Abstract
The persistence of an invasive species is influenced by its reproductive ecology, and a successful control program must operate on this premise. However, the reproductive ecology of invasive species may be enigmatic due to factors that also limit their management, such as cryptic coloration and behavior. We explored the mating and reproductive ecology of the invasive Brown Treesnake (BTS: Boiga irregularis) by reconstructing a multigenerational genomic pedigree based on 654 single nucleotide polymorphisms for a geographically closed population established in 2004 on Guam (N = 426). The pedigree allowed annual estimates of individual mating and reproductive success to be inferred for snakes in the study population over a 14‐year period. We then employed generalized linear mixed models to gauge how well phenotypic and genomic data could predict sex‐specific annual mating and reproductive success. Average snout–vent length (SVL), average body condition index (BCI), and trappability were significantly related to annual mating success for males, with average SVL also related to annual mating success for females. Male and female annual reproductive success was positively affected by SVL, BCI, and trappability. Surprisingly, the degree to which individuals were inbred had no effect on annual mating or reproductive success. When juxtaposed with current control methods, these results indicate that baited traps, a common interdiction tool, may target fecund BTS in some regards but not others. Our study emphasizes the importance of reproductive ecology as a focus for improving BTS control and promotes genomic pedigree reconstruction for such an endeavor in this invasive species and others.
Keywords: Boiga irregularis, Brown Treesnake, ddRAD, parentage, reproductive ecology, single nucleotide polymorphisms
Our study details the use of genomic tools (i.e., double‐digest restriction site‐associated DNA sequencing) to develop novel genomic resources (>6,000 SNPs) for a high‐profile and invasive vertebrate pest (i.e., Brown Treesnake). We used a subset of these SNPs to reconstruct a multigenerational genomic pedigree for a wild population of Brown Treesnakes from which we inferred patterns and predictors of annual mating and reproductive success, and we juxtaposed phenotypes of individuals with high annual mating and reproductive success against those of commonly trapped individuals to assess existing management efficacy. Our results promote the use of genomic pedigree reconstruction to elucidate the reproductive ecology of invasive species with cryptic coloration and behaviors, so as to inform and improve management.

1. INTRODUCTION
Species declines and extinctions are driven by multiple factors, the most egregious being anthropogenic habitat alteration (Travis, 2003), species introductions (Vitousek, Mooney, Lubchenco, & Melillo, 1997), and climate change (Thomas et al., 2004). Introduced species, in particular, have provoked negative responses in a variety of ecological contexts: community assembly (Sanders, Gotelli, Heller, & Gordon, 2003), competitive exclusion/niche displacement (Mooney & Cleland, 2001), interspecific hybridization/introgression (Muhlfeld et al., 2009), and even natural selection (Strauss, Lau, & Carroll, 2006). Introductions are deemed second only to human‐induced habitat loss as a major cause of species endangerment (Simberloff, 2001), yet are the primary cause of global avian extinctions (Clavero & García‐Berthou, 2005). Within a more social context, invasive species also impact global economies (Olson, 2006) and human health (Juliano & Lounibos, 2005).
Reproductive ecology is a key element in the establishment and persistence of an invasive species. Those that exhibit high fecundity not only increase their probability of establishment but also mitigate the potential for an Allee effect and/or issues that stem from demographic and environmental stochasticity (Lockwood, Hoopes, & Marchetti, 2013). Following invasion, a species also must be able to persist and cope with changes in an alien environment. These responses are mediated through the mating system (e.g., selfing, monogamy, promiscuity), its characteristics (e.g., traits that promote mating and reproductive success), and associated reproductive phenomena (e.g., inbreeding, multiple paternity) that influence genetic variation and evolutionary potential (Ellegren & Galtier, 2016).
Two reproductive ecology parameters critical to development of successful invasive species control are the number of offspring that an individual produces annually (referred to here as annual reproductive success; ARS) and the number of mates with which an individual produces offspring annually (referred to here as annual mating success; AMS). Quantification of average ARS in an invasive species yields an important estimate of the annual ability of a population to replenish itself. Further, when phenotypes associated with high ARS can be identified, these can be juxtaposed with phenotypes of individuals removed by existing control methods to gauge the efficacy of management and identify areas for improvement. Differently, the number of mates with which an individual produces offspring can have implications for maintenance of genetic diversity (Ellegren & Galtier, 2016), as production of offspring with multiple mating partners increases the overall genetic variation present in the resulting offspring (Foerster, Delhey, Johnsen, Lifjeld, & Kempenaers, 2003). In this regard, juxtaposition of phenotypes associated with high AMS against those of individuals targeted by control can also provide information regarding the potential effect of control methods on genetic variation and evolutionary potential over time.
Here, we applied these concepts to explore the reproductive ecology of the Brown Treesnake (Boiga irregularis; BTS), introduced to Guam from the island of Manus in the Admiralty Archipelago during or shortly after World War II and subsequently deemed one of the “world's worst” invasive species (Lowe, Browne, Boudjelas, & De Poorter, 2000). Despite limited propagule pressure (≤10 individuals; Richmond, Wood, Stanford, & Fisher, 2014), its population size reached two million by the 1980s (Fritts & Rodda, 1998). BTS have since caused considerable ecological changes, to include extirpation/extinction of 10 native bird species (Savidge, 1987) and population declines of endemic nonavian vertebrates (Rodda, Fritts, & Chiszar, 1997). Not surprisingly, this decline in biodiversity has had a cascading effect on community dynamics and structure (Caves, Lambers, Tewksbury, & Rogers, 2013; Mortensen, Dupont, & Olesen, 2008; Rogers, Lambers, Miller, & Tewksbury, 2012). The BTS invasion has also been detrimental to the economy (Perry & Vice, 2009) and has implications for human health (Fritts, McCoid, & Haddock, 1990).
Birth rate is a demographic parameter fundamental to population persistence (Cole, 1954), and thus, an in‐depth understanding of reproductive ecology should be a focus of control efforts for BTS and other invasive species. Not surprisingly, this theme has been amplified in the BTS literature (e.g., Engbring & Fritts, 1988; Jordan & Rodda, 1994; Moore et al., 2005; Rodda, Fritts, McCoid, & Campbell, 1999; Siegel, Aldridge, Clark, Poldemann, & Gribbins, 2009). However, the cryptic behavior of BTS constrains field studies and stymies in‐depth research on its reproduction in the wild (Greene & Mason, 1998; Kahl, Henke, Hall, & Britton, 2012; Mathies, Franklin, & Miller, 2004; Trembath & Fearn, 2008). This is unfortunate in that successful control and eradication hinges on the ability to eliminate breeding individuals more rapidly than they are replenished (Rodda et al., 2002). If phenotypes associated with elevated mating and reproductive success can be so targeted, then the potential for management to achieve this goal is enhanced considerably.
Fortunately, ARS and AMS can be inferred from genomic pedigrees, as reconstructed from DNA samples. However, genetic markers for BTS are limited, particularly with regard to fine‐grained estimates of relatedness [but see Richmond et al. (2014) and Unger et al. (2015)]. This capacity has been expanded as of late through derivation of single nucleotide polymorphisms (SNPs) that are not only cost‐effective but also highly applicable to nonmodel organisms (Ekblom & Galindo, 2011).
Our central goal was to reconstruct a multigenerational genomic pedigree for BTS that would allow patterns of mating and reproductive success to be inferred in the wild. To do so, we juxtaposed genome‐wide SNPs identified from double‐digest restriction site‐associated DNA (ddRAD) libraries against phenotypic and genomic data collected over a 14‐year period for 426 BTS from a geographically closed population on Guam. Predictors of AMS and ARS were identified and the genetic mating system of BTS characterized. The results of this study can be applied to assess the efficacy of existing BTS control and to improve management. In a more general sense, this study highlights the importance of understanding invasive species reproductive ecology in the context of management action and promotes the use of genomic pedigree reconstruction to achieve this goal.
1.1. Hypotheses
We postulated that both sexes are promiscuous (i.e., producing offspring with more than one mating partner per year) as this is the most common snake mating system (Rivas & Burghardt, 2005). We predicted that four factors, each focusing on a different aspect of BTS ecology, were related to AMS and ARS. Specifically, we predicted that individual AMS and ARS would be influenced by the following: (a) snout–vent length (SVL; the length from the tip of the snout to the cloacal vent), (b) body condition index (BCI; a measure of body mass relative to SVL), (c) trappability (a measure of the propensity to enter baited traps; Le Cœur et al., 2015), and (d) the degree to which an individual is inbred.
We predicted an influence of SVL on AMS and ARS because SVL is a trait correlated with AMS (Shine et al., 2000) and ARS in other snakes (Levine et al., 2015). Male snakes with larger SVLs have a competitive advantage over smaller males in combat for gaining priority‐of‐access to females (Duvall & Schuett, 1997; Madsen & Shine, 1993, 1994), and such “combat dances” have been observed among male BTS in a laboratory setting (Greene & Mason, 2000). Differently, females with larger SVLs may appear more attractive to males (Blouin‐Demers, Gibbs, & Weatherhead, 2005), resulting in greater AMS, and their larger body cavities may correlate with an increased capacity to produce eggs (Blouin‐Demers et al., 2005; Brown & Shine, 2007). We expected body condition to be influential for AMS and ARS in that underweight individuals in either sex may lack sufficient energy reserves to search for mates (Lind & Beaupré, 2015), engage in mating and mating‐related activities [e.g., male combat (Shine et al., 2000)], and/or produce offspring (Aubret, Bonnet, Shine, & Lourdais, 2002).
We also postulated that trappability [i.e., the propensity to enter baited traps (Le Cœur et al., 2015)] would impact AMS and ARS for both sexes for two reasons. First, trappability may serve as a proxy for risk‐taking behaviors (Boyer, Réale, Marmet, Pisanu, & Chapuis, 2010; Réale, Gallant, Leblanc, & Festa‐Bianchet, 2000; Wilson, Coleman, Clark, & Biederman, 1993). In this sense, positive correlations exist between boldness/exploratory behavior and trappability across taxa (Biro & Dingemanse, 2009). We predicted that individuals with high trappability would display greater values for AMS and ARS in that they would be more likely to take risks regarding mate searching and/or acquisition. Second, those individuals may also enter traps more often due to enhanced olfactory capabilities that improve their capacity to find baited traps (Shivik, 1998; Shivik & Clark, 1997). This should also promote mating and reproductive success in that olfaction influences mate finding in male BTS (Greene, Stark, & Mason, 2001; Mathies, Levine, Engeman, & Savidge, 2013). Importantly, we recognize the complexity of the relationship between trappability, AMS, and ARS, and thus simply offer a rationale for their association. Finally, we predicted that a negative relationship would be found between AMS/ARS and the degree to which the focal individual is inbred. This would represent an echo of the founder effects manifested by BTS on Guam (Richmond et al., 2014).
2. MATERIALS AND METHODS
2.1. Study site
The study site is a 5‐ha enclosure on Andersen Air Force Base (northern Guam) that was fenced in 2004 to prevent immigration/emigration of BTS (Rodda, Savidge, Tyrrell, Christy, & Ellingson, 2007; Tyrrell et al., 2009). We collected tissue samples (blood, tail clips, and ventral scale clips) from 426 unique individuals (217 females, 207 males, and two of unknown sex) over an eight‐year span (2009–2017). We back‐extrapolated median estimated hatch dates (assuming a hatch size of 350 mm SVL) for each individual from sex‐specific growth rates and SVL at first capture that ranged from years 2002 to 2016. From 2004 to 2018, we captured BTS in baited traps or by hand during nocturnal visual searches along maintained transects. Baited traps deployed in the enclosure were similar to those used in operational control on the island. Over the course of the study, data including SVL, mass, and method of capture were collected from individuals each time they were encountered.
2.2. ddRAD library preparation
We extracted genomic DNA using the QIAamp Fast DNA Tissue Kit (QIAGEN©) and quantified concentrations with a Qubit 2.0 Fluorometer (Invitrogen, Inc.), following manufacturer protocols. We verified the presence of high‐quality genomic DNA (i.e., molecular weight > 10kb) by separating a 5‐µl aliquot of each extract on a 2% agarose gel for 50 m at 100 mV, with visualization via GelGreen on a blue‐light transilluminator (Gel Doc™ EZ Imager; Bio‐Rad). We prepared extracted DNA samples using a ddRAD protocol (Peterson, Weber, Kay, Fisher, & Hoekstra, 2012) subsequently modified in Bangs, Douglas, Mussmann, and Douglas (2018; Appendix S1) for single‐end sequencing (100 bp length) on an Illumina HiSeq 4000.
2.3. Bioinformatics
We inspected fastq files for quality using FastQC (Andrews, 2014). We used the process_radtags module of Stacks 2.0 (Catchen, Amores, Hohenlohe, Cresko, & Postlethwait, 2011; Catchen, Hohenlohe, Bassham, Amores, & Cresko, 2013) to demultiplex reads by individual barcode with default values for score limit (s = 10) and sliding window size (w = 0.15). We clustered raw reads into loci using Stacks 2.0, which uses three main parameters to cluster reads: minimum number of identical sequencing reads to be considered a putative locus (=m), maximum number of nucleotide differences within each locus (stack) per individual (=M), and maximum number of nucleotide differences between individuals at a locus (=n; Catchen et al., 2011). We determined the correct parameters for clustering reads into loci by following published protocols (Paris, Stevens, & Catchen, 2017; Rochette & Catchen, 2017). The correct values of these parameters were revealed by parameter optimization to be as follows: m = 3; M = 2; n = 2 (Appendix S1).
We then used Stacks 2.0 to cluster raw reads from all samples, with 75 selected for catalog construction (Rochette & Catchen, 2017) to span the entire sampling period, include only high‐coverage individuals (mean number of reads/locus = 26.67 ± 9.69), and minimize potential batch effects that could stem from digestion, ligation, and sequencing procedures. We excluded those individuals sequenced more than once for quality control (Appendix S1). Upon completion of the core modules (ustacks, cstacks, sstacks, tsv2bam, gstacks), we used the populations module to retain only those loci present in at least 95% of individuals (r = 0.95). To minimize linkage disequilibrium, we also only retained the first SNP at each locus (‐‐write_single_snp).
2.4. Pedigree reconstruction
We used the R package Sequoia (Huisman, 2017) to iteratively reconstruct a maximum‐likelihood multigenerational pedigree from SNP genotypes, sex data, and estimated birth years. Sequoia is optimized for SNP data sets, jointly considers a variety of alternative relationship categories (e.g., grandparents/grand‐offspring in additional to parents/offspring), and allows the consideration of more than two generations at a time. Due to the longitudinal nature of our study and the presence of multiple overlapping generations of BTS in the study population, this last point is essential as multiple generations of snakes can feasibly produce offspring in the same cohort, thereby making it impossible to declare a priori a clear candidate parent group for each cohort [as is required by other parentage analysis software such as Colony (Jones & Wang, 2010)]. To prepare the SNP data set for pedigree reconstruction, we first used plink 1.9 (Purcell et al., 2007) to test for and discard loci in linkage disequilibrium (LD) and out of Hardy–Weinberg equilibrium (HWE). For LD and HWE tests, we considered all individuals (n = 24) born from 2002 to 2004 (2004 = year of fence construction) as founders. We tested for LD with the ‐‐indep function, evaluating 50 SNP windows, five SNPs at a time, and with a variance inflation factor (VIF) cutoff = 2.
The Sequoia user manual recommends retaining only those loci with a minor allele frequency (MAF) ≥0.3 in the population, and to tweak this parameter value and the level of LD tolerated until a set of 300–700 SNPs is achieved. Rather than compromising the stringency of LD that was tolerated in our data set and risking potential nonindependence of SNPs, we chose a MAF = 0.3 for the population which resulted in 654 SNPs for pedigree reconstruction.
We accomplished initial parentage assignments with the genotype file and a life history file with MaxSibiter = 0. This allowed us to scan the pedigree for obvious errors, as well as for duplicates accidentally retained. We altered the parameter data frame (=Specs) to increase MaxSibshipSize = 100, MaxSibiter = 40, and UseAge=“Extra.” The agepriors file was modified to prevent impossible parentage assignments (e.g., one‐year‐old parents; J. Huisman, personal communication, 9 April 2018). This was accomplished by changing the one‐year‐old prior for maternity from 0.035 to 0.000 and the one‐year‐old prior for paternity from 0.001 to 0.000. All other parameters were kept at default. We constructed the full pedigree by setting the altered parameter file as the SeqList. We ran Sequoia using R v. 3.4.3 (R Core Team, 2013).
We assessed the accuracy of our reconstructed pedigree and the ability of our SNP data set to correctly identify familial relationships in three ways. First, we used Sequoia's EstConf function to calculate confidence probabilities of parentage assignments for dams and sires of known ID (Table S1). We ran simulations for 50 iterations (nSim = 50) and assumed that 40% of parents were not sampled (ParMis = 0.4), and we found all probabilities of parentage assignments to known individuals ranged from 0.93 to 0.99. Second, we regressed pairwise genomic relatedness [estimated from the entire set of SNPs (N = 6,180) using the QG89_avg estimator in the R package irelr (Gonçalves da Silva & Russello, 2011)] on to pairwise pedigree relatedness [estimated from the reconstructed Sequoia pedigree including dummy individuals using the R package Pedantics (Morrissey & Wilson, 2010)]. We found a strong correlation between estimated pairwise genomic relatedness and estimated pairwise pedigree relatedness (Pearson correlation coefficient = 0.71; Figure S1) that was also within the range of correlations previously reported for data sets analyzed with Sequoia (=0.47–0.81; Huisman, 2017). Third, we tested the ability of Sequoia to correctly identify duplicate individuals by running it on a data set with genotypes of 114 individuals that were sequenced twice in different lanes following separate library preparations (Appendix S1), and found that all duplicates were correctly flagged. We ran Sequoia and irelr with R v. 3.4.3, and we ran Pedantics in RStudio (RStudio Team, 2015) with R v. 3.5.0.
We parsed the Pedigree file generated by Sequoia to calculate AMS and ARS for each individual for each calendar year that they were known to be alive, and we included dummy parents assigned by Sequoia in estimates of AMS. Breeding is largely aseasonal in BTS on Guam (Savidge, Qualls, & Rodda, 2007), although there is some evidence that peak copulation occurs in the dry season with hatching occurring in the following wet season (least rain occurs in March; Rodda & Savidge, 2007). Using calendar years as cutoffs for AMS and ARS is thus somewhat arbitrary but consistent among individuals, a fact which should make our analyses more conservative. Fourteen individuals were assigned a known individual as one parent but neither a known nor dummy individual as their other parent. We included these assignments in estimates of AMS and ARS so as to not underestimate ARS, and preliminary analyses without these assignments demonstrated that their inclusion had no effect on the results of downstream analyses.
2.5. Statistical analyses
We fit generalized linear mixed models (GLMMs) for each sex separately to test predictors of AMS and ARS using the glmer function of the R package lme4 (Bates, Maechler, Bolker, & Walker, 2015). We excluded from analyses records for two individuals of unknown sex, as well as annual records for which AMS and ARS > 0 but also for which phenotypic data were incomplete or lacking (=11 annual female records and 10 annual male records). These exclusions removed data collected during year 2014 from analyses. The final data set comprised records of AMS and ARS for years 2004–2013 and 2015–2018.
For both males and females, we analyzed a complete data set that included all annual data records for individuals (hereafter “complete”; data collected during years 2004–2013 and 2015–2018) and a data set filtered by mean annual SVL to only include data collected when individuals were likely adults (hereafter “SVL‐filtered”; data collected during years 2005–2013 and 2015–2018). BTS can mature over a large size range (Savidge et al., 2007), so drawing a clear cutoff by age or SVL at which snakes are deemed sexually mature is, at best, difficult and, at worst, risks biasing the data set by excluding from analyses legitimate adults that matured earlier than is average or including juveniles that matured later. Nevertheless, we recognize that our inclusion of juvenile records in the complete data sets may be problematic, particularly if traits undergo ontogenetic shifts [e.g., change in trappability as BTS mature (Rodda et al., 2007)]. Therefore, we also analyzed sex‐specific data sets that we filtered to include only data collected when individuals were likely adults (=“SVL‐filtered”). Savidge et al. (2007) reported that most females and males reach sexual maturity at SVLs between 910 and 1,025 mm and between 940 and 1,030 mm, respectively. Thus, these SVL‐filtered data sets only included annual records that exceeded these cutoffs (i.e., mean female SVL ≥ 910 mm, mean male SVL ≥ 940 mm). Filtering the data set by mean annual SVL also ensured that only fully trappable snakes were included in analyses of these data (Tyrrell et al., 2009).
We used plots to visualize the structure and distribution of all data sets prior to fitting the models (complete data sets shown in Figures 1, 2, 3, 4). We assayed for the potential presence of interactions among explanatory variables by generating coplots and tested for collinearity among explanatory variables (Zuur, Ieno, & Elphick, 2010). To do so, we visually inspected correlation matrices and calculated VIFs with the R package MCtest (Imdadullah, Aslam, & Altaf, 2016), with no evidence of significant collinearity among explanatory variables [all VIFs < 2 (Zuur et al., 2010)].
Figure 1.
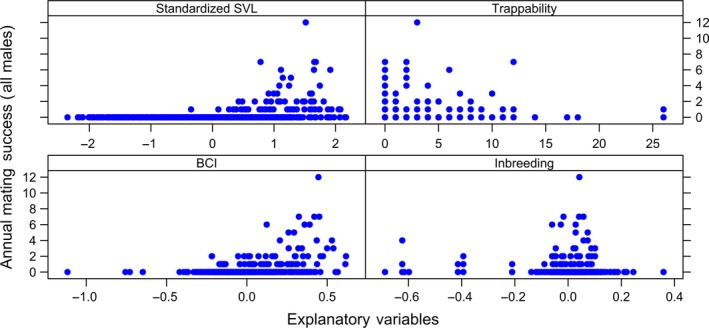
Distribution of four annual measurements with respect to annual mating success (i.e., number of mates with which an individual produced offspring over the course of a calendar year) for male Brown Treesnakes (Boiga irregularis) collected over a 14‐year period from a geographically closed 5‐hectare population on Guam. Annual measurements include standardized average snout–vent length (“Standardized SVL”), number of times the individual was captured in a baited trap (“Trappability”), average body condition index (=BCI), and degree of genome‐wide inbreeding (“Inbreeding”). There were a total of 661 annual observations of 207 males
Figure 2.
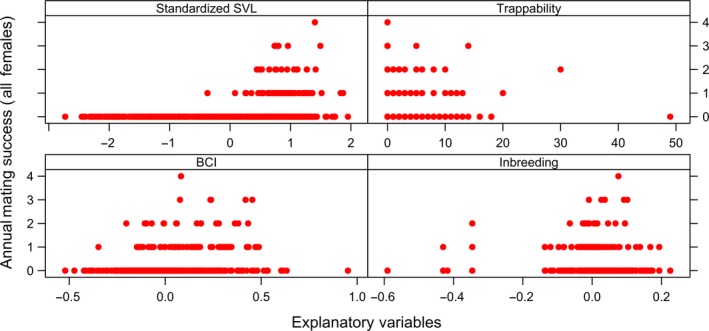
Distribution of four annual measurements with respect to annual mating success (i.e., number of mates with which an individual produced offspring over the course of a calendar year) for female Brown Treesnakes (Boiga irregularis) collected over a 14‐year period from a geographically closed 5‐hectare population on Guam. Annual measurements include standardized average snout–vent length (“Standardized SVL”), number of times the individual was captured in a baited trap (“Trappability”), average body condition index (“BCI”), and degree of genome‐wide inbreeding (“Inbreeding”). There were a total of 735 annual observations of 217 females
Figure 3.
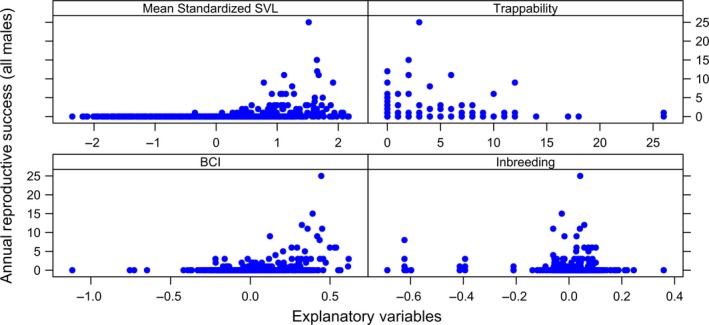
Distribution of four annual measurements with respect to annual reproductive success (i.e., number of offspring an individual produced over the course of a calendar year) for male Brown Treesnakes (Boiga irregularis) collected over a 14‐year period from geographically closed 5‐hectare population on Guam. Annual measurements include standardized average snout–vent length (“Standardized SVL”), number of times the individual was captured in a baited trap (“Trappability”), average body condition index (“BCI”), and degree of genome‐wide inbreeding (“Inbreeding”). There were a total of 661 annual observations of 207 males
Figure 4.
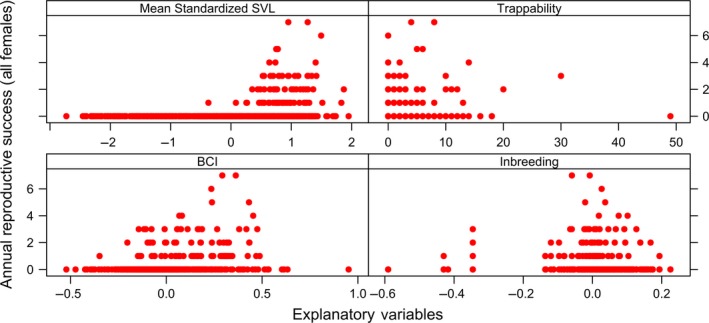
Distribution of four annual measurements with respect to annual reproductive success (i.e., number of offspring an individual produced over the course of a calendar year) for female Brown Treesnakes (Boiga irregularis) collected over a 14‐year period from a geographically closed 5‐hectare population on Guam. Annual measurements include standardized average snout–vent length (“Standardized SVL”), number of times the individual was captured in a baited trap (“Trappability”), average body condition index (“BCI”), and degree of genome‐wide inbreeding (“Inbreeding”). There were a total of 735 annual observations of 217 females
We did not model interactions among explanatory variables in sex‐specific GLMMs of AMS and ARS for several reasons. First, we had no a priori expectation that explanatory variables would interact to influence AMS or ARS (Harrison et al., 2018). Second, visualization of potential interactions with coplots did not indicate strong interactions among predictor variables (Bolker et al., 2009; Zuur & Ieno, 2016; Zuur et al., 2010). Third, we avoided overparameterization of our models by not including interaction terms (Harrison et al., 2018).
We specified a Poisson error distribution with a log‐link for GLMM analyses of the complete and SVL‐filtered sex‐specific data sets because of its utility for count data (Zuur, Ieno, Walker, Savaliev, & Smith, 2009) and the means of our response variables (i.e., µ < 5; DuVal, 2012). We chose not to convert our data to zeros (=AMS/ARS = 0) and ones (=AMS/ARS > 0) for modeling with a binomial distribution instead as this resulted in extreme parameter estimates and nonrandom patterns within our residuals. We validated our choice of distribution and method of modeling our data for final reduced models by testing for overdispersion (all p > .05), generating Q‐Q plots, and plotting scaled residuals against predicted values [simulated with the DHARMa package in R (Hartig, 2017)], with all models compliant. A Poisson distribution has also been used previously to model predictors of numbers of offspring produced in other organisms [e.g., male eastern chipmunks (Patterson & Schulte‐Hostedde, 2011)].
We modeled sex‐specific AMS and ARS as linear functions of four fixed and two random effects. Fixed effects included annual mean BCI, annual mean SVL, annual trappability, and a genome‐wide estimate of inbreeding [F hat3 (Yang, Lee, Goddard, & Visscher, 2011)]. Random effects included individual and year (Bolker et al., 2009). We included the “individual” random effect to account for repeated measurements of individuals and the “year” random effect to accommodate temporal variation over the course of the study (e.g., variable numbers of traps deployed per night each year).
We estimated annual mean BCIs by taking the residuals of the regression of natural log‐transformed annual mean body mass versus natural log‐transformed annual mean SVL separately for each sex (Schulte‐Hostedde, Zinner, Mllar, & Hickling, 2005). We assessed annual trappability by summing the number of times an individual was caught in a baited trap in a given year [see Réale et al. (2000) and Le Cœur et al. (2015)]. Average annual SVL was calculated for each individual, and this variable was standardized to have a mean of zero and a standard deviation of one to facilitate convergence of GLMMs (Harrison et al., 2018). Finally, we used PLINK 1.9 to derive the genome‐wide estimate of inbreeding (F hat3, Yang et al., 2011) for each individual from the final filtered set of SNPs (N = 654). After running the global models, we used the drop1{stats} function in R (R Core Team, 2013) to test the significance of fixed effects using likelihood ratio tests of the global model against a null model lacking the predictor of interest (χ 2, α = .05; see Nystrand, Cassidy, and Dowling (2018) and Sales et al. (2018) for similar approaches). To avoid possible bias of effect sizes, we only report estimates and standard errors for parameters for the global models (Harrison et al., 2018; Tables 1 and 2). Statistical analyses were conducted in RStudio (R v. 3.5.0; RStudio Team, 2015).
Table 1.
Results for sex‐specific generalized linear mixed models (GLMMs) of annual mating success (AMS) for Brown Treesnakes (Boiga irregularis) from a geographically closed population on Guam. GLMMs were run for (A) complete sex‐specific data sets (male = 661 records; female = 735 records) and (B) sex‐specific data sets filtered by SVL to include only likely adult records (male = 312 records; female = 367 records). Sex‐specific AMS was modeled as a linear function of four annual fixed effects (“Parameter”): average body condition index (“BCI”), standardized average snout–vent length (“SVL”), the number of times the individual was captured in a baited trap (“Trappability”), and the individual's genome‐wide estimate of inbreeding (“Inbreeding”). GLMMs also included individual and year of sampling as random effects (not shown). GLMMs employed a Poisson error distribution with a log‐link
| Fixed effect | Male AMS | Female AMS | ||||||
|---|---|---|---|---|---|---|---|---|
| Estimate | SE | LRT | p | Estimate | SE | LRT | p | |
| (A) Complete | ||||||||
| BCI | 3.717 | 0.910 | 15.496 | <.001 | 1.184 | 0.738 | 2.613 | .106 |
| SVL | 1.643 | 0.275 | 65.820 | <.001 | 1.483 | 0.245 | 63.988 | <.001 |
| Trappability | 0.095 | 0.033 | 8.635 | .003 | 0.034 | 0.020 | 2.640 | .104 |
| Inbreeding | 0.368 | 1.168 | 0.099 | .753 | −0.607 | 1.073 | 0.310 | .578 |
| (B) SVL‐filtered | ||||||||
| BCI | 3.909 | 0.974 | 15.108 | <.001 | 0.868 | 0.736 | 1.408 | .235 |
| SVL | 1.416 | 0.394 | 13.105 | <.001 | 0.794 | 0.351 | 5.000 | .025 |
| Trappability | 0.091 | 0.034 | 7.441 | .006 | 0.028 | 0.020 | 1.921 | .166 |
| Inbreeding | 0.510 | 1.188 | 0.185 | .667 | −0.525 | 1.038 | 0.248 | .618 |
Significance of fixed effects was assessed with likelihood ratio tests of the global model containing the effect of interest against a null model without the effect, with significant p‐values in bold (χ 2, α = .05). Estimates and standard errors are reported for effects in the global model to avoid overestimation of effect sizes.
Abbreviations: Estimate, statistical value; LRT, log‐likelihood ratio; p, p‐value; SE, standard error.
Table 2.
Results for sex‐specific generalized linear mixed models (GLMMs) of annual reproductive success (ARS) for Brown Treesnake (Boiga irregularis) from a geographically closed population on Guam. GLMMs were run for (A) complete sex‐specific data sets (male = 661 records; female = 735 records) and (B) sex‐specific data sets filtered by SVL to include only likely adult records (male = 312 records; female = 367 records). Sex‐specific ARS was modeled as a linear function of four annual fixed effects (“Parameter”): average body condition index (“BCI”), standardized average snout–vent length (“SVL”), the number of times the individual was captured in a baited trap (“Trappability”), and the individual's genome‐wide estimate of inbreeding (“Inbreeding”). GLMMs also included individual and year of sampling as random effects (not shown). GLMMs employed a Poisson error distribution with a log‐link
| Fixed effect | Male ARS | Female ARS | ||||||
|---|---|---|---|---|---|---|---|---|
| Estimate | SE | LRT | p | Estimate | SE | LRT | p | |
| (A) Complete | ||||||||
| BCI | 4.194 | 0.938 | 19.240 | <0.001 | 1.723 | 0.736 | 5.616 | 0.018 |
| SVL | 1.755 | 0.290 | 68.804 | <0.001 | 1.683 | 0.262 | 78.852 | <0.001 |
| Trappability | 0.113 | 0.034 | 11.730 | <0.001 | 0.049 | 0.020 | 5.623 | 0.018 |
| Inbreeding | 0.404 | 1.388 | 0.085 | 0.771 | −0.582 | 1.219 | 0.223 | 0.636 |
| (B) SVL‐filtered | ||||||||
| BCI | 4.365 | 1.004 | 18.393 | <0.001 | 1.369 | 0.734 | 3.548 | 0.060 |
| SVL | 1.601 | 0.415 | 14.971 | <0.001 | 1.013 | 0.365 | 7.556 | 0.006 |
| Trappability | 0.113 | 0.036 | 10.810 | 0.001 | 0.044 | 0.020 | 4.664 | 0.031 |
| Inbreeding | 0.621 | 1.439 | 0.187 | 0.666 | −0.505 | 1.138 | 0.194 | 0.660 |
Significance of fixed effects was assessed with likelihood ratio tests of the global model containing the effect of interest against a null model without the effect, with significant p‐values in bold (χ 2, α = 0.05). Estimates and standard errors are reported for effects in the global model to avoid overestimation of effect sizes.
Abbreviations: Estimate, statistical value; LRT, log‐likelihood ratio; p, p‐value; SE, standard error.
3. RESULTS
3.1. ddRAD sequencing and bioinformatic processing
Illumina sequencing of ddRAD libraries resulted in 1,590,917,152 raw reads, with mean number/individual = 3,734,547 (standard deviation [SD] = ±1,114,685.18). When considering samples duplicated for quality control, sequencing resulted in 1,971,406,088 raw reads (µ = 3,650,752 reads ± 1,105,292.25). Mean sequencing coverage/individual = 25.97x (±8.26), while mean coverage/individual = 24.80x (±8.32), including duplicates.
After clustering raw reads into loci and filtering with the populations module of Stacks 2.0, we identified 6,180 SNPs, each present in at least 95% of sequenced BTS (N = 426). Of the 6,180 SNPs that passed initial filtering, we discarded 217 due to departures from HWE (p < .05), 482 with evidence of LD (VIF > 2), and 4,827 that had allele frequencies < MAF threshold (=<0.3). We used all remaining loci (n = 654) for pedigree reconstruction.
3.2. Pedigree reconstruction
We were able to assign 69 known females as dams to 199 known individuals and 51 known males as sires to 257 known individuals with confidence probabilities ranging from 0.93 to 0.99 (Table S1). A promiscuous mating system was evident in that both sexes produced offspring via multiple mates each year. Of individuals that reproduced, males produced offspring with an average of 2.28 mates/year (±2.06), whereas females produced offspring with an average of 1.33 mates/year (±0.64). Of these individuals, mean male ARS = 3.38 (±4.03) offspring/year, while mean female ARS = 2.01 (±1.33).
3.3. GLMMs of AMS and ARS
We analyzed 1,396 complete annual data records, to include 661 records for males and 735 for females collected during years 2004–2013 and 2015–2018. The SVL‐filtered data sets included 312 records for likely adult males and 367 records for likely adult females collected during years 2005–2013 and 2015–2018. Mean age of reproduction was 3.68 (±1.09) and 3.94 (±1.20) years for males and females, respectively. Mean F hat3 was 0.02 (±0.11) and ranged from −0.69 to 0.36.
We found BCI, SVL, and trappability to have significant positive effects on male AMS when considering both the complete data set and the SVL‐filtered data set for males (Table 1A,B). When considering the complete and SVL‐filtered data sets for females, only SVL had a significant positive effect on AMS (Table 1A,B). BCI, SVL, and trappability had significant positive effects on ARS when analyzing both the complete and SVL‐filtered data sets for males (Table 2A,B). SVL, BCI, and trappability also had positive significant effects on female ARS when considering the complete female data set (Table 2A), with only significant positive effects of SVL and trappability on female ARS remaining when analyzing the SVL‐filtered data set (Table 2B).
4. DISCUSSION
4.1. Characterization of the BTS mating system
Our expectation of a promiscuous mating system for BTS on Guam was supported; both sexes produced offspring with multiple mates each year. Promiscuity is the most common type of mating system in snakes (Rivas & Burghardt, 2005), although polygyny (multiple mating only by males) and polyandry (multiple mating only by females) are also prevalent (Duvall, Schuett, & Arnold, 1993; Kissner, Weatherhead, & Gibbs, 2005).
For individuals that produced offspring in a given year, we found mean ARS for males and females to be 3.38 (±4.03) and 2.01 (±1.33), respectively, and these data provide important insight into annual BTS reproductive capacity. Mean clutch size has been a difficult parameter to estimate on Guam, with few wild clutches located, and low annual recruitment (as estimated from appearance of hatchlings) has been reported for the study population [i.e., 0.5 female offspring per female per year (Rodda & Savidge, 2007)]. Gravid females have previously been found to contain 3–12 eggs when palpated (Rodda & Savidge, 2007), and Savidge et al. (2007) estimated mean clutch size to be 4.3 eggs (±2.2). Yet, this value was based off of counts of ovarian follicles and oviductal eggs rather than eggs laid by females. A few gravid females have been collected in the wild on Guam and taken into the laboratory for parturition, resulting in clutches ranging from 3 to 11 eggs (Savidge et al., 2007). However, successful hatching of eggs can be variable. For example, of three Guam females induced to reproduce in a laboratory setting, the number of eggs hatched was 0 (out of 3 eggs laid), 2 (out of 8 eggs laid), and 7 (out of 10 eggs; Mathies & Miller, 2003). In this regard, our estimate of female ARS in particular may be a more informative metric for BTS reproductive ecology than clutch size, as it represents the realized annual reproductive success of females and not a count that may include unfertilized or unhatched eggs. Importantly, these estimates represent minimum mean AMS and ARS for males and females as it is possible that some hatchlings died before they could be sampled.
4.2. Predictors of AMS and ARS
We predicted that AMS and ARS in both sexes would be influenced by SVL, BCI, trappability, and degree of genome‐wide inbreeding. We found significant effects of SVL, BCI, and trappability on AMS for males, and a significant effect of SVL on AMS for females regardless of the data set analyzed (Table 1A,B). Further, we found significant effects of SVL, BCI, and trappability on both male and female ARS when analyzing the complete data sets, with significance remaining for these effects when the SVL‐filtered data sets were analyzed, save the effect of BCI on female ARS (Table 2A,B).
The effect of SVL on AMS and ARS for males and females was present regardless of the data set analyzed (i.e., complete vs. SVL‐filtered). Larger SVLs have previously been shown in male snakes to correlate with greater AMS (Shine et al., 2000) and ARS (Levine et al., 2015) due to larger males having a competitive advantage over smaller males for gaining priority‐of‐access to females (Duvall & Schuett, 1997; Madsen & Shine, 1993, 1994). While male–male combat for priority‐of‐access to females has not been observed in wild BTS, it has been observed in captivity (Greene & Mason, 2000). Larger females, on the other hand, may reflect adequate energy reserves for production of offspring (Aubret et al., 2002) and, given this, appear more attractive to males (Blouin‐Demers et al., 2005), thereby facilitating offspring production with more partners. Furthermore, larger females have larger body cavities which may correlate with an increased capacity to produce offspring (Blouin‐Demers et al., 2005; Brown & Shine, 2007). An additional factor that may contribute to our results is that males and females with larger SVLs are older, and therefore are more likely to have reached sexual maturity and to participate and in mating and reproduction. This is supported by a stronger effect of SVL on AMS and ARS for both males and females when analyzing the complete (as opposed to the SVL‐filtered) data sets (Tables 1 and 2).
We predicted that BCI would influence AMS and ARS, and found this to be true for males regardless of the data set analyzed. Males with better body condition have greater energy reserves, and this should support a variety of mating‐related behaviors. For example, larger male BTS gain access to females by being successful in confrontations with smaller males (Greene & Mason, 2000). Sufficient energy reserves also permit mate searching by males (Lind & Beaupré, 2015) and allow more time for mate searching in that competing activities such as foraging are less mandatory (Beaupré, 2008). Although the energetic status of male snakes often influences their ability to mate [e.g., Red‐Sided Garter Snake (Shine & Mason, 2005), Timber Rattlesnake (Lind & Beaupré, 2015)], the relationship between BCI and male reproductive success remains somewhat nebulous (Shine & Mason, 2005). Male snakes seemingly contribute little energy to actual reproductive success, in that gamete production requires limited energy (Aubret et al., 2002). However, they do expend considerable energy during related activities (Lind & Beaupré, 2015; Shine & Mason, 2005). It is possible that the significant effect of BCI on ARS we identified is due to factors associated with mate acquisition, or even the physical ability of males to mate [e.g., adequate plasma testosterone levels (Bonnet & Naulleau, 1996); elevated corticosterone levels due to food stress (Waye & Mason, 2008)]. Indeed, chronic stress and elevated corticosterone levels are associated with low BCI in BTS (Waye & Mason, 2008), whereas elevated male BCIs are related to higher levels of plasma testosterone (Mathies, Cruz, Lance, & Savidge, 2010), and both elevated corticosterone and reduced plasma testosterone negatively affect reproduction in male BTS (Aldridge, Siegel, Bufalino, Wisniewski, & Jellen, 2010; Moore et al., 2005). Male BCI may also have the capacity to directly influence reproductive success. Male body size is associated with testes mass, such that larger male BTS may in fact have greater rates of sperm production, and thus an increased capacity for fertilization (Mathies et al., 2010).
We did not find a significant effect of BCI on female AMS regardless of the data set analyzed. Further, there was a significant effect of BCI on female ARS only when analyzing the complete data set and not the SVL‐filtered data set. We found the inconsistent effect of BCI on female AMS and ARS surprising as adequate energy reserves should be necessary for females to participate in mating and produce offspring. However, females may actively forage while vitellogenic, so as to acquire energy for reproduction [i.e., income breeding (Bonnet, Bradshaw, & Shine, 1998)]. In support of this hypothesis, Savidge et al. (2007) found that 79% of female BTS captured on Guam with vitellogenic follicles had prey in their stomachs. Therefore, a lack of a consistent effect of BCI on female AMS and ARS could be due to the presence of income breeding in BTS. Future work will be required to determine why BCI is important to male AMS and ARS, but seemingly less so to female AMS and ARS.
We also predicted that trappability would influence AMS and ARS in both sexes. With regard to AMS, we only found a significant positive effect for males (although these estimates were small for both the complete and SVL‐filtered data sets; Table 1A,B). These results indicate that males with a greater propensity to enter baited traps mate with more partners to produce offspring. Importantly, although juveniles are not considered trappable by baited trap (Rodda et al., 2007), the significant effect of trappability on male AMS remained when the male records were filtered by SVL to retain only those for likely adults. Trappability has often been used as a proxy for propensity of individuals to engage in risk‐taking behaviors (Biro & Dingemanse, 2009). Presumably, males bold enough to enter baited traps will also take risks to acquire mates. For instance, movement by bold individuals in search for mates may increase their risk of predation, while those less bold would not, with the likelihood of encountering potential mates subsequently diminished (Sih, Bell, & Johnson, 2004).
Trappability is also related to the ability of an individual to detect chemical stimuli from baited traps. BTS use olfactory cues to find prey in baited traps (Shivik, 1998), and male BTS also use olfactory cues (i.e., female pheromones) to find mates (Greene et al., 2001; Parker, Patel, Zachry, & Kimball, 2018) and elicit courtship (Greene & Mason, 1998). Therefore, it is likely that males with better chemosensory abilities will not only find baited traps more frequently but also have greater success in finding and acquiring mates. This may be the reason that we found a relationship between trappability and AMS in males but not females. The use of olfactory cues to detect prey in baited traps may also contribute to both males and females with greater trappability having greater ARS. Individuals that are better able to detect prey in baited traps may also be better at energy acquisition which would result in greater energy availability for mating and reproduction. However, additional research will be required to untangle the complicated relationship between trappability and ARS.
We found the degree to which an individual was inbred to have no effect on AMS or ARS for males or females, and these results were consistent regardless of the data set analyzed. This finding was surprising because most individuals (68.4%) in our study had estimates of genome‐wide inbreeding (F hat3) > 0, with several (8.3% females; 6.8% males) consistent with having half‐sibling parental relationships (F hat3 > 0.125). In other systems, mating success in the wild is negatively impacted by the degree to which focal individuals are inbred (Janicke, Vellnow, Lamy, Chapuis, & David, 2014; Joron & Brakefield, 2003).
Although we found no significant effect on AMS or ARS for either sex, inbreeding effects may instead manifest indirectly by influencing other traits that, in turn, impact AMS and ARS [e.g., by affecting propensity for risk‐taking behavior (Richardson & Smiseth, 2017) or motivation to mate (De Boer, Eens, & Müller, 2018)]. The effects of inbreeding may also be context‐dependent, with negative impacts more pronounced under stressful conditions (Armbruster & Reed, 2005). Guam's relatively constant environment (Rodda et al., 1999) may counter environmental stress and thus act to dampen negative effects of inbreeding on AMS and ARS. Further, individuals may avoid inbreeding effects through behavioral plasticity (Lucia‐Simmons & Keane, 2015). Finally, inbreeding may simply exert minimal effects on AMS and ARS (Gooley, Hogg, Belov, & Grueber, 2017). Future work will be required to determine why inbreeding seemingly does not significantly affect AMS and ARS in Guam BTS, particularly considering the small size of the founding population (Richmond et al., 2014).
4.3. Implications for control
A variety of control efforts have been implemented to reduce or eliminate BTS on Guam and to prevent its dispersal to other areas (Clark, Clark, & Siers, 2018; Engbring & Fritts, 1988; Rodda & Savidge, 2007). Baited traps are a primary method of control, and improving trap success will likewise promote BTS management. Here, we interpret our results with regard to common phenotypes found in trapped individuals.
First, several studies have explored the relationship between body size and trappability (Boyarski, Savidge, & Rodda, 2008; Lardner et al., 2013; Rodda et al., 2007; Tyrrell et al., 2009), with larger individuals trapped more often than those smaller. Given this finding, our results indicate that baited traps are effective at capturing males and females that produce more offspring and males that produce offspring with more mating partners.
However, while several studies have also evaluated the relationship between trapping success and BCI, results have been variable. For example, Tyrrell et al. (2009) found a minimal positive effect of BCI on female trappability and a negative effect of BCI on male trappability, such that males with low BCIs were caught in baited traps more often. Differently, Lardner et al. (2013) found a positive correlation between BCI and accession of bait tubes for both sexes. Our study demonstrated that BCI has a positive effect on AMS and ARS for males, with a similar effect of BCI on ARS supported for females. Given these relationships between BCI and AMS/ARS, our incomplete understanding of how BCI is related to trappability (by either baited traps or bait tubes) is troubling.
Finally, it is promising that overall trappability is related to AMS for males and ARS for both males and females, although the estimates for the effects of trappability on AMS and ARS were small. Males and females with higher trappability produce more offspring annually and males with higher trappability produce offspring with more mates; importantly, these effects remain when removing likely juveniles (untrappable) from analyses. Removal of trapped individuals thus has the potential to depress the birth rate of the population by eliminating more fecund males and females. However, significant unexplained heterogeneity in trappability exists among individuals (Clark, Savarie, Shivik, Breck, & Dorr, 2012; Mason, Savidge, Rodda, & Yackel Adams, 2011; Rodda et al., 2002; Tyrrell et al., 2009), prompting concerns that trappability may have a heritable genetic component (Tyrrell et al., 2009), as documented in fishes (Cooke, Suski, Ostrand, Wahl, & Philipp, 2007). If BTS trappability is indeed heritable, then selection may yield a population with overall lower ARS, but also one that is trap‐shy (Tyrrell et al., 2009). We are currently evaluating the heritability of being trappable to gauge the potential for inadvertent adaptive responses of reproductive ecology to management action.
5. CONCLUSION
An understanding of the reproductive ecology of invasive species is critical for the development of effective control. Phenotypes associated with enhanced annual mating and reproductive success can be targeted to maximally impact the birth rate of the population. Furthermore, by identifying correlates of mating and reproductive success and juxtaposing them with “controllable” phenotypes, the long‐term efficacy of control can be gauged, particularly considering eco‐evo dynamics generated by the control methods themselves (Závorka et al., 2018). Here, we demonstrated the use of multigenerational genomic pedigree reconstruction as an avenue for identification of predictors of AMS and ARS in an invasive vertebrate and compared phenotypes associated with elevated AMS and ARS with those targeted by control. We did so using genomic markers that are widely applicable to nonmodel organisms (Peterson et al., 2012). These results will serve to promote similar endeavors for other invasive species.
CONFLICT OF INTEREST
The authors declare no competing interests.
AUTHOR CONTRIBUTIONS
All authors contributed to the conceptualization and design of the study. AAYA, BAL, BL, RNR, and JAS collected field data. AAYA and BL organized and maintained the U.S. Geological Survey Brown Treesnake database. BAL conducted all laboratory work and statistical analyses, with guidance from MRD and MED. Most editing was accomplished by MRD and MED, but all authors contributed editorial feedback.
Supporting information
ACKNOWLEDGMENTS
This work was supported by two grants [U.S. Geological Survey RWO42 (MRD and MED); Sigma Xi GIAR (BAL)] and two University of Arkansas endowments [Bruker Professor in Life Sciences (MRD); 21st Century Chair in Global Change (MED)]. Funding for the USGS agreement was provided by the Department of Interior's Office of Insular Affairs and by the USGS Invasive Species Program. We collected all samples and data with approvals from Colorado State University IACUC (protocol #15‐5892A) and University of Arkansas IACUC (protocol #16063). We thank the Arkansas High Performance Computing Center (AHPCC) for providing computational resources to accomplish all bioinformatic analyses and the University of Oregon Genomic and Cell Characterization Core Facility (GC3F) and University of Wisconsin Biotechnology Center (UWBC) for sequencing our ddRAD libraries. We also thank Dr. David Krementz, Dr. Warren Booth, and three anonymous reviewers for helpful comments that improved this manuscript. This project would not have been possible without the sampling efforts of past and present USGS Brown Treesnake Project team members. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. government.
Levine BA, Douglas MR, Yackel Adams AA, et al. Genomic pedigree reconstruction identifies predictors of mating and reproductive success in an invasive vertebrate. Ecol Evol. 2019;9:11863–11877. 10.1002/ece3.5694
DATA AVAILABILITY STATEMENT
Raw sequencing reads are archived with the DRYAD data repository at https://doi.org/10.5061/dryad.47ss1b4. Data analyzed in the study are available through ScienceBase at https://doi.org/10.5066/P9X1AKVJ.
REFERENCES
- Aldridge, R. D. , Siegel, D. S. , Bufalino, A. P. , Wisniewski, S. S. , & Jellen, B. C. (2010). A multiyear comparison of the male reproductive biology of the brown treesnake (Boiga irregularis) from Guam and the native range. Australian Journal of Zoology, 58(1), 24–32. 10.1071/ZO09068 [DOI] [Google Scholar]
- Andrews, S. (2014). A quality control tool for high throughput sequence data. Retrieved from http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- Armbruster, P. , & Reed, D. H. (2005). Inbreeding depression in benign and stressful environments. Heredity, 95(3), 235–242. 10.1038/sj.hdy.6800721 [DOI] [PubMed] [Google Scholar]
- Aubret, F. , Bonnet, X. , Shine, R. , & Lourdais, O. (2002). Fat is sexy for females but not males: The influence of body reserves on reproduction in snakes (Vipera aspis). Hormones and Behavior, 42(2), 135–147. 10.1006/hbeh.2002.1793 [DOI] [PubMed] [Google Scholar]
- Bangs, M. R. , Douglas, M. R. , Mussmann, S. M. , & Douglas, M. E. (2018). Unraveling historical introgression and resolving phylogenetic discord within Catostomus (Osteichthys: Catostomidae). BMC Evolutionary Biology, 18(1), 10.1186/s12862-018-1197-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates, D. , Maechler, M. , Bolker, B. M. , & Walker, S. C. (2015). Fitting linear mixed‐effects models using lme4. Journal of Statistical Software, 67, 1–48. [Google Scholar]
- Beaupré, S. J. (2008). Annual variation in time‐energy allocation by timber rattlesnakes (Crotalus horridus) in relation to food acquisition In Hayes W., Beaman K., Cardwell M., & Bush S. (Eds.), The biology of rattlesnakes (pp. 111–122). Loma Linda, CA: Loma Linda University Press. [Google Scholar]
- Biro, P. A. , & Dingemanse, N. J. (2009). Sampling bias resulting from animal personality. Trends in Ecology and Evolution, 24(2), 66–67. 10.1016/j.tree.2008.10.004 [DOI] [PubMed] [Google Scholar]
- Blouin‐Demers, G. , Gibbs, H. L. , & Weatherhead, P. J. (2005). Genetic evidence for sexual selection in black rat snakes, Elaphe obsoleta . Animal Behaviour, 69, 225–234. 10.1016/j.anbehav.2004.03.012 [DOI] [Google Scholar]
- Bolker, B. M. , Brooks, M. E. , Clark, C. J. , Geange, S. W. , Poulsen, J. R. , Stevens, M. H. H. , & White, J.‐S.‐S. (2009). Generalized linear mixed models: A practical guide for ecology and evolution. Trends in Ecology and Evolution, 24(3), 127–135. 10.1016/j.tree.2008.10.008 [DOI] [PubMed] [Google Scholar]
- Bonnet, X. , Bradshaw, D. , & Shine, R. (1998). Capital versus income breeding: An ectothermic perspective. Oikos, 83(2), 333–342. 10.2307/3546846 [DOI] [Google Scholar]
- Bonnet, X. , & Naulleau, G. (1996). Are body reserves important for reproduction in male dark green snakes (Colubridae: Coluber viridiflavus)? Herpetologica, 52(2), 137–146. [Google Scholar]
- Boyarski, V. L. , Savidge, J. A. , & Rodda, G. H. (2008). Brown treesnake (Boiga irregularis) trappability: Attributes of the snake, environment and trap. Applied Herpetology, 5, 47–61. 10.1163/157075408783489239 [DOI] [Google Scholar]
- Boyer, N. , Réale, D. , Marmet, J. , Pisanu, B. , & Chapuis, J. L. (2010). Personality, space use and tick load in an introduced population of Siberian chipmunks Tamias sibiricus . Journal of Animal Ecology, 79(3), 538–547. 10.1111/j.1365-2656.2010.01659.x [DOI] [PubMed] [Google Scholar]
- Brown, G. P. , & Shine, R. (2007). Repeatability and heritability of reproductive traits in free‐ranging snakes. Journal of Evolutionary Biology, 20, 588–596. 10.1111/j.1420-9101.2006.01256.x [DOI] [PubMed] [Google Scholar]
- Catchen, J. M. , Amores, A. , Hohenlohe, P. , Cresko, W. , & Postlethwait, J. H. (2011). Stacks: Building and genotyping loci de novo from short‐read sequences. Genes ‐ Genomes ‐ Genetics, 1(3), 171–182. 10.1534/g3.111.000240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen, J. , Hohenlohe, P. A. , Bassham, S. , Amores, A. , & Cresko, W. A. (2013). Stacks: An analysis tool set for population genomics. Molecular Ecology, 22(11), 3124–3140. 10.1111/mec.12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caves, E. M. , Jennings, S. B. , HilleRisLambers, J. , Tewksbury, J. J. , & Rogers, H. S. (2013). Natural experiment demonstrates that bird loss leads to cessation of dispersal of native seeds from intact to degraded forests. PLoS ONE, 8(5), e65618 10.1371/journal.pone.0065618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, L. , Clark, C. , & Siers, S. (2018). Brown tree snakes – methods and approaches for control In Pitt W., Beasley J., & Witmer G. (Eds.), Ecology and management of terrestrial vertebrate invasive species in the United States (pp. 107–134). Boca Raton, FL: CRC Press. [Google Scholar]
- Clark, L. , Savarie, P. J. , Shivik, J. A. , Breck, S. W. , & Dorr, B. S. (2012). Efficacy, effort, and cost comparisons of trapping and acetaminophen‐baiting for control of brown treesnakes on Guam. Human‐Wildlife Interactions, 6(2), 222–236. [Google Scholar]
- Clavero, M. , & García‐Berthou, E. (2005). Invasive species are a leading cause of animal extinctions. Trends in Ecology and Evolution, 20(3), 110 10.1016/j.tree.2005.01.003 [DOI] [PubMed] [Google Scholar]
- Cole, L. C. (1954). The population consequences of life history phenomena. The Quarterly Review of Biology, 29(2), 103–137. 10.1086/400074 [DOI] [PubMed] [Google Scholar]
- Cooke, S. J. , Suski, C. D. , Ostrand, K. G. , Wahl, D. H. , & Philipp, D. P. (2007). Physiological and behavioral consequences of long‐term artificial selection for vulnerability to recreational angling in a teleost fish. Physiological and Biochemical Zoology, 80(5), 480–490. 10.1086/520618 [DOI] [PubMed] [Google Scholar]
- De Boer, R. A. , Eens, M. , & Müller, W. (2018). An experimental study: Does inbreeding increase the motivation to mate? PLoS ONE, 13(6), 1–13. 10.1371/journal.pone.0199182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuVal, E. H. (2012). Variation in annual and lifetime reproductive success of lance‐tailed manakins: Alpha experience mitigates effects of senescence on siring success. Proceedings of the Royal Society B: Biological Sciences, 279(1733), 1551–1559. 10.1098/rspb.2011.1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall, D. , & Schuett, G. W. (1997). Straight‐line movement and competitive mate searching in prairie rattlesnakes. Animal Behaviour, 54, 329–334. [DOI] [PubMed] [Google Scholar]
- Duvall, D. , Schuett, G. W. , & Arnold, S. J. (1993). Ecology and evolution of snake mating systems In Seigel R. A., & Collins J. (Eds.), Snakes: Ecology and behavior (pp. 165–200). New York, NY: McGraw‐Hill. [Google Scholar]
- Ekblom, R. , & Galindo, J. (2011). Applications of next generation sequencing in molecular ecology of non‐model organisms. Heredity, 107(1), 1–15. 10.1038/hdy.2010.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren, H. , & Galtier, N. (2016). Determinants of genetic diversity. Nature Reviews Genetics, 17(7), 422–433. 10.1038/nrg.2016.58 [DOI] [PubMed] [Google Scholar]
- Engbring, J. , & Fritts, T. H. (1988). Demise of an insular avifauna: The brown tree snake on Guam. Transactions of the Western Section of the Wildlife Society, 24, 31–37. [Google Scholar]
- Foerster, K. , Delhey, K. , Johnsen, A. , Lifjeld, J. T. , & Kempenaers, B. (2003). Females increase offspring heterozygosity and fitness through extra‐pair matings. Nature, 425(6959), 714–717. 10.1038/nature01969 [DOI] [PubMed] [Google Scholar]
- Fritts, T. H. , McCoid, M. J. , & Haddock, R. L. (1990). Risks to infants on Guam from bites of the brown tree snake (Boiga irregularis). The American Journal of Tropical Medicine and Hygiene, 42(6), 607–611. 10.4269/ajtmh.1990.42.607 [DOI] [PubMed] [Google Scholar]
- Fritts, T. H. , & Rodda, G. H. (1998). The role of introduced species in the degradation of island ecosystems: A case history of Guam. Annual Review of Ecology and Systematics, 29, 113–140. 10.1146/annurev.ecolsys.29.1.113 [DOI] [Google Scholar]
- Gonçalves da Silva, A. , & Russello, M. A. (2011). iREL: Software for implementing pairwise relatedness estimators and evaluating their performance. Conservation Genetics Resources, 3(1), 69–71. 10.1007/s12686-010-9292-4 [DOI] [Google Scholar]
- Gooley, R. , Hogg, C. J. , Belov, K. , & Grueber, C. E. (2017). No evidence of inbreeding depression in a Tasmanian devil insurance population despite significant variation in inbreeding. Scientific Reports, 7(1), 1–11. 10.1038/s41598-017-02000-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene, M. J. , & Mason, R. T. (1998). Chemically mediated sexual behavior of the brown tree snake, Boiga irregularis . Ecoscience, 5(3), 405–409. [Google Scholar]
- Greene, M. J. , & Mason, R. T. (2000). Courtship, mating, and male combat of the brown tree snake, Boiga irregularis . Herpetologica, 56(2), 166–175. [Google Scholar]
- Greene, M. J. , Stark, S. L. , & Mason, R. T. (2001). Pheromone trailing behavior of the brown tree snake, Boiga irregularis . Journal of Chemical Ecology, 27(11), 2193–2201. [DOI] [PubMed] [Google Scholar]
- Harrison, X. A. , Donaldson, L. , Correa‐Cano, M. E. , Evans, J. , Fisher, D. N. , Goodwin, C. E. D. , … Inger, R. (2018). A brief introduction to mixed effects modelling and multi‐model inference in ecology. PeerJ, 6, 1–32. 10.7717/peerj.4794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig, F. (2017). DHARMa: Residual diagnostics for hierarchical (multi‐level/mixed) regression models. R package version 0.2.0. Retrieved from https://cran.r-project.org/package=DHARMa [Google Scholar]
- Huisman, J. (2017). Pedigree reconstruction from SNP data: Parentage assignment, sibship clustering and beyond. Molecular Ecology, 17, 1009–1024. 10.1111/1755-0998.12665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imdadullah, M. , Aslam, M. , & Altaf, S. (2016). mctest: An R package for detection of collinearity among regressors. The R Journal, 8(2), 495–505. 10.32614/RJ-2016-062 [DOI] [Google Scholar]
- Janicke, T. , Vellnow, N. , Lamy, T. , Chapuis, E. , & David, P. (2014). Inbreeding depression of mating behavior and its reproductive consequences in a freshwater snail. Behavioral Ecology, 25(2), 288–299. 10.1093/beheco/art122 [DOI] [Google Scholar]
- Jones, O.R. , & Wang, J. (2010). COLONY: A program for parentage and sibship inference from multilocus genotype data. Molecular Ecology Resources, 10(3), 551–555. 10.1111/j.1755-0998.2009.02787.x [DOI] [PubMed] [Google Scholar]
- Jordan, M. A. , & Rodda, G. H. (1994). Identification of sex in Boiga irregularis: Implications for understanding population dynamics in Guam. Journal of Herpetology, 28(3), 381–384. 10.2307/1564541 [DOI] [Google Scholar]
- Joron, M. , & Brakefield, P. M. (2003). Captivity masks inbreeding effects on male mating success in butterflies. Nature, 424(6945), 191–194. 10.1038/nature01713 [DOI] [PubMed] [Google Scholar]
- Juliano, S. A. , & Lounibos, P. (2005). Ecology of invasive mosquitoes: Effects on resident species and on human health. Ecology Letters, 8(5), 558–574. 10.1109/TMI.2012.2196707.Separate [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahl, S. S. , Henke, S. E. , Hall, M. A. , & Britton, D. K. (2012). Brown treesnakes: A potential invasive species for the United States. Human‐Wildlife Interactions, 6(2), 181–203. [Google Scholar]
- Kissner, K. J. , Weatherhead, P. J. , & Gibbs, H. L. (2005). Experimental assessment of ecological and phenotypic factors affecting male mating success and polyandry in northern watersnakes, Nerodia sipedon . Behavioral Ecology and Sociobiology, 59(2), 207–214. 10.1007/s00265-005-0026-7 [DOI] [Google Scholar]
- Lardner, B. , Yackel Adams, A. A. , Savidge, J. A. , Rodda, G. H. , Reed, R. N. , & Clark, C. S. (2013). Effectiveness of bait tubes for brown treesnake control on Guam. Wildlife Society Bulletin, 37(3), 664–673. 10.1002/wsb.297 [DOI] [Google Scholar]
- Le Cœur, C. , Thibault, M. , Pisanu, B. , Thibault, S. , Chapuis, J. L. , & Baudry, E. (2015). Temporally fluctuating selection on a personality trait in a wild rodent population. Behavioral Ecology, 26(5), 1285–1291. 10.1093/beheco/arv074 [DOI] [Google Scholar]
- Levine, B. A. , Smith, C. F. , Schuett, G. W. , Douglas, M. S. , Davis, M. A. , & Douglas, M. E. (2015). Bateman‐Trivers in the 21st century: Sexual selection in a North American pitviper. Biological Journal of the Linnean Society, 114(1948), 436–445. 10.1111/bij.12434 [DOI] [Google Scholar]
- Lind, C. M. , & Beaupré, S. J. (2015). Male snakes allocate time and energy according to individual energetic status: Body condition, steroid hormones, and reproductive behavior in timber rattlesnakes, Crotalus horridus . Physiological and Biochemical Zoology, 88(6), 624–633. 10.1086/683058 [DOI] [PubMed] [Google Scholar]
- Lockwood, J. , Hoopes, M. , & Marchetti, M. (2013). Invasion ecology (2nd ed.). Chichester, UK: Wiley‐Blackwell. [Google Scholar]
- Lowe, S. , Browne, M. , Boudjelas, S. , & De Poorter, M. (2000). 100 of the world's worst invasive alien species. A selection from the Global Invasive Species Database (pp. 4). Auckland, New Zealand: The Invasive Species Specialist Group (ISSG) a specialist group of the Species Survival Commission (SSC) of the World Conservation Union (IUCN). [Google Scholar]
- Lucia‐Simmons, K. E. , & Keane, B. (2015). Behavioral plasticity in nest residency compensates for inbreeding depression in male prairie voles. Behavioral Ecology, 26(4), 1060–1070. 10.1093/beheco/arv053 [DOI] [Google Scholar]
- Madsen, T. , & Shine, R. (1993). Temporal variability in sexual selection acting on reproductive tactics and body size in male snakes. American Naturalist, 141, 167–171. 10.1086/285467 [DOI] [PubMed] [Google Scholar]
- Madsen, T. , & Shine, R. (1994). Components of lifetime reproductive success in adders, Vipera berus . Journal of Applied Ecology, 63, 561–568. 10.2307/5222 [DOI] [Google Scholar]
- Mason, L. C. , Savidge, J. A. , Rodda, G. H. , & Yackel Adams, A. A. (2011). Scented guide ropes as a method to enhance brown treesnake (Boiga irregularis) trap capture success on Guam. Journal of Herpetology, 45(3), 308–312. 10.1670/10-026.1 [DOI] [Google Scholar]
- Mathies, T. , Cruz, J. , Lance, V. , & Savidge, J. (2010). Reproductive biology of male brown treesnakes (Boiga irregularis) on Guam. Journal of Herpetology, 44(2), 209–221. 10.1670/06-069.1 [DOI] [Google Scholar]
- Mathies, T. , Franklin, E. A. , & Miller, L. A. (2004). Proximate cues for ovarian recrudescence and ovulation in the brown treesnake (Boiga irregularis) under laboratory conditions. Herpetological Review, 35(1), 46–49. [Google Scholar]
- Mathies, T. , Levine, B. , Engeman, R. , & Savidge, J. A. (2013). Pheromonal control of the invasive brown treesnake: Potency of female sexual attractiveness pheromone varies with ovarian state. International Journal of Pest Management, 59(2), 141–149. 10.1080/09670874.2013.784374 [DOI] [Google Scholar]
- Mathies, T. , & Miller, L. A. (2003). Cool temperatures elicit reproduction in a biologically invasive predator, the brown treesnake (Boiga irregularis). Zoo Biology, 22, 227–238. 10.1002/zoo.10084 [DOI] [Google Scholar]
- Mooney, H. A. , & Cleland, E. E. (2001). The evolutionary impact of invasive species. Proceedings of the National Academy of Sciences of the United States of America, 98(10), 5446–5451. 10.1073/pnas.091093398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, I. T. , Greene, M. J. , Lerner, D. T. , Asher, C. E. , Krohmer, R. W. , Hess, D. L. , … Mason, R. T. (2005). Physiological evidence for reproductive suppression in the introduced population of brown tree snakes (Boiga irregularis) on Guam. Biological Conservation, 121, 91–98. 10.1016/j.biocon.2004.04.012 [DOI] [Google Scholar]
- Morrissey, M. B. , & Wilson, A. J. (2010). Pedantics: An R package for pedigree‐based genetic simulation and pedigree manipulation, characterization and viewing. Molecular Ecology Resources, 10(4), 711–719. 10.1111/j.1755-0998.2009.02817.x [DOI] [PubMed] [Google Scholar]
- Mortensen, H. S. , Dupont, Y. L. , & Olesen, J. M. (2008). A snake in paradise: Disturbance of plant reproduction following extirpation of bird flower‐visitors on Guam. Biological Conservation, 141(8), 2146–2154. 10.1016/j.biocon.2008.06.014 [DOI] [Google Scholar]
- Muhlfeld, C. C. , Kalinowski, S. T. , McMahon, T. E. , Taper, M. L. , Painter, S. , Leary, R. F. , & Allendorf, F. W. (2009). Hybridization rapidly reduces fitness of a native trout in the wild. Biology Letters, 5(3), 328–331. 10.1098/rsbl.2009.0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystrand, M. , Cassidy, E. J. , & Dowling, D. K. (2018). The effects of a bacterial challenge on reproductive success of fruit flies evolved under low or high sexual selection. Ecology and Evolution, 8, 9341–9352. 10.1002/ece3.4450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, L. J. (2006). The economics of terrestrial invasive species: A review of the literature. Agricultural and Resource Economics, 35, 178–194. 10.1017/S1068280500010145 [DOI] [Google Scholar]
- Paris, J. R. , Stevens, J. R. , & Catchen, J. M. (2017). Lost in parameter space: A road map for stacks. Methods in Ecology and Evolution, 8(10), 1360–1373. 10.1111/2041-210X.12775 [DOI] [Google Scholar]
- Parker, M. R. , Patel, S. M. , Zachry, J. E. , & Kimball, B. A. (2018). Feminization of male brown treesnake methyl ketone expression via steroid hormone manipulation. Journal of Chemical Ecology, 44(2), 189–197. 10.1007/s10886-018-0935-3 [DOI] [PubMed] [Google Scholar]
- Patterson, L. D. , & Schulte‐Hostedde, A. I. (2011). Behavioural correlates of parasitism and reproductive success in male eastern chipmunks, Tamias striatus . Animal Behaviour, 81, 1129–1137. 10.1016/j.anbehav.2011.02.016 [DOI] [Google Scholar]
- Perry, G. , & Vice, D. (2009). Forecasting the risk of brown tree snake dispersal from Guam: A mixed transport‐establishment model. Conservation Biology, 23(4), 992–1000. 10.1111/j.1523-1739.2009.01169.x [DOI] [PubMed] [Google Scholar]
- Peterson, B. K. , Weber, J. N. , Kay, E. H. , Fisher, H. S. , & Hoekstra, H. E. (2012). Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non‐model species. PLoS ONE, 7(5), 1–11. 10.1371/journal.pone.0037135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell, S. , Neale, B. , Todd‐Brown, K. , Thomas, L. , Ferreira, M. A. R. , Bender, D. , … Sham, P. C. (2007). PLINK: A tool set for whole‐genome association and population‐based linkage analyses. The American Journal of Human Genetics, 81(3), 559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2013). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; Retrieved from http://www.r-project.org/ [Google Scholar]
- Réale, D. , Gallant, B. Y. , Leblanc, M. , & Festa‐Bianchet, M. (2000). Consistency of temperament in bighorn ewes and correlates with behaviour and life history. Animal Behaviour, 60(5), 589–597. 10.1006/anbe.2000.1530 [DOI] [PubMed] [Google Scholar]
- Richardson, J. , & Smiseth, P. T. (2017). Intraspecific competition and inbreeding depression: Increased competitive effort by inbred males is costly to outbred opponents. The American Naturalist, 189(5), 539–548. 10.1086/691328 [DOI] [PubMed] [Google Scholar]
- Richmond, J. Q. , Wood, D. A. , Stanford, J. W. , & Fisher, R. N. (2014). Testing for multiple invasion routes and source populations for the invasive brown treesnake (Boiga irregularis) on Guam: Implications for pest management. Biological Invasions, 17(1), 337–349. 10.1007/s10530-014-0733-y [DOI] [Google Scholar]
- Rivas, J. A. , & Burghardt, G. M. (2005). Snake mating systems, behavior, and evolution: The revisionary implications of recent findings. Journal of Comparative Psychology, 119(4), 447–454. 10.1037/0735-7036.119.4.447 [DOI] [PubMed] [Google Scholar]
- Rochette, N. C. , & Catchen, J. M. (2017). Deriving genotypes from RAD‐seq short‐read data using Stacks. Nature Protocols, 12, 2640 10.1038/nprot.2017.123 [DOI] [PubMed] [Google Scholar]
- Rodda, G. , Fritts, T. , Campbell, E. W. I. , Dean‐Bradley, K. , Perry, G. , & Qualls, C. (2002). Practical concerns in the eradication of island snakes In Veitch C., & Clout M. (Eds.), Turning the tide: The eradication of invasive species (27th ed., pp. 260–265). Gland, Switzerland: IUCN Species Survival Commission. [Google Scholar]
- Rodda, G. H. , Fritts, T. H. , & Chiszar, D. (1997). The disappearance of Guam's wildlife. BioScience, 47(9), 565–574. 10.2307/1313163 [DOI] [Google Scholar]
- Rodda, G. H. , Fritts, T. H. , McCoid, M. J. , & Campbell, E. W. I. (1999). An overview of the biology of the brown treesnake (Boiga irregularis), a costly introduced pest on Pacific Islands In Rodda G. H., Sawai Y., Chiszar D., & Tanaka H. (Eds.), Problem snake management: The habu and the brown treesnake (pp. 44–80). Ithaca, NY: Cornell University Press. [Google Scholar]
- Rodda, G. H. , & Savidge, J. A. (2007). Biology and impacts of Pacific Island invasive species. 2. Boiga irregularis, the brown tree snake (Reptilia: Colubridae). Pacific Science, 61(3), 307–324. [Google Scholar]
- Rodda, G. H. , Savidge, J. A. , Tyrrell, C. , Christy, M. T. , & Ellingson, A. R. (2007). Size bias in visual searches and trapping of brown treesnakes on Guam. Journal of Wildlife Management, 71(2), 656–661. 10.2193/2005-742 [DOI] [Google Scholar]
- Rogers, H. , Lambers, J. H. R. , Miller, R. , & Tewksbury, J. J. (2012). “Natural experiment” demonstrates top‐down control of spiders by birds on a landscape level. PLoS ONE, 7(9), e43446 10.1371/journal.pone.0043446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio Team (2015). RStudio: Integrated development for R. Boston, MA: RStudio, Inc; Retrieved from http://www.rstudio.com/ [Google Scholar]
- Sales, K. , Vasudeva, R. , Dickinson, M. E. , Godwin, J. L. , Lumley, A. J. , Michalczyk, Ł. , … Gage, M. J. G. (2018). Experimental heatwaves compromise sperm function and cause transgenerational damage in a model insect. Nature Communications, 9, 4771 10.1038/s41467-018-07273-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, N. J. , Gotelli, N. J. , Heller, N. E. , & Gordon, D. M. (2003). Community disassembly by an invasive species. Proceedings of the National Academy of Sciences of the United States of America, 100(5), 2474–2477. 10.1073/pnas.0437913100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savidge, J. A. (1987). Extinction of an island forest avifauna by an introduced snake. Ecology, 68(3), 660–668. 10.2307/1938471 [DOI] [Google Scholar]
- Savidge, J. A. , Qualls, F. J. , & Rodda, G. H. (2007). Reproductive biology of the brown tree snake, Boiga irregularis (Reptilia: Colubridae), during colonization of Guam and comparison with that in their native range. Pacific Science, 61(2), 191–199. 10.2984/1534-6188(2007)61[191:RBOTBT]2.0.CO;2 [DOI] [Google Scholar]
- Schulte‐Hostedde, A. I. , Zinner, B. , Mllar, J. S. , & Hickling, G. J. (2005). Restitution of mass‐size residuals: Validating body condition indices. Ecology, 86(1), 155–163. 10.1890/04-0232 [DOI] [Google Scholar]
- Shine, R. , & Mason, R. T. (2005). Do a male garter snake's energy stores limit his reproductive effort? Canadian Journal of Zoology, 83(10), 1265–1270. 10.1139/z05-119 [DOI] [Google Scholar]
- Shine, R. , Olsson, M. M. , Moore, I. T. , Lemaster, M. P. , Greene, M. , & Mason, R. T. (2000). Body size enhances mating success in male garter snakes. Animal Behaviour, 59, F4–F11. 10.1006/anbe.1999.1338 [DOI] [PubMed] [Google Scholar]
- Shivik, J. A. (1998). Brown tree snake response to visual and olfactory cues. The Journal of Wildlife Management, 62(1), 105–111. 10.2307/3802268 [DOI] [Google Scholar]
- Shivik, J. A. , & Clark, L. (1997). Carrion seeking in brown tree snakes: Importance of olfactory and visual cues. Journal of Experimental Zoology, 279(6), 549–553. [DOI] [Google Scholar]
- Siegel, D. S. , Aldridge, R. D. , Clark, C. S. , Poldemann, E. H. , & Gribbins, K. M. (2009). Stress and reproduction in Boiga irregularis with notes on the ultrastructure of the sexual segment of the kidney in squamates. Canadian Journal of Zoology, 87, 1138–1146. 10.1139/Z09-103 [DOI] [Google Scholar]
- Sih, A. , Bell, A. , & Johnson, J. C. (2004). Behavioral syndromes: An ecological and evolutionary overview. Trends in Ecology and Evolution, 19(7), 372–378. 10.1016/j.tree.2004.04.009 [DOI] [PubMed] [Google Scholar]
- Simberloff, D. (2001). Biological invasions – How are they affecting us, and what can we do about them? Western North American Naturalist, 61(3), 308–315. [Google Scholar]
- Strauss, S. Y. , Lau, J. A. , & Carroll, S. P. (2006). Evolutionary responses of natives to introduced species: What do introductions tell us about natural communities? Ecology Letters, 9(3), 357–374. 10.1111/j.1461-0248.2005.00874.x [DOI] [PubMed] [Google Scholar]
- Thomas, C. D. , Cameron, A. , Green, R. E. , Bakkenes, M. , Beaumont, L. J. , Collingham, Y. C. , … Williams, S. E. (2004). Extinction risk from climate change. Nature, 427, 145–148. 10.1038/nature02121 [DOI] [PubMed] [Google Scholar]
- Travis, J. M. J. (2003). Climate change and habitat destruction: A deadly anthropogenic cocktail. Proceedings of the Royal Society B: Biological Sciences, 270(1514), 467–473. 10.1098/rspb.2002.2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trembath, D. F. , & Fearn, S. (2008). Body sizes, activity times, food habits and reproduction of brown tree snakes (Boiga irregularis) (Serpentes: Colubridae) from tropical north Queensland, Australia. Australian Journal of Zoology, 56(3), 173–178. 10.1071/ZO08008 [DOI] [Google Scholar]
- Tyrrell, C. L. , Christy, M. T. , Rodda, G. H. , Yackel Adams, A. A. , Ellingson, A. R. , Savidge, J. A. , … Bischof, R. (2009). Evaluation of trap capture in a geographically closed population of brown treesnakes on Guam. Journal of Applied Ecology, 46, 128–135. 10.1111/j.1365-2664.2008.01591.x [DOI] [Google Scholar]
- Unger, S. D. , Abernethy, E. F. , Lance, S. L. , Beasley, R. R. , Kimball, B. A. , McAuliffe, T. W. , … Rhodes, O. E. (2015). Development and characterization of 33 novel polymorphic microsatellite markers for the brown tree snake Boiga irregularis . BMC Research Notes, 8(1), 8–11. 10.1186/s13104-015-1620-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitousek, P. M. , Mooney, H. A. , Lubchenco, J. , & Melillo, J. M. (1997). Human domination of Earth's ecosystems. Science, 277, 494–499. 10.1126/science.277.5325.494 [DOI] [Google Scholar]
- Waye, H. L. , & Mason, R. T. (2008). A combination of body condition measurements is more informative than conventional condition indices: Temporal variation in body condition and corticosterone in brown tree snakes (Boiga irregularis). General and Comparative Endocrinology, 155(3), 607–612. 10.1016/j.ygcen.2007.08.005 [DOI] [PubMed] [Google Scholar]
- Wilson, D. S. , Coleman, K. , Clark, A. B. , & Biederman, L. (1993). Shy‐bold continuum in pumpkinseed sunfish (Lepomis gibbosus): An ecological study of a psychological trait. Journal of Comparative Psychology, 107(3), 250–260. 10.1037/0735-7036.107.3.250 [DOI] [Google Scholar]
- Yang, J. , Lee, S. H. , Goddard, M. E. , & Visscher, P. M. (2011). GCTA: A tool for genome‐wide complex trait analysis. American Journal of Human Genetics, 88(1), 76–82. 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Závorka, L. , Lang, I. , Raffard, A. , Evangelista, C. , Britton, J. R. , Olden, J. D. , & Cucherousset, J. (2018). Importance of harvest‐driven trait changes for invasive species management. Frontiers in Ecology and the Environment, 16(6), 317–318. 10.1002/fee.1922 [DOI] [Google Scholar]
- Zuur, A. F. , & Ieno, E. N. (2016). A protocol for conducting and presenting results of regression‐type analyses. Methods in Ecology and Evolution, 7(6), 636–645. 10.1111/2041-210X.12577 [DOI] [Google Scholar]
- Zuur, A. F. , Ieno, E. N. , & Elphick, C. S. (2010). A protocol for data exploration to avoid common statistical problems. Methods in Ecology and Evolution, 1(1), 3–14. 10.1111/j.2041-210X.2009.00001.x [DOI] [Google Scholar]
- Zuur, A. F. , Ieno, E. N. , Walker, N. , Savaliev, A. , & Smith, G. (2009). Chapter 8: Meet the exponential family Gail M., Krickeberg K., Samet J. M., Tsiatis A., & Wong W. (Eds.), Mixed effects models and extensions in R (p. 205). New York, NY: Springer. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing reads are archived with the DRYAD data repository at https://doi.org/10.5061/dryad.47ss1b4. Data analyzed in the study are available through ScienceBase at https://doi.org/10.5066/P9X1AKVJ.


