Main Text
Rogers et al.1 provide a detailed study of a novel class of clade F adeno-associated virus (AAV) capsids (AAV hematopoietic stem cells [AAVHSCs])2 to facilitate homologous recombination in the presence or absence of site-specific DNA cleavage in human CD34+ hematopoietic stem and progenitor cells (HSPCs). The recent publication that clade F AAVs can facilitate high-efficiency homologous recombination in the absence of a targeted DNA break3 was surprising due to the high sequence homology of AAVHSC variants to the relatively inefficient AAV9 and the long-standing knowledge in the field that AAV-based viral vectors do not undergo high frequency-targeted integration (1% or less).4, 5, 6 Rogers et al.1 provide a direct comparison with AAV6, the serotype most commonly utilized for this method and cell type and demonstrate that long-lived (>14 days) expression of a GFP-reporter encoding donor template is only observed after specific induction of a DNA break using zinc-finger nucleases (ZFNs). They additionally report that the highest efficiency of editing is observed using AAV6. Here we provide additional independently generated confirmation of the findings that AAV6 is a superior vector for homologous recombination-based genome editing and discuss potential technical differences in the presented studies that may influence the data interpretation and conflicting interpretation of these studies. As gene therapy and gene editing become more widely applied therapeutics, transparency and reproducibility is critical for the successful development and implementation as these therapies transition into the clinic.
Genome editing-based gene therapies are being developed as a class of molecular therapeutics that have the potential to cure previously untreatable diseases through permanent correction of a disease-causing genetic mutation.7 The development of ZFNs,8, 9 transcription activator-like effector nuclease (TALENs),10, 11, 12 and Cas913, 14-based gene editing methodologies have allowed specific manipulation of a DNA sequence of interest by targeted insertion, deletion, or single base editing with high efficiency. Recombinant AAV is an ideal delivery vehicle to provide DNA templates for gene correction through homology-directed repair after induction of a specific DNA break because it is a non-integrating virus and thus has the key property for a “hit and run” process in dividing cells. The combination of a nuclease-induced break with transduction of AAV6 to deliver the donor has been efficient in a wide variety of primary cell types, including HSPCs and T cells, and genetic loci.15, 16, 17, 18, 19, 20
Smith et al.3 claimed that a novel set of AAV capsid variants, which they called AAVHSC, were able to mediate high frequencies of targeted integration into CD34+ human HSPCs without the need for a double-strand break (DSB). These clade F AAVHSC variants are a highly related subgroup of AAV capsids that are most closely related to AAV9, most of which differ by only one or two single amino acid substitutions in the capsid. Since 1998, when Russell and Hirata4 published their seminal paper showing that AAV could integrate in a targeted fashion at much higher frequencies than naked plasmid DNA, AAV has been used in this manner for research purposes. The frequency without selection, however, has been 1% or lower.21, 22, 23 We demonstrated that AAV6 mediates targeted integration without a break in HSCs, for example, at a frequency of ∼0.5%.24 The frequency of targeted integration without a break by AAV is dose dependent.23, 25
Rogers et al.1 directly compared one variant, AAV9-G505R (AAVHSC13), to AAV9 and AAV6 in transducing and serving as a template for HDR genome editing. They demonstrate that, although AAV6, AAV9, and AAV9-G505R can all transduce HSPCs as shown by GFP expression from an exogenous promoter (phosphoglycerate kinase promoter [PGK]), high efficiency targeted integration at multiple loci (CCR5 and AAVS1) can only occur in the presence of a site-specific ZFN-induced DNA break. This was demonstrated using flow cytometry to demonstrate sustained high-level expression of GFP after 14 days as well as by molecular characterization of genomic integration by digital droplet PCR (ddPCR) primers spanning the targeted integration site. Importantly, GFP expression observed from HSPC transduction by any of the three tested serotypes encapsidating the CCR5-PGK-GFP construct at day 2 was lost by day 14 in the absence of a site-specific break. These data suggest that AAVs, including the novel class of clade F AAVHSCs, are unable to facilitate long-lasting genomic integration in HSPCs without a DNA DSB.
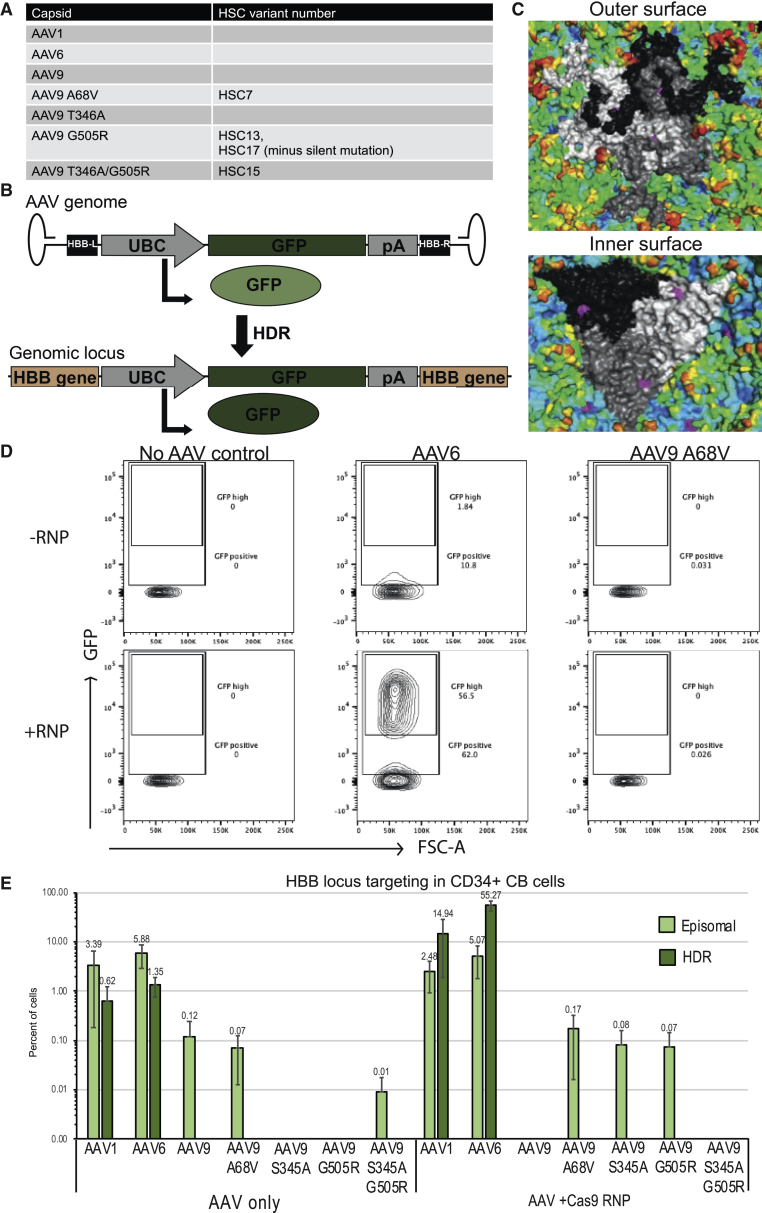
We have independently confirmed that AAV6 is a superior vector for homology-directed genome editing in HSPCs as compared to AAV9-G505R as well as a panel of other AAVHSC viral vectors by using an additional method for generating DNA DSBs (CRISPR/Cas9 nuclease) and an additional therapeutically relevant genomic locus (HBB). Using site-directed mutagenesis, we generated a small panel of AAVHSC variants (Figure 1A) that were previously characterized to have high-level AAV production as single-stranded AAV (ssAAV) and high transduction in HSPCs.2 We characterized the ability of these capsids to facilitate homology-directed repair (HDR) in the presence and absence of a site-specific Cas9/ribonucleoprotein (RNP) complex in comparison to their parental capsid AAV9 as a control as well as the commonly used AAV6 and highly related AAV1. This panel of capsids was used to encapsidate a homology donor construct containing GFP driven from ubiquitin C (UBC) promoter with homology arms compatible with the HBB gene (Figure 1B).26 Episomal expression of this construct demonstrates low intensity fluorescence, which is lost over time as the AAV genome is diluted through cell division, or high intensity fluorescence after site-specific genomic integration following HDR.27 The reported capsid sequence of each of these variants was reported to differ by only one (AAVHSC7, HSC13) or two (AAVHSC7) amino acids relative to AAV9 (Figure 1A). The location of AAV9-A68 cannot be visualized in the AAV9 crystal structure28 due to its location in the VP1 unique region, which is unresolved in AAV structures: the physical locations of G505 (Figure 1C, top) and T346 (Figure 1C, bottom) are highlighted in magenta mapped on the AAV9 crystal structure, with individual VP3 monomers highlighted in white, gray, and black. G505 is a surface-exposed residue located at the interface of two adjacent monomers, while T346 is located on the inner surface of the capsid on individual monomers. The highly distinct locations of these two different point mutations does not suggest an obvious underlying mechanism by which the two amino acid variants may be specifically influencing the homologous recombination (HR) activity of these capsids.
Figure 1.
AAV6 Is Superior to AAVHSCs in Facilitating HDR in CD34+ Cells and Requires a Double-Strand Break
(A) Table of tested AAV capsid variants and corresponding AAVHSC variant name from Smith et al.2 (B) Diagram of donor template packaged into the tested AAV capsids, containing GFP driven from UBC promoter with flanking human beta-globin homology arms and AAV ITRs. Episomal expression from the AAV genome facilitates low-intensity GFP fluorescence (top), while expression after HDR facilitates long-lived high-intensity GFP fluorescence (bottom).27 (C) Molecular modeling of location of AAVHSC variants in relation of parental capsid AAV9. Outer (top) and inner (bottom) surface representation of AAV9 capsid crystal structure: individual viral protein (VP) monomers are highlighted in white, black, or gray and HSC point mutations are highlighted magenta. (D) Gating strategy used for quantification of episomal or HDR-mediated GFP expression in CD34+ HSPCs. Samples from one representative experiment are shown. (E) Quantification of episomal or HDR-mediated GFP expression with standard error measurements from three individual donors, quantified 12–14 days post-transduction. Episomal expression was calculated by subtracting GFP high (HDR) cells from total GFP-positive cells after using AAV6+RNP as gating control for cells that underwent HDR.
Quantification of the total number of GFP-positive or highly GFP expressing cells was used as a surrogate readout for HDR, with no virus and no RNP conditions as the gating controls (Figure 1D). Transgene expression levels and the amount of targeted integration using this donor has been previously characterized with AAV6 and is consistent with our observations.15 While some low intensity GFP expression can be seen after transduction with each vector, an appreciable number of cells demonstrating high intensity GFP expression after HDR is only observed in the AAV6+RNP condition (Figure 1D). These data are consistent with previous reports that AAV6 is more efficient at transducing HSPCs than other AAV serotypes.29, 30 Long-term GFP expression was quantified at 12 to 14 days post-transduction in three independent experiments using cord blood-derived CD34+ HSPCs from three different individual donors (Figure 1D). The percentage of cells with episomal or HDR GFP expression is presented on a log scale to highlight the large differences in efficacy of these vectors. Our data are in agreement with Rogers et al.,1 which highlights AAV6 as the superior vector for delivery of HDR donors and the requirement of a site-specific nuclease for this reaction to occur at high efficiency and maintain long-term expression.
While the results presented here and in Rogers et al.1 are in conflict with the previous reports of high efficiency HSPC gene editing by clade F AAVs, Rogers et al.1 point out that there are potential technical differences that exist between the studies that may have influenced the conflicting results or interpretation of data. Here we discuss the technical differences between the studies in hopes that further clarification of the methodology will reconcile these conflicting results. The sequences between flanking inverted terminal repeats (ITRs) are the same in Rogers et al.1 and Smith et al.:3 the backbone containing the ITR sequences is from different plasmid constructs (pAAV-MCS or pSaiLuc AAV2, respectively) and the potential for sequence differences in the ITR may alter transgene expression. Sequences downstream of the right homology arm may influence episomal GFP expression, as a minimal enhancer-promoter element has been described in this region,31 although it is unclear whether this sequence would be functional in HSPCs. Conversely, other studies suggest that ITR sequences may have a negative impact on transgene expression.32 AAV production protocols differ between the studies, as we and Rogers et al.1 used a helper virus-free system in which helper functions are provided by co-transfection of a helper plasmid,33 whereas vector used by Smith et al.3 produced AAV using replication-competent herpes simplex virus type 1 (HSV1) as a helper.34 Although the described purification method cites either heat inactivation or cesium chloride purification for inactivation of HSV1, there is still the potential for contamination of live helper virus or incomplete heat inactivation, which may boost the signal observed from AAV transduction experiments. Interestingly, inactivated HSV has been shown to cause induction of expression of endogenous retroviruses (ERVs),35 which are observed at the AAVS1 locus (USC Genome Browser), and could therefore lead to stalled replication forks, inducing double-stranded breaks, and subsequent integration being detected at this locus. Of note, the target loci used in Smith et al.3 have been shown to undergo nuclease-free recombination,36 which may occur at higher frequency in the presence of heat inactivated HSV1. Additionally, BRCA2−/− cells have been shown to have aberrant immune activation,37 which may lead to nuclease-free recombination or altered cellular responses to AAV, especially in the context of an HSV1 helper. When considered in aggregate, it is clear that more mechanistic characterization of the recombination induced by this novel class of AAVs will lead to a better understanding of their utility as vectors for gene editing.
An additional important consideration when comparing different capsids is the method of titration and dilution buffer used for titration. Both groups used TaqMan qPCR for AAV genome titration, either using GFP-, AAVS1-, or Luc-specific primer-probes (Smith et al.)3 or ITR-specific primer-probe with AAV2 reference standard material from American Type Culture Collection (ATCC).1 Neither group mentions whether surfactant is used in vector dilutions for qPCR titration, which is an important consideration when comparing different capsids, as the lack of surfactant can increase capsid adherence to plastic and cause under-titration by up to 10-fold for some capsids.38 The reported genome dose used by Rogers et al.1 is 104 vector genomes (vg)/cell, while Smith et al.3 report using up to 4 × 105 vg/cell. We and others have observed a cytotoxic effect from high vg/cell transduction of extremely efficient AAV capsids and, as such, the high dose of AAV may be causing apoptosis in this sensitive cell population. This would be consistent with previous reports that high levels of AAV cause p53-induced apoptosis in other stem cells such as human embryonic stem cells (hESCs)39 and relates back to the method of genome titration for the different capsids. For a more in-depth discussion of AAV-mediated toxicity in stem cells, we refer readers to a review by Brown et al.40
The recent publication of high efficiency genome editing induced by AAVHSC variants in the absence of a DNA break and the conflicting report presented by Rogers et al.1 highlight the importance of a better mechanistic understanding of how HDR occurs in the context of these gene editing and gene delivery technologies. A thorough examination of HDR induced by AAV9-G505R suggests that the previously reported DSB independent mechanism of site-specific recombination facilitated by capsid may need revision or at least further exploration of technical differences leading to such drastically conflicted studies. We have discussed potential technical differences that may influence experimental outcomes presented by these studies with the goal of aiding in the transparency and reproducibility of novel gene therapy methodologies as an effort to consolidate our understanding of how to develop gene editing-based therapeutics.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.09.005.
Supplemental Information
References
- 1.Rogers G.L., Chen H.-Y., Morales H., Cannon P.M. Homologous recombination-based genome editing by clade F AAVs is inefficient in the absence of a targeted DNA break. Mol. Ther. 2019;27:1726–1736. doi: 10.1016/j.ymthe.2019.08.019. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith L.J., Ul-Hasan T., Carvaines S.K., Van Vliet K., Yang E., Wong K.K., Jr., Agbandje-McKenna M., Chatterjee S. Gene transfer properties and structural modeling of human stem cell-derived AAV. Mol. Ther. 2014;22:1625–1634. doi: 10.1038/mt.2014.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith L.J., Wright J., Clark G., Ul-Hasan T., Jin X., Fong A., Chandra M., St Martin T., Rubin H., Knowlton D. Stem cell-derived clade F AAVs mediate high-efficiency homologous recombination-based genome editing. Proc. Natl. Acad. Sci. USA. 2018;115:E7379–E7388. doi: 10.1073/pnas.1802343115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russell D.W., Hirata R.K. Human gene targeting by viral vectors. Nat. Genet. 1998;18:325–330. doi: 10.1038/ng0498-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller D.G., Trobridge G.D., Petek L.M., Jacobs M.A., Kaul R., Russell D.W. Large-Scale Analysis of Adeno-Associated Virus Vector Integration Sites in Normal Human Cells. J Virol. 2005;79:11434–11442. doi: 10.1128/JVI.79.17.11434-11442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porteus M.H. Mammalian gene targeting with designed zinc finger nucleases. Mol Ther. 2006;13:438–446. doi: 10.1016/j.ymthe.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Porteus M.H. A New Class of Medicines through DNA Editing. N. Engl. J. Med. 2019;380:947–959. doi: 10.1056/NEJMra1800729. [DOI] [PubMed] [Google Scholar]
- 8.Durai S., Mani M., Kandavelou K., Wu J., Porteus M.H., Chandrasegaran S. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005;33:5978–5990. doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Porteus M.H., Carroll D. Gene targeting using zinc finger nucleases. Nat. Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 10.Christian M., Cermak T., Doyle E.L., Schmidt C., Zhang F., Hummel A., Bogdanove A.J., Voytas D.F. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li T., Huang S., Jiang W.Z., Wright D., Spalding M.H., Weeks D.P., Yang B. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011;39:359–372. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller J.C., Tan S., Qiao G., Barlow K.A., Wang J., Xia D.F., Meng X., Paschon D.E., Leung E., Hinkley S.J. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 13.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho S.W., Kim S., Kim J.M., Kim J.-S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 15.Dever D.P., Bak R.O., Reinisch A., Camarena J., Washington G., Nicolas C.E., Pavel-Dinu M., Saxena N., Wilkens A.B., Mantri S. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature. 2016;539:384–389. doi: 10.1038/nature20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pavel-Dinu M., Wiebking V., Dejene B.T., Srifa W., Mantri S., Nicolas C.E., Lee C., Bao G., Kildebeck E.J., Punjya N. Gene correction for SCID-X1 in long-term hematopoietic stem cells. Nat. Commun. 2019;10:1634. doi: 10.1038/s41467-019-09614-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eyquem J., Mansilla-Soto J., Giavridis T., van der Stegen S.J.C., Hamieh M., Cunanan K.M., Odak A., Gönen M., Sadelain M. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543:113–117. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J., Exline C.M., DeClercq J.J., Llewellyn G.N., Hayward S.B., Li P.W.-L., Shivak D.A., Surosky R.T., Gregory P.D., Holmes M.C., Cannon P.M. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat. Biotechnol. 2015;33:1256–1263. doi: 10.1038/nbt.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen S., Sun S., Moonen D., Lee C., Lee A.Y.-F., Schaffer D.V., He L. CRISPR-READI: Efficient Generation of Knockin Mice by CRISPR RNP Electroporation and AAV Donor Infection. Cell Rep. 2019;27:3780–3789.e4. doi: 10.1016/j.celrep.2019.05.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Ravin S.S., Reik A., Liu P.-Q., Li L., Wu X., Su L., Raley C., Theobald N., Choi U., Song A.H. Targeted gene addition in human CD34(+) hematopoietic cells for correction of X-linked chronic granulomatous disease. Nat. Biotechnol. 2016;34:424–429. doi: 10.1038/nbt.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiramoto T., Li L.B., Funk S.E., Hirata R.K., Russell D.W. Nuclease-free Adeno-Associated Virus-Mediated Il2rg Gene Editing in X-SCID Mice. Mol. Ther. 2018;26:1255–1265. doi: 10.1016/j.ymthe.2018.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barzel A., Paulk N.K., Shi Y., Huang Y., Chu K., Zhang F., Valdmanis P.N., Spector L.P., Porteus M.H., Gaensler K.M., Kay M.A. Promoterless gene targeting without nucleases ameliorates haemophilia B in mice. Nature. 2015;517:360–364. doi: 10.1038/nature13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirata R.K., Russell D.W. Design and packaging of adeno-associated virus gene targeting vectors. J. Virol. 2000;74:4612–4620. doi: 10.1128/jvi.74.10.4612-4620.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bak R.O., Porteus M.H. CRISPR-Mediated Integration of Large Gene Cassettes Using AAV Donor Vectors. Cell Rep. 2017;20:750–756. doi: 10.1016/j.celrep.2017.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Porteus M.H., Cathomen T., Weitzman M.D., Baltimore D. Efficient gene targeting mediated by adeno-associated virus and DNA double-strand breaks. Mol. Cell. Biol. 2003;23:3558–3565. doi: 10.1128/MCB.23.10.3558-3565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vakulskas C.A., Dever D.P., Rettig G.R., Turk R., Jacobi A.M., Collingwood M.A., Bode N.M., McNeill M.S., Yan S., Camarena J. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med. 2018;24:1216–1224. doi: 10.1038/s41591-018-0137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charlesworth C.T., Camarena J., Cromer M.K., Vaidyanathan S., Bak R.O., Carte J.M., Potter J., Dever D.P., Porteus M.H. Priming Human Repopulating Hematopoietic Stem and Progenitor Cells for Cas9/sgRNA Gene Targeting. Mol. Ther. Nucleic Acids. 2018;12:89–104. doi: 10.1016/j.omtn.2018.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DiMattia M.A., Nam H.J., Van Vliet K., Mitchell M., Bennett A., Gurda B.L., McKenna R., Olson N.H., Sinkovits R.S., Potter M. Structural insight into the unique properties of adeno-associated virus serotype 9. J. Virol. 2012;86:6947–6958. doi: 10.1128/JVI.07232-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellis B.L., Hirsch M.L., Barker J.C., Connelly J.P., Steininger R.J., 3rd, Porteus M.H. A survey of ex vivo/in vitro transduction efficiency of mammalian primary cells and cell lines with Nine natural adeno-associated virus (AAV1-9) and one engineered adeno-associated virus serotype. Virol. J. 2013;10:74. doi: 10.1186/1743-422X-10-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veldwijk M.R., Sellner L., Stiefelhagen M., Kleinschmidt J.A., Laufs S., Topaly J., Fruehauf S., Zeller W.J., Wenz F. Pseudotyped recombinant adeno-associated viral vectors mediate efficient gene transfer into primary human CD34(+) peripheral blood progenitor cells. Cytotherapy. 2010;12:107–112. doi: 10.3109/14653240903348293. [DOI] [PubMed] [Google Scholar]
- 31.Logan G.J., Dane A.P., Hallwirth C.V., Smyth C.M., Wilkie E.E., Amaya A.K., Zhu E., Khandekar N., Ginn S.L., Liao S.H.Y. Identification of liver-specific enhancer-promoter activity in the 3′ untranslated region of the wild-type AAV2 genome. Nat. Genet. 2017;49:1267–1273. doi: 10.1038/ng.3893. [DOI] [PubMed] [Google Scholar]
- 32.Yan Z., Sun X., Feng Z., Li G., Fisher J.T., Stewart Z.A., Engelhardt J.F. Optimization of Recombinant Adeno-Associated Virus-Mediated Expression for Large Transgenes, Using a Synthetic Promoter and Tandem Array Enhancers. Hum. Gene Ther. 2015;26:334–346. doi: 10.1089/hum.2015.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao X., Li J., Samulski R.J. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 1998;72:2224–2232. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chatterjee S., Wong J.K.K. Adeno-Associated Viral Vectors for the Delivery of Antisense RNA. Methods. 1993;5:51–59. [Google Scholar]
- 35.Brudek T., Lühdorf P., Christensen T., Hansen H.J., Møller-Larsen A. Activation of endogenous retrovirus reverse transcriptase in multiple sclerosis patient lymphocytes by inactivated HSV-1, HHV-6 and VZV. J Neuroimmunol. 2007;187:147–155. doi: 10.1016/j.jneuroim.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 36.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 37.Reisländer T., Lombardi E.P., Groelly F.J., Miar A., Porru M., Di Vito S., Wright B., Lockstone H., Biroccio A., Harris A. BRCA2 abrogation triggers innate immune responses potentiated by treatment with PARP inhibitors. Nat. Commun. 2019;10:3143. doi: 10.1038/s41467-019-11048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanmiguel J., Gao G., Vandenberghe L.H. Springer New York; 2019. Quantitative and Digital Droplet-Based AAV Genome Titration; pp. 51–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirsch M.L., Fagan B.M., Dumitru R., Bower J.J., Yadav S., Porteus M.H., Pevny L.H., Samulski R.J. Viral single-strand DNA induces p53-dependent apoptosis in human embryonic stem cells. PLoS ONE. 2011;6:e27520. doi: 10.1371/journal.pone.0027520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown N., Song L., Kollu N.R., Hirsch M.L. Adeno-Associated Virus Vectors and Stem Cells: Friends or Foes? Hum. Gene Ther. 2017;28:450–463. doi: 10.1089/hum.2017.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.