Abstract
Berberine (BBR) is a natural active principle with potential antitumor activity. The compound targets multiple cell signaling pathways, including proliferation, differentiation, and epithelial–mesenchymal transition. The aim of this study was to elucidate the mechanisms behind the anticancer activity of BBR by comparing the effects of purified BBR with those of the extract of Tinospora cordifolia, a medicinal plant that produces this metabolite. The expression levels of a panel of 44 selected genes in human colon adenocarcinoma (HCA-7) cell line were quantified by real-time polymerase chain reaction (PCR). BBR treatment resulted in a time- and dose-dependent down regulation of 33 genes differently involved in cell cycle, differentiation, and epithelial–mesenchymal transition. The trend was confirmed across the two types of treatment, the two time points, and the different absolute dosage of BBR. These findings suggest that the presence of BBR in T. cordifolia extract significantly contributes to its antiproliferative activity.
Keywords: berberine, colorectal cancer, gene expression, liquid chromatography–mass spectrometry, Tinospora cordifolia
Introduction
The anticancer activity of natural origin compounds has been recently focused by several studies.1 In this context, plants used in the traditional medicine systems can be considered a valuable source of active compounds and important supplements for chemotherapy/treatment.2
Tinospora cordifolia (Willd.) Miers is a widely used plant in Ayurvedic medicine, and it has been found endowed with several biological activities, including anticancer.3
Among the secondary metabolites produced by this plant, isoquinoline alkaloids such as berberine (BBR) are claimed to be responsible for most of the beneficial effects attributed to the plant source. For instance, BBR has been used to treat different types of diarrhea,4,5 and more recently, it has been found effective in the treatment of gastroenteritis, diabetes, hyperlipidemia, cardiovascular diseases, inflammatory conditions, hyperglycemia, and obesity.6–9 Moreover, this compound is endowed with anticancer activity on different cell lines.10
In particular, regarding colon cancer cells, previous studies have shown the ability of BBR to induce cells death through different mechanisms, such as: activation of JNK/p38 apoptotic pathway, suppression of the Wnt/β-catenin signaling pathway and inhibition of growth/proliferation processes for example by inducing G1/S and G2/M cell cycle phase arrest.11,12
Nevertheless, only few studies have been focused on the gene expression effects of BBR in colon cancer cells. Specifically, Hu et al.13 reported a strong dose-dependent antiproliferative BBR activity on human colon cancer cells (HT-29), together with a gradual increasing of apoptotic cells proportion associated with a reduction of the expression of the survivin gene. Moreover, BBR has been reported to increase nonsteroidal anti-inflammatory drug-activated gene-1 (NAG-1) expression in HCT-116 and CaCo-2 cells. This gene has been investigated for its proapoptotic and anti-tumorigenic activity, since it is highly expressed in colon but significantly reduced in most human tumor biopsies.14
With the aim to explore the potential chemopreventive activity of T. cordifolia and BBR, we tested their effect on the expression of a panel of genes selected because of their involvement in self-renewal, tumor progression, malignant transformation, cell mobility, and invasiveness.
This evaluation could lead to the identification of genes and cellular pathways involved in colon cancer facilitating the identification of possible therapeutic targets.
Materials and methods
Plant material and preparation of extracts
Dried and powdered T. cordifolia Willd. stem was supplied by Maharishi Ayurveda Product, Italy (Verona, Italy). The plant material was collected from Ram Bagh (Rajasthan, India) and authenticated by Dr M.R. Uniyal (Maharishi Ayurveda Product Ltd, Noida, India).
Extraction was performed by treating 300 g of plant with 2 L of MeOH/H2O (1:1) and sonication for 1 h. The extract was filtered on Büchner funnel and the solvents were evaporated under vacuum at 40°C. The procedure was repeated for three times, obtaining 28.4 g of crude extract.
BBR analysis
T. cordifolia extract was preliminarily analyzed by ultraviolet (UV) spectrophotometry (Jasco, Tokyo, Japan) and ultraviolet high-powered liquid chromatography (HPLC-UV; Jasco, Tokyo, Japan). BBR UV spectrum shows the most intense absorbance at the wavelengths of 228, 266, and 342 nm, among which the work wavelength chosen for HPLC-UV analysis was 266 nm.
The screening data obtained by UV and HPLC-UV were subsequently confirmed by MS and LC-MS (Waters, Milford, USA). The triple quadrupole mass analyzer was operating in positive mode electrospray ionization (ESI+), and acquisitions were performed in multiple reaction monitoring (MRM).
Both HPLC-UV and liquid chromatography–mass spectrometry (LC-MS) methods were original, developed, and validated ad hoc for the application to T. cordifolia extract.
T. cordifolia extract was subjected to standardization, using BBR pure powder as a reference standard (Sigma-Aldrich, Inc, St. Louis, MO, USA).
The title of BBR in the extract of T. cordifolia was 120 mg/g dry extract.
Cell culture
Human colon adenocarcinoma (HCA-7) cells line, kindly provided by Dr Mariella Chiricolo (University of Bologna, Italy) were initially grown in standard Dulbecco’s Modified Eagle’s Medium (DMEM) (Sigma-Aldrich, Inc) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 0.6 mg/mL glutamine, and 200 U/mL penicillin/streptomycin (Sigma-Aldrich, Inc).
Cell cultures were replicated for subsequent experiments and maintained in water saturated atmosphere at 37°C and 5% CO2.
Cell viability test
A stock solution of T. cordifolia was prepared dissolving 0.0167 g of dry extract in 1 mL of dimethyl sulfoxide (DMSO; Sigma-Aldrich, Inc). The pure BBR purchased from Sigma-Aldrich was re-suspended in a stock solution 5.4 mM. In both solutions, the BBR is considered to have the same concentration. Both solutions were prepared freshly.
Further dilutions were made with the culture medium to the desired concentrations just before use.
HCA-7 cells were seeded into 96-well plates at a density of 104 cells per well, in 100 µL of cell culture medium, and incubated for 24 h to allow cell adherence.
Serial dilutions of T. cordifolia solution (30.9, 92.7, 154.5, and 309.0 µg/mL) and BBR solution (10, 30, 50, and 100 µM) were added (three wells for each concentration). The vehicle DMSO and cell culture medium alone were used as positive and negative control, respectively. After 48 h of incubation, cell viability was measured using PrestoBlue™ Reagent Protocol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Briefly, the PrestoBlue solution (10×) was added into each well containing 100 µL of treatment solution. Plates were then placed back into the incubator for 1 h, after which absorbance was measured at wavelengths of 570 nm excitation and 620 nm emission by an automated microplate reader (Sunrise™, Tecan Trading AG, Switzerland). The percentage of viable cells was determined by comparing the average absorbance in drug-treated wells with average absorbance in control wells exposed to vehicle alone. The results are presented as the mean ± standard deviation of three measures.
Cells treatment
Once determined, the most appropriate concentration for the treatments, HCA-7 cells were seeded at a density of 105 cells/mL into 9 cm2 (3 mL) wells. Cells were washed two times with phosphate buffered serum (PBS) and incubated for 18 h at 37°C with serum-free DMEM.
After serum starvation, cells were treated with the following solutions: (a) 30.9 and 92.7 µg/mL of T. cordifolia methanol extract; (b) 10 and 30 µM of BBR solution, and (c) DMSO solution at the same concentration used to dissolve the extract. This treatment was the negative control.
All treatments were performed for three biological replicates in DMEM supplemented with 2% FBS, antibiotics, and amino acids.
After the end of the exposure time (24 and 48 h), cells were trypsinized and processed for RNA extraction.
RNA isolation, reverse transcription, and quantitative RT-PCR
Total RNA was isolated from cells using GenElute mammalian total RNA purification mini prep kit (Sigma-Aldrich, Inc) according to manufacturer’s instructions. Total RNA concentration and quality were measured using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). Complementary cDNA was obtained using PrimeScript RT Master Mix (Takara Bio Inc, Shiga, Japan) starting from 500 ng of total RNA. cDNA was amplified by real-time quantitative polymerase chain reaction (PCR) using the Power SYBR® Green Master Mix™ (Life Technologies, Foster City, CA, USA) and specific forward and reverse pre-designed assays (Sigma-Aldrich, Inc). The selected genes grouped by functional pathway are listed in Table 1.
Table 1.
Selected genes used in real-time PCR grouped by functional pathway.
| Pathway | Gene |
|---|---|
| Cell cycle | ANAPC2 (anaphase-promoting complex subunit 2) |
| CCNA2 (cyclin A2) | |
| CCND1 (cyclin D1) | |
| CCNE1 (cyclin E1) | |
| CCNT1 (cyclin T1) | |
| CDC6 (cell division cycle 6) | |
| CDK4 (cyclin-dependent kinase 4) | |
| MCM2 (minichromosome maintenance complex component 2) | |
| WEE1 (WEE1 G2 checkpoint kinase) | |
| Cell regulation | ABCB5 (ATP binding cassette subfamily B member 5) |
| ALDH1A1 (aldehyde dehydrogenase 1 family member A1) | |
| TP63 (tumor protein p63) | |
| MAPK1 (mitogen-activated protein kinase 1) | |
| Differentiation | AKT1 (v-AKT murine thymoma viral oncogene homolog 1) |
| BMP2 (bone morphogenetic protein 2) | |
| BMP7 (bone morphogenetic protein 7) | |
| EGFR (epidermal growth factor receptor) | |
| FZD7 (frizzled class receptor 7) | |
| NOTCH1 (notch 1) | |
| PTP4A1 (protein tyrosine phosphatase type IVA, member 1) | |
| SMAD2 (SMAD family member 2) | |
| TGFB1 (transforming growth factor beta 1) | |
| TGFB2 (transforming growth factor beta 2) | |
| TIMP1 (TIMP metallopeptidase inhibitor 1) | |
| Epithelial–mesenchymal transition | AHNAK (AHNAK nucleoprotein) |
| BMP1 (bone morphogenetic protein 1) | |
| CALD1 (caldesmon 1) | |
| CaMKIIG (calcium/calmodulin dependent protein kinase II gamma) | |
| CDH1 (cadherin 1) | |
| CDH2 (cadherin 2) | |
| COL1A2 (collagen type I alpha 2) | |
| CTNNB1 (catenin beta 1) | |
| DSP (desmoplakin) | |
| ITGA5 (integrin subunit alpha 5) | |
| ROCK1 (Rho associated coiled-coil containing protein kinase 1) | |
| ROCK2 (Rho associated coiled-coil containing protein kinase 2) | |
| SNAI2 (snail family zinc finger 2) | |
| TCF4 (transcription factor 4) | |
| VCAN (versican) | |
| VIM (vimentin) | |
| KRT19 (keratin 19) | |
| OCLN (occluding) | |
| TSPAN13 (tetraspanin 13) | |
| TSPAN8 (tetraspanin 8) |
PCR reactions were performed in 20 µL of final volume using the ABI PRISM 7500 (Applied Biosystems, Foster City, CA, USA). Each reaction contained 10 µL of 2× Power SYBR Green Master Mix, 400 nM concentration of each primer and 300 nM of cDNA.
After an initial denaturing step at 95°C for 10 min, the amplification proceeded with 40 cycles of a two-step profile of 15 s at 95°C and 60 s at 60°C. As final step, a melt curve dissociation analysis was performed. All experiments included non-template controls in order to check for the presence of contamination.
Statistical analysis
A preliminary test was performed to select the reference gene among three housekeeping genes. RPL13 expression appeared most consistent with the amount of RNA input. Gene expression quantification was conducted with the delta/delta Ct calculation method,15 and RPL13 was used as reference gene to normalize the gene expression levels.
After normalization, the delta Ct of treated cells, and control were compared by a paired sample t-test. Mean expression levels of treated cells were calculated as fold changes relative to the expression of untreated cells with the delta/delta calculation method.
Results
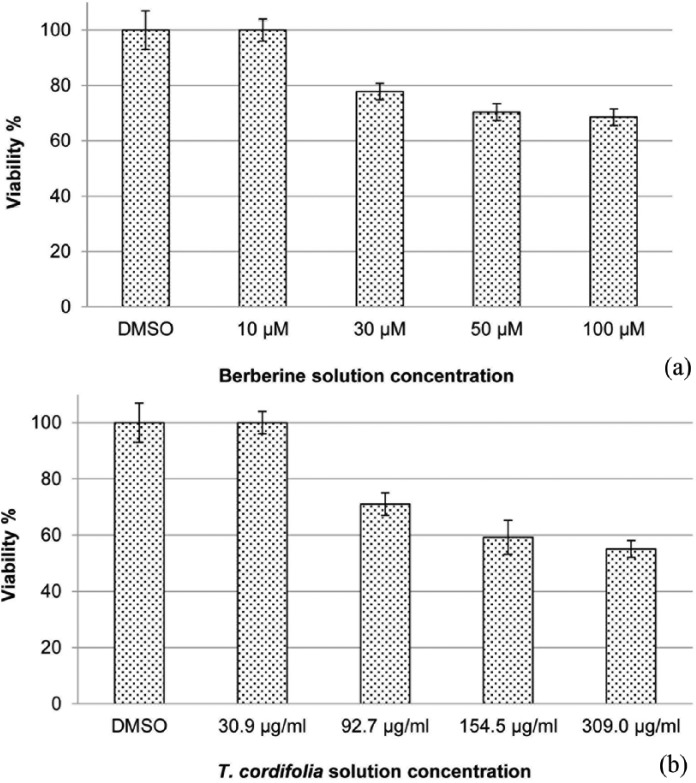
The assay based on vital dye PrestoBlue showed that both T. cordifolia extract and BBR had no effect on HCA-7 cell viability at 30.9 µg/mL and 10 µM, respectively (Figure 1(a) and (b)). An evident decrease of viability was observed at concentration starting from 92.7 µg/mL and 30 µM, respectively. The number of viable cells at these concentrations was still adequate to investigate the biologic effects of treatments, such as modulation of gene expression and cellular pathways.
Figure 1.
(a) Cell viability of berberine solution in HCA-7 cell line assessed by PrestoBlue reagent protocol. Cells were treated with different concentrations of drug (10, 30, 50, and 100 µM) for 48 h and (b) cell viability of T. cordifolia solution in HCA-7 cell line assessed by PrestoBlue Reagent Protocol. Cells were treated with various concentrations of dry extract solution (30.9, 92.7, 154.5, 309 µg/ml) for 48 h.
Data are means ± SD.
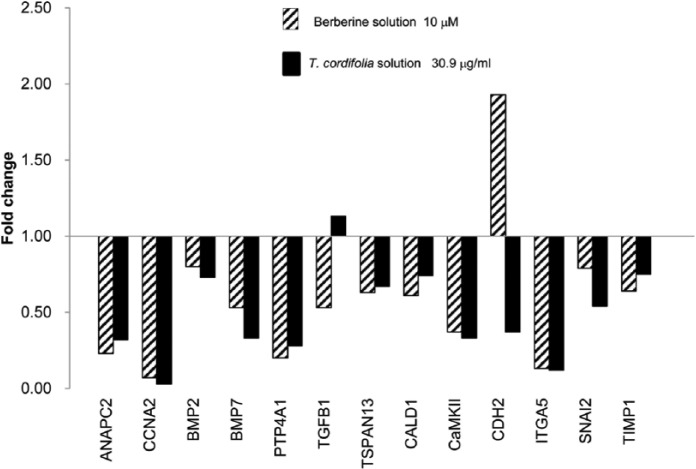
Figure 2.
Gene expression profile of HCA-7-treated cells (30.9 µg/mL of T. cordifolia and 10 µM of berberine) vs control at 48 h.
Gene expression of 44 genes belonging to different pathways involved in tumor progression and cell differentiation was investigated in HCA-7 cell line treated with T. cordifolia and BBR solutions at two different concentrations and time points. Tables 2 and 3 report the effects of 24 and 48 h treatment on gene expression. Bold fonts indicate significant variation of gene expression level: fold change ⩾2 and P value ⩽ 0.05 for up-regulated genes, and fold change ⩽0.5 and P value ⩽ 0.05 for significantly down-regulated genes.
Table 2.
Significantly de-regulated genes after 24 h of treatment in HCA-7 cell line.
| Gene | Berberine 10 µM | Berberine 30 µM | T. cordifolia solution | T. cordifolia solution | Pathway | ||||
|---|---|---|---|---|---|---|---|---|---|
| 30.9 µg/mL | 92.7 µg/mL | ||||||||
| fold change | P value | fold change | P value | fold change | P value | fold change | P value | ||
| ANAPC2 | 0.37 | 0.007 | 0.23 | 0.005 | 1.41 | 0.214 | 1.4 | 0.231 | Cell cycle |
| CCNA2 | 0.63 | 0.004 | 0.46 | 0.007 | 0.97 | 0.774 | 1.65 | 0.073 | Cell cycle |
| CCNE1 | 0.6 | 0.030 | 0.75 | 0.083 | 1.42 | 0.090 | 2.03 | 0.001 | Cell cycle |
| CDC6 | 0.9 | 0.532 | 0.78 | 0.409 | 0.37 | 0.004 | 0.6 | 0.056 | Cell cycle |
| CDK4 | 0.94 | 0.727 | 0.48 | 0.008 | 2.02 | 0.010 | 1.36 | 0.127 | Cell cycle |
| LGR5 | 1.14 | 0.548 | 2.71 | 0.005 | 1.33 | 0.218 | 3.17 | 0.005 | Cell cycle |
| MCM2 | 0.06 | <0.001 | 0.06 | <0.001 | 0.18 | <0.001 | 0.15 | 0.001 | Cell cycle |
| WEE1 | 1.06 | 0.830 | 1.77 | 0.007 | 2.57 | 0.011 | 3.03 | 0.001 | Cell cycle |
| ABCB5 | 0.19 | 0.013 | 4.86 | 0.010 | 0.25 | 0.001 | 4.15 | 0.015 | Cell regulation |
| AKT1 | 0.83 | 0.326 | 0.34 | 0.003 | 1.58 | 0.061 | 1.62 | 0.005 | Differentiation |
| BMP2 | 0.98 | 0.949 | 0.82 | 0.536 | 2.56 | 0.022 | 2.37 | 0.014 | Differentiation |
| BMP7 | 0.6 | 0.025 | 0.69 | 0.172 | 1.14 | 0.330 | 3.12 | 0.002 | Differentiation |
| FZD7 | 0.32 | <0.001 | 0.49 | 0.005 | 1.3 | 0.130 | 1.09 | 0.561 | Differentiation |
| PTP4A1 | 0.25 | 0.001 | 0.34 | 0.003 | 1.21 | 0.375 | 1.47 | 0.091 | Differentiation |
| SMAD2 | 13.8 | <0.001 | 12.32 | <0.001 | 1.3 | 0.278 | 1.99 | 0.001 | Differentiation |
| TGFB1 | 0.64 | 0.065 | 0.41 | 0.002 | 1.43 | 0.179 | 1.1 | 0.424 | Differentiation |
| TSPAN13 | 0.44 | 0.016 | 0.56 | 0.014 | 0.76 | 0.195 | 0.92 | 0.568 | Differentiation |
| TSPAN8 | 0.94 | 0.799 | 2.22 | 0.009 | 2.21 | 0.001 | 3.06 | 0.006 | Differentiation |
| CALD1 | 0.87 | 0.456 | 0.76 | 0.075 | 1.58 | 0.061 | 2.02 | 0.002 | EMT |
| CaMKII | 1.3 | 0.011 | 1.26 | 0.079 | 2.83 | <0.001 | 3.86 | <0.001 | EMT |
| CDH2 | 0.08 | <0.001 | 0.36 | 0.003 | 0.11 | 0.006 | 0.66 | 0.078 | EMT |
| DSP | 0.37 | 0.015 | 0.8 | 0.569 | 1.56 | 0.142 | 3.68 | 0.009 | EMT |
| ITGA5 | 0.22 | <0.001 | 0.2 | <0.001 | 0.52 | 0.007 | 0.48 | 0.006 | EMT |
| KRT19 | 0.18 | <0.001 | 0.18 | <0.001 | 0.73 | 0.055 | 0.86 | 0.482 | EMT |
| OCLN | 0.89 | 0.686 | 1.34 | 0.191 | 2.15 | 0.045 | 1.86 | 0.020 | EMT |
| ROCK1 | 0.3 | 0.005 | 0.53 | <0.001 | 1.06 | 0.611 | 1.96 | 0.002 | EMT |
| SNAI2 | 0.23 | 0.005 | 0.73 | 0.355 | 0.58 | 0.061 | 1.86 | 0.113 | EMT |
| VCAN | 1.14 | 0.631 | 2.99 | <0.001 | 1.25 | 0.425 | 2.19 | 0.046 | EMT |
| VIM | 0.62 | 0.034 | 2.17 | 0.007 | 1.39 | 0.098 | 2.15 | 0.007 | EMT |
EMT: epithelial–mesenchymal transition.
In bold significant gene expression level.
Significantly up-regulated gene: fold change ⩾2 and P value ⩽ 0.05.
Significantly down-regulated gene: fold change ⩽0.5 and P value ⩽ 0.05.
Table 3.
Significantly de-regulated genes after 48 h of treatment in HCA-7 cell line.
| Gene | Berberine 10 µM | Berberine 30 µM | T. cordifolia solution | T. cordifolia solution | Pathway | ||||
|---|---|---|---|---|---|---|---|---|---|
| 30.9 µg/mL | 92.7 µg/mL | ||||||||
| fold change | P value | fold change | P value | fold change | P value | fold change | P value | ||
| ANAPC2 | 0.23 | 0.001 | 0.3 | 0.021 | 0.32 | 0.010 | 0.32 | 0.024 | Cell cycle |
| CCNA2 | 0.07 | <0.001 | 0.05 | <0.001 | 0.03 | <0.001 | 0.04 | <0.001 | Cell cycle |
| BMP2 | 0.8 | 0.290 | 0.3 | 0.014 | 0.73 | 0.358 | 0.3 | 0.010 | Differentiation |
| BMP7 | 0.53 | 0.002 | 0.55 | 0.007 | 0.33 | <0.001 | 0.22 | <0.001 | Differentiation |
| PTP4A1 | 0.2 | <0.001 | 0.27 | 0.020 | 0.28 | 0.002 | 0.32 | 0.001 | Differentiation |
| TGFB1 | 0.53 | 0.006 | 0.3 | <0.001 | 1.13 | 0.445 | 0.71 | 0.125 | Differentiation |
| TSPAN13 | 0.63 | 0.026 | 0.37 | 0.001 | 0.67 | 0.050 | 0.46 | 0.003 | Differentiation |
| CALD1 | 0.61 | 0.005 | 0.27 | 0.005 | 0.74 | 0.038 | 0.51 | 0.007 | EMT |
| CaMKII | 0.37 | <0.001 | 0.26 | 0.002 | 0.33 | <0.001 | 0.24 | <0.001 | EMT |
| CDH2 | 1.93 | 0.028 | 1.24 | 0.283 | 0.37 | 0.005 | 0.52 | 0.034 | EMT |
| ITGA5 | 0.13 | <0.001 | 0.18 | 0.001 | 0.12 | <0.001 | 0.14 | <0.001 | EMT |
| SNAI2 | 0.79 | 0.177 | 0.44 | 0.008 | 0.54 | 0.014 | 0.22 | <0.001 | EMT |
| TIMP1 | 0.64 | 0.156 | 0.22 | 0.001 | 0.75 | 0.187 | 0.38 | 0.005 | EMT |
EMT: epithelial–mesenchymal transition.
In bold significant gene expression level.
Significantly up-regulated gene: fold change ⩾2 and P value ⩽ 0.05.
Significantly down-regulated gene: fold change ⩽0.5 and P value ⩽ 0.05.
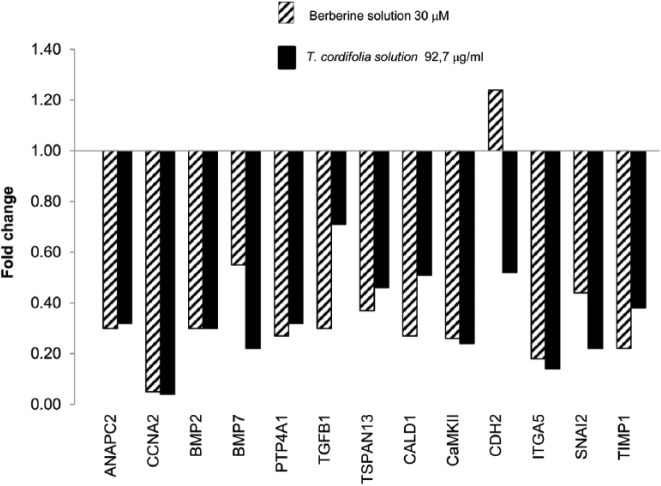
The most relevant effects were observed after 48 h (Figures 2 and 3).
Figure 3.
Gene expression profile of HCA-7-treated cells (92.7 µg/mL of T. cordifolia and 30 µM of berberine) vs control at 48 h.
A total of 13 genes of the 44 studied were significantly over- and down-regulated and belong essentially to three pathways: cell cycle and regulation, differentiation, epithelial–mesenchymal transition (EMT).
Interestingly, both the plant extract and the BBR showed the same effects on the expression profiling of treated cells. In particular, the genes that maintained the same expression trend following all the treatments were ANAPC2 and CCNA2, involved in cycle regulation; BMP7 and PTP4A1 belonging to cell differentiation pathway; CaMKII and ITGA5 involved in EMT. This set of genes was significantly down-regulated in treated cells with respect to the control.
Discussion
Plant-based drug discovery has played an important role in the development of anticancer drugs currently in clinical use.1
Several medicinal plants used in different traditional medicine systems showed anti-tumorigenic activity.16–18 Specifically, plants containing BBR might be promising suppressing agents.
In this investigation, we observed the effect of T. cordifolia extract on the HCA-7 colon cancer cell line as compared with purified BBR. The content of BBR in the extract of T. cordifolia was evaluated by LC-MS analysis in order to treat the cells with comparable concentration of BBR. Two different concentrations of BBR were chosen for the assay based on the result of a viability test.
Considering the quality of the mRNA extracted from treated cells the highest concentration of T. cordifolia (92.7 µg/mL) and of BBR used (30 µM) was supposed not lethal but sufficient to induce a significant response. In fact, at the second time point (48 h from the treatment), a similar trend of down regulation of selected genes was confirmed both for the treatment with T. cordifolia extract and for the one with pure BBR for the two different concentrations (30.9 µg/mL, 92.7 µg/mL, and 10 µM, 30 µM, respectively).
These observations let hypothesize the involvement of BBR in the activity showed by the T. cordifolia.
The transcriptional level of 44 genes known to be involved in specific pathways such as cell cycle, differentiation, or EMT was monitored.
Among the genes modulated by the treatments, a couple of genes of cell cycle pathway resulted significantly down-regulated: ANAPC2 and CCNA2. The first one codes for a subunit of the anaphase-promoting complex (APC), involved in the progression of the cell cycle. Recently, the chemopreventive and chemotherapeutic potential of curcumin, contained in the Asian spice Curcuma longa, has been related to APC inhibition and apoptosis induction.19 The gene CCNA2 codes for cyclin A2, that is able to bind to CDK2 and plays critical roles in cells proliferation; the over expression of CCNA2 has been linked to colorectal carcinogenesis and tumor invasion.20
Alteration of epithelial cells homeostasis and acquisition of migratory mesenchymal phenotype are crucial steps in progression of epithelial cancers. In this study, it was shown that the expression level of several genes involved in differentiation and EMT was modulated by T. cordifolia and BBR treatments.
Bone morphogenetic proteins (BMPs) are responsible for tumorigenesis and metastatic progression of many types of cancer, inducing EMT and activating the tumorigenic potential of cancer stem cells in human colorectal cancer.21 The up-regulation of BMP7 in colorectal cancer determines a poor prognosis caused by the increased aggressiveness of the tumor and onset of liver metastases.22 We found significantly down-regulated both BMP2 and BMP7 in our system.
The expression of PTP4A1 was found four times lower in treated cells compared to control. Its product is localized preferentially in the cytoplasm of colon adenocarcinoma cells, endothelial cells, and smooth muscle cells in proximity of vessels surrounding lymph node metastases.23,24
Another down-regulated gene in treated cells was TSPAN13 that encodes a transmembrane protein involved in cell motility. Up-regulation of this gene has been reported at the progression stage of many cancers such as ovarian adenocarcinoma, breast cancer, bladder carcinoma, and acute lymphoblastic leukemia.25
Two Ca2+/calmodulin-dependent proteins, CALD1 and CAMKIIG, were down-regulated in HCA-7 cells after exposure to T. cordifolia and BBR. CALD1 is involved in migration, invasion, and proliferation of many tumors, including colorectal cancer. Overexpression of CALD1 in peritoneal cancer cells has been associated to reduced tumor prognosis,19 while inhibition of CAMKIIG through synthetic compounds suppressed the growth of cancer cells, inducing apoptosis.26
TGFB1 increases the invasive properties of colon cancer cells increasing the expression of CDH2 coding for the N-cadherin, located at the tips of myofibroblast filopodia.27 In our investigation, CDH2 was significantly down-regulated at the higher concentration of both treatments.
The integrin α5, ITGA5, was strongly down-regulated in treated cells compared to control with an expression decreased by 10 times. The altered expression of ITGA5 during tumor progression has been largely demonstrated in colon cancer cells line with highly invasive potential.28
Two genes involved in EMT and down-regulated only at the higher concentration of T. cordifolia and BBR were SNAI2 and TIMP1. SNAI2 mediates resistance to 5-fluorouracil chemotherapy in colorectal cancer, representing a new therapeutic target to improve current treatment strategies.29 Serum and tissue levels of TIMP1 are increased in patients with colon cancer. TIMP1 has anti-apoptotic role through the pathway of Bcl-2, stimulating cell growth and promoting the migration and metastatic invasion.30
Conclusion
Our study demonstrated that extract of T. cordifolia is able to inhibit the expression of several genes involved in colon cancer development and progression. The effects of T. cordifolia treatment were well resembled by pure BBR; this evidence suggested that the main effects of T. cordifolia were mediated by BBR. The ability to strongly reduce expression of genes involved in proliferation, differentiation, cell motility, and EMT suggests further research efforts to explore the use of these substances as chemotherapeutic agent in colorectal cancer.
Acknowledgments
The authors are grateful to Dr Paolo Scartezzini (Maharishi Ayurveda Product, Italy) for providing T. cordifolia samples and for technical assistance during the early steps of this project.
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iDs: Luca Scapoli  https://orcid.org/0000-0003-4006-9910
https://orcid.org/0000-0003-4006-9910
Laura Mercolini  https://orcid.org/0000-0002-0644-9461
https://orcid.org/0000-0002-0644-9461
References
- 1. Khazir J, Mir BA, Pilcher L, et al. (2014) Role of plants in anticancer drug discovery. Phytochemistry Letters 7: 9. [Google Scholar]
- 2. Balachandran P, Govindarajan R. (2005) Cancer—An ayurvedic perspective. Pharmacological Research 51(1): 19–30. [DOI] [PubMed] [Google Scholar]
- 3. Bala M, Pratap K, Verma PK, et al. (2015) Validation of ethnomedicinal potential of Tinospora cordifolia for anticancer and immunomodulatory activities and quantification of bioactive molecules by HPTLC. Journal of Ethnopharmacology 175: 131–137. [DOI] [PubMed] [Google Scholar]
- 4. Lahiri SC, Dutta NK. (1967) Berberine and chloramphenicol in the treatment of cholera and severe diarrhoea. Journal of the Indian Medical Association 48(1): 1–11. [PubMed] [Google Scholar]
- 5. Sun D, Abraham SN, Beachey EH. (1988) Influence of berberine sulfate on synthesis and expression of Pap fimbrial adhesin in uropathogenic Escherichia coli. Antimicrobial Agents and Chemotherapy 32(8): 1274–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gibbs PJ, Seddon KR. (2000) Berberine. Altern Med Rev 5: 175–177. [PubMed] [Google Scholar]
- 7. Chen C, Yu Z, Li Y, et al. (2014) Effects of berberine in the gastrointestinal tract—A review of actions and therapeutic implications. The American Journal of Chinese Medicine 42(5): 1053–1070. [DOI] [PubMed] [Google Scholar]
- 8. Liu C, Wang Z, Song Y, et al. (2015) Effects of berberine on amelioration of hyperglycemia and oxidative stress in high glucose and high fat diet-induced diabetic hamsters in vivo. Biomed Research International 2015: 313808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang WL, Zhu L, Jiang JG. (2014) Active ingredients from natural botanicals in the treatment of obesity. Obesity Reviews 15(12): 957–967. [DOI] [PubMed] [Google Scholar]
- 10. Tillhon M, Guaman Ortiz LM, Lombardi P, et al. (2012) Berberine: New perspectives for old remedies. Biochemical Pharmacology 84(10): 1260–1267. [DOI] [PubMed] [Google Scholar]
- 11. Albring KF, Weidemuller J, Mittag S, et al. (2013) Berberine acts as a natural inhibitor of Wnt/beta–catenin signaling: Identification of more active 13-arylalkyl derivatives. Biofactors 39(6): 652–662. [DOI] [PubMed] [Google Scholar]
- 12. Chidambara Murthy KN, Jayaprakasha GK, Patil BS. (2012) The natural alkaloid berberine targets multiple pathways to induce cell death in cultured human colon cancer cells. European Journal of Pharmacology 688(1–3): 14–21. [DOI] [PubMed] [Google Scholar]
- 13. Hu W, Yu L, Wang MH. (2011) Antioxidant and antiproliferative properties of water extract from Mahonia bealei (Fort.) Carr. leaves. Food and Chemical Toxicology 49(4): 799–806. [DOI] [PubMed] [Google Scholar]
- 14. Eling TE, Baek SJ, Shim M, et al. (2006) NSAID activated gene (NAG-1): A modulator of tumorigenesis. Journal of Biochemistry and Molecular Biology 39(6): 649–655. [DOI] [PubMed] [Google Scholar]
- 15. Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25(4): 402–408. [DOI] [PubMed] [Google Scholar]
- 16. Mondal S, Bandyopadhyay S, Ghosh MK, et al. (2012) Natural products: Promising resources for cancer drug discovery. Anticancer Agents in Medicinal Chemistry 12(1): 49–75. [DOI] [PubMed] [Google Scholar]
- 17. Fadeyi SA, Fadeyi OO, Adejumo AA, et al. (2013) In vitro anticancer screening of 24 locally used Nigerian medicinal plants. BMC Complementary and Alternative Medicine 13: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang CY, Bai XY, Wang CH. (2014) Traditional Chinese medicine: A treasured natural resource of anticancer drug research and development. The American Journal of Chinese Medicine 42(3): 543–559. [DOI] [PubMed] [Google Scholar]
- 19. Lee SJ, Langhans SA. (2012) Anaphase-promoting complex/cyclosome protein Cdc27 is a target for curcumin-induced cell cycle arrest and apoptosis. BMC Cancer 12: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li JQ, Miki H, Wu F, et al. (2002) Cyclin A correlates with carcinogenesis and metastasis, and p27(kip1) correlates with lymphatic invasion, in colorectal neoplasms. Human Pathology 33(10): 1006–1015. [DOI] [PubMed] [Google Scholar]
- 21. Kim BR, Oh SC, Lee DH, et al. (2015) BMP-2 induces motility and invasiveness by promoting colon cancer stemness through STAT3 activation. Tumour Biology 36(12): 9475–9486. [DOI] [PubMed] [Google Scholar]
- 22. Motoyama K, Tanaka F, Kosaka Y, et al. (2008) Clinical significance of BMP7 in human colorectal cancer. Annals of Surgical Oncology 15(5): 1530–1537. [DOI] [PubMed] [Google Scholar]
- 23. Wang Y, Li ZF, He J, et al. (2007) Expression of the human phosphatases of regenerating liver (PRLs) in colonic adenocarcinoma and its correlation with lymph node metastasis. International Journal of Colorectal Disease 22(10): 1179–1184. [DOI] [PubMed] [Google Scholar]
- 24. Fiordalisi JJ, Keller PJ, Cox AD. (2006) PRL tyrosine phosphatases regulate rho family GTPases to promote invasion and motility. Cancer Research 66(6): 3153–3161. [DOI] [PubMed] [Google Scholar]
- 25. Arencibia JM, Martin S, Perez-Rodriguez FJ, et al. (2009) Gene expression profiling reveals overexpression of TSPAN13 in prostate cancer. International Journal of Oncology 34(2): 457–463. [PubMed] [Google Scholar]
- 26. Wang C, Li N, Liu X, et al. (2008) A novel endogenous human CaMKII inhibitory protein suppresses tumor growth by inducing cell cycle arrest via p27 stabilization. The Journal of Biological Chemistry 283(17): 11565–11574. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27. De Wever O, Westbroek W, Verloes A, et al. (2004) Critical role of N-cadherin in myofibroblast invasion and migration in vitro stimulated by colon-cancer-cell-derived TGF-beta or wounding. J Cell Sci 117: 4691–4703. [DOI] [PubMed] [Google Scholar]
- 28. Janouskova H, Ray AM, Noulet F, et al. (2013) Activation of p53 pathway by Nutlin-3a inhibits the expression of the therapeutic target alpha5 integrin in colon cancer cells. Cancer Letters 336(2): 307–318. [DOI] [PubMed] [Google Scholar]
- 29. Findlay VJ, Wang C, Nogueira LM, et al. (2014) SNAI2 modulates colorectal cancer 5-fluorouracil sensitivity through miR145 repression. Molecular Cancer Therapeutics 13(11): 2713–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Niewiarowska K, Pryczynicz A, Dymicka-Piekarska V, et al. (2014) Diagnostic significance of TIMP-1 level in serum and its immunohistochemical expression in colorectal cancer patients. Polish Journal of Pathology 65(4): 296–304. [DOI] [PubMed] [Google Scholar]