Ethanol tolerance is essential for L. plantarum strains living in substances with more than 9% ethanol, such as wine and beer. The details regarding how L. plantarum adapts to ethanol are still lacking. This study demonstrates that AcrR regulates the de novo synthesis of fatty acids in L. plantarum adapting to toxic levels of ethanol. We also identified the ability of the TetR/AcrR family regulator to bind to the fatty acid biosynthesis gene promoter, PfabZ1, in L. plantarum and defined the binding sites. This finding facilitates the induction of the adaptation of L. plantarum strains to ethanol for food fermentation applications.
KEYWORDS: Lactobacillus plantarum, AcrR, fatty acid de novo synthesis, ethanol tolerance
ABSTRACT
Lactobacillus plantarum is a versatile bacterium with significant adaptability to harsh habitats containing excessive ethanol concentrations. It was found that the L. plantarum NF92-TetR/AcrR family regulator, AcrR, significantly enhanced the growth rate of this lactic acid bacterium in the presence of ethanol. Through screening 172 ethanol-resistant related genes by electrophoretic mobility shift and quantitative reverse transcription-PCR (RT-qPCR) assays, six genes were identified to be regulated by AcrR under ethanol stress. Among these was a gene coding for a 3-hydroxyacyl-ACP dehydratase (fabZ1) regulated by AcrR under ethanol stress. AcrR regulated fabZ1 under ethanol stress by binding to its promoter, PfabZ1. DNase I footprinting analysis indicated that there were two specific AcrR binding sites on PfabZ1. RT-PCR results showed fabZ1 could cotranscribe with its downstream 12 genes and conform a fatty acid de novo biosynthesis (fab) gene cluster under the control of PfabZ1. Both RT-qPCR of the fab gene cluster in acrR knockout and overexpression strains and fatty acid methyl ester analysis of the acrR knockout strain showed that AcrR could promote fatty acid synthesis in L. plantarum NF92. Membrane fluorescence anisotropy analysis of acrR knockout and overexpression strains showed that AcrR could increase membrane fluidity under ethanol stress. Thus, AcrR could regulate fatty acid synthesis and membrane fluidity to promote the adaption of L. plantarum NF92 to a high ethanol concentration.
IMPORTANCE Ethanol tolerance is essential for L. plantarum strains living in substances with more than 9% ethanol, such as wine and beer. The details regarding how L. plantarum adapts to ethanol are still lacking. This study demonstrates that AcrR regulates the de novo synthesis of fatty acids in L. plantarum adapting to toxic levels of ethanol. We also identified the ability of the TetR/AcrR family regulator to bind to the fatty acid biosynthesis gene promoter, PfabZ1, in L. plantarum and defined the binding sites. This finding facilitates the induction of the adaptation of L. plantarum strains to ethanol for food fermentation applications.
INTRODUCTION
Lactic acid bacteria (LAB) are widely distributed in a range of niches, including vegetables, meat, dairy products, and gastrointestinal tracts, as well as numerous fermented foods and beverages (1–3). Sometimes, these environments are not very suitable for the survival of LAB, and many stress factors in these habitats, such as toxic alcoholic content, suboptimal growth temperature, acidic pH, starvation, and growth-inhibitory compounds originating from bacterial metabolism, are detrimental to LAB. Numerous studies aimed at elucidating the mechanisms of LAB to sense stress signals and protect themselves against acid, oxidative chemical agents, bile, and heat stresses have been reported (4–9). These studies have shown that LAB respond rapidly to the habitat by modulating different cellular processes, including stress response pathways, amino acid metabolism, carbohydrate metabolism, cell envelope composition, and others (10).
Lactobacillus plantarum, some representatives of which have been characterized as probiotics, has been used to improve food fermentations, as well as human health (11, 12). L. plantarum can survive in many different ecological niches and is able to tolerate varied stressors. Ethanol is a common stress factor in food fermentations, especially in the wine industry, and represents a hurdle for the survival and viability of L. plantarum. The toxicity of ethanol is generally attributed to the disturbance of the plasma, resulting in a loss of membrane integrity and an increase in membrane permeability (13, 14). L. plantarum is widely distributed in the early, middle, and late stages of fermented grains and has a good adaptability to this environment (15). In the fermentation process, L. plantarum plays an important role in the acidification of red grapes, improving flavor and inhibiting pathogens or spoilage bacteria (16–19). It has been reported that L. plantarum tolerates up to 13% (vol/vol) ethanol at 18°C (20), and several studies have been conducted to explore the response of L. plantarum to this solvent (21–27). For example, overproduction of Hsp 18.55 and Hsp 19.3 can enhance the survival of L. plantarum in the presence of butanol (1% [vol/vol]) or ethanol (12% [vol/vol]) (26). Overexpression of the L. plantarum peptidoglycan biosynthesis associated gene, murA2, can increase the tolerance of Escherichia coli to alcohols and enhance ethanol production (24). However, these studies did not reveal a detailed molecular basis for how L. plantarum adapts to ethanol stress.
The TetR/AcrR family is a large and important family of regulators and is widespread among bacteria and archaea (28). Regulators in the TetR/AcrR family are always one-component signal transduction systems, which contain a conserved helix-turn-helix DNA-binding domain and a C-terminal ligand regulatory domain. These regulators play important roles in a wide range of cellular activities, including osmotic stress response, biosynthesis of antibiotics, multidrug efflux pumping, catabolic pathways, secondary metabolite biosynthesis, and the pathogenicity of bacteria (29–33). However, the role of AcrR in ethanol tolerance is not documented. It has been reported that AcrR has a very slight negative impact on the ethanol tolerance of E. coli (34); however, the exact regulation mechanism is unknown.
TetR/AcrR family regulators not only have various functions in different kinds of microorganisms but also can regulate many cellular activities in the same bacteria (34–36). Our previous report showed that the TetR/AcrR family regulator AcrR could participate in sorbitol or mannitol utilization by regulating the aldehyde-alcohol dehydrogenase encoding gene adhE in L. plantarum NF92 isolated from the Chinese liquor fermented grains (37). In this study, we found that AcrR could positively regulate fatty acid biosynthesis to enhance the ethanol resistance of L. plantarum NF92. The mechanism by which AcrR regulates L. plantarum NF92 tolerance to ethanol and adaptation was explored.
RESULTS
AcrR is involved in ethanol tolerance in L. plantarum NF92.
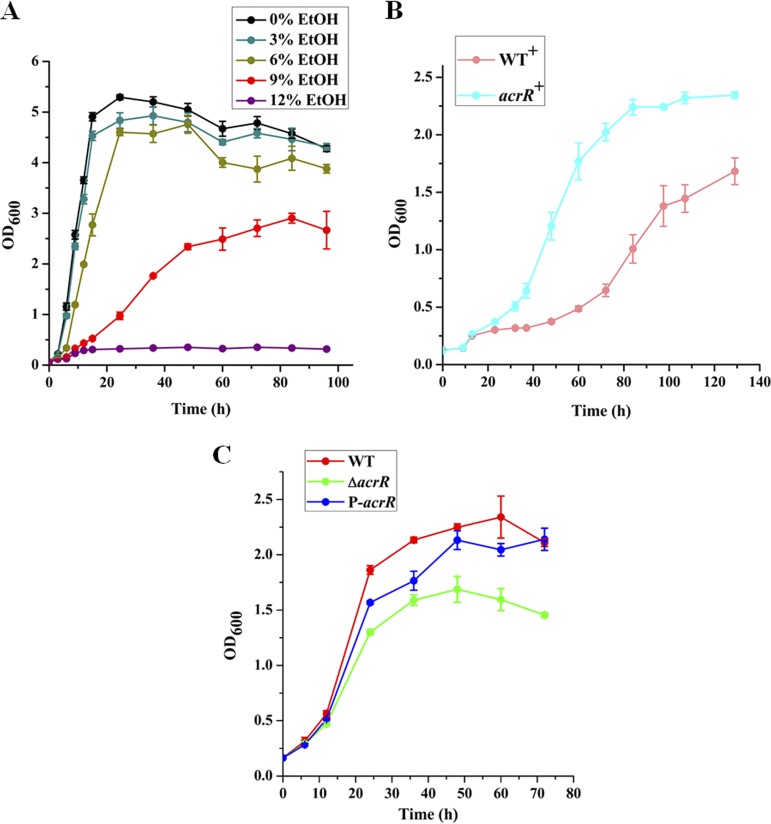
L. plantarum NF92 is a strain that was isolated from Chinese liquor fermented grains. We measured the growth of L. plantarum NF92 in MRS supplemented with ethanol. The results showed that the growth of L. plantarum NF92 was not affected obviously when grown in 3% (vol/vol) and 6% (vol/vol) ethanol but was completely repressed by 12% (vol/vol) ethanol. Meanwhile, 9% (vol/vol) ethanol obviously repressed the growth of L. plantarum NF92; however, the cell density of NF92 could still reach half that of the control. As a result, we chose 9% (vol/vol) as the ethanol stress concentration in this study (Fig. 1A). In our previous report, we found that AcrR could promote the expression of AdhE, which could catalyze acetyl coenzyme A (acetyl-CoA) to ethanol (37). At the same time, we found that it might be also involved in ethanol tolerance of L. plantarum NF92. As shown in Fig. 1B, the acrR overexpression strain of L. plantarum NF92 (acrR+) had a significantly shorter growth time and higher cell densities in 9% (vol/vol) ethanol than that of the empty vector containing wild-type strain (WT+), suggesting that AcrR might be involved in ethanol resistance in L. plantarum NF92. Further, the growth of the L. plantarum NF92 wild-type strain, the acrR knockout (ΔacrR) strain, and the acrR complemented strain P-acrR was tested in MRS supplemented with 9% (vol/vol) ethanol. Compared to the wild-type strain, the growth of the ΔacrR strain was significantly repressed. The growth of the acrR complemented strain P-acrR was much stronger than that of ΔacrR strain in MRS supplemented with 9% (vol/vol) ethanol and partly recovered to the level of wild-type strain (Fig. 1C). These results demonstrate that AcrR plays an important role in ethanol tolerance in L. plantarum NF92.
FIG 1.
Growth curves of L. plantarum NF92. (A) Growth curve of L. plantarum NF92 in the presence of various concentrations (vol/vol) of ethanol (EtOH). (B) Effects of acrR overexpression on the growth of L. plantarum NF92 in MRS supplemented with 9% (vol/vol) ethanol. (C) Effects of acrR disruption on the growth of L. plantarum NF92 in MRS supplemented with 9% (vol/vol) ethanol. Values and standard deviations were calculated from three repeated samples.
Genes that may be regulated by AcrR during ethanol tolerance in L. plantarum NF92.
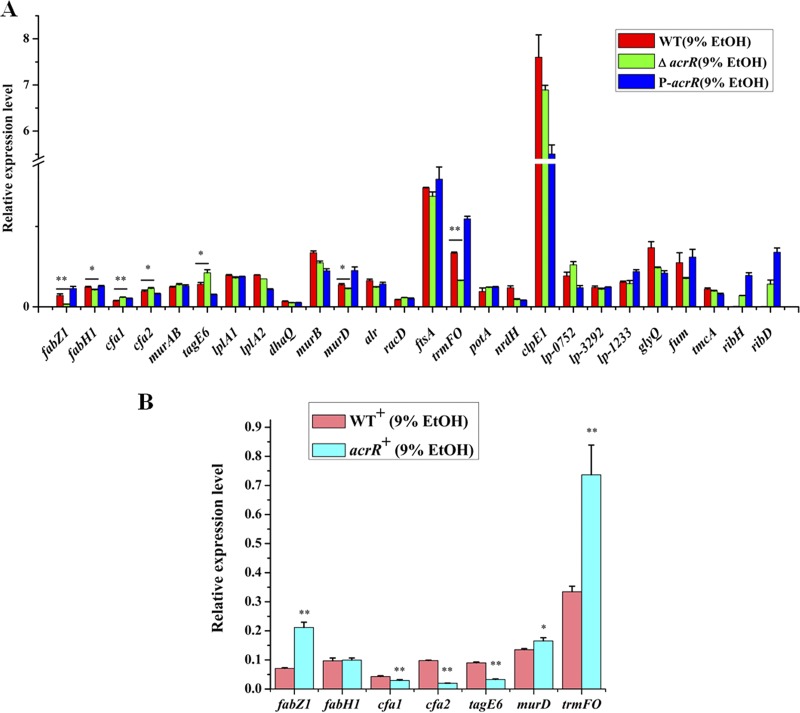
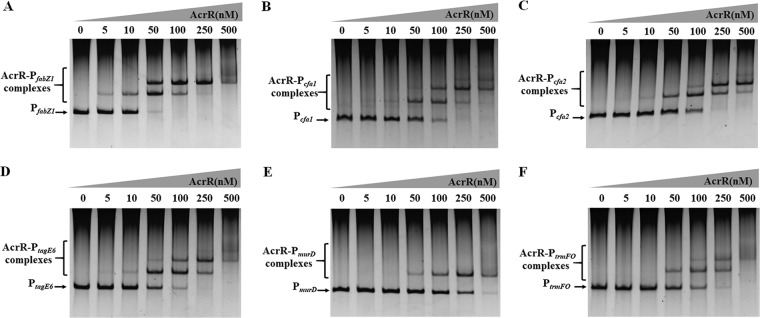
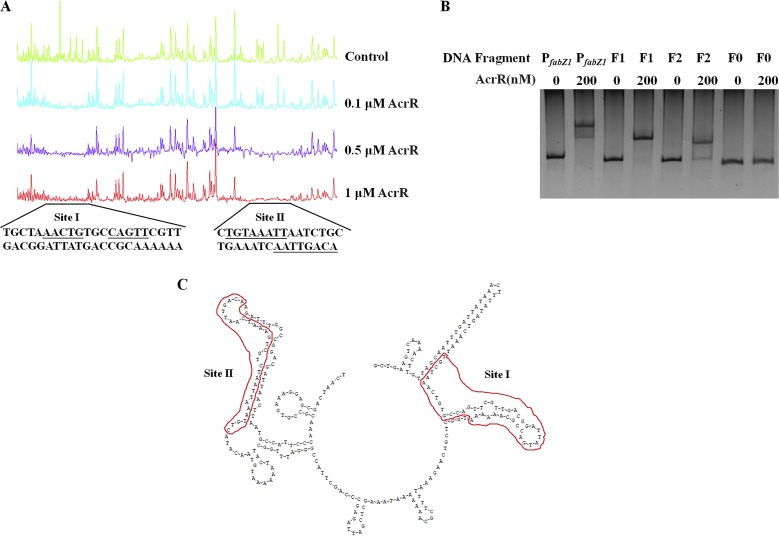
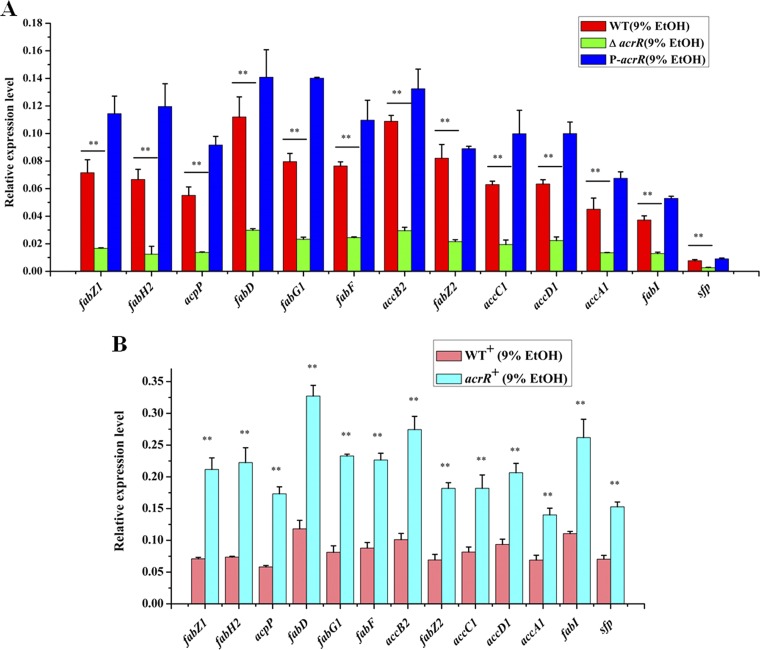
In order to identify ethanol tolerance related genes regulated by AcrR in L. plantarum NF92, we checked the binding capabilities of AcrR to 88 promoters. These promoters were referred to 172 genes that are differently expressed after 8% ethanol treatment in L. plantarum WCFS1 (21) and the main genes of peptidoglycan and wall teichoic acid biosynthesis related to stress response in LAB (10, 38). These genes are mainly involved in stress response, cell division, cell envelope synthesis, citrate metabolism, pyrimidine synthesis, and so on. The related genes and promoters were found in L. plantarum NF92 and are listed in Table S1 in the supplemental material. Electrophoretic mobility shift assays (EMSAs) were used to detect the binding of AcrR to 88 promoters. As shown in Table S1, 51 of these promoters were detected to be bound by AcrR, and among them, 25 promoters showed stronger binding abilities. The expressions of the first genes controlled by these 25 promoters in WT, ΔacrR, and P-acrR strains grown in 9% (vol/vol) ethanol were further analyzed by quantitative reverse transcription-PCR (RT-qPCR). The results showed that the expression levels of seven genes, namely, fabZ1 (3-hydroxyacyl-ACP dehydratase, controlled by PfabZ1), fabH1 (ketoacyl-ACP synthase III, controlled by PfabH1), cfa1 (cyclopropane-fatty-acyl-phospholipid synthase, controlled by Pcfa1), cfa2 (cyclopropane-fatty-acyl-phospholipid synthase, controlled by Pcfa2), murD (UDP-N-acetylmuramoyl-l-alanine-d-glutamate ligase, controlled by PmurD), trmFO {[FADH(2)-oxidizing methylenetetrahydrofolate-tRNA-(uracil(54)-C(5)]-methyltransferase, controlled by PtrmFO}, and tagE6 (glycosyltransferase, controlled by PtagE6), were obviously changed in the ΔacrR strain compared to those in the WT and recovered in P-acrR. In the ΔacrR strain grown in 9% ethanol, the expression levels of fabZ1, fabH1, murD, and trmFO were significantly downregulated, whereas cfa1, cfa2, and tagE6 were significantly upregulated, compared to those in the WT strain grown in 9% ethanol. In the acrR+ strain grown in 9% (vol/vol) ethanol, the transcriptional levels of fabZ1, murD, and trmFO were significantly upregulated and cfa1, cfa2, and tagE6 were significantly downregulated compared to those in the WT strain grown in 9% ethanol, whereas the expression of fabH1 did not show obvious differences before or after acrR was overexpressed (Fig. 2B). These results demonstrate that fabZ1, murD, and trmFO may be positively regulated by AcrR, while cfa1, cfa2, and tagE6 may be negatively regulated by AcrR, when L. plantarum NF92 is grown in 9% (vol/vol) ethanol. The EMSA results also showed that AcrR had strong binding abilities to the promoters of these six genes (Fig. 3). AcrR could bind to PfabZ1, Pcfa1, Pcfa2, and PtagE6 at a concentration of 10 nM and to PmurD and PtrmFO at a concentration of 50 nM. Among these genes, fabZ1, which is related to fatty acid de novo biosynthesis, was most obviously affected by AcrR under ethanol stress. The expression of fabZ1 decreased 3.28-fold when acrR was knocked out and increased 2-fold when acrR was overexpressed. Moreover, the fabZ1 promoter PfabZ1 had the strongest binding ability to AcrR, and a clear binding band could be seen when the concentration of AcrR was 5 nM (Fig. 3A). The possible binding sites of AcrR to PfabZ1 were identified by a DNase I footprinting assay. Two regions located on PfabZ1 at bp –221 to bp –178 (site I) and bp –92 to bp –62 (site II) upstream of the fabZ1 start codon ATG were identified (Fig. 4A). The site I (TGCTAAACTGTGCCAGTTCGTTGACGGATTATGACCGCAAAAAA, 44 bp) and site II (CTGTAAATTAATCTGCTGAAATCAATTGACA, 31 bp) fragments were inserted into a nonspecific DNA fragment (fragment 0 [F0]), respectively creating fragment 1 (F1) and fragment 2 (F2). EMSA showed that AcrR could bind to F1 and F2 but not to F0 (Fig. 4B), indicating that AcrR can bind to the site I and site II regions of PfabZ1. The second structure of PfabZ1 was predicted by DNAMAN, and both site I and site II were included in the hairpin structures in the structure of PfabZ1 (Fig. 4C). Site I contains a 5-bp (AACTG) repeat sequence and site II contains an 8-bp (TGTAAATT) imperfect repeat sequence. We proposed that fabZ1 and the possible genes controlled by fabZ1 promoter might be regulated by AcrR to participate in ethanol tolerance in L. plantarum NF92.
FIG 2.
RT-qPCR transcriptional analysis of genes that may be regulated by AcrR under ethanol stress. (A) Transcriptional levels of genes that may be regulated by AcrR in WT, ΔacrR, and P-acrR strains under 9% (vol/vol) ethanol. (B) Transcriptional levels of genes that may be regulated by AcrR in the WT+ and acrR overexpression (acrR+) strains under 9% (vol/vol) ethanol. Relative values were obtained using the transcription of gyrB as reference. cDNA used as template in this experiment was diluted 1,000-fold. The x axis represents different genes. Values and standard deviations were calculated from three repeated samples. **, P < 0.01; *, P < 0.05.
FIG 3.
Binding ability analysis of AcrR to promoters by EMSA. (A) EMSA of AcrR binding to the upstream region of fabZ1 (PfabZ1). (B) EMSA of AcrR binding to the upstream region of cfa1 (Pcfa1). (C) EMSA of AcrR binding to the upstream region of cfa2 (Pcfa2). (D) EMSA of AcrR binding to the upstream region of tagE6 (PtagE6). (E) EMSA of AcrR binding to the upstream region of murD (PmurD). (F) EMSA of AcrR binding to the upstream region of trmFO (PtrmFO). Each lane contained 20 ng of DNA probe. Concentrations of AcrR ranged from 0 to 500 nM.
FIG 4.
Identification of AcrR binding site on PfabZ1. (A) Determination of AcrR binding sites on PfabZ1 by DNase I footprinting. Each lane contained 1.5 μg of DNA probe, which was incubated with increasing concentrations of AcrR protein (0.1, 0.5, and 1 μM). DNA probe incubated with 7 μM bovine serum albumin was used as a negative control. (B) Binding of AcrR to the 44-bp sequence (site I) and 31-bp sequence (site II). F1 contained the 44-bp sequence; F2 contained the 31-bp sequence. F0, nonspecific DNA as a negative control; PfabZ1, the positive control. Each lane was loaded with a 20-ng DNA fragment and either 0 or 200 nM AcrR, as indicated in the figure. (C) Second structure of a part of PfabZ1, which was predicted by DNAMAN. Red lines indicate the positions of site I and site II.
Fatty acid de novo biosynthesis gene cluster is regulated by AcrR under ethanol.
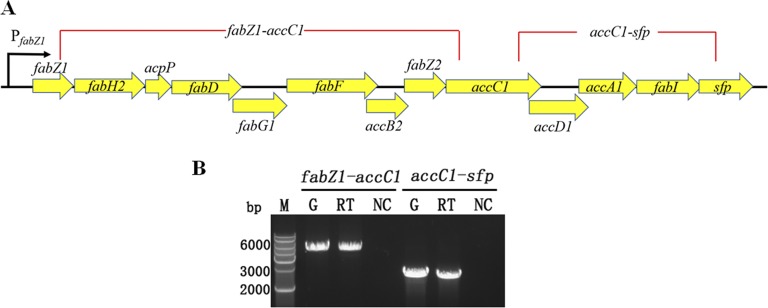
FabZ1 is a 3-hydroxyacyl-ACP dehydratase that is involved in the dehydration of 3-hydroxyacyl-ACPs of all chain lengths in the basic type II fatty acid biosynthetic pathway (39). The location of fabZ1 in the genome of L. plantarum NF92 was analyzed, and another 12 genes were located downstream of it. Twelve of these thirteen genes are involved in type II fatty acid synthesis, and another is necessary for the activity of ACP during fatty acid biosynthesis; details are listed in Table S1 in the supplemental material. These 13 genes composed the fatty acid biosynthesis (fab) gene cluster, with a total length of 9,890 bp. The transcriptional units of the fab gene cluster were determined by RT-PCR, which detected two parts of the fab gene cluster, namely, the fragments from fabZ1 to accC1 and from accC1 to sfp. As shown in Fig. 5, single bands of the expected size were obtained for genes from fabZ1 to accC1 and from accC1 to sfp. This indicated that these 13 genes could form a cotranscribed unit that was controlled by the same promoter PfabZ1. It is proposed that the fab gene cluster might be regulated by AcrR during ethanol stress. RT-qPCR analysis was performed using total RNAs from WT, ΔacrR, P-acrR, WT+, and acrR+ strains. The results showed that the expression levels of these 13 genes were all significantly reduced in the acrR knockout strain (ΔacrR) and increased in the overexpression strain (acrR+) compared to the wild type (Fig. 6). These results suggest AcrR is a positive regulator of fab gene cluster and may promote fatty acid de novo biosynthesis in L. plantarum NF92 under ethanol stress.
FIG 5.
Organization and transcription unit of fab gene cluster in L. plantarum NF92. (A) Organization of the fab gene cluster in L. plantarum NF92. The gene cluster was divided into two parts (red lines) for RT-PCR to determine potential cotranscription. fabZ1, 3-hydroxyacyl-ACP dehydratase FabZ; fabH2, ketoacyl-ACP synthase III; acpP, acyl carrier protein; fabD, ACP S-malonyltransferase; fabG1, SDR family oxidoreductase; fabF, β-ketoacyl-[acyl-carrier-protein] synthase II; accB2, acetyl-CoA carboxylase biotin carboxyl carrier protein subunit; fabZ2, β-hydroxyacyl-ACP dehydratase; accC1, acetyl-CoA carboxylase biotin carboxylase subunit; accD1, acetyl-CoA carboxylase carboxyl transferase subunit beta; accA1, acetyl-CoA carboxylase carboxyl transferase subunit alpha; fabI, enoyl-[acyl-carrier-protein] reductase FabI; sfp, 4′-phosphopantetheinyl transferase superfamily protein. (B) RT-PCR for cotranscription analysis of genes in fab gene cluster. fabZ1-accC1, 5,354 bp; accC1 to sfp, 2,610 bp. RNA was extracted from the WT strain. Lanes: G, positive controls with genomic DNA; RT, cDNA from the RNA sample; NC, negative controls consisting of DNase I-treated RNA sample; M, DNA marker.
FIG 6.
RT-qPCR transcriptional analysis of fab gene cluster in WT, ΔacrR, P-acrR, WT+, and acrR+ strains under ethanol stress. (A) Effects of acrR disruption on fab transcription in WT, ΔacrR, and P-acrR strains under 9% (vol/vol) ethanol stress. (B) Effects of acrR overexpression on fab transcription in WT+ and acrR+ strains under 9% (vol/vol) ethanol stress. Relative values were obtained using the transcription of gyrB as a reference. cDNA used as template in this experiment was diluted 1,000-fold. The x axis represents different genes. Values and standard deviations were calculated from three repeated samples. **, P < 0.01.
AcrR promotes fatty acid synthesis under ethanol stress.
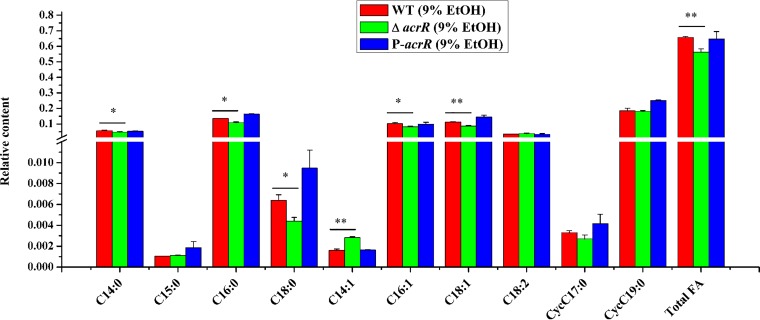
In order to test whether AcrR affect the fatty acid contents of L. plantarum NF92 under ethanol stress, fatty acid methyl ester (FAME) analysis was used to detect the fatty acid contents in WT, ΔacrR, and P-acrR strains grown in 9% (vol/vol) ethanol. Analysis of the fatty acid composition of L. plantarum NF92 during ethanol stress revealed that mainly ten kinds of fatty acids were detected, including saturated fatty acid (SFA; C14:0, C15:0, C16:0, and C18:0), unsaturated fatty acid (UFA; C14:1, C16:1, C18:1, and C18:2), and cyclopropane fatty acid (CFA; cycC17:0 and cycC19:0). Compared to the WT and complemented P-acrR strains, most of the fatty acids, especially C18:1, were decreased in the acrR disruption strain. The relative content of total fatty acid was also significantly decreased in the ΔacrR strain (Fig. 7). This demonstrates that AcrR can promote fatty acid synthesis under ethanol stress, which is consistent with its influence on fab gene cluster.
FIG 7.
Effects of AcrR on composition and contents of fatty acids of L. plantarum NF92 under 9% (vol/vol) ethanol stress. Fatty acids of WT, ΔacrR, and P-acrR strains were determined in logarithmic cultures grown in MRS containing 9% (vol/vol) ethanol. The y axis represents the relative content of fatty acid per mg of dried cells. Values and standard deviations were calculated from three repeated samples. *, P < 0.05; **, P < 0.01.
AcrR increases membrane fluidity of L. plantarum NF92 under ethanol stress.
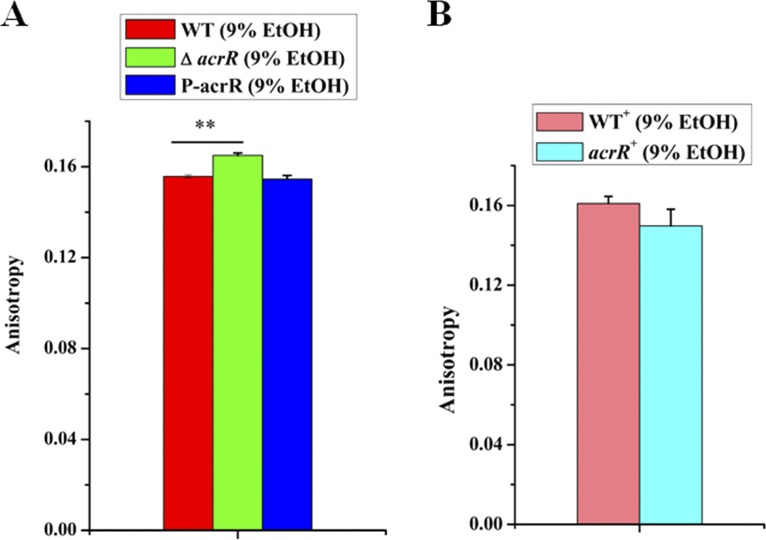
Changes in fatty acids always influence cell membrane fluidity. We hypothesized that AcrR might be involved in regulating membrane fluidity of L. plantarum NF92 under ethanol stress. In order to elucidate the possible effects of AcrR on membrane-related alterations, membrane fluidity was evaluated by fluorescence anisotropy using TMA-DPH {1-[4 (trimethylamino)phenyl]-6-phenyl-1,3,5-hexatriene} as a probe. Samples were obtained from the exponential phase of WT, ΔacrR, P-acrR, WT+, and acrR+ strains under 9% (vol/vol) ethanol. Higher fluorescence anisotropy values were observed in the ΔacrR strain than in the WT and P-acrR strains (Fig. 8A), which meant that disruption of acrR reduced the membrane fluidity of L. plantarum NF92. Furthermore, the fluorescence anisotropy values in the acrR+ strain were lower than in the WT+ strain (Fig. 8B), indicating that overexpression of acrR increased the membrane fluidity of L. plantarum NF92. These results demonstrate that AcrR may enhance membrane fluidity through upregulation of fatty acid biosynthesis in L. plantarum NF92 under ethanol stress.
FIG 8.
Effects of AcrR on membrane fluidity of L. plantarum NF92 under 9% (vol/vol) ethanol stress. (A) Effects of acrR disruption on membrane fluidity of L. plantarum NF92 under 9% (vol/vol) ethanol stress. (B) Effects of acrR overexpression on membrane fluidity of L. plantarum NF92 under 9% (vol/vol) ethanol stress. The y axis represents the fluorescence anisotropy. Values and standard deviations were calculated from three repeated samples. **, P < 0.01.
DISCUSSION
AcrR, as a member of the TetR/AcrR family, was proved to participate in sorbitol or mannitol utilization in our previous report (37). In the present study, we found that AcrR could enhance the tolerance of L. plantarum NF92 to ethanol through regulating fatty acid de novo biosynthesis. We show a novel regulation mechanism of AcrR in L. plantarum that has not been reported.
Cell envelope acts as a physical barrier between the cell and its surroundings, and it is the first barrier of bacteria to defend environmental stress. The ability to adjust lipid composition to adapt to different environments is crucially important for bacterial survival. Fatty acids are central to microbial environmental adaptation. For example, Bacillus species are able to adapt to a wide range of habitat changes, including changes in the growth medium, temperature, and pH, by modifying their fatty acid patterns (40). L. plantarum also changes fatty acid contents to survive in harsh environmental conditions, including heat, acid, and ethanol (27, 41). It is essential for the cell to keep a tight regulation of fatty acid biosynthesis to efficiently respond to stresses. Regulation of fatty acid de novo synthesis has been studied in several species, such as Escherichia coli, Bacillus subtilis, mycobacteria, Lactococcus lactis, and Streptococcus pneumoniae (42–48). However, there is little report about the regulation of fatty acid synthesis in Lactobacillus. Here, we proved that AcrR could regulate fatty acid synthesis in L. plantarum NF92 under ethanol stress. In the previous reports, several regulators have been identified to regulate fatty acid synthesis, such as FabR in E. coli, FapR in B. subtilis, FabT in Streptococcus pneumoniae, and FasR in Streptococcus coelicolor, Mycobacterium tuberculosis, or Corynebacterium glutamicum. Among these regulators, FasR from C. glutamicum, FasR from Mycobacterium tuberculosis, and FabR from E. coli are all TetR family regulators. These findings indicate that several TetR family regulators are involved in regulating fatty acid synthesis, which supports our conclusion in L. plantarum. However, AcrR showed only 10.8, 12.61, and 13.19% identities with FasR from C. glutamicum, FasR from Mycobacterium tuberculosis, and FabR from E. coli, respectively. We analyzed the binding sites on the promoters that were recognized by AcrR, FasR, and FabR, and no conserved sequences were found among them (data not shown). Meanwhile, AcrR in L. plantarum NF92 could regulate all of the genes related to the type II fatty acid de novo biosynthesis pathway, whereas FabR from E. coli and FasR from C. glutamicum could only regulate parts of related genes. For example, FabR functions as a repressor controlling the expression of fabA (β-hydroxydecanoyl-ACP dehydratase) and fabB (β-ketoacyl-ACP synthase I) from E. coli. The function and regulation of AcrR in L. plantarum NF92 are different from the reported TetR family regulators involving in fatty acid synthesis.
Ethanol tolerance has been associated with high plasma membrane fluidity (49–51). Changes of fatty acids always lead to the fluidization response based on the hypothesis of “homeoviscous adaptation” (52). In our study, we found when acrR was knocked out, the CFA/SFA ratio and the CFA/UFA ratio were both significantly increased, and the UFA/SFA ratio was decreased (Table S2), which meant the membrane fluidity would be decreased. This was consistent with the membrane fluidity evaluation results that acrR knockout strain showed a lower membrane fluidity. Thus, AcrR could enhance membrane fluidity under ethanol stress. It has been reported that the increase of membrane fluidity may help to protect Lactobacillus hilgardii avoid rigidity disruption induced by ethanol (51). We proposed that in L. plantarum NF92, the membrane fluidity was increased through regulating fatty acid biosynthesis by AcrR to adapt to the ethanol stress. In addition, in the ΔacrR strain, the percentage of cycC19:0 was significantly increased, and C18:1 was significantly decreased (Table S2). Both C18:1 and cycC19:0 are important in overcoming the toxic effects of ethanol (53, 54). Methylenoctadecenoic acid cycC19:0 could be synthesized in situ by the transfer of a methylene group from S-adenosyl-l-methionine to the double bond of unsaturated fatty acid by the Cfa synthase enzymes (55), whose transcriptional levels were increased in the ΔacrR strain under ethanol stress. AcrR may also have a central role in maintaining the balance between C18:1 and cycC19:0 during ethanol stress.
TetR family regulators are known to regulate numerous aspects of bacterial physiology and some of them are global regulators (28). In our study, we found that AcrR could bind to promoters of various genes. In addition to the fab gene cluster, AcrR could regulate murD, trmFO, and tagE6 in L. plantarum NF92 under ethanol stress. Peptidoglycan (PG) is the main component of the Gram-positive cell wall. MurD, which could catalyze the addition of d-Glu and help to form UDP-MurNAc-pentapeptide, has a central role in PG synthesis (38). RNA modification plays a key role in regulating the cellular response to various stressors (56). Methylation is one of the most common chemical modifications in RNA; for example, tRNA contains abundant methylated nucleotides. These methylated nucleotides can stabilize the L-shaped tRNA structure and improve molecular recognition. One such modification involves a conserved uridine at position 54 (U54) in the T-loop of tRNA, which is often modified to 5-methyluridine (m5U; ribothymidine) by the folate-dependent tRNA methyltransferase TrmFO (57, 58). TagE6 is a glycosyltransferase, which is required for protein O-glycosylation. Protein glycosylation is a highly ubiquitous protein modification in nature and is considered to be one of the posttranslational modifications involved in protein stability, signaling and response to stress, adaptation to changing environments, regulation of toxic and damaged proteins, protein localization, and host-pathogen interactions (59, 60). In L. plantarum strains, protein glycosylation is a common feature, and the cell wall teichoic acid biosynthesis could also require the activity of specific TagE (teichoic acid glycosylation) proteins (61). In L. plantarum NF92 there is another glycosyltransferase-encoding gene, tagE5, located downstream of tagE6. Both TagE6 and TagE5 are required for the glycosylation of proteins, especially those O-glycosylated with N-acetylhexosamine, likely N-acetylglucosamine (GlcNAc) in L. plantarum WCFS1 (61). In L. plantarum NF92, the amino acid sequences of TagE6 and TagE5 showed identities of 99.81 and 99%, respectively, with those in L. plantarum WCFS1. They might also have a central role in protein glycosylation. Above all, in addition to regulating fatty acid synthesis, AcrR might participate in ethanol tolerance by promoting cell wall synthesis, enhancing tRNA stabilization and molecular recognition ability and influencing protein glycosylation. Much more additional research on the relationship of these genes with ethanol tolerance is needed.
Conclusion.
AcrR is a TetR/AcrR family regulator that controls the expression of fatty acid synthesis in L. plantarum NF92. AcrR can bind to multiple gene promoters, among which the promoter of the fab gene cluster (PfabZ1) shows the strongest ability to bind AcrR. All the genes on fab gene cluster are strongly regulated by AcrR in L. plantarum NF92 under ethanol stress. AcrR enhances fatty acid synthesis by directly binding to site I (TGCTAAACTGTGCCAGTTCGTTGACGGATTATGACCGCAAAAAA) and site II (CTGTAAATTAATCTGCTGAAATCAATTGACA) of PfabZ1 in L. plantarum NF92 under ethanol stress. The adaption of L. plantarum NF92 to ethanol is mainly regulated by AcrR through promoting fatty acid synthesis and improving membrane fluidity.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
L. plantarum NF92 and its derivatives—the ΔacrR (the acrR knockout strain), P-acrR (the acrR complemented strain), WT+ (the empty vector-containing strain), and acrR+ (the acrR overexpressed strain) strains—were grown in De Man-Rogosa-Sharp (MRS) medium at 30°C. Chloramphenicol (20 μg/ml) was used for the WT+ and acrR+ strains. The growth of L. plantarum NF92 in MRS supplemented with ethanol was determined by measuring the cell optical density at 600 nm with a spectrophotometer (model 680; Bio-Rad). The AcrR E. coli expression strain (acrR) was grown in Luria broth (LB) with aeration and 50 μg/ml kanamycin at 37°C. All the bacterial strains used in this study were constructed as described in our previous study (37).
Protein expression and purification.
The expression and purification of AcrR were conducted according to a previous report (37). Briefly, AcrR was induced and expressed using IPTG (isopropyl-β-d-thiogalactopyranoside), purified on an immobilized metal affinity chromatography Ni2+ column (GE Healthcare Life Science), confirmed by SDS-PAGE, and quantified using a Pierce BCA protein assay kit (Thermo Scientific).
Screening genes that may be regulated by AcrR during ethanol tolerance.
According to the microarray data of L. plantarum WCFS1 (21) and the peptidoglycan and wall teichoic acid biosynthesis in LAB (10, 38), 172 genes whose transcription was differently influenced by ethanol or may be related to stress response were chosen to screen the genes that might be regulated by AcrR under ethanol stress. The main screening methods and processes were as follows. For step 1, the promoters of these genes were deduced according to the functions and locations of these genes in the genome of L. plantarum NF92 and predicted by BDGP (http://www.fruitfly.org/seq_tools/promoter.html). An EMSA was used to detect the binding abilities of promoters of these genes to AcrR. A high binding ability was defined as promoters that could bind when the concentration of AcrR was ≤50 nM. For step 2, the transcriptional levels of genes whose promoters had a high affinity for AcrR were analyzed in WT, ΔacrR, and P-acrR strains under ethanol stress by RT-qPCR. For step 3, the transcriptional levels of the selected genes in step 2 were confirmed in WT+ [containing an empty vector pMG36c(M)] and acrR+ strains under ethanol stress by RT-qPCR. Genes that were differentially expressed in both steps 2 and 3 were considered to be regulated by AcrR under ethanol stress. EMSA and RT-qPCR were performed according to a previous report, with some modifications (37). As DNA probes, promoters of these genes were amplified by PCR from the genomic DNA of L. plantarum NF92. Total RNAs were extracted from the cells of L. plantarum NF92 and its derivative strains (ΔacrR, P-acrR, WT+, and acrR+), which were cultured to exponential phase in 9% (vol/vol) ethanol containing MRS broth in triplicate. cDNAs were synthesized by using a RevertAid first-strand cDNA synthesis kit (Thermo Scientific) and used as templates for the target genes and the internal control (i.e., the DNA gyrase B-encoding gene gyrB) (62). H2O was used as a negative control. The primers used in this study are described in Table 1.
TABLE 1.
Primers used in this study
| Primera | Function | Nucleotide sequence (5′–3′) |
|---|---|---|
| P(cfa1)-F | Cloning of Pcfa1 for EMSA | GCTGAATAGTCTCGATGTCTGGCAACAGAC |
| P(cfa1)-R | CCTGAATTTGATTTTCTGGAAGATTGATTGTC | |
| P(cfa2)-F | Cloning of Pcfa2 for EMSA | ACAGCGATAATTACTGCAAACACGATC |
| P(cfa2)-R | GGTGTGGTAAAAGGTTTTTTCTAGCATTG | |
| P(tagE6)-F | Cloning of PtagE6 for EMSA | GTTGTGTTGTTATTTCGATTGAATGGTGAT |
| P(tagE6)-R | TATTACTCGCTCCCTTACACGACGTTCTG | |
| P(fabZ1)-F | Cloning of PfabZ1 for EMSA | GCGTGCATCTAAGCTAGTGACAGTTGAAC |
| P(fabZ1)-R | AGTTAGTCGCTCCTTTCACGGCGTTTG | |
| P(trmFO)-F | Cloning of PtrmFO for EMSA | ATGAAAAAAAGGTCATCTACGGATGATCT |
| P(trmFO)-R | CCAGAATTCCTCTTTTCTTGTTGTCATACC | |
| P(murD)-F | Cloning of PmurD for EMSA | CATTCTTGTCCGAGTTAGCTGTACATAATTG |
| P(murD)-R | AAACCAAGATCTCCCTATAATGTTTGTAGATG | |
| QgyrB-F2 | qRT-PCR analysis of gyrB | AGGTGGGACGCATGAAGAAG |
| QgyrB-R2 | CCGTCATCCCTTCACGAACA | |
| Q-0752-F | qRT-PCR analysis of lp_0752 | ACCCATGGCTTTGGTGTTTTG |
| Q-0752-R | GCTTGCGGTTTAGCTTGAGT | |
| Q-murA1-F | qRT-PCR analysis of murA1 | CGCTAAAATTCGGGGTGCTG |
| Q-murA1-R | TCGAAGCAGCCAGGGATAGA | |
| Q-3292-F | qRT-PCR analysis of lp_3292 | CATCATCAATCGCGTTATCGGT |
| Q-3292-R | GCAACAGCCACATCGTTACC | |
| Q-1233-F | qRT-PCR analysis of lp_1233 | AGATTGGGCGATTCATCCGT |
| Q-1233-R | TCGACTTCTCGTGGCAATGG | |
| Q-nrdH-F | qRT-PCR analysis of nrdH | GTTCCTCACGGCGCACAATA |
| Q-nrdH-R | TTTCAACGACCGGAACAGCA | |
| Q-glyQ-F | qRT-PCR analysis of glyQ | GGAACGATGAGCCCCTACAC |
| Q-glyQ-R | GGGTTCTCACCATACCGACC | |
| Q-potA-F | qRT-PCR analysis of potA | CCCACGATCAGGAAGAAGCC |
| Q-potA-R | GCCACGAAATGGTTGATCGG | |
| Q-alr-F | qRT-PCR analysis of alr | GCGATTCTAGCGAAGGGACT |
| Q-alr-R | CGCCCTGCCATAAACTCGAT | |
| Q-cfa1-F2 | qRT-PCR analysis of cfa1 | TTTCCCAGGTGGCTACGTTC |
| Q-cfa1-R2 | CCCAGATTTCAGTCGTCCGT | |
| Q-cfa2-F | qRT-PCR analysis of cfa2 | AGGCATCCGTCGTTTGATGA |
| Q-cfa2-R | CAGCAAATTCCAACGCCACA | |
| Q-ftsA-F | qRT-PCR analysis of ftsA | GACGGAAGCCGAATTTGACG |
| Q-ftsA-R | CCACGTTGTGCCATCACTTC | |
| Q-trmFO-F | qRT-PCR analysis of trmFO | CAGCGGGTCAGCTTAAAACC |
| Q-trmFO-R | CGCGCTTGAATCTTGGTCTG | |
| Q-murB-F | qRT-PCR analysis of murB | GCCGGGCGTGTTGATTATTG |
| Q-murB-R | TTTTGCGGAACTTGGCATCC | |
| Q-fabZ1-F | qRT-PCR analysis of fabZ1 | GGTCATTACGCGACGTTTGG |
| Q-fabZ1-R | AAGGCCCAGCGGTTAGAATC | |
| Q-fabH2-F | qRT-PCR analysis of fabH2 | CTGCGACGGCTTGTTTAGTG |
| Q-fabH2-R | TTTGGTATTGCCCACTGCGA | |
| Q-fabH1-F | qRT-PCR analysis of fabH1 | CGCGACAGGGTTATCAGCAA |
| Q-fabH1-R | TCAGTCTCCGCAACTATGCT | |
| Q-accB1-F | qRT-PCR analysis of accB1 | TCCTGCTAAGATGGTCACGC |
| Q-accB1-R | TAGTCGTTGGCCGTTCACTG | |
| Q-tagE6-F | qRT-PCR analysis of tagE6 | ACCCATGGCTTTGGTGTTTTG |
| Q-tagE6-R | GCTTGCGGTTTAGCTTGAGT | |
| Q-fum-F | qRT-PCR analysis of fum | CCATCAAGACCCTAGCAGCC |
| Q-fum-R | GGGTTCGTTAGCCGGAATGT | |
| Q-tmcA-F | qRT-PCR analysis of tmcA | TTCCAAAAGCCCACTCCGTT |
| Q-tmcA-R | GCGGTTATAGCGGTCTGGTT | |
| Q-ribD-F | qRT-PCR analysis of ribD | GTGGCGACGTACCAAAATCC |
| Q-ribD-R | ACATCCACCTCAGCATGGTC | |
| Q-lplA1-F | qRT-PCR analysis of lplA1 | CTCGGTTGATGACGGCTTGA |
| Q-lplA1-R | TTTAGCGATGGCCTCATGGT | |
| Q-lplA2-F | qRT-PCR analysis of lplA2 | TTAACGCCCCCTGCATCATT |
| Q-lplA2-R | ATAAACCGCTCCACCACCAG | |
| Q-racD-F | qRT-PCR analysis of racD | GACCGTTACCCATCGTGACC |
| Q-racD-R | TGCACATCCGCTTCCAATGA | |
| Q-clpE1-F | qRT-PCR analysis of clpE1 | GGTGAAGCTGGGGTTGGTAA |
| Q-clpE1-R | TGAACAAGGGAAGCCACGTC | |
| Q-murD-F | qRT-PCR analysis of murD | CGCAAATCCTAGCGGGAGAG |
| Q-murD-R | AGCTTTACCGGCATTTGTGC | |
| Q-dak1A-F | qRT-PCR analysis of dak1A | GGAGTTGCTGGCACGATTTT |
| Q-dak1A-R | AGCAACCCCAATCGTGTGAA | |
| Q-acpP-F | qRT-PCR analysis of acpP | ACGATGACAACGAACTTTACCG |
| Q-acpP-R | ATCACCAACCGTGGCTAACG | |
| Q-fabD-F | qRT-PCR analysis of fabD | ACCCCCTTTAGCCAGGAAAC |
| Q-fabD-R | AAACTATGCCCCGGTCCAAC | |
| Q-fabG1-F | qRT-PCR analysis of fabG1 | GGCAAGCATTAACGGCTGAA |
| Q-fabG1-R | GAGTCCATCCAAGCGACCAA | |
| Q-fabF-F | qRT-PCR analysis of fabF | GTATTGCAGGGTTTGCGTCC |
| Q-fabF-R | TAAGACTAACGTCGCACCCC | |
| Q-accB2-F | qRT-PCR analysis of accB2 | ACGGATACGGAAGCGATGAC |
| Q-accB2-R | TGTGAACGCGCTGAATTTGT | |
| Q-FabZ2-F | qRT-PCR analysis of fabZ2 | CCGTATCGTAGCGGTCCAAC |
| Q-FabZ2-R | GCCGAATCATCGTTCTTCCG | |
| Q-accC1-F | qRT-PCR analysis of accC1 | CCGGCGAAGCATATCGAAGT |
| Q-accC1-R | GACCGCACATGGACTTTCCT | |
| Q-accD1-F | qRT-PCR analysis of accD1 | TATGTTGAGCGAGCCACGAG |
| Q-accD1-R | CCAATAAGGTTTCCGCCCGT | |
| Q-accA1-F | qRT-PCR analysis of accA1 | TCATGGTGATCGTCAACGGG |
| Q-accA1-R | GCCAAAATGCCGAGCTTGAT | |
| Q-FabI-F | qRT-PCR analysis of fabI | CAGTATTGCCTGGGGGTGTA |
| Q-FabI-R | CACAGGCAATTAACGGCACA | |
| Q-sfp-F | qRT-PCR analysis of sfp | CGGTGCAAGATCGTTATCGG |
| Q-sfp-R | TCAAGAACCTCAACACGGCT | |
| murD-Q-F | Transcriptional unit analysis of murD, murG, and ftsQ gene cluster | GCTCTAGAGATGAAGTCGGTTGAGCAGTATCGTA |
| murD-Q-R | AAAACTGCAGTCAATTATTCTGATTCTTGAACGG | |
| tagE6-5-F | Transcriptional unit analysis of tagE6 and tagE5 gene cluster | GCTCTAGAGATGTATTATTTTGTGAATACCAGCATT |
| tagE6-5-R | AAAACTGCAGTCACTGAACAACCGGCTGCCACT | |
| fabZ1-accC1-F | Transcriptional unit analysis of fab gene cluster | TCCAACATGGGAACGGTGAGTTGTCAG |
| fabZ1-accC1-R | CACTGAATAGACAGCGACCGCTTTAATCC | |
| accC1-sfp-F | GTTCAAACGAACCGGATCTTCTTAGCAGG | |
| accC1-sfp-R | TTGCTGAATTAATTGGCGACTCACGGT |
F, forward; R, reverse.
DNase I footprinting assays.
DNase I footprinting assays were conducted according to a previous report (37), with some modifications. As a DNA probe, the spacer sequence between fabZ1 and its upstream gene (PfabZ1) was amplified by PCR using the fluorescently labeled primers FAM-PfabZ1-F [5′-(6-FAM)-AGCGTTGCCGTTTAGGTTAAAAGTTATGGG-3′] and HEX–PfabZ1-R [5′-(HEX)-AGTTAGTCGCTCCTTTCACGGCGTTTG-3′]. To confirm the binding sites, two binding sequences from PfabZ1 were inserted into the fragment of the 16S rRNA gene (F0), generating fragment 1 (F1) and fragment 2 (F2). Their binding abilities with AcrR were determined by EMSA.
RT-PCR.
RT-PCR was performed to analyze transcription units using primers (see Table 1) designed to amplify across the intergenic regions of neighboring genes. RNA samples were extracted from cells of L. plantarum NF92. cDNA was used as the template for PCR amplification. Genomic DNA was used as the positive control to verify the amplicon size, and DNase I-treated total RNA was used as a negative control to rule out possible contamination of the RNA sample with genomic DNA.
FAME analysis.
Preparation and analysis of FAMEs were performed according to a previous report (63), with some modifications. L. plantarum NF92 and its derivatives (the ΔacrR and P-acrR strains) were cultured in MRS with 9% (vol/vol) ethanol, independently. Exponential-phase cells were harvested and washed with phosphate-buffered saline twice. After freezing for 24 h at –80°C, the samples were immediately freeze-dried in a freeze dryer with a condenser temperature at −96°C and a chamber pressure of <0.20 hPa for 48 h. Portions (150 mg) of dried cells were saponified with 3 ml of 1 mol/liter NaOH-methanol (MeOH) at 70°C for 10 min and methylated with 6 ml of 10% H2SO4-MeOH at 70°C for 15 min, and then FAMEs were extracted by using n-hexane. A gas chromatograph (Agilent, 7890A) and a 5975C mass spectrometer with an HP-5 MS capillary column (30 m by 0.25 mm by 0.25-μm film) were used to analyze the FAMEs. Fatty acids were identified according to the NIST database. Relative quantification of each peak was performed by using benzoic acid as an internal standard (IS). For quantification, the area ratios of a protonated molecule of a given FAME versus the IS were calculated.
Measurement of membrane fluidity.
The membrane fluidity of L. plantarum NF92 was investigated by fluorescence anisotropy as previously described (64), with some modifications. Briefly, cells from WT, ΔacrR, P-acrR, WT+, and acrR+ strains grown in MRS containing 9% (vol/vol) ethanol were harvested at exponential phase. The samples were incubated for 1 h at 37°C with TMA-DPH at a final concentration of 5 μM. The fluorescence anisotropy was measured at 37°C using an F-7000 spectrofluorometer (Hitachi, Tokyo, Japan) with excitation at 360 nm and emission at 430 nm using 5- and 5-nm slits, respectively, and a 3-s integration time. Anisotropy values (r) were calculated according to the following equation:
where I is the corrected fluorescence intensity and subscripts V and H indicate the values obtained with vertical or horizontal orientation, respectively, of the excitation polarizer and emission analyzer (in that order). In these experiments, decreases in the degree of fluorescence anisotropy reflected increases in the fluidity of the lipid bilayer, which controls or alters the mobility of TMA-DPH in the membrane.
Statistical analysis.
Determinations were performed from three independent cultures of each bacterial strain. A one-way analysis of variance was performed by OriginPro 8.0 to test whether there were any significant differences in RT-qPCR results, fatty acid compositions, and membrane fluidities. If the P value was <0.05, the differences were considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (grant 31570114), the Special Fund for Agro-scientific Research in the Public Interest (grant 201503134), and the Guangxi Major Science and Technology Project (grant AA18118041).
J. Zhong and K. Teng were involved in the conception and design of the study and manuscript revision. X. Yang drafted the manuscript. X. Yang, L. Li, R. Su, and J. Zhang were involved in the acquisition, analysis, and interpretation of the data. G. Ai analyzed the fatty acid methyl esters. All authors discussed the results and reviewed the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01690-19.
REFERENCES
- 1.Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N, Shakhova V, Grigoriev I, Lou Y, Rohksar D, Lucas S, Huang K, Goodstein DM, Hawkins T, Plengvidhya V, Welker D, Hughes J, Goh Y, Benson A, Baldwin K, Lee JH, Díaz-Muñiz I, Dosti B, Smeianov V, Wechter W, Barabote R, Lorca G, Altermann E, Barrangou R, Ganesan B, Xie Y, Rawsthorne H, Tamir D, Parker C, Breidt F, Broadbent J, Hutkins R, O’Sullivan D, Steele J, Unlu G, Saier M, Klaenhammer T, Richardson P, Kozyavkin S, Weimer B, Mills D. 2006. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci U S A 103:15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rhee SJ, Lee JE, Lee CH. 2011. Importance of lactic acid bacteria in Asian fermented foods. Microb Cell Fact 10(Suppl 1):S5. doi: 10.1186/1475-2859-10-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miranda-Castilleja DE, Martínez-Peniche RÁ, Aldrete-Tapia JA, Soto-Muñoz L, Iturriaga MH, Pacheco-Aguilar JR, Arvizu-Medrano SM. 2016. Distribution of native lactic acid bacteria in wineries of Queretaro, Mexico, and their resistance to wine-like conditions. Front Microbiol 7:1769. doi: 10.3389/fmicb.2016.01769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang C, Cui Y, Qu X. 2018. Mechanisms and improvement of acid resistance in lactic acid bacteria. Arch Microbiol 200:195–201. doi: 10.1007/s00203-017-1446-2. [DOI] [PubMed] [Google Scholar]
- 5.Su J, Wang T, Li YY, Li J, Zhang Y, Wang Y, Wang H, Li H. 2015. Antioxidant properties of wine lactic acid bacteria: Oenococcus oeni. Appl Microbiol Biotechnol 99:5189–5202. doi: 10.1007/s00253-015-6425-4. [DOI] [PubMed] [Google Scholar]
- 6.Miyoshi A, Rochat T, Gratadoux J-J, Le Loir Y, Oliveira SC, Langella P, Azevedo V. 2003. Oxidative stress in Lactococcus lactis. Genet Mol Res 2:348–359. [PubMed] [Google Scholar]
- 7.Rincé A, Le Breton Y, Verneuil N, Giard J-C, Hartke A, Auffray Y. 2003. Physiological and molecular aspects of bile salt response in Enterococcus faecalis. Int J Food Microbiol 88:207–213. doi: 10.1016/s0168-1605(03)00182-x. [DOI] [PubMed] [Google Scholar]
- 8.Chen MJ, Tang HY, Chiang ML. 2017. Effects of heat, cold, acid and bile salt adaptations on the stress tolerance and protein expression of kefir-isolated probiotic Lactobacillus kefiranofaciens M1. Food Microbiol 66:20–27. doi: 10.1016/j.fm.2017.03.020. [DOI] [PubMed] [Google Scholar]
- 9.Bron PA, Molenaar D, de Vos WM, Kleerebezem M. 2006. DNA micro-array-based identification of bile-responsive genes in Lactobacillus plantarum. J Appl Microbiol 100:728–738. doi: 10.1111/j.1365-2672.2006.02891.x. [DOI] [PubMed] [Google Scholar]
- 10.Papadimitriou K, Alegria A, Bron PA, de Angelis M, Gobbetti M, Kleerebezem M, Lemos JA, Linares DM, Ross P, Stanton C, Turroni F, van Sinderen D, Varmanen P, Ventura M, Zuniga M, Tsakalidou E, Kok J. 2016. Stress physiology of lactic acid bacteria. Microbiol Mol Biol Rev 80:837–890. doi: 10.1128/MMBR.00076-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shan Y, Man CX, Han X, Li L, Guo Y, Deng Y, Li T, Zhang LW, Jiang YJ. 2015. Evaluation of improved gamma-aminobutyric acid production in yogurt using Lactobacillus plantarum NDC75017. J Dairy Sci 98:2138–2149. doi: 10.3168/jds.2014-8698. [DOI] [PubMed] [Google Scholar]
- 12.Seddik HA, Bendali F, Gancel F, Fliss I, Spano G, Drider D. 2017. Lactobacillus plantarum and its probiotic and food potentialities. Probiotics Antimicrob Proteins 9:111–122. doi: 10.1007/s12602-017-9264-z. [DOI] [PubMed] [Google Scholar]
- 13.Olguin N, Champomier-Verges M, Anglade P, Baraige F, Cordero-Otero R, Bordons A, Zagorec M, Reguant C. 2015. Transcriptomic and proteomic analysis of Oenococcus oeni PSU-1 response to ethanol shock. Food Microbiol 51:87–95. doi: 10.1016/j.fm.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Auesukaree C. 2017. Molecular mechanisms of the yeast adaptive response and tolerance to stresses encountered during ethanol fermentation. J Biosci Bioeng 124:133–142. doi: 10.1016/j.jbiosc.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 15.Pang X-N, Han B-Z, Huang X-N, Zhang X, Hou L-F, Cao M, Gao L-J, Hu G-H, Chen J-Y. 2018. Effect of the environment microbiota on the flavour of light-flavour Baijiu during spontaneous fermentation. Sci Rep 8:3396. doi: 10.1038/s41598-018-21814-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cappello MS, Zapparoli G, Logrieco A, Bartowsky EJ. 2017. Linking wine lactic acid bacteria diversity with wine aroma and flavour. Int J Food Microbiol 243:16–27. doi: 10.1016/j.ijfoodmicro.2016.11.025. [DOI] [PubMed] [Google Scholar]
- 17.Onetto CA, Bordeu E. 2015. Pre-alcoholic fermentation acidification of red grape must using Lactobacillus plantarum. Antonie Van Leeuwenhoek 108:1469–1475. doi: 10.1007/s10482-015-0602-4. [DOI] [PubMed] [Google Scholar]
- 18.Rouse S, van Sinderen D. 2008. Bioprotective potential of lactic acid bacteria in malting and brewing. J Food Prot 71:1724–1733. doi: 10.4315/0362-028x-71.8.1724. [DOI] [PubMed] [Google Scholar]
- 19.Hong X, Chen J, Liu L, Wu H, Tan H, Xie G, Xu Q, Zou H, Yu W, Wang L, Qin N. 2016. Metagenomic sequencing reveals the relationship between microbiota composition and quality of Chinese rice wine. Sci Rep 6:26621. doi: 10.1038/srep26621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.G-Alegría EL, Ruiz JI, Sáenz J, Fernández E, Zarazaga M, Dizy M, Torres C, Ruiz-Larrea F. 2004. High tolerance of wild Lactobacillus plantarum and Oenococcus oeni strains to lyophilization and stress environmental conditions of acid pH and ethanol. FEMS Microbiol Lett 230:53–61. doi: 10.1016/S0378-1097(03)00854-1. [DOI] [PubMed] [Google Scholar]
- 21.van Bokhorst-van de Veen H, Abee T, Tempelaars M, Bron PA, Kleerebezem M, Marco ML. 2011. Short- and long-term adaptation to ethanol stress and its cross-protective consequences in Lactobacillus plantarum. Appl Environ Microbiol 77:5247–5256. doi: 10.1128/AEM.00515-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SG. 2012. Proteomic analysis of proteins increased or reduced by ethanol of Lactobacillus plantarum ST4 isolated from Makgeolli, traditional Korean rice wine. J Microbiol Biotechnol 22:516–525. doi: 10.4014/jmb.1109.09012. [DOI] [PubMed] [Google Scholar]
- 23.Bravo-Ferrada BM, Brizuela N, Gerbino E, Gomez-Zavaglia A, Semorile L, Tymczyszyn EE. 2015. Effect of protective agents and previous acclimation on ethanol resistance of frozen and freeze-dried Lactobacillus plantarum strains. Cryobiology 71:522–528. doi: 10.1016/j.cryobiol.2015.10.154. [DOI] [PubMed] [Google Scholar]
- 24.Yuan Y, Bi C, Nicolaou SA, Zingaro KA, Ralston M, Papoutsakis ET. 2014. Overexpression of the Lactobacillus plantarum peptidoglycan biosynthesis murA2 gene increases the tolerance of Escherichia coli to alcohols and enhances ethanol production. Appl Microbiol Biotechnol 98:8399. doi: 10.1007/s00253-014-6004-0. [DOI] [PubMed] [Google Scholar]
- 25.Bravo-Ferrada BM, Gonçalves S, Semorile L, Santos NC, Tymczyszyn EE, Hollmann A. 2015. Study of surface damage on cell envelope assessed by AFM and flow cytometry of Lactobacillus plantarum exposed to ethanol and dehydration. J Appl Microbiol 118:1409–1417. doi: 10.1111/jam.12796. [DOI] [PubMed] [Google Scholar]
- 26.Fiocco D, Capozzi V, Goffin P, Hols P, Spano G. 2007. Improved adaptation to heat, cold, and solvent tolerance in Lactobacillus plantarum. Appl Microbiol Biotechnol 77:909–915. doi: 10.1007/s00253-007-1228-x. [DOI] [PubMed] [Google Scholar]
- 27.Bravo-Ferrada BM, Gomez-Zavaglia A, Semorile L, Tymczyszyn EE. 2015. Effect of the fatty acid composition of acclimated oenological Lactobacillus plantarum on the resistance to ethanol. Lett Appl Microbiol 60:155–161. doi: 10.1111/lam.12350. [DOI] [PubMed] [Google Scholar]
- 28.Cuthbertson L, Nodwell JR. 2013. The TetR family of regulators. Microbiol Mol Biol Rev 77:440–475. doi: 10.1128/MMBR.00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang M, Gao CH, Hu J, Zhao L, Huang Q, He ZG. 2015. InbR, a TetR family regulator, binds with isoniazid and influences multidrug resistance in Mycobacterium bovis BCG. Sci Rep 5:13969. doi: 10.1038/srep13969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou JH, Hu YH, Zhang M, Sun L. 2009. Identification and characterization of the AcrR/AcrAB system of a pathogenic Edwardsiella tarda strain. J Gen Appl Microbiol 55:191–199. doi: 10.2323/jgam.55.191. [DOI] [PubMed] [Google Scholar]
- 31.Gristwood T, Fineran PC, Everson L, Salmond GP. 2008. PigZ, a TetR/AcrR family repressor, modulates secondary metabolism via the expression of a putative four-component resistance-nodulation-cell-division efflux pump, ZrpADBC, in Serratia sp. ATCC 39006. Mol Microbiol 69:418–435. doi: 10.1111/j.1365-2958.2008.06291.x. [DOI] [PubMed] [Google Scholar]
- 32.Deng W, Li C, Xie J. 2013. The underling mechanism of bacterial TetR/AcrR family transcriptional repressors. Cell Signal 25:1608–1613. doi: 10.1016/j.cellsig.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 33.Chen M, Wang P, Xie Z. 2018. A complex mechanism involving LysR and TetR/AcrR that regulates iron scavenger biosynthesis in Pseudomonas donghuensis HYS. J Bacteriol 200:e00087-18. doi: 10.1128/JB.00087-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luhe AL, Gerken H, Tan L, Wu J, Zhao H. 2012. Alcohol tolerance of Escherichia coli acrR and marR regulatory mutants. J Mol Catal B Enzym Enzymatic 76:89–93. doi: 10.1016/j.molcatb.2011.11.013. [DOI] [Google Scholar]
- 35.Lee JO, Cho KS, Kim OB. 2014. Overproduction of AcrR increases organic solvent tolerance mediated by modulation of SoxS regulon in Escherichia coli. Appl Microbiol Biotechnol 98:8763–8773. doi: 10.1007/s00253-014-6024-9. [DOI] [PubMed] [Google Scholar]
- 36.Ma D, Alberti M, Lynch C, Nikaido H, Hearst JE. 1996. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol Microbiol 19:101–112. doi: 10.1046/j.1365-2958.1996.357881.x. [DOI] [PubMed] [Google Scholar]
- 37.Yang X, Teng K, Su R, Li L, Zhang T, Fan K, Zhang J, Zhong J. 2019. AcrR and Rex control mannitol and sorbitol utilization through their cross-regulation of aldehyde-alcohol dehydrogenase (AdhE) in Lactobacillus plantarum. Appl Environ Microbiol 85:e02035-18. doi: 10.1128/AEM.02035-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chapot-Chartier MP, Kulakauskas S. 2014. Cell wall structure and function in lactic acid bacteria. Microb Cell Fact 13(Suppl 1):S9. doi: 10.1186/1475-2859-13-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujita Y, Matsuoka H, Hirooka K. 2007. Regulation of fatty acid metabolism in bacteria. Mol Microbiol 66:829–839. doi: 10.1111/j.1365-2958.2007.05947.x. [DOI] [PubMed] [Google Scholar]
- 40.Diomande SE, Nguyen-The C, Guinebretiere MH, Broussolle V, Brillard J. 2015. Role of fatty acids in Bacillus environmental adaptation. Front Microbiol 6:813. doi: 10.3389/fmicb.2015.00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haddaji N, Mahdhi AK, Ismaiil MB, Bakhrouf A. 2017. Effect of environmental stress on cell surface and membrane fatty acids of Lactobacillus plantarum. Arch Microbiol 199:1243–1250. doi: 10.1007/s00203-017-1395-9. [DOI] [PubMed] [Google Scholar]
- 42.Zhang YM, Marrakchi H, Rock CO. 2002. The FabR (YijC) transcription factor regulates unsaturated fatty acid biosynthesis in Escherichia coli. J Biol Chem 277:15558–15565. doi: 10.1074/jbc.M201399200. [DOI] [PubMed] [Google Scholar]
- 43.Schujman GE, Paoletti L, Grossman AD, de Mendoza D. 2003. FapR, a bacterial transcription factor involved in global regulation of membrane lipid biosynthesis. Dev Cell 4:663–672. doi: 10.1016/S1534-5807(03)00123-0. [DOI] [PubMed] [Google Scholar]
- 44.Eckhardt TH, Skotnicka D, Kok J, Kuipers OP. 2013. Transcriptional regulation of fatty acid biosynthesis in Lactococcus lactis. J Bacteriol 195:1081–1089. doi: 10.1128/JB.02043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu YJ, Rock CO. 2006. Transcriptional regulation of fatty acid biosynthesis in Streptococcus pneumoniae. Mol Microbiol 59:551–566. doi: 10.1111/j.1365-2958.2005.04951.x. [DOI] [PubMed] [Google Scholar]
- 46.Arabolaza A, D’Angelo M, Comba S, Gramajo H. 2010. FasR, a novel class of transcriptional regulator, governs the activation of fatty acid biosynthesis genes in Streptomyces coelicolor. Mol Microbiol 78:47–63. doi: 10.1111/j.1365-2958.2010.07274.x. [DOI] [PubMed] [Google Scholar]
- 47.Mondino S, Gago G, Gramajo H. 2013. Transcriptional regulation of fatty acid biosynthesis in mycobacteria. Mol Microbiol 89:372–387. doi: 10.1111/mmi.12282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nickel J, Irzik K, van Ooyen J, Eggeling L. 2010. The TetR-type transcriptional regulator FasR of Corynebacterium glutamicum controls genes of lipid synthesis during growth on acetate. Mol Microbiol 78:253–265. doi: 10.1111/j.1365-2958.2010.07337.x. [DOI] [PubMed] [Google Scholar]
- 49.Sharma SC, Raj D, Forouzandeh M, Bansal MP. 1996. Salt-induced changes in lipid composition and ethanol tolerance in Saccharomyces cerevisiae. Appl Biochem Biotechnol 56:189–195. doi: 10.1007/BF02786949. [DOI] [PubMed] [Google Scholar]
- 50.Alexandre H, Rousseaux I, Charpentier C. 1994. Relationship between ethanol tolerance, lipid composition and plasma membrane fluidity in Saccharomyces cerevisiae and Kloeckera apiculata. FEMS Microbiol Lett 124:17–22. doi: 10.1111/j.1574-6968.1994.tb07255.x. [DOI] [PubMed] [Google Scholar]
- 51.Couto JA, Rozès N, Hogg T. 1996. Ethanol-induced changes in the fatty acid composition of Lactobacillus hilgardii, its effects on plasma membrane fluidity and relationship with ethanol tolerance. J Appl Bacteriol 81:126–132. doi: 10.1111/j.1365-2672.1996.tb04489.x. [DOI] [Google Scholar]
- 52.Sinensky M. 1974. Homeoviscous adaptation: a homeostatic process that regulates the viscosity of membrane lipids in Escherichia coli. Proc Natl Acad Sci U S A 71:522–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.You KM, Rosenfield CL, Knipple DC. 2003. Ethanol tolerance in the yeast Saccharomyces cerevisiae is dependent on cellular oleic acid content. Appl Environ Microbiol 69:1499–1503. doi: 10.1128/aem.69.3.1499-1503.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grandvalet C, Assad-Garcia JS, Chu-Ky S, Tollot M, Guzzo J, Gresti J, Tourdot-Marechal R. 2008. Changes in membrane lipid composition in ethanol- and acid-adapted Oenococcus oeni cells: characterization of the cfa gene by heterologous complementation. Microbiology 154:2611. doi: 10.1099/mic.0.2007/016238-0. [DOI] [PubMed] [Google Scholar]
- 55.Velly H, Bouix M, Passot S, Penicaud C, Beinsteiner H, Ghorbal S, Lieben P, Fonseca F. 2015. Cyclopropanation of unsaturated fatty acids and membrane rigidification improve the freeze-drying resistance of Lactococcus lactis subsp. lactis TOMSC161. Appl Microbiol Biotechnol 99:907–918. doi: 10.1007/s00253-014-6152-2. [DOI] [PubMed] [Google Scholar]
- 56.Popis MC, Blanco S, Frye M. 2016. Posttranscriptional methylation of transfer and ribosomal RNA in stress response pathways, cell differentiation, and cancer. Curr Opin Oncol 28:65–71. doi: 10.1097/CCO.0000000000000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamagami R, Yamashita K, Nishimasu H, Tomikawa C, Ochi A, Iwashita C, Hirata A, Ishitani R, Nureki O, Hori H. 2012. The tRNA recognition mechanism of folate/FAD-dependent tRNA methyltransferase (TrmFO). J Biol Chem 287:42480–42494. doi: 10.1074/jbc.M112.390112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hamdane D, Argentini M, Cornu D, Myllykallio H, Skouloubris S, Hui-Bon-Hoa G, Golinelli-Pimpaneau B. 2011. Insights into folate/FAD-dependent tRNA methyltransferase mechanism: role of two highly conserved cysteines in catalysis. J Biol Chem 286:36268–36280. doi: 10.1074/jbc.M111.256966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martinez MR, Dias TB, Natov PS, Zachara NE. 2017. Stress-induced O-GlcNAcylation: an adaptive process of injured cells. Biochem Soc Trans 45:237–249. doi: 10.1042/BST20160153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mehaffy C, Belisle JT, Dobos KM. 2019. Mycobacteria and their sweet proteins: an overview of protein glycosylation and lipoglycosylation in M. tuberculosis. Tuberculosis (Edinb) 115:1–13. doi: 10.1016/j.tube.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 61.Lee I-C, van Swam II, Tomita S, Morsomme P, Rolain T, Hols P, Kleerebezem M, Bron PA. 2014. GtfA and GtfB are both required for protein O-glycosylation in Lactobacillus plantarum. J Bacteriol 196:1671–1682. doi: 10.1128/JB.01401-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shao Y, Zhang W, Guo H, Pan L, Zhang H, Sun T. 2015. Comparative studies on antibiotic resistance in Lactobacillus casei and Lactobacillus plantarum. Food Control 50:250–258. doi: 10.1016/j.foodcont.2014.09.003. [DOI] [Google Scholar]
- 63.Yang X, Teng K, Zhang J, Wang F, Zhang T, Ai G, Han P, Bai F, Zhong J. 2017. Transcriptome responses of Lactobacillus acetotolerans F28 to a short and long term ethanol stress. Sci Rep 7:2650. doi: 10.1038/s41598-017-02975-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen C, Zhao G, Chen W, Guo B. 2015. Metabolism of fructooligosaccharides in Lactobacillus plantarum ST-III via differential gene transcription and alteration of cell membrane fluidity. Appl Environ Microbiol 81:7697–7707. doi: 10.1128/AEM.02426-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.