Abstract
Fundamental metabolic pathways are essential for mammalian cells to provide energy, precursors for biosynthesis of macromolecules, and reducing power for redox regulation. While dysregulated metabolism (e.g., aerobic glycolysis also known as the Warburg effect) has long been recognized as a hallmark of cancer, recent discoveries of metabolic reprogramming in immune cells during their activation and differentiation have led to an emerging concept of “immunometabolism.” Considering the recent success of cancer immunotherapy in the treatment of several cancer types, increasing research efforts are being made to elucidate alterations in metabolic profiles of cancer and immune cells during their interplays in the setting of cancer progression and immunotherapy. In this review, we summarize recent advances in studies of metabolic reprogramming in cancer as well as differentiation and functionality of various immune cells. In particular, we will elaborate how distinct metabolic pathways in the tumor microenvironment cause functional impairment of immune cells and contribute to immune evasion by cancer. Lastly, we highlight the potential of metabolically reprogramming the tumor microenvironment to promote effective and long-lasting antitumor immunity for improved immunotherapeutic outcomes.
1. Introduction
The profound effect of metabolic alterations in cancer cells on disease development is well established and metabolic reprogramming is now considered one of the hallmarks of cancer (Cairns, Harris, & Mak, 2011; DeBerardinis & Thompson, 2012; Galluzzi, Kepp, Vander Heiden, & Kroemer, 2013; Hanahan & Weinberg, 2011). However, the metabolic modulation of the immune system is not well defined. There is growing interest in the emerging role of “immunometabolism” as an important regulator of the fate and function of immune cells (Barton & Medzhitov, 2002; Ganeshan & Chawla, 2014; Grohmann & Bronte, 2010; Lochner, Berod, & Sparwasser, 2015; Pearce & Pearce, 2013). The changes in key metabolic programs within immune cells are now known to be triggered not only by nutrients or oxygen conditions, but also by immune signals (O’Neill & Pearce, 2016). It is apparent that, other than energy production and biosynthesis, distinct metabolic pathways can govern the phenotype and function of immune cells.
Recent advances in the field of cancer immunotherapy have generated new powerful modalities for cancer management (e.g., immune checkpoint blockade, T cell therapy, and cancer vaccines) and are beginning to re-shape the landscape of cancer therapy (Guo et al., 2013; Hodi et al., 2010; Kantoff et al., 2010; Pardoll, 2012; Wang, Zuo, Sarkar, & Fisher, 2011). The immune checkpoint inhibitors (ICIs) that bolster antitumor immunity are now FDA approved for the treatment of a broad spectrum of cancers, culminating in unprecedented responses in patients with several types of advanced diseases (Ribas & Wolchok, 2018). However, a substantial number of patients fail to respond to these clinically approved immune-modulating drugs. Multiple mechanisms (e.g., elevation of immune checkpoint molecules, recruitment of immunosuppressive cells or factors, impaired antigen presentation) may contribute to immune escape of cancer cells and prevent effective antitumor immunity (Chen & Mellman, 2013; Dunn, Old, & Schreiber, 2004; Hanahan & Coussens, 2012; Motz & Coukos, 2013). Increasing evidence suggests that the deregulation of energy metabolism could be responsible for the failure of cancer immunotherapy (Martinez-Outschoorn, Peiris-Pages, Pestell, Sotgia, & Lisanti, 2017).
Complex and dynamic metabolic reprogramming is a common feature of cancer cells, which accommodates the biosynthetic and bioenergetic demands for growth and adaptation to the “stressful” tumor microenvironment (TME) (Viale & Draetta, 2016). Beyond the “Warburg effect,” i.e., preferential use of glycolysis by cancer cells for ATP generation, hypoxia and pH also play a major role in defining the metabolic TME (Cairns et al., 2011; Kareva & Hahnfeldt, 2013; Warburg, 1956; Ward & Thompson, 2012; Xie & Simon, 2017). Metabolic activity of cancer cells can shape the immune compartment by actively competing for key nutrients (e.g., glucose, glutamine, lipids, and amino acids) or producing metabolic by-products, which directly or indirectly impairs activation, fitness, and effector function of immune cells (Ben-Shoshan, Maysel-Auslender, Mor, Keren, & George, 2008; Biswas, 2015; Cairns & Mak, 2017; Chang et al., 2015; Fischer et al., 2007; Lochner et al., 2015). As a consequence, these dysfunctional immune cells not only fail to eradicate cancer cells, but also may transition into tumor-supporting cells to facilitate cancer progression and invasion. However, our knowledge of the fundamental impact of metabolic reprogramming on immune cells within the TME or during cancer immunotherapy is relatively limited. In this review, we describe our current understanding of metabolic reprogramming in cancer cells as well as immune cells during their activation and differentiation. We will also expand on the intrinsic and extrinsic metabolic pathways involved in cancer-induced immune dysfunction and potential development of novel strategies to metabolically reprogram the cancer-immune interface, thereby enhancing or optimizing existing immunotherapies.
2. Cell metabolism: Overview
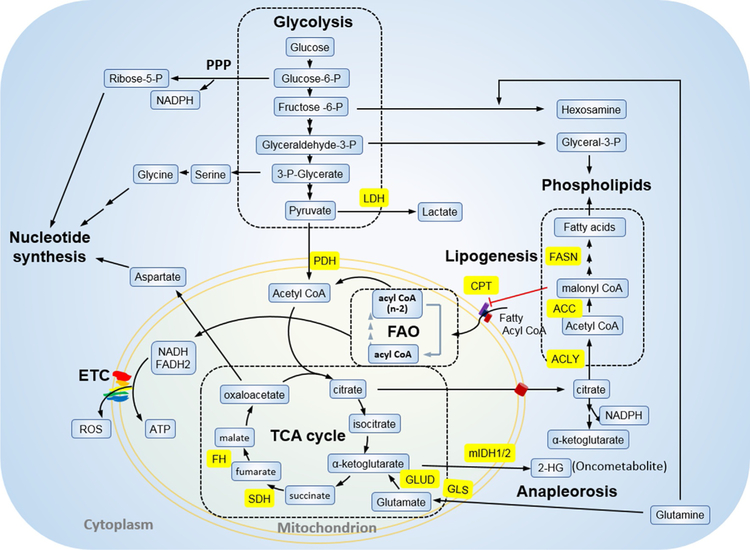
Mammalian cells rely on fundamental catabolic pathways to generate energy, precursors for biosynthesis of macromolecules, and reducing power (NADPH) for redox regulation (Vander Heiden, Cantley, & Thompson, 2009). Glucose, amino acids (primarily glutamine), and fatty acids (FAs) are the main nutrients supporting these processes (Fig. 1) (Cantor & Sabatini, 2012). Glucose is the preferred carbon source of energy via glycolytic metabolism to provide acetyl-CoA for oxidation through the tricarboxylic acid (TCA) cycle in the mitochondrion. The pentose phosphate pathway that branches off glycolysis provides an alternative pathway for glucose breakdown to generate NADPH and ribose-5-phosphate for biosynthesis of nucleic acids. When blood sugar levels drop, FAs from stored fat are split into acetyl-CoA through fatty acid β oxidation (FAO) in the mitochondrion. This process is associated with electron transfers to electron transport chain (ETC) to generate ATP. The resulting acetyl-CoA joins the TCA cycle or when in excess in the liver, is processed to form ketone bodies for use by extrahepatic tissues. Recent studies demonstrate that FAO-generated acetyl-CoA enters the TCA cycle to form citrate, which can be exported to the cytoplasm to engage NADPH-producing oxidation of isocitrate to α-ketoglutarate by isocitrate dehydrogenase (Carracedo, Cantley, & Pandolfi, 2013; Palmieri et al., 2015; Williams & O’Neill, 2018). Hence, FAO is also involved in replenishing cytosolic NADPH.
Fig. 1.
Overview of cell metabolism. The metabolism of glucose, fatty acids, and glutamine generates intermediates to support cell survival and proliferation. Glycolysis and FAO provide acetyl-CoA for oxidation through TCA cycle. The pentose phosphate pathway branching off glycolysis generates NADPH and ribose-5-phosphate for nucleotide synthesis. Cytosolic aspartate and acetyl-CoA generated by TCA cycle metabolites oxaloacetate and citrate are used for nucleotide and FAS, respectively. The intermediate malonyl CoA from the FAS pathway blocks CPT1, the rate-setting enzyme of FAO. CPT1, carnitine palmitoyltransferase I; ETC, electron transport chain; FAO, fatty acid oxidation; FAS, fatty acid synthesis; FASN, fatty acid synthase; FH, fumarate hydratase; GLUD, glutamate dehydrogenase; GLS, glutaminase; mIDH1/2, mutant isocitrate dehydrogenases 1 and 2; LDH, lactate dehydrogenase; SDH, succinate dehydrogenase; TCA, tricarboxylic acid.
The TCA cycle coordinates oxidation of acetyl-CoA from diverse nutrients to generate energy, and exit of intermediates from the cycle to support multiple biosynthetic pathways. Typically, oxaloacetate (OAA) serves as a vehicle to carry the two carbons from acetyl-CoA, first forming citrate followed by a series of isomerization and oxidation steps to generate reducing equivalents to drive ATP production via oxidative phosphorylation. After the two carbons are completely oxidized and released in the form of CO2 through the cycle, OAA will be recovered to load acetyl group again for the next cycle. However, in growing cells that require biomass accumulation, TCA intermediates are drawn off as precursors for biosynthesis of amino acids, lipids and other biomolecules. Under these conditions, OAA would become limited and must be replenished by OAA-producing pathways termed anaplerosis, which enables the TCA cycle to function as a biosynthetic pathway as opposed to a purely bioenergetic process (Owen, Kalhan, & Hanson, 2002).
The most significant anaplerostic contribution is conferred by glutaminolysis, a series of biochemical reactions to lyse the non-essential amino acid glutamine to form glutamate, aspartate, pyruvate, lactate, alanine, asparagine, serine, proline, and citrate (DeBerardinis et al., 2007). Glutamate is further converted to α-ketoglutarate, which enters the TCA cycle to generate ATP. In addition to fueling the TCA cycle, glutamine serves as an inter-organ shuttle of carbon and nitrogen and is the major source of nitrogen for non-essential amino acids, nucleotides, and hexosamines (Hensley, Wasti, & DeBerardinis, 2013). In rapidly dividing cells such as lymphocytes and enterocytes of the small intestine, glutamine is vigorously consumed and utilized for both bioenergy production and biomass accumulation (Windmueller & Spaeth, 1974).
3. Metabolic reprogramming in cancer
To meet the demands of a highly proliferative state and survival in various unfavorable microenvironments, tumors undergo fundamental alterations in their metabolism of carbohydrates, lipids, and glutamine (DeBerardinis & Chandel, 2016). Cancer cells are subsequently metabolically transformed, as characterized by increased glucose and glutamine consumption, dependence on glycolytic ATP as a major energetic pathway, and elevated de novo lipid synthesis to provide building blocks for membrane biosynthesis (Pavlova & Thompson, 2016).
3.1. The glycolytic phenotype
The most prominent aspect of metabolic transformation is the glycolytic phenotype or the Warburg effect, whereby cancer cells exhibit high glycolytic activity under aerobic conditions (Warburg, Posener, & Negelein, 1924). Because Warburg also observed increased aerobic glycolysis in normal proliferating cells and noted that it was reversible under normal conditions, he postulated that aerobic glycolysis was a consequence of irreversible damage to respiration in tumors (Warburg et al., 1924). Although later studies found mutations in some mitochondrial DNA-encoded ETC genes and genomic DNA-encoded genes of the TCA cycle (Baysal et al., 2000; Ward et al., 2010; Wu, Akhtari, & Alachkar, 2018), damaged respiration as an intrinsic property or “origin” of cancer is not established ( Ju et al., 2014; Stewart et al., 2015). In addition, hypoxia has long been known to stimulate glycolytic metabolism (Semenza, 2012). However, hypoxia is apparently not a causal factor of cancer glycolytic phenotype that occurs in both hypoxic and oxygenated regions of a tumor. Tumor cells also undergo glycolysis when cultured in vitro under normoxic conditions (Maxwell et al., 1999; Pollard et al., 2005).
In the last decade, advances in molecular biology and cancer genetics provide definite evidence for a mechanistic link between dysfunction of oncogenic proteins or tumor suppressors and hyperactive glycolysis in cancer (Levine & Puzio-Kuter, 2010). Ras, Akt, c-Myc, EGFR, BCR-ABL and ALK promote glycolysis via upregulation of or interaction with various glycolytic enzymes or intermediates including glucose transporter I (GLUT1) (Yun et al., 2009), hexokinase 2 (HK2) (Ma et al., 2016), lactate dehydrogenase A (LDHA) (Miller, Thomas, Islam, Muench, & Sedoris, 2012), and fructose-1,6-bisphosphate (F1,6BP) (Lim et al., 2016). Many oncoproteins activate hypoxia inducible factor (HIF) via hypoxia-independent mechanisms or a pseudohypoxic state to enhance tumor glycolysis (Lee et al., 2008; Ma et al., 2016). A multitude of glycolytic enzymes including HK2, phosphofructokinase 2 (PFK2), pyruvate kinase M2 (PKM2), LDHA, and pyruvate dehydrogenase kinase (PDK) have been identified to be HIF-targeted genes (Marin-Hernandez, Gallardo-Perez, Ralph, Rodriguez-Enriquez, & Moreno-Sanchez, 2009). The c-Myc transcription factor that commonly amplified and overexpressed in numerous tumors drives expression of GLUT1, HK2, PFKM, and LDHA (Miller et al., 2012). Loss of the tumor suppressor p53 inhibits the mitochondrial respiratory chain via suppression of SCO2 (the synthesis of cytochrome c oxidase protein) and promotes glycolysis via TIGAR, a TP53-induced glycolysis and apoptosis regulator (Won et al., 2012).
Despite the complex mechanisms underlying the cancer glycolytic phenotype, the potential advantages of glycolysis versus respiration in cancer cells are evident (Lunt & Vander Heiden, 2011; Vander Heiden et al., 2009). Although it yields a lower amount of ATP than mitochondrial oxidative phosphorylation (OXPHOS), direct phosphorylation of ADP during glycolysis is a more expedient way of ATP production. The enhanced rate of energy production may be more important than ATP yield to cancer cells. A preference for glycolysis is also evident in normal proliferating cells. Secondly, active OXPHOS is associated with generation of reactive oxygen species (ROS), byproduct production, and risk of oxidative stress. By diverting to glycolysis, actively proliferating cancer cells can avoid robust ROS production and oxidative damage to membranes, DNA and proteins, thereby favoring long-term survival of cancer cells. In addition to a bioenergetic role, glycolytic intermediates serve as precursors for biosynthesis of specific amino acids (e.g., serine, glycine) (Amelio, Cutruzzola, Antonov, Agostini, & Melino, 2014; Locasale, 2013) and particularly lipids that are essential building blocks of biological membranes, which are indispensable to increase tumor mass (Currie, Schulze, Zechner, Walther, & Farese, 2013). These key benefits inherent with aerobic glycolysis drive cancer cells to favor glycolysis over mitochondrial oxidation (Liberti & Locasale, 2016).
Although aerobic glycolysis as a signature of cancer cell metabolism has been successfully exploited in FDG-PET diagnosis of tumors in clinical oncology (Alavi & Reivich, 2002), the efforts in targeting glycolysis for treatment of cancer have not yet translated into clinical benefits. Inhibitors of HK2 (Maschek et al., 2004; Robustelli della Cuna & Pedrazzoli, 1991), PFK2 (Clem et al., 2013; Mondal et al., 2018), PKM2 (Dayton, Jacks, & Vander Heiden, 2016), LDHA (Le et al., 2010), and PDK (Michelakis et al., 2010) have antitumor effects both in vitro and in vivo. However, clinical trial failures with the HK2 inhibitors, 2-deoxyglucose and lonidamine have brought into question the feasibility of glycolysis-targeted therapies (Sborov, Haverkos, & Harris, 2015). Apparently, a more comprehensive understanding of functionally relevant mediators of tumor glycolysis and development of their specific inhibitors are critical to the successful applications of anti-glycolysis strategies as cancer therapies.
3.2. The lipogenic phenotype
Another prominent metabolic change in tumor cells compared with their normal counterparts is intensified de novo lipid synthesis, irrespective of the lipid precursors in the circulation (Currie et al., 2013). This lipogenic phenotype occurs at early stages of tumorigenesis including precursor lesions and becomes more pronounced in advanced stages of cancer (Menendez & Lupu, 2007; Wang, Rajput, Watabe, Liao, & Cao, 2010). Tumor cells depend heavily on or are “addicted” to the lipogenic phenotype as inhibition of de novo FA synthesis interferes with growth or survival of malignant cells.
In both normal and neoplastic cells, FAs are synthesized from glucose-derived acetyl-CoA. Acetyl-CoA in the mitochondrial matrix condenses with oxaloacetate to synthesize citrate as the first step of the TCA cycle. Once moved to the cytosol by citrate transporter on the mitochondrial membrane, citrate releases acetyl-CoA by the action of ATP citrate lyase (ACLY). Acetyl-CoA is then carboxylated by acetyl-CoA carboxylase (ACC) to generate malonyl-CoA, an intermediate for synthesis of palmitate by multimeric fatty acid synthase (FASN) (Mashima, Seimiya, & Tsuruo, 2009). Palmitate is further modified to increase the length of fatty acyl chain by Elongase or to add double bonds by Desaturase. FAs are esterified to synthesize triacylglycerides (TAG) as storage lipid droplets or phosphoglycerolipids as essential biological membrane components (Currie et al., 2013). It is evident that acquisition of an accelerated lipid synthesis rate independent of uncertain external lipid supplies confers a significant advantage to rapidly growing tumor cells (Medes, Thomas, & Weinhouse, 1953; Menendez, Colomer, & Lupu, 2005).
Hyperactive lipogenesis in cancer is mediated by increased expression and/or activity of key lipogenic enzymes, including ACLY, ACC and FASN (Hatzivassiliou et al., 2005; Swinnen, Brusselmans, & Verhoeven, 2006; Yoon et al., 2007). Each of these enzymes is over-expressed in different types of cancer. ACC1 is the rate-limiting enzyme of de novo FA synthesis under normal physiological conditions. This enzyme is activated by high glucose and insulin-induced dephosphorylation at Ser-79 and inhibited by glucagon and PKA-mediated phosphorylation (Fullerton et al., 2013). There are also other serine residues targeted for phosphorylation and inhibition by the energy sensor AMPK (Davies, Sim, & Hardie, 1990). Recent investigations show links of certain tumor suppressors or oncoproteins to dysregulation of ACC activity in cancer. For example, breast cancer type 1 susceptibility protein (BRCA1), a tumor suppressor, promotes phosphorylation of ACC (Moreau et al., 2006). Loss of BRCA1 function leads to dephosphorylation and activation of ACC1. Aldo-keto reductase family 1 B10 (AKR1B10), a novel protein overexpressed in human hepatocellular carcinoma and non-small cell lung carcinoma, can protect ACC from degradation through the ubiquitination-proteasome pathway (Ma et al., 2008). At the transcription level, ACC is regulated by the transcription factors carbohydrate response element binding protein (ChERBP) (Postic, Dentin, Denechaud, & Girard, 2007) and sterol response element binding protein 1c (SREBP-1c) (Yahagi et al., 2005). In cancer cells, the phosphoinositide 3-kinase (PI3K)-AKT (i.e., protein kinase B or PKB) pathway activates SREBP-1c and expression of SREBP-1c-dependent ACC and other cholesterol synthesis related genes (Campa et al., 2009). A recent study of breast cancer pathogenesis showed that the ACC gene is present in recurrent amplicons associated with reduced patient survival (Chin et al., 2006).
Compared to ACC, more evidence and therapeutic studies have attributed the cancer lipogenic phenotype to FASN hyperactivity. Originally found as a tumor antigen in the blood of cancer patients (Kuhajda, 2000), the large multimeric protein is abundantly expressed in multiple types of cancer, representing one of the most frequent phenotypic alterations in cancer (Flavin, Peluso, Nguyen, & Loda, 2010). Overexpression of FASN correlates with cancer progression, poor patient prognosis (Gansler, Hardman, Hunt, Schaffel, & Hennigar, 1997), and drug resistance (Wu, Qin, Fako, & Zhang, 2014). Many studies with FASN inhibitors of diverse mechanisms of action demonstrate that FASN hyperactivity is indispensable for maintaining the malignant phenotype of cancer (Alwarawrah et al., 2016; Lupu & Menendez, 2006a; Puig et al., 2009).
The precise mechanisms responsible for tumor-associated FASN over-expression remain poorly understood. FASN gene amplification does not appear to be common in cancer. A number of studies on transcriptional regulation of FASN have highlighted the importance of signaling through receptor tyrosine kinases (Bandyopadhyay et al., 2005; Mashima et al., 2009) or G protein-coupled receptors (Mukherjee, Wu, Barbour, & Fang, 2012). EGF/EGFR and ERBB2 have been shown to induce SREBP-dependent FASN expression via the PI3K-AKT and MAPK/ERK pathways (Bandyopadhyay et al., 2005; Mashima et al., 2009). Consistent with the regulatory role of the PI3K-AKT signaling, mutation of the PTEN tumor suppressor gene in prostate cancer is coupled to FASN overexpression as a result of constitutive PI3K-AKT activation (Van de Sande, De Schrijver, Heyns, Verhoeven, & Swinnen, 2002). In hormonally responsive cancers, FASN overexpression seems to be upregulated by steroid hormones as exemplified by the effects of estrogen E2 and androgen in breast carcinoma and prostate cancer, respectively (Santolla et al., 2012; Swinnen, Esquenet, Goossens, Heyns, & Verhoeven, 1997). Similarly, the stimulatory effects of hormones also appear to involve activation of the PI3K-AKT and MAPK/ERK pathways (Lupu & Menendez, 2006b). There is also evidence for cancer-specific inhibition of ubiquitin proteasome-mediated degradation of FASN (Yu et al., 2013).
FASN consists of two identical multifunctional polypeptides, each containing seven functional domains (Asturias et al., 2005; Chirala & Wakil, 2004; Maier, Jenni, & Ban, 2006), generating an X-shaped structure with a full set of active sites amenable for drug development. A number of FASN inhibitors targeting different catalytic domains have been developed as anti-obesity drugs or potential anti-cancer agents (Buckley et al., 2017). These include Orlistat (FDA approved anti-obesity drug) and C75, the most frequently used FASN inhibitor in culture and in experimental animals (Kridel, Axelrod, Rozenkrantz, & Smith, 2004; Kuhajda et al., 2000). However, their applications to treatment of cancer are limited by the fact that they are nonselective suppressors of FASN in both normal and malignant tissues, which could promote weight loss, anorexia, fatigue, and other cancer-associated complications ( Jones & Infante, 2015). To specifically block lipid synthesis in cancer cells, it will be necessary to develop FASN inhibitors that are more accessible to cancer cells or exhibit differential inhibitory activities toward abundantly expressed FASN in cancer cells versus limited FASN expression in normal tissues.
3.3. The glutaminolytic phenotype
As a source for nitrogen and carbon in an array of growth-promoting pathways, glutamine is particularly crucial for proliferating cells (Rubin, 1990). Mirroring these critical metabolic functions of glutamine in the growing normal cells, glutaminolysis is highly active in cancer (Hensley et al., 2013). Multiple types of cancer cells exhibit increased glutamine consumption and glutamine addiction (Wise & Thompson, 2010). The biochemical basis for the strict dependence of cancer cells on glutaminolysis is now well understood. First and probably most importantly, glutamine lies in the central position to produce the most non-essential amino acids for protein synthesis. The constant supply of these amino acids maintains cellular activity of mTORC1, also known as mammalian target of rapamycin complex 1 (Duran et al., 2012), another mode for glutamine supply to promote protein synthesis. Indeed, tracer experiments have demonstrated that more than 50% of non-essential amino acids used in protein synthesis are directly derived from glutamine in growing cancer cells in culture (Alberghina & Gaglio, 2014). Glutamine supply is critical for cell survival by preventing activation of amino acid-sensing kinase and the subsequent integrated stress response (ISR) (Harding et al., 2003).
Glutamine is a substrate for glutamine fructose-6-phosphate aminotransferase (GFAT), a rate-limiting step leading to biosynthesis of hexosamine and the downstream uridine diphosphate N-acetylglucosamine (UDP-GlcNAc) (McKnight et al., 1992). UDP-GlcNAc is required for O-linked protein glycosylation. Accordingly, glutaminolysis is required for protein synthesis, protein glycosylation, and glycosylation-dependent protein folding and trafficking, and suppression of the endoplasmic reticulum (ER) stress (Wellen et al., 2010).
Besides regulation of protein metabolism and functional modifications, glutaminolysis is involved in nucleotide biosynthesis that is essential for cell division. Glutamine is a nitrogen reservoir for de novo purine and pyrimidine biosynthesis (Lane & Fan, 2015). Glutamine-derived aspartate serves as a crucial carbon source for both purine and pyrimidine (Birsoy et al., 2015). In agreement with an essential role of glutamine in nucleotide biosynthesis, glutamine-deprived cancer cells fail to enter the S phase of the cell cycle, which can be rescued specifically by exogenous nucleotides or supplementation with aspartate (Gaglio, Soldati, Vanoni, Alberghina, & Chiaradonna, 2009).
In addition to amino acids, proteins, and nucleotides, glutamine metabolism is responsible for production of most glutathione, an antioxidant vital to a cell’s immune defense and other cellular functions (Welbourne, 1979). Glutamine can generate glutamate and glycine, and regulates transport of cysteine as substrates to synthesize the tri-peptide (Glu-Cys-Gly) (Welbourne, 1979). Recent studies also suggest that glutamine may participate in the production of the reducing equivalent of NADPH to decrease glutathione (Botman, Tigchelaar, & Van Noorden, 2014). This is most likely accomplished via the link from α-ketoglutarate to malate and oxidation of malate by malic enzymes (DeBerardinis et al., 2007). Together, the glutamine regulation of glutathione and NADPH production neutralizes ROS activity to protect cancer cells from oxidative stress.
Mechanistically, glutaminolysis is re-wired in cancer as a consequence of oncogenic insults, loss of tumor suppressors or compensation for other cancer-specific metabolic programs (Altman, Stine, & Dang, 2016). Many proteins/enzymes in glutamine metabolism are dysregulated by oncogenes such as c-Myc (Gao et al., 2009), Ras (Gaglio et al., 2011) and YAP (Edwards et al., 2017). In particular, the c-Myc oncoprotein upregulates expression of glutamine transporters and glutaminase 1 (GSL1) to generate glutamate and further flux to α-ketoglutarate for the TCA cycle and glutathione production (Gao et al., 2009; Wise et al., 2008). c-Myc also mediates glutaminolysis activation induced by other oncogenic pathways (Csibi et al., 2014). K-Ras is another example of oncoprotein-mediated activation of glutamine metabolism. The K-Ras harboring G12C or G12D mutations induces activity of aminotransferases, glutamine-dependent production of nucleotides and NADPH (Son et al., 2013).
Glutamine metabolism could be stimulated by energy status of cancer cells. As discussed earlier, the primary source of ATP in cancer cells are derived from glycolysis, a quick yet inefficient way to generate ATP (Vander Heiden et al., 2009). Although this may be sufficient to support cancer cell physiology, it is possible that overall ATP/ADP ratios are relatively low in the context of cancer cells. Glutamate dehydrogenase (GLUD), the key enzyme to convert glutamate to α-ketoglutarate, is activated allosterically by ADP or low energetic state of cancer cells (Yang et al., 2009). Furthermore, hyperactive glycolysis and glutaminolysis may be driven by common transcription factors such as HIF-1α that can be activated through pseudohypoxia. Glutamine itself can increase stabilization of HIF-1α that, in turn, directs glutamine toward biosynthetic products (Drogat et al., 2007; Kwon & Lee, 2005; Zhdanov, Waters, Golubeva, & Papkovsky, 2015).
Many classes of compounds that target glutamine metabolism, from initial transport to conversion to α-ketoglutarate, have been evaluated for their anti-tumor potential in preclinical models and clinical trials (Altman et al., 2016). Some of these compounds such as CB-839 (an inhibitor of GLS) showed promising preclinical results and are currently moving into clinical trials (Gross et al., 2014; Jacque et al., 2015). A general concern with glutaminolysis-targeted compounds is their lack of selectivity toward cancer cells. None of the glutamine-metabolizing enzymes has been found to be mutated or exclusively expressed in cancer. The question is further complicated by dysregulation of multiple glutaminolytic enzymes in cancer. No validated target has been identified for the glutaminolysis pathway that is of general acceptance by the scientists in the field. Moreover, functional redundancy or metabolic compensation from other isoforms could prevent or minimize the dependence of cancer on a single glutaminolytic enzyme.
3.4. Mutations in TCA cycle enzymes in cancer
Although metabolic reprogramming is a well-recognized hallmark of cancer, a causal role for metabolic dysfunction in cancer pathogenesis remains controversial, mainly due to the lack of genetic mutations of key metabolic enzymes of the major metabolic pathways discussed above. This view, however, has changed with the discovery of cancer-associated mutations in three enzymes of the TCA cycle: isocitrate dehydrogenase (IDH), succinate dehydrogenase (SDH) and fumarate hydratase (FH) (Laurenti & Tennant, 2016). These mutations predispose carriers to a number of hereditary and sporadic malignancies. FDA has recently approved an IDH1 mutant-targeted inhibitor Tibsovo (ivosidenib) to treat elapsed or refractory acute myeloid leukemia (AML) (DiNardo et al., 2018).
Due to the central importance of the TCA cycle in cell metabolism, IDH, SDH or FH mutations cause metabolic perturbations and re-wiring to synthesize anabolic building blocks such as amino acids, FAs and nucleotides (Metallo et al., 2011; Tong et al., 2011). The truncated TCA cycle is accompanied by increases in glycolysis to supply ATP, glutaminolysis to maintain TCA intermediates and amino acid synthesis, and lipogenesis from excess acetyl-CoA. Most clearly, these mutations are associated with the accumulation of metabolites that are able to influence many aspects of cancer. For example, mutation in IDH1 or IDH2 leads to loss of the enzyme’s oxidative decarboxylation activity while concomitantly acquiring a neomorphic activity characterized by an ability to catalyze conversion of a-KG to 2-hydroxyglutarate (2-HG), a prototype oncometabolite (Dang et al., 2009). Due to its structural similarity to α-KG, 2-HG can interact and inhibit α-KG-dependent dioxygenase enzymes including the HIF prolyl hydroxylase (PHD) (Zhao et al., 2009). PHD induces hydroxylation of HIF-1α in normoxia, allowing ubiquitination by the E3 ubiquitin ligase pVHL (product of von Hippel-Lindau tumor suppressor gene) and subsequent proteasomal degradation. Thus accumulation of 2-HG in IDH defective cells mimics hypoxia to stabilize HIF-1α, another mode of pseudohypoxia (Yang, Soga, & Pollard, 2013). Mutations of SDH or FH also appear to inhibit PHD activity via abnormal accumulation of succinate or fumarate (Isaacs et al., 2005; Selak et al., 2005). Other common consequences of these mutations of the TCA cycle are inhibition of NADPH and glutathione, and increases in ROS production and oxidative stress (Yang, Soga, & Pollard, 2013). In sum, mutations in TCA cycle enzymes likely promote tumorigenesis through inducing cancer-associated metabolic phenotypes such as increased dependence on glycolytic and glutaminolytic metabolism, production of oncometabolites to activate HIF-1α, and increased intracellular ROS.
4. Metabolic regulation of immune functions
The main function of the host immune system is removal of invading pathogens and maintenance of tissue homeostasis. This highly interactive and complex immune network, which comprises lymphoid organs and various types of immune cells together with immune modulating factors (e.g., cytokines, chemokines, surface molecules), can be mobilized to mount a properly controlled immune response to eliminate pathogens or transformed cells, while avoiding massive tissue destruction. However, the failure of this system can result in lethal infections, cancer, autoimmunity, and many other disorders (Parkin & Cohen, 2001; Villani, Sarkizova, & Hacohen, 2018). There are two major arms of the immune system. The innate immune system, e.g., natural killer (NK) cells, macrophages/monocytes, or neutrophils, can provide immediate immunity as a first-line of defense. The adaptive immune system, e.g., T lymphocytes, B lymphocytes, can be activated to generate an antigen-specific long-lasting immune response (Delves & Roitt, 2000). In recent years, an improved appreciation of the complex interplay between metabolic reprogramming and immune cell development or functionality has emerged. A large body of research in the field of immunometabolism is focusing on elucidating metabolic changes associated with phenotypic differentiation and activation of major immune cell subsets (e.g., macrophages, T cells), and more importantly, the functional outcomes of metabolic reprogramming in these immune cells. These findings have advanced our fundamental understanding of distinct metabolic alterations in defining the diversity and functional plasticity of immune cells.
4.1. Dendritic cells
Dendritic cells (DCs) are specialized antigen-presenting cells (APCs) that play an essential role in innate and adaptive immunity. These cells can sense a wide range of inflammatory stimuli derived from microbial pathogens or transformed cells through pattern recognition receptors and initiate protective immune responses (Akira & Takeda, 2004; Banchereau & Steinman, 1998; Barton & Medzhitov, 2002). It is well documented that DCs at different differentiation or activation stages are buttressed by diverse types of cellular metabolism to fulfill their bioenergetic and biosynthetic demands.
Upon exposure to granulocyte-macrophage colony-stimulating factor (GM-CSF) and interleukin 4 (IL-4), human monocytes differentiate into DCs, a process that requires lipid metabolism and mitochondrial biogenesis (Ishikawa et al., 2007; Le Naour et al., 2001). Blockade of FA synthesis markedly diminishes human DC development from peripheral blood mononuclear cell precursors (Rehman et al., 2013). In contrast, DCs consume more glucose and produce higher amounts of lactate after activation by toll-like receptor (TLR) ligands (Everts et al., 2012; Jantsch et al., 2008; Krawczyk et al., 2010). This metabolic shift toward glycolysis is dependent on inhibition of OXPHOS by nitrite oxide (NO) produced during DC activation. Activation and survival of DCs after TLR stimulation are severely reduced by glucose restriction (Everts et al., 2012; Krawczyk et al., 2010). In addition to glucose, DCs also express specific isoforms of enzymes essential for glycogen metabolism to support their maturation and effector function, particularly at an early stage of activation or under glucoserestricted conditions (Thwe et al., 2017). The engagement of TLRs triggers a marked glycolytic flux, which is essential for de novo synthesis of FAs. FA synthesis can then support expansion of the ER and Golgi for the expression of co-stimulatory molecules and secretion of inflammatory cytokines (Everts et al., 2014). In contrast to activated or mature DCs, tolerogenic DCs display high mitochondrial oxidative activity and manifest substantially higher FAO rate. As a result, inhibition of FAO can partially enhance T cell-stimulating capacity of tolerogenic DCs (Malinarich et al., 2015). However, the molecular and biochemical switch from glycolysis to OXPHOS in tumor-associated DCs that are known to be functionally impaired or tolerogenic remains to be defined.
4.2. Macrophages
Macrophages are multifunctional innate immune cells involved in diverse physiological and pathological states, including host defense, tissue homeostasis, tumor promotion or destruction, and regulation of T cell immunity. Initial studies using in vitro stimuli have led to identification of two main phenotypes of macrophage activation or polarization. Inflammatory stimuli, such as IFN-γ plus LPS, can induce classical activation of macrophages (i.e., M1 phenotype), which is characterized by production of inflammatory cytokines (e.g., TNF-α, IL-12) and reactive nitrogen and oxygen intermediates (RNIs, ROIs), as well as enhanced microbicidal functions. In contrast, M2 polarization or alternative activation of macrophages is often induced by anti-inflammatory stimuli (e.g., IL-4, IL-13, glucocorticoids), which is associated with a functional shift toward tissue recovery or immunosuppression (Biswas & Mantovani, 2010; Wynn, Chawla, & Pollard, 2013; Xue et al., 2014).
M1/M2 polarization of macrophages displays distinctive metabolic programs. While M1 macrophages preferentially consume glucose (Rodriguez-Prados et al., 2010), alternatively activated macrophages mainly use FAO and mitochondrial biogenesis (Vats et al., 2006). LPS stimulation of macrophages enhances the levels of succinate, an intermediate of the TCA cycle, followed by upregulation of glycolytic genes and reduced expression of mitochondrial biogenesis genes. Inhibition of glycolysis in macrophages with 2-deoxyglucose decreases IL-1β production induced by LPS (Tannahill et al., 2013). In response to IL-4, signal transducer and activator of transcription 6 (STAT6) and PPARγ-coactivator-1β (PGC-1β), which link cellular programs of inflammation and lipid homeostasis, initiate metabolic reprogramming in macrophages toward FAO and mitochondrial biogenesis (Lin, Handschin, & Spiegelman, 2005; Puigserver et al., 1998; Vats et al., 2006). M1 macrophages upregulate the glucose transporter GLUT1 (Fukuzumi, Shinomiya, Shimizu, Ohishi, & Utsumi, 1996), whereas lipoprotein lipase and CD36, which regulate uptake and transport of triacylglycerol, are induced in M2 macrophages (Feng et al., 2000; Huang et al., 2014). Uptake and lipolysis of exogenous triacylglycerols generate FAs for FAO and promote the expression of genes that define M2 macrophages (Huang et al., 2014). However, the role of FAO in M2 macrophages needs to be investigated further. It was recently reported that, while the CPT2-deficient macrophages showed a defect in FAO, these cells were properly polarized to a M2 phenotype in vitro and in vivo (Nomura et al., 2016). Additionally, inhibition of FAO in human macrophages with etomoxir has no effect on IL-4-induced M2 polarization (Namgaladze & Brune, 2014).
Regulation of macrophage function by cholesterol metabolism was also documented recently. Engaging type I interferon (IFN) signaling in macrophages results in metabolic shift, reflected by a decrease in cholesterol biosynthesis and an increase in cholesterol import. Artificially promoting this metabolic shift in macrophages induces the expression of IFN-inducible genes and enhances the anti-viral immune response (York et al., 2015). Intriguingly, limiting flux via the cholesterol biosynthetic pathway engages a STING (i.e., stimulator of interferon genes)-dependent type I IFN response, which not only delineates a metabolic-inflammatory circuit involving lipid biosynthesis and innate immunity, but also provides supporting evidence for involving metabolic reprogramming to modulate the host immunity.
Amino acid metabolism has been implicated in functional modulation of macrophages. IFN-γ and LPS stimulation can upregulate inducible nitric oxide synthase (iNOS or NOS2) that promotes the conversion of l-arginine into NO in macrophages. However, IL-4 treatment of macrophages induces arginase 1 that catalyzes the conversion of l-arginine to l-ornithine (Modolell, Corraliza, Link, Soler, & Eichmann, 1995). Indeed, distinctiveness of arginine metabolism has been shown to correlate with diverse functions of macrophages, including elimination of microbial pathogens and cancer cells, tissue remodeling and, in some cases, support of tumor growth (Chang, Liao, & Kuo, 2001; Rath, Muller, Kropf, Closs, & Munder, 2014). A recent study showed that glutamine catabolism and UDP-GlcNAc-associated modules are required during M2 polarization of macrophages. Glutamine deprivation or inhibition of N-glycosylation can suppress M2 polarization and production of the chemokine CCL22 ( Jha et al., 2015).
The NOD-like receptor family pyrin domain containing 3 (NLRP3) inflammasome is a widely studied intracellular pattern recognition multiprotein complex that tightly regulates the innate immune response and the production of pro-inflammatory cytokines such as IL-1β and IL-18 (Guo, Callaway, & Ting, 2015; Latz, Xiao, & Stutz, 2013; Martinon, Burns, & Tschopp, 2002). Lipid synthesis stimulated by mitochondrial uncoupling protein-2 (UCP2) has been shown to regulate activation of the NLRP3 inflammasome in macrophages, which is supported by reduced production of IL-1β and IL-18 in LPS challenged UCP2-deficient mice. Defective lipid synthesis in the absence of UCP2 resulted from decreased expression of FASN (Moon, Lee, et al., 2015). Strikingly, FAO promoted activation of the NLRP3 inflammasome in macrophages through NADPH oxidase 4 (NOX4)-dependent upregulation of carnitine palmitoyltransferase 1A (CPT1A), a rate limiting enzyme that controls mitochondrial FAO (Moon et al., 2016). Additionally, saturated FA palmitate, not unsaturated oleate, can trigger the activation of the NLRP3 inflammasome in macrophages (Wen et al., 2011). Stimulation of a glycolytic phenotype in macrophages is also linked to NLRP3 inflammasome activation, evidenced by the observation that inhibition of glycolysis in macrophages suppresses both caspase-1 activation and IL-1β maturation in response to LPS and ATP (Moon, Hisata, et al., 2015). Together, metabolic reprogramming is essential for the phenotypic polarization and functional activation of macrophages.
4.3. NK cells
NK cells are innate immune lymphocytes that function as a key element of the first line of defense against infection and cancer cells. These cells are actively involved in host defense either through cytotoxicity and cytokine secretion (i.e., IFN-γ) or through modulating the functions of other immune cells (i.e., DCs, T cells) (Biron & Brossay, 2001; French & Yokoyama, 2003; Long, Kim, Liu, Peterson, & Rajagopalan, 2013; Malmberg et al., 2017; Vivier, Tomasello, Baratin, Walzer, & Ugolini, 2008). NK cells use mitochondrial OXPHOS primarily in a resting state. Upon NK cell activation with cytokines (e.g., IL-2), dramatic metabolic reprogramming occurs, indicated by elevated rates of glycolysis. Inhibition of glycolysis is sufficient to reduce IFN-γ and granzyme B production by activated NK cells (Donnelly et al., 2014). The transcription factor Srebp is essential for cytokine-induced metabolic reprogramming of NK cells. During elevated glycolysis and OXPHOS in NK cells, Srebp is believed to promote metabolization of glucose to cytosolic citrate via the citratemalate shuttle. Blocking Srebp activation dampens NK cell activation as well as cytotoxicity against cancer cells (Assmann et al., 2017). Accordingly, metabolic reprogramming toward glycolysis is a prerequisite for NK cell effector function. Differences in the metabolic requirements for IFN-γ production by murine NK cells also depend upon the nature of the activation signal. While activation of NK cells by IL-12 plus IL-18 is independent of glycolysis or mitochondrial OXPHOS, stimulation of IFN-γ production via activating NK receptors requires glucose-driven OXPHOS (Keppel, Saucier, Mah, Vogel, & Cooper, 2015), suggesting that metabolism can serve as a second signal for IFN-γ production resulting from NK cell receptor activation.
4.4. T cells
T cells are essential adaptive immune cells required for host defense, homeostasis, immune tolerance, and immune memory. T-cell receptors (TCRs) are engaged to specifically recognize various antigens derived from invading pathogens, malignant cells or the environment. Based on lineage markers and functionality, T cells are classified into two major subsets, i.e., CD8+ T cells and CD4+ T cells. CD8+ T cells can be activated and differentiated into cytotoxic T lymphocytes (CTLs) after recognition of antigenic peptides in complex with MHC class I molecules. In contrast, CD4+ T cells can be primed by antigen in the context of MHC class II molecules present on professional APCs, including DCs, macrophages, and B cells. Upon activation, naïve CD4+ T cells differentiate into effector helper T cells depending on the cytokine milieu of the microenvironment ( Jenkins et al., 2001; Kumar, Connors, & Farber, 2018; Tao, Constant, Jorritsma, & Bottomly, 1997; Zhu & Paul, 2008). CTLs can mediate direct cytotoxic effects on target cells (e.g., infected or transformed cells), whereas helper CD4+ T cells mostly provide help for CTL function. Although the majority of effector T cells die after eradication of pathogens or cancer cells, a heterogeneous pool of memory T cells survive and remain in the body, which can be mobilized to mount a more rapid and stronger immune response upon encountering secondary infection from an antigenically similar pathogen (Gebhardt et al., 2009; Mueller, Gebhardt, Carbone, & Heath, 2013; Sallusto, Lenig, Forster, Lipp, & Lanzavecchia, 1999). Accumulating studies demonstrate that T cells undergo metabolic reprogramming during activation, which leads to imprinting of their distinct fates and functions.
While naïve T cells mainly utilize OXPHOS of various nutrients such as glucose and amino acids to generate energy, differentiating T effector cells following antigen encounter and activation shift toward glycolysis to support their rapid proliferation and function, e.g., production of cytokines (Chang et al., 2013; Frauwirth & Thompson, 2004; Rathmell, Vander Heiden, Harris, Frauwirth, & Thompson, 2000). Several signaling pathways govern the metabolic reprogramming of activated T cells. In addition to initiating intracellular signals that drive cell proliferation, engaging CD3 and CD28 on T cells triggers activation of signaling pathways involving PI3K and Akt (Appleman, van Puijenbroek, Shu, Nadler, & Boussiotis, 2002; Pages et al., 1994; Siska & Rathmell, 2015; Ward, Westwick, Hall, & Sansom, 1993). Akt is well-established in supporting cytokine production and cell survival, both of which are required for activated T cells (Datta, Brunet, & Greenberg, 1999; Jones et al., 2000; Kane, Andres, Howland, Abbas, & Weiss, 2001; Rathmell, Elstrom, Cinalli, & Thompson, 2003). These signaling events promote glycolysis by inducing glucose transporters (e.g., GLUT1) and rate limiting enzymes of glycolysis (e.g., HK2), and amino acid transporters (Barthel et al., 1999; Frauwirth et al., 2002; Rathmell et al., 2003). It is worth noting that many of these signaling and metabolic changes occurring in activated T cells resemble alterations observed in cancer cells.
In addition to co-stimulation signals, activation of T cells is tightly regulated through immune checkpoint molecules or co-inhibition signals to avoid aberrant immune responses, such as those promoting autoimmunity. Cytotoxic T-lymphocyte associated protein 4 (CTLA-4), an immune checkpoint molecule that displays much higher affinity for the co-stimulation B7 molecules on APCs, is induced during T cell activation. The competition for binding to B7 molecules blocks co-stimulation signaling through CD28 and abrogates T cell activation (Krummel & Allison, 1995; Walunas et al., 1994). Programmed cell death-1 (PD-1) is another immune checkpoint molecule that restrains T cell response (Freeman et al., 2000), which can cause exhaustion of effector T cells during chronic infections and cancer immune evasion (Crawford et al., 2014; Wherry, 2011; Wherry et al., 2007). It was recently shown that engagement of PD-1 signaling in human CD4+ T cells impairs their capability to uptake and utilize glucose, amino acids, and glutamine. Instead, these T cells display an increased rate of FAO, which correlates with elevation of the key enzyme CPT1A (Patsoukis et al., 2015). Similar to PD-1 ligation, CTLA-4 also reduces glucose uptake in T lymphocytes following co-stimulation (Frauwirth et al., 2002; Parry et al., 2005). However, molecular studies reveal that CTLA-4 inhibits glycolysis through down-regulation of GLUT1 without impacting CPT1A level and FAO rate (Patsoukis et al., 2015), implicating a role of CTLA-4 in maintaining the metabolic profile of non-stimulated cells. Additional studies showed that both PD-1 and CTLA-4 signaling block CD28-mediated activation of PI3K and Akt (Frauwirth et al., 2002; Parry et al., 2005, 1997), suggesting that Akt represents a conserved mechanism used by immune checkpoint molecules to attenuate T cell activation.
Activated T cells also increase uptake of FAs and promote lipid synthesis (Lochner et al., 2015). ACC1 is an enzyme that converts acetyl coenzyme A to malonyl coenzyme A, which is required for long-chain FA synthesis. T cell-specific deletion of ACC1 impairs expansion of antigen-specific CD8+ T cells due to increased cell death. Supplying exogenous FAs can rescue the survival of ACC1-deficient CD8+ T cells, supporting a critical role of FA synthesis during the activation and expansion of CD8+ T cells (Lee et al., 2014). Differentiation of inflammatory Th17 cells also depends on de novo FA synthesis through ACC1. Inhibition of ACC1 suppresses the differentiation of Th17 cells by promoting the polarization of anti-inflammatory Foxp3+ regulatory T cells or Tregs (Berod et al., 2014), accentuating the importance of selectivity for FA sources in dictating the development of different T cell lineages.
As opposed to the activated effector T cells that preferentially use glycolysis, energy metabolism in Tregs or CD8+ memory T cells depends largely on OXPHOS and FAO. In an asthma model, highly activated AMP-activated protein kinase in Tregs decreases GLUT1 expression and supports high lipid oxidation rates during Treg development (Michalek et al., 2011). CD8+ memory T cells with longevity post pathogen clearance (Beura & Masopust, 2014) carry a metabolic signature of enhanced mitochondrial FAO. In contrast to effector CD8+ T cells, CD8+ memory T cells use extracellular glucose to support OXPHOS and FAO, indicating lipid synthesis is needed to provide substrates for FAO (O’Sullivan et al., 2014). Several lines of evidence show that memory T cells require IL-7 and IL-15 for survival and self-renewal (Becker et al., 2002; Kaech et al., 2003; Kieper et al., 2002). During infection with Listeria monocytogenes, CD8+ memory T cells possess substantial mitochondrial spare respiratory capacity (SRC) in order to maintain cellular survival. IL-15 was found to promote the expression of CPT1A (van der Windt et al., 2012). TAG synthesis was shown to be a central component of IL-7-supported survival of human and mouse CD8+ memory T cells (Cui et al., 2015). IL-7 can induce the expression of the mammalian glycerol channel aquaporin 9 (AQP9) in CD8+ memory T cells. Lack of AQP9 impairs glycerol transportation into memory CD8+ T cells for TAG synthesis and storage (Carbrey et al., 2003; Cui et al., 2015; Rojek et al., 2007). These observations enrich our understanding of how cytokines define distinct T cell functions through mitochondrial metabolic regulation.
Metabolism of amino acids, which serve as a fuel source as well as precursors for synthesis of macromolecules, is another critical determinant for T cell development and function. Protein synthesis requires metabolism of l-arginine, which is also a precursor of immuno-modulating metabolites, such as polyamines and NO (Grohmann & Bronte, 2010). Elevation of l-arginine sensed by transcription factors BAZ1B, PSIP1, and TSN in activated T cells induces a metabolic shift from glycolysis to OXPHOS, which facilitates the generation of central memory T cells with enhanced survival capacity (Geiger et al., 2016), underscoring a role of l-arginine for reprogramming metabolism as well as functionality of T cells. Moreover, glutamine, the most abundant amino acid in blood, is required for energy production to support rapid division of activated T cells (Sinclair et al., 2013). TCR engagement can trigger amino acid transporter ASCT2-mediated uptake of glutamine and leucine, resulting in the differentiation of Th1 and Th17 cells (Nakaya et al., 2014). Glutamine also appears to be critical for the fitness of CD8+ CTLs and development of CD8+ memory T cells (Blagih et al., 2015). Taken together, these extensive studies elucidate the complexity of metabolic changes that can impact on the activation and differentiation of immune cell types.
5. Metabolic alterations in immune cells during cancer progression
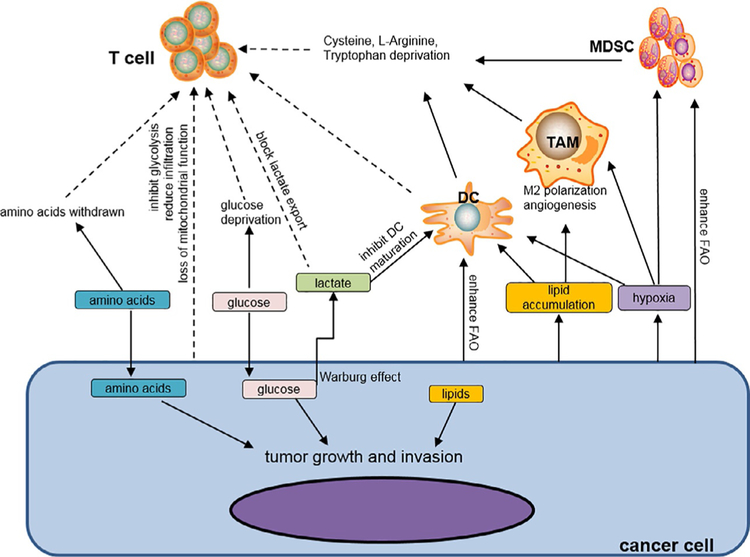
Metabolic reprogramming is a hallmark of cancer during tumorigenesis and progression (Hanahan & Weinberg, 2011; Ward & Thompson, 2012). Cancer cells preferentially use glucose to produce energy (i.e., Warburg effect) for supporting cell growth and proliferation (Cairns et al., 2011; Vander Heiden et al., 2009; Warburg, 1956). Growth-factor signaling within the TME activates PI3K-Akt, which in turn promotes the elevation of a number of glycolytic genes (i.e., GLUT1, HK2, PFKFB3 and LDHA) and PDK that suppresses the TCA cycle (Cairns et al., 2011; Semenza, 2003; Ward & Thompson, 2012). Consequently, these molecular events culminate in deprivation of glucose and amino acids in the TME, which directly causes dysfunction of tumor-infiltrating immune cells. In addition to supporting cancer cell growth, the metabolic reprogramming in cancer cells generates waste products, such as lactate, phosphoenolpyruvate, arginine and tryptophan by-products, which can further contribute to an immune suppressive environment and shape the fate or function of immune cells.
5.1. Immune evasion through metabolic competition
Although NK cells have intrinsic tumoricidal activity, they generally fail to control tumor progression due to their functional defects in the TME (Carrega et al., 2008; Mamessier et al., 2011; Platonova et al., 2011; Smyth, Hayakawa, Takeda, & Yagita, 2002). In a Kras-driven lung cancer transgenic model, upregulation of fructose-1,6-bisphosphatase (FBP1) in NK cells inhibits glycolysis, thereby impairing their tumor-destroying capability (Cong et al., 2018). In addition to glucose, cancer cells also have a high demand for glutamine (Still & Yuneva, 2017; Zhang, Pavlova, & Thompson, 2017). Glutamine withdrawal or systemic blockade of l-amino acid transport inhibits induction of the transcription factor c-Myc during NK cell activation. The loss of c-Myc reduces cell growth and tumor cytotoxicity of NK cells (Loftus et al., 2018), suggesting an essential role of glutamine and amino acids for NK cell function.
Increasing evidence indicates that glucose deprivation caused by the tumor Warburg effect suppresses effector functions of tumor-reactive T cells. Cancer cells can escape from CD4+ T cell-mediated immune surveillance by increasing glycolysis via expression of HK2. Insufficient glycolytic metabolite phosphoenolpyruvate (PEP) in CD4+ T cells due to metabolic competition results in an increase in the SERCA-mediated Ca2+ uptake and a defective T cell activation (Ho et al., 2015). In a mouse sarcoma model, it was demonstrated that competitive glucose consumption by tumors blocks mTOR activity, glycolytic capacity, and IFN-γ production within T cells, consequently disabling the tumor-protective ability of T cells (Chang et al., 2015). Moreover, a substantial elevation of glycolysis-related genes is commonly seen in human melanoma and lung cancer samples that are poorly infiltrated by T cells, indicating that tumor glycolysis likely creates a barrier for T cell infiltration (Cascone et al., 2018). The enhanced glycolysis of cancer cells causes excessive production of lactic acid (LA) in the TME, which can further disturb T cell metabolism and function by blocking export of LA from T cells. This is consistent with a positive correlation between serum levels of lactate and tumor burden in cancer patients (Fischer etal., 2007). Therefore, it is conceivablethat Tcellresponsivenessismodulated, at least partially, by tumor-triggered glucose restrictions and glucose consumption may serve as an additional mechanism underlying tumor immune evasion. Intriguingly, it was recently found that T cells infiltrating murine and human tumors experience a loss of mitochondrial mass and function. This impaired mitochondrial biogenesis in tumor-specific T cells resulted from defective Akt-PPAR-γ coactivator 1α (PGC1α) signaling (Scharping et al., 2016), indicating that repression of oxidative metabolism in tumor-infiltrating T cells abrogates their effector function by limiting metabolic requirements.
5.2. Metabolic reprogramming and dysfunction of tumor-associated APCs
Active glycolysis in cancer cells and stromal cells (e.g., cancer-associated fibroblasts or CAFs) are known to cause accumulation of LA, an end-product of glycolysis (Ghesquiere, Wong, Kuchnio, & Carmeliet, 2014). Tumor-derived LA is a key factor involved in tumor escape from immune surveillance, supported by its effect on inhibition of IL-12 production by DCs and induction of a phenotype similar as tumor-associated DC during DC differentiation (Gottfried et al., 2006).
Enhanced FA synthesis in DCs in response to TLR stimulation increases lipid storage (Everts et al., 2014; Maroof, English, Bedford, Gabrilovich, & Knight, 2005). However, high lipid content in tumor-associated DCs have been associated with dysfunction of DCs in the TME. Prostaglandin E2 (PGE2) is a prostanoid lipid that can enhance cancer cell survival, growth, migration, angiogenesis, and immunosuppression. Cyclooxygenase (COX)-1 and 2, the catalytic enzymes for production of PGE2, are highly upregulated in colorectal, breast, stomach, lung, and pancreatic cancers (Dannenberg & Subbaramaiah, 2003; Wang & DuBois, 2010). PGE2 produced by melanoma cells was reported to prevent accumulation and activation CD103+ DCs and to downregulate the expression of molecules essential for anti-tumor type I immunity, such as IFN-γ, T-bet, and IL-12 (Zelenay et al., 2015). DCs from tumor-bearing mice or cancer patients also accumulate higher amounts of triglycerides as compared with DCs from healthy hosts. Lipid accumulation in tumor-associated DCs is believed to be largely caused by upregulation of scavenger receptor A (i.e., CD204), a negative regulator of DC immunogenicity (Guo, Yi, Yu, Hu, et al., 2012; Guo, Yi, Yu, Zuo, et al., 2012; Qian et al., 2011; Wang, Facciponte, Chen, Subjeck, & Repasky, 2007; Yi et al., 2011; Yu, Guo, Fisher, Subjeck, & Wang, 2015). Indeed, lipid laden DCs exhibit reduced capacity to process antigen and stimulate allogenic T cells. Use of an acetyl-CoA carboxylase inhibitor or anti-SRA antibodies to reduce lipid content in DCs can substantially enhance the anti-tumor potency of DC vaccination (Herber et al., 2010). Tumor-derived oxidized neutral lipids, such as triglycerides, cholesterol esters and FAs, but not non-oxidized lipids, decreased the cell surface expression of peptide-MHC class I complexes and block the tumor antigen cross-presentation activity of DCs (Ramakrishnan et al., 2014). The X-box binding protein 1 (XBP1) is a major transcription factor mediating an ER stress response, which has been documented to directly support tumorigenesis by promoting cancer cell survival and metastasis (Chen et al., 2014; Lee, Iwakoshi, Anderson, & Glimcher, 2003; Tang et al., 2014). Constitutive activation of XBP1 in ovarian tumor-associated DCs can also induce a triglyceride biosynthetic program resulting in abnormal lipid accumulation, which subsequently blunts antitumor immunity (Cubillos-Ruiz et al., 2015), suggesting that targeting the XBP-1-mediated ER stress response may offer a unique strategy to metabolically enhance the immune-genicity of DCs.
While it is well recognized that aberrant β-catenin signaling plays an important role in driving cancer development and metastasis (Klaus & Birchmeier, 2008; Reya & Clevers, 2005), its involvement in functional impairment of immune cells is beginning to be clarified. Cancer cells can activate β-catenin signaling in DCs, resulting in inhibition of the cross-priming of T cells (Liang et al., 2014). Tumor-infiltrating DCs also metabolize vitamin A to produce retinoic acid via β-catenin signaling, which subsequently facilitates a regulatory T-cell response and immune tolerance. Deletion of β-catenin or blocking of the β-catenin pathway in DCs can effectively reduce regulatory T cell response and inhibit the growth of mouse melanoma (Hong et al., 2015). A recent study reported that melanoma establishes an immune privileged environment via a paracrine Wnt5a-β-catenin-peroxisome proliferator-activated receptor-γ (PPAR-γ) pathway, which upregulates the expression of CPT1A for increased FAO in DCs. This metabolic shift toward FAO promotes the induction of indoleamine 2,3-dioxygenase-1 (IDO1) while decreasing the production of immunostimulatory cytokines (e.g., IL-12), culminating in a tolerogenic phenotype of DCs and expansion of Tregs (Zhao et al., 2018). Based on these considerations, it is suggested that manipulation of lipid accumulation in DCs can improve anti-tumor immunity and the therapeutic efficacy of DC-based cancer vaccinations.
5.3. Metabolic reprogramming in suppressive immune cells
Although the metabolic conditions in the TME dampen antitumor function of tumor-specific effector T cells, Tregs can continue to exert their immunosuppressive effect due to their metabolic reliance on FAO (MacIver, Michalek, & Rathmell, 2013; Michalek et al., 2011). Production of lactate and amino acids by the hypermetabolic cancer cells as well as induction of HIF-1α in tumor can support the expansion of Tregs, which inhibit activation and cytolytic function of effector T cells in the TME (Ben-Shoshan et al., 2008; Siska et al., 2017).
The interactions between cancer cells and tumor-associated macrophages (TAMs) or monocytes through metabolic pathways or signals may also play a role in tumor development. Tumor produced LA can modulate TAM function to amplify tumor-promoting inflammation in the TME. In the presence of tumor-derived LA, human and mouse monocytes/macrophages activated by TLR ligands display enhanced transcription of IL-23p19 (Shime et al., 2008). IL-23 promotes cancer supporting inflammation through upregulation of the matrix metalloprotease MMP-9, expansion of inflammatory Th17 cells, and by preventing tumor infiltration of CTLs (Langowski et al., 2006; Numasaki et al., 2003). Activation of human monocytes by LPS triggers the glycolytic flux and the secretion of lactate. The abundance of tumor-derived LA can also influence activation of monocytes because addition of LA to reduce lactate export or blocking of glycolysis by 2-deoxyglucose strongly inhibits monocyte/macrophages activation-associated TNF production (Dietl et al., 2010). Further evidence for the contribution of LA to dysfunction of TAMs is provided by the observation that LA from cancer cells induces HIF-1α-dependent vascular endothelial growth factor and arginase 1, which accelerate tumor growth in syngeneic murine cancer models (Colegio et al., 2014).
Accumulating data indicate that TAMs undergo significant alterations in lipid profile. A reprogramming of lipid metabolism in macrophages was observed in an orthotropic lung cancer model, indicated by changes of several genes involved in lipid signaling. Compared with healthy controls, TAMs exhibited elevated levels of COX-2, which positively correlated with increased tumor angiogenesis (Nakao et al., 2005; Poczobutt et al., 2016). Expression of high 15-lipoxygenase 2 (15-LOX2) and its lipid product called the 15(S)-hydroxyeicosatetraenoic acid in renal cell carcinoma associated macrophages also correlated with the amounts of the chemokine CCL2 as well as the immunosuppressive cytokine IL-10, which facilitates cancer-supporting inflammation and immune escape (Daurkin et al., 2011). Cancer cells also regulate FA metabolism in TAMs to fulfill their tumor-promoting function. Lewis lung carcinoma cells actively produce macrophage colony-stimulating factor (M-CSF) to induce macrophage-intrinsic FASN and IL-10 production to facilitate tumor cell invasion (Park et al., 2015). Mechanistic studies revealed that FASN acts upstream of the nuclear receptor PPARβ/δ, a key regulator of tumor angiogenesis (Abdollahi et al., 2007; Muller-Brusselbach et al., 2007). FA binding proteins (FABPs) are lipid chaperones that can reversibly bind hydrophobic ligands (i.e., saturated or unsaturated long-chain FAs, eicosanoids and other lipids) and dictate their biological functions (Haunerland & Spener, 2004; Makowski & Hotamisligil, 2005). In the stroma of mousemammarytumor, epidermal FABP (E-FABP) is highly upregulated in M1-like macrophages, whereas macrophages with low expression of E-FABP are more M2 oriented, evidenced by differential production of IFN-β. Agonistic activation of E-FABP strongly enhances the antitumor activity of macrophages (Rao et al., 2015; Zhang et al., 2014). Therefore, the pro-tumor phenotype of TAMs may be defined by their intracellular metabolic lipid profiles.
A potential link is implied between amino acid or nutrient depletion and dysfunction of tumor-associated myeloid cells. Cysteine, which is required for protein synthesis and cell proliferation, can be acquired by cells through importing extracellular disulfide-bonded cystine via membrane cystine transporter and reducing them to cysteine (Arner & Holmgren, 2000; Mansoor, Svardal, & Ueland, 1992) or converting intracellular methionine to cysteine through cystathionase (Gout, Buckley, Simms, & Bruchovsky, 2001; Ishii et al., 2004). Myeloid-derived suppressor cells (MDSCs) consist of heterogeneous populations of immature cells phenotypically characterized as CD11b+Gr1+ cells in tumor-bearing mice and CD11b+CD14− CD33+ cells in cancer patients (Gabrilovich, Ostrand-Rosenberg, & Bronte, 2012). Expansion of MDSCs represents a major mechanism of cancer evasion of antitumor immunity. Due to the absence of Alanine-Serine-Cysteine (ASC) transporter, export of cysteine is blocked in MDSCs. However, MDSCs can import extracellular cysteine through transporter. As a result, MDSCs sequester cysteine in the TME and limit the availability of cysteine to T cells (Srivastava, Sinha, Clements, Rodriguez, & Ostrand-Rosenberg, 2010). Since T cells do not express cystathionase or the transporter, their proliferation and activation require cysteine mainly from other cells (Eagle, Washington, & Friedman, 1966), lack of cysteine in the TME leads to impaired T cell activation (Srivastava et al., 2010). Similarly, MDSCs suppress activation of T cells by depletion of arginine through upregulation of arginase I, which decreases the expression of CD3ζ on T cells (Rodriguez et al., 2009). In addition to MDSCs, arginase I is also highly expressed in mature myeloid cells infiltrating mouse lung carcinoma or human non-small cell carcinoma. These myeloid cells, not the tumor cells or infiltrating lymphocytes, are believed to be the primary source of intra-tumor arginase I and can efficiently deplete extracellular l-arginine via cationic amino acid transporter 2B. Similar to MDSCs, depletion of l-arginine by tumor-associated mature myeloid cells inhibit the activation of antigen-specific T cells by downregulating CD3ζ expression (Rodriguez et al., 2004). Tumor-infiltrating myeloid cells, such as MDSCs, DCs or TAMs also catalyze tryptophan metabolism in the kynurenine pathway through upregulation of IDO, which can inhibit T cell activation through depletion of tryptophan and expansion of Tregs (Martinez, Gordon, Locati, & Mantovani, 2006; Munn & Mellor, 2007, 2016). Accordingly, amino acid deprivation by tumor-associated myeloid cells and resultant impairment of TCR signaling or T cell activation represents a major mechanism by which cancer cells avoid immune recognition and/or attack.
Several recent studies also highlight involvement of metabolic pathways that impact on the suppressive functions of MDSCs. Tumor-infiltrating MDSCs sorted from mouse and human display a metabolic characteristic of activated FAO (Hossain et al., 2015). The increased FAO and FA uptake are associated with elevated arginase I as well as an enhanced capacity of MDSCs to inhibit T cells. In response to high level of FAO, MDSCs produce cytokines that can sustain the expansion of MDSC (e.g., G-CSF, GM-CSF, IL-1β, IL-6 and IL-10), which is blocked by inhibition of FAO (Hossain et al., 2015). Cancer cell-derived cytokines including G-CSF and GM-CSF can signal through STAT3 and STAT5 in a paracrine manner to induce expression of lipid transporters and lipid uptake in MDSCs (Al-Khami et al., 2017). Intracellular accumulation of lipids increases further oxidative metabolism and strengthens immunosuppressive function of MDSCs, which can be reversed by blocking STAT3/5 signaling or ablation of the FA translocase CD36. Strikingly, lipid transport proteins are highly upregulated in human tumor-infiltrating and peripheral blood MDSCs (Al-Khami et al., 2017). Lectin-type oxidized LDL receptor 1 (LOX-1) is specifically expressed on polymorphonuclear MDSCs (PMN-MDSCs) from cancer patients, but not on those from healthy donors. LOX-1 expressing PMN-MDSCs display a gene profile associated with immune suppression (Condamine et al., 2016). Consequently, tumor-derived lipids cause profound metabolic changes in MDSCs, which can modulate their ability to counteract the effector function of antitumor T cells.
6. Reprogramming metabolic pathways to potentiate cancer immunotherapy
There is support for the notion that metabolic changes alter the phenotype and function of immune cells within the TME, which is believed to at least partially contribute to the failure of current cancer immunotherapy. A major goal of immunotherapy is to overcome immune suppression for efficient mobilization of tumor-specific T effector cells and generation of T memory cells, which will allow for long-lasting immune-mediated elimination or control of cancer. Since metabolic programing drives immune cell development and function, use of metabolism-targeting drugs could offer new opportunities to improve cancer immunotherapies. Evidence is beginning to emerge that metabolic regulation, e.g., inhibition of cholesterol esterification (Yang, Bai, et al., 2016), blockade of conversion of adenosine monophosphate (Allard, Pommey, Smyth, & Stagg, 2013), and targeting of the unbalanced lipid accumulation in tumor-infiltrating DCs (Cubillos-Ruiz et al., 2015; Herber et al., 2010) can be harnessed to strengthen or normalize immune functions for optimized treatment of cancers.
6.1. Overcoming immune suppression by metabolic reprogramming of myeloid cells
An effective DC vaccination or DC-targeted immunotherapy is dependent on optimized processing and presentation of tumor antigens by DCs as well as induction of co-stimulatory molecules on the cell surface. mTOR regulates a variety of cellular responses (e.g., metabolism and survival) through nutrient sensing thereby representing a potential metabolic target for modulating DC functionality (Kapahi et al., 2010; Meijer & Codogno, 2008; Sengupta, Peterson, & Sabatini, 2010). Inhibition of mTOR prevents TLR activation-induced metabolic switch to glycolysis in DCs (Amiel et al., 2012; Krawczyk et al., 2010). Blocking mTOR expression or function also significantly prolongs the lifespan of activated DCs, increases the levels of co-stimulatory molecules, and enhances their T cell-priming capacity. Lastly, treatment of DCs with mTOR inhibitors during their activation improves the therapeutic efficacy of DC vaccination to control the progression of mouse B16 melanoma (Amiel et al., 2012). Mechanistic studies show that mTOR inhibition reduces the dependence of activated DCs on glycolysis to generate ATP and shifts the metabolic pathway toward OXPHOS. This metabolic change is primarily attributed to the downregulation of iNOS and NO (Amiel et al., 2014). These findings provide supporting evidence for strategic targeting of the mTOR pathway in DCs to enhance their immunostimulatory potential to improve immuno-therapeutic outcomes.
Small-molecule inhibitors have also been tested to repress the FAO cycle in myeloid cells, e.g., etomoxir that irreversibly blocks CPT1A and ranolazine that targets the trifunctional enzyme HADHA (Bressler et al., 1989; MacInnes et al., 2003). In support of the role of FAO in the suppressive function of MDSCs, administration of etomoxir or ranolazine effectively reduces the growth of mouse 3LL lung carcinoma and MCA-38 colon carcinoma cells, which is dependent on T cells (Hossain et al., 2015). FAO inhibition confers an antitumor effect that appears to involve MDSCs, since etomoxir treatment did not directly affect cancer cells or T cell functions (e.g., proliferation, cytokine production, cytolytic activity) (Hossain et al., 2015). Several additional studies also demonstrate that FAO inhibition significantly increases antitumor potency of low-dose chemotherapy or T cell therapy by targeting MDSC-associated immune suppression (Coussens, Zitvogel, & Palucka, 2013; DeNardo et al., 2011; Hossain et al., 2015).
6.2. Metabolic reprogramming of T cells for enhanced therapeutic efficacy
Adoptive cell therapy (ACT) is a form of immunotherapy that utilizes tumor-reactive T cells to eradicate cancers (Chen & Mellman, 2013; Goff et al., 2016). However, a glucose-poor tumor microenvironment limits aerobic glycolysis in tumor-infiltrating T cells, resulting in suppression of their tumoricidal functions. Among melanoma patients that are refractory to ACT, there is a high elevation of glycolytic activity in cancer cells, implicating a potential role of tumor-intrinsic glycolysis in dampening the therapeutic potency of adoptively transferred T cells (Cascone et al., 2018). The glycolytic metabolite PEP was recently shown to serve as new metabolic checkpoint for sustaining TCR-mediated Ca2+-NFAT signaling and effector functions of T cells. Tumor-specific CD4+ and CD8+ T cells can be metabolically reprogrammed by increasing PEP production through overexpression of phosphoenolpyruvate carboxykinase 1 (PCK1), which improves their effector functions. PCK1 overexpression enhances the antitumor efficacy of adoptively transferred melanoma-reactive T cells, which is associated with increased production of IFN-γ and CD40L in TILs, but not in T cells within peripheral lymphoid organs where glucose is abundant (Ho et al., 2015). This finding indicates that metabolic reprogramming of antitumor T cells for enhanced glycolysis may be potentially used to overcome glucose deprivation in the TME and rejuvenate functionally impaired TILs.
While enhanced glycolysis can promote effector T cell functions as well as their terminal differentiation, inhibition of glycolysis generates memory cell-like CD8+ T cells that display longevity and superior antitumor function (Sukumar et al., 2013, 2016). Inhibition of glycolysis during ex vivo expansion of antigen-specific T cells promotes a transcriptional program embodying characteristics of memory cells (Sukumar et al., 2013). Activation of CD8+ T cells in the presence of glycolytic inhibition enhances the generation of long-lived memory T cells and improves their antitumor activity after transfer into B16 tumor bearing mice (Sukumar et al., 2013). It is feasible that glycolysis-active effector T cells can lead to a rapid therapeutic effect, however, long-lasting memory T cells are essential for control of relapsed tumors. Therefore, a challenge remains relative to the appropriate timing of metabolic reprogramming of T cells to achieve successful treatment outcomes in the setting of ACT.
In mouse and human melanoma, hypoglycemia and hypoxia in the TME enhance PPAR-α signaling and catabolism of FAs in vaccination-induced CD8+ T cells, which can partially preserve the effector functions of TILs. Such a metabolic switch associates with enhanced uptake of FAs and exacerbated FAO. Metabolic reprogramming of CD8+ T cells by increasing FA catabolism enhanced the antitumor efficacy of ACT, which can also synergize immune checkpoint inhibition therapy for improved tumor elimination (Zhang, Kurupati, et al., 2017). This result is also buttressed by other studies demonstrating that memory CD8+ T cells, which rely on FAO and OXPHOS for energy production, are more efficient in delaying tumor growth than effector cells (Crompton et al., 2015; Sukumar et al., 2013).
The PI3K-Akt-mTOR pathway is implicated in regulation of the formation of memory CD8+ T cells (Araki et al., 2009; Macintyre et al., 2011; Okkenhaug et al., 2002). Pharmacologic inhibition of Akt facilitates the expansion and differentiation of human tumor-specific lymphocytes into memory T cells by enhancing FAO and SRC. Upon Akt inhibition, human CTLs exhibit prolonged persistence after adoptive transfer into NSG mice. Transfer of mouse B16 melanoma-reactive CTLs after treatment with the Akt inhibitor also show a superior antitumor activity, indicated by reduced tumor growth and improved animal survival (Crompton et al., 2015). However, sustained and strong activation of Akt was shown to play a non-redundant role in CTLs to coordinate the TCR- and IL-2-induced transcriptional programs that control expression of key cytolytic effector molecules, adhesion molecules, cytokine and chemokine receptors that distinguish effector T cells from memory or naïve T cells (Laplante & Sabatini, 2012; Macintyre et al., 2011). Therefore, despite the fact that Akt is dispensable for metabolism, the strength and duration of Akt activity can dictate transcriptional programs in CTLs that determine their fate.
l-arginine can increase the survival of human and mouse T cells and favor their central memory phenotype, which is superior to effector T cells in eradicating mouse tumors (Geiger et al., 2016; Klebanoff et al., 2005). Supplement with l-arginine provides OT-I cells with a higher survival capacity. l-arginine treatment of OT-I cells prior to adoptive transfer into B16-OVA bearing tumor mice mounts an enhanced therapeutic effect and prolongs the lifespan of mice. Alternatively, feeding B16-OVA tumor bearing mice with l-arginine also renders more potent OT-I T cell activation primed in vivo by OVA plus Alum immunization as well as optimized tumor control (Geiger et al., 2016). These observations collectively demonstrate that increasing l-arginine level in CD8+ T cells enhances their antitumor activity.
Manipulating cholesterol metabolism may also be exploited to potentiate the tumor cytotoxicity of CD8+ T cells, which is often suppressed in the TME. Genetic ablation or pharmacological inhibition of ACAT1, a key cholesterol esterification enzyme (Chang, Chang, Ohgami, & Yamauchi, 2006), specifically enhances the proliferation and effector function of CD8+ T cells, not CD4+ T cells (Yang, Bai, et al., 2016). This is achieved via increasing plasma membrane cholesterol levels, causing enhanced TCR clustering and signaling as well as formation of the immunological synapse. Selective ablation of ACAT1 in CD8+ T cells reduces the progression and metastasis of mouse melanoma in vivo. This improved immunotherapeutic outcome can be recapitulated by using the ACAT inhibitor avasimibe, which was previously tested in clinical trials for treatment of atherosclerosis (Yang, Bai, et al., 2016).
Chimeric antigen receptor engineered T cell (CAR-T) therapy is an immunotherapeutic strategy that combines the effector functions of T cells with extracellular variable regions of an antibody to allow specific recognition of cancer cells independent of antigen cross-presentation (Eshhar, Waks, Gross, & Schindler, 1993; Kuwana et al., 1987). CAR-T therapy has achieved remarkable clinical responses in hematological malignancies, e.g., chronic lymphocytic leukemia (CLL), acute lymphoblastic leukemia (ALL), and diffused large B cell lymphoma. The new generation CAR-T therapy incorporates the cytoplasmic domains of co-stimulatory molecules, i.e., CD28, 4–1BB (CD137), ICOS or OX40, either individually or in combination for enhanced proliferation and function of CAR-T cells (Maus et al., 2014; Sadelain, Brentjens, & Riviere, 2013). Although similar responses were observed in ALL patients receiving CAR-T therapy that has incorporated signaling domain of CD28 or 4–1BB (Brentjens et al., 2013; Lee et al., 2015; Maude et al., 2014), the presence of the 4–1BB signaling domain results in superior treatment outcomes in CLL as compared to that of CD28 domain, which is due to increased persistence of CAR-T cells and their resistance to exhaustion (Brentjens et al., 2013, 2011; Long et al., 2015; Porter et al., 2015). Intriguingly, incorporation of CD28 or 4–1BB domains causes activation of distinct metabolic pathways within CAR-T cells. CAR-T cells equipped with 4–1BBζ display a central memory phenotype and preferentially use metabolic pathways such as FAO to achieve their bioenergetic demands for long term survival. In contrast, CD28ζ CAR-T cells consume a greater amount of glucose and show increased aerobic glycolysis via activation of the PI3K-Akt pathway during effector cell differentiation (Frauwirth et al., 2002; Kawalekar et al., 2016; Sabbagh, Pulle, Liu, Tsitsikov, & Watts, 2008). These observations are consistent with a previous report of a critical role of the 4–1BB signaling in long-term survival of T cells (Sabbagh et al., 2008). Consequently, metabolic reprogramming represents a critical mechanism for determining T cell responses during cancer progression and T cell-based immunotherapies. Strategical engineering of CAR-T cells with altered metabolic activities that favor the persistence and functionality of T cells in the hostile TME can lead to improved therapeutic outcomes.
6.3. Metabolic reprogramming and immune checkpoint blockade therapy
The immune checkpoint proteins, which play a crucial role in regulation of the immune response to maintain self-tolerance and prevent excessive immune activation, are often dysregulated in tumors and immune cells, thereby resulting in immune evasion by cancer cells (Keir, Butte, Freeman, & Sharpe, 2008). ICIs, which reinvigorate antitumor immunity by restoring T cell function while preventing their exhaustion, have shown striking and durable clinical responses in several types of cancer (Brahmer et al., 2012; Hamid et al., 2013; Hodi et al., 2010; Pardoll, 2012; Wei, Duffy, & Allison, 2018). Accumulating evidence indicate that these immune check-point signals can cause metabolic reprogramming of both cancer cells and immune cells. An early study showed that ligation of CTLA-4 and PD-1 signaling reduced CD3/CD28 stimulation triggered elevation of glucose metabolism and Akt activity in T cells (Parry et al., 2005). Indeed, tumor PD-L1 expression can promote tumor-intrinsic glycolysis and Akt-mTOR activation while suppressing mTOR activity in T cells through glucose competition within the tumor niche. In a mouse sarcoma model, treatment with ICIs (i.e., anti-PD-1/L1, anti-CTLA-4 antibodies) inhibited glucose uptake in tumor cells and enhanced glucose uptake and glycolysis as well as mTOR activity in T cells, which overlapped with increased IFN-γ production by T cells (Chang et al., 2015). In support of this finding, an increased glucose rate in TME and improved glycolysis as well as cytolytic function of T cells is also observed in a mouse melanoma model following treatment with ICIs (Zhao et al., 2016), suggesting blockade of immune checkpoint molecules can influence the metabolic profile in the TME by favoring T cell functions.
Recent studies by Patsoukis et al showed that activating the PD-L1/PD-1 signaling prevents the development of effector T cells by inhibiting glycolytic reprogramming, promoting FAO of endogenous lipids through overexpression of CPT1A, and inducing lipolysis (Patsoukis et al., 2015). Given the impact of lipid metabolism on the proliferation and function of T cells, use of metabolic modulator to improve therapeutic outcomes in the setting of immune checkpoint blockade has been attempted. Anti-PD-1 antibodies combined with avasimibe, which targets cholesterol esterification for treatment of atherosclerosis, demonstrated better efficacy than monotherapies in controlling tumor growth (Yang, Bai, et al., 2016).
Considering that production of LA and resultant low-pH TME impair proliferation and cytolytic activity of CTLs, neutralization of the TME has been evaluated for its therapeutic impact on cancer immunotherapy (Choi, Collins, Gout, & Wang, 2013; Romero-Garcia, Moreno-Altamirano, Prado-Garcia, & Sanchez-Garcia, 2016). Buffering LA with oral bicarbonate improves the pH of TME, resulting in enhanced CTL activity and improved inhibition of tumor growth when combined with anti-PD-1 therapy (Pilon-Thomas et al., 2016). Recently, 4–1BB costimulatory signaling was shown to provide metabolic support to TILs in an immunosuppressive and metabolically challenged TME, which is known to impose metabolic restrictions on T cell function (Menk et al., 2018). A short course of 4–1BB treatment is adequate to improve metabolic sufficiency in endogenous or adoptive transferred CD8+ T cells through enhanced mitochondrial capacity and by engaging PGC1α pathways. Strikingly, 4–1BB stimulation combined with PD-1 inhibitor results in robust antitumor immunity, further highlighting the potential of metabolic reprogramming to enhance the fitness of CTLs for immunotherapy (Menk et al., 2018).
6.4. Targeting immunometabolism to ameliorate ICIs-associated side effects
While ICIs are revolutionizing the treatment of cancer, accumulating evidence indicates that these immunotherapeutic agents are associated with a diverse spectrum of autoimmune or inflammatory complications (e.g., colitis, hepatitis, rheumatoid arthritis or RA, inflammatory bowel disease, pneumonia), referred to as immune-related adverse events (irAEs) (Postow, Sidlow, & Hellmann, 2018). A number of irAEs that emerge with ICIs have been observed, ranging from severe and potentially life-threatening pneumonitis (Barjaktarevic, Qadir, Suri, Santamauro, & Stover, 2013; Topalian et al., 2012), hepatitis (Hamid et al., 2013; Robert et al., 2015), and colitis (Beck et al., 2006), to autoimmune arthritis (Cappelli et al., 2017) and dermatitis (Sibaud et al., 2016). Although the exact pathways mediating irAEs are not completely understood, it is conceivable that blockade of immune checkpoint molecules, such as CTLA-4 or PD-1, could cause aberrant T cell activation and loss of self-tolerance with a myriad of off-target inflammatory and autoimmune manifestations (Calabrese, Calabrese, & Cappelli, 2018; Calabrese & Velcheti, 2017; Sandigursky & Mor, 2018). As observed in cancer, metabolic alterations have been implicated in abnormal immune activation in autoimmune disorders (Perl, 2017). In patients with RA, CD4+ T cells hyper-proliferate and differentiate into cytokine-producing effector cells involved in disease pathology. In contrast to those from healthy controls, glycolytic breakdown of glucose in T cells in RA is reduced as a result of diminished activity of the regulatory enzyme PFKFB3, which impairs ATP production, autophagy, and redox balance (Yang, Fujii, Mohan, Goronzy, & Weyand, 2013). Impaired glycolysis in RA T cells is also caused by up-regulation of glucose-6-phosphate dehydrogenase (G6PD) (Yang, Bai, et al., 2016). Excess G6PD diverts glucose into the pentose phosphate pathway, resulting in the accumulation of NADPH and consumption of ROS. Restoring oxidant signaling by replenishing intracellular ROS can correct the pro-arthritogenic activity of T cells and alleviate synovial inflammation (Yang, Shen, et al., 2016). Additionally, podosome scaffolding protein TKS5, i.e., tyrosine kinase substrate with five Src homology 3 domains, is critically involved in the pro-inflammatory and tissue-invasive behavior of T cells in RA (Shen et al., 2017). Low glycolysis in RA T cells drives the overexpression of TKS5 by reducing pyruvate and promoting FA biosynthesis. Pharmacologically restoring pyruvate production or inhibiting FA synthesis suppresses the tissue-invasive and proinflammatory functions of RA T cells in vivo (Shen et al., 2017).
Macrophages, a dominant cell population in the inflamed joint in RA, also undergo metabolic reprogramming that drives their pro-inflammatory phenotype. Intestinally, as opposed to a phenotype of ATPlowNADPHhighROSlow in RA T cells, RA macrophages gorge on glucose due to upregulation of glucose transporters and display increased ROS production. Subsequent post-translational modifications of the glycolytic enzyme pyruvate kinase further amplifies STAT3-dependent production of IL-1β and IL-6 (Weyand, Zeisbrich, & Goronzy, 2017). Given the metabolic alterations in these hyper-inflammatory pathogenic cells, it is possible to target cell-type-exclusive metabolic set points to alleviate irAEs. However, a remaining challenge of metabolic interference as a means of immunomodulation is the complexity of metabolic programing in different immune cell types, e.g., coexistence of energy-deprived T-cells and glucose-overloaded macrophages as cytokine super-producers in autoimmune disorders. Understanding how specific ICIs functionally and metabolically modulate tumor-specific and self-attacking immune cells will be essential for rational development of metabolism modifying approaches to optimize immune checkpoint blockade therapy.
7. Concluding remarks
The immune system needs to constantly adapt and respond to diverse environments, which requires coordination of the bioenergetic machinery within the highly interactive immune cell network to achieve efficient and appropriate effector functions. The emerging field of immunometabolism highlights the importance of metabolism in the activation, differentiation, and function of immune cells (O’Neill, Kishton, & Rathmell, 2016), which also provides a molecular and biochemical basis for understanding metabolic changes in immune cells during tumor progression or in the setting of immunotherapy. While dysregulated cellular energetics in cancer cells is established as a means of conferring growth advantage, recent studies are beginning to shed light onto the immunological impact of metabolic events in cancer cells and consequent tumor escape from immune surveillance. Metabolic regulation provides a key determinant of the behavior of cancer cells as well as the functional fate of immune cells. The interplay between cancer cells and immune cells promotes metabolic reprogramming in both entities, which is exemplified by altered glycolysis, glutaminolysis, lipid metabolism, mitochondrial biogenesis, etc. Cancer cells are believed to evade the immune system metabolically through multiple mechanisms (Fig. 2), including nutrient competition, release of metabolites that impair immune cell activity or induce expansion of suppressive immune cells (e.g., M2 macrophage, MDSC, Treg). The functionalities of these suppressive or regulatory immune cells are adapted to the hostile TME and can facilitate tumor progression or invasion. Mechanistic understanding of the metabolic reprogramming in the TME may provide a means for developing novel approaches to enhance the antitumor potential of immune effector cells. Indeed, several glycolysis inhibitors designed to target tumor glucose metabolism to reverse Warburg effect have been investigated preclinically or are currently in clinical trials. However, the specific effects of those inhibitors on immune cells, especially in cancer immunotherapy, remain largely unknown. Identifying metabolic pathways shared by cancer and immune cells will allow the selection of appropriate metabolism-targeting drugs as potential immune modulators. Although immunotherapy is emerging as one of the most promising therapeutic modalities for cancer, long-term clinical benefits are not achieved in the majority of patients. Thus, pharmacological inhibition of metabolic pathways may be exploited for combinatorial therapy with other types of immunotherapies for improved treatment outcomes.
Fig. 2.
Metabolic evasion of the immune system by cancer. Cancer cells metabolically alter the tumor immune compartment through nutrient competition, release of immunosuppressive metabolites that can also facilitate the expansion and function of suppressive immune cells, such as M2 macrophages, myeloid-derived suppressor cells (MDSCs). Tumors impair effector function of T cells by limiting the nutrient availability in the tumor microenvironment (TME), e.g., glucose or amino acids deprivation. This can cause a mitochondrial function loss in T cells and poor tumor infiltration by T cells. Accumulation of lactate due to Warburg effects also inhibits glycolysis in T cells. Tumorinfiltrating dendritic cells (DCs) display a defect in antigen cross-presentation and T cell priming due to the presence of lactate, lipid accumulation, and enhanced FAO. Suppressive immune cells, including tumor-associated macrophages (TAMs) and MDSCs, can adapt to the metabolically challenged TME via their metabolic reliance on FAO, which lead to amplified immune suppression and tumor promotion. The hypoxia TME further enforces the activity of infiltrating myeloid cells in restricting the availability of amino acids that are essential for T cell activation. Targeting metabolic pathways in both tumor and immune cell can be therapeutically exploited to overcome immune suppression and reinvigorate immune functions for improved antitumor immunity.
Despite the recent progress in the field, several critical questions remain to be addressed. Further research is required to better understand the timing of the major metabolic switches during immune cell activation and differentiation. In addition to pharmacological agents, there is a pressing need to use a broader spectrum of genetic modifications including transgenic mouse models, which target diverse metabolic regulators to precisely determine their immune modulating effects. Likewise, it is not well understood as to how antitumor immune cells (e.g., TILs) and tumor-promoting cells (e.g., MDSCs) adapt to the metabolic constrains within the TME and to what degree this impacts on their functionality. Considering that memory T cells provide superior antitumor immunity, it is critical to determine how metabolic pathways support their proliferative capacity and function in the TME. Additional work is also needed to define the relative contribution of metabolic targeting to tumor inhibition and immune activation. For instance, despite the safety of the glucose analog 2-deoxyglucose (2-DG) and its efficiency in cancer cells, 2-DG has also been shown to impair the metabolism of T cells, resulting in reduced antitumor function (Gu et al., 2017; Sukumar et al., 2013). The overlapping nature of many metabolic pathways calls for caution in target selection and drug design. Novel platforms that can deliver metabolic reprogramming agents in a cell type-specific manner will be necessary to achieve selective targeting. Future studies should address the mechanistic connection between immunotherapy and the metabolism of cancer or immune cells. These include defining a therapeutic window to be exploited for optimized metabolic interventions, elucidation of the metabolic contributions mediated through immune checkpoint molecules for rational targeting to maximize a durable antitumor response, and potential involvement of metabolic reprogramming in tumor resistance to immunotherapies, e.g., ICIs ( Jenkins, Barbie, & Flaherty, 2018). Answering these questions will expand our comprehension of the interplay between metabolic regulation and the antitumor immune response, potentially providing new avenues to improve cancer immunotherapy. It is anticipated that strategically targeting metabolic vulnerabilities of cancer cells and concurrent metabolic strengthening of antitumor fitness or function of immune cells should translate the current success of immunotherapy to additional cancer types and extend clinical immunotherapeutic benefits to a larger proportion of patients.
Acknowledgments
This study was supported in part by National Institutes of Health (NIH) Grants CA175033, CA CA229812, CA099326, the Department of Defense Award W81XWH-17–1-0317, the National Foundation for Cancer Research, NCI Cancer Center Support Grant to VCU Massey Cancer Center (MCC) P30CA16059, and MCC Pilot Project Fund. X.-Y.W. is the Harrison Endowed Scholar in Cancer Research in the VCU Massey Cancer Center. P.B.F. holds the Thelma Newmeyer Corman Chair in Cancer Research in the VCU Massey Cancer Center.
Footnotes
Competing interests
None declared.
References
- Abdollahi A, Schwager C, Kleeff J, Esposito I, Domhan S, Peschke P, et al. (2007). Transcriptional network governing the angiogenic switch in human pancreatic cancer. Proceedings of the National Academy of Sciences of the United States of America, 104, 12890–12895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S, & Takeda K (2004). Toll-like receptor signalling. Nature Reviews. Immunology, 4, 499–511. [DOI] [PubMed] [Google Scholar]
- Alavi A, & Reivich M (2002). Guest editorial: The conception of FDG-PET imaging. Seminars in Nuclear Medicine, 32, 2–5. [DOI] [PubMed] [Google Scholar]
- Alberghina L, & Gaglio D (2014). Redox control of glutamine utilization in cancer. Cell Death & Disease, 5, e1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khami AA, Zheng L, Del Valle L, Hossain F, Wyczechowska D, Zabaleta J, et al. (2017). Exogenous lipid uptake induces metabolic and functional reprogramming of tumor-associated myeloid-derived suppressor cells. Oncoimmunology, 6, e1344804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard B, Pommey S, Smyth MJ, & Stagg J (2013). Targeting CD73 enhances the antitumor activity of anti-PD-1 and anti-CTLA-4 mAbs. Clinical Cancer Research, 19, 5626–5635. [DOI] [PubMed] [Google Scholar]
- Altman BJ, Stine ZE, & Dang CV (2016). From Krebs to clinic: Glutamine metabolism to cancer therapy. Nature Reviews. Cancer, 16, 619–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alwarawrah Y, Hughes P, Loiselle D, Carlson DA, Darr DB, Jordan JL, et al. (2016). Fasnall, a selective FASN inhibitor, shows potent anti-tumor activity in the MMTV-neu model of HER2(+) breast cancer. Cell Chemical Biology, 23, 678–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amelio I, Cutruzzola F, Antonov A, Agostini M, & Melino G (2014). Serine and glycine metabolism in cancer. Trends in Biochemical Sciences, 39, 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiel E, Everts B, Freitas TC, King IL, Curtis JD, Pearce EL, et al. (2012). Inhibition of mechanistic target of rapamycin promotes dendritic cell activation and enhances therapeutic autologous vaccination in mice. The Journal of Immunology, 189, 2151–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiel E, Everts B, Fritz D, Beauchamp S, Ge B, Pearce EL, et al. (2014). Mechanistic target of rapamycin inhibition extends cellular lifespan in dendritic cells by preserving mitochondrial function. The Journal of Immunology, 193, 2821–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleman LJ, van Puijenbroek AAFL, Shu KM, Nadler LM, & Boussiotis VA (2002). CD28 costimulation mediates down-regulation of p27kip1 and cell cycle progression by activation of the PI3K/PKB signaling pathway in primary human T cells. The Journal of Immunology, 168, 2729–2736. [DOI] [PubMed] [Google Scholar]
- Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, et al. (2009). mTOR regulates memory CD8 T-cell differentiation. Nature, 460, 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arner ES, & Holmgren A (2000). Physiological functions of thioredoxin and thioredoxin reductase. European Journal of Biochemistry, 267, 6102–6109. [DOI] [PubMed] [Google Scholar]
- Assmann N, O’Brien KL, Donnelly RP, Dyck L, Zaiatz-Bittencourt V, Loftus RM, et al. (2017). Srebp-controlled glucose metabolism is essential for NK cell functional responses. Nature Immunology, 18, 1197–1206. [DOI] [PubMed] [Google Scholar]
- Asturias FJ, Chadick JZ, Cheung IK, Stark H, Witkowski A, Joshi AK, et al. (2005). Structure and molecular organization of mammalian fatty acid synthase. Nature Structural & Molecular Biology, 12, 225–232. [DOI] [PubMed] [Google Scholar]
- Banchereau J, & Steinman RM (1998). Dendritic cells and the control of immunity. Nature, 392, 245–252. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S, Pai SK, Watabe M, Gross SC, Hirota S, Hosobe S, et al. (2005). FAS expression inversely correlates with PTEN level in prostate cancer and a PI 3-kinase inhibitor synergizes with FAS siRNA to induce apoptosis. Oncogene, 24, 5389–5395. [DOI] [PubMed] [Google Scholar]
- Barjaktarevic IZ, Qadir N, Suri A, Santamauro JT, & Stover D (2013). Organizing pneumonia as a side effect of ipilimumab treatment of melanoma. Chest, 143, 858–861. [DOI] [PubMed] [Google Scholar]
- Barthel A, Okino ST, Liao J, Nakatani K, Li J, Whitlock JP Jr., et al. (1999). Regulation of GLUT1 gene transcription by the serine/threonine kinase Akt1. The Journal of Biological Chemistry, 274, 20281–20286. [DOI] [PubMed] [Google Scholar]
- Barton GM, & Medzhitov R (2002). Control of adaptive immune responses by Toll-like receptors. Current Opinion in Immunology, 14, 380–383. [DOI] [PubMed] [Google Scholar]
- Baysal BE, Ferrell RE, Willett-Brozick JE, Lawrence EC, Myssiorek D, Bosch A, et al. (2000). Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science, 287, 848–851. [DOI] [PubMed] [Google Scholar]
- Beck KE, Blansfield JA, Tran KQ, Feldman AL, Hughes MS, Royal RE, et al. (2006). Enterocolitis in patients with cancer after antibody blockade of cytotoxic T-lymphocyte-associated antigen 4. Journal of Clinical Oncology, 24, 2283–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker TC, Wherry EJ, Boone D, Murali-Krishna K, Antia R, Ma A, et al. (2002). Interleukin 15 is required for proliferative renewal of virus-specific memory CD8 T cells. The Journal of Experimental Medicine, 195, 1541–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shoshan J, Maysel-Auslender S, Mor A, Keren G, & George J (2008). Hypoxia controls CD4 +CD25 + regulatory T-cell homeostasis via hypoxia-inducible factor-1alpha. European Journal of Immunology, 38, 2412–2418. [DOI] [PubMed] [Google Scholar]
- Berod L, Friedrich C, Nandan A, Freitag J, Hagemann S, Harmrolfs K, et al. (2014). De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nature Medicine, 20, 1327–1333. [DOI] [PubMed] [Google Scholar]
- Beura LK, & Masopust D (2014). SnapShot: Resident memory T cells. Cell, 157, 1488–1488.e1. [DOI] [PubMed] [Google Scholar]
- Biron CA, & Brossay L (2001). NK cells and NKT cells in innate defense against viral infections. Current Opinion in Immunology, 13, 458–464. [DOI] [PubMed] [Google Scholar]
- Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, & Sabatini DM (2015). An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell, 162, 540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas SK (2015). Metabolic reprogramming of immune cells in cancer progression. Immunity, 43, 435–449. [DOI] [PubMed] [Google Scholar]
- Biswas SK, & Mantovani A (2010). Macrophage plasticity and interaction with lymphocyte subsets: Cancer as a paradigm. Nature Immunology, 11, 889–896. [DOI] [PubMed] [Google Scholar]
- Blagih J, Coulombe F, Vincent EE, Dupuy F, Galicia-Vazquez G, Yurchenko E, et al. (2015). The energy sensor AMPK regulates T cell metabolic adaptation and effector responses in vivo. Immunity, 42, 41–54. [DOI] [PubMed] [Google Scholar]
- Botman D, Tigchelaar W, & Van Noorden CJ (2014). Determination of glutamate dehy-drogenase activity and its kinetics in mouse tissues using metabolic mapping (quantitative enzyme histochemistry). The Journal of Histochemistry and Cytochemistry, 62, 802–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. (2012). Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. The New England Journal of Medicine, 366, 2455–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brentjens RJ, Davila ML, Riviere I, Park J, Wang X, Cowell LG, et al. (2013). CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Science Translational Medicine, 5, 177ra138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brentjens RJ, Riviere I, Park JH, Davila ML, Wang X, Stefanski J, et al. (2011). Safety and persistence of adoptively transferred autologous CD19-targeted T cells in patients with relapsed or chemotherapy refractory B-cell leukemias. Blood, 118, 4817–4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressler R, Gay R, Copeland JG, Bahl JJ, Bedotto J, & Goldman S (1989). Chronic inhibition of fatty acid oxidation: New model of diastolic dysfunction. Life Sciences, 44, 1897–1906. [DOI] [PubMed] [Google Scholar]
- Buckley D, Duke G, Heuer TS, O’Farrell M, Wagman AS, McCulloch W, et al. (2017). Fatty acid synthase—Modern tumor cell biology insights into a classical oncology target. Pharmacology & Therapeutics, 177, 23–31. [DOI] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, & Mak TW (2011). Regulation of cancer cell metabolism. Nature Reviews. Cancer, 11, 85–95. [DOI] [PubMed] [Google Scholar]
- Cairns RA, & Mak TW (2017). Fire and water: Tumor cell adaptation to metabolic conditions. Experimental Cell Research, 356, 204–208. [DOI] [PubMed] [Google Scholar]
- Calabrese LH, Calabrese C, & Cappelli LC (2018). Rheumatic immune-related adverse events from cancer immunotherapy. Nature Reviews. Rheumatology, 14, 569–579. [DOI] [PubMed] [Google Scholar]
- Calabrese L, & Velcheti V (2017). Checkpoint immunotherapy: Good for cancer therapy, bad for rheumatic diseases. Annals of the Rheumatic Diseases, 76, 1–3. [DOI] [PubMed] [Google Scholar]
- Campa D, McKay J, Sinilnikova O, Husing A, Vogel U, Hansen RD, et al. (2009). Genetic variation in genes of the fatty acid synthesis pathway and breast cancer risk. Breast Cancer Research and Treatment, 118, 565–574. [DOI] [PubMed] [Google Scholar]
- Cantor JR, & Sabatini DM (2012). Cancer cell metabolism: One hallmark, many faces. Cancer Discovery, 2, 881–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappelli LC, Gutierrez AK, Baer AN, Albayda J, Manno RL, Haque U, et al. (2017). Inflammatory arthritis and sicca syndrome induced by nivolumab and ipilimumab. Annals of the Rheumatic Diseases, 76, 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbrey JM, Gorelick-Feldman DA, Kozono D, Praetorius J, Nielsen S, & Agre P (2003). Aquaglyceroporin AQP9: Solute permeation and metabolic control of expression in liver. Proceedings of the National Academy of Sciences of the United States of America, 100, 2945–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carracedo A, Cantley LC, & Pandolfi PP (2013). Cancer metabolism: Fatty acid oxidation in the limelight. Nature Reviews. Cancer, 13, 227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrega P, Morandi B, Costa R, Frumento G, Forte G, Altavilla G, et al. (2008). Natural killer cells infiltrating human nonsmall-cell lung cancer are enriched in CD56 bright CD16(−) cells and display an impaired capability to kill tumor cells. Cancer, 112, 863–875. [DOI] [PubMed] [Google Scholar]
- Cascone T, McKenzie JA, Mbofung RM, Punt S, Wang Z, Xu C, et al. (2018). Increased tumor glycolysis characterizes immune resistance to adoptive T cell therapy. Cell Metabolism, 27, 977–987.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TY, Chang CC, Ohgami N, & Yamauchi Y (2006). Cholesterol sensing, trafficking, and esterification. Annual Review of Cell and Developmental Biology, 22, 129–157. [DOI] [PubMed] [Google Scholar]
- Chang CH, Curtis JD, Maggi LB Jr., Faubert B, Villarino AV, O’Sullivan D, et al. (2013). Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell, 153, 1239–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CI, Liao JC, & Kuo L (2001). Macrophage arginase promotes tumor cell growth and suppresses nitric oxide-mediated tumor cytotoxicity. Cancer Research, 61, 1100–1106. [PubMed] [Google Scholar]
- Chang C-H, Qiu J, O’Sullivan D, Buck MD, Noguchi T, Curtis JD, et al. (2015). Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell, 162, 1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Iliopoulos D, Zhang Q, Tang Q, Greenblatt MB, Hatziapostolou M, et al. (2014). XBP1 promotes triple-negative breast cancer by controlling the HIF1α pathway. Nature, 508, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DS, & Mellman I (2013). Oncology meets immunology: The cancer-immunity cycle. Immunity, 39, 1–10. [DOI] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, et al. (2006). Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell, 10, 529–541. [DOI] [PubMed] [Google Scholar]
- Chirala SS, & Wakil SJ (2004). Structure and function of animal fatty acid synthase. Lipids, 39, 1045–1053. [DOI] [PubMed] [Google Scholar]
- Choi SY, Collins CC, Gout PW, & Wang Y (2013). Cancer-generated lactic acid: A regulatory, immunosuppressive metabolite? The Journal of Pathology, 230, 350–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clem BF, O’Neal J, Tapolsky G, Clem AL, Imbert-Fernandez Y, Kerr DA 2nd, et al. (2013). Targeting 6-phosphofructo-2-kinase (PFKFB3) as a therapeutic strategy against cancer. Molecular Cancer Therapeutics, 12, 1461–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colegio OR, Chu NQ, Szabo AL, Chu T, Rhebergen AM, Jairam V, et al. (2014). Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature, 513, 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condamine T, Dominguez GA, Youn JI, Kossenkov AV, Mony S, Alicea-Torres K, et al. (2016). Lectin-type oxidized LDL receptor-1 distinguishes population of human polymorphonuclear myeloid-derived suppressor cells in cancer patients. Science Immunology, 1, pii: aaf8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong J, Wang X, Zheng X, Wang D, Fu B, Sun R, et al. (2018). Dysfunction of natural killer cells by FBP1-induced inhibition of glycolysis during lung cancer progression. Cell Metabolism, 28, 243–255.e5. [DOI] [PubMed] [Google Scholar]
- Coussens LM, Zitvogel L, & Palucka AK (2013). Neutralizing tumor-promoting chronic inflammation: A magic bullet? Science (New York, N.Y.), 339, 286–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford A, Angelosanto JM, Kao C, Doering TA, Odorizzi PM, Barnett BE, et al. (2014). Molecular and transcriptional basis of CD4(+) T cell dysfunction during chronic infection. Immunity, 40, 289–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crompton JG, Sukumar M, Roychoudhuri R, Clever D, Gros A, Eil RL, et al. (2015). Akt inhibition enhances expansion of potent tumor-specific lymphocytes with memory cell characteristics. Cancer Research, 75, 296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csibi A, Lee G, Yoon SO, Tong H, Ilter D, Elia I, et al. (2014). The mTORC1/S6K1 pathway regulates glutamine metabolism through the eIF4B-dependent control of c-Myc translation. Current Biology, 24, 2274–2280. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cubillos-Ruiz JR, Silberman PC, Rutkowski MR, Chopra S, Perales-Puchalt A, Song M, et al. (2015). ER stress sensor XBP1 controls anti-tumor immunity by disrupting dendritic cell homeostasis. Cell, 161, 1527–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G, Staron MM, Gray SM, Ho PC, Amezquita RA, Wu J, et al. (2015). IL-7-induced glycerol transport and TAG synthesis promotes memory CD8 + T cell longevity. Cell, 161, 750–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie E, Schulze A, Zechner R, Walther TC, & Farese RV Jr. (2013). Cellular fatty acid metabolism and cancer. Cell Metabolism, 18, 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. (2009). Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature, 462, 739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg AJ, & Subbaramaiah K (2003). Targeting cyclooxygenase-2 in human neoplasia: Rationale and promise. Cancer Cell, 4, 431–436. [DOI] [PubMed] [Google Scholar]
- Datta SR, Brunet A, & Greenberg ME (1999). Cellular survival: A play in three Akts. Genes & Development, 13, 2905–2927. [DOI] [PubMed] [Google Scholar]
- Daurkin I, Eruslanov E, Stoffs T, Perrin GQ, Algood C, Gilbert SM, et al. (2011). Tumor-associated macrophages mediate immunosuppression in the renal cancer microenvironment by activating the 15-lipoxygenase-2 pathway. Cancer Research, 71, 6400–6409. [DOI] [PubMed] [Google Scholar]
- Davies SP, Sim AT, & Hardie DG (1990). Location and function of three sites phosphorylated on rat acetyl-CoA carboxylase by the AMP-activated protein kinase. European Journal of Biochemistry, 187, 183–190. [DOI] [PubMed] [Google Scholar]
- Dayton TL, Jacks T, & Vander Heiden MG (2016). PKM2, cancer metabolism, and the road ahead. EMBO Reports, 17, 1721–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, & Chandel NS (2016). Fundamentals of cancer metabolism. Science Advances, 2, e1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, et al. (2007). Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proceedings of the National Academy of Sciences of the United States of America, 104, 19345–19350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, & Thompson CB (2012). Cellular metabolism and disease: What do metabolic outliers teach us? Cell, 148, 1132–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delves PJ, & Roitt IM (2000). The Immune System. New England Journal of Medicine, 343, 108–117. [DOI] [PubMed] [Google Scholar]
- DeNardo DG, Brennan DJ, Rexhepaj E, Ruffell B, Shiao SL, Madden SF, et al. (2011). Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discovery, 1, 54–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietl K, Renner K, Dettmer K, Timischl B, Eberhart K, Dorn C, et al. (2010). Lactic acid and acidification inhibit TNF secretion and glycolysis of human monocytes. Journal of Immunology (Baltimore, Md.: 1950), 184, 1200–1209. [DOI] [PubMed] [Google Scholar]
- DiNardo CD, Stein EM, de Botton S, Roboz GJ, Altman JK, Mims AS, et al. (2018). Durable remissions with Ivosidenib in IDH1-mutated relapsed or refractory AML. The New England Journal of Medicine, 378, 2386–2398. [DOI] [PubMed] [Google Scholar]
- Donnelly RP, Loftus RM, Keating SE, Liou KT, Biron CA, Gardiner CM, et al. (2014). mTORC1-dependent metabolic reprogramming is a prerequisite for NK cell effector function. Journal of Immunology (Baltimore, Md.: 1950), 193, 4477–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drogat B, Bouchecareilh M, North S, Petibois C, Deleris G, Chevet E, et al. (2007). Acute L-glutamine deprivation compromises VEGF-a upregulation in A549/8 human carcinoma cells. Journal of Cellular Physiology, 212, 463–472. [DOI] [PubMed] [Google Scholar]
- Dunn GP, Old LJ, & Schreiber RD (2004). The three Es of cancer immunoediting. Annual Review of Immunology, 22, 329–360. [DOI] [PubMed] [Google Scholar]
- Duran RV, Oppliger W, Robitaille AM, Heiserich L, Skendaj R, Gottlieb E, et al. (2012). Glutaminolysis activates Rag-mTORC1 signaling. Molecular Cell, 47, 349–358. [DOI] [PubMed] [Google Scholar]
- Eagle H, Washington C, & Friedman SM (1966). The synthesis of homocystine, cystathionine, and cystine by cultured diploid and heteroploid human cells. Proceedings of the National Academy of Sciences of the United States of America, 56, 156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DN, Ngwa VM, Wang S, Shiuan E, Brantley-Sieders DM, Kim LC, et al. (2017). The receptor tyrosine kinase EphA2 promotes glutamine metabolism in tumors by activating the transcriptional coactivators YAP and TAZ. Science Signaling, 10, pii: eaan4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshhar Z, Waks T, Gross G, & Schindler DG (1993). Specific activation and targeting of cytotoxic lymphocytes through chimeric single chains consisting of antibody-binding domains and the gamma or zeta subunits of the immunoglobulin and T-cell receptors. Proceedings of the National Academy of Sciences of the United States of America, 90, 720–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts B, Amiel E, Huang SC, Smith AM, Chang CH, Lam WY, et al. (2014). TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKvarepsilon supports the anabolic demands of dendritic cell activation. Nature Immunology, 15, 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts B, Amiel E, van der Windt GJW, Freitas TC, Chott R, Yarasheski KE, et al. (2012). Commitment to glycolysis sustains survival of NO-producing inflammatory dendritic cells. Blood, 120, 1422–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Han J, Pearce SF, Silverstein RL, Gotto AM Jr., Hajjar DP, et al. (2000). Induction of CD36 expression by oxidized LDL and IL-4 by a common signaling pathway dependent on protein kinase C and PPAR-gamma. Journal of Lipid Research, 41, 688–696. [PubMed] [Google Scholar]
- Fischer K, Hoffmann P, Voelkl S, Meidenbauer N, Ammer J, Edinger M, et al. (2007). Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood, 109, 3812–3819. [DOI] [PubMed] [Google Scholar]
- Flavin R, Peluso S, Nguyen PL, & Loda M (2010). Fatty acid synthase as a potential therapeutic target in cancer. Future Oncology (London, England), 6, 551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauwirth KA, Riley JL, Harris MH, Parry RV, Rathmell JC, Plas DR, et al. (2002). The CD28 signaling pathway regulates glucose metabolism. Immunity, 16, 769–777. [DOI] [PubMed] [Google Scholar]
- Frauwirth KA, & Thompson CB (2004). Regulation of T lymphocyte metabolism. Journal of Immunology (Baltimore, Md.: 1950), 172, 4661–4665. [DOI] [PubMed] [Google Scholar]
- Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, et al. (2000). Engagement of the Pd-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. The Journal of Experimental Medicine, 192, 1027–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French AR, & Yokoyama WM (2003). Natural killer cells and viral infections. Current Opinion in Immunology, 15, 45–51. [DOI] [PubMed] [Google Scholar]
- Fukuzumi M, Shinomiya H, Shimizu Y, Ohishi K, & Utsumi S (1996). Endotoxin-induced enhancement of glucose influx into murine peritoneal macrophages via GLUT1. Infection and Immunity, 64, 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton MD, Galic S, Marcinko K, Sikkema S, Pulinilkunnil T, Chen ZP, et al. (2013). Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin-sensitizing effects of metformin. Nature Medicine, 19, 1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrilovich DI, Ostrand-Rosenberg S, & Bronte V (2012). Coordinated regulation of myeloid cells by tumours. Nature Reviews. Immunology, 12, 253–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio D, Metallo CM, Gameiro PA, Hiller K, Danna LS, Balestrieri C, et al. (2011). Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Molecular Systems Biology, 7, 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio D, Soldati C, Vanoni M, Alberghina L, & Chiaradonna F (2009). Glutamine deprivation induces abortive s-phase rescued by deoxyribonucleotides in k-ras transformed fibroblasts. PLoS One, 4, e4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Kepp O, Vander Heiden MG, & Kroemer G (2013). Metabolic targets for cancer therapy. Nature Reviews. Drug Discovery, 12, 829–846. [DOI] [PubMed] [Google Scholar]
- Ganeshan K, & Chawla A (2014). Metabolic regulation of immune responses. Annual Review of Immunology, 32, 609–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gansler TS, Hardman W 3rd, Hunt DA, Schaffel S, & Hennigar RA (1997). Increased expression of fatty acid synthase (OA-519) in ovarian neoplasms predicts shorter survival. Human Pathology, 28, 686–692. [DOI] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, et al. (2009). c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature, 458, 762–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt T, Wakim LM, Eidsmo L, Reading PC, Heath WR, & Carbone FR (2009). Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nature Immunology, 10, 524–530. [DOI] [PubMed] [Google Scholar]
- Geiger R, Rieckmann JC, Wolf T, Basso C, Feng Y, Fuhrer T, et al. (2016). Larginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell, 167, 829–842.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghesquiere B, Wong BW, Kuchnio A, & Carmeliet P (2014). Metabolism of stromal and immune cells in health and disease. Nature, 511, 167–176. [DOI] [PubMed] [Google Scholar]
- Goff SL, Dudley ME, Citrin DE, Somerville RP, Wunderlich JR, Danforth DN, et al. (2016). Randomized, prospective evaluation comparing intensity of lymphodepletion before adoptive transfer of tumor-infiltrating lymphocytes for patients with metastatic melanoma. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology, 34, 2389–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottfried E, Kunz-Schughart LA, Ebner S, Mueller-Klieser W, Hoves S, Andreesen R, et al. (2006). Tumor-derived lactic acid modulates dendritic cell activation and antigen expression. Blood, 107, 2013–2021. [DOI] [PubMed] [Google Scholar]
- Gout PW, Buckley AR, Simms CR, & Bruchovsky N (2001). Sulfasalazine, a potent suppressor of lymphoma growth by inhibition of the x(c)-cystine transporter: A new action for an old drug. Leukemia, 15, 1633–1640. [DOI] [PubMed] [Google Scholar]
- Grohmann U, & Bronte V (2010). Control of immune response by amino acid metabolism. Immunological Reviews, 236, 243–264. [DOI] [PubMed] [Google Scholar]
- Gross MI, Demo SD, Dennison JB, Chen L, Chernov-Rogan T, Goyal B, et al. (2014). Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Molecular Cancer Therapeutics, 13, 890–901. [DOI] [PubMed] [Google Scholar]
- Gu L, Yi Z, Zhang Y, Ma Z, Zhu Y, & Gao J (2017). Low dose of 2-deoxy-D-glucose kills acute lymphoblastic leukemia cells and reverses glucocorticoid resistance via N-linked glycosylation inhibition under normoxia. Oncotarget, 8, 30978–30991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Callaway JB, & Ting JPY (2015). Inflammasomes: Mechanism of action, role in disease, and therapeutics. Nature Medicine, 21, 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Manjili MH, Subjeck JR, Sarkar D, Fisher PB, & Wang XY (2013). Therapeutic cancer vaccines: Past, present, and future. Advances in Cancer Research, 119, 421–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Yi H, Yu X, Hu F, Zuo D, Subjeck JR, et al. (2012). Absence of scavenger receptor A promotes dendritic cell-mediated cross-presentation of cell-associated antigen and antitumor immune response. Immunology and Cell Biology, 90, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Yi H, Yu X, Zuo D, Qian J, Yang G, et al. (2012). In situ vaccination with CD204 gene-silenced dendritic cell, not unmodified dendritic cell, enhances radiation therapy of prostate cancer. Molecular Cancer Therapeutics, 11, 2331–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, et al. (2013). Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. The New England Journal of Medicine, 369, 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, & Coussens LM (2012). Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell, 21, 309–322. [DOI] [PubMed] [Google Scholar]
- Hanahan D, & Weinberg RA (2011). Hallmarks of cancer: The next generation. Cell, 144, 646–674. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, et al. (2003). An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Molecular Cell, 11, 619–633. [DOI] [PubMed] [Google Scholar]
- Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, et al. (2005). ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell, 8, 311–321. [DOI] [PubMed] [Google Scholar]
- Haunerland NH, & Spener F (2004). Fatty acid-binding proteins—insights from genetic manipulations. Progress in Lipid Research, 43, 328–349. [DOI] [PubMed] [Google Scholar]
- Hensley CT, Wasti AT, & DeBerardinis RJ (2013). Glutamine and cancer: Cell biology, physiology, and clinical opportunities. The Journal of Clinical Investigation, 123, 3678–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herber DL, Cao W, Nefedova Y, Novitskiy SV, Nagaraj S, Tyurin VA, et al. (2010). Lipid accumulation and dendritic cell dysfunction in cancer. Nature Medicine, 16, 880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho P-C, Bihuniak JD, Macintyre AN, Staron M, Liu X, Amezquita R, et al. (2015). Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell, 162, 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. (2010). Improved survival with ipilimumab in patients with metastatic melanoma. The New England Journal of Medicine, 363, 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Manoharan I, Suryawanshi A, Majumdar T, Angus-Hill ML, Koni PA, et al. (2015). β-catenin promotes regulatory T-cell responses in tumors by inducing vitamin A metabolism in dendritic cells. Cancer Research, 75, 656–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain F, Al-Khami AA, Wyczechowska D, Hernandez C, Zheng L, Reiss K, et al. (2015). Inhibition of fatty acid oxidation modulates immunosuppressive functions of myeloid-derived suppressor cells and enhances cancer therapies. Cancer Immunology Research, 3, 1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SC, Everts B, Ivanova Y, O’Sullivan D, Nascimento M, Smith AM, et al. (2014). Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nature Immunology, 15, 846–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs JS, Jung YJ, Mole DR, Lee S, Torres-Cabala C, Chung YL, et al. (2005). HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: Novel role of fumarate in regulation of HIF stability. Cancer Cell, 8, 143–153. [DOI] [PubMed] [Google Scholar]
- Ishii I, Akahoshi N, Yu XN, Kobayashi Y, Namekata K, Komaki G, et al. (2004). Murine cystathionine gamma-lyase: Complete cDNA and genomic sequences, promoter activity, tissue distribution and developmental expression. The Biochemical Journal, 381, 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa F, Niiro H, Iino T, Yoshida S, Saito N, Onohara S, et al. (2007). The developmental program of human dendritic cells is operated independently of conventional myeloid and lymphoid pathways. Blood, 110, 3591–3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacque N, Ronchetti AM, Larrue C, Meunier G, Birsen R, Willems L, et al. (2015). Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood, 126, 1346–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch J, Chakravortty D, Turza N, Prechtel AT, Buchholz B, Gerlach RG, et al. (2008). Hypoxia and hypoxia-inducible factor-1 alpha modulate lipopolysaccharide-induced dendritic cell activation and function. Journal of Immunology (Baltimore, Md.: 1950), 180, 4697–4705. [DOI] [PubMed] [Google Scholar]
- Jenkins RW, Barbie DA, & Flaherty KT (2018). Mechanisms of resistance to immune checkpoint inhibitors. British Journal of Cancer, 118, 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins MK, Khoruts A, Ingulli E, Mueller DL, McSorley SJ, Reinhardt RL, et al. (2001). In vivo activation of antigen-specific CD4 T cells. Annual Review of Immunology, 19, 23–45. [DOI] [PubMed] [Google Scholar]
- Jha AK, Huang SC-C, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, et al. (2015). Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity, 42, 419–430. [DOI] [PubMed] [Google Scholar]
- Jones SF, & Infante JR (2015). Molecular pathways: Fatty acid synthase. Clinical Cancer Research, 21, 5434–5438. [DOI] [PubMed] [Google Scholar]
- Jones RG, Parsons M, Bonnard M, Chan VSF, Yeh W-C, Woodgett JR, et al. (2000). Protein kinase B regulates T lymphocyte survival, nuclear factor κb activation, and Bcl-Xl levels in vivo. The Journal of Experimental Medicine, 191, 1721–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju YS, Alexandrov LB, Gerstung M, Martincorena I, Nik-Zainal S, Ramakrishna M, et al. (2014). Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. Elife, 3 10.7554/eLife.02935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, & Ahmed R (2003). Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nature Immunology, 4, 1191. [DOI] [PubMed] [Google Scholar]
- Kane LP, Andres PG, Howland KC, Abbas AK, & Weiss A (2001). Akt provides the CD28 costimulatory signal for up-regulation of IL-2 and IFN-gamma but not TH2 cytokines. Nature Immunology, 2, 37–44. [DOI] [PubMed] [Google Scholar]
- Kantoff PW, Higano CS, Shore ND, Berger ER, Small EJ, Penson DF, et al. (2010). Sipuleucel-T immunotherapy for castration-resistant prostate cancer. The New England Journal of Medicine, 363, 411–422. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Chen D, Rogers AN, Katewa SD, Li PW, Thomas EL, et al. (2010). With TOR, less is more: A key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metabolism, 11, 453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kareva I, & Hahnfeldt P (2013). The emerging “hallmarks” of metabolic reprogramming and immune evasion: Distinct or linked? Cancer Research, 73, 2737–2742. [DOI] [PubMed] [Google Scholar]
- Kawalekar OU, O’Connor RS, Fraietta JA, Guo L, McGettigan SE, Posey AD Jr., et al. (2016). Distinct signaling of coreceptors regulates specific metabolism pathways and impacts memory development in CAR T cells. Immunity, 44, 380–390. [DOI] [PubMed] [Google Scholar]
- Keir ME, Butte MJ, Freeman GJ, & Sharpe AH (2008). PD-1 and its ligands in tolerance and immunity. Annual Review of Immunology, 26, 677–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppel MP, Saucier N, Mah AY, Vogel TP, & Cooper MA (2015). Activation-specific metabolic requirements for NK Cell IFN-γ production. The Journal of Immunology, 194, 1954–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieper WC, Tan JT, Bondi-Boyd B, Gapin L, Sprent J, Ceredig R, et al. (2002). Overexpression of interleukin (IL)-7 Leads to IL-15-independent generation of memory phenotype CD8+ T cells. The Journal of Experimental Medicine, 195, 1533–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus A, & Birchmeier W (2008). Wnt signalling and its impact on development and cancer. Nature Reviews. Cancer, 8, 387–398. [DOI] [PubMed] [Google Scholar]
- Klebanoff CA, Gattinoni L, Torabi-Parizi P, Kerstann K, Cardones AR, Finkelstein SE, et al. (2005). Central memory self/tumor-reactive CD8+ T cells confer superior antitumor immunity compared with effector memory T cells. Proceedings of the National Academy of Sciences of the United States of America, 102, 9571–9576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk CM, Holowka T, Sun J, Blagih J, Amiel E, DeBerardinis RJ, et al. (2010). Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood, 115, 4742–4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kridel SJ, Axelrod F, Rozenkrantz N, & Smith JW (2004). Orlistat is a novel inhibitor of fatty acid synthase with antitumor activity. Cancer Research, 64, 2070–2075. [DOI] [PubMed] [Google Scholar]
- Krummel MF, & Allison JP (1995). CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. The Journal of Experimental Medicine, 182, 459–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhajda FP (2000). Fatty-acid synthase and human cancer: New perspectives on its role in tumor biology. Nutrition, 16, 202–208. [DOI] [PubMed] [Google Scholar]
- Kuhajda FP, Pizer ES, Li JN, Mani NS, Frehywot GL, & Townsend CA (2000). Synthesis and antitumor activity of an inhibitor of fatty acid synthase. Proceedings of the National Academy of Sciences of the United States of America, 97, 3450–3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar BV, Connors TJ, & Farber DL (2018). Human T cell development, localization, and function throughout life. Immunity, 48, 202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwana Y, Asakura Y, Utsunomiya N, Nakanishi M, Arata Y, Itoh S, et al. (1987). Expression of chimeric receptor composed of immunoglobulin-derived V regions and T-cell receptor-derived C regions. Biochemical and Biophysical Research Communications, 149, 960–968. [DOI] [PubMed] [Google Scholar]
- Kwon SJ, & Lee YJ (2005). Effect of low glutamine/glucose on hypoxia-induced elevation of hypoxia-inducible factor-1alpha in human pancreatic cancer MiaPaCa-2 and human prostatic cancer DU-145 cells. Clinical Cancer Research, 11, 4694–4700. [DOI] [PubMed] [Google Scholar]
- Lane AN, & Fan TW (2015). Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Research, 43, 2466–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langowski JL, Zhang X, Wu L, Mattson JD, Chen T, Smith K, et al. (2006). IL-23 promotes tumour incidence and growth. Nature, 442, 461–465. [DOI] [PubMed] [Google Scholar]
- Laplante M, & Sabatini DM (2012). mTOR signaling in growth control and disease. Cell, 149, 274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latz E, Xiao TS, & Stutz A (2013). Activation and regulation of the inflammasomes. Nature Reviews. Immunology, 13, 379–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti G, & Tennant DA (2016). Isocitrate dehydrogenase (IDH), succinate dehydrogenase (SDH), fumarate hydratase (FH): Three players for one phenotype in cancer? Biochemical Society Transactions, 44, 1111–1116. [DOI] [PubMed] [Google Scholar]
- Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, et al. (2010). Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proceedings of the National Academy of Sciences of the United States of America, 107, 2037–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Naour F, Hohenkirk L, Grolleau A, Misek DE, Lescure P, Geiger JD, et al. (2001). Profiling changes in gene expression during differentiation and maturation of monocyte-derived dendritic cells using both oligonucleotide microarrays and proteomics. The Journal of Biological Chemistry, 276, 17920–17931. [DOI] [PubMed] [Google Scholar]
- Lee A-H, Iwakoshi NN, Anderson KC, & Glimcher LH (2003). Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proceedings of the National Academy of Sciences of the United States of America, 100, 9946–9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BL, Kim WH, Jung J, Cho SJ, Park JW, Kim J, et al. (2008). A hypoxia-independent up-regulation of hypoxia-inducible factor-1 by AKT contributes to angiogenesis in human gastric cancer. Carcinogenesis, 29, 44–51. [DOI] [PubMed] [Google Scholar]
- Lee DW, Kochenderfer JN, Stetler-Stevenson M, Cui YK, Delbrook C, Feldman SA, et al. (2015). T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: A phase 1 dose-escalation trial. Lancet (London, England), 385, 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Walsh MC, Hoehn KL, James DE, Wherry EJ, & Choi Y (2014). Reg-ulator of fatty acid metabolism, acetyl coenzyme a carboxylase 1, controls T cell immunity. Journal of Immunology (Baltimore, Md.: 1950), 192, 3190–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, & Puzio-Kuter AM (2010). The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science, 330, 1340–1344. [DOI] [PubMed] [Google Scholar]
- Liang X, Fu C, Cui W, Ober-Blobaum JL, Zahner SP, Shrikant PA, et al. (2014). beta-catenin mediates tumor-induced immunosuppression by inhibiting cross-priming of CD8(+) T cells. Journal of Leukocyte Biology, 95, 179–190. [DOI] [PubMed] [Google Scholar]
- Liberti MV, & Locasale JW (2016). The warburg effect: How does it benefit cancer cells? Trends in Biochemical Sciences, 41, 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SO, Li CW, Xia W, Lee HH, Chang SS, Shen J, et al. (2016). EGFR signaling enhances aerobic glycolysis in triple-negative breast cancer cells to promote tumor growth and immune escape. Cancer Research, 76, 1284–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Handschin C, & Spiegelman BM (2005). Metabolic control through the PGC-1 family of transcription coactivators. Cell Metabolism, 1, 361–370. [DOI] [PubMed] [Google Scholar]
- Locasale JW (2013). Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nature Reviews. Cancer, 13, 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochner M, Berod L, & Sparwasser T (2015). Fatty acid metabolism in the regulation of T cell function. Trends in Immunology, 36, 81–91. [DOI] [PubMed] [Google Scholar]
- Loftus RM, Assmann N, Kedia-Mehta N, O’Brien KL, Garcia A, Gillespie C, et al. (2018). Amino acid-dependent cMyc expression is essential for NK cell metabolic and functional responses in mice. Nature Communications, 9, 2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long AH, Haso WM, Shern JF, Wanhainen KM, Murgai M, Ingaramo M, et al. (2015). 4–1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nature Medicine, 21, 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long EO, Kim HS, Liu D, Peterson ME, & Rajagopalan S (2013). Controlling natural killer cell responses: Integration of signals for activation and inhibition. Annual Review of Immunology, 31, 227–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt SY, & Vander Heiden MG (2011). Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annual Review of Cell and Developmental Biology, 27, 441–464. [DOI] [PubMed] [Google Scholar]
- Lupu R, & Menendez JA (2006a). Pharmacological inhibitors of Fatty Acid Synthase (FASN)—Catalyzed endogenous fatty acid biogenesis: A new family of anti-cancer agents? Current Pharmaceutical Biotechnology, 7, 483–493. [DOI] [PubMed] [Google Scholar]
- Lupu R, & Menendez JA (2006b). Targeting fatty acid synthase in breast and endometrial cancer: An alternative to selective estrogen receptor modulators? Endocrinology, 147, 4056–4066. [DOI] [PubMed] [Google Scholar]
- Ma J, Yan R, Zu X, Cheng JM, Rao K, Liao DF, et al. (2008). Aldo-keto reductase family 1 B10 affects fatty acid synthesis by regulating the stability of acetyl-CoA carboxylase-alpha in breast cancer cells. The Journal of Biological Chemistry, 283, 3418–3423. [DOI] [PubMed] [Google Scholar]
- Ma Y, Yu C, Mohamed EM, Shao H, Wang L, Sundaresan G, et al. (2016). A causal link from ALK to hexokinase II overexpression and hyperactive glycolysis in EML4-ALK-positive lung cancer. Oncogene, 35, 6132–6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacInnes A, Fairman DA, Binding P, Rhodes J, Wyatt MJ, Phelan A, et al. (2003). The antianginal agent trimetazidine does not exert its functional benefit via inhibition of mitochondrial long-chain 3-ketoacyl coenzyme A thiolase. Circulation Research, 93, e26–e32. [DOI] [PubMed] [Google Scholar]
- Macintyre AN, Finlay D, Preston G, Sinclair LV, Waugh CM, Tamas P, et al. (2011). Protein kinase B controls transcriptional programs that direct cytotoxic T cell fate but is dispensable for T cell metabolism. Immunity, 34, 224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIver NJ, Michalek RD, & Rathmell JC (2013). Metabolic regulation of T lymphocytes. Annual Review of Immunology, 31, 259–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier T, Jenni S, & Ban N (2006). Architecture of mammalian fatty acid synthase at 4.5 A resolution. Science, 311, 1258–1262. [DOI] [PubMed] [Google Scholar]
- Makowski L, & Hotamisligil GS (2005). The role of fatty acid binding proteins in metabolic syndrome and atherosclerosis. Current Opinion in Lipidology, 16, 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinarich F, Duan K, Hamid RA, Bijin A, Lin WX, Poidinger M, et al. (2015). High mitochondrial respiration and glycolytic capacity represent a metabolic phenotype of human tolerogenic dendritic cells. The Journal of Immunology, 194, 5174–5186. [DOI] [PubMed] [Google Scholar]
- Malmberg KJ, Carlsten M, Bjorklund A, Sohlberg E, Bryceson YT, & Ljunggren HG (2017). Natural killer cell-mediated immunosurveillance of human cancer. Seminars in Immunology, 31, 20–29. [DOI] [PubMed] [Google Scholar]
- Mamessier E, Sylvain A, Thibult ML, Houvenaeghel G, Jacquemier J, Castellano R, et al. (2011). Human breast cancer cells enhance self tolerance by promoting evasion from NK cell antitumor immunity. The Journal of Clinical Investigation, 121, 3609–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansoor MA, Svardal AM, & Ueland PM (1992). Determination of the in vivo redox status of cysteine, cysteinylglycine, homocysteine, and glutathione in human plasma. Analytical Biochemistry, 200, 218–229. [DOI] [PubMed] [Google Scholar]
- Marin-Hernandez A, Gallardo-Perez JC, Ralph SJ, Rodriguez-Enriquez S, & Moreno-Sanchez R (2009). HIF-1alpha modulates energy metabolism in cancer cells by inducing over-expression of specific glycolytic isoforms. Mini Reviews in Medicinal Chemistry, 9, 1084–1101. [DOI] [PubMed] [Google Scholar]
- Maroof A, English NR, Bedford PA, Gabrilovich DI, & Knight SC (2005). Developing dendritic cells become ‘lacy’ cells packed with fat and glycogen. Immunology, 115, 473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez FO, Gordon S, Locati M, & Mantovani A (2006). Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: New molecules and patterns of gene expression. Journal of Immunology (Baltimore, Md: 1950), 177, 7303–7311. [DOI] [PubMed] [Google Scholar]
- Martinez-Outschoorn UE, Peiris-Pages M, Pestell RG, Sotgia F, & Lisanti MP (2017). Cancer metabolism: A therapeutic perspective. Nature Reviews. Clinical Oncology, 14, 11–31. [DOI] [PubMed] [Google Scholar]
- Martinon F, Burns K, & Tschopp J (2002). The Inflammasome: A molecular platform triggering activation of inflammatory caspase and processing of proIL-beta. Molecular Cell, 10, 417–426. [DOI] [PubMed] [Google Scholar]
- Maschek G, Savaraj N, Priebe W, Braunschweiger P, Hamilton K, Tidmarsh GF, et al. (2004). 2-deoxy-D-glucose increases the efficacy of adriamycin and paclitaxel in human osteosarcoma and non-small cell lung cancers in vivo. Cancer Research, 64, 31–34. [DOI] [PubMed] [Google Scholar]
- Mashima T, Seimiya H, & Tsuruo T (2009). De novo fatty-acid synthesis and related pathways as molecular targets for cancer therapy. British Journal of Cancer, 100, 1369–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, et al. (2014). Chimeric antigen receptor T cells for sustained remissions in leukemia. The New England Journal of Medicine, 371, 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maus MV, Fraietta JA, Levine BL, Kalos M, Zhao Y, & June CH (2014). Adoptive immunotherapy for cancer or viruses. Annual Review of Immunology, 32, 189–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, et al. (1999). The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature, 399, 271–275. [DOI] [PubMed] [Google Scholar]
- McKnight GL, Mudri SL, Mathewes SL, Traxinger RR, Marshall S, Sheppard PO, et al. (1992). Molecular cloning, cDNA sequence, and bacterial expression of human glutamine: Fructose-6-phosphate amidotransferase. The Journal of Biological Chemistry, 267, 25208–25212. [PubMed] [Google Scholar]
- Medes G, Thomas A, & Weinhouse S (1953). Metabolism of neoplastic tissue. IV. A study of lipid synthesis in neoplastic tissue slices in vitro. Cancer Research, 13, 27–29. [PubMed] [Google Scholar]
- Meijer AJ, & Codogno P (2008). Nutrient sensing: TOR’s Ragtime. Nature Cell Biology, 10, 881–883. [DOI] [PubMed] [Google Scholar]
- Menendez JA, Colomer R, & Lupu R (2005). Why does tumor-associated fatty acid synthase (oncogenic antigen-519) ignore dietary fatty acids? Medical Hypotheses, 64, 342–349. [DOI] [PubMed] [Google Scholar]
- Menendez JA, & Lupu R (2007). Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nature Reviews. Cancer, 7, 763–777. [DOI] [PubMed] [Google Scholar]
- Menk AV, Scharping NE, Rivadeneira DB, Calderon MJ, Watson MJ, Dunstane D, et al. (2018). 4–1BB costimulation induces T cell mitochondrial function and biogenesis enabling cancer immunotherapeutic responses. The Journal of Experimental Medicine, 215, 1091–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, et al. (2011). Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature, 481, 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, et al. (2011). Cutting edge: Distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4 + T cell subsets. Journal of Immunology, 186, 3299–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelakis ED, Sutendra G, Dromparis P, Webster L, Haromy A, Niven E, et al. (2010). Metabolic modulation of glioblastoma with dichloroacetate. Science Translational Medicine, 2, 31ra34. [DOI] [PubMed] [Google Scholar]
- Miller DM, Thomas SD, Islam A, Muench D, & Sedoris K (2012). c-Myc and cancer metabolism. Clinical Cancer Research, 18, 5546–5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modolell M, Corraliza IM, Link F, Soler G, & Eichmann K (1995). Reciprocal regulation of the nitric oxide synthase/arginase balance in mouse bone marrow-derived macrophages by TH1 and TH2 cytokines. European Journal of Immunology, 25, 1101–1104. [DOI] [PubMed] [Google Scholar]
- Mondal S, Roy D, Sarkar Bhattacharya S, Jin L, Jung D, Zhang S, et al. (2018). Therapeutic targeting of PFKFB3 with a novel glycolytic inhibitor PFK158 promotes lipophagy and chemosensitivity in gynecologic cancers. International Journal of Cancer. 10.1002/ijc.31868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JS, Hisata S, Park MA, DeNicola GM, Ryter SW, Nakahira K, et al. (2015). mTORC1-induced HK1-dependent glycolysis regulates NLRP3 inflammasome activation. Cell Reports, 12, 102–115. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Moon JS, Lee S, Park MA, Siempos II, Haslip M, Lee PJ, et al. (2015). UCP2-induced fatty acid synthase promotes NLRP3 inflammasome activation during sepsis. The Journal of Clinical Investigation, 125, 665–680. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Moon JS, Nakahira K, Chung KP, DeNicola GM, Koo MJ, Pabon MA, et al. (2016). NOX4-dependent fatty acid oxidation promotes NLRP3 inflammasome activation in macrophages. Nature Medicine, 22, 1002–1012. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Moreau K, Dizin E, Ray H, Luquain C, Lefai E, Foufelle F, et al. (2006). BRCA1 affects lipid synthesis through its interaction with acetyl-CoA carboxylase. The Journal of Biological Chemistry, 281, 3172–3181. [DOI] [PubMed] [Google Scholar]
- Motz GT, & Coukos G (2013). Deciphering and reversing tumor immune suppression. Immunity, 39, 61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SN, Gebhardt T, Carbone FR, & Heath WR (2013). Memory T cell subsets, migration patterns, and tissue residence. Annual Review of Immunology, 31, 137–161. [DOI] [PubMed] [Google Scholar]
- Mukherjee A, Wu J, Barbour S, & Fang X (2012). Lysophosphatidic acid activates lipogenic pathways and de novo lipid synthesis in ovarian cancer cells. The Journal of Biological Chemistry, 287, 24990–25000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Brusselbach S, Komhoff M, Rieck M, Meissner W, Kaddatz K, Adamkiewicz J, et al. (2007). Deregulation of tumor angiogenesis and blockade of tumor growth in PPARbeta-deficient mice. The EMBO Journal, 26, 3686–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn DH, & Mellor AL (2007). Indoleamine 2,3-dioxygenase and tumor-induced tolerance. The Journal of Clinical Investigation, 117, 1147–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn DH, & Mellor AL (2016). IDO in the tumor microenvironment: Inflammation, counter-regulation, and tolerance. Trends in Immunology, 37, 193–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao S, Kuwano T, Tsutsumi-Miyahara C, Ueda S, Kimura YN, Hamano S, et al. (2005). Infiltration of COX-2-expressing macrophages is a prerequisite for IL-1 beta-induced neovascularization and tumor growth. The Journal of Clinical Investigation, 115, 2979–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya M, Xiao Y, Zhou X, Chang JH, Chang M, Cheng X, et al. (2014). Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity, 40, 692–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namgaladze D, & Brune B (2014). Fatty acid oxidation is dispensable for human macrophage IL-4-induced polarization. Biochimica et Biophysica Acta, 1841, 1329–1335. [DOI] [PubMed] [Google Scholar]
- Nomura M, Liu J, Rovira II, Gonzalez-Hurtado E, Lee J, Wolfgang MJ, et al. (2016). Fatty acid oxidation in macrophage polarization. Nature Immunology, 17, 216–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numasaki M, Fukushi J-I, Ono M, Narula SK, Zavodny PJ, Kudo T, et al. (2003). Interleukin-17 promotes angiogenesis and tumor growth. Blood, 101, 2620–2627. [DOI] [PubMed] [Google Scholar]
- Okkenhaug K, Bilancio A, Farjot G, Priddle H, Sancho S, Peskett E, et al. (2002). Impaired B and T cell antigen receptor signaling in p110δ PI 3-kinase mutant mice. Science (New York, N.Y.), 297, 1031–1034. [DOI] [PubMed] [Google Scholar]
- O’Neill LA, Kishton RJ, & Rathmell J (2016). A guide to immunometabolism for immunologists. Nature Reviews. Immunology, 16, 553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill LA, & Pearce EJ (2016). Immunometabolism governs dendritic cell and macrophage function. The Journal of Experimental Medicine, 213, 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan D, van der Windt GJ, Huang SC, Curtis JD, Chang CH, Buck MD, et al. (2014). Memory CD8(+) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity, 41, 75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen OE, Kalhan SC, & Hanson RW (2002). The key role of anaplerosis and cataplerosis for citric acid cycle function. The Journal of Biological Chemistry, 277, 30409–30412. [DOI] [PubMed] [Google Scholar]
- Pages F, Ragueneau M, Rottapel R, Truneh A, Nunes J, Imbert J, et al. (1994). Binding of phosphatidylinositol-3-OH kinase to CD28 is required for T-cell signalling. Nature, 369, 327–329. [DOI] [PubMed] [Google Scholar]
- Palmieri EM, Spera I, Menga A, Infantino V, Porcelli V, Iacobazzi V, et al. (2015). Acetylation of human mitochondrial citrate carrier modulates mitochondrial citrate/malate exchange activity to sustain NADPH production during macrophage activation. Biochimica et Biophysica Acta, 1847, 729–738. [DOI] [PubMed] [Google Scholar]
- Pardoll DM (2012). The blockade of immune checkpoints in cancer immunotherapy. Nature Reviews. Cancer, 12, 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Lee SE, Hur J, Hong EB, Choi JI, Yang JM, et al. (2015). M-CSF from cancer cells induces fatty acid synthase and PPARbeta/delta activation in tumor myeloid cells, leading to tumor progression. Cell Reports, 10, 1614–1625. [DOI] [PubMed] [Google Scholar]
- Parkin J, & Cohen B (2001). An overview of the immune system. The Lancet, 357, 1777–1789. [DOI] [PubMed] [Google Scholar]
- Parry RV, Chemnitz JM, Frauwirth KA, Lanfranco AR, Braunstein I, Kobayashi SV, et al. (2005). CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Molecular and Cellular Biology, 25, 9543–9553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry RV, Reif K, Smith G, Sansom DM, Hemmings BA, & Ward SG (1997). Ligation of the T cell co-stimulatory receptor CD28 activates the serine-threonine protein kinase protein kinase B. European Journal of Immunology, 27, 2495–2501. [DOI] [PubMed] [Google Scholar]
- Patsoukis N, Bardhan K, Chatterjee P, Sari D, Liu B, Bell LN, et al. (2015). PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nature Communications, 6, 6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova NN, & Thompson CB (2016). The emerging hallmarks of cancer metabolism. Cell Metabolism, 23, 27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, & Pearce EJ (2013). Metabolic pathways in immune cell activation and quiescence. Immunity, 38, 633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl A (2017). Review: Metabolic control of immune system activation in rheumatic diseases. Arthritis & Rhematology, 69, 2259–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon-Thomas S, Kodumudi KN, El-Kenawi AE, Russell S, Weber AM, Luddy K, et al. (2016). Neutralization of tumor acidity improves antitumor responses to immunotherapy. Cancer Research, 76, 1381–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platonova S, Cherfils-Vicini J, Damotte D, Crozet L, Vieillard V, Validire P, et al. (2011). Profound coordinated alterations of intratumoral NK cell phenotype and function in lung carcinoma. Cancer Research, 71, 5412–5422. [DOI] [PubMed] [Google Scholar]
- Poczobutt JM, De S, Yadav VK, Nguyen TT, Li H, Sippel TR, et al. (2016). Expression profiling of macrophages reveals multiple populations with distinct biological roles in an immunocompetent orthotopic model of lung cancer. The Journal of Immunology, 196, 2847–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard PJ, Briere JJ, Alam NA, Barwell J, Barclay E, Wortham NC, et al. (2005). Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutations. Human Molecular Genetics, 14, 2231–2239. [DOI] [PubMed] [Google Scholar]
- Porter DL, Hwang WT, Frey NV, Lacey SF, Shaw PA, Loren AW, et al. (2015). Chimeric antigen receptor T cells persist and induce sustained remissions in relapsed refractory chronic lymphocytic leukemia. Science Translational Medicine, 7, 303ra139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postic C, Dentin R, Denechaud PD, & Girard J (2007). ChREBP, a transcriptional regulator of glucose and lipid metabolism. Annual Review of Nutrition, 27, 179–192. [DOI] [PubMed] [Google Scholar]
- Postow MA, Sidlow R, & Hellmann MD (2018). Immune-related adverse events associated with immune checkpoint blockade. The New England Journal of Medicine, 378, 158–168. [DOI] [PubMed] [Google Scholar]
- Puig T, Turrado C, Benhamu B, Aguilar H, Relat J, Ortega-Gutierrez S, et al. (2009). Novel inhibitors of fatty acid synthase with anticancer activity. Clinical Cancer Research, 15, 7608–7615. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, & Spiegelman BM (1998). A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell, 92, 829–839. [DOI] [PubMed] [Google Scholar]
- Qian J, Yi H, Guo C, Yu X, Zuo D, Chen X, et al. (2011). CD204 suppresses large heat shock protein-facilitated priming of tumor antigen gp100-specific T cells and chaperone vaccine activity against mouse melanoma. Journal of Immunology (Baltimore, Md.: 1950), 187, 2905–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan R, Tyurin VA, Veglia F, Condamine T, Amoscato A, Mohammadyani D, et al. (2014). Oxidized lipids block antigen cross-presentation by dendritic cells in cancer. Journal of Immunology (Baltimore, Md.: 1950), 192, 2920–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao E, Singh P, Zhai X, Li Y, Zhu G, Zhang Y, et al. (2015). Inhibition of tumor growth by a newly-identified activator for epidermal fatty acid binding protein. Oncotarget, 6, 7815–7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath M, Muller I, Kropf P, Closs EI, & Munder M (2014). Metabolism via arginase or nitric oxide synthase: Two competing arginine pathways in macrophages. Frontiers in Immunology, 5, 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathmell JC, Elstrom RL, Cinalli RM, & Thompson CB (2003). Activated Akt promotes increased resting T cell size, CD28-independent T cell growth, and development of autoimmunity and lymphoma. European Journal of Immunology, 33, 2223–2232. [DOI] [PubMed] [Google Scholar]
- Rathmell JC, Vander Heiden MG, Harris MH, Frauwirth KA, & Thompson CB (2000). In the absence of extrinsic signals, nutrient utilization by lymphocytes is insufficient to maintain either cell size or viability. Molecular Cell, 6, 683–692. [DOI] [PubMed] [Google Scholar]
- Rehman A, Hemmert KC, Ochi A, Jamal M, Henning JR, Barilla R, et al. (2013). Role of fatty-acid synthesis in dendritic cell generation and function. Journal of Immunology (Baltimore, Md.: 1950), 190, 4640–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, & Clevers H (2005). Wnt signalling in stem cells and cancer. Nature, 434, 843–850. [DOI] [PubMed] [Google Scholar]
- Ribas A, & Wolchok JD (2018). Cancer immunotherapy using checkpoint blockade. Science, 359, 1350–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert C, Schachter J, Long GV, Arance A, Grob JJ, Mortier L, et al. (2015). Pembrolizumab versus ipilimumab in advanced melanoma. The New England Journal of Medicine, 372, 2521–2532. [DOI] [PubMed] [Google Scholar]
- Robustelli della Cuna G, & Pedrazzoli P (1991). Toxicity and clinical tolerance of lonidamine. Seminars in Oncology, 18, 18–22. [PubMed] [Google Scholar]
- Rodriguez PC, Ernstoff MS, Hernandez C, Atkins M, Zabaleta J, Sierra R, et al. (2009). Arginase I-producing myeloid-derived suppressor cells in renal cell carcinoma are a subpopulation of activated granulocytes. Cancer Research, 69, 1553–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez PC, Quiceno DG, Zabaleta J, Ortiz B, Zea AH, Piazuelo MB, et al. (2004). Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Research, 64, 5839–5849. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Prados JC, Traves PG, Cuenca J, Rico D, Aragones J, Martin-Sanz P, et al. (2010). Substrate fate in activated macrophages: A comparison between innate, classic, and alternative activation. Journal of Immunology (Baltimore, Md.: 1950), 185, 605–614. [DOI] [PubMed] [Google Scholar]
- Rojek AM, Skowronski MT, Füchtbauer E-M, Füchtbauer AC, Fenton RA, Agre P, et al. (2007). Defective glycerol metabolism in aquaporin 9 (AQP9) knockout mice. Proceedings of the National Academy of Sciences of the United States of America, 104, 3609–3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Garcia S, Moreno-Altamirano MM, Prado-Garcia H, & Sanchez-Garcia FJ (2016). Lactate contribution to the tumor microenvironment: Mechanisms, effects on immune cells and therapeutic relevance. Frontiers in Immunology, 7, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin AL (1990). Suppression of transformation by and growth adaptation to low concentrations of glutamine in NIH-3T3 cells. Cancer Research, 50, 2832–2839. [PubMed] [Google Scholar]
- Sabbagh L, Pulle G, Liu Y, Tsitsikov EN, & Watts TH (2008). ERK-dependent Bim modulation downstream of the 4–1BB-TRAF1 signaling axis is a critical mediator of CD8 T cell survival in vivo. Journal of Immunology (Baltimore, Md.: 1950), 180, 8093–8101. [DOI] [PubMed] [Google Scholar]
- Sadelain M, Brentjens R, & Riviere I (2013). The basic principles of chimeric antigen receptor design. Cancer Discovery, 3, 388–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F, Lenig D, Forster R, Lipp M, & Lanzavecchia A (1999). Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature, 401, 708–712. [DOI] [PubMed] [Google Scholar]
- Sandigursky S, & Mor A (2018). Immune-related adverse events in cancer patients treated with immune checkpoint inhibitors. Current Rheumatology Reports, 20, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolla MF, Lappano R, De Marco P, Pupo M, Vivacqua A, Sisci D, et al. (2012). G protein-coupled estrogen receptor mediates the up-regulation of fatty acid synthase induced by 17beta-estradiol in cancer cells and cancer-associated fibroblasts. The Journal of Biological Chemistry, 287, 43234–43245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sborov DW, Haverkos BM, & Harris PJ (2015). Investigational cancer drugs targeting cell metabolism in clinical development. Expert Opinion on Investigational Drugs, 24, 79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharping NE, Menk AV, Moreci RS, Whetstone RD, Dadey RE, Watkins SC, et al. (2016). The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity, 45, 374–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, et al. (2005). Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell, 7, 77–85. [DOI] [PubMed] [Google Scholar]
- Semenza GL (2003). Targeting HIF-1 for cancer therapy. Nature Reviews. Cancer, 3, 721–732. [DOI] [PubMed] [Google Scholar]
- Semenza GL (2012). Hypoxia-inducible factors: Mediators of cancer progression and targets for cancer therapy. Trends in Pharmacological Sciences, 33, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, & Sabatini DM (2010). Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Molecular Cell, 40, 310–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Wen Z, Li Y, Matteson EL, Hong J, Goronzy JJ, et al. (2017). Metabolic control of the scaffold protein TKS5 in tissue-invasive, proinflammatory T cells. Nature Immunology, 18, 1025–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shime H, Yabu M, Akazawa T, Kodama K, Matsumoto M, Seya T, et al. (2008). Tumor-secreted lactic acid promotes IL-23/IL-17 proinflammatory pathway. Journal of Immunology (Baltimore, Md.: 1950), 180, 7175–7183. [DOI] [PubMed] [Google Scholar]
- Sibaud V, Meyer N, Lamant L, Vigarios E, Mazieres J, & Delord JP (2016). Dermatologic complications of anti-PD-1/PD-L1 immune checkpoint antibodies. Current Opinion in Oncology, 28, 254–263. [DOI] [PubMed] [Google Scholar]
- Sinclair LV, Rolf J, Emslie E, Shi YB, Taylor PM, & Cantrell DA (2013). Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nature Immunology, 14, 500–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siska PJ, Beckermann KE, Mason FM, Andrejeva G, Greenplate AR, Sendor AB, et al. (2017). Mitochondrial dysregulation and glycolytic insufficiency functionally impair CD8 T cells infiltrating human renal cell carcinoma. JCI Insight, 2, pii: 93411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siska PJ, & Rathmell JC (2015). T cell metabolic fitness in antitumor immunity. Trends in Immunology, 36, 257–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth MJ, Hayakawa Y, Takeda K, & Yagita H (2002). New aspects of natural-killer-cell surveillance and therapy of cancer. Nature Reviews. Cancer, 2, 850–861. [DOI] [PubMed] [Google Scholar]
- Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, et al. (2013). Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature, 496, 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava MK, Sinha P, Clements VK, Rodriguez P, & Ostrand-Rosenberg S (2010). Myeloid-derived suppressor cells inhibit T-cell activation by depleting cystine and cysteine. Cancer Research, 70, 68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JB, Alaei-Mahabadi B, Sabarinathan R, Samuelsson T, Gorodkin J, Gustafsson CM, et al. (2015). Simultaneous DNA and RNA mapping of somatic mitochondrial mutations across diverse human cancers. PLoS Genetics, 11, e1005333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Still ER, & Yuneva MO (2017). Hopefully devoted to Q: Targeting glutamine addiction in cancer. British Journal of Cancer, 116, 1375–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumar M, Liu J, Ji Y, Subramanian M, Crompton JG, Yu Z, et al. (2013). Inhibiting glycolytic metabolism enhances CD8 + T cell memory and antitumor function. The Journal of Clinical Investigation, 123, 4479–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumar M, Liu J, Mehta GU, Patel SJ, Roychoudhuri R, Crompton JG, et al. (2016). Mitochondrial membrane potential identifies cells with enhanced stemness for cellular therapy. Cell Metabolism, 23, 63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinnen JV, Brusselmans K, & Verhoeven G (2006). Increased lipogenesis in cancer cells: New players, novel targets. Current Opinion in Clinical Nutrition and Metabolic Care, 9, 358–365. [DOI] [PubMed] [Google Scholar]
- Swinnen JV, Esquenet M, Goossens K, Heyns W, & Verhoeven G (1997). Androgens stimulate fatty acid synthase in the human prostate cancer cell line LNCaP. Cancer Research, 57, 1086–1090. [PubMed] [Google Scholar]
- Tang C-HA, Ranatunga S, Kriss CL, Cubitt CL, Tao J, Pinilla-Ibarz JA, et al. (2014). Inhibition of ER stress–associated IRE-1/XBP-1 pathway reduces leukemic cell survival. The Journal of Clinical Investigation, 124, 2585–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, et al. (2013). Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature, 496, 238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X, Constant S, Jorritsma P, & Bottomly K (1997). Strength of TCR signal determines the costimulatory requirements for Th1 and Th2 CD4 + T cell differentiation. Journal of Immunology (Baltimore, Md.: 1950), 159, 5956–5963. [PubMed] [Google Scholar]
- Thwe PM, Pelgrom L, Cooper R, Beauchamp S, Reisz JA, D’Alessandro A, et al. (2017). Cell-intrinsic glycogen metabolism supports early glycolytic reprogramming required for dendritic cell immune responses. Cell Metabolism, 26, 558–567.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong WH, Sourbier C, Kovtunovych G, Jeong SY, Vira M, Ghosh M, et al. (2011). The glycolytic shift in fumarate-hydratase-deficient kidney cancer lowers AMPK levels, increases anabolic propensities and lowers cellular iron levels. Cancer Cell, 20, 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. (2012). Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. The New England Journal of Medicine, 366, 2443–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Sande T, De Schrijver E, Heyns W, Verhoeven G, & Swinnen JV (2002). Role of the phosphatidylinositol 3’-kinase/PTEN/Akt kinase pathway in the over-expression of fatty acid synthase in LNCaP prostate cancer cells. Cancer Research, 62, 642–646. [PubMed] [Google Scholar]
- van der Windt GJW, Everts B, Chang C-H, Curtis JD, Freitas TC, Amiel E, et al. (2012). Mitochondrial respiratory capacity is a critical regulator of CD8 + T cell memory development. Immunity, 36, 68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, & Thompson CB (2009). Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science (New York, N.Y.), 324, 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vats D, Mukundan L, Odegaard JI, Zhang L, Smith KL, Morel CR, et al. (2006). Oxidative metabolism and PGC-1β attenuate macrophage-mediated inflammation. Cell Metabolism, 4, 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viale A, & Draetta GF (2016). Metabolic features of cancer treatment resistance. Recent Results in Cancer Research, 207, 135–156. [DOI] [PubMed] [Google Scholar]
- Villani AC, Sarkizova S, & Hacohen N (2018). Systems immunology: Learning the rules of the immune system. Annual Review of Immunology, 36, 813–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier E, Tomasello E, Baratin M, Walzer T, & Ugolini S (2008). Functions of natural killer cells. Nature Immunology, 9, 503–510. [DOI] [PubMed] [Google Scholar]
- Walunas TL, Lenschow DJ, Bakker CY, Linsley PS, Freeman GJ, Green JM, et al. (1994). CTLA-4 can function as a negative regulator of T cell activation. Immunity, 1, 405–413. [DOI] [PubMed] [Google Scholar]
- Wang D, & DuBois RN (2010). Eicosanoids and cancer. Nature Reviews. Cancer, 10, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XY, Facciponte J, Chen X, Subjeck JR, & Repasky EA (2007). Scavenger receptor-A negatively regulates antitumor immunity. Cancer Research, 67, 4996–5002. [DOI] [PubMed] [Google Scholar]
- Wang C, Rajput S, Watabe K, Liao DF, & Cao D (2010). Acetyl-CoA carboxylase-a as a novel target for cancer therapy. Frontiers in Bioscience (Scholar Edition), 2, 515–526. [DOI] [PubMed] [Google Scholar]
- Wang XY, Zuo D, Sarkar D, & Fisher PB (2011). Blockade of cytotoxic T-lymphocyte antigen-4 as a new therapeutic approach for advanced melanoma. Expert Opinion on Pharmacotherapy, 12, 2695–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O (1956). On the origin of cancer cells. Science (New York, NY), 123, 309–314. [DOI] [PubMed] [Google Scholar]
- Warburg O, Posener K, & Negelein E (1924). u€eber den Stoffwechsel der Tumoren. Biochemische Zeitschrift, 152, 319–344. [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. (2010). The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell, 17, 225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, & Thompson CB (2012). Metabolic reprogramming: A cancer hallmark even warburg did not anticipate. Cancer Cell, 21, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward SG, Westwick J, Hall ND, & Sansom DM (1993). Ligation of CD28 receptor by B7 induces formation of D-3 phosphoinositides in T lymphocytes independently of T cell receptor/CD3 activation. European Journal of Immunology, 23, 2572–2577. [DOI] [PubMed] [Google Scholar]
- Wei SC, Duffy CR, & Allison JP (2018). Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discovery, 8, 1069–1086. [DOI] [PubMed] [Google Scholar]
- Welbourne TC (1979). Ammonia production and glutamine incorporation into glutathione in the functioning rat kidney. Canadian Journal of Biochemistry, 57, 233–237. [DOI] [PubMed] [Google Scholar]
- Wellen KE, Lu C, Mancuso A, Lemons JM, Ryczko M, Dennis JW, et al. (2010). The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes & Development, 24, 2784–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Gris D, Lei Y, Jha S, Zhang L, Huang MT, et al. (2011). Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nature Immunology, 12, 408–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyand CM, Zeisbrich M, & Goronzy JJ (2017). Metabolic signatures of T-cells and macrophages in rheumatoid arthritis. Current Opinion in Immunology, 46, 112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ (2011). T cell exhaustion. Nature Immunology, 12, 492–499. [DOI] [PubMed] [Google Scholar]
- Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, et al. (2007). Molecular signature of CD8 + T cell exhaustion during chronic viral infection. Immunity, 27, 670–684. [DOI] [PubMed] [Google Scholar]
- Williams NC, & O’Neill LAJ (2018). A role for the Krebs cycle intermediate citrate in metabolic reprogramming in innate immunity and inflammation. Frontiers in Immunology, 9, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windmueller HG, & Spaeth AE (1974). Uptake and metabolism of plasma glutamine by the small intestine. The Journal of Biological Chemistry, 249, 5070–5079. [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, et al. (2008). Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proceedings of the National Academy of Sciences of the United States of America, 105, 18782–18787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, & Thompson CB (2010). Glutamine addiction: A new therapeutic target in cancer. Trends in Biochemical Sciences, 35, 427–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won KY, Lim SJ, Kim GY, Kim YW, Han SA, Song JY, et al. (2012). Regulatory role of p53 in cancer metabolism via SCO2 and TIGAR in human breast cancer. Human Pathology, 43, 221–228. [DOI] [PubMed] [Google Scholar]
- Wu S, Akhtari M, & Alachkar H (2018). Characterization of mutations in the mitochondrial encoded electron transport chain complexes in acute myeloid leukemia. Science Reporter, 8, 13301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Qin L, Fako V, & Zhang JT (2014). Molecular mechanisms of fatty acid synthase (FASN)-mediated resistance to anti-cancer treatments. Advances in Biological Regulation, 54, 214–221. [DOI] [PubMed] [Google Scholar]
- Wynn TA, Chawla A, & Pollard JW (2013). Macrophage biology in development, homeostasis and disease. Nature, 496, 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, & Simon MC (2017). Oxygen availability and metabolic reprogramming in cancer. The Journal of Biological Chemistry, 292, 16825–16832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I, et al. (2014). Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity, 40, 274–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahagi N, Shimano H, Hasegawa K, Ohashi K, Matsuzaka T, Najima Y, et al. (2005). Co-ordinate activation of lipogenic enzymes in hepatocellular carcinoma. European Journal of Cancer, 41, 1316–1322. [DOI] [PubMed] [Google Scholar]
- Yang W, Bai Y, Xiong Y, Zhang J, Chen S, Zheng X, et al. (2016). Potentiating the antitumour response of CD8(+) T cells by modulating cholesterol metabolism. Nature, 531, 651–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Fujii H, Mohan SV, Goronzy JJ, & Weyand CM (2013). Phosphofructokinase deficiency impairs ATP generation, autophagy, and redox balance in rheumatoid arthritis T cells. The Journal of Experimental Medicine, 210, 2119–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Shen Y, Oishi H, Matteson EL, Tian L, Goronzy JJ, et al. (2016). Restoring oxidant signaling suppresses proarthritogenic T cell effector functions in rheumatoid arthritis. Science Translational Medicine, 8, 331ra338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Soga T, & Pollard PJ (2013). Oncometabolites: Linking altered metabolism with cancer. The Journal of Clinical Investigation, 123, 3652–3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Sudderth J, Dang T, Bachoo RM, McDonald JG, & DeBerardinis RJ (2009). Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer Research, 69, 7986–7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H, Guo C, Yu X, Gao P, Qian J, Zuo D, et al. (2011). Targeting the immunoregulator SRA/CD204 potentiates specific dendritic cell vaccine-induced T-cell response and antitumor immunity. Cancer Research, 71, 6611–6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S, Lee MY, Park SW, Moon JS, Koh YK, Ahn YH, et al. (2007). Upregulation of acetyl-CoA carboxylase alpha and fatty acid synthase by human epidermal growth factor receptor 2 at the translational level in breast cancer cells. The Journal of Biological Chemistry, 282, 26122–26131. [DOI] [PubMed] [Google Scholar]
- York AG, Williams KJ, Argus JP, Zhou QD, Brar G, Vergnes L, et al. (2015). Limiting cholesterol biosynthetic flux spontaneously engages type I IFN signaling. Cell, 163, 1716–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Deng R, Zhu HH, Zhang SS, Zhu C, Montminy M, et al. (2013). Modulation of fatty acid synthase degradation by concerted action of p38 MAP kinase, E3 ligase COP1, and SH2-tyrosine phosphatase Shp2. The Journal of Biological Chemistry, 288, 3823–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Guo C, Fisher PB, Subjeck JR, & Wang XY (2015). Scavenger receptors: Emerging roles in cancer biology and immunology. Advances in Cancer Research, 128, 309–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, et al. (2009). Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science, 325, 1555–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelenay S, van der Veen AG, Böttcher JP, Snelgrove KJ, Rogers N, Acton SE, et al. (2015). Cyclooxygenase-dependent tumor growth through evasion of immunity. Cell, 162, 1257–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kurupati R, Liu L, Zhou XY, Zhang G, Hudaihed A, et al. (2017). Enhancing CD8(+) T cell fatty acid catabolism within a metabolically challenging tumor microenvironment increases the efficacy of melanoma immunotherapy. Cancer Cell, 32, 377–391.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Pavlova NN, & Thompson CB (2017). Cancer cell metabolism: The essential role of the nonessential amino acid, glutamine. The EMBO Journal, 36, 1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sun Y, Rao E, Yan F, Li Q, Zhang Y, et al. (2014). Fatty acid-binding protein E-FABP restricts tumor growth by promoting IFN-beta responses in tumor-associated macrophages. Cancer Research, 74, 2986–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, et al. (2009). Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science, 324, 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao E, Maj T, Kryczek I, Li W, Wu K, Zhao L, et al. (2016). Cancer mediates effector T cell dysfunction by targeting microRNAs and EZH2 via glycolysis restriction. Nature Immunology, 17, 95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Xiao C, Evans KS, Theivanthiran T, DeVito N, Holtzhausen A, et al. (2018). Paracrine Wnt5a-beta-catenin signaling triggers a metabolic program that drives dendritic cell tolerization. Immunity, 48, 147–160.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhdanov AV, Waters AH, Golubeva AV, & Papkovsky DB (2015). Differential contribution of key metabolic substrates and cellular oxygen in HIF signalling. Experimental Cell Research, 330, 13–28. [DOI] [PubMed] [Google Scholar]
- Zhu J, & Paul WE (2008). CD4 T cells: Fates, functions, and faults. Blood, 112, 1557–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]