Abstract
Emerging evidence has revealed that excessive activation of macrophages may result in an adverse lung inflammation involved in sepsis-related acute lung injury (ALI). However, it has never been clearly identified whether peripheral circulating serum exosomes participate in the pathogenesis of sepsis-related ALI. Therefore, the purposes of our study were to investigate the effect of serum exosomes on macrophage activation and elucidate a novel mechanism underlying sepsis-related ALI. Here we found that exosomes were abundant in the peripheral blood from ALI mice and selectively loaded microRNAs (miRNAs), such as miR-155. In vivo experiments revealed that intravenous injection of serum exosomes harvested from ALI mice, but not control mice, increased the number of M1 macrophages in the lung, and it caused lung inflammation in naive mice. In vitro, we demonstrated that serum exosomes from ALI mice delivered miR-155 to macrophages, stimulated nuclear factor κB (NF-κB) activation, and induced the production of tumor necrosis factor alpha (TNF-α) and interleukin (IL)-6. Furthermore, we also showed that serum exosome-derived miR-155 promoted macrophage proliferation and inflammation by targeting SHIP1 and SOCS1, respectively. Collectively, our data suggest the important role of circulating exosomes secreted into peripheral blood as a key mediator of septic lung injury via exosome-shuttling miR-155.
Keywords: exosomes, miRNAs, inflammation, macrophages, lung injury
Excessive activation of macrophages may result in severe lung injury. Deng et al. demonstrate that peripheral circulating serum exosomes could transfer miR-155 into lung macrophages and then activate macrophages during sepsis. The present report reveals the importance of peripheral circulating exosomes in the development of septic lung injury.
Introduction
MicroRNAs (miRNAs), a class of non-coding single-stranded RNAs, negatively regulate gene expression by inhibiting translation or degrading target mRNAs.1 Due to their post-transcriptional regulatory abilities, miRNAs downregulate the expression of proteins involved in a wide array of biological processes, including cellular apoptosis and proliferation, immune response, and metabolism.2, 3, 4 An increasing body of evidence indicates that miRNAs are implicated in the pathogenesis of various inflammatory diseases and emerged as novel targets for intervention therapy.5, 6, 7 For instance, miR-155 is a highly conserved miRNA among mammals, and it has been generally recognized as a critical regulator of cell proliferation.8 Additionally, miR-155 has been recently reported to regulate macrophage-mediated inflammation during atherogenesis.9
In general, miRNAs are thought to act within the cells where they are generated. However, recent studies have suggested that miRNAs are present in circulating exosomes.10, 11 Exosomes are nanometer-sized membranous vesicles (30–150 nm) released by many types of cells, including epithelial cells,12 tumor cells,13 and macrophages,14 and they are thought to play a key role in cell-to-cell communications.15 It has been reported that exosomes contain lipids, proteins, mRNAs, and miRNAs derived from parent cells that are transferred between cells and, thus, regulate the functions of recipient cells.16 Recently, exosomally transferred miRNAs have emerged as important modulators of cellular function.17 A growing number of studies have confirmed that some specific miRNAs can be transferred via exosomes between immune cells or other cell types.10, 18 For example, hepatocyte-derived exosomes can transfer miR-122 and sensitize monocytes to endotoxin,19 while monocyte-derived miR-150-containing exosomes enhance endothelial cell migration.20 Thus, exosomes can serve as a novel therapeutic target and biomarker for a variety of disorders, including lung diseases.
Acute lung injury (ALI) is a common clinical syndrome occurring in critically ill patients.21 ALI is usually associated with the development of a systemic inflammatory response, such as sepsis and trauma, and sepsis is one of the most common causes of ALI in humans.22 Despite decades of experimental studies, the mortality rate of ALI remains relatively high (35%–45%).23 Histologically, ALI features an acute pulmonary inflammatory response that critically impairs lung function.24 Emerging evidence has revealed that excessive activation of inflammatory cells such as macrophages may result in an adverse lung inflammatory response, which could cause ALI.25
Macrophages, an essential effector cell of host defense against foreign stimuli, play a critical role in the pathogenesis of lung inflammation involved in ALI.4 Once activated (M1 activation), macrophages regulate leukocyte influx by the release of inflammatory cytokines and chemokines.26, 27 Previous studies have put a lot more emphasis on the involvement of cytokines and chemokines in the progression of ALI.28 However, it has never been clearly identified whether blood-borne exosomes participate in ALI associated with systemic inflammatory response. In fact, macrophages have been shown to be the recipient cells for exogenous exosomes. For instance, human mononuclear macrophages can be stimulated to produce pro-inflammatory factors upon taking up gastric cancer cell-derived exosomes.29 Macrophages are in direct contact with peripheral serum exosomes, raising the interesting possibility that macrophages may exhibit a series of biological functions upon taking up serum exosomes. Therefore, we hypothesized that serum exosomes deliver their cargo miRNAs to macrophages and they are involved in the activation of macrophages during sepsis-related ALI.
Results
miR-155 Expression Is Increased in the Mouse Lung Tissues after ALI
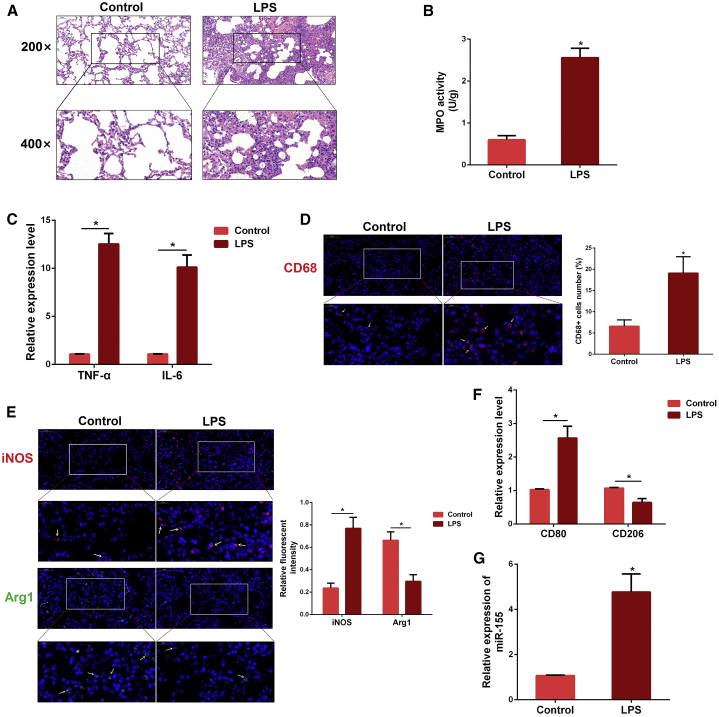
To validate the successful establishment of a mouse model of sepsis-related ALI, we performed H&E staining. Compared with the control group, the lung tissues from the lipopolysaccharide (LPS) group showed severe pathological changes, including interstitial edema, alveolar hyperemia, and inflammatory cell infiltration (Figure 1A). Myeloperoxidase (MPO) is a crucial indicator of inflammatory cell accumulation.30 The severity of pulmonary inflammation was further confirmed by subsequent MPO results (Figure 1B). Besides, qPCR assay also showed an increased production of pro-inflammatory cytokines interleukin (IL)-6 and tumor necrosis factor alpha (TNF-α) after ALI (Figure 1C).
Figure 1.
Macrophages Were Activated in the Lung Tissues of ALI Mice
(A) Histopathological analysis of lung tissues. Mice were intraperitoneally (i.p.) injected with LPS for 12 h, and the degree of lung inflammation was assessed with H&E staining (n = 3 per group). (B) Infiltration of inflammatory cells into the lung tissues was measured by MPO activity (n = 4 per group). (C) The levels of pro-inflammatory cytokines TNF-α and IL-6 were detected by qPCR (n = 3 per group). GAPDH was used as an endogenous control. (D) Sectioned tissues were stained with anti-CD68 antibody to identify the number of macrophages in lung tissues (n = 3 per group). The CD68-positive macrophages were normalized to DAPI-positive cells. (E) Immunofluorescence intensity of M1 and M2 macrophages in lung tissues (n = 3 per group). iNOS (M1, green) or Arg1 (M2, red). (F) The expression of CD80 and CD206 was determined by qPCR. GAPDH was used as an endogenous control (n = 3 per group). (G) The miR-155 expression was detected in the lung tissues of LPS-treated mice by qPCR (n = 4 per group). U6 snRNA was used as an endogenous control. Data are expressed as the mean ± SEM. *p < 0.05.
The macrophage is an important effector cell of host defense against foreign stimuli, and its activation promotes pulmonary inflammation involved in ALI.31 Thus, we measured the expression of macrophage marker CD68, and we found that the number of macrophages was significantly increased after exposure to LPS (Figure 1D). The activated phenotype (M1 or M2) of macrophage was also analyzed by inducible nitric oxide (iNOS) (M1 marker) and Arg1 (M2 marker) expressions. The immunofluorescence results showed that the fluorescent intensity of pro-inflammatory M1 macrophages was obviously increased while anti-inflammatory M2 macrophages were decreased in the LPS group (Figure 1E). Meanwhile, an increased mRNA level of the specific M1 marker CD80 was found with LPS stimulation, with decreased expression of another M2 indicator, CD206, further verifying the immunofluorescence results (Figure 1F). To investigate whether miR-155 is involved in the pulmonary inflammation during ALI, we detected its expression, and we found that miR-155 expression was markedly increased in the lung tissues of ALI mice (Figure 1G).
Isolation and Characterization of Serum Exosomes
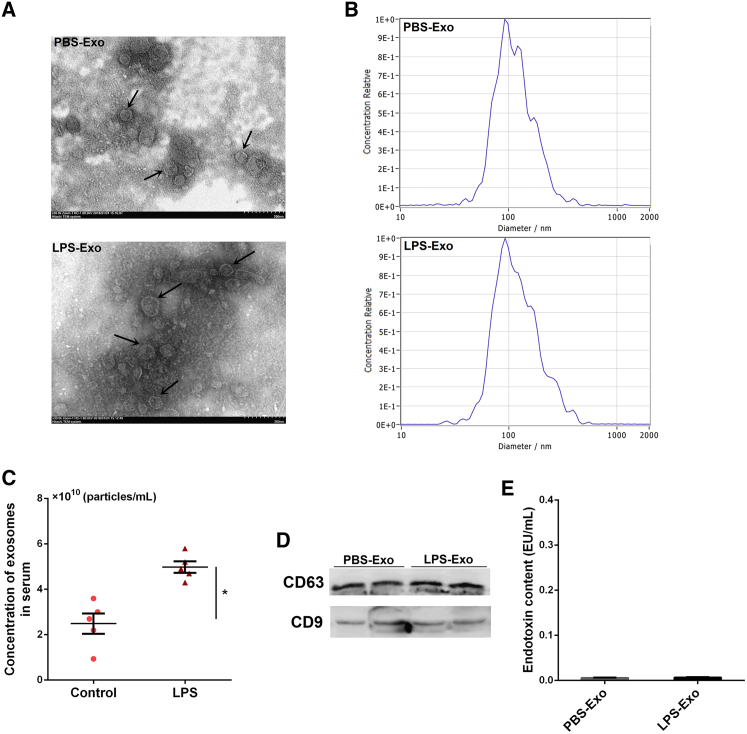
We isolated exosomes from the sera of mice by ultracentrifugation, and we observed the ultrastructure of exosomes using transmission electron microscopy (TEM). The TEM images showed the cup-shaped morphology of the exosomes, with a size range of 40–150 nm (Figure 2A). To further identify the size distribution profiles of isolated exosomes, nanoparticle-tracking analysis (NTA) was performed by a ZetaView Nanoparticle Tracking Analyzer (Particle Metrix, Germany). As shown in Figure 2B, the peak diameters of exosomes were not different between groups, which is in line with a previous report.32 Besides, the concentration of serum exosomes was significantly higher in ALI mice (Figure 2C), indicating that the secretion of exosomes is enhanced under inflammatory conditions. Western blot analysis also indicated that the exosome markers CD63 and CD9 were enriched in the exosomal fraction (no loading control) (Figure 2D). In addition, endotoxin test results showed that endotoxin (LPS) contamination was barely detectable in these serum exosomes (Figure 2E).
Figure 2.
Isolation and Characterization of Serum Exosomes
(A) Transmission electron micrographs of serum exosomes isolated from control and ALI mice. Scale bar, 200 nm. (B) The size distribution profiles of isolated serum exosomes. (C) NTA showing the concentration of serum exosomes isolated from control and ALI mice (n = 5 per group). (D) Western blot analysis of exosome markers CD63 and CD9 in the exosomal preparations. (E) Endotoxin levels in the exosomal preparations. PBS-Exo and LPS-Exo represent serum exosomes isolated from control and ALI mice, respectively. Data are expressed as the mean ± SEM. *p < 0.05.
Injected Serum Exosomes Reach the Lung and Induce Pulmonary Inflammation In Vivo
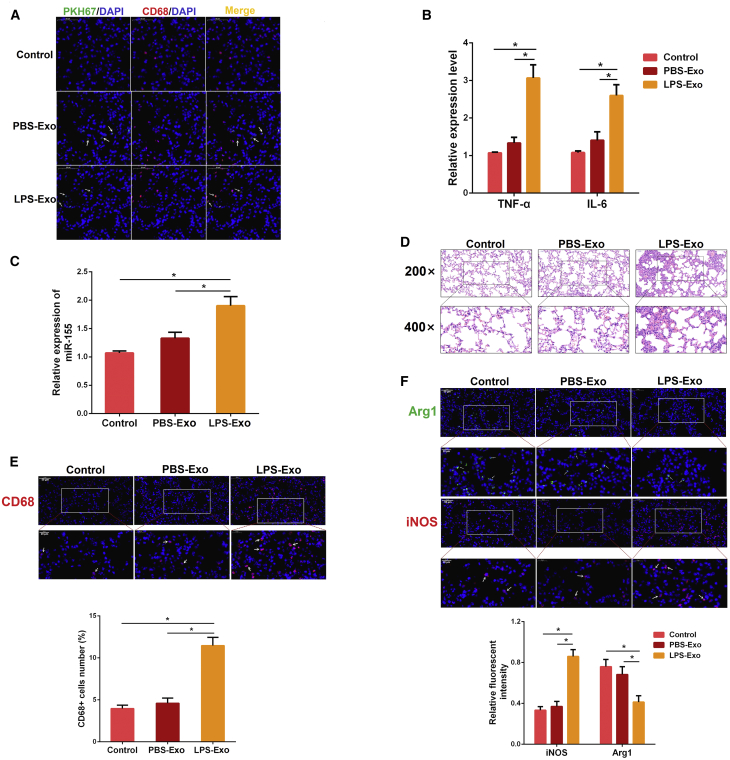
To explore whether circulating serum exosomes mediate sepsis-related ALI, an equal amount of serum exosomes from mice treated with PBS or LPS was intravenously injected into naive mice. Interestingly, the frozen lung sections showed that the PKH67-labeled exosomes were taken up into the lung parenchyma 6 h after injection (Figure 3A), which is consistent with a previous report.33 Subsequently, we determined the lung inflammation at 24 h after exosome injection. The expressions of pro-inflammatory cytokines (IL-6 and TNF-α) and miR-155 were increased after the injection of serum exosomes from LPS-treated mice (LPS-Exo). In contrast, injection of serum exosomes from PBS-treated mice (PBS-Exo) failed to cause a lung inflammatory response (Figures 3B and 3C). Histologic examination also revealed a significant increase in inflammatory cell infiltration into the alveolar space by the injection of LPS-Exo (Figure 3D). To further confirm the ability of LPS-Exo to cause lung inflammation in naive mice, we observed a significant increase in the number of lung macrophages (CD68-positive cells) after the injection of LPS-Exo (Figure 3E). Furthermore, we also found that the intensity of M1 macrophages was markedly increased and M2 macrophages were significantly decreased in the LPS-Exo group (Figure 3F). Collectively, these data indicate that circulating serum exosomes activate lung macrophages and then induce pulmonary inflammation in vivo.
Figure 3.
Injection of Serum Exosomes from ALI Mice Induces Lung Inflammation in Naive Mice
(A) Serum exosomes from each experimental group (control and ALI) and PBS as a control were labeled with green fluorescent dye and injected into naive mice via tail vein. After 6 h, the lung tissues were stained with anti-CD68 antibody (macrophages, red) and DAPI (nucleus, blue) (n = 3 per group). Pictures were obtained by fluorescence microscopy. (B) The levels of TNF-α and IL-6 were detected by qPCR 24 h after exosome injection (n = 3 per group). GAPDH was used as an endogenous control. (C) The miR-155 expression was measured by qPCR (n = 4 per group). U6 snRNA was used as an endogenous control. (D) Representative images of lung histology stained with H&E (n = 3 per group). (E) Sectioned tissues were stained with anti-CD68 antibody to identify the number of macrophages in lung tissues. The CD68-positive macrophages were normalized to DAPI-positive cells (n = 3 per group). (F) Immunofluorescence intensity of M1 and M2 macrophages in lung tissues (n = 3 per group). iNOS (M1, green) or Arg1 (M2, red). PBS-Exo and LPS-Exo represent serum exosomes isolated from control and ALI mice, respectively. Data are expressed as the mean ± SEM. *p < 0.05.
Serum Exosomes from ALI Mice Selectively Load miR-155
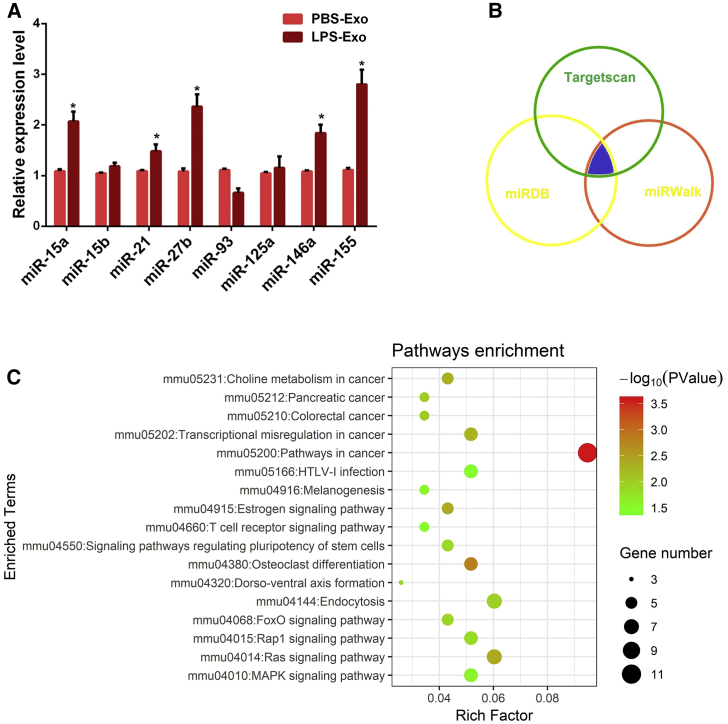
Recent studies have suggested that exosomes exert important regulatory effects on the physiological functions of recipient cells via shuttling miRNAs.34 Thus, we selected a group of inflammation-related miRNAs (miR-15a, miR-15b, miR-21, miR-27b, miR-93, miR-125a, miR-146a, and miR-155), according to others’ reports,32, 35 and we analyzed the differential expressions of these miRNAs in serum exosomes using qPCR. Among them, the expression of miR-155 in LPS-Exo was highest, and it was significantly higher than that in PBS-Exo (Figure 4A), revealing that serum exosomes from ALI mice selectively loaded miR-155.
Figure 4.
Serum Exosomes from ALI Mice Selectively Load miR-155
(A) The expression levels of inflammatory miRNAs in the serum exosomes from control and ALI mice were measured by qPCR. (B) The potential targets of miR-155 were predicted by integrating the results of three databases (TargetScan, miRDB, and miRWalk). (C) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was performed on the predicted target genes of miR-155. PBS-Exo and LPS-Exo represent serum exosomes isolated from control and ALI mice, respectively. Data are expressed as the mean ± SEM. *p < 0.05.
To unravel the role of exosome-contained miR-155 in the pathogenesis of ALI, the TargetScan, miRDB, and miRWalk databases were applied to predict the target genes of miR-155 (Figure 4B). Thereafter, the pathway analysis of the predicted target genes was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation system. As shown in Figure 4C, the predicted target genes were significantly enriched in immune- and proliferation-related pathways, such as the mitogen-activated protein kinase (MAPK)-signaling pathway, T cell receptor-signaling pathway, Ras-signaling pathway, and FOXO-signaling pathway. Thus, we hypothesized that serum exosomes may transfer miR-155 into lung macrophages and then activate macrophages during sepsis-related ALI.
Serum Exosome-Derived miR-155 Promotes Macrophage Proliferation and Inflammation In Vitro
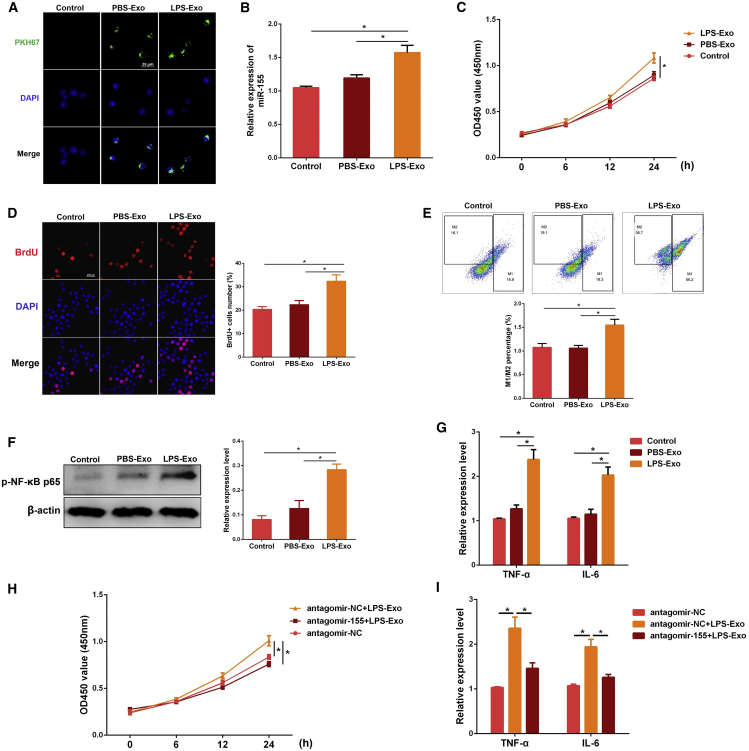
To test the above hypothesis, we labeled serum exosomes with PKH67 and incubated them with macrophages for the indicated time. Intriguingly, we observed that macrophages could uptake these exosomes in vitro (Figure 5A). Meanwhile, the miR-155 level increased significantly when the cells were treated with LPS-Exo (Figure 5B), indicating that serum exosomes from ALI mice can transfer miR-155 into macrophages. Next, the proliferation of macrophages was determined by cell counting kit-8 (CCK-8) and bromodeoxyuridine (BrdU) assays. The results of CCK-8 and BrdU assays showed that LPS-Exo, but not PBS-Exo, promoted the proliferation of macrophages (Figures 5C and 5D).
Figure 5.
Serum Exosome-Derived miR-155 Promotes Macrophage Proliferation and Inflammation In Vitro
(A) The PKH67-labeled exosomes (green) were co-cultured with macrophages for the indicated time. The uptake of exosomes by macrophages was observed under a confocal microscope. (B–E) Macrophages were treated with the serum exosomes from control or ALI mice (PBS-Exo or LPS-Exo) for the indicated time. (B) The miR-155 expression was determined by qPCR. U6 snRNA was used as an endogenous control. The effect of serum exosomes on the macrophage proliferation was measured by CCK-8 (C) and BrdU (D) assays, respectively. The values of OD (450 nm) represent cell viability. The BrdU-positive cells were normalized to DAPI-positive cells. (E) The effect of serum exosomes on the macrophage polarization was analyzed by flow cytometry. (F and G) The pro-inflammatory effect of serum exosomes on macrophages was assessed by measuring the levels of NF-κB p65 (F), TNF-α, and IL-6 (G). (H) Macrophages were transfected with antagomir-155 or antagomir-NC for 24 h and then treated with PBS-Exo or LPS-Exo for the indicated time. The proliferation of macrophages was assessed by CCK-8 assay. (I) Cells were treated as in (H), and the levels of TNF-α and IL-6 were detected by qPCR. GAPDH was used as an endogenous control. PBS-Exo and LPS-Exo represent serum exosomes isolated from control and ALI mice, respectively. Data are expressed as the mean ± SEM. *p < 0.05.
The M1:M2 ratio of macrophages was also obviously increased after LPS-Exo treatment, as evidenced by flow cytometry, which implied that the promoted proliferation of macrophages by LPS-Exo might be due to the increase in M1 macrophages (Figure 5E). Besides, we also evaluated the effect of serum exosomes on the inflammation of macrophages. We demonstrated that pro-inflammatory cytokines IL-6 and TNF-α were remarkably increased in LPS-Exo-stimulated cells compared to those stimulated with PBS-Exo (Figure 5G). Nuclear factor κB (NF-κB) is a nuclear transcription factor that regulates downstream inflammatory cytokines involved in the inflammation process.30 Our findings also revealed higher NF-κB p65 expression levels in LPS-Exo-stimulated cells than those in PBS-Exo-stimulated cells (Figure 5F).
To further verify the role of exosome-contained miR-155 in macrophage proliferation and inflammation, macrophages were incubated with LPS-Exo in the presence of antagomir-155 (a miR-155 inhibitor). Compared with the antagomir-negative control (NC), inhibition of miR-155 reversed the increased proliferation and inflammation of macrophages induced by LPS-Exo (Figures 5H and 5I). The above results strongly suggest that serum exosomes from ALI mice are a kind of pro-inflammatory exosome and can promote macrophage proliferation and inflammation by shuttling miR-155.
miR-155 Promotes Macrophage Proliferation by Targeting SHIP1
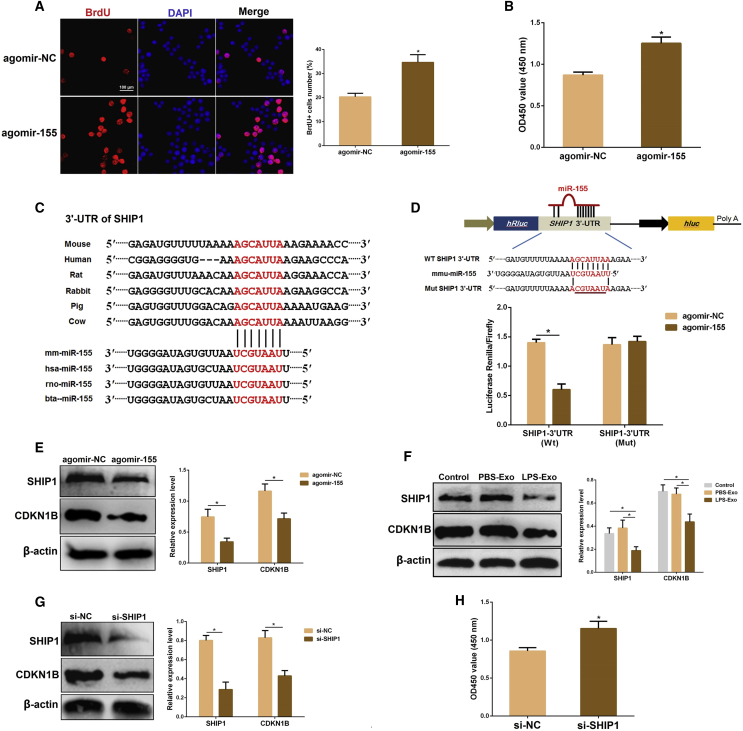
To figure out whether miR-155 promotes macrophage proliferation, we transfected macrophages with agomir-155. Overexpression of miR-155 enhanced macrophage proliferation, as detected by BrdU (Figure 6A) and CCK-8 (Figure 6B) assays. Subsequently, we analyzed the possible targets of miR-155 involved in cell proliferation to uncover the molecular mechanism through which miR-155 affects macrophage proliferation. The bioinformatics analysis revealed that the 3′ UTR of SHIP1 mRNA contains a complementary binding site for the seed region of miR-155, which is conserved across species (Figure 6C).
Figure 6.
miR-155 Promotes Macrophage Proliferation by Targeting SHIP1
(A and B) Cell viability was examined using BrdU (A) and CCK-8 (B) assays after transfection with agomir-155 or agomir-NC. (C) Conservation of the miR-155 target sequence in SHIP1 3′ UTR among different species and conservation of the miR-155 sequence among different species. (D) The dual-luciferase reporter assay was performed in 293T cells. Cells were co-transfected with the wild- or mutant-type SHIP1 3′ UTR luciferase reporter plasmids, as well as agomir-155 or agomir-NC. The ratio of Renilla activity:Firefly activity represents luciferase activity. (E) The protein levels of SHIP1 and CDKN1B were detected using western blot analysis after transfection with agomir-155 or agomir-NC. β-actin was used as an internal control. (F) The protein levels of SHIP1 and CDKN1B were determined using western blot analysis after incubation with PBS-Exo or LPS-exo. β-actin was used as an internal control. (G) The protein levels of SHIP1 and CDKN1B were detected using western blot analysis after transfection with si-SHIP1 or si-NC. β-actin was used as an internal control. (H) The proliferation of macrophages was assessed by the CCK-8 assay after co-transfection with si-SHIP1 or si-NC. GAPDH was used as an endogenous control. PBS-Exo and LPS-Exo represent serum exosomes isolated from control and ALI mice, respectively. Data are expressed as the mean ± SEM. *p < 0.05.
SHIP1 has been reported to negatively regulate the proliferation of osteoclast precursors through upregulating cyclin-dependent kinase inhibitors 1B (CDKN1B).36 To provide direct evidence that SHIP1 is a target of miR-155, we carried out a luciferase reporter assay. The results showed that agomir-155 remarkably reduced the luciferase activity for the wild-type 3′ UTR of SHIP1 but showed no inhibition effect for the mutated 3′ UTR of SHIP1 (Figure 6D). SHIP1 and CDKN1B proteins were also measured in macrophages transfected with agomir-155 or agomir-NC. As expected, the protein levels of SHIP1 and CDKN1B were strongly depressed by the overexpression of miR-155 (Figure 6E). In conclusion, miR-155 inhibits SHIP1 expression in macrophages by directly targeting the 3′ UTR of SHIP1 mRNA.
Next, we assessed the effects of serum exosomes on SHIP1 and CDKN1B in macrophages. Surprisingly, treatment with LPS-Exo resulted in decreased expressions of SHIP1 and CDKN1B compared with the PBS-Exo group (Figure 6F). Furthermore, knockdown of SHIP1 also reduced CDKN1B expression (Figure 6G) and then induced macrophage proliferation (Figure 6H). Overall, these data demonstrate that serum exosome-derived miR-155 promotes cellular proliferation by suppressing SHIP1 in macrophages.
miR-155 Induces Macrophage Inflammation by Targeting SOCS1
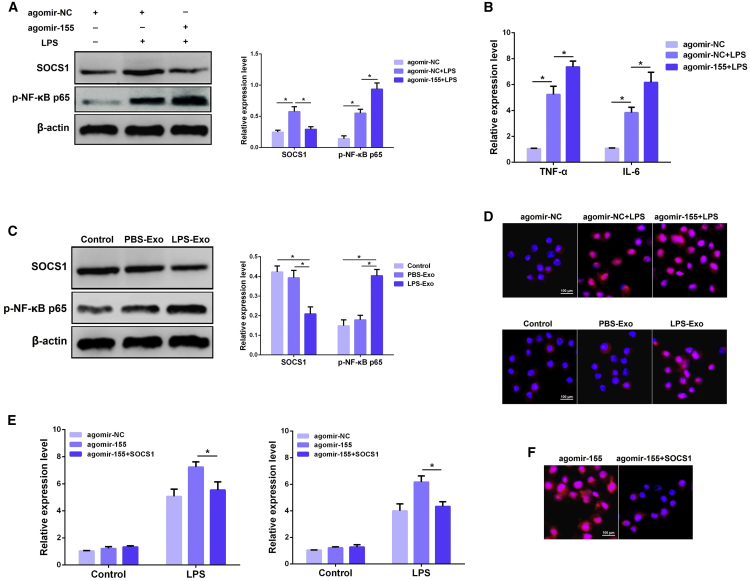
Recent studies have highlighted the importance of miR-155 as a regulatory target of immune responses in a wide range of inflammatory diseases.37, 38 SOCS1, a known target of miR-155,39 can suppress NF-κB activity by reducing p65 stability.40 To further obtain direct evidence that miR-155 targets SOCS1 in macrophages, we measured the protein level of SOCS1 in macrophages transfected with agomir-155 after LPS stimulation. Overexpression of miR-155 significantly decreased the expression of SOCS1, displaying an inverse relationship between SOCS1 and miR-155 (Figure 7A). Moreover, we also observed an obvious increase in the levels of NF-κB p65 (Figure 7A) and pro-inflammatory cytokines TNF-α and IL-6 (Figure 7B) in macrophages overexpressing miR-155.
Figure 7.
miR-155 Promotes Macrophage Inflammation by Targeting SOCS1
(A) The protein levels of SOCS1 and NF-κB p65 were detected using western blot analysis after transfection with agomir-155 or agomir-NC. β-actin was used as an internal control. (B) The levels of TNF-α and IL-6 were detected by qPCR after transfection with agomir-155 or agomir-NC. (C) The protein levels of SOCS1 and NF-κB p65 were determined using western blot analysis after incubation with PBS-Exo or LPS-Exo. β-actin was used as an internal control. (D) Translocation of the NF-κB p65 subunit from the cytoplasm into the nucleus was evaluated by immunofluorescence. Blue spots represent cell nuclei and red spots represent NF-κB p65 staining. (E) The levels of TNF-α and IL-6 were detected by qPCR after co-transfection with agomir-155 and pCDNA.3.1-SOCS1. GAPDH was used as an endogenous control. (F) Translocation of the NF-κB p65 subunit from the cytoplasm into the nucleus was evaluated by immunofluorescence. Blue spots represent cell nuclei and red spots represent NF-κB p65 staining. PBS-Exo and LPS-Exo represent serum exosomes isolated from control and ALI mice, respectively. Data are expressed as the mean ± SEM. *p < 0.05.
Additionally, we further determined the effects of serum exosomes on SOCS1 and NF-κB p65 in macrophages. Compared with the PBS-Exo group, treatment with LPS-Exo decreased the expression of SOCS1 and increased the NF-κB p65 expression (Figure 7C). Similarly, immunofluorescence results confirmed that miR-155 or LPS-Exo induced an increase in the nuclear translocation of NF-κB p65 (Figure 7D). Mechanically, overexpression of SOCS1 markedly inversed the pro-inflammatory effect of miR-155 on macrophages, as evidenced by the inhibition of TNF-α, IL-6 (Figure 7E), and NF-κB p65 nuclear translocation (Figure 7F). Collectively, these results suggest that serum exosome-derived miR-155 induces macrophage inflammation by targeting SOCS1.
Discussion
Patients with sepsis have a higher risk of multiple organ dysfunction syndrome (MODS), including ALI, which is thought to be crucial for their prognosis.41 Although neutrophil recruitment and activation within the lung are key contributors to the pathogenesis of ALI,42 overwhelming evidence confirms that the activation of macrophages also contributes to the initiation of inflammatory responses and resultant lung injury.43, 44 However, it remains poorly understood how lung macrophages are activated during the development of sepsis-related lung injury. LPS is a gram-negative bacterial endotoxin that is implicated in the initiation of ALI, and the administration of LPS has been widely used as a model of lung injury in several species.45, 46, 47 Thus, we employed a mouse model of LPS-induced, sepsis-related ALI to delineate the possible mechanisms underlying macrophage activation and lung inflammation during sepsis. Here we showed that the macrophages in lung tissues were noticeably activated by intraperitoneally administered LPS.
As detailed in the Introduction, exosomes are actively secreted into various body fluids, including blood; modulate both physiological and pathological processes in an organism; and mediate diverse biological functions.15 It has been shown that blood-borne exosomes can be internalized by dendritic cells (DCs) and then induce donor-specific transplant tolerance.48 On the other hand, serum exosomes from diabetic mice contribute to aortic endothelial dysfunction in nondiabetic mice.49 Notably, blood-borne microparticles (containing exosomes) from patients with sepsis have been reported to induce endothelial cell apoptosis through peroxynitrite generation.50 However, little has been reported about whether serum exosomes can affect macrophages during septic lung injury. Thus, we hypothesized that peripheral circulating serum exosomes may affect macrophages and induce lung inflammation during sepsis-related lung injury.
In this study, we reported for the first time an increased number of serum exosomes isolated from samples collected after ALI. To ascertain whether serum exosomes mediate the lung injury, we labeled these exosomes using PKH67 dye and transfused them into naive mice via tail vein injection. Intriguingly, we found that these labeled serum exosomes were mobilized to the lung parenchyma and some of them were taken up by lung macrophages. Indeed, the finding is in agreement with a previously published report that confirmed that exosomes from mesenteric lymph could reach the lung parenchyma and be internalized by macrophages after intravenous injection.33 Thereafter, we observed that serum exosomes from ALI mice resulted in a significant lung inflammatory response, as manifested by increased expressions of pro-inflammatory cytokines in the lung tissues. In addition, the number of pro-inflammatory M1 macrophages also increased following exosome transfusion, implying a potential role of serum exosomes in septic lung injury. Similarly, a recent paper by Bonjoch et al.51 also demonstrated that circulating exosomes are involved in the lung inflammation associated with experimental acute pancreatitis. Based on previous studies and our present results, circulating serum exosomes from ALI mice are a kind of pro-inflammatory exosome, and they can activate lung macrophages and then induce pulmonary inflammation.
Although exosomes contain abundant bioactive molecules, studies have suggested that their biological effects on recipient cells are partly dependent on the loaded miRNA content.14, 35 Exosomal miRNAs play pivotal roles in the development of cancer and inflammatory diseases. For example, adipose tissue macrophage-derived exosomal miRNAs can modulate in vivo and in vitro insulin sensitivity.52 Transfer of miR-365 in macrophage-derived exosomes induces the drug resistance of pancreatic adenocarcinoma.53 Some exosomal miRNAs have also been identified as valuable biomarkers for disease diagnosis in certain clinical settings.54 Noteworthily, Azevedo et al.55 demonstrated that exosomes from septic shock patients carry miRNAs, which are novel players in the pathogenesis of sepsis. Consequently, we determined a group of the most common miRNAs associated with inflammation (miR-15a, miR-15b, miR-21, miR-27b, miR-93, miR-125a, miR-146a, and miR-155) in serum exosomes based on others’ studies.32, 35
We showed that serum exosomes harvested from ALI mice exhibited differentially expressed miRNAs. Although some miRNAs (such as miR-15a, miR-27b, and miR-146a) were also upregulated in the serum exosomes, the miR-155 expression was highest, suggesting that miR-155 may be a significant contributor to the biological activities of these inflammatory exosomes. Indeed, miR-155 is a highly conserved and important miRNA, which is not only closely related to the occurrence of cancer but also can regulate the inflammatory response. It was reported that miR-155 could regulate macrophage-mediated inflammation during atherogenesis.9 More notably, some recent studies have revealed that miR-155 is implicated in the development of inflammatory lung disease, such as Staphylococcal Enterotoxin B-induced lung injury.56 Meanwhile, in our pilot experiments, we also found that treatment with miR-155 inhibitors in vivo could significantly alleviate the inflammatory lung injury in septic mice (Figures S1A and S1B), further confirming that miR-155 plays a crucial role in the pathogenesis of septic-related ALI. Taken together, these findings prompted us to perform in vitro experiments to investigate the role of exosomal miR-155 in macrophage activation during septic-related ALI.
Macrophages are the known recipient cells for exogenous exosomes. For instance, human macrophages can be stimulated to produce pro-inflammatory factors upon taking up gastric cancer cell-derived exosomes.29 Breast cancer-derived exosomes are capable of inducing IL-6 secretion and a pro-survival phenotype in bone marrow-derived macrophages.57 We provided the first evidence that serum exosomes from ALI mice increased the expression of pro-inflammatory cytokines in vitro in macrophages by activating NF-κB signaling. Besides, the proliferation rate of macrophages was also enhanced after incubation with these inflammatory exosomes, which is consistent with the in vivo data. More importantly, administration with inflammatory exosomes also increased the miR-155 expression in macrophages compared with the control cells, implying a transfer of miR-155 between peripheral circulating exosomes and macrophages.
To identify whether exosomal miR-155 mediated the effects of serum exosomes, we incubated macrophages with these inflammatory exosomes upon exposure to miR-155 inhibitors (antagomir-155). Intriguingly, the pro-inflammatory and pro-proliferative activities of serum exosomes were reversed following miR-155 knockdown. To further corroborate exosomal miR-155 as a determinant of the bioactivity of these inflammatory exosomes, we isolated serum exosomes from miR-155-knockdown mice. Knocking down miR-155 via in vivo inhibitor transduction lowered the expression of serum exosomal miR-155 (Figure S1C). Importantly, these exosomes with low miR-155 expression resulted in a weaker inflammatory response in macrophages compared with the exosomes from wild-type mice (Figure S1D). Based on our observations, we speculated that circulating serum exosomes could transfer miR-155 into lung macrophages and then activate macrophages, thus leading to lung injury. This finding accords with an earlier study, which showed that macrophage miR-155 promotes LPS-induced lung injury in mice.7
miRNAs are thought to regulate many intricate physiological processes by inhibiting the expressions of target genes having related functions.58 KEGG analysis revealed that the target genes of miR-155 were significantly enriched in immune- and proliferation-related pathways. Through the databases, we found that SHIP1 and SOCS1 are the potential targets of miR-155. SHIP1, a tumor suppressor, has been widely recognized to inhibit cell proliferation in many types of tumor cells.8, 59 It has been reported that SHIP1 inhibits the proliferation of osteoclast precursors via upregulating CDKN1B.36 CDKN1B controls cell progression and causes cell-cycle arrest in a wide range of tumors.60 However, the roles of SHIP1 and CDKN1B in macrophage proliferation have been rarely reported. SOCS1 acts as a negative feedback regulator to inhibit JAK2/STAT3 signaling and control inflammation during ischemic stroke.61 Macrophage deletion of SOCS1 gene increases sensitivity to LPS and results in systemic inflammation.62 SOCS1 can also restrain NF-κB by decreasing p65 stability within the cell nucleus and then repress the release of inflammatory cytokines.40 Based on a series of functional experiments, we further demonstrated exosomal miR-155 promoted the proliferation and inflammation of macrophages through downregulating SHIP1 and SOCS1, respectively.
Due to technical limitations, it remains difficult to identify the precise origins of these lung-targeting serum exosomes. Recent studies have shown that alveolar epithelial cells respond to hyperoxia stress and then activate macrophages through the release of extracellular vesicles.63 Thus, we speculate that a proportion of serum exosomes are probably from lung cells and so they can reach the lung to induce lung inflammation. Identification of the sources of serum exosomes will serve as a subject for further research in the near future. In addition, our study cannot rule out the possibility that other exosomal miRNAs with altered levels, such as miR-15a and miR-27b, might exert minor effects in serum exosome-induced inflammation, which requires future study.
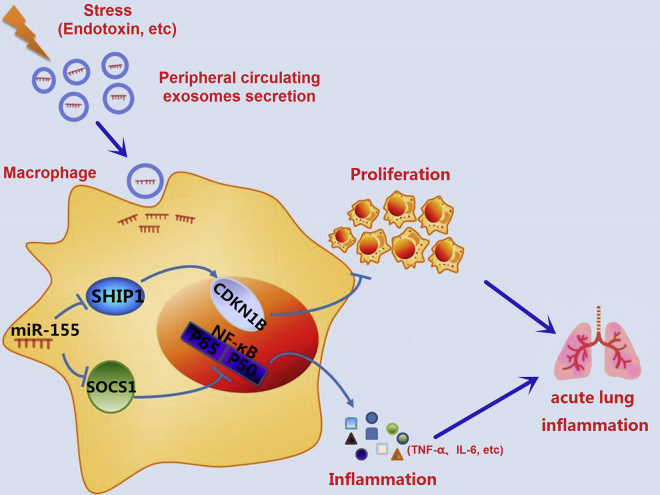
In summary, here we demonstrate that serum exosomes are abundant in the peripheral blood of ALI mice and these exosomes selectively load miRNAs, such as miR-155. In vivo, serum exosomes are able to reach the lung macrophages and cause lung injury (Figure 8). In vitro, serum exosome-derived miR-155 promotes macrophage proliferation and inflammation by targeting SHIP1 and SOCS1, respectively. The present report unravels the importance of circulating serum exosomes in the regulation of macrophage activation during sepsis. This novel route for intercellular communication may contribute to a better understanding for the mechanism of ALI after exposure to various stimuli such as sepsis.
Figure 8.
The Effects of miR-155-Containing Serum Exosomes on Macrophage Proliferation and Inflammation
During sepsis, miR-155-enriched exosomes are secreted into peripheral blood and then able to reach the lung macrophages. Exosome-derived miR-155 promotes macrophage proliferation and inflammation, which leads to acute lung inflammation.
Materials and Methods
Mice
Adult male BALB/c mice were obtained from the Experimental Animal Center of Huazhong Agricultural University (Wuhan, China). All of the protocols involving animals in this study were approved by the Ethical Committee on Animal Research at Huazhong Agricultural University, and they were carried out in accordance with the approved guidelines.
Mouse Model of LPS-Induced ALI
The method for establishing the LPS-induced ALI model was performed as previously described.47 Briefly, the mice were intraperitoneally (i.p.) injected with LPS (20 mg/kg, Sigma), and the control mice were given an equal amount of PBS. At 12 h after LPS injection, the serum samples and lung tissues were collected for further analysis.
Lung Histological Analysis
For histological analysis, lung tissues were excised and fixed with 4% paraformaldehyde for 24 h. Thereafter, the fixed tissues were embedded in paraffin, sliced to a thickness of 4–6 μm, and stained with H&E.
MPO Evaluation
The infiltration of inflammatory cells into lung tissues was evaluated by MPO activity. Lung tissues were homogenized with reaction buffer (1/9 w/v), and the MPO activity was measured with an MPO test kit (Nanjing Jiancheng Bioengineering Institute, China), according to the manufacturer’s instructions.
Serum Exosome Isolation, Identification, Labeling, and Uptake
The mouse blood was collected in 1.5-mL tubes and allowed to clot for 1 h at room temperature, and then serum was obtained by centrifugation at 2,000 × g for 10 min at 4°C. Serum exosomes were isolated according to the protocol previously described.49 Briefly, the serum was centrifuged at 3,000 × g for 10 min at 4°C. The supernatant was then diluted with an equal volume of sterile PBS (pH 7.4) and centrifuged again at 10,000 × g for 30 min at 4°C. The resultant supernatant fluid was filtered through a 0.22-μm filter (Millipore, USA), transferred to a sterile ultracentrifuge tube, and then centrifuged at 200,000 × g for 2 h at 4°C (Beckman Optima XE-90, Beckman Coulter, USA). For exosome purification, the pellets were washed in a large volume of PBS and centrifuged at 200,000 × g for 1 h at 4°C. Following this step, the pellets containing the exosomes were collected and resuspended in filtrated PBS for subsequent studies.
After isolation, exosomes were analyzed by electron microscopy and NTA in terms of ultrastructure, concentration, and size distribution. Protein markers such as CD63 and CD9 were assessed by western blot.
To determine whether macrophages can uptake serum exosomes, we labeled exosomes using a PKH67 green fluorescent labeling kit (Sigma, MINI67), as previously described.64 Subsequently, we injected these labeled exosomes into mice or co-cultured them with macrophages. At 6 h post-injection, the lung tissues from mice were excised and then observed with a laser-scanning confocal microscope (Zeiss LSM 800, Zeiss, Germany) to track whether exosomes could reach the lung. After the indicated time of co-culture, we stained macrophages with DAPI and observed them with a laser-scanning confocal microscope.
Administration of LPS-Stimulated Serum Exosomes from Donor to Recipient Mice
Serum exosomes from mice treated either with or without LPS were isolated and resuspended in filtrated PBS. A total of 200 μL exosomes was intravenously injected into the recipient mice. Mice injected with the same amount of PBS were used as a control group. Mice were all euthanized at 24 h after the injection, followed up by the examination of inflammatory markers.
TEM
Isolated exosome pellets were fixed in 2% paraformaldehyde, loaded onto formvar carbon-coated grids, and then left to settle for 15 min. Thereafter, the samples were negatively stained with uranyl oxalate for 5 min and then visualized using a Hitachi H-600 transmission electron microscope (Hitachi, Schaumburg, IL, USA).
Cell Culture
The mouse RAW264.7 macrophages and HEK293T cells were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in DMEM (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Sigma, St. Louis, MO, USA), streptomycin (50 μg/mL), and penicillin (50 U/mL) at 37°C in a humidified atmosphere of 5% CO2. Cells for exosome treatment were cultured in DMEM with 10% exosome-depleted FBS (System Biosciences, San Francisco, CA, USA).
Immunofluorescent Staining
Cells or excised tissues were fixed in 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and then blocked with 10% BSA. Slides were incubated with the indicated primary antibodies overnight at 4°C and then incubated with fluorescently labeled secondary antibodies for 1 h at room temperature. Nuclei were stained using DAPI for 10 min. Six images per animal were captured under a fluorescence microscope (AX70, Olympus, Japan), and the average value from the six images was used to represent the value from one mouse lung. Antibodies used included the following: anti-CD68, anti-iNOS, anti-Arg-1, anti-BrdU (Abcam, Cambridge, MA, USA), and anti-NF-κB p65 (Cell Signaling Technology, MA, USA).
Western Blot Analysis
Proteins were extracted from exosomes or cells using radioimmunoprecipitation (RIPA) lysis buffer (BioSharp, China), and their concentrations were measured using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Rockford, IL, USA). Samples with equal amounts of protein were separated by SDS-PAGE gels, transferred onto polyvinylidene fluoride (PVDF) membranes, and probed with the indicated primary antibodies overnight. Membranes were washed and incubated with horseradish peroxidase-conjugated secondary antibodies and then visualized using the ImageQuant LAS4000 mini (GE Healthcare, Piscataway, NJ). Primary antibodies against CD63 and CD9 (1:1,500) were purchased from Abcam (Cambridge, MA, USA); primary antibodies against SHIP1, CDKN1B, and SOCS1 (1:800) were from Santa Cruz Biotechnology (Dallas, TX, USA); and primary antibodies against β-actin and phospho-NF-κB p65 (1:1,000) were from Cell Signaling Technology (Beverly, MA, USA). Secondary antibodies (1:4,000) were from Cell Signaling Technology (Beverly, MA, USA). The ray values of the protein bands were measured by Image-Pro Plus 6.0 software (Media Cybernetics, Silver Spring, MD, USA), and the relative expressions of the indicated proteins were normalized to an internal control β-actin.
RNA Extraction and qPCR Analysis
Total RNA was extracted using the TRIzol reagent (Invitrogen, USA), following the manufacturer’s instructions, and then reverse transcribed into cDNA using the M-MLV Reverse Transcriptase (Takara, Dalian, China) or Hairpin-it miRNAs qPCR Quantitation Kit (GenePharma, Shanghai, China). Then, qPCR was performed using the SYBR Green Master Mix and run on a StepOne Plus Real-Time PCR System (Applied Biosystems, USA). The relative expression levels of miRNAs or mRNAs were normalized to U6 small nuclear RNA (snRNA) or GAPDH following the 2–ΔΔCt comparative method. The primers are listed in Table S1.
Cell Transfection
Macrophages were transfected with the indicated miRNA agomirs, antagomirs, small interfering RNAs (siRNAs), or NCs using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), following the manufacturer’s instructions. The cells were harvested for subsequent studies 24 h after transfection. agomir-NC, agomir-155, antagomir-NC, antagomir-155, siRNAs, and the corresponding siRNA NC (si-NC) were designed and synthesized by GenePharma (Shanghai, China).
Plasmid Construction and Luciferase Reporter Assay
The possible miRNA-binding sites on the SHIP1 gene were predicted using the TargetScan, miRDB, and miRWalk databases. The luciferase reporter plasmids were constructed as described previously. Briefly, the miR-155-binding sites in the SHIP1 3′ UTR sequence were cloned into a psiCHECK-2 vector (Promega, Madison, WI, USA). For the luciferase reporter assay, HEK293T cells were co-transfected with the luciferase reporter plasmids combined with agomir-155 or controls, respectively. After 24 h of transfection, the cells were lysed and measured for luciferase activities using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA), following the manufacturer’s protocols.
Cell Proliferation Assays
'Cell proliferation was measured using the CCK-8 (Dojindo Laboratories, Japan) and BrdU incorporation assays, respectively. For the CCK-8 assay, cells were plated at a density of 4 × 104 cells/well in 96-well plates for 1–2 h. After treatment, 10 μL CCK-8 solution was added to each well, and the wells were incubated continuously for 3 h. The optical density (OD) was measured at a wavelength of 450 nm on a microplate reader (Bio-Rad, Hercules, CA, USA). Alternatively, the BrdU incorporation assay was carried out as previously described.65 Cells were incubated with BrdU (100 nM; Sigma) for 4 h and then subjected to BrdU immunostaining. Fluorescent images were obtained using a fluorescence microscope (AX70, Olympus, Japan).
Flow Cytometry Analysis
The cells were scraped from the culture dish and then washed three times with cold PBS. The polarization of macrophages was assessd by multi-color flow cytometry, as described previously.66 The data were obtained and further analyzed using FlowJo software (version 10.0.7; Tree Star, Ashland, OR, USA).
Statistical Analysis
All values were expressed as the means ± SEM. The results were analyzed using a two-tailed Student’s t test for comparison of two groups, and one-way ANOVA followed by Dunnett’s multiple comparison test when three groups were compared. For all analyses, a p value of <0.05 was considered to be statistically significant.
Author Contributions
K.J. and G.D. conceived and designed the experiments. K.J., J.Y., and S.G. carried out the experiments. K.J., J.Y., H.W., and G.Z. analyzed the data. K.J., J.Y., and G.D. wrote the manuscript. All authors agreed to be responsible for the content of the work.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (31772816).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.07.003.
Supplemental Information
References
- 1.Wu X.M., Liu M.Y., Xu Q., Guo W.W. Identification and characterization of microRNAs from citrus expressed sequence tags. Tree Genet. Genomes. 2011;7:117–133. [Google Scholar]
- 2.Li S., Wang L., Fu B., Berman M.A., Diallo A., Dorf M.E. TRIM65 regulates microRNA activity by ubiquitination of TNRC6. Proc. Natl. Acad. Sci. USA. 2014;111:6970–6975. doi: 10.1073/pnas.1322545111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rottiers V., Näär A.M. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang K., Guo S., Zhang T., Yang Y., Zhao G., Shaukat A., Wu H., Deng G. Downregulation of TLR4 by miR-181a Provides Negative Feedback Regulation to Lipopolysaccharide-Induced Inflammation. Front. Pharmacol. 2018;9:142. doi: 10.3389/fphar.2018.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai R., Ahmed S.A. MicroRNA, a new paradigm for understanding immunoregulation, inflammation, and autoimmune diseases. Transl. Res. 2011;157:163–179. doi: 10.1016/j.trsl.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalal S.R., Kwon J.H. The Role of MicroRNA in Inflammatory Bowel Disease. Gastroenterol. Hepatol. (N. Y.) 2010;6:714–722. [PMC free article] [PubMed] [Google Scholar]
- 7.Wang W., Liu Z., Su J., Chen W.S., Wang X.W., Bai S.X., Zhang J.Z., Yu S.Q. Macrophage microRNA-155 promotes lipopolysaccharide-induced acute lung injury in mice and rats. Am. J. Physiol. Lung Cell. Mol. Physiol. 2016;311:L494–L506. doi: 10.1152/ajplung.00001.2016. [DOI] [PubMed] [Google Scholar]
- 8.Banerjee N., Talcott S., Safe S., Mertens-Talcott S.U. Cytotoxicity of pomegranate polyphenolics in breast cancer cells in vitro and vivo: potential role of miRNA-27a and miRNA-155 in cell survival and inflammation. Breast Cancer Res. Treat. 2012;136:21–34. doi: 10.1007/s10549-012-2224-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Z., Liang K., Zou G., Chen X., Shi S., Wang G., Zhang K., Li K., Zhai S. Inhibition of miR-155 attenuates abdominal aortic aneurysm in mice by regulating macrophage-mediated inflammation. Biosci. Rep. 2018;38 doi: 10.1042/BSR20171432. BSR20171432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kosaka N., Iguchi H., Yoshioka Y., Takeshita F., Matsuki Y., Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexander M., O’Connell R. Exosomal miRNAs regulate inflammatory responses (IRM11P.624) J. Immunol. 2015;194(1 Suppl):132.3. [Google Scholar]
- 12.Borges F.T., Melo S.A., Özdemir B.C., Kato N., Revuelta I., Miller C.A., Gattone V.H., 2nd, LeBleu V.S., Kalluri R. TGF-β1-containing exosomes from injured epithelial cells activate fibroblasts to initiate tissue regenerative responses and fibrosis. J. Am. Soc. Nephrol. 2013;24:385–392. doi: 10.1681/ASN.2012101031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andre F., Schartz N.E., Movassagh M., Flament C., Pautier P., Morice P., Pomel C., Lhomme C., Escudier B., Le Chevalier T. Malignant effusions and immunogenic tumour-derived exosomes. Lancet. 2002;360:295–305. doi: 10.1016/S0140-6736(02)09552-1. [DOI] [PubMed] [Google Scholar]
- 14.Wang C., Zhang C., Liu L., A X., Chen B., Li Y., Du J. Macrophage-Derived mir-155-Containing Exosomes Suppress Fibroblast Proliferation and Promote Fibroblast Inflammation during Cardiac Injury. Mol. Ther. 2017;25:192–204. doi: 10.1016/j.ymthe.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li M., Zeringer E., Barta T., Schageman J., Cheng A., Vlassov A.V. Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369:20130502. doi: 10.1098/rstb.2013.0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bretz N.P., Ridinger J., Rupp A.K., Rimbach K., Keller S., Rupp C., Marmé F., Umansky L., Umansky V., Eigenbrod T. Body fluid exosomes promote secretion of inflammatory cytokines in monocytic cells via Toll-like receptor signaling. J. Biol. Chem. 2013;288:36691–36702. doi: 10.1074/jbc.M113.512806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexander M., Hu R., Runtsch M.C., Kagele D.A., Mosbruger T.L., Tolmachova T., Seabra M.C., Round J.L., Ward D.M., O’Connell R.M. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat. Commun. 2015;6:7321. doi: 10.1038/ncomms8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montecalvo A., Larregina A.T., Shufesky W.J., Stolz D.B., Sullivan M.L., Karlsson J.M., Baty C.J., Gibson G.A., Erdos G., Wang Z. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119:756–766. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Momen-Heravi F., Bala S., Kodys K., Szabo G. Exosomes derived from alcohol-treated hepatocytes horizontally transfer liver specific miRNA-122 and sensitize monocytes to LPS. Sci. Rep. 2015;5:9991. doi: 10.1038/srep09991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Liu D., Chen X., Li J., Li L., Bian Z., Sun F., Lu J., Yin Y., Cai X. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol. Cell. 2010;39:133–144. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 21.Jin Y., Choi A.M. Cytoprotection of heme oxygenase-1/carbon monoxide in lung injury. Proc. Am. Thorac. Soc. 2005;2:232–235. doi: 10.1513/pats.200503-023AC. [DOI] [PubMed] [Google Scholar]
- 22.Wheeler A.P., Bernard G.R. Acute lung injury and the acute respiratory distress syndrome: a clinical review. Lancet. 2007;369:1553–1564. doi: 10.1016/S0140-6736(07)60604-7. [DOI] [PubMed] [Google Scholar]
- 23.Phua J., Badia J.R., Adhikari N.K., Friedrich J.O., Fowler R.A., Singh J.M., Scales D.C., Stather D.R., Li A., Jones A. Has mortality from acute respiratory distress syndrome decreased over time?: A systematic review. Am. J. Respir. Crit. Care Med. 2009;179:220–227. doi: 10.1164/rccm.200805-722OC. [DOI] [PubMed] [Google Scholar]
- 24.Mura M., Han B., Andrade C.F., Seth R., Hwang D., Waddell T.K., Keshavjee S., Liu M. The early responses of VEGF and its receptors during acute lung injury: implication of VEGF in alveolar epithelial cell survival. Crit. Care. 2006;10 doi: 10.1186/cc5042. R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han S., Mallampalli R.K. The acute respiratory distress syndrome: from mechanism to translation. J. Immunol. 2015;194:855–860. doi: 10.4049/jimmunol.1402513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matute-Bello G., Frevert C.W., Martin T.R. Animal models of acute lung injury. Am. J. Physiol. Lung Cell. Mol. Physiol. 2008;295:L379–L399. doi: 10.1152/ajplung.00010.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopf M., Schneider C., Nobs S.P. The development and function of lung-resident macrophages and dendritic cells. Nat. Immunol. 2015;16:36–44. doi: 10.1038/ni.3052. [DOI] [PubMed] [Google Scholar]
- 28.Ward P.A. Role of complement, chemokines, and regulatory cytokines in acute lung injury. Ann. N Y Acad. Sci. 1996;796:104–112. doi: 10.1111/j.1749-6632.1996.tb32572.x. [DOI] [PubMed] [Google Scholar]
- 29.Wu L., Zhang X., Zhang B., Shi H., Yuan X., Sun Y., Pan Z., Qian H., Xu W. Exosomes derived from gastric cancer cells activate NF-κB pathway in macrophages to promote cancer progression. Tumour Biol. 2016;37:12169–12180. doi: 10.1007/s13277-016-5071-5. [DOI] [PubMed] [Google Scholar]
- 30.Jiang K.F., Zhao G., Deng G.Z., Wu H.C., Yin N.N., Chen X.Y., Qiu C.W., Peng X.L. Polydatin ameliorates Staphylococcus aureus-induced mastitis in mice via inhibiting TLR2-mediated activation of the p38 MAPK/NF-κB pathway. Acta Pharmacol. Sin. 2017;38:211–222. doi: 10.1038/aps.2016.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang K., Guo S., Yang C., Yang J., Chen Y., Shaukat A., Zhao G., Wu H., Deng G. Barbaloin protects against lipopolysaccharide (LPS)-induced acute lung injury by inhibiting the ROS-mediated PI3K/AKT/NF-κB pathway. Int. Immunopharmacol. 2018;64:140–150. doi: 10.1016/j.intimp.2018.08.023. [DOI] [PubMed] [Google Scholar]
- 32.Li J.J., Wang B., Kodali M.C., Chen C., Kim E., Patters B.J., Lan L., Kumar S., Wang X., Yue J., Liao F.F. In vivo evidence for the contribution of peripheral circulating inflammatory exosomes to neuroinflammation. J. Neuroinflammation. 2018;15:8. doi: 10.1186/s12974-017-1038-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kojima M., Gimenes-Junior J.A., Chan T.W., Eliceiri B.P., Baird A., Costantini T.W., Coimbra R. Exosomes in postshock mesenteric lymph are key mediators of acute lung injury triggering the macrophage activation via Toll-like receptor 4. FASEB J. 2017;32:97–110. doi: 10.1096/fj.201700488R. [DOI] [PubMed] [Google Scholar]
- 34.Ti D., Hao H., Tong C., Liu J., Dong L., Zheng J., Zhao Y., Liu H., Fu X., Han W. LPS-preconditioned mesenchymal stromal cells modify macrophage polarization for resolution of chronic inflammation via exosome-shuttled let-7b. J. Transl. Med. 2015;13:308. doi: 10.1186/s12967-015-0642-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poon K.S., Palanisamy K., Chang S.S., Sun K.T., Chen K.B., Li P.C., Lin T.C., Li C.Y. Plasma exosomal miR-223 expression regulates inflammatory responses during cardiac surgery with cardiopulmonary bypass. Sci. Rep. 2017;7:10807. doi: 10.1038/s41598-017-09709-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou P., Kitaura H., Teitelbaum S.L., Krystal G., Ross F.P., Takeshita S. SHIP1 negatively regulates proliferation of osteoclast precursors via Akt-dependent alterations in D-type cyclins and p27. J. Immunol. 2006;177:8777–8784. doi: 10.4049/jimmunol.177.12.8777. [DOI] [PubMed] [Google Scholar]
- 37.Béres N.J., Szabó D., Kocsis D., Szűcs D., Kiss Z., Müller K.E., Lendvai G., Kiss A., Arató A., Sziksz E. Role of Altered Expression of miR-146a, miR-155, and miR-122 in Pediatric Patients with Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2016;22:327–335. doi: 10.1097/MIB.0000000000000687. [DOI] [PubMed] [Google Scholar]
- 38.Bala S., Petrasek J., Ward J., Alao H., Levin I., Szabo G. Serum MicroRNA-122 and miR-155 as Biomarkers of Liver Injury and Inflammation in Models of Acute and Chronic Liver Disease. Gastroenterology. 2011;140(Suppl 1):S-906. [Google Scholar]
- 39.Chen Y., Liu W., Sun T., Huang Y., Wang Y., Deb D.K., Yoon D., Kong J., Thadhani R., Li Y.C. 1,25-Dihydroxyvitamin D promotes negative feedback regulation of TLR signaling via targeting microRNA-155-SOCS1 in macrophages. J. Immunol. 2013;190:3687–3695. doi: 10.4049/jimmunol.1203273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strebovsky J., Walker P., Lang R., Dalpke A.H. Suppressor of cytokine signaling 1 (SOCS1) limits NFkappaB signaling by decreasing p65 stability within the cell nucleus. FASEB J. 2011;25:863–874. doi: 10.1096/fj.10-170597. [DOI] [PubMed] [Google Scholar]
- 41.Alberti C., Brun-Buisson C., Goodman S.V., Guidici D., Granton J., Moreno R., Smithies M., Thomas O., Artigas A., Le Gall J.R., European Sepsis Group Influence of systemic inflammatory response syndrome and sepsis on outcome of critically ill infected patients. Am. J. Respir. Crit. Care Med. 2003;168:77–84. doi: 10.1164/rccm.200208-785OC. [DOI] [PubMed] [Google Scholar]
- 42.Aggarwal A., Baker C.S., Evans T.W., Haslam P.L. G-CSF and IL-8 but not GM-CSF correlate with severity of pulmonary neutrophilia in acute respiratory distress syndrome. Eur. Respir. J. 2000;15:895–901. doi: 10.1034/j.1399-3003.2000.15e14.x. [DOI] [PubMed] [Google Scholar]
- 43.Kooguchi K., Hashimoto S., Kobayashi A., Kitamura Y., Kudoh I., Wiener-Kronish J., Sawa T. Role of alveolar macrophages in initiation and regulation of inflammation in Pseudomonas aeruginosa pneumonia. Infect. Immun. 1998;66:3164–3169. doi: 10.1128/iai.66.7.3164-3169.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marriott H.M., Dockrell D.H. The role of the macrophage in lung disease mediated by bacteria. Exp. Lung Res. 2007;33:493–505. doi: 10.1080/01902140701756562. [DOI] [PubMed] [Google Scholar]
- 45.Esbenshade A.M., Newman J.H., Lams P.M., Jolles H., Brigham K.L. Respiratory failure after endotoxin infusion in sheep: lung mechanics and lung fluid balance. J. Appl. Physiol. 1982;53:967–976. doi: 10.1152/jappl.1982.53.4.967. [DOI] [PubMed] [Google Scholar]
- 46.Emery D.A., Nagaraja K.V., Sivanandan V., Lee B.W., Zhang C.L., Newman J.A. Endotoxin lipopolysaccharide from Escherichia coli and its effects on the phagocytic function of systemic and pulmonary macrophages in turkeys. Avian Dis. 1991;35:901–909. [PubMed] [Google Scholar]
- 47.Takaoka Y., Goto S., Nakano T., Tseng H.P., Yang S.M., Kawamoto S., Ono K., Chen C.L. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) prevents lipopolysaccharide (LPS)-induced, sepsis-related severe acute lung injury in mice. Sci. Rep. 2014;4:5204. doi: 10.1038/srep05204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morelli A.E., Larregina A.T., Shufesky W.J., Sullivan M.L., Stolz D.B., Papworth G.D., Zahorchak A.F., Logar A.J., Wang Z., Watkins S.C. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood. 2004;104:3257–3266. doi: 10.1182/blood-2004-03-0824. [DOI] [PubMed] [Google Scholar]
- 49.Zhang H., Liu J., Qu D., Wang L., Wong C.M., Lau C.W., Huang Y., Wang Y.F., Huang H., Xia Y. Serum exosomes mediate delivery of arginase 1 as a novel mechanism for endothelial dysfunction in diabetes. Proc. Natl. Acad. Sci. USA. 2018;115:E6927–E6936. doi: 10.1073/pnas.1721521115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gambim M.H., do Carmo Ade.O., Marti L., Veríssimo-Filho S., Lopes L.R., Janiszewski M. Platelet-derived exosomes induce endothelial cell apoptosis through peroxynitrite generation: experimental evidence for a novel mechanism of septic vascular dysfunction. Crit. Care. 2007;11:R107. doi: 10.1186/cc6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bonjoch L., Casas V., Carrascal M., Closa D. Involvement of exosomes in lung inflammation associated with experimental acute pancreatitis. J. Pathol. 2016;240:235–245. doi: 10.1002/path.4771. [DOI] [PubMed] [Google Scholar]
- 52.Ying W., Riopel M., Bandyopadhyay G., Dong Y., Birmingham A., Seo J.B., Ofrecio J.M., Wollam J., Hernandez-Carretero A., Fu W. Adipose Tissue Macrophage-Derived Exosomal miRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell. 2017;171:372–384.e12. doi: 10.1016/j.cell.2017.08.035. [DOI] [PubMed] [Google Scholar]
- 53.Binenbaum Y., Fridman E., Yaari Z., Milman N., Schroeder A., Ben-David G., Shlomi T., Gil Z. Transfer of miRNA in macrophage-derived exosomes induces drug resistance in pancreatic adenocarcinoma. Cancer Res. 2018;78:5287–5299. doi: 10.1158/0008-5472.CAN-18-0124. [DOI] [PubMed] [Google Scholar]
- 54.Nicholas J. A new diagnostic tool with the potential to predict tumor metastasis. J. Natl. Cancer Inst. 2013;105:371–372. doi: 10.1093/jnci/djt048. [DOI] [PubMed] [Google Scholar]
- 55.Azevedo L.C.P., Real J.M., Bezerra J.E., Machado F.R., Salomao R. Microparticles from septic shock patients contain microRNA and messenger RNA: new players in the pathogenesis of sepsis? Crit. Care. 2013;17(Suppl 4):P96. [Google Scholar]
- 56.Rao R., Rieder S.A., Nagarkatti P., Nagarkatti M. Staphylococcal enterotoxin B-induced microRNA-155 targets SOCS1 to promote acute inflammatory lung injury. Infect. Immun. 2014;82:2971–2979. doi: 10.1128/IAI.01666-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ham S., Lima L.G., Chai E.P.Z., Muller A., Lobb R.J., Krumeich S., Wen S.W., Wiegmans A.P., Möller A. Breast Cancer-Derived Exosomes Alter Macrophage Polarization via gp130/STAT3 Signaling. Front. Immunol. 2018;9:871. doi: 10.3389/fimmu.2018.00871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nowek K., Sun S.M., Bullinger L., Bindels E.M., Exalto C., Dijkstra M.K., van Lom K., Döhner H., Erkeland S.J., Löwenberg B., Jongen-Lavrencic M. Aberrant expression of miR-9/9* in myeloid progenitors inhibits neutrophil differentiation by post-transcriptional regulation of ERG. Leukemia. 2016;30:229–237. doi: 10.1038/leu.2015.183. [DOI] [PubMed] [Google Scholar]
- 59.Condé C., Gloire G., Piette J. Enzymatic and non-enzymatic activities of SHIP-1 in signal transduction and cancer. Biochem. Pharmacol. 2011;82:1320–1334. doi: 10.1016/j.bcp.2011.05.031. [DOI] [PubMed] [Google Scholar]
- 60.Koff A. How to decrease p27Kip1 levels during tumor development. Cancer Cell. 2006;9:75–76. doi: 10.1016/j.ccr.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 61.Wang X.L., Qiao C.M., Liu J.O., Li C.Y. Inhibition of the SOCS1-JAK2-STAT3 Signaling Pathway Confers Neuroprotection in Rats with Ischemic Stroke. Cell. Physiol. Biochem. 2017;44:85–98. doi: 10.1159/000484585. [DOI] [PubMed] [Google Scholar]
- 62.Sachithanandan N., Graham K.L., Galic S., Honeyman J.E., Fynch S.L., Hewitt K.A., Steinberg G.R., Kay T.W. Macrophage deletion of SOCS1 increases sensitivity to LPS and palmitic acid and results in systemic inflammation and hepatic insulin resistance. Diabetes. 2011;60:2023–2031. doi: 10.2337/db11-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee H., Zhang D., Zhu Z., Dela Cruz C.S., Jin Y. Epithelial cell-derived microvesicles activate macrophages and promote inflammation via microvesicle-containing microRNAs. Sci. Rep. 2016;6:35250. doi: 10.1038/srep35250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bang C., Batkai S., Dangwal S., Gupta S.K., Foinquinos A., Holzmann A., Just A., Remke J., Zimmer K., Zeug A. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J. Clin. Invest. 2014;124:2136–2146. doi: 10.1172/JCI70577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Calkins M.J., Reddy P.H. Assessment of newly synthesized mitochondrial DNA using BrdU labeling in primary neurons from Alzheimer’s disease mice: Implications for impaired mitochondrial biogenesis and synaptic damage. Biochim. Biophys. Acta. 2011;1812:1182–1189. doi: 10.1016/j.bbadis.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang J., Li S., Wang L., Du F., Zhou X., Song Q., Zhao J., Fang R. Ginsenoside Rg3 Attenuates Lipopolysaccharide-Induced Acute Lung Injury via MerTK-Dependent Activation of the PI3K/AKT/mTOR Pathway. Front. Pharmacol. 2018;9:850. doi: 10.3389/fphar.2018.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.