Abstract
Cystic fibrosis (CF) is an autosomal recessive monogenic disease caused by mutations in the CFTR gene. Therapeutic approaches that are focused on correcting CFTR protein face the challenge of the heterogeneity in CFTR mutations and resulting defects. Thus, while several small molecules directed at CFTR show benefit in the clinic for subsets of CF patients, these drugs cannot treat all CF patients. Additionally, the clinical benefit from treatment with these modulators could be enhanced with novel therapies.
To address this unmet need, we utilized an approach to increase CFTR protein levels through antisense oligonucleotide (ASO)-mediated steric inhibition of 5′ UTR regulatory elements. We identified ASOs to upregulate CFTR protein expression and confirmed the regulatory role of the sites amenable to ASO-mediated upregulation. Two ASOs were investigated further, and both increased CFTR protein expression and function in cell lines and primary human bronchial epithelial cells with distinct CF genotypes. ASO treatment further increased CFTR function in almost all CF genotypes tested on top of treatment with the FDA approved drug Symdeko (ivacaftor and tezacaftor). Thus, we present a novel approach to CFTR therapeutic intervention, through ASO-mediated modulation of translation.
In this issue, Sasaki et al. identified antisense oligonucleotides (ASOs) to increase CFTR protein expression through steric inhibition of 5′ UTR regulatory elements. CFTR function was also increased in several models of cystic fibrosis. Thus, the authors present a novel approach to CFTR therapeutic intervention, through ASO-mediated modulation of translation.
Introduction
Cystic fibrosis (CF) is an autosomal recessive monogenic disease caused by mutations in the CFTR gene that result in dysfunctional CFTR protein production, folding, trafficking, and ion channel activity.1 CFTR is an epithelial anion channel critical for chloride and bicarbonate transport and airway hydration in the lung. CFTR defects impair its function in many tissues, including the lungs, liver, intestine, and pancreas. Airway dehydration and impaired mucociliary clearance occur in the absence of functional CFTR, enabling persistent infection of the airways and progressive lung disease, which is the primary cause of morbidity and mortality in CF patients.2
Therapeutic approaches for CF have focused both on restoring airway hydration and correcting defective CFTR protein. Symptomatic treatments such as mucolytic agents and enzyme replacement therapies have substantially increased patient lifespan, but they do not address the underlying cause of disease. A significant challenge is the considerable number of CFTR mutations that give rise to CF, which have heterogeneous effects on CFTR protein. Disease-causing mutations have been classified into six groups, based on the resulting effect on CFTR protein: mutations (splicing or nonsense mutations) that result in no protein (I), protein that does not traffic to the cell membrane (II), protein that is not functional due to gating defects (III), protein that has limited function (IV), lower protein expression (V), and protein that is not stable at the cell surface (VI). In the past decade, several small molecule modifiers of CFTR protein have shown benefit in the clinic for subsets of CF patients. For example, ivacaftor (Kalydeco) is a potentiator that can specifically improve outcomes for patients with gating-defective CFTR (class III mutations—G551D, etc.). Similarly, trafficking corrector compounds, such as lumacaftor and the more potent compound tezacaftor, increase the amount of CFTR at the cell membrane for patients with improperly trafficked CFTR (class II mutations—F508del, N1303K, etc.). Unfortunately, even when used in combination, these drugs cannot treat all CF patients, due to the heterogeneity of CFTR defects. Additionally, the clinical benefit from treatment with these modulators is heterogeneous and could be increased with novel therapies. Thus, there remains a significant unmet need for a therapy that would benefit all CF patients.
We hypothesized that an approach to increase CFTR protein levels, in a mutation-agnostic manner, would be beneficial either in combination with existing therapies for patients with mutations resulting in defective CFTR function or as a stand-alone treatment for patients with mutations resulting in diminished CFTR protein expression. Previous reports have demonstrated that CFTR is regulated at the level of translation, through an upstream open reading frame (uORF) and putative secondary structure element in the 5′ UTR and through microRNA (miRNA) and RNA-binding protein sites in the 3′ UTR.3, 4, 5 Thus, we sought to increase CFTR translation through the use of antisense oligonucleotides (ASOs).
ASOs act on RNA to modulate splicing, elicit degradation of the transcript, or activate or inhibit translation through highly specific hybridization to a target RNA. There are several approved ASO drugs for indications such as spinal muscular atrophy, familial hypercholesterolemia, and hereditary transthyretin amyloidosis, validating the clinical application of this class of therapeutics.6, 7 ASOs are synthetic single strands of 16–20 nucleotides and are chemically modified to increase pharmacological properties and nuclease resistance. They are engineered to regulate mRNA and protein expression with nucleotide specificity through several distinct mechanisms. RNase H1-mediated endonucleolytic cleavage of a target transcript is the most commonly used mechanism of ASO action, which results in degradation of the target mRNA.6, 8 ASOs to recruit RNase H1 are designed with a DNA-like “gap” lacking sugar modification that results in recognition by RNase H1 upon hybridization to the target mRNA. The “gap” is flanked by “wings” that have sugar modifications to enhance stability and affinity for the target. ASOs that are modified uniformly, without an unmodified “gap,” can be used to mask regulatory elements in a transcript via steric blocking. This is exemplified by ASOs that modulate splicing, such as the approved drugs Spinraza and etiplirsen. More recently, uniformly modified ASOs have been used to increase translation of specific transcripts through steric inhibition of 5′ UTR regulatory elements, such as uORFs or secondary structure elements.9, 10, 11
We sought to increase CFTR expression levels through ASO-mediated steric inhibition of 5′ UTR regulatory elements. We designed ASOs to tile the 5′ UTR at single-nucleotide resolution, resulting in the identification of ASOs that increased CFTR protein expression and function in relevant cell lines and preclinical CF models. Our data exemplifies a novel approach to CF drug discovery and represents the first application of this ASO mechanism to a therapeutic target.
Results
Identification of Putative Translation Regulatory Elements in the 5′ UTR of CFTR mRNA
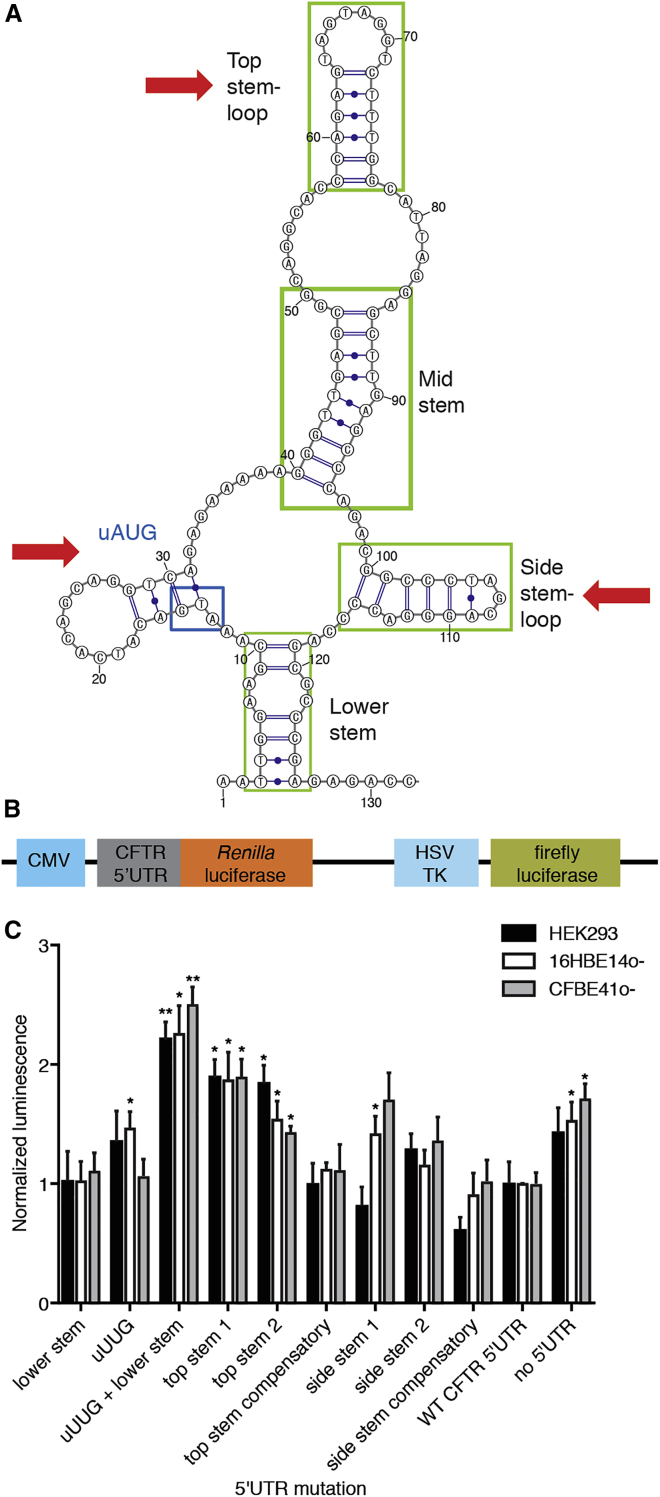
We performed bioinformatic analysis of the 5′ UTR to identify sites of potential translational regulation. Upon structure prediction analysis of the 5′ UTR, we identified several stem loops that we hypothesized may contribute to regulation of CFTR translation, indicated in green boxes (Figure 1A). The “top stem loop” and “mid stem” also had significant overlap with a previously identified structural element, which in reporter assays was shown to co-regulate reporter mRNA translation in combination with the uORF.3 We also identified additional structured regions of the 5′ UTR that have not been previously reported (Figure 1A, “side stem loop” and “lower stem”).
Figure 1.
Identification of Regulatory Elements in the CFTR 5′ UTR
(A) Structure prediction of the human CFTR 5′ UTR. Structures of potential interest are boxed in green; the uAUG site is boxed in blue. Red arrows indicate regions of overlap with the ASO screen. (B) Schematic of reporter construct. CMV, CMV promoter; HSV TK, HSV TK promoter. (C) Representative reporter assay data in three cell lines for various mutations introduced into the CFTR 5′ UTR. Data are mean + SEM. *p < 0.05, **p < 0.005 by Holm-Sidak method, α = 0.05.
To determine if inhibition of elements in the 5′ UTR could increase CFTR protein expression, ASOs were designed to tile the full 132-nucleotide 5′ UTR at single-nucleotide resolution. An ELISA was developed to detect CFTR protein changes in a high-throughput manner for use in ASO screens (Figure S1). This assay was validated in several cell lines with varying levels of CFTR expression. 16HBE14o− cells were selected for ASO screens, as they had easily detectable levels of CFTR protein expression and have previously been demonstrated to be amenable to CFTR functional assays.12
An initial screen of the 5′ UTR was performed using 16-nucleotide ASOs with phosphorothioate backbone and 2′ methoxy modification, based on previously demonstrated upregulation of other targets with this chemistry.10 Three regions emerged as most amenable to upregulation, with CFTR protein expression increased by 3- to 4-fold. These regions were consistent with two stem loops in the predicted structure (Figure 1A, “top stem loop” and “side stem”). Additionally, the previously reported upstream AUG site (Figure 1A, outlined in blue box), which is predicted to initiate translation of an uORF,3 was blocked by several ASOs that increased CFTR expression.
Confirmation of a Regulatory Role for uAUG and Predicted Structure in the 5′ UTR
To test the regions of interest for potential regulatory effects on translation, we engineered a dual luciferase reporter assay system. The Renilla luciferase open reading frame was placed immediately downstream of the CFTR 5′ UTR, and firefly luciferase was inserted after a distinct promoter to serve as a control for transfection efficiency of this reporter construct (Figure 1B). We performed mutational analysis of the 5′ UTR and evaluated effects on translation of the reporter in three distinct cell lines: HEK293, 16HBE14o−, and CFBE41o−. HEK293 cells are commonly used for reporter assays and were also used for ease of transfection. 16HBE14o− and CFBE41o− cells are well characterized human bronchial epithelial cells used for CFTR research, homozygous for wild-type or F508del-CFTR, respectively.
The wild-type 5′ UTR (“WT CFTR 5′ UTR”) on its own exhibited decreased reporter translation compared to a construct lacking the CFTR 5′ UTR (Figure 1C, “no 5′ UTR”). Mutation of the uAUG to uUUG paired with mutations that disrupted some 5′ structure (Figure 1C, “uUUG + lower stem”) significantly enhanced translation compared to a construct with the wild-type 5′ UTR. Similar increases in reporter translation were observed upon mutation of either side of the top stem (Figure 1C, “top stem 1” and “top stem 2”). Restoration of base-pairing interactions through compensatory mutation fully reduced reporter translation back to wild-type levels (Figure 1C, “top stem compensatory”). Mutation of the side stem loop resulted in a similar effect, though to a lesser degree than the top stem or uUUG mutations (Figure 1C, “side stem 1,” “side stem 2,” and “side stem compensatory”). All three cell lines tested showed similar trends in reporter translation in response to the different mutations. These data support a translation regulatory role for the uAUG site and top and side stem loops that were identified as amenable to upregulation via ASO treatment.
ASOs Increase CFTR Surface-Localized Protein and Function in Wild-Type Cell Lines
Based on the reporter assay results, we selected two ASOs for additional analysis in wild-type CFTR cells, one that blocks the uAUG site (“uORF ASO”) and one that blocks the top stem of the predicted structure (“TSE ASO”). ASOs were delivered to cells without transfection (free uptake13, 14). A chemistry-matched scrambled ASO that has no human transcript targets was used as a control (“control ASO”).
The uORF ASO demonstrated a dose-responsive increase in CFTR protein expression in 16HBE14o− cells (Figure 2A). The translation suppression element (TSE) ASO increased CFTR protein expression to a lesser extent (3-fold) and did not exhibit a clear dose response. The duration of effect was 24–36 h for both uORF and TSE ASOs.
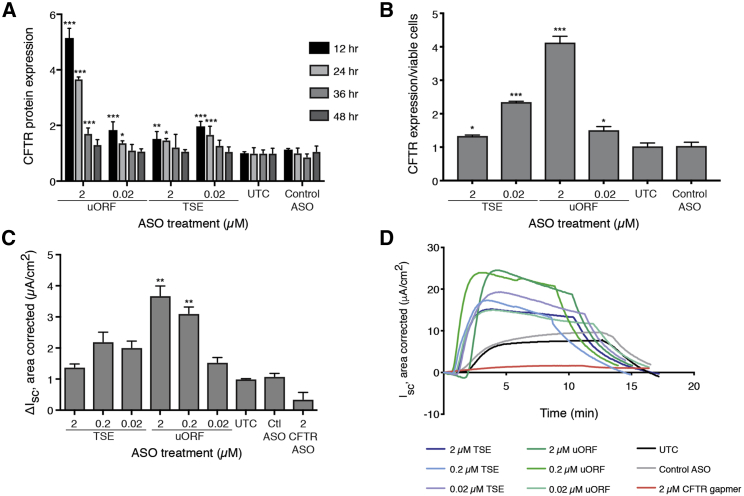
Figure 2.
Effect of ASOs to Upregulate CFTR in 16HBE14o− Cells
(A) CFTR protein expression after indicated ASO treatments, normalized to untreated control (UTC). *p < 0.05, **p < 0.005, ***p < 0.0005 by two-way ANOVA with Dunnett’s multiple-comparison test. (B) CFTR surface-localized protein expression. *p < 0.05, ***p < 0.0005 by Holm-Sidak method. (C) CFTR function, measured by Ussing chamber and quantified relative to untreated control. Mean electrophysiological data for ASO-treated 16HBE14o− cells. Data is normalized to untreated control (UTC). Ctl, Control ASO; CFTR ASO, CFTR gapmer ASO. Amiloride (100 μM) was added to block the epithelial sodium channel, followed by forskolin (10 μM) to stimulate CFTR and the inhibitor 172 (CFTRinh-172, 50 μM) to inhibit CFTR. Data are mean + SEM. *p < 0.05, **p < 0.005 by Holm-Sidak method, α = 0.05. (D) Representative traces of functional response in Ussing chambers after the indicated ASO treatments in 16HBE14o− cells.
CFTR requires trafficking to the cell surface to function as a chloride channel. Thus, we sought to determine the effect of ASO treatment on CFTR cell surface expression. We developed an assay to specifically detect surface-localized CFTR through use of a CFTR antibody that recognizes an extracellular loop of the protein (Figure S2). Upon ASO treatment, surface-localized CFTR levels were increased up to 4-fold with uORF ASO treatment and about 2-fold with TSE ASO treatment (Figure 2B), similar to the increases seen in the CFTR ELISA.
We reasoned that since both ASOs could increase surface-localized CFTR protein levels, they should also increase CFTR channel activity; therefore, we next sought to determine the effect of TSE and uORF ASO treatment on chloride transport in 16HBE14o− cells. After basolateral delivery of the ASOs to cells grown on transwell inserts, Ussing chamber assays were performed to measure the CFTR-specific ion current. CFTR was activated with forskolin, potentiated with the modulator VX-770, and inhibited with CFTR-Inh 172. Treatment with TSE and uORF ASOs increased CFTR function by 2- to 4-fold, compared to the untreated control. uORF ASO treatment had dose-responsive effects on CFTR function (Figure 2C), consistent with the CFTR protein expression effects (Figure 2A). The control ASO had no effect on CFTR function, as expected. A “gapmer” ASO to knockdown CFTR through RNase H1 recruitment was used as a positive control for ASO delivery to the cells. As expected, the gapmer ASO reduced CFTR function significantly. This is consistent with mRNA and protein reductions in CFTR with this ASO (Figures S3C and S3D).
ASOs Are Effective at Increasing CFTR Expression in Cell Lines with Trafficking-Defective CFTR
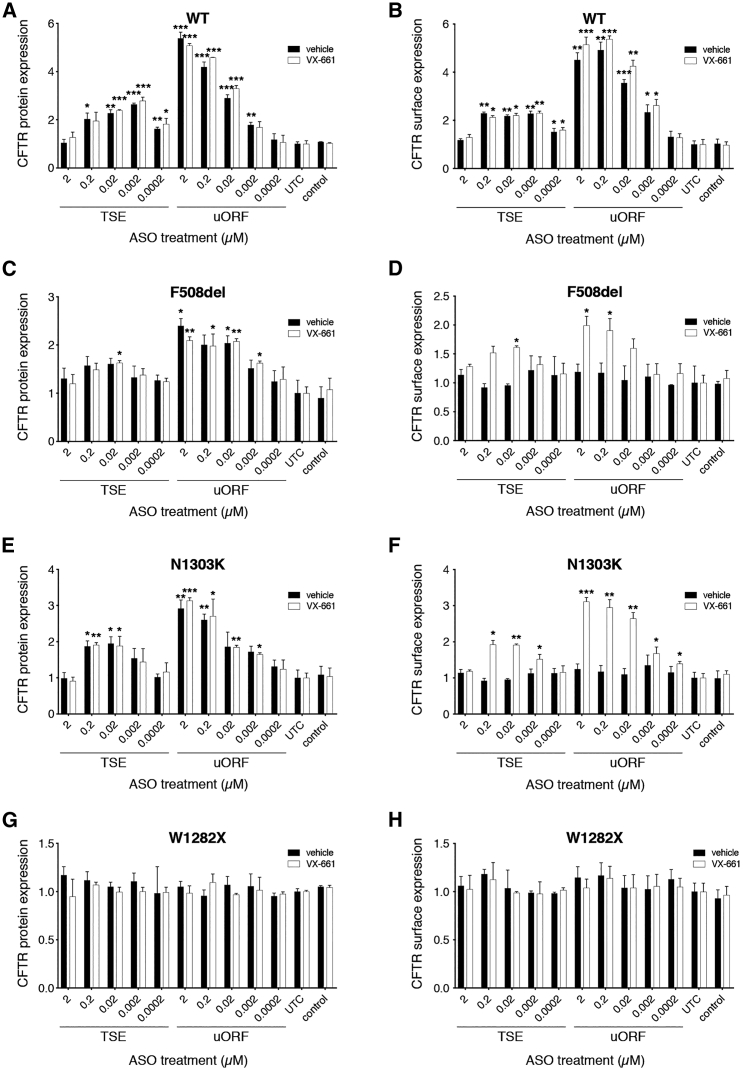
To determine the effect of ASO treatment on mutated CFTR, we utilized gene-edited 16HBE14o− cell lines homozygous for distinct CFTR mutations, generated by Cystic Fibrosis Foundation Therapeutics (CFF-16HBEge cells).12 We first tested CFTR mRNA expression, protein expression, and function in the relevant assays and confirmed that W1282X cells showed reduced CFTR mRNA and protein, F508del and N1303K cells had similar CFTR mRNA levels relative to the parental 16HBE14o− cells but decreased CFTR protein, and all demonstrated defective CFTR function (Figure S4). Treatment with either the uORF or TSE ASO had no effect on CFTR mRNA levels in W1282X, N1303K, or F508del cells (Figure S5). Combination of ASO with VX-661 (tezacaftor), a clinically effective trafficking corrector compound,15 also did not affect mRNA levels. These data indicate that the ASOs increase CFTR protein without affecting mRNA stability, in agreement with the effect on translation reported for these types of ASOs.10, 11 Total CFTR protein levels were increased in a dose-responsive manner in WT, N1303K, and F508del cells with uORF ASO treatment, with similar degrees of upregulation observed with and without VX-661 treatment (Figures 3A, 3C, and 3E). W1282X-CFTR levels were not affected by uORF or TSE ASO treatment (Figures 3G and 3H). TSE ASO treatment had very similar effects on CFTR mRNA and total protein, to a lesser extent and without dose response. Surface-localized CFTR was also increased with uORF and TSE ASO treatment, but only in combination with VX-661 in mutant cells (Figures 3B, 3D, and 3F). Taken together, these data indicate that uORF and TSE ASOs increase trafficking-defective CFTR protein, but not nonsense-mutated CFTR, at the relevant cellular location.
Figure 3.
Effect of ASO Combination with Corrector Compounds in Cell Lines with Defective CFTR
CFTR protein expression with and without the corrector compound VX-661 (18 μM) after indicated ASO treatments in 16HBE14o− (WT) cells (A), CFF-16HBEge-F508del cells (C), CFF-16HBEge-N1303K cells (E), and CFF-16HBEge-W1282X cells (G). Surface-localized CFTR protein expression for each cell line is represented in (B), (D), (F), and (H). All data are normalized to untreated control (UTC). Data are mean + SEM. *p < 0.05, **p < 0.005, ***p < 0.0005 by Holm-Sidak method, α = 0.05.
ASOs Increase Defective CFTR Function in Combination with Correctors in Cell Lines
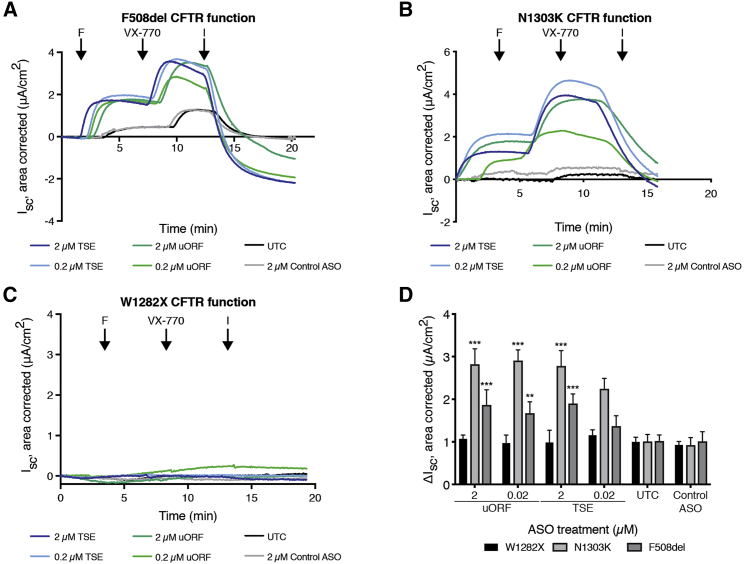
We sought to determine the effect of ASO-mediated upregulation on CFTR function in the CFF-16HBEge cell lines. We treated cells with ASO through the basolateral route three times prior to Ussing chamber measurement. Representative tracings for F508del-CFTR demonstrate a significant 2- to 3-fold improvement in forskolin and VX-770 activated current after uORF or TSE ASO treatment, with VX-661 combination (Figures 4A and 4D). The same conditions were tested for W1282X and N1303K cells and quantified relative to VX-661 treatment alone (Figures 4B–4D). Both uORF and TSE ASOs increased N1303K and F508del function but had no effect on W1282X function.
Figure 4.
Functional Effect of ASO Combination with Corrector Compounds in Cell Lines with Defective CFTR
(A–C) Representative traces of functional response in Ussing chambers after ASO treatment in CFF-16HBEge-F508del (A), CFF-16HBEge-N1303K (B), and CFF-16HBEge-W1282X (C) cells. Amiloride (100 μM) was added to block the epithelial sodium channel, followed by forskolin (10 μM) to stimulate CFTR, VX-770 (10 μM) to potentiate CFTR, and the inhibitor 172 (CFTRinh-172, 50 μM) to inhibit CFTR. F, forskolin; I, CFTR inhibitor 172. (D) Mean electrophysiological data for all CFF-16HBEge cell lines tested. Data is normalized to untreated control (UTC). Two-way ANOVA analysis was conducted with Dunnett’s multiple-comparison test between each mean and the mean of the untreated control. Data are mean + SEM. ***p < 0.0005, **p < 0.005.
ASOs Increase Defective CFTR in Combination with Correctors in Clinically Relevant Models of CF
Patient-derived primary human bronchial epithelial (hBE) cells cultured at the air-liquid interface are a clinically relevant model of a pseudostratified airway epithelium, with detectable cilia beating and airway surface liquid, as well as measurable CFTR function that correlates well with spirometry measures in patients (FEV1).16 Thus, we next tested ASO effects in this model. As these cells form tight monolayers, we first confirmed ASO delivery through use of the RNaseH1 ASO to knockdown CFTR (Figure S6). CFTR mRNA levels were reduced dose-dependently, compared to the control ASO and relative to the untreated control in cells from a healthy donor (Figure S6A). Dose-dependent loss of CFTR function was observed in Ussing chamber assays conducted after treatment with the RNaseH1 ASO (Figure S6B), indicating that ASOs are effectively delivered to primary hBE cells and can modulate CFTR function in this model.
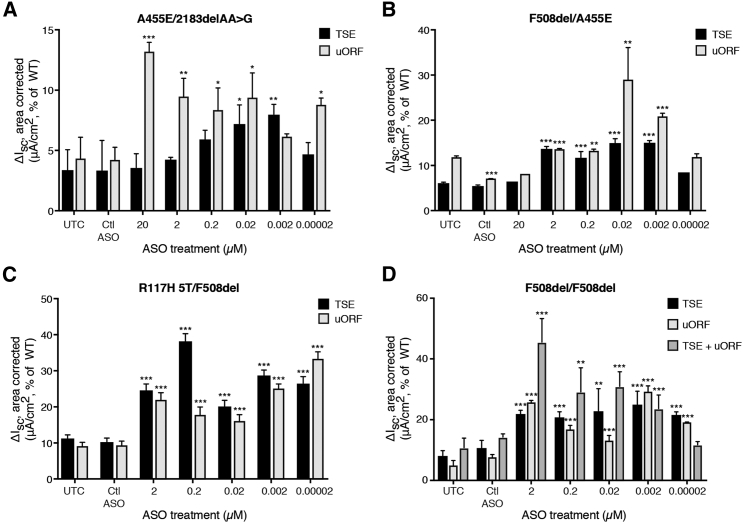
Next, uORF and TSE ASOs were delivered to cells with four distinct CF genotypes to test the effect of ASO treatment on CFTR from different mutation classes. We tested the effect of upregulation ASO treatment in cells from one F508del/R117H 5T/9T donor, one A455E/2183delAA > G donor, one F508del/A455E donor, and two F508del/F508del donors.
In A455E/2183AA→G cells, VX-661/770-corrected CFTR function was increased with ASO treatment by 2- to 4-fold relative to VX-661/770 treatment alone (UTC, Figure 5A). A455E is a common missense mutation in a highly conserved nucleotide-binding domain of CFTR that results in both reduced protein expression and stability as well as impaired function.17, 18 2183AA→G is a rare frameshift mutation in exon 13 that leads to premature termination of CFTR translation.19, 20, 21 This mutation has a prevalence of ≤ 1% and on its own is not responsive to current modulator therapies. Thus, for these cells, basal function was quite low relative to the wild-type donor cells, likely representative of A455E-CFTR function alone. ASO treatment therefore only restored defective CFTR function to 15% of WT (Figure 5A). Similar effects were observed with ASO treatment to F508del/A455E cells (Figure 5B).
Figure 5.
Effect of ASOs on CFTR Function in CF Patient-Derived Primary Cells
Mean electrophysiological data for ASO-treated primary hBE cells from A455E/2183delAA > G (A), F508del/A455E (B), R117H 5T/F508del (C), and F508del/F508del (D) donors. Data are represented as percent of WT donor function. n ≥ 5 per donor. Data are mean + SEM. *p < 0.05, **p < 0.005, ***p < 0.0005 by Holm-Sidak method, α = 0.05.
In R117H 5T/F508del cells, we again observed 2- to 4-fold increases in CFTR function with either uORF or TSE ASO treatment (Figure 5C). R117H results in a conductance defect that can be rescued by VX-770 treatment. F508del is a mutation in NBD1 that causes improper CFTR post-translational processing and therefore does not traffic to the cell surface. F508del trafficking can be restored with VX-661 treatment, which results in CFTR channels with a gating defect. Thus, VX-661 and VX-770 are co-administered to patients with at least one F508del allele. Surprisingly, even with both VX-661 and VX-770 treatment, function was only about 10% of WT (Figure 5C). Thus, ASO treatment resulted in increases of up to ∼30%–40% of WT function.
In F508del homozygous cells, both ASOs increased VX-661/770-corrected CFTR function by at least 2-fold (Figure 5D). Interestingly, the combination of both uORF and TSE ASOs at equal concentrations resulted in a more robust and dose-responsive effect on F508del-CFTR function. Compared to cells from a wild-type donor, function was restored to almost 45%.
Discussion
Here, we demonstrate the first application of ASOs to increase the expression of a therapeutic target in multiple cell lines and CF patient-derived primary cells, a clinically relevant disease model. We identified ASOs to upregulate CFTR protein expression through steric inhibition of both previously reported and novel regulatory sites. Two ASOs were selected for further analysis, and both increased CFTR protein expression and function in cell lines and hBE cells with distinct CF genotypes. Importantly, ASO treatment further increased CFTR function in almost all CF genotypes tested on top of ivacaftor and tezacaftor (Symdeko) treatment. Our data suggest a novel approach for CFTR modulation at the level of translation, which may provide benefit in the clinic, particularly when combined with existing CFTR modulator therapies.
The reporter assay revealed multiple sites of regulation within the 5′ UTR of the primary CFTR transcript. The uORF was previously reported to inhibit translation of a reporter RNA in HT29 and HEK293 cells.3 Here, we show that it contributes to decreased translation of a reporter in 16HBE14o− and CFBE41o− cells, suggestive of a conserved mechanism of translational control of CFTR mRNA. We also demonstrate that blocking the uAUG and structured sites increases CFTR protein levels in CFTR-defective cell lines and primary cells, without an effect on CFTR steady-state mRNA levels, supportive of the expected effect on translation. Further mechanistic analysis will be helpful in determining whether that is indeed the case and which steps of translation are altered. Other studies have demonstrated that ASOs to block 5′ UTR elements affect protein expression through increased ribosome occupancy, dependent on helicase activity.10, 11 It is likely that the ASOs that upregulate CFTR protein act through a similar mechanism, but additional experiments are required to test this hypothesis. Our data builds on and supports previous studies and indicates a path for therapeutic development.
CFTR is an attractive target, as mutations in CFTR give rise to the monogenic disease CF. There are existing drugs for CF patients, but many of them are either symptomatic or specific to subsets of patients. The strategy reported here would enable increased CFTR translation for most CFTR mutation classes. For mutations that result in CFTR that cannot traffic to the membrane or transport ion effectively, this strategy would have to be paired with an approach to correct the defect. Combination of 5′ UTR-binding ASOs with trafficking corrector compounds increases CFTR function greater than corrector compound treatment alone for F508del and N1303K-CFTR. Mutations that are subject to nonsense-mediated mRNA decay (NMD) may also require combination with other pharmacological agents, most likely including NMD inhibitors. The NMD-targeted mutation W1282X-CFTR did not respond to 5′ UTR-binding ASO treatment. The W1282X-CFTR mRNA may be too unstable for 5′ UTR ASOs to act on, or the effect on W1282X-CFTR protein and function levels may have been too minor to detect, due to the sensitivity of the assays. Future studies to combine NMD inhibition with 5′ UTR ASOs would clarify the lack of effect and may result in increased W1282X-CFTR protein and function. Taken together, our data suggest that increasing the pool of defective CFTR that is available for correction would provide clinically meaningful benefit, for mutations from four of the five classes tested in this study. Of the six mutation classes, only the gating-defective class of CFTR mutants was not evaluated. Future studies may provide insight into the application of this approach for gating-defective CFTR.
While this mechanism shows significant therapeutic potential, there remain some critical challenges. The dose dependence of both uORF and TSE ASOs is unclear, particularly in primary cell models. While the protein expression and chloride transport data are very consistent, both ASOs exhibit upregulation effects at a very large range of doses. The primary cells were treated with ASO multiple times to the basolateral compartment over the course of 1 week, and these cells are no longer proliferating. Thus, there could be significant accumulation of ASO that may account for observed activity at very low doses. Ribosome occupancy and CFTR mRNA structure analyses after ASO treatment have been challenging due to low CFTR mRNA levels; therefore, a more complete mechanistic understanding of this activity remains elusive. Improved detection of CFTR with these methods may yield better understanding of the mechanism and the dose response.
Additionally, in vivo efficacy of upregulation ASOs remains to be evaluated. It will be critical for therapeutic development to determine if the ASOs achieve similar efficacy in a CF or CF-like lung as in primary hBE models and to determine the pharmacokinetic properties in vivo. Prior experience with gapmer ASOs to inhibit ENaC in animal models suggests that ASOs can distribute effectively to the lung and maintain similar activity and pharmacokinetics even in models that recapitulate the significant mucus accumulation seen in CF.22 It remains to be seen whether the same would hold true for ASOs that do not recruit RNase H1, as differences in chemical modification of the ASOs might influence their tissue distribution. Additionally, the dose response of uniformly modified ASOs for CFTR is distinct in cell models from that of RNase H1 ASOs that target CFTR. However, in vivo studies of ASOs to upregulate other targets have demonstrated similar effects to those observed in vitro.10 Studies in animal models possessing the human CFTR 5′ UTR, which have been intractable thus far, will help to elucidate this further.
Despite these challenges, this is an important extension of previously reported application of ASOs to increase translation.10, 11 Liang et al. demonstrated that ASOs could be used to block 5′ UTR elements to increase ribosome association with the start codon and thereby augment protein production for a number of candidate transcripts.9, 10, 11 Here, we applied this novel ASO strategy to a therapeutic target, CFTR, and showed potential for this mechanism in therapeutic development. This strategy can be extended to inhibition of other regulatory elements in the CFTR transcript, such as miRNA sites in the 3′ UTR, which have been shown to negatively regulate CFTR expression.4, 5 Additionally, this mechanism has broad applicability for therapeutic development. uORFs have been identified in many transcripts, some of which are implicated in disease.23 However, it remains to be seen how many of these uORFs negatively regulate translation of the downstream primary ORF and would therefore be viable therapeutic targets.
In summary, we demonstrate a novel application of ASO-mediated translational control to increase CFTR expression in clinically relevant models. Our data suggests that ASOs can effectively inhibit elements that have a regulatory effect on translation to increase protein expression of a therapeutic target. While this strategy still needs additional in vivo and clinical validation, it is an attractive and tractable alternative to mRNA replacement and gene therapy that can be applied to other loss-of-function diseases.
Materials and Methods
Structure Prediction
Secondary structure of human CFTR 5′ UTR (positions 1–132 of NM_000492.3) was predicted using RNAfold 2.1.8.24 Standard parameters were used for the prediction. The predicted 5′ UTR structure was visualized using VARNA.25
Cell Culture and Lysis
Calu-3, T84, and HeLa cells were cultured per ATCC guidelines.16HBE14o− and CFBE41o− cells (from Dieter Gruenert) and CFF-16HBEge CFTR cells (from the Cystic Fibrosis Foundation Therapeutics [CFFT] lab) were cultured as described.26 Cells in 96-well plates were washed three times with 150 μL cold PBS. 100 μL cold IP lysis buffer (Pierce immunoprecipitation [IP] lysis buffer with Halt protease inhibitors [Thermo Fisher, Carlsbad, CA, USA]) was added, and plates were shaken (4°C for 30 min) and transferred to a −80°C freezer for 12 h. After freeze-thaw, lysates were mixed by pipetting and protein was quantified (Pierce BCA).
hBE cells prepared as described previously were provided by Drs. Chris Penland (the Cystic Fibrosis Foundation) and Scott H. Randell (Marsico Lung Institute, The University of North Carolina at Chapel Hill, Chapel Hill, NC, USA).27 The cells were obtained under protocol #03-1396 approved by the University of North Carolina at Chapel Hill Biomedical Institutional Review Board. Cells were thawed and expanded in Pneumacult EX basal media (STEMCELL Technologies, Vancouver, Canada), supplemented with antibiotics, and then plated onto 3T3-conditioned media-coated transwell inserts at a density of 0.55 × 106 cells. Upon reaching confluence, apical media was removed. Cells were cultured at the air-liquid interface in Pneumacult air-liquid interface media (STEMCELL Technologies, Vancouver, Canada), with basal compartment media changes 2 to 3 times per week, for 4 to 5 weeks until fully differentiated. For mRNA and protein extraction, cells were washed apically with sodium bicarbonate to remove mucus. PBS washes to both compartments were conducted prior to application of 50 μL cold IP lysis buffer (for protein extraction) or 100 μL RLT buffer (QIAGEN, Hilden, Germany) for RNA extraction. Protein and RNA extraction were performed as for other cells.
ASO Delivery to Cells
ASOs were delivered to cells by free uptake. For the cell lines, cells were seeded in fresh media for 24 h and were treated with ASOs diluted in media to the final concentration indicated for each experiment. Primary hBE cells were treated, after reaching full differentiation (24–28 days after seeding), by replacing media in the basolateral compartment with ASO diluted in media. Media changes were conducted three times with ASO prior to Ussing chamber or other analysis.
mRNA Extraction and qRT-PCR
mRNA extraction and qRT-PCR were performed as described.28 Primer and probe sequences were as follows: forward, 5′-TCCTTTCCAACAACCTGAAC-3′; reverse, 5′-CAAGTCCACAGAAGGCAGAC-3′; probe, 5′-Fam-CCCCATGAGGAGTGCCACTTGC-Iowa Black-3′.
CFTR ELISA
All incubation steps were performed with shaking at 700 rpm. Between incubations, wells were washed three times with 150 μL PBS + 0.05% Tween-20 before addition of the next reagent. Small-spot streptavidin plates (L45SA, MesoScale Diagnostics [MSD], Rockville, MD, USA) were blocked for 1 h at room temperature with 150 μL/well 5% blocker A in PBS and coated with 0.075 pmol of Abcam CFTR antibody (ab181782) biotinylated with Pierce EZ-link sulfo-NHS-LC-biotin (Thermo Fisher, Carlsbad, CA, USA). After 1 h at room temperature, 25 μL sample was added to each well and incubated overnight at 4°C, followed by 1 h room temperature incubation in UNC596 antibody (1:1,000, in 1% block A + 0.1% block B + 0.1% block DR, all in PBS). Sulfo-tag α-mouse antibody was added at 1 μg/mL (in 1% block A in PBS). After 1 h at room temperature, plates were read in 1× MSD read buffer on a MSD Sector plate reader. Samples were analyzed in triplicate.
CFTR Surface Expression Assay
Cells were seeded onto high-bind plates (L15XB, MesoScale Diagnostics, Rockville, MD, USA) 16 h prior to ASO treatment and/or assay. Incubations were at 37°C, and wells were washed three times with PBS between incubations. Viability was measured by CellTiterFluor assay (Promega, Madison, WI, USA). Cells were incubated for 1 h with 10% fetal bovine serum (FBS) in PBS, followed by 1:500 CF3 antibody (Thermo Fisher, Carlsbad, CA, USA) for 1 h. MSD Sulfo-tag conjugated α-mouse secondary was added at 1:10,000 for 1 h prior to detection in 1× MSD read buffer without detergent on a MSD Sector plate reader.
Surface Biotinylation and Pulldown
Surface biotinylation and pulldown were performed as described, except that biotin labeling was conducted for 1 h.29
Western Blot
Lysates (25 μg) were separated on 3%–8% NuPage Tris-acetate gels (Thermo Fisher, Carlsbad, CA, USA) and transferred to polyvinylidene fluoride (PVDF) (Thermo Fisher, Carlsbad, CA, USA). Membranes were probed with UNC596 antibody (1:2,000, provided by John Riordan and CFFT) and α-tubulin (1:10,000, Cell Signaling Technologies, Danvers, MA, USA) or α-Na/K ATPase (1:100,000, Abcam, Cambridge, MA, USA) followed by α-rabbit IRDye680 (1:15,000) and α-mouse IRDye800 (1:15,000) and imaged on a LI-COR Odyssey.
Ussing Chamber Assay
Air-liquid interface cultures were mounted into Ussing chambers (Physiologic Instruments, San Diego, CA, USA) and voltage clamped. Buffers to establish a chloride gradient were as described.30 Short-circuit current and resistance were measured through Acquire and Analyze (Physiologic Instruments, San Diego, CA, USA). The following compounds were added acutely: 100 μM amiloride (Sigma, St. Louis, MO, USA) to the mucosal side, to inhibit the epithelial sodium channel; 10 μM forskolin (Sigma, St. Louis, MO, USA) to both sides to activate CFTR; 10 μM VX-770 (Selleck Chemicals, Houston, TX, USA) to the mucosal side to further potentiate CFTR; and 50 μM CFTR inhibitor 172 (received from Robert Bridges through CFFT) to the mucosal side to inhibit CFTR.
Reporter Assay
HEK293, 16HBE14o−, or CFBE41o− cells were seeded in triplicate wells in 96-well plates. 24 h later, cells were transfected with reporter plasmid using Lipofectamine 2000 per manufacturer’s instructions. After 24 h, reporter assay was conducted using the Dual-Glo luciferase reporter assay system (Promega, Madison, WI, USA) according to manufacturer’s instructions.
Cloning of Reporter Plasmids
The CFTR 5′ UTR (positions 1–132 of NM_000492.3) was ordered as a gBlock (IDT, Coralville, IA, USA) and inserted via Gibson assembly cloning,31 using a Gibson assembly kit (NEB, Ipswich, MA, USA), into the pcDNA5/FRT plasmid (Thermo Fisher, Carlsbad, CA, USA). A subsequent Gibson assembly was conducted to insert the dual luciferase reporter (amplified out of psiCHECK-2 [Promega, Madison, WI, USA]) immediately downstream of the CFTR 5′ UTR. A dual luciferase reporter control plasmid without the 5′ UTR was constructed as well. Site-directed mutagenesis was used to generate the mutations of interest using the Q5 site-directed mutagenesis kit (NEB, Ipswich, MA, USA).
Author Contributions
Conceptualization, S.S., J.R.C., B.P.M., S.G.; Methodology, S.S., R.S.; Investigation, S.S., R.S.; Writing – Original Draft, S.S.; Writing – Review & Editing, S.S. and S.G.; Funding Acquisition, J.R.C, B.P.M., S.G.; Resources, H.-H.B.; Supervision, J.R.C., B.P.M., S.G.
Conflicts of Interest
S.S., H.-H.B., J.R.C., B.P.M., and S.G. are employees of Ionis Pharmaceuticals.
Acknowledgments
We thank Chenguang Zhao, Dong Bai, and Min Zhang for help establishing assays and cell models and Tracy Reigle for help preparing the figures. We also thank Dieter Gruenert for the parental 16HBE14o− and CFBE41o− cell lines; Chris Penland and Scott Randell for the primary hBE cells; Cystic Fibrosis Foundation Therapeutics (CFFT) lab for the CFF-16HBEge cells; and Xue-hai Liang, Michael Migawa, Sue Freier, Melissa Keenan, and Lulu Huang for helpful discussions. Provision of cells by Dr. Randell was supported by a grant and contract from the Cystic Fibrosis Foundation (BOUCHE15R0 and CFFT Cell Resource). We thank Robert Bridges, PhD (Rosalind Franklin University of Medicine and Science and Cystic Fibrosis Foundation Therapeutics) for providing CFTR inhibitor 172 as well as John Riordan, PhD (University of North Carolina at Chapel Hill and Cystic Fibrosis Foundation Therapeutics) for providing the UNC596 antibody. This project was supported by a Cystic Fibrosis Foundation fellowship to S.S. (IONIS16X1-SC).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.06.016.
Supplemental Information
References
- 1.Mall M.A., Hartl D. CFTR: cystic fibrosis and beyond. Eur. Respir. J. 2014;44:1042–1054. doi: 10.1183/09031936.00228013. [DOI] [PubMed] [Google Scholar]
- 2.Ratjen F., Bell S.C., Rowe S.M., Goss C.H., Quittner A.L., Bush A. Cystic fibrosis. Nat. Rev. Dis. Primers. 2015;1:15010. doi: 10.1038/nrdp.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lukowski S.W., Rothnagel J.A., Trezise A.E. CFTR mRNA expression is regulated by an upstream open reading frame and RNA secondary structure in its 5′ untranslated region. Hum. Mol. Genet. 2015;24:899–912. doi: 10.1093/hmg/ddu501. [DOI] [PubMed] [Google Scholar]
- 4.Viart V., Bergougnoux A., Bonini J., Varilh J., Chiron R., Tabary O., Molinari N., Claustres M., Taulan-Cadars M. Transcription factors and miRNAs that regulate fetal to adult CFTR expression change are new targets for cystic fibrosis. Eur. Respir. J. 2015;45:116–128. doi: 10.1183/09031936.00113214. [DOI] [PubMed] [Google Scholar]
- 5.Lutful Kabir F., Ambalavanan N., Liu G., Li P., Solomon G.M., Lal C.V., Mazur M., Halloran B., Szul T., Gerthoffer W.T. MicroRNA-145 Antagonism Reverses TGF-β Inhibition of F508del CFTR Correction in Airway Epithelia. Am. J. Respir. Crit. Care Med. 2018;197:632–643. doi: 10.1164/rccm.201704-0732OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett C.F., Baker B.F., Pham N., Swayze E., Geary R.S. Pharmacology of Antisense Drugs. Annu. Rev. Pharmacol. Toxicol. 2017;57:81–105. doi: 10.1146/annurev-pharmtox-010716-104846. [DOI] [PubMed] [Google Scholar]
- 7.Benson M.D., Pandey S., Witchell D., Jazayeri A., Siwkowski A., Monia B., Kluve-Beckerman B. Antisense oligonucleotide therapy for TTR amyloidosis. Amyloid. 2011;18(Suppl 1):60. doi: 10.3109/13506129.2011.574354021. [DOI] [PubMed] [Google Scholar]
- 8.Crooke S.T. Molecular Mechanisms of Antisense Oligonucleotides. Nucleic Acid Ther. 2017;27:70–77. doi: 10.1089/nat.2016.0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang X.H., Shen W., Crooke S.T. Specific Increase of Protein Levels by Enhancing Translation Using Antisense Oligonucleotides Targeting Upstream Open Frames. Adv. Exp. Med. Biol. 2017;983:129–146. doi: 10.1007/978-981-10-4310-9_9. [DOI] [PubMed] [Google Scholar]
- 10.Liang X.H., Shen W., Sun H., Migawa M.T., Vickers T.A., Crooke S.T. Translation efficiency of mRNAs is increased by antisense oligonucleotides targeting upstream open reading frames. Nat. Biotechnol. 2016;34:875–880. doi: 10.1038/nbt.3589. [DOI] [PubMed] [Google Scholar]
- 11.Liang X.H., Sun H., Shen W., Wang S., Yao J., Migawa M.T., Bui H.H., Damle S.S., Riney S., Graham M.J. Antisense oligonucleotides targeting translation inhibitory elements in 5′ UTRs can selectively increase protein levels. Nucleic Acids Res. 2017;45:9528–9546. doi: 10.1093/nar/gkx632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valley H.C., Bukis K.M., Bell A., Cheng Y., Wong E., Jordan N.J., Allaire N.E., Sivachenko A., Liang F., Bihler H. Isogenic cell models of cystic fibrosis-causing variants in natively expressing pulmonary epithelial cells. J. Cyst. Fibros. 2019;18:476–483. doi: 10.1016/j.jcf.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Crooke S.T., Wang S., Vickers T.A., Shen W., Liang X.H. Cellular uptake and trafficking of antisense oligonucleotides. Nat. Biotechnol. 2017;35:230–237. doi: 10.1038/nbt.3779. [DOI] [PubMed] [Google Scholar]
- 14.Wang S., Sun H., Tanowitz M., Liang X.H., Crooke S.T. Annexin A2 facilitates endocytic trafficking of antisense oligonucleotides. Nucleic Acids Res. 2016;44:7314–7330. doi: 10.1093/nar/gkw595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor-Cousar J.L., Munck A., McKone E.F., van der Ent C.K., Moeller A., Simard C., Wang L.T., Ingenito E.P., McKee C., Lu Y. Tezacaftor-Ivacaftor in Patients with Cystic Fibrosis Homozygous for Phe508del. N. Engl. J. Med. 2017;377:2013–2023. doi: 10.1056/NEJMoa1709846. [DOI] [PubMed] [Google Scholar]
- 16.Awatade N.T., Wong S.L., Hewson C.K., Fawcett L.K., Kicic A., Jaffe A., Waters S.A. Human Primary Epithelial Cell Models: Promising Tools in the Era of Cystic Fibrosis Personalized Medicine. Front. Pharmacol. 2018;9:1429. doi: 10.3389/fphar.2018.01429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Goor F., Yu H., Burton B., Hoffman B.J. Effect of ivacaftor on CFTR forms with missense mutations associated with defects in protein processing or function. J. Cyst. Fibros. 2014;13:29–36. doi: 10.1016/j.jcf.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 18.Cebotaru L., Rapino D., Cebotaru V., Guggino W.B. Correcting the cystic fibrosis disease mutant, A455E CFTR. PLoS ONE. 2014;9:e85183. doi: 10.1371/journal.pone.0085183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheppard D.N., Rich D.P., Ostedgaard L.S., Gregory R.J., Smith A.E., Welsh M.J. Mutations in CFTR associated with mild-disease-form Cl- channels with altered pore properties. Nature. 1993;362:160–164. doi: 10.1038/362160a0. [DOI] [PubMed] [Google Scholar]
- 20.Welsh M.J., Smith A.E. Molecular mechanisms of CFTR chloride channel dysfunction in cystic fibrosis. Cell. 1993;73:1251–1254. doi: 10.1016/0092-8674(93)90353-r. [DOI] [PubMed] [Google Scholar]
- 21.Bozon D., Zielenski J., Rininsland F., Tsui L.C. Identification of four new mutations in the cystic fibrosis transmembrane conductance regulator gene: I148T, L1077P, Y1092X, 2183AA-->G. Hum. Mutat. 1994;3:330–332. doi: 10.1002/humu.1380030329. [DOI] [PubMed] [Google Scholar]
- 22.Crosby J.R., Zhao C., Jiang C., Bai D., Katz M., Greenlee S., Kawabe H., McCaleb M., Rotin D., Guo S., Monia B.P. Inhaled ENaC antisense oligonucleotide ameliorates cystic fibrosis-like lung disease in mice. J. Cyst. Fibros. 2017;16:671–680. doi: 10.1016/j.jcf.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 23.Barbosa C., Peixeiro I., Romão L. Gene expression regulation by upstream open reading frames and human disease. PLoS Genet. 2013;9:e1003529. doi: 10.1371/journal.pgen.1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorenz R., Bernhart S.H., Höner Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darty K., Denise A., Ponty Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cozens A.L., Yezzi M.J., Kunzelmann K., Ohrui T., Chin L., Eng K., Finkbeiner W.E., Widdicombe J.H., Gruenert D.C. CFTR expression and chloride secretion in polarized immortal human bronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 1994;10:38–47. doi: 10.1165/ajrcmb.10.1.7507342. [DOI] [PubMed] [Google Scholar]
- 27.Fulcher M.L., Randell S.H. Human nasal and tracheo-bronchial respiratory epithelial cell culture. Methods Mol. Biol. 2013;945:109–121. doi: 10.1007/978-1-62703-125-7_8. [DOI] [PubMed] [Google Scholar]
- 28.Hung G., Xiao X., Peralta R., Bhattacharjee G., Murray S., Norris D., Guo S., Monia B.P. Characterization of target mRNA reduction through in situ RNA hybridization in multiple organ systems following systemic antisense treatment in animals. Nucleic Acid Ther. 2013;23:369–378. doi: 10.1089/nat.2013.0443. [DOI] [PubMed] [Google Scholar]
- 29.Cheng J., Wang H., Guggino W.B. Modulation of mature cystic fibrosis transmembrane regulator protein by the PDZ domain protein CAL. J. Biol. Chem. 2004;279:1892–1898. doi: 10.1074/jbc.M308640200. [DOI] [PubMed] [Google Scholar]
- 30.Fischer H., Illek B., Sachs L., Finkbeiner W.E., Widdicombe J.H. CFTR and calcium-activated chloride channels in primary cultures of human airway gland cells of serous or mucous phenotype. Am. J. Physiol. Lung Cell. Mol. Physiol. 2010;299:L585–L594. doi: 10.1152/ajplung.00421.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., 3rd, Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.