Abstract
Background: Dysregulated expression of long non-coding RNA gastric carcinoma high expressed transcript 1 (lncRNA GHET1) has been observed in several cancers, however, definite conclusion on the prognostic value of lncRNA GHET1 expression in human cancers has not been determined. The aim of this meta-analysis was to evaluate the prognostic significance of lncRNA GHET1 expression in cancers. Methods: PubMed, Web of Science and Embase were comprehensively searched for relevant studies. Meta-analyses of overall survival (OS) and clinicopathological features were conducted. Results: Ten studies were finally analyzed in the present study. High lncRNA GHET1 expression was associated with shorter OS than low lncRNA GHET1 expression in cancers (hazard ratio (HR) = 2.59, 95% CI = 1.93–3.47, P<0.01). Online cross-validation using The Cancer Genome Atlas (TCGA) data observed similar results (HR = 1.10, P<0.05). When compared with low lncRNA GHET1 expression, high lncRNA GHET1 expression was related to larger tumor size (P<0.01), worse differentiation (P<0.01), earlier distant metastasis (P=0.02), earlier lymph node metastasis (P<0.01) and more advanced clinical stage (P<0.01). Conclusion: High lncRNA GHET1 expression is associated with worse cancer prognosis and can serve as a promising prognostic factor of human cancers.
Keywords: Cancer, LncRNA GHET1, Meta-analysis, Prognosis
Background
Cancer has become a leading cause of death and a vital public health problem worldwide [1,2]. Although great advancements have been achieved in the diagnosis and treatment of cancers in recent years, many people suffer from disappointing results [2]. The lack of efficient biomarkers to supervise the clinical outcomes and predict the prognosis is supposed to be an important reason for the poor prognosis of cancer patients [3–6].
Long non-coding RNA (lncRNA), longer than 200 nucleotides, is an important member of non-coding RNA family [7]. A great number of studies have found that lncRNAs play a crucial role in the development of human diseases although lncRNA is short of the ability to code proteins [7,8]. Recently, accumulating evidence shows lncRNA is involved with tumor tumorigenesis, invasion and metastasis [9]. Several lncRNAs have been identified as prognostic factors in cancers, such as metastasis-associated lung adenocarcinoma transcript 1 (MAlAT1) [10] and cancer susceptibility 2 (CASC2) [5]. Gastric carcinoma high expressed transcript 1 (GHET1), a kind of lncRNA with the length of 1913 nt, is located at chromosome 7q36.1 position in the human genome [11]. Recently, many studies found that lncRNA GHET1 contributed to the cancer progression and had the potential ability to predict the cancer prognosis [12–21]. However, definite conclusion has not been obtained for contradictory results among different studies. For instance, Xia et al. [19] study showed there was no obvious relationship between lncRNA GHET1 expression and lymph node metastasis (P=0.41), similar results were observed in Yang et al. study (P=0.20) [20]. Nevertheless, Liu et al. [15] detected the significant association between lncRNA GHET1 expression and lymph node metastasis (P<0.01), similarly, Shen et al. [17] also discovered the evident connection between high lncRNA GHET1 expression and earlier lymph node metastasis (P<0.01). In view of these conflicting data, for the first time, we performed this systematic review and meta-analysis to evaluate the prognostic significance of lncRNA GHET1 expression in cancers.
Materials and methods
Literature search and selection
We searched PubMed, Web of Science and Embase using the following strategy: (‘long non-coding RNA’ OR ‘lncRNA’) AND (‘gastric carcinoma high expressed transcript 1’ OR ‘GHET1’) AND (‘tumor’ OR ‘cancer’ OR ‘carcinoma’). The last literature search was conducted on 5 May 2019. Literature selection was performed according to inclusion and exclusion standards. Two authors completed the literature search and selection independently, and any disagreement was solved by group discussion.
Inclusion standards and exclusion standards
The study would be included into this research if it met the following inclusion standards: (i) patients were diagnosed with cancers; (ii) patients with high lncRNA GHET1 expression were divided into research group; (iii) patients with low lncRNA GHET1 expression were divided into control group; (iv) association of lncRNA GHET1 expression with overall survival (OS), progression-free survival (PFS) or clinicopathological parameters was reported; (v) studies contained retrospective cohorts or perspective cohorts. The following studies were directly excluded from this meta-analysis: reviews, comments, letters, animal experiments, cell experiments, duplicated publications or studies without sufficient data.
Data extraction and quality assessment
We extracted the following items using a prepared template: first author, publication year, country, sample size, gender, lncRNA GHET1 expression, cut-off value, detection methods, type of cancer and outcomes. Especially, hazard ratio (HR) and 95% confidence interval (CI) of OS were directly obtained from published studies. If HR and corresponding 95% CI were not directly reported, both of them could be indirectly extracted from survival curves used Engauge Digitizer 4.1 [22]. Quality of each included study was assessed using Newcastle–Ottawa Scale (NOS) [23]. The study with NOS ≥ 6 was considered as high-quality study. The process of data extraction and quality assessment was completed by two authors independently. Any disagreement was solved by group discussion.
Online cross-validation
We conducted online cross-validation to validate the prognostic role of lncRNA GHET1 expression in human cancers using Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/index.html) based on The Cancer Genome Atlas (TCGA) data (https://cancergenome.nih.gov/).
Statistical analysis
HR and corresponding 95% CI were pooled to determine the association between lncRNA GHET1 expression and OS. Odds ratio (OR) and 95% CI were used to assess the relationship between lncRNA GHET1 expression and clinicopathological features, such as age, gender and tumor size. Heterogeneity was assessed via chi-square-based Q and I2 tests across studies. A fixed-effect model was used when the heterogeneity was obvious (I2 > 50 or P<0.05). Otherwise, a random-effect model was applied (I2 ≤ 50 or P≥0.05). Forest plot was applied to show the overall effects. Funnel plot, Begg’s test and Egger’s test were generated to evaluate the publication bias. Sensitivity analysis was conducted to check the robustness of results by omitting one study at a time. All analyses were performed using Review Manager 5.3 (The Cochrane Collaboration, Copenhagen, Denmark) and Stata 12.0 (Stata, College Station, TX, U.S.A.). All P-values were two-sided and a P-value less than 0.05 indicated the results were statistically significant.
Results
Literature search and selection
As shown in Figure 1, a total of 81 papers were retrieved from three common databases. After removal of duplicates, 27 papers remained for further evaluation. Then, 13 papers were directly excluded by scanning titles or abstracts. The remaining 14 papers were further checked for eligibility by reading full-texts, and then 4 papers were removed. Ultimately, ten studies were included into this systematic review and meta-analysis.
Figure 1. Literature search and selection.
Characteristics of included studies
Characteristics of included studies were listed in Table 1. A total of 654 patients (350 males and 304 females) were included in this research [12–21]. All studies were conducted in China and sample size varied from 42 to 105 [12–21]. There were 323 and 331 patients in high lncRNA GHET1 expression group and low lncRNA GHET1 expression group, respectively. The expression level of lncRNA GHET1 was evaluated using quantitative real-time polymerase chain reaction (qRT-PCR) in all studies [12–21]. Besides, nine studies used median value [12,14–21] and one study used mean value [13] as the cut-off value. Eight kinds of cancer were investigated, including non-small-cell lung carcinoma (NSCLC) [12,17], hepatocellular carcinoma (HCC) [13], bladder cancer [14], esophageal squamous cell carcinoma (ESCC) [16], head and neck cancer (HNC) [15], breast cancer [18], gastric cancer [19,20] and pancreatic cancer [21]. Moreover, all studies reported clinicopathological parameters [12–21], seven studies reported OS [12–15,17,18,20] and one study reported PFS [17]. NOS was equal to or greater than six in all studies, which suggested all studies were of high quality [12–21].
Table 1. Characteristics of included studies.
| Study | Country | Sample size (n) | Gender (M/F) (n) | GHET1 expression (H/L) (n) | Detection method | Cut-off value | Cancer | Outcomes | NOS |
|---|---|---|---|---|---|---|---|---|---|
| Guan (2017) [12] | China | 52 | 40/12 | 25/27 | qRT-PCR | Median | NSCLC | CP, OS | 7 |
| Jin (2017) [13] | China | 68 | 35/33 | 27/41 | qRT-PCR | Mean | HCC | CP, OS | 7 |
| Li (2014) [14] | China | 80 | 43/37 | 39/41 | qRT-PCR | Median | Bladder cancer | CP, OS | 7 |
| Liu (2017) [16] | China | 55 | 34/21 | 28/27 | qRT-PCR | Median | ESCC | CP | 6 |
| Liu (2018) [15] | China | 86 | 61/25 | 43/43 | qRT-PCR | Median | HNC | CP,OS | 7 |
| Shen (2018) [17] | China | 105 | 44/61 | 53/52 | qRT-PCR | Median | NSCLC | CP, OS, PFS | 7 |
| Song (2018) [18] | China | 60 | 0/60 | 30/30 | qRT-PCR | Median | Breast cancer | CP, OS | 7 |
| Xia (2018) [19] | China | 42 | 28/14 | 21/21 | qRT-PCR | Median | Gastric cancer | CP | 6 |
| Yang (2014) [20] | China | 42 | 31/11 | 21/21 | qRT-PCR | Median | Gastric cancer | CP, OS | 7 |
| Zhou (2017) [21] | China | 64 | 34/30 | 36/28 | qRT-PCR | Median | Pancreatic cancer | CP | 6 |
Abbreviations: CP, clinicopathological parameter; F, female; H, high GHET1 expression; L, low GHET1 expression; M, male.
Meta-analysis of OS
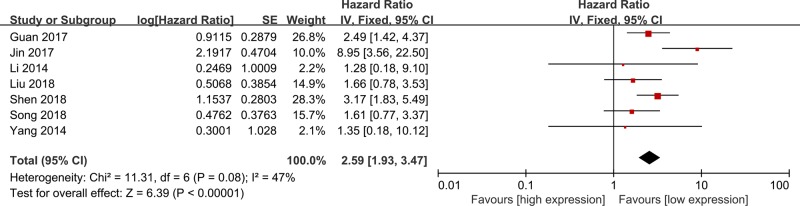
Seven studies were included in the meta-analysis of OS (Figure 2) [12–15,17,18,20]. A fixed-effect model was used for mild heterogeneity across included studies (I2 = 47%, P=0.08), and results showed high lncRNA GHET1 expression was significantly associated with shorter OS than low lncRNA GHET1 expression in cancers (HR = 2.59, 95% CI = 1.93–3.47, P<0.01). Subgroup analyses also indicated the obvious relationship between high lncRNA GHET1 expression and shorter OS in cancers (P<0.05) (Table 2).
Figure 2. Meta-analysis of OS.
Table 2. Subgroup analysis of OS.
| Variables | Studies (n) | HR, 95%CI | P | Heterogeneity | Model | |
|---|---|---|---|---|---|---|
| I2 (%) | P | |||||
| Sample size (n) | ||||||
| ≤60 | 3 | 2.07 (1.34, 3.21) | <0.01* | 0 | 0.60 | Fixed |
| >60 | 4 | 3.10 (1.47, 6.55) | <0.01* | 65 | 0.04 | Random |
| Cut-off value | ||||||
| Median | 6 | 2.26 (1.66, 3.07) | <0.01* | 0 | 0.61 | Fixed |
| Mean | 1 | 8.95 (3.56, 22.50) | <0.01* | NA | NA | Fixed |
| Cancer type | ||||||
| NSCLC | 2 | 2.82 (1.90, 4.18) | <0.01* | 0 | 0.55 | Fixed |
| Others | 5 | 2.34 (1.06, 5.16) | 0.03* | 62 | 0.03 | Random |
Abbreviation: NA, not available.
*P<0.05 indicating significant association between GHET1 expression and OS.
Online cross-validation
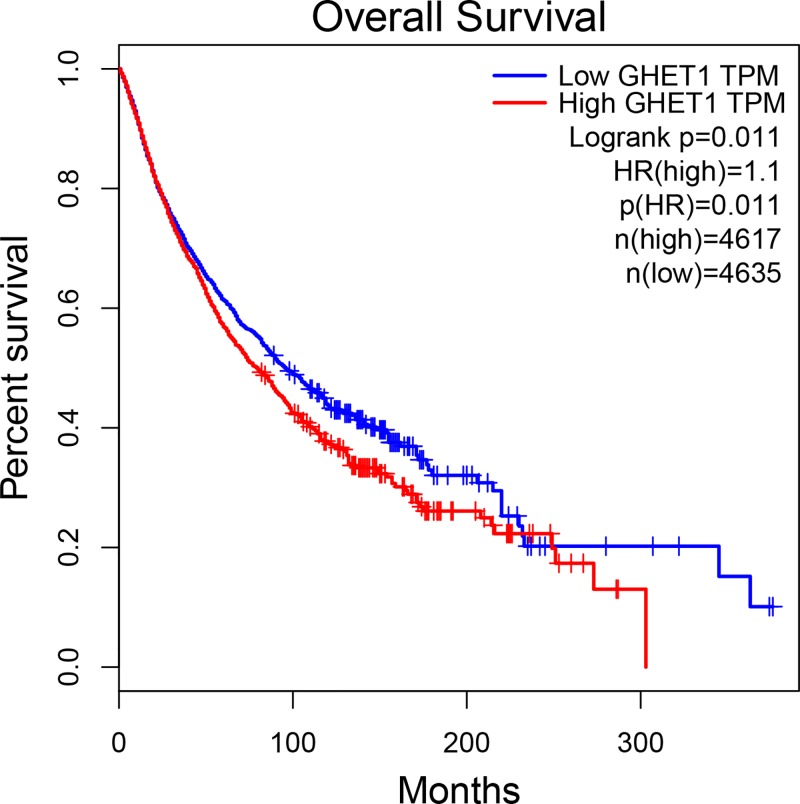
As shown in Figure 3, online cross-validation using TCGA data showed patients with high lncRNA GHET1 expression tended to have shorter OS compared with those with low lncRNA GHET1 expression (HR = 1.10, P<0.05).
Figure 3. Online cross-validation using TCGA data.
Meta-analysis of clinicopathological features
As listed in Table 3, there was no distinct relationship between lncRNA GHET1 expression and age (P=0.70) or gender (P=0.74). Nevertheless, high lncRNA GHET1 expression was obviously related to larger tumor size (P<0.01), worse differentiation (P<0.01), earlier distant metastasis (P=0.02), earlier lymph node metastasis (P<0.01) and more advanced clinical stage (P<0.01) than low lncRNA GHET1 expression in cancers.
Table 3. Association between GHET1 expression and clinicopathological features.
| Variables | Studies (n) | Patients (n) | High expression group (%) | Low expression group (%) | OR 95% CI | P | Heterogeneity | Model | |
|---|---|---|---|---|---|---|---|---|---|
| I2 (%) | P | ||||||||
| Age (old versus young) | 9 | 574 | 50.7 versus 49.3 | 52.1 versus 47.9 | 0.94 (0.67, 1.31) | 0.70 | 0 | 0.88 | Fixed |
| Gender (male versus female) | 8 | 514 | 59.1 versus 40.9 | 60.4 versus 39.6 | 0.94 (0.65, 1.35) | 0.74 | 0 | 0.81 | Fixed |
| Tumor size (large versus small) | 8 | 522 | 62.9 versus 37.1 | 35.7 versus 64.3 | 3.06 (2.14, 4.38) | <0.01* | 45 | 0.08 | Fixed |
| Tumor differentiation (poor versus well) | 6 | 345 | 63.6 versus 36.4 | 44.4 versus 55.6 | 2.32 (1.48, 3.64) | <0.01* | 27 | 0.23 | Fixed |
| Distant metastasis (yes versus no) | 3 | 148 | 15.4 versus 84.6 | 2.9 versus 97.1 | 4.63 (1.23, 17.38) | 0.02* | 0 | 0.59 | Fixed |
| Lymph node metastasis (yes versus no) | 7 | 442 | 59.7 versus 40.3 | 30.3 versus 69.7 | 3.81 (2.51, 5.77) | <0.01* | 49 | 0.07 | Fixed |
| Clinical stage (III/IV versus I/II) | 6 | 422 | 62.8 versus 37.2 | 30.4 versus 69.6 | 3.92 (2.60, 5.91) | <0.01* | 0 | 0.92 | Fixe |
*P<0.05 indicating significant association between GHET1 expression and clinicopathological features.
Publication bias and sensitivity analysis
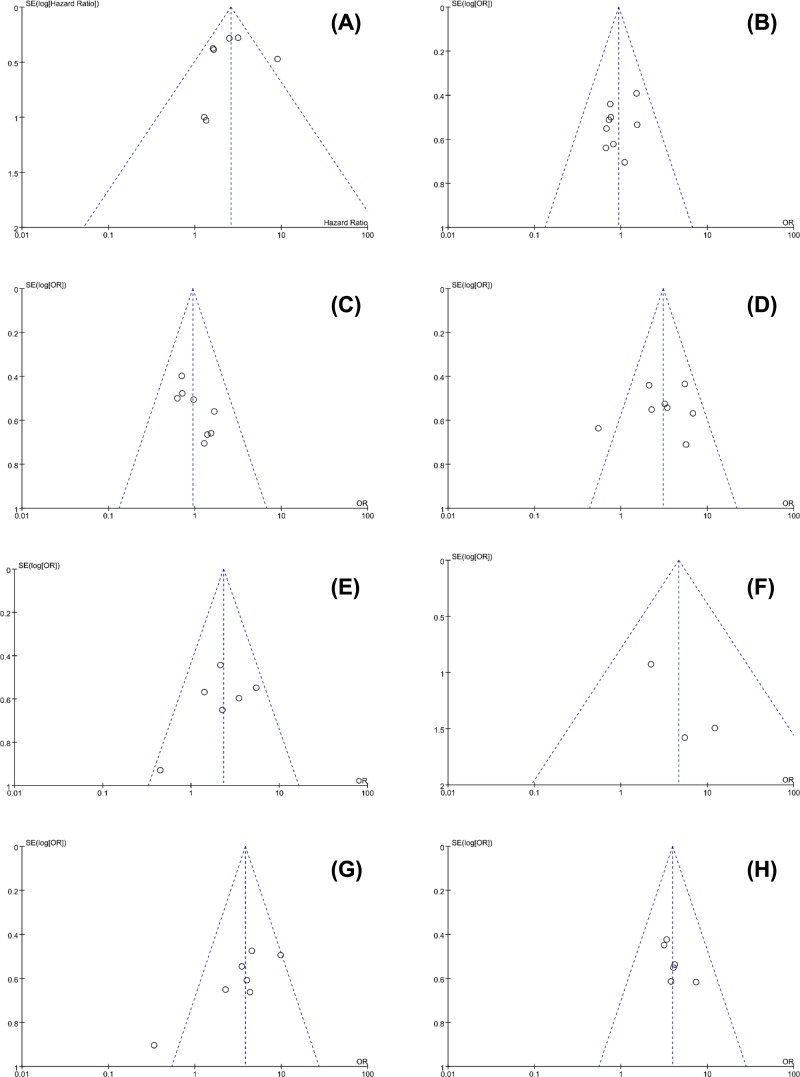
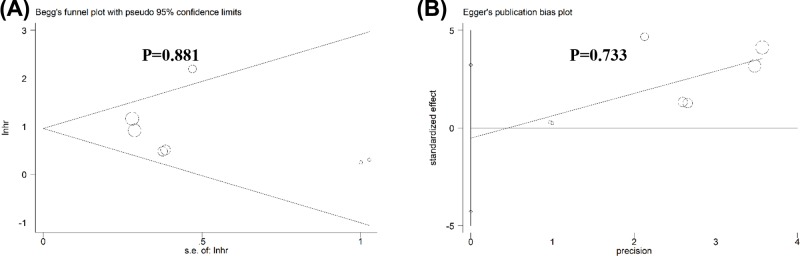
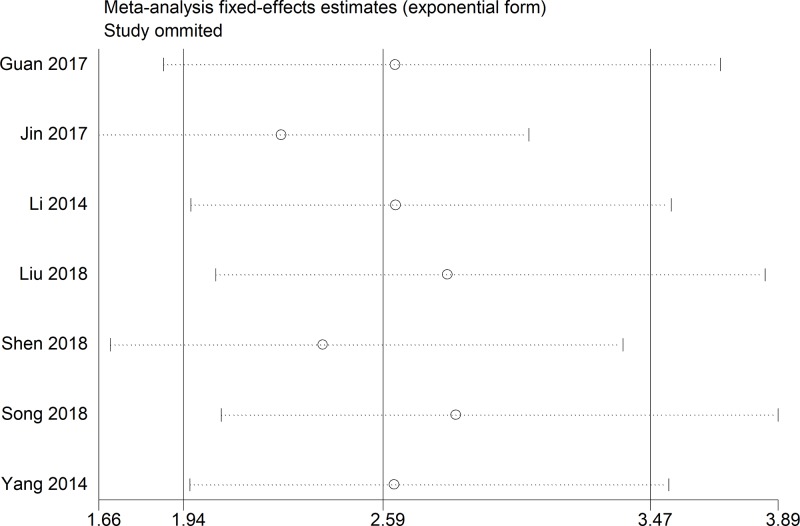
No obvious publication bias across included studies was observed in all analyses (Figure 4). Especially, as for the meta-analysis of OS, publication bias was also assessed using Begg’s test (P=0.881) and Egger’s test (P=0.733), and no distinct publication bias was found (Figure 5). Sensitivity analysis for the meta-analysis of OS was conducted, and results were not altered after removal of any included study (Figure 6).
Figure 4. Funnel plots for all meta-analyses.
Figure 5. Begg’s test and Egger’s test for the meta-analysis of OS.
Figure 6. Sensitivity analysis for the meta-analysis of OS.
Discussion
LncRNA has been proved to associate with cancer tumorigenesis, invasion, differentiation and metastasis [24]. Several lncRNAs have been demonstrated as prognostic biomarkers of human cancers [5,25]. Although accelerating evidence indicates lncRNA GHET1 may have the potential ability to predict the cancer prognosis, clear mechanism has not been obtained. Guan et al. [12] found that knockdown of lncRNA GHET1 could suppress the proliferation and invasion capacity of NSCLC cells by suppressing LATS1/YAP pathway signaling pathway in NSCLC cells. Xia et al. [19] discovered that down-regulation of lncRNA GHET1 inhibited the migration, invasion and proliferation of gastric cancer cells via up-regulating P21 expression and down-regulating cyclin and CDK expression to inhibit the G0/G1 to S phase transition. Song et al. [18] found lncRNA GHET1 promoted the cancer progression via EMT in breast cancer. Jin et al. [13] found lncRNA GHET1 facilitated the HCC cell proliferation by silencing KLF2 and further caused disappointing results. Ding et al. [26] study showed overexpression of ATF1 reversed the lncRNA GHET1 knockdown-mediated inhibition on the progression of HCC cells. Yang et al. [20] observed that lncRNA GHET1 promoted cancer cell proliferation by increasing c-Myc mRNA stability in gastric cancer.
In order to determine the prognostic significance of lncRNA GHET1 expression in human cancers, we performed this meta-analysis by integrating the current evidence [12–21]. To our knowledge, the present study was the first meta-analysis to evaluate the association between lncRNA GHET1 expression and cancer prognosis. We discovered that, compared with low lncRNA GHET1 expression, high lncRNA GHET1 expression was associated with worse OS and several clinicopathological features, including tumor size, differentiation, distant metastasis, lymph node metastasis and clinical stage. Besides, online-cross validation also indicated that high lncRNA GHET1 expression was an unfavorable prognostic factor of cancer. Overall, lncRNA GHET1 expression could serve as a potential prognostic biomarker for human cancers.
Some limitations should be considered when elucidating our data. First, only ten studies were included into this meta-analysis, and the relatively small sample size might lower the stringency of results. To eliminate this limitation, we have used TCGA data with a large population to validate our results, hence, we believe our study can provide reliable conclusion. Second, although we do not impose restrictions on the country when performing literature search and selection, all included studies are performed in China, which generates a region bias. However, as aforesaid, we have used TCGA data to validate the results, therefore, we believe our conclusion can be extended into other countries. Third, the cut-off value of lncRNA GHET1 expression varies a lot among different studies, as a result, definite cut-off value has not be obtained, which may limit the clinical use of our conclusion. Nevertheless, as aforesaid, this meta-analysis is a preliminary study to acknowledge the prognostic significance of lncRNA GHET1 expression in cancers, and more researches should be carried out to identify the optimal cut-off value of lncRNA GHET1 expression in future.
Conclusion
High lncRNA GHET1 expression is associated with worse OS and clinicopathological features compared with low lncRNA GHET1 expression in human cancers. LncRNA GHET1 expression can serve as a promising prognostic factor of cancers.
Abbreviations
- CI
confidence interval
- GHET1
gastric carcinoma high expressed transcript 1
- HCC
hepatocellular carcinoma
- HR
hazard ratio
- LncRNA
long non-coding RNA
- NOS
Newcastle–Ottawa scale
- NSCLC
non-small-cell lung carcinoma
- OS
overall survival
- PFS
progression-free survival
- qRT-PCR
quantitative real-time polymerase chain reaction
- TCGA
The Cancer Genome Atlas
Author Contribution
Study concepts and design: Haiyan Sun and Jing Ye. Literature search: Jing Ye, Zhengquan Feng and Qiqing Zhang. Data extraction: Jing Ye, Qiqin Zhang and Yongliang Xia. Manuscript preparation and revision: Jing Ye, Qiqing Zhang and Yunxi Ji. All authors have participated sufficiently in the study and approved the final version.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This study is supported by Construction Program of Chen Yi National Famous Old TCM Experts Inheritance Studio [grant number G.TCM.R.J.F.20110041].
References
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J.and Jemal A. (2015) Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.Miller K.D., Siegel R.L., Lin C.C., Mariotto A.B., Kramer J.L.and Rowland J.H. (2016) Cancer treatment and survivorship statistics, 2016. CA Cancer J. Clin. 66, 271–289 10.3322/caac.21349 [DOI] [PubMed] [Google Scholar]
- 3.Zhou Y., Cheng S., Chen S.and Zhao Y. (2018) Prognostic and clinicopathological value of SIRT3 expression in various cancers: a systematic review and meta-analysis. OncoTargets Ther. 11, 2157–2167 10.2147/OTT.S157836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou Y., Cheng S., Fathy A.H., Qian H.and Zhao Y. (2018) Prognostic value of platelet-to-lymphocyte ratio in pancreatic cancer: a comprehensive meta-analysis of 17 cohort studies. OncoTargets Ther. 11, 1899–1908 10.2147/OTT.S154162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu L., Chen S., Bao H., Zhang W., Liao M.and Liang Q. (2018) The role of lncRNA CASC2 on prognosis of malignant tumors: a meta-analysis and bioinformatics. OncoTargets Ther. 11, 4355–4365 10.2147/OTT.S166132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang T., Wu D.M., Deng S.H., Han R., Liu T.and Li J. (2018) Integrated analysis reveals that long non-coding RNA TUBA4B can be used as a prognostic biomarker in various cancers. Cell. Physiol. Biochem. 49, 530–544 10.1159/000492991 [DOI] [PubMed] [Google Scholar]
- 7.Espinosa J.M. (2017) On the origin of lncRNAs: missing link found. Trends Genet. 33, 660 10.1016/j.tig.2017.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jandura A.and Krause H.M. (2017) The new RNA world: growing evidence for long noncoding RNA functionality. Trends Genet. 33, 665 10.1016/j.tig.2017.08.002 [DOI] [PubMed] [Google Scholar]
- 9.Yang G., Lu X.and Yuan L. (2014) LncRNA: a link between RNA and cancer. Biochim. Biophys. Acta 1839, 1097–1109 [DOI] [PubMed] [Google Scholar]
- 10.Li J., Cui Z., Li H., Lv X., Gao M.and Yang Z. (2018) Clinicopathological and prognostic significance of long noncoding RNA MALAT1 in human cancers: a review and meta-analysis. Cancer Cell Int. 18, 109 10.1186/s12935-018-0606-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ni W., Luo L., Zuo P., Li R.P., Xu X.B.and Wen F. (2018) lncRNA GHET1 down-regulation suppresses the cell activities of glioma. Cancer Biomark. 10.3233/CBM-171002 [DOI] [PubMed] [Google Scholar]
- 12.Guan Z.B., Cao Y.S., Li Y., Tong W.N.and Zhuo A.S. (2018) Knockdown of lncRNA GHET1 suppresses cell proliferation, invasion and LATS1/YAP pathway in non small cell lung cancer. Cancer Biomark. 21, 557–563 10.3233/CBM-170431 [DOI] [PubMed] [Google Scholar]
- 13.Jin L., He Y., Tang S.and Huang S. (2018) LncRNA GHET1 predicts poor prognosis in hepatocellular carcinoma and promotes cell proliferation by silencing KLF2. J. Cell. Physiol. 233, 4726–4734 10.1002/jcp.26257 [DOI] [PubMed] [Google Scholar]
- 14.Li L.J., Zhu J.L., Bao W.S., Chen D.K., Huang W.W.and Weng Z.L. (2014) Long noncoding RNA GHET1 promotes the development of bladder cancer. Int. J. Clin. Exp. Pathol. 7, 7196–7205 [PMC free article] [PubMed] [Google Scholar]
- 15.Liu H.and Wu Y. (2018) Long non-coding RNA gastric carcinoma highly expressed transcript 1 promotes cell proliferation and invasion in human head and neck cancer. Oncol. Lett. 15, 6941–6946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu H., Zhen Q.and Fan Y. (2017) LncRNA GHET1 promotes esophageal squamous cell carcinoma cells proliferation and invasion via induction of EMT. Int. J. Biol. Mark. 32, e403 10.5301/ijbm.5000304 [DOI] [PubMed] [Google Scholar]
- 17.Shen Q.M., Wang H.Y.and Xu S. (2018) LncRNA GHET1 predicts a poor prognosis of the patients with non-small cell lung cancer. Eur. Rev. Med. Pharmacol. Sci. 22, 2328–2333 [DOI] [PubMed] [Google Scholar]
- 18.Song R., Zhang J., Huang J.and Hai T. (2018) Long non-coding RNA GHET1 promotes human breast cancer cell proliferation, invasion and migration via affecting epithelial mesenchymal transition. Cancer Biomark. 22, 565–573 10.3233/CBM-181250 [DOI] [PubMed] [Google Scholar]
- 19.Xia Y., Yan Z., Wan Y., Wei S., Bi Y.and Zhao J. (2018) Knockdown of long noncoding RNA GHET1 inhibits cellcycle progression and invasion of gastric cancer cells. Mol. Med. Rep. 18, 3375–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang F., Xue X., Zheng L., Bi J., Zhou Y.and Zhi K. (2014) Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. FEBS J. 281, 802–813 10.1111/febs.12625 [DOI] [PubMed] [Google Scholar]
- 21.Zhou H.Y., Zhu H., Wu X.Y., Chen X.D., Qiao Z.G.and Ling X. (2017) Expression and clinical significance of long-non-coding RNA GHET1 in pancreatic cancer. Eur. Rev. Med. Pharmacol. Sci. 21, 5081–5088 [DOI] [PubMed] [Google Scholar]
- 22.Tierney J.F., Stewart L.A., Ghersi D., Burdett S.and Sydes M.R. (2007) Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 8, 16 10.1186/1745-6215-8-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stang A. (2010) Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 25, 603–605 10.1007/s10654-010-9491-z [DOI] [PubMed] [Google Scholar]
- 24.Slaby O., Laga R.and Sedlacek O. (2017) Therapeutic targeting of non-coding RNAs in cancer. Biochem. J. 474, 4219 10.1042/BCJ20170079 [DOI] [PubMed] [Google Scholar]
- 25.Liu X., Ming X., Jing W., Luo P., Li N.and Zhu M. (2018) Long non-coding RNA XIST predicts worse prognosis in digestive system tumors: a systemic review and meta-analysis. Biosci. Rep. 38, 10.1042/BSR20180169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding G., Li W., Liu J., Zeng Y., Mao C.and Kang Y. (2017) LncRNA GHET1 activated by H3K27 acetylation promotes cell tumorigenesis through regulating ATF1 in hepatocellular carcinoma. Biomed. Pharmacother. 94, 326–331 [DOI] [PubMed] [Google Scholar]