Abstract
Menin is encoded by multiple endocrine neoplasia type 1 (MEN1) gene, the germ line mutations of which are the main cause of pancreatic neuroendocrine tumors (PNETs). To date, a large number of frameshift, nonsense and missense mutations of MEN1 have been identified to be responsible for part of MEN1-defficient PNETs patients due to truncation or rapid degradation of menin protein. However, the stability of the wild-type (WT) menin in PNETs is totally unknown. In the present study, we observed ubiquitination of WT menin in 293T cells by transfection of ectopic WT menin and HA-ubiquitin. As expected, either endogenous or ectopic WT menin is stable in 293T cells, whereas in INS-1 cells, a rat insulinoma cell line derived from PNETs, either endogenous or ectopic WT menin is rapidly degraded through ubiquitin-proteasome pathway. Furthermore, the degradation of WT menin is more rapid in the presence of serum. Our findings suggest that in part of PNETs patients with WT MEN1, a ubiquitin-proteasome system targeting menin is untimely activated.
Keywords: INS-1 cell, Insulinoma, Menin, Ubiquitin-proteasome
Introduction
Pancreatic neuroendocrine tumors (PNETs) arise from the endocrine cells of the pancreas which are also known as the islets of Langerhans [1]. Some PNETs occur in patients with multiple endocrine neoplasia type 1 (MEN1) [2], identified by the presence of neoplasms in two or more different endocrine tissues such as parathyroid glands, endocrine pancreas and anterior pituitary gland [3]. The MEN1 gene encodes for the protein menin and individuals with MEN1 mutations will develop MEN1 [1]. As reported by Papadopoulos et al. in 2011, 44% of the PNETs had somatic inactivating mutations in MEN1 [4], indicating a strong correlation between menin-deficiency and the development/progression/initiation of PNETs.
Menin, an evolutionarily conserved and ubiquitously distributed scaffold protein, has functions in multiple physiological processes. Menin has been shown to function in DNA repair, cell cycle and cell structure through interacting with FANCD2 [5], the 32-kDa subunit (RPA2) of replication protein A [6] and GFAP and vemintin [7]. In addition, Menin can interact with a number of transcriptional activators including protein-energy malnutrition (Pem) [8], Hlxb9 [9] and mixed lineage leukemia proteins (MLL) [10] to activate the expression of tissue-specific target genes and recruit histone modifiers including histone deacetylase (HDAC)1/2 [11], Sirt1 [12], the histone H3 lysine 27 methyltransferase EZH2 [13], and protein arginine methyltransferase (PRMT) 5 [14] to repress the transcription of target genes. As a tumor suppressor, the deletion of MEN1 in mouse pancreatic β cells results in the formation of functional PNETs (insulinomas) after 6 months of age [15], suggesting that menin is essential for the control of β-cells proliferation.
Although majority of the deficiency of encoded protein menin in PNETs is caused by abnormal transcripts from inactivating mutations of MEN1 [16] or the rapid degradation of missense mutants via the ubiquitin-proteasome pathway [17,18], some PNETs with wild-type (WT) sequence of MEN1 also showed a lower level of menin expression in menin immunohistochemical staining [19]. Furthermore, it is unknown that whether the menin in PNETs with WT MEN1 is also unstable. In the present study, we observed ubiquitination of WT menin in vivo and first reported the rapid degradation of endogenous or ectopic WT menin in one of PNET-derived cell line.
Materials and methods
Cell lines and cell culture
Stable Flag-Menin-expressing INS-1 cells were established by transduction with pMX-puro-Menin and RetroQ-puro-Shmen1-derived retroviruses, as previously reported by Feng et al. [20]. 293T cells were cultured in Dulbecco’s Modified Eagle’s Medium (HyClone) supplemented with 10% FBS and 1% Pen/Strep. INS-1 cells were cultured in RPMI 1640 (Gibco) supplemented with 10% FBS, 1 mol/l Hepes, 0.2 mol/l l-glutamine, 0.1 mol/l Sodium pyruvate, 55 mmol/l β-Mercaptoethanol.
Plasmids
WT full-length menin was amplified by PCR and cloned into the BamHI/NotI site of pCDNA3.1. Retroviral plasmid pMX-puro-menin was constructed by inserting PCR-amplified Menin cDNA into the BamHI/NotI site of the retroviral vector pMX-puro.
Plasmids transfection
Transfection of Flag-Menin and/or HA-Ubiquitin was performed according to the typical Lipofectamine 2000 transfection procedure. Briefly, diluted 10 μg DNA with 1 ml Opti-MEM Medium and mixed with 1 ml diluted Lipofectamine 2000 Reagent. The mixture was incubated for 5 min at room temperature and DNA–lipid complex was added into 293T cells afterward.
Immunoprecipitation
For immunoprecipitation, cells were suspended in lysis buffer (50 mmol/l Tris/Cl, pH 7.4, 150 mmol/l NaCl, 5% glycerol, 1% NP-40, 1 mmol/l EDTA), supplemented with 1 mM PMSF, 4 μg/ml protease inhibitor cocktail (Sigma). Lysates were centrifuged at 13000×g for 10 min, and the supernatant was added to 2 μl indicated antibodies and 100 μl Protein A agarose (Invitrogen) to incubate for 4 h at 4°C. Afterward, Protein A agarose was washed by 250 mmol NaCl for four-times.
Western blotting
For Western blotting, cells were collected at indicated time points and then were lysed by RIPA lysis buffer (Beyotime, Nantong, China). Cell lysates (90 μl) were mixed with 30 μl SDS loading buffer and boiled for 5 min at 100°C for SDS/PAGE. Primary antibodies were diluted according to instructions. HRP-labeled secondary antibody was used at a dilution of 1:3000. Immuno-reactive bands were revealed by enhanced chemiluminescence (Clarity™ Western ECL Substrate, Bio-Rad) and visualized by the Image Quant LAS 4000 mini (GE). Band intensities were quantified and analyzed with ImageJ and normalized against the level of β-actin.
Chemicals and inhibitors
MG132 was purchased from Sigma–Aldrich and dissolved in DMSO. Cycloheximide (CHX) was purchased from AMRESCO and dissolved in DMSO.
Antibodies
Antibodies used were Rabbit polyclonal anti-menin (Bethyl, A300-105A), mouse monoclonal anti-HA (CWbiotech, CW0092A) and mouse monoclonal anti-β-actin (Santa Cruz, SC47778).
Statistical analysis
Data are presented as mean ± S.E.M. for the indicated number of experiments (n). Statistical significance was evaluated using the Student’s t test. Data were considered significant when P<0.05.
Results
WT menin can be ubiquitinated in vivo
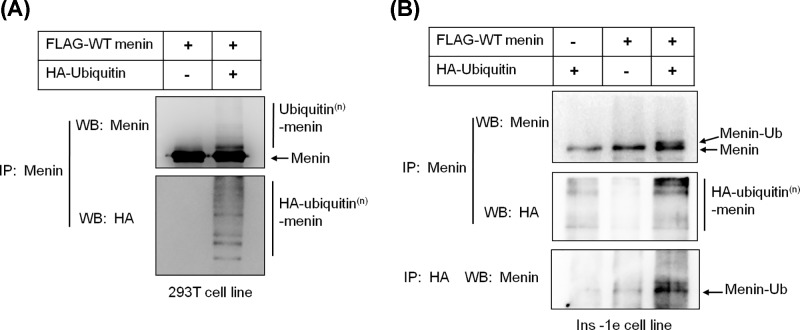
To determine whether WT menin could be ubiquitinated, we performed an in vivo ubiquitination assay. 293T cells were co-transfected with constructs expressing HA-ubiquitin and FLAG-tagged WT menin or WT menin only, and lysates were immunoprecipitated with an anti-menin antibody. Western blotting with anti-menin and anti-HA antibodies respectively detected menin-ubiquitin bands (Figure 1A), indicating WT menin is ubiquitinated in vivo. Given that the mutant menin could be degraded in MEN1, we also performed an in vivo ubiquitination assay of WT menin in an insulinoma cell line, INS-1. The result showed that WT menin was also ubiquitinated in the INS-1 cells (Figure 1B).
Figure 1. WT Menin can be modified with ubiquitin in 293T cells.
(A) Equal amounts of Menin and/or HA-ubquitin constructs were transfected into 293T cells, followed by MG132 (20 μmol) treatment for 4 h and lysed for IP and Western blot with the indicated antibodies. (B) Equal amounts of Menin and/or HA-ubquitin constructs were transfected into INS-1 cells, followed by MG132 (20 μmol) treatment for 4 h and lysed for IP and Western blot with the indicated antibodies.
WT menin is stable in 293T cells
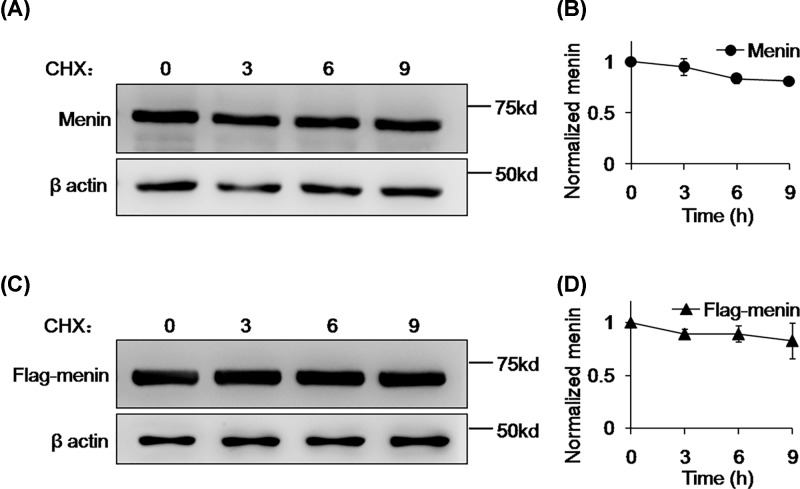
As the WT menin could be modified with ubiquitin in vivo, we next estimated the stability of WT menin in 293T cells with CHX treatment. Whole-cell extracts from 293T cells treated with CHX for indicated times were subjected to Western blotting and the clearance rate of WT menin was monitored by Western blot analysis. The protein level of WT menin from indicated CHX treatment exhibited no obvious difference in Western blot bands (Figure 2A) and the clearance rate was no more than 20% after 9-h CHX treatment (Figure 2B). This result was confirmed by CHX chase analysis of the ectopic FLAG-tagged WT menin. 293T cells transfected with FLAG-tagged WT menin were treated with CHX for indicated times and subjected to Western blotting. As shown in Figure 2C,D, FLAG-tagged WT menin was also stable at protein level and clearance rate in 293T cells.
Figure 2. Menin is stable in 293T cells.
(A) 293T cells were treated with CHX (20 μg/ml) for the indicated times and lysed for Western blot. (B) Quantitation of menin protein level from (A). Gray analysis was executed with ImageJ and menin protein levels were quantified and normalized to β-actin. (C) 293T cells transfected with Falg-menin were treated with CHX (20 μg/ml) for the indicated times and lysed for Western blot. (D) Quantitation of the Flag-menin protein level from (C). Gray analysis was executed with ImageJ and Flag-menin protein levels were quantified and normalized to β-actin.
WT menin is degraded rapidly in INS-1 cells
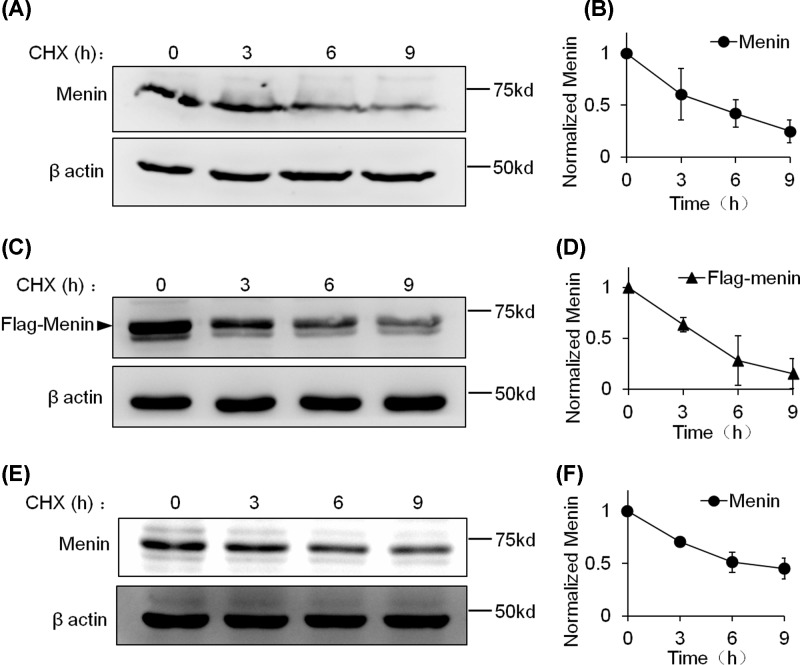
Given that the WT menin is stable in 293T cells, we turned to investigating the stability of WT menin in pathological cells, such as PNETs. To this end, we employed INS-1 cell, a rat insulinoma cell line, to estimate the degradation rate of WT menin. Whole-cell extracts from INS-1 cells treated with CHX for indicated times were subjected to Western blotting and the clearance rate of WT menin was monitored by Western blot analysis. In contrast with the stability in 293T cells, we observed rapid degradation of WT menin in INS-1 cells (Figure 3A). After CHX treatment for 9 h, nearly 80 percent of WT menin was degraded and the half-life of WT menin was estimated to be approximately 4 h in INS-1 cells (Figure 3B). To confirm the rapid degradation of WT menin in INS-1 cells, we constructed INS-1 cell line stably expressing ectopic FLAG-tagged WT menin which was the same as the one we used in 293T cells. Similarly, we observed significant degradation of FLAG-tagged WT menin in ectopic menin-expressing INS-1 cells (Figure 3C,D). To confirm that the WT menin can be degraded in insulinoma cells, we employed another insulinoma cell line, TGP-61, to detect the stability of WT menin. The result showed that WT menin was also significantly degraded in TGP-61 cell line (Figure 3E,F). These findings suggest that WT menin reduction resulting from rapid degradation is the possible cause of PNETs in part of patients who have normal MEN1 gene.
Figure 3. WT menin degrades rapidly in the presence of CHX in INS-1 cells.
(A) INS-1 cells were treated with CHX (20 μg/ml) for the indicated times and lysed for Western blot. (B) Quantitation of menin protein level from (A). Gray analysis was executed with ImageJ and menin protein levels were quantified and normalized to β-actin. (C) INS-1 cells expressing ectopic Falg-menin were treated with CHX (20 μg/ml) for the indicated times and lysed for Western blot. (D) Quantitation of the Flag-menin protein level from (C). Gray analysis was executed with ImageJ and Flag-menin protein levels were quantified and normalized to β-actin. (E) TGP-61 cells were treated with CHX (20 μg/ml) for the indicated times and lysed for Western blot. (F) Quantitation of the menin protein level from (E). Gray analysis was executed with ImageJ and menin protein levels were quantified and normalized to β-actin.
WT menin is targeted for degradation via the ubiquitin-proteasome pathway in INS-1 cells
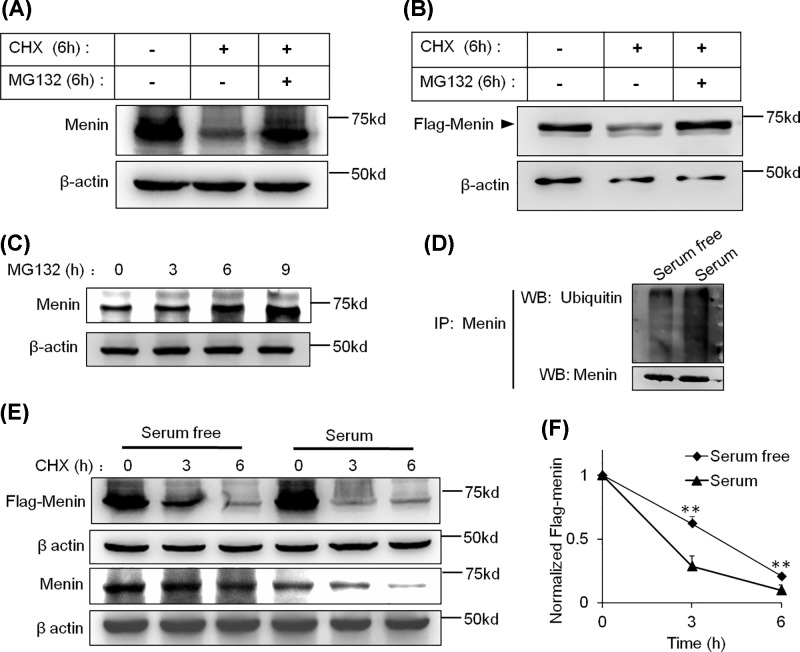
In eukaryotic cells, the ubiquitin-proteasome pathway is one of the major mechanisms for the targeted degradation of unstable proteins. Therefore, we investigated the effects of the proteasome inhibitor MG132 on the stability of WT menin during CHX addition to confirm whether WT menin is targeted for degradation via the ubiquitin-proteasome pathway in INS-1 cells. As shown in Figure 4, whether the amount of WT menin (Figure 4A) or FLAG-tagged WT menin (Figure 4B) could be restored by MG132 treatment in the presence of CHX in INS-1 cells. Especially, MG132 treatment alone significantly accumulated the amount of WT menin (Figure 4C). These data suggest that the ubiquitin-proteasome pathway was implicated in the rapid degradation of WT menin.
Figure 4. Degradation of WT menin could be blocked by MG132 in INS-1 cells.
(A) INS-1 cells were treated with CHX (20 μg/ml) and MG132 (20 μmol) or CHX (20 μg/ml) only for 6 h and lysed for Western blot. (B) INS-1 cells expressing ectopic Flag-menin were treated with CHX (20 μg/ml) and MG132 (20 μmol) or CHX (20 μg/ml) only for 6 h and lysed for Western blot. (C) INS-1 cells were treated with MG132 (20 μmol) for the indicated times and lysed for Western blot. (D) INS-1 cells were treated with MG132 (20 μmol) for 4 h and lysed for IP and Western blot with the indicated antibodies. (E) INS-1 cells or INS-1 cells expressing ectopic Flag-menin were treated with CHX (20 μg/ml) for the indicated times in the presence or absence of serum and lysed for Western blot. (F) Quantitation of Flag-menin protein level from (E). Gray analysis was executed with ImageJ and Flag-menin protein levels were quantified and normalized to β-actin. **, P<0.01.
WT menin is degraded slowly in serum-free medium
To further investigate the regulation of WT menin degradation in INS-1 cells, we performed CHX chase analysis with removal of the serum from medium. INS-1 cells expressing ectopic WT menin were serum-starved overnight and then incubated with or without 10% FBS and CHX for indicated times. Whole-cell extracts were analyzed by IP with menin antibody and Western blotting with menin and ubiquitin antibodies. The result showed that there was more ubiquitination of menin in the presence of serum than that in the absence of serum (Figure 4D). For further confirmation, the clearance rate of WT menin was monitored by Western blot analysis. As shown in Figure 4E, the protein level of WT menin (Flag-menin or endogenous menin) at 3-h CHX treatment in serum-free medium was obviously higher than that in serum medium, which was also confirmed in clearance rate (62 vs 29%; Figure 4F). These results were further confirmed by the fact that serum-free culture led to a little reduction in menin mRNA, but with no significance (Supplementary Figure S1A), indicating that some factors in serum might contribute to the ubiquitin-proteasome pathway mediated degradation of WT menin.
Discussion
To date, more than 1500 different somatic and germline MEN1 gene mutations have been identified [21,22]. Most of the studies focused on the truncated forms of nonsense mutations and the rapid degradation of missense mutations of MEN1 [17,18], whereas few researchers investigate the stability of WT menin even though some PNETs with WT MEN1 gene show lower protein level of menin. Here, we reported that the WT menin can be ubiquitinated but is very stable in 293T cells (Figures 1 and 2) and for the first time, we reported the rapid degradation of WT menin mediated by ubiquitin-proteasome pathway in a PNET-derived cell line, INS-1 (Figures 3 and 4).
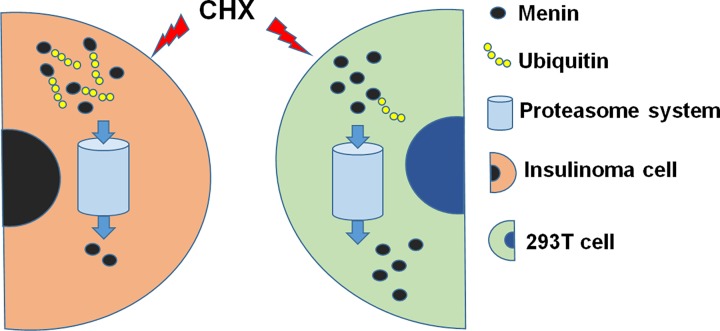
The finding that WT menin is very stable in 293T cells is also consistent with the results reported by Tsukada et al. [17]. In their studies, whether transiently expressed FLAG-tagged WT menin or endogenous menin is stable in 293T cells [17]. In some PNETs with WT MEN1, the protein level of menin in nucleus was reduced and the cytoplasmic staining was also absent, which excluded the possibility that menin was transported from nucleus to cytoplasm [19]. This finding indicates a potential transcriptional repression or protein degradation system targeting WT menin in PNETs with WT MEN1 gene. As a rat insulinoma cell line, INS-1 cell is quite an ideal cell model to study the stability of WT menin in PNETs. With this cell model, we observed obvious degradation of WT menin in vivo. Similar to the degradation of missense mutations reported previously [17,18], the degradation of WT menin is also mediated by ubiquitin-proteasome system. However, unlike the interaction of missense mutations with CHIP, one of the E3 ligases, to initiate the degradation, Co-IP assay showed no interaction of WT menin with CHIP (Supplementary Figure S1B) in the INS-1 cells, suggesting that the ubiquitin-proteasome mediated degradation of WT menin in INS-1 is dependent on another E3 ligase. This E3 ligase might mediated ubiquitin-proteasome system to degraded WT menin only in the insulinoma cells (for example, INS-1 or TGP-61), but not non-insulinoma cells (for example, 293T cells). In a word, our studies propose a new regulation pathway of WT menin in PNETs (Figure 5).
Figure 5. Schematic diagram demonstrates a new regulatory pathway of WT menin in insulinoma cells.
.
Supplementary Material
Abbreviations
- CHIP
carboxylterminus of the hsp70 interacting protein
- CHX
cycloheximide
- Co-IP
co-immunoprecipitation
- EZH2
enhancer of zeste homolog 2
- FANCD2
fanconi anemia group D2
- GFAP
glial fibrillary acidic protein
- HA
hemagglutinin
- Hlxb9
H2.0-like homeobox b9
- HRP
horseradish peroxidase
- IP
immunoprecipitation
- MEN1
multiple endocrine neoplasia type 1
- Opti-MEM
optimal minimal essential medium
- PNET
pancreatic neuroendocrine tumor
- RIPA
radio immunoprecipitation assay
- RPA2
replication protein A2
- WT
wild-type
Author Contribution
Zongzhe Jiang designed the experiments and wrote the manuscript. Shengrong Wan and Bowen Xing performed the experiments.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported in part by the National Natural Science Foundation of China [grant numbers 81600597, 81600596].
References
- 1.Zhang J., Francois R., Iyer R., Seshadri M., Zajac-Kaye M.and Hochwald S.N. (2013) Current understanding of the molecular biology of pancreatic neuroendocrine tumors. J. Natl. Cancer Inst. 105, 1005–1017 10.1093/jnci/djt135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang X., Cao Y., Li F., Su Y., Li Y., Peng Y. et al. (2014) Targeting beta-catenin signaling for therapeutic intervention in MEN1-deficient pancreatic neuroendocrine tumours. Nat. Commun. 5, 5809 10.1038/ncomms6809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gracanin A., Dreijerink K.M., van der Luijt R.B., Lips C.J.and Hoppener J.W. (2009) Tissue selectivity in multiple endocrine neoplasia type 1-associated tumorigenesis. Cancer Res. 69, 6371–6374 10.1158/0008-5472.CAN-09-0678 [DOI] [PubMed] [Google Scholar]
- 4.Jiao Y., Shi C., Edil B.H., de Wilde R.F., Klimstra D.S., Maitra A. et al. (2011) DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 331, 1199–1203 10.1126/science.1200609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin S., Mao H., Schnepp R.W., Sykes S.M., Silva A.C., D’Andrea A.D. et al. (2003) Menin associates with FANCD2, a protein involved in repair of DNA damage. Cancer Res. 63, 4204–4210 [PubMed] [Google Scholar]
- 6.Sukhodolets K.E., Hickman A.B., Agarwal S.K., Sukhodolets M.V., Obungu V.H., Novotny E.A. et al. (2003) The 32-kilodalton subunit of replication protein A interacts with menin, the product of the MEN1 tumor suppressor gene. Mol. Cell. Biol. 23, 493–509 10.1128/MCB.23.2.493-509.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopez-Egido J., Cunningham J., Berg M., Oberg K., Bongcam-Rudloff E.and Gobl A. (2002) Menin’s interaction with glial fibrillary acidic protein and vimentin suggests a role for the intermediate filament network in regulating menin activity. Exp. Cell Res. 278, 175–183 10.1006/excr.2002.5575 [DOI] [PubMed] [Google Scholar]
- 8.Lemmens I.H., Forsberg L., Pannett A.A., Meyen E., Piehl F., Turner J.J. et al. (2001) Menin interacts directly with the homeobox-containing protein Pem. Biochem. Biophys. Res. Commun. 286, 426–431 10.1006/bbrc.2001.5405 [DOI] [PubMed] [Google Scholar]
- 9.Shi K., Parekh V.I., Roy S., Desai S.S.and Agarwal S.K. (2013) The embryonic transcription factor Hlxb9 is a menin interacting partner that controls pancreatic beta-cell proliferation and the expression of insulin regulators. Endocr. Relat. Cancer 20, 111–122 10.1530/ERC-12-0077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokoyama A., Wang Z., Wysocka J., Sanyal M., Aufiero D.J., Kitabayashi I. et al. (2004) Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol. Cell. Biol. 24, 5639–5649 10.1128/MCB.24.13.5639-5649.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim H., Lee J.E., Cho E.J., Liu J.O.and Youn H.D. (2003) Menin, a tumor suppressor, represses JunD-mediated transcriptional activity by association with an mSin3A-histone deacetylase complex. Cancer Res. 63, 6135–6139 [PubMed] [Google Scholar]
- 12.Gang D., Hongwei H., Hedai L., Ming Z., Qian H.and Zhijun L. (2013) The tumor suppressor protein menin inhibits NF-kappaB-mediated transactivation through recruitment of Sirt1 in hepatocellular carcinoma. Mol. Biol. Rep. 40, 2461–2466 10.1007/s11033-012-2326-0 [DOI] [PubMed] [Google Scholar]
- 13.Gao S.B., Feng Z.J., Xu B., Wu Y., Yin P., Yang Y. et al. (2009) Suppression of adenocarcinoma through menin and polycomb gene-mediated repression of growth factor pleiotrophin. Oncogene 28, 4095–4104 10.1038/onc.2009.273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gurung B., Feng Z., Iwamoto D.V., Thiel A., Jin G., Fan C.M. et al. (2013) Menin epigenetically represses Hedgehog signaling in MEN1 tumor syndrome. Cancer Res. 73, 2650–2658 10.1158/0008-5472.CAN-12-3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertolino P., Tong W.M., Herrera P.L., Casse H., Zhang C.X.and Wang Z.Q. (2003) Pancreatic beta-cell-specific ablation of the multiple endocrine neoplasia type 1 (MEN1) gene causes full penetrance of insulinoma development in mice. Cancer Res. 63, 4836–4841 [PubMed] [Google Scholar]
- 16.Jiang X.H., Lu J.L., Cui B., Zhao Y.J., Wang W.Q., Liu J.M. et al. (2007) MEN1 mutation analysis in Chinese patients with multiple endocrine neoplasia type 1. Endocr. Relat. Cancer 14, 1073–1079 10.1677/ERC-07-0015 [DOI] [PubMed] [Google Scholar]
- 17.Yaguchi H., Ohkura N., Takahashi M., Nagamura Y., Kitabayashi I.and Tsukada T. (2004) Menin missense mutants associated with multiple endocrine neoplasia type 1 are rapidly degraded via the ubiquitin-proteasome pathway. Mol. Cell. Biol. 24, 6569–6580 10.1128/MCB.24.15.6569-6580.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Canaff L., Vanbellinghen J.F., Kanazawa I., Kwak H., Garfield N., Vautour L. et al. (2012) Menin missense mutants encoded by the MEN1 gene that are targeted to the proteasome: restoration of expression and activity by CHIP siRNA. J. Clin. Endocrinol. Metab. 97, E282–E291 10.1210/jc.2011-0241 [DOI] [PubMed] [Google Scholar]
- 19.Corbo V., Dalai I., Scardoni M., Barbi S., Beghelli S., Bersani S. et al. (2010) MEN1 in pancreatic endocrine tumors: analysis of gene and protein status in 169 sporadic neoplasms reveals alterations in the vast majority of cases. Endocr. Relat. Cancer 17, 771–783 10.1677/ERC-10-0028 [DOI] [PubMed] [Google Scholar]
- 20.Feng Z., Wang L., Sun Y., Jiang Z., Domsic J., An C. et al. (2017) Menin and Daxx interact to suppress neuroendocrine tumors through epigenetic control of the membrane metallo-endopeptidase. Cancer Res. 77, 401–411 10.1158/0008-5472.CAN-16-1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemos M.C.and Thakker R.V. (2008) Multiple endocrine neoplasia type 1 (MEN1): analysis of 1336 mutations reported in the first decade following identification of the gene. Hum. Mutat. 29, 22–32 10.1002/humu.20605 [DOI] [PubMed] [Google Scholar]
- 22.Concolino P., Costella A.and Capoluongo E. (2016) Multiple endocrine neoplasia type 1 (MEN1): an update of 208 new germline variants reported in the last nine years. Cancer Genet. 209, 36–41 10.1016/j.cancergen.2015.12.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.