Abstract
Background
Euglena gracilis, a unicellular flagellated microalga, is regarded as one of the most promising species as microalgal feedstock for biofuels. Its lipids (mainly wax esters) are suitable for biodiesel and jet fuel. Culture of E. gracilis using wastewater effluent will improve the economics of E. gracilis biofuel production. Enhancement of the productivity of E. gracilis biomass is critical to creating a highly efficient biofuels production system. Certain bacteria have been found to promote microalgal growth by creating a favorable microenvironment. These bacteria have been characterized as microalgae growth-promoting bacteria (MGPB). Co-culture of microalgae with MGPB might offer an effective strategy to enhance microalgal biomass production in wastewater effluent culture systems. However, no MGPB has been identified to enhance the growth of E. gracilis. The objectives of this study were, therefore, to isolate and characterize the MGPB effective for E. gracilis and to demonstrate that the isolated MGPB indeed enhances the production of biomass and lipids by E. gracilis in wastewater effluent culture system.
Results
A bacterium, Emticicia sp. EG3, which is capable of promoting the growth of microalga E. gracilis, was isolated from an E. gracilis-municipal wastewater effluent culture. Biomass production rate of E. gracilis was enhanced 3.5-fold and 3.1-fold by EG3 in the co-culture system using a medium of heat-sterilized and non-sterilized wastewater effluent, respectively, compared to growth in the same effluent culture but without EG3. Two-step culture system was examined as follows: E. gracilis was cultured with or without EG3 in wastewater effluent in the first step and was further grown in wastewater effluent in the second step. Production yields of biomass and lipids by E. gracilis were enhanced 3.2-fold and 2.9-fold, respectively, in the second step of the system in which E. gracilis was co-cultured with EG3 in the first step.
Conclusion
Emticicia sp. EG3 is the first MGPB for E. gracilis. Growth-promoting bacteria such as EG3 will be promising agents for enhancing E. gracilis biomass/biofuel productivities.
Keywords: Euglena gracilis, Growth-promoting bacteria, Biomass production, Lipid production, Wastewater effluent, Wastewater effluent
Background
Microalgae are widely recognized as alternative, renewable, and sustainable feedstocks for biofuels. The use of wastewater effluent as a medium for microalgal culture will no doubt improve the economics and sustainability of microalgal biomass production [1, 2]. Microalgal biomass production using wastewater effluent decreases the costs for fertilization and irrigation. Coupling microalgal production with wastewater treatment would transform a wastewater treatment plant into a biorefinery plant [3].
Euglena gracilis, a unicellular flagellated microalga, is regarded as one of the most promising species as microalgal feedstock for functional foods and biofuels [4, 5]. Depending on culture conditions, E. gracilis can synthesize and accumulate a variety of valuable products such as wax esters [6], paramylon (β-1,3-glucan) [7], β-carotene, and vitamins C and E [8]. Those products have numerous commercial applications. Wax esters—which consist mainly of C14:0 saturated fatty acid, myristic acid, C14:0 saturated fatty alcohols, and myristyl alcohols—are considered high-value biofuels [9, 10]. These wax esters of E. gracilis are suitable for biodiesel and jet fuel [5, 10, 11].
Enhancement of the productivity of E. gracilis biomass is critical to creating a highly efficient biofuels production system and to reducing the price of the biofuel. Genetic recombination [12], selective breeding [13], optimization of growth conditions [14, 15], and supplementation with additives, including nutrients [14] and plant hormones [16], have been investigated to enhance the production of biomass and lipids by E. gracilis. However, to realize a highly efficient E. gracilis’s biofuel producing system, development of synergistic technologies is needed to further enhance the growth of E. gracilis.
In both natural aquatic environments and microalgal cultures, certain bacteria have been found to promote microalgal growth by creating a favorable microenvironment [17–19], such as providing nutrients [20], vitamins [21], phytohormones [22], chelators [23], or volatile organic compounds [24]. These bacteria have been characterized as microalgae growth-promoting bacteria (MGPB). MGPB have been reported for various microalgae. For example, Azospirillum brasilense for Chlorella vulgaris [25]; Rhizobium sp. for Botryococcus braunii [26]; Pelagibaca bacilliformis and Stappia sp. for Tetraselmis striata [27]; and Rhizobium sp. for Chlamydomonas reinhardtii, C. vulgaris, Scenedesmus sp., and B. braunii [28]. The use of MGPB as inoculant in co-cultures with microalgae might offer an alternative and promising strategy to effectively enhance microalgal biomass production. To the best of our knowledge, however, no MGPB has been identified to enhance the growth of E. gracilis. In addition, no studies have shown that MGPB enhance microalgal biomass production in wastewater effluent under non-sterile culture conditions where exist indigenous bacteria. There is thus high demand for obtaining a novel MGPB that is capable of enhancing biomass/lipid production by E. gracilis in wastewater effluent.
The objectives of this study were, therefore, to (i) isolate and characterize the MGPB effective for E. gracilis and (ii) demonstrate that the isolated MGPB indeed enhances the production of biomass and lipids by E. gracilis in wastewater effluent culture system.
Methods
E. gracilis material and culturing of E. gracilis
Axenic E. gracilis (NIES-48) was obtained from the Microbial Culture Collection, National Institute for Environmental Studies, Tsukuba, Japan. E. gracilis was cultured in C medium supplemented with 400 mg/L yeast extract and 600 mg/L polypeptone, namely CYP medium. C medium contained per liter 150 mg Ca(NO3)2·4H2O, 100 mg KNO3, 50 mg β-Na2 glycerophosphate·5H2O, 40 mg MgSO4·7H2O, 500 mg tris(hydroxymethyl)aminomethane, 0.1 μg vitamin B12, 0.1 μg biotin, 10 μg thiamine HCl, and 3 mL PIV metals (1000 mg/L Na2EDTA·H2O, 196 mg/L FeCl3·6H2O, 36 mg/L MnCl2·4H2O, 10.4 mg/L ZnCl2, 4 mg/L CoCl2·6H2O, and 2.5 mg/L Na2MoO4·H2O). pH was adjusted to 7.5 by adding NaOH. The axenic E. gracilis culture was incubated in a growth chamber (28 ± 1 °C with fluorescent lamps at a photosynthetic photon flux density of 80 μmol/m2/s and 16-h photoperiod) for 1 week. Thereafter, subculture was started by routine transfer of E. gracilis cells into fresh CYP medium every week. E. gracilis was harvested by centrifugation (3000×g, room temperature, 5 min), washed with sterile C medium, and re-suspended in aliquots of sterile C medium. Such an E. gracilis cell suspension was used as the inoculum for each subculture experiment.
Wastewater effluent samples
Municipal wastewater effluent was collected from the final sedimentation process of a conventional activated sludge municipal wastewater treatment plant in Kofu City, Yamanashi, Japan. The effluent sample was first passed through a glass microfiber filter (pore size, 1 μm; GF/B grade; GE Healthcare UK Ltd, Buckinghamshire, England) and then through a membrane filter (pore size, 0.8 μm; mixed cellulose esters membrane; Merck Millipore Ltd, Cork, Ireland) to remove suspended solids and organisms larger than bacteria—including microalgae—from the effluent sample. The effluent filtrate, therefore, included the indigenous bacterial community. For each of the following experiments, a fresh effluent sample was collected and used each day. Table 1 shows the water quality characteristics (total organic carbon [TOC], ammonium-nitrogen [NH4-N], nitrite-N [NO2-N], nitrate-N [NO3-N], phosphate [PO4-P]), and total bacteria concentrations). TOC was measured using TOC-LCSH (Shimadzu, Kyoto, Japan). For NH4-N, the indophenol method was used; for NO2-N, the N-(1-naphthyl) ethylenediamine method was used; for NO3-N, the reduction-N-(1-naphthyl) ethylenediamine method and UV adsorption (at 220 and 275 nm) method was used; and for PO4-P, the molybdenum blue method was used. Total culturable bacteria in effluent were quantified using R2A agar plates (0.5 g/L Peptone, 0.5 g/L Yeast extract, 0.5 g/L Casamino acid, 0.5 g/L Glucose, 0.5 g/L Soluble starch, 0.3 g/L K2HPO4, 0.05 g/L MgSO4·7H2O, 0.3 g/L Sodium pyruvate; pH 7.0; Agar 15 g/L). In some experiments, sterile effluent samples were prepared by autoclaving (121 °C, 20 min) for heat sterilization.
Table 1.
Initial concentrations of total organic carbon (TOC), inorganic nitrogen (NH4-N, NO2-N, and NO3-N), phosphate (PO4-P), and total bacteria in effluent samples for each experiment
| TOC (mg/L) | NH4-N (mg/L) | NO2-N (mg/L) | NO3-N (mg/L) | PO4-P (mg/L) | Total bacteria (CFU/mL) | |
|---|---|---|---|---|---|---|
| Experiment of section 2.3 | 15.4 | 7.2 | 0.0 | 6.1 | 3.3 | 4.5 ± 0.2 × 105 |
| Experiment of section 2.4 | 13.9 | 6.8 | 0.0 | 6.9 | 3.1 | 1.9 ± 0.1 × 105 |
| Experiment of section 2.7 | 14.2 | 6.1 | 0.0 | 5.8 | 3.4 | 3.6 ± 0.2 × 105 |
| Experiment of section 2.8 | 16.5 | 6.6 | 0.0 | 6.6 | 3.3 | 6.4 ± 0.5 × 105 |
Culturing of E. gracilis in wastewater effluent and isolation of MGPB candidates for E. gracilis
A cell suspension (10 mL) of E. gracilis prepared by the above method was added to 100 mL of (non-sterilized) effluent filtrate in a 200-mL glass flask. The flask was incubated in the growth chamber (28 ± 1 °C with fluorescent lamps at 80 μmol photons m−2 s−1 and 16-h photoperiod) for 10 days, and then, about 10 mL of this culture was transferred to 100 mL of fresh effluent and incubated for another 10 days. This transfer and 10-day batch culture growth was repeated once more. During the three repeated growth periods, the flasks were shaken for 1 min three times a day to disperse and aerate the E. gracilis. The chlorophyll concentration in each flask was measured daily. E. gracilis was also cultured in 100 mL of autoclaved effluent without living indigenous bacteria. The culture was used as a control treatment to determine the effect of the indigenous bacteria in the effluent on the growth of E. gracilis. The initial chlorophyll a + b concentration of each culture was adjusted to about 0.5 μg/mL.
After the third 10-day batch culture growth period, 20 mL of the non-sterile E. gracilis-effluent culture was transferred into a 50-mL tube, vortexed at maximum speed for 3 min, ultrasonicated (40 kHz) for 1 min, and vortexed again to disperse the E. gracilis and bacterial cells. The sample was then filtered through a GF/B glass microfiber filter to remove E. gracilis cells. The filtrate-containing bacteria was serially diluted and spread on R2A agar plates. The plates were then incubated at 28 °C. A total of ten bacterial strains were obtained from the isolation culture. A pure culture of each strain was maintained on an R2A agar plate.
Screening of MGPB
Six isolated strains grew well in R2A liquid medium. Each strain was cultured in R2A liquid medium for 24 h at 28 °C. Cells were harvested by centrifugation (10,000×g, room temperature, 5 min) and washed twice with 50 mM potassium phosphate buffer (pH 7.5). E. gracilis was inoculated into 100 mL of autoclaved wastewater effluent in 200-mL glass flasks. Optical densities of cultures of isolated bacterial cells were measured at 600 nm (OD600), and each bacterial cell suspension was then added into the E. gracilis culture at bacterial cell density at OD600 of 0.05. The co-cultures of E. gracilis and each isolated strain were incubated in a growth chamber (28 ± 1 °C with fluorescent lamps at 80 μmol photons/m2/s and 16-h photoperiod) for 7 days. At 7th day, concentrations of chlorophyll a + b were measured. E. gracilis was also cultured in autoclaved wastewater effluent without a bacterial inoculation. The growth-promoting ability of MGPB was assessed by comparing the chlorophyll a + b concentration at the end of 7-day cultures with and without bacterial inoculation.
Identification and characterization of strain EG3
Among the isolated bacterial strains, strain EG3 showed the highest growth-promoting ability in the screening test. Strain EG3 was thus characterized and identified by physiological and phylogenetic analyses. The physiological characterization was performed with an API 20NE kit according to the manufacturer’s instructions (BioMérieux Japan, Tokyo, Japan). A comparative 16S rRNA gene sequence analysis was performed as follows. Partial 16S rRNA genes were amplified by PCR using primers 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) [29] and 1510R (5′-GGTTACCTTGTTACGACTT-3′) [30]. A genus-level identification was carried out based on 16S rRNA gene sequence similarity with that of a type strain sequence in NCBI-GenBank using BLAST. The 16S rRNA sequence data (1431 bp) of isolated strain EG3 have been submitted to the DDBJ/EMBL/GenBank databases under accession number LC441033.
Culture conditions of EG3 and preparation of cell suspension
Strain EG3 was grown in R2A liquid medium for 24 h at 28 °C with shaking at 150 rpm. Cells were harvested by centrifugation (10,000×g, room temperature, 5 min), washed twice with phosphate buffer, and then suspended in the phosphate buffer. Highly concentrated EG3 cell suspension in phosphate buffers (DO600 = 5, 10, or 20) were prepared for the following experiments. For each experiment, 1% (v/v) of the EG3 cell suspension was inoculated into the experimental culture (sterilized or non-sterilized effluent). Thus, the initial EG3 cell density of each experiment was adjusted as OD600 = 0.05, 0.1, or 0.2. To convert OD600 value into EG3 cell dry weight concentration (mg dry weight/mL) and cell count (CFU/mL), dry weight of cells was measured after drying at 90 °C for 3 h, and CFU of EG3 suspensions (OD600 = 0.05, 0.1, and 0.2) was measured by R2A agar plate method in triplicate. An OD600 = 1.0 of EG3 cells was equivalent to about 0.45 ± 0.03 mg dry weight/mL. Also, OD600 = 0.05, 0.1, and 0.2 of EG3 cells were equivalent to about 2 × 106, 8 × 106, and 4 × 107 CFU/mL on R2A agar plate. In this manuscript, cell density of EG3 was described using CFU/mL.
Co-culturing of E. gracilis with EG3 in effluent under several conditions
Euglena gracilis was inoculated into 1 L of autoclave sterilized wastewater effluent in a 2-L glass bottle. To determine the effect of the EG3 cell density on the growth of E. gracilis, EG3 was inoculated into 1 L of an E. gracilis culture in sterile wastewater effluent at an initial cell density of 2 × 106, 8 × 106, and 4 × 107 CFU/mL. The co-cultures of E. gracilis with EG3 were incubated in a growth chamber (28 ± 1 °C with fluorescent lamps at 80 μmol photons/m2/s and 16-h photoperiod) for 7 days. E. gracilis was also cultured without EG3 in 1 L of autoclave sterilized wastewater effluent under the above conditions as the control experiment.
Both sterilized and non-sterilized effluent samples were used to examine the effects of co-existing indigenous bacteria in wastewater effluent on the ability of EG3 to promote E. gracilis growth. E. gracilis was inoculated into 1 L of wastewater effluent in 2-L glass bottles. EG3 was then inoculated into the E. gracilis culture at 8 × 106 CFU/mL, because the growth-promoting ability of EG3 was highest at this inoculation cell density. The co-cultures of E. gracilis with EG3 were incubated in the growth chamber (28 ± 1 °C with fluorescent lamps at 80-μmol photons/m2/s and 16-h photoperiod) for 7 days.
During the experiments, chlorophyll a + b concentration was monitored daily. After 7 days, the dry weight of E. gracilis was measured. The experiments were conducted in triplicate.
Experiment to demonstrate EG3-enhanced production of biomass and lipids by E. gracilis in wastewater effluent: two-step enhanced E. gracilis biomass/lipids production system
In this part of our study, possibility of a two-step enhanced biomass production system was evaluated. The first step was co-culture of E. gracilis with strain EG3 to promote the growth of E. gracilis. The E. gracilis cells produced in this first step were used as the E. gracilis inoculum for the second step. In the second step, the productions of biomass and lipids by the E. gracilis inoculum from the first step were measured (Fig. 1).
Fig. 1.
Schematic design of two-step enhanced E. gracilis biomass/lipid production system
The experimental design for the co-culture of E. gracilis and EG3 in the first step was as follows. E. gracilis was inoculated into 5 L of non-sterilized wastewater effluent in a 15-L polycarbonate culture vessel with an inner diameter of about 290 mm and a height of about 430 mm. EG3 was inoculated into the E. gracilis culture at 8 × 106 CFU/mL. The co-culture of E. gracilis and EG3 in the first step was bubbled with atmospheric air by an air pump at a flow rate 0.5 L/min for 7 days in a growth chamber (28 ± 1 °C with fluorescent lamps at 80-μmol photons/m2/s and 16-h photoperiod). After 7 days, 2–4 L of culture was collected, and E. gracilis was harvested by low gravity centrifugation (3000×g, 15 min). The E. gracilis cells were washed twice with distilled water to remove EG3 cells and harvested by centrifugation. We confirmed that the collected E. gracilis pellets contained only little EG3 cells (Additional file 1: Fig. S1). The collected E. gracilis pellet was used for the second step culture. E. gracilis was also cultured without EG3 in 5 L of wastewater effluent under the above conditions, and the control E. gracilis pellet from this experiment was also used for the second step.
The production of biomass and lipids in the second step culture was performed in 15-L polycarbonate culture vessels. Fifteen liters of unautoclaved wastewater effluent was added to the culture vessel. The E. gracilis co-cultured with EG3 or the control E. gracilis only culture was inoculated into the unautoclaved wastewater effluent. The initial cell density of E. gracilis was about 80 mg dry weight/L. The biomass/lipid production step was conducted in a growth chamber (28 ± 1 °C with fluorescent lamps at 80 μmol photons/m2/s and 16-h photoperiod) for 7 days. Aeration and mixing were achieved by bubbling with atmospheric air at a flow rate 1 L/min. Chlorophyll a + b and biomass were measured every day. The lipid content of E. gracilis was measured at the beginning and end of the second step.
E. gracilis growth measurement and E. gracilis lipid measurement
In this study, chlorophyll concentration was monitored as an indicator of E. gracilis growth. Chlorophyll concentration was measured spectrophotometrically after extraction in 100% methanol for 30 min [31]. Absorbance of the extract was measured at 665 nm (A665) and 650 nm (A650) with a spectrophotometer (UVmini-1240; Shimadzu Co. Ltd., Kyoto, Japan). The total chlorophyll (chlorophyll a + chlorophyll b: Chl a + b) concentration (μg/mL) was calculated as follows [31]:
| 1 |
The biomass (dry weight) of E. gracilis was measured as follows. Fifty milliliters of the culture was collected and vortexed for 30 s to uniformly suspend the bacterial and microalgal cells. The mixture was centrifuged (3000×g, 5 min); the pellet was washed with 20 mL of distilled water; and the pellet was then suspended in 20 mL of distilled water. The E. gracilis cells in the suspension were collected on a pre-weighed GF/B filter (pore size, 1 μm), dried (90 °C, 3 h), and then weighed. We confirmed that this method could collect E. gracilis cells with little interference from co-existing EG3 cells (Additional file 1: Fig. S1).
The lipid content of the E. gracilis cells was quantified in terms of the percent of dry biomass accounted for by lipids. E. gracilis was harvested by centrifugation (3000×g, 5 min), washed with distilled water, dried, and then powdered. The E. gracilis powder (20 mg) was crushed with 1 mL of n-hexane in a BioMasher (Takara Bio, Kusatsu, Japan). The E. gracilis sample was transferred into a 50-mL tube, and 9 mL of n-hexane and 6-mL isopropanol were added to the tube. The tube was shaken at 225 rpm for 24 h. Thirty milliliters of distilled water was then added to the tube. The tube was shaken for 1 min and then centrifuged (10,000×g, 5 min). The n-hexane layer containing the lipids was collected on a pre-weighed aluminum tray, dried at room temperature overnight, and then dried at 90 °C for 3 h. Finally, the collected lipids were weighed.
Biomass production rate (mg/L/day) and lipid production rate (mg/L/day) were calculated as follows:
| 2 |
| 3 |
Statistical analysis
Each value used in the statistical analysis represents the results from three samples (n = 3 replicates) per experiment. All results are expressed as mean ± SD. Significance (p < 0.05) was analyzed using the t test in SPSS Statistics v. 22.0 (IBM, Armonk, NY, USA).
Results
Isolation and identification of the MGPB Emticicia sp. EG3 for E. gracilis
To test the hypothesis that indigenous bacteria in wastewater effluent could enhance the growth of E. gracilis, E. gracilis was grew in wastewater effluent with (non-sterilized effluent) or without living bacteria (autoclave heat-sterilized effluent) in three sequencing batch cultures. After 2 days of each batch culture, the chlorophyll a + b concentration was higher (p < 0.05) in the E. gracilis-effluent culture with living indigenous bacteria than in the culture without living bacteria (Fig. 2). The results suggested that MGPB were present in the E. gracilis-effluent culture with living indigenous bacteria. Ten bacterial strains were isolated from the E. gracilis culture in non-sterilized effluent. Six of the ten strains grew well in R2A medium and were examined for their ability to enhance E. gracilis growth. Four strains were found to enhance the growth of E. gracilis. Among them, strain EG3 showed the highest growth-promoting ability (Additional file 2: Fig. S2). In addition, strain EG3 showed high biomass yield in R2A liquid medium culture.
Fig. 2.

Changes in chlorophyll a + b content in E. gracilis-municipal wastewater effluent culture with indigenous living bacteria (non-sterilized effluent, white circle) and without living indigenous bacteria (autoclave heat-sterilized effluent, black circle). Values are means ± SDs (n = 3)
Colonies of strain EG3 on R2A agar were circular, smooth, convex, and orange color. EG3 showed positive for oxidase, glucose fermentation, and aesculin hydrolysis, but negative for nitrate reduction, indole production, arginine dihydrolase, urease, gelatinase, and β-galactosidase activities. EG3 utilized glucose, d-mannose, or maltose as sole carbon sources but did not utilize l-arabinose, d-mannitol, N-acetyl-d-glucosamine, gluconate, n-caprate, adipate, d,l-malate, citrate, or phenylacetate. Almost the entire sequence of the 16S rRNA gene (1431 bp) of EG3 was very similar to the analogous sequences of Emticicia fontis strain IMCC1731T (97.9% sequence similarity), Emticicia ginsengisoli strain Gsoil 085T (97.6%), Emticicia soli strain ZZ-4T (97.5%), Emticicia oligotrophica strain GPTSA100-15T (94.4%), Emticicia aquatica strain HMF2925T (93.9%), Emticicia sediminis strain JBR12T (93.7%), and Emticicia aquatilis strain THG-DN6.14T (93.7%). We, therefore, identified strain EG3 as Emticicia sp.
Growth promotion of E. gracilis by strain EG3 at different cell densities
To determine the cell density of EG3 needed to promote the growth of E. gracilis in wastewater effluent, E. gracilis was grew in heat-sterilized wastewater effluent with various EG3 cell densities, 0 (control), 2 × 106, 8 × 106, and 4 × 107 CFU/mL. The chlorophyll a + b concentrations and biomass production rate (mg/L/day) of E. gracilis were significantly higher (p < 0.05) in E. gracilis cultures with EG3 at 2 × 106, 8 × 106, and 4 × 107 CFU/mL than in the E. gracilis control culture (Fig. 3). The growth-promoting effect of EG3 was higher at cell density of 8 × 106 and 4 × 107 CFU/mL than at 2 × 106 CFU/mL. The EG3 initial inoculation cell density was an important determinant of whether EG3 greatly enhanced E. gracilis biomass production rate. We decided to use 8 × 106 CFU/mL for follow-up experiments, because the difference in the ability of EG3 to promote the growth of E. gracilis between initial inocula 8 × 106 and 4 × 107 CFU/mL was not significant.
Fig. 3.
Effects of EG3 on the growth of E. gracilis co-cultured with various initial cell densities of EG3—0 (control), 2 × 106, 8 × 106, and 4 × 107 CFU/mL—in heat-sterilized wastewater effluent for 7 days. a Changes in chlorophyll a + b content in E. gracilis co-cultured with EG3 at initial cell density of 0 (control, black triangle), 2 × 106 CFU/mL (white diamond), 8 × 106 CFU/mL (white square), or 4 × 107 CFU/mL (white circle). b Biomass production rate of E. gracilis in co-culture with EG3 at initial cell density of 0 (control), 2 × 106, 8 × 106, and 4 × 107 CFU/mL during 7 days. Values are means ± SDs (n = 3). *Significant difference (p < 0.05) from control
Effect of indigenous bacteria in wastewater effluent on the ability of EG3 to promote the growth of E. gracilis
To examine the effect of bacteria that co-existed with strain EG3, E. gracilis and EG3 were co-cultured in heat-sterilized or non-sterilized wastewater effluent. The chlorophyll a + b concentrations of E. gracilis-EG3 co-cultures in both sterilized and non-sterilized wastewater effluent were significantly higher (p < 0.05) than those of E. gracilis cultures without EG3 (Fig. 4). During the 7-day culture experiment, the biomass production rate (mg/L/day) of E. gracilis co-cultured with EG3 were 3.7-fold and 3.1-fold in sterilized and non-sterilized wastewater effluent, respectively, those of E. gracilis cultured without EG3 (Fig. 4). These results clearly indicated that EG3 promoted the growth of E. gracilis in real wastewater effluent, which harbors a 3.6 ± 0.2 × 105 CFU/mL complex indigenous bacterial community.
Fig. 4.
Effects of EG3 on the growth of E. gracilis co-cultured with initial EG3 cell density of 8 × 106 CFU/mL in heat-sterilized and non-sterilized wastewater effluents for 7 days. a Changes in chlorophyll a + b content in E. gracilis cultured without (black square) and with EG3 (white square) in heat-sterilized effluent; changes in chlorophyll a + b content in E. gracilis cultured without (black circle) and with EG3 (white circle) in non-sterilized effluent. b Biomass production rate of E. gracilis cultured without or with EG3 in sterilized or non-sterilized effluent during 7 days. Values are means ± SDs (n = 3). *Significant difference (p < 0.05) from control
Enhanced production of biomass and lipids by E. gracilis in effluent culture using strain EG3 in a two-step culture system
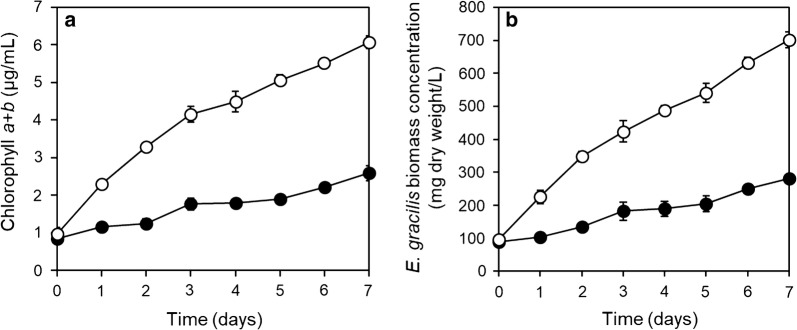
We operated a two-step E. gracilis culture system to enhance production of biomass and lipids by E. gracilis (Fig. 1). First, E. gracilis was co-cultured with EG3 to promote E. gracilis growth. The E. gracilis produced in this first step was used as the inoculum for the second culture system, in which production of biomass and lipids by E. gracilis was monitored in 15 L of wastewater effluent for 7 days. In the control culture, E. gracilis was grown without EG3 in the first step, and then, the E. gracilis produced in the first step was used to inoculate a second culture system containing 15 L of wastewater effluent. The chlorophyll a + b concentration and biomass (dry weight) concentration of E. gracilis in the second culture system were significantly higher (p < 0.05) in the system inoculated with E. gracilis that had been co-cultured with EG3 in the first step than in the system inoculated with the control E. gracilis culture (Fig. 5). During the 7 days of second culture inoculated with the E. gracilis-EG3, E. gracilis biomass concentration reached the highest level of 702 ± 23 mg/L at the end of 7-day culture (Fig. 5). Figure 6 compares the rates of biomass and lipid production and the lipid content of E. gracilis during the second step in the systems inoculated with the E. gracilis-EG3 and control E. gracilis cultures. The rate of biomass production by E. gracilis-EG3 (87 ± 2.8 mg/L/day) was 3.2 times of that of the control E. gracilis (27 ± 2.7 mg/L/day). The lipid content of E. gracilis-EG3 (28.4% ± 0.7%) was slightly but significantly lower than that of the control E. gracilis (30.9% ± 0.8%). The lipid production rate of E. gracilis-EG3 (25 ± 0.8 mg/L/day) was 2.9 times that of the control E. gracilis (8.5 ± 0.8 mg/L/day).
Fig. 5.
Time-course of chlorophyll a + b content and biomass (dry weight) concentration of E. gracilis in second culture of two-step E. gracilis culture systems. a Changes in chlorophyll a + b content in control culture of E. gracilis without co-culture with EG3 (black circle) and with co-culture with EG3 (white circle) in non-sterilized effluent during period of 7 days. b Changes in biomass concentration of E. gracilis in control culture without co-culture with EG3 (black circle) and with co-culture with EG3 (white circle) in non-sterilized effluent during period of 7 days. Values are means ± SDs (n = 3)
Fig. 6.
Effects of co-culturing E. gracilis with EG3 in first culture of a two-step E. gracilis biomass/lipid production system. a Biomass production rate of E. gracilis in second culture of two-step E. gracilis culture systems after co-culturing with EG3 and without (control) in the first step. b Lipid content of E. gracilis in second culture of two-step E. gracilis culture systems after co-culturing with EG3 and control. c Lipid production rate of E. gracilis in second culture of two-step E. gracilis culture systems after co-culturing with EG3 and control. Values are means ± SDs (n = 3). *Significant difference (p < 0.05) from control
Discussion
Co-culture with MGPB is useful for enhancing microalgal growth in mass culture systems. For example, “Candidatus Phycosocius bacilliformis” BOTRYCO-2 and P. bermudensis KCTC 13073BP increase the rate of biomass production by B. braunii and T. striata 1.8-fold [32] and twofold [27], respectively. To make this strategy most successful, it is necessary to use the MGPB suitable for the host microalgal species. Use of the MGPB isolated from an E. gracilis-wastewater effluent culture was important for enhancing the biomass/lipid productivity of E. gracilis in a wastewater effluent culture system. The MGPB for three microalgae—C. reinhardtii, C. vulgaris, and E. gracilis—have recently been found to be present in common wastewater effluent [33]. In this study, an MGPB for E. gracilis, Emticicia sp. EG3, was isolated from an E. gracilis-municipal wastewater effluent culture and was found to be capable of promoting the growth of E. gracilis. Strain EG3 is the first MGPB for E. gracilis. Members of the genus Emticicia have been isolated from various environments, including freshwater [34–37], wetlands [38], seawater [39], the sediments of a shallow stream [40], and soil [41, 42]. Although the ecological role of Emticicia and its potential for industrial use are still unclear, our results reveal that one of its ecosystem services may be to function in a symbiotic relationship that enhances the growth of microalgae such as E. gracilis.
In co-cultures of E. gracilis with strain EG3 at 8 × 106 CFU/mL, EG3 enhanced the production rate of E. gracilis biomass 3.7-fold and 3.1-fold in heat-sterilized and non-sterilized wastewater effluent, respectively (Fig. 4). The growth-promoting effect of EG3 on E. gracilis was similar to the effect of other MGPB on their host microalgae. These effects have previously been studied under single-MGPB and single-microalga axenic culture conditions without other bacterial communities. In natural field conditions, the introduction of plant growth-promoting bacteria often fails to show beneficial effects on the crops because of competition with other indigenous microorganisms [43, 44]. It is, therefore, noteworthy that strain EG3 significantly increased biomass productivity of E. gracilis in non-sterilized wastewater effluent, which harbors a complex community of indigenous bacteria. This ability of EG3 may, therefore, be of great practical value.
Some MGPB can increase the content of lipids [24, 45–47], fatty acids [47], starch [48], or hydrocarbons [32] in microalgal cells. A. brasilense Cd can significantly increase the accumulation of fatty acids and lipids in C. vulgaris [45, 47]. In this study, however, strain EG3 did not have any positive effect on the accumulation of lipids in E. gracilis (Fig. 6). The effects of EG3 on the production of other cellular components such as paramylon, β-carotene, starch, and proteins, or on lipid composition were not considered. Elucidation of those effects and that of the growth-promoting mechanisms of strain EG3 remain to be investigated.
Co-immobilization of a microalga-MGPB symbiosis has been proposed as a promising and effective strategy for enhancing microalgal abilities and for use in wastewater treatment and biomass production [49]. C. vulgaris has been co-immobilized with A. brasilense Cd in alginate beads, and the symbiosis has resulted in (1) enhanced nutrient removal in synthetic wastewater compared to immobilized C. vulgaris alone, and (2) enhanced biomass, lipid, and starch productivities in synthetic media [45, 48, 50]. In this study, we proposed a two-step culture system as another application of MGPB to enhance biomass production of E. gracilis. E. gracilis was first co-cultured with strain EG3 at 8 × 106 CFU/mL to promote the growth of E. gracilis, and the production of biomass and lipids was then enhanced by growing the stimulated E. gracilis in wastewater effluent culture. In the second E. gracilis culture step, E. gracilis rapidly grew over a period of 7 days (Fig. 5). Biomass and lipid production rates during the 7-day culture period enhanced 3.2-fold and 2.9-fold, respectively, in real wastewater effluent compared to control E. gracilis cultures without EG3 inoculation (Fig. 6).
Our study is the first to demonstrate the potential of the MGPB Emticicia sp. EG3 to increase biomass/lipid productivities in wastewater-E. gracilis cultures. Promotion of E. gracilis growth has been achieved by genetic engineering or by supplements of exogenous nutrients, plant hormones, and ferulic acid. Ogawa et al. [12] were able to enhance E. gracilis biomass (dry weight) production rate twofold in CM medium under high-light and high CO2 conditions by overexpressing the cyanobacterial fructose-1,6-/sedoheptulose-1,7-bisphosphatase gene, which is involved in the Calvin cycle. The highest biomass concentration of the transgenic E. gracilis was 631.1 ± 89.9 mg/L. Zhu and Wakisaka [51] have reported that the addition of ferulic acid made from rice bran into an E. gracilis culture at 500 mg/L can increase E. gracilis cell density 2.5-fold and E. gracilis biomass (dry weight) production rate 2.2-fold. The maximum E. gracilis biomass concentration in CM medium with 500 mg/L ferulic acid was 670 ± 40 mg/L. Noble et al. [16] have reported that exogenous phytohormones stimulate the growth of E. gracilis; a combination of trans-zeatin (10−7 M) and abscisic acid (10−9 M) produced optimal conditions for growth of E. gracilis and increased growth rate about threefold compared to an E. gracilis culture without any phytohormones. Enhancement of biomass production rate (3.2-fold) and highest biomass concentration (702 ± 23 mg/L) of E. gracilis by EG3 co-culture at 8 × 106 CFU/mL in real wastewater effluent were comparable to or higher than the results of these previous enhancement methods. Our results indicate the potential utility of a two-step E. gracilis culture system with EG3—the first step being co-culture of E. gracilis with EG3, and the second step being a large biomass production culture system for enhanced productivities of biomass and lipids by E. gracilis in wastewater effluent. A schematic design of the two-step E. gracilis culture system coupling with wastewater treatment plant is shown in Fig. 7. In combination with the other methods noted above, our strategy would greatly enhance the biomass/lipid productivities of E. gracilis. Optimization of the two-step culture system and better understanding of the growth-promoting effect of EG3 are the next challenges for commercial application of this strategy.
Fig. 7.
Coupling of E. gracilis culture with wastewater treatment. Enhanced biomass and lipid production by a two-step E. gracilis culture system using growth-promoting bacterium EG3
Conclusions
Emticicia sp. EG3, which is capable of promoting the growth of E. gracilis, was isolated from an E. gracilis-municipal wastewater effluent culture. In a co-culture of EG3 and E. gracilis, EG3 significantly increased the biomass productivity of E. gracilis, not only in heat-sterilized wastewater effluent, but also in non-sterilized effluent that contained an indigenous wastewater effluent bacterial community. A two-step E. gracilis culture system was proposed to enhance E. gracilis biomass/lipid productivities by exploiting the stimulatory effect of EG3 on E. gracilis. First, E. gracilis was co-cultured with EG3 to promote E. gracilis growth in 5 L of effluent. Second, biomass and lipids were produced using the promoted E. gracilis in a 15-L wastewater effluent culture. Biomass and lipid productivities of E. gracilis were increased 3.2-fold and 2.9-fold, respectively, compared to a culture without EG3. Growth-promoting bacteria such as EG3 are promising agents for enhancing microalgal biomass/biofuels productivities.
Supplementary information
Additional file 1: Figure S1. Summary of E. gracilis collection method and fate of EG3 cells during this method.
Additional file 2: Figure S2. Effects of isolated strains EG3, EG5, EG6, EG8, EG9, and EG10 on E. gracilis growth in autoclave heat-sterilized wastewater effluent.
Acknowledgements
Not applicable.
Abbreviations
- MGPB
microalgae growth-promoting bacteria
- TOC
total organic carbon
- NH4-N
ammonium-nitrogen
- NO2-N
nitrite-nitrogen
- NO3-N
nitrate-nitrogen
- PO4-P
phosphate
- CFU
colony-forming units
- OD
optical density
- rRNA
ribosomal ribonucleic acid
- PCR
polymerase chain reaction
- NCBI
National Center for Biotechnology Information
- BLAST
Basic Local Alignment Search Tool
- DDBJ
DNA Data Bank of Japan
- EMBL
European Molecular Biology Laboratory
- SD
standard deviation
Authors’ contributions
TT designed and performed the experiments, interpreted the results, and drafted the manuscript. TH performed the experiments and interpreted the results. KY, KS, YT, MM, and KM interpreted the results and critically revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported in part by the Advanced Low Carbon Technology Research and Development Program of the Japan Science and Technology Agency (Grant Number JPMJAL1108), and by a Grant-in-Aid for Challenging Exploratory Research (No. 15K12243) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Availability of data and materials
The data sets used and/or analyzed in this study are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable. There are no any individual person’s data.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13068-019-1544-2.
References
- 1.Pittman JK, Dean AP, Osundeko O. The potential of sustainable algal biofuel production using wastewater resources. Bioresour Technol. 2011;102:17–25. doi: 10.1016/j.biortech.2010.06.035. [DOI] [PubMed] [Google Scholar]
- 2.Uggetti E, Garcia J, Alvarez JA, Garcia-Galan MJ. Start-up of a microalgae-based treatment system within the biorefinery concept: from wastewater to bioproducts. Water Sci Technol. 2018;78:114–124. doi: 10.2166/wst.2018.195. [DOI] [PubMed] [Google Scholar]
- 3.Menger-Krug E, Niederste-Hollenberg J, Hillenbrand T, Hiessl H. Integration of microalgae systems at municipal wastewater treatment plants: implications for energy and emission balances. Environ Sci Technol. 2012;46:11505–11514. doi: 10.1021/es301967y. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki K. Large-scale cultivation of Euglena. In: Schwartzhach SD, Shigeoka S, editors. Euglena: biochemistry, cell and molecular biology. Berlin: Springer; 2017. pp. 285–293. [Google Scholar]
- 5.Gissibl A, Sun A, Care A, Nevalainen H, Sunna A. Bioproducts from Euglena gracilis: synthesis and application. Front Bioeng Biotechnol. 2019;7:108. doi: 10.3389/fbioe.2019.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inui H, Miyatake K, Nakano Y, Kitaoka S. Wax ester fermentation in Euglena gracilis. FEBS Lett. 1982;150:89–93. doi: 10.1016/0014-5793(82)81310-0. [DOI] [Google Scholar]
- 7.Calvayrac R, Laval-Martin D, Briand J, Farineau J. Paramylon synthesis by Euglena gracilis photoheterotrophically grown under low O2 pressure. Planta. 1981;153:6–13. doi: 10.1007/BF00385311. [DOI] [PubMed] [Google Scholar]
- 8.Takeyama H, Kanamaru A, Yoshino Y, Kakuta H, Kawamura Y, Matsunaga T. Production of antioxidant vitamins, β-carotene, vitamin C, and vitamin E, by two-step culture of Euglena gracilis Z. Biotechnol Bioeng. 1997;53:185–190. doi: 10.1002/(SICI)1097-0290(19970120)53:2<185::AID-BIT8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 9.Hulanicka D, Erwin J, Bloch K. Lipid metabolism of Euglena gracilis. J Biol Chem. 1964;239:2778–2787. [PubMed] [Google Scholar]
- 10.Furuhashi T, Ogawa T, Nakai R, Nakazawa M, Okazawa A, Padermschoke A, Nishio K, Hirai MY, Arita M, Ohta D. Wax ester and lipophilic compound profiling of Euglena gracilis by gas chromatography–mass spectrometry: toward understanding of wax ester fermentation under hypoxia. Metabolomics. 2015;11:175–183. doi: 10.1007/s11306-014-0687-1. [DOI] [Google Scholar]
- 11.Inui H, Ishikawa T, Tamoi M. Wax ester fermentation and its application for biofuel production. In: Schwartzhach SD, Shigeoka S, editors. Euglena: biochemistry, cell and molecular biology. Berlin: Springer; 2017. pp. 269–283. [DOI] [PubMed] [Google Scholar]
- 12.Ogawa T, Tamoi M, Kimura A, Mine A, Sakuyama H, Yoshida E, Maruta T, Suzuki K, Ishikawa T, Shigeoka S. Enhancement of photosynthetic capacity in Euglena gracilis by expression of cyanobacterial fructose-1,6-/sedoheptulose-1,7-bisphosphatase leads to increases in biomass and wax ester production. Biotechnol Biofuels. 2015;8:80. doi: 10.1186/s13068-015-0264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamada K, Suzuki H, Takeuchi T, Kazama Y, Mitra S, Abe T, Goda K, Suzuki K, Iwata O. Efficient selective breeding of live oil-rich Euglena gracilis with fluorescence-activated cell sorting. Sci Rep. 2016;6:26327. doi: 10.1038/srep26327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogbonna JC, Tomiyamal S, Tanaka H. Heterotrophic cultivation of Euglena gracilis Z for efficient production of α-tocopherol. J Appl Phycol. 1998;10:67–74. doi: 10.1023/A:1008011201437. [DOI] [Google Scholar]
- 15.Chae SR, Hwang EJ, Shin HS. Single cell protein production of Euglena gracilis and carbon dioxide fixation in an innovative photo-bioreactor. Bioresour Technol. 2006;97:322–329. doi: 10.1016/j.biortech.2005.02.037. [DOI] [PubMed] [Google Scholar]
- 16.Noble A, Kisiala A, Galer A, Clysdale D, Emery RJN. Euglena gracilis (Euglenophyceae) produces abscisic acid and cytokinins and responds to their exogenous application singly and in combination with other growth regulators. Eur J Phycol. 2014;49:244–254. doi: 10.1080/09670262.2014.911353. [DOI] [Google Scholar]
- 17.Amin SA, Parker MS, Armbrust EV. Interactions between diatoms and bacteria. Microbiol Mol Biol Rev. 2012;76:667–684. doi: 10.1128/MMBR.00007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuentes JL, Garbayo I, Cuaresma M, Montero Z, Gonzalez-Del-Valle M, Vilchez C. Impact of microalgae-bacteria interactions on the production of algal biomass and associated compounds. Mar Drugs. 2016;14:100. doi: 10.3390/md14050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramanan R, Kim BH, Cho DH, Oh HM, Kim HS. Algae-bacteria interactions: evolution, ecology and emerging applications. Biotechnol Adv. 2016;34:14–29. doi: 10.1016/j.biotechadv.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Villa JA, Ray EE, Barney BM. Azotobacter vinelandii siderophore can provide nitrogen to support the culture of the green algae Neochloris oleoabundans and Scenedesmus sp. BA032. FEMS Microbiol Lett. 2014;351:70–77. doi: 10.1111/1574-6968.12347. [DOI] [PubMed] [Google Scholar]
- 21.Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature. 2005;438:90–93. doi: 10.1038/nature04056. [DOI] [PubMed] [Google Scholar]
- 22.Amin SA, Hmelo LR, Tol HM, Durham BP, Carlson LT, Heal KR, Morales RL, Berthiaume CT, Parker MS, Djunaedi B, Ingalls AE, Parsek MR, Moran MA, Armbrust EV. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature. 2015;522:98–101. doi: 10.1038/nature14488. [DOI] [PubMed] [Google Scholar]
- 23.Amin SA, Green DH, Hart MC, Küpper FC, Sunda WG, Carrano CJ. Photolysis of iron-siderophore chelates promotes bacterial–algal mutualism. Proc Natl Acad Sci USA. 2009;106:17071–17076. doi: 10.1073/pnas.0905512106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amavizca E, Bahan Y, Ryu CM, Farag MA, Behout BM, de-Bashan LE. Enhanced performance of the microalga Chlorella sorokiniana remotely induced by the plant growth-promoting bacteria Azospirillum brasilense and Bacillus pumilus. Sci Rep. 2017;7:41310. doi: 10.1038/srep41310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez LE, Bashan Y. Increased growth of the microalga Chlorella vulgaris when co-immobilized and cocultured in alginate beads with the plant-growth-promoting bacterium Azospirillum brasilense. Appl Environ Microbiol. 2000;66:1527–1531. doi: 10.1128/AEM.66.4.1527-1531.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rivas MO, Vargas P, Riquelme CE. Interactions of Botryococcus braunii cultures with bacterial biofilms. Microb Ecol. 2010;60:628–635. doi: 10.1007/s00248-010-9686-6. [DOI] [PubMed] [Google Scholar]
- 27.Park J, Park BS, Wang P, Patidar SK, Kim JH, Kim SH, Han MS. Phycospheric native bacteria Pelagibaca bermudensis and Stappia sp. ameliorate biomass productivity of Tetraselmis striata (KCTC1432BP) in co-cultivation system through mutualistic interaction. Front Plant Sci. 2017;8:289. doi: 10.3389/fpls.2017.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim BH, Ramanan R, Cho DH, Oh HM, Kim HS. Role of Rhizobium, a plant growth promoting bacterium, in enhancing algal biomass through mutualistic interaction. Biomass Bioenergy. 2014;69:95–105. doi: 10.1016/j.biombioe.2014.07.015. [DOI] [Google Scholar]
- 29.Edwards U, Rogall T, Blöcker H, Emde M, Böttger EC. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reysenbach AL, Wickham GS, Pace NR. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl Environ Microbiol. 1994;60:2113–2119. doi: 10.1128/aem.60.6.2113-2119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hipkins MF, Baker NR. Spectroscopy. In: Hipkins MF, Baker NR, editors. Photosynthesis, energy transduction: a practical approach. Oxford: IRL Press; 1986. pp. 51–101. [Google Scholar]
- 32.Tanabe Y, Okazaki Y, Yoshida M, Matsuura H, Kai A, Shiratori T, Ishida K, Nakano S, Watanabe MM. A novel alphaproteobacterial ectosymbiont promotes the growth of the hydrocarbon-rich green alga Botryococcus braunii. Sci Rep. 2015;5:10467. doi: 10.1038/srep10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toyama T, Kasuya M, Hanaoka T, Kobayashi N, Tanaka Y, Inoue D, Sei K, Morikawa M, Mori K. Growth promotion of three microalgae, Chlamydomonas reinhardtii, Chlorella vulgaris and Euglena gracilis, by in situ indigenous bacteria in wastewater effluent. Biotechnol Biofuels. 2018;11:176. doi: 10.1186/s13068-018-1174-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joung Y, Hong S, Kim H, Kang H, Farrance CE, Joh K. Taeseokella kangwonensis gen. nov., sp. nov., isolated from a freshwater reservoir. Int J Syst Evol Microbiol. 2015;65:4309–4314. doi: 10.1099/ijsem.0.000582. [DOI] [PubMed] [Google Scholar]
- 35.Joung Y, Seo M-A, Kang H, Kim H, Ahn T-S, Cho J-C, Joh K. Emticicia aquatica sp. nov., a species of the family Cytophagaceae isolated from fresh water. Int J Syst Evol Microbiol. 2015;65:4358–4362. doi: 10.1099/ijsem.0.000577. [DOI] [PubMed] [Google Scholar]
- 36.Nam GG, Joung Y, Song J, Lim Y, Cho J-C. Emticicia fontis sp. nov., isolated from a freshwater pond. Int J Syst Evol Microbiol. 2016;66:5161–5166. doi: 10.1099/ijsem.0.001490. [DOI] [PubMed] [Google Scholar]
- 37.Ngo HTT, Trinh H, Yang J-E, Won K-H, Chu D-H, Kook M, Yi T-H. Emticicia aquatilis sp. nov., isolated from a freshwater sample. Int J Syst Evol Microbiol. 2017;67:1703–1708. doi: 10.1099/ijsem.0.001595. [DOI] [PubMed] [Google Scholar]
- 38.Kang H, Kim H, Joung Y, Joh K. Emticicia paludis sp. nov., isolated from wetland freshwater. Int J Syst Evol Microbiol. 2016;66:3383–3387. doi: 10.1099/ijsem.0.001205. [DOI] [PubMed] [Google Scholar]
- 39.Li D-D, Peng M, Wang N, Wang X-J, Zhang X-Y, Chen X-L, Su H-N, Zhang Y-Z, Shi M. Arcticibacterium luteifluviistationis gen. nov., sp. nov., isolated from Arctic seawater. Int J Syst Evol Microbiol. 2017;67:664–669. doi: 10.1099/ijsem.0.001690. [DOI] [PubMed] [Google Scholar]
- 40.Park S-E, Lee S, Kim H, Ahn T-Y. Emticicia sediminis sp. nov. isolated from sediment of a shallow stream. Int J Syst Evol Microbiol. 2015;65:2496–2499. doi: 10.1099/ijs.0.000292. [DOI] [PubMed] [Google Scholar]
- 41.Liu Q-M, Ten LN, Yu H-S, Jin F-X, Im W-T, Lee S-T. Emticicia ginsengisoli sp. nov., a species of the family ‘Flexibacteraceae’ isolated from soil of a ginseng field. Int J Syst Evol Microbiol. 2008;58:1100–1105. doi: 10.1099/ijs.0.65386-0. [DOI] [PubMed] [Google Scholar]
- 42.Chen Q, Zang X-X, Hang X, Wang H-M, Jia W-B, Li S-P, Jiang J-D, Chen K. Emticicia soli sp. nov., a novel member of the family ‘Flexibacteraceae’, isolated from tetrabromobisphenol A-contaminated soil. Int J Syst Evol Microbiol. 2017;67:2885–2890. doi: 10.1099/ijsem.0.002041. [DOI] [PubMed] [Google Scholar]
- 43.Lucy M, Reed E, Glick BR. Applications of free living plant growth-promoting rhizobacteria. Antonie Van Leeuwenhoek. 2004;86:1–25. doi: 10.1023/B:ANTO.0000024903.10757.6e. [DOI] [PubMed] [Google Scholar]
- 44.Liu W, Yang C, Shi S, Shu W. Effects of plant growth-promoting bacteria isolated from copper tailings on plants in sterilized and non-sterilized tailings. Chemosphere. 2014;97:47–53. doi: 10.1016/j.chemosphere.2013.10.089. [DOI] [PubMed] [Google Scholar]
- 45.Leyva LA, Bashan Y, Mendoza A, de Bashan LE. Accumulation fatty acids of in Chlorella vulgaris under heterotrophic conditions in relation to activity of acetyl-CoA carboxylase, temperature, and co-immobilization with Azospirillum brasilense. Naturwissenschaften. 2014;101:819–830. doi: 10.1007/s00114-014-1223-x. [DOI] [PubMed] [Google Scholar]
- 46.Cho DH, Ramanan R, Heo J, Lee J, Kim BH, Oh HM, Kim HS. Enhanced microalgal biomass productivity by engineering a microalgal–bacterial community. Bioresour Technol. 2015;175:578–585. doi: 10.1016/j.biortech.2014.10.159. [DOI] [PubMed] [Google Scholar]
- 47.Xue L, Shang H, Ma P, Wang X, He X, Niu J, Wu J. Analysis of growth and lipid production characteristics of Chlorella vulgaris in artificially constructed consortia with symbiotic bacteria. J Basic Microbiol. 2018;58:358–367. doi: 10.1002/jobm.201700594. [DOI] [PubMed] [Google Scholar]
- 48.Palacios OA, Choix FJ, Bashan Y, de Bashan LE. Influence of tryptophan and indole-3-acetic acid on starch accumulation in the synthetic mutualistic Chlorella sorokiniana–Azospirillum brasilense system under heterotrophic conditions. Res Microbiol. 2016;167:367–379. doi: 10.1016/j.resmic.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 49.Shen Y, Gao J, Li L. Municipal wastewater treatment via co-immobilized microalgal–bacterial symbiosis: microorganism growth and nutrients removal. Bioresour Technol. 2017;243:905–913. doi: 10.1016/j.biortech.2017.07.041. [DOI] [PubMed] [Google Scholar]
- 50.Palacios OA, Lopez BR, Bashan Y, de Bashan LE. Early changes in nutritional conditions affect formation of synthetic mutualism between Chlorella sorokiniana and the bacterium Azospirillum brasilense. Microb Ecol. 2019;77:980–992. doi: 10.1007/s00248-018-1282-1. [DOI] [PubMed] [Google Scholar]
- 51.Zhu J, Wakisaka M. Growth promotion of Euglena gracilis by ferulic acid from rice bran. AMB Express. 2018;8:16. doi: 10.1186/s13568-018-0547-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Summary of E. gracilis collection method and fate of EG3 cells during this method.
Additional file 2: Figure S2. Effects of isolated strains EG3, EG5, EG6, EG8, EG9, and EG10 on E. gracilis growth in autoclave heat-sterilized wastewater effluent.
Data Availability Statement
The data sets used and/or analyzed in this study are available from the corresponding author upon reasonable request.