Abstract
This study investigated whether overexpression of paired-related homeobox 1 (prrx1) can successfully induce differentiation of brown adipose-derived stem cells (BADSCs) into sinus node-like cells. The experiments were performed in two groups: adenovirus–green fluorescent protein (Ad-GFP) group and Ad-prrx1 group. After 5–7 days of adenoviral transfection, the expression levels of sinus node cell-associated pacing protein (hyperpolarization-activated cyclic nucleotide-gated potassium channel 4 [HCN4]) and ion channel (calcium channel, voltage-dependent, T type, alpha 1G subunit [Cacna1g]), as well as transcription factors (T-box 18 [TBX18], insulin gene enhancer binding protein 1 [ISL-1], paired-like homeodomain transcription factor 2 [pitx2], short stature homeobox 2 [shox2]), were detected by western blot and reverse transcription–quantitative polymerase chain reaction. Immunofluorescence assay was carried out to detect whether prrx1 was coexpressed with HCN4, TBX18, and ISL-1. Finally, whole-cell patch-clamp technique was used to record pacing current hyperpolarization-activated inward current (If). The isolated cells were CD90+, CD29+, and CD45−, indicating that pure BADSCs were successfully isolated. After 5–7 days of Ad transfection into cells, the mRNA levels and protein levels of pacing-related factors (TBX18, ISL-1, HCN4, shox2, and Cacna1g) in Ad-prrx1 group were significantly higher than those in Ad-GFP group. However, the expression level of pitx2 was decreased. Immunofluorescence analysis showed that prrx1 was coexpressed with TBX18, ISL-1, and HCN4 in the Ad-prrx1 group, which did not appear in the Ad-GFP group. Whole-cell patch clamps were able to record the If current in the experimental group rather than in the Ad-GFP group. Overexpression of prrx1 can successfully induce sinus node-like cells.
Keywords: biological pacing, sinus node-like cells, prrx1, TBX18
Introduction
The clinical manifestations of sick sinus syndrome (SSS) are diverse. Those milder symptoms include fatigue, dizziness, memory loss, whereas severe symptoms show as stun syncope, transient black sputum, Aspen synthesis, and so on. Electronic pacemaker is the mainstay technique used to improve the quality of life of SSS patients and to prolong their life expectancy. However, electronic pacemakers have some limitations, especially for children, such as lead malfunction, finite battery life, and device-related infections. To solve this problem, some experts have proposed that a biological pacemaker is likely to be a more effective technology (Cingolani et al., 2018).
At present, four methods are used to produce the sinus pacemaker-like cells. The earliest approach involved a direct knockout or introduction of various ion channel genes, for instance, inhibition of KCNJ2 gene (encoding inward rectifier potassium ion channel IK1 to reduce the outward current) (Miake et al., 2002) or advancement of a hyperpolarized-activated cyclic nucleotide (HCN)-gated channel (encoding hyperpolarization-activated inward current [If] current to increase inward current) (Qu et al., 2003). The mechanism of this approach was to improve the depolarizing current and to simplify automatic depolarization, which is actually helpful to increase the heart rate. This means that it can induce sinus node-like cells functionally and not morphologically. The second approach is the application of stem cell transplantation with an aim of imitating embryonic heart development (Kehat et al., 2004). In this way, the progenitor stem cells are induced to differentiate into the sinus junction cells. The other approach is the fusion of cells and genes. The discovery of induced pluripotent stem cells (iPSCs) has enabled to introduce biological pacemakers factors such as T-box 18 (TBX18) into iPSCs to induce sinoatrial node (SAN)-like cells (Gorabi et al., 2019a). Finally, somatic reprogramming is one of the most effective and promising technology. Scientists directly constructed a virus carrying TBX18, which was injected into the left ventricular myocytes of adult pigs (Kapoor et al., 2013). The successfully produced node-like cells exhibit both desired morphology and function.
The methods mentioned aim at inducing sinus node-like cells with biological characteristics that can be comparable with the natural SAN cells (both morphological and functional). The construction of sinus node-like cells by reprogramming or injecting adenovirus-TBX18 (Ad-TBX18) in vivo has put researchers' focus into the embryonic process of sinus node. It has been found that many factors regulate the development of the sinus node, such as Tbx18 (Gorabi et al., 2019b), T-box 3 (Tbx3) (Choudhury et al., 2018), short stature homeobox 2 (Shox2) (Hu et al., 2018), insulin gene enhancer binding protein 1 (ISL-1) (Liang et al., 2015; Zhang et al., 2019), NK2 transcription factor-related locus 5 (Nkx2.5) (Protze et al., 2017) and other transcription factors, HCN-gated potassium channel 4 (HCN4) (D'Souza et al., 2017), L-type calcium channel (Howarth et al., 2018), sodium calcium exchange plasma channel, cell surface adhesion molecule—cluster of differentiation 166 (CD166) (Scavone et al., 2013), connexion (Cx) protein—Cx45 and Cx43 (Martinez et al., 2002), and others. This study proposed a new transcription factor that may induce pacemaker-like cells.
During embryonic development, primary biological pacemaker is first generated on the left side. Along with the development of the embryonic phase, the origin of heartbeats shifts to the right and the pulsing site on the left side gradually disappears (Wang et al., 2014). The nodal-paired-like homeodomain transcription factor 2 (pitx2) axis plays an key role in this process. Recent studies have shown that a cable-like structure of paired-related homeobox 1 (prrx1)-positive cells at the wall of the right vitelline vein and right sinus venosus horns in control embryos, compatible with the displacement of the posterior pole to the left side. The loss of Left/Right morphological asymmetry after prrx1 silencing highly expresses in the right lateral mesoderm. It means that the expressions of prrx1 are significantly higher in the right circulation of the zebrafish heart than those on the left side (Ocana et al., 2017). In addition, prrx1 gene knockout impairs the development of right side of the venous sinus and leads to a small atrial volume. However, arteries are not significantly affected. Ocana et al. also demonstrated that ISL-1 colocalized with prrx1 in the posterior pole and its expression was downregulated with prrx1 RNAi. Of interest, Tbx18 is repressed when Prrx1 is downregulated. Based on the findings described previously, we propose that prrx1 might be a upstream transcription factors of TBX18 and ISL-1 to control the development of sinus node. Therefore, overexpression of prrx1 may induce the formation of the sinus-like node cells.
Materials and Methods
Animals
Adult male Sprague-Dawley (SD) rats (n = 6; age, 3–4 weeks; weight, 40–80 g) were purchased from the Center for Disease Control and Prevention of Hubei Province (Hubei, China). The SD rats were kept in four per cage with standard laboratory chow and given sterilized water. The room environment was controlled under a constant temperature (22 ± 2°C), constant humidity (55 ± 5%), and a 12:12 h light/dark cycle. This study was approved by the Experimental Animal Committee of Wuhan University (Hubei, China; no. WDRM20180807). All experimental procedures were approved by the Ethics Committee of Animal Research, Wuhan University Health Science Center, and the investigation conformed to the Guide for the Care and Use of Laboratory Animals published by the U.S. National Institutes of Health (NIH Publication, 8th Edition, 2011).
Isolation and culture of brown adipose-derived stem cells
All experimental procedures were conducted in accordance with the Institutional Guidelines for the Care and Use of Laboratory Animals at Wuhan University (Wuhan, China) and conformed to the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Ethics approval was provided by the Ethics Committee of Wuhan University. Brown adipose-derived stem cells (BADSCs) were obtained using a previously described method modification (Hao et al., 2017). In brief, SD rat was anesthetized with an intraperitoneal injection of 3% pentobarbital sodium (30 mg/kg). Brown adipose tissue was bilaterally obtained from the area of scapula and washed with sterile phosphate-buffered saline (PBS). The brown adipose tissue was then cut into 1 × 1 mm3 pieces after the careful removal of blood vessels and fascia. The tissue pieces were digested by incubation with 1 mg/mL collagenase type I (Sigma, St. Louis, MO) for 1 h at 37°C with constant shaking. The cell suspension was centrifuged at 1000 × g for 10 min. The pellet was resuspended in Dulbecco's modified Eagle's medium (DMEM)/F12 supplemented with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA) and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA). After 48 h of culture in an incubator at 37°C with a 5% CO2 atmosphere, the medium was removed and replaced with fresh medium. When the cells reached 80–90% confluence, 0.25% trypsin (Genom, Hangzhou, China) was used to digest and passage BADSCs. Cells of passages 3–5 were used for all subsequent analyses.
Ad construction and purification
The adenoviral vector expressing GV315Ad-MSC-GFP-prrx1 was constructed by inserting the human prrx1 gene (positive clone sequence: ACCGGCGCCACCATGACCTCCAGCTACGGGCACGTTCTGGAGCGGCAACCGGCGCTGGGCGGCCGCTTGGACAGCCCGGGCAACCTCGACACCCTGCAGGCGAAAAAGAACTTCTCCGTCAGTCACCTGCTAGACCTGGAGGAAGCCGGGGACATGGTGGCGGCACAGGCGGATGAGAACGTGGGCAGGCTGGCCGGAGCCTGCTGGAGTCGCCGGGACTCACCAGCGGCAGCGACACCCCGCAGCAGGACAATGACCAGCTGAACTCAGAAGAAAAAAAGAAGAGAAAGCAGCGAAGGAATAGGACAACCTTCAATAGCAGCCAGCTGCAGGCTTTGGAGCGTGTCTTTGAGCGGACACACTATCCTGATGCTTTTGTGCGAGAAGACCTTGCCCGCCGGGTGAACCTCACCGAGGCGAGAGTGCAGGTGTGGTTTCAGAACCGAAGAGCCAAGTTCCGCAGGAATGAGAGAGCCATGCTAGCCAATAAAAACGCTTCCCTCCTCAAATCCTACTCAGGAGACGTGACTGCTGTGGAGCAGCCCATCGTACCTCGTCCTGCTCCGAGACCCACCGATTATCTCTCCTGGGGGACAGCGTCTCCGTACAGATCCTCGTCCCTCCCAAGATGTTGTTTACACGAGGGGCTTCATAACGGATTCTAACTAGC) into GV315Ad-MSC-GFP vector (Shanghai Genechem Co., Ltd., Shanghai, China) using AgeI/NheI (cat. no. CON267) restriction sites, all obtained from Shanghai Genechem Co., Ltd. Ad–green fluorescent protein (Ad-GFP) and Ad-GFP-prrx1 were measured as 5 × 1010 PFU/mL and 6 × 1010 PFU/mL respectively, which were preserved at −80°C.
BADSCs transfected with Ad-prrx1 and Ad-GFP
BADSCs of passages 3–5 were removed from the culture dishes by digestion. A cell suspension was prepared and then inoculated onto six-well plates. When cell confluence reached 70–80%, Ad-prrx1 in DMEM/F12 was added to the cells at different MOI values. The control group was treated with Ad-GFP. After a 12-h incubation period, the medium was replaced with fresh complete medium. The cells were observed under a light microscope and a fluorescent microscope (BX51 systems; Olympus Corporation, Tokyo, Japan).
Reverse transcription–quantitative polymerase chain reaction
Total RNA was extracted from the transfected BADSCs after 1 week using TRIzol® reagent (Invitrogen). Quantitative PCR (qPCR) was performed to evaluate the mRNA expression of human prrx1, rat shox2, rat ISL-1, rat TBX18, rat HCN4, rat pitx2 and rat calcium channel, voltage-dependent, T type, alpha 1G subunit (Cacna1g). Isolated RNA (2 μg) was converted into cDNA using the First Strand cDNA Synthesis kit (Toyobo, Tokyo, Japan). The primers used for PCR amplification were synthesized by Invitrogen Biotechnology (Shanghai, China) and are given in Table 1. Reverse transcription–qPCR (RT-qPCR) was performed using the StepOne™ Real-time PCR system (Life Technologies, Carlsbad, CA). The reactions were then conducted using the SYBR® Premix Ex Taq TM II (Takara Bio, Japan). Semilog amplification curves were analyzed using the 2−ΔΔCt comparative quantification method and the expression of each gene was normalized to β-actin. PCR analyses were repeated at least three times to verify results.
Table 1.
Polymerase Chain Reaction Primers Used in This Study
| Gene | Accession no. | Primer (5′–3′) | Size (bp) | |
|---|---|---|---|---|
| R-β-actin | NM_031144.3 | Forward | CGTTGACATCCGTAAAGACCTC | 110 |
| Reverse | TAGGAGCCAGGGCAGTAATCT | |||
| H-prrx1 | NM_006902.4 | Forward | TTTGTGCGAGAAGACCTTGC | 142 |
| Reverse | AGTAGGATTTGAGGAGGGAAGC | |||
| R-shox2 | NM_013028.1 | Forward | AGGTGTCCCCTGAACTGAAGG | 121 |
| Reverse | AGCTCGTTGAGTTGTTCCAGG | |||
| R-ISL1 | NM_017339.3 | Forward | TGCGGAGTGTAATCAGTATTTGG | 136 |
| Reverse | GTCGTTCTTGCTGAAGCCTATG | |||
| R-TBX18 | NM_001108173.1 | Forward | GGAGACTTGGATGAGACAAGTGAT | 282 |
| Reverse | TTGGCAAATGGATTCCTGTCT | |||
| R-HCN4 | NM_021658.1 | Forward | CACTAAGGGCAACAAGGAGACC | 281 |
| Reverse | GGTAGTTGAAGACGCCTGAGTTG | |||
| R-Cacnalg | NM_00130830 | Forward | AGAAACCGCTACCCAGACATG | 185 |
| Reverse | TACGAGTAGCCGGGGTACATG | |||
| R-Pitx2 | NM_001042505.1 | Forward | AGAAACCGCTACCCAGACATG | 212 |
| Reverse | TACGAGTAGCCGGGGTACATG | |||
Cacna1g, calcium channel, voltage-dependent, T type, alpha 1G subunit; HCN4, hyper-polarization activated cyclic nucleotide gated potassium channel 4; ISL-1, insulin gene enhancer binding protein 1; Pitx2, paired-like homeodomain transcription factor 2; Prrx1, paired-related homeobox 1; R-shox2, short stature homeobox 2; TBX18, T-box 18.
Western blot analysis
After transfected with Ad-prrx1 and Ad-GFP for 5–7 days, BADSCs were digested from six-well culture dishes. The cells were harvested using RIPA lysis buffer (Beyotime Institute of Biotechnology, Haimen, China). Equal amounts of protein were loaded onto a gel for sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and the separated proteins were transferred to a nitrocellulose membrane, and then incubated with the primary antibodies against HCN4 (ab32675; Abcam, Cambridge, MA), TBX18 (ab115262; Abcam), ISL-1 (abs132916; Abcam), and shox2 (ab55740; Abcam) overnight at 4°C. The primary antibodies were detected by incubating the membrane with horseradish peroxidase-conjugated secondary antibodies (KPL, 14-16-06, 074-1506) raised in the appropriate species, and then performing enhanced chemiluminescence detection (Beyotime Institute of Biotechnology). The level of β-actin was used to normalize the signal intensities. Western blot were repeated at least three times to verify results.
Immunostaining studies
BADSCs transfected with Ad-Prrx1 and Ad-GFP were plated on gelatin-coated coverslips in six-well culture dishes. The cell cultures were washed with PBS and fixed with 4% paraformaldehyde. After permeabilization with 0.1% Triton X-100, the cells were incubated with the primary antibody anti-HCN4 or anti-TBX18 or anti-ISL-1 overnight at 4°C. The secondary antibodies Cy3-conjugated Affinipure goat anti-rabbit IgG (111-165-003) and FITC-AffiniPure F(ab′)2 fragment goat anti-mouse IgG (115-096-006) (both from Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) were used to detect cTnI. 4′,6-Diamidino-2-phenylindole (DAPI) was used to visualize the nuclei. The cells were observed under a fluorescent microscope (BX51 systems; Olympus Corporation). Three randomly selected visual fields were examined for each treatment.
Electrophysiological recordings
Whole-cell electrophysiology recordings were performed as described (Kapoor et al., 2013). Cells were plated on gelatin-coated coverslips in 24-well culture dishes. The whole-cell patch-clamp technique was used to record the funny current (If) after 5–7 days of transfection. We selected smooth and plump cells to record current. The impedance of the fluid-filled electrode was 3–4 MΩ. The experiments were performed using an Axon patch-clamp amplifier 700B (Molecular Devices, Sunnyvale, CA). A digital 700AD/DA converter and 6.0.4 pClamp (both from Axon Instruments, Union City, CA) were used for recording and analyzing the data.
If bath solution contained the following reagents (in mM): 140 NaCl, 5.4 KCl, 1.0 MgCl2, 1.8 CaCl2, 1.0 BaCl2, 5.5 HEPES, 5.0 glucose (pH 7.4); the pipette solution (in mM): 20 KCl, 125 K-gluconate, 1.0 MgCl2, 5.0 NaCl, 10 HEPES, 5 K2ATP (pH 7.2). The whole-cell recording mode was used to record If. The Clampex program was applied to the sample. The sampling frequency was 10 kHz, and the filtering rate was 5 kHz. If currents were recorded by holding the resting membrane potential at −35 mV, then stepping to a test voltage of −140 mV for 2 s with 20 mV step in each sweep. Each test potential was followed by a step to −140 mV for 1 s to examine its activation kinetics, and then brought to the holding potential of −35 mV. CsCl (4 mM) was used to inhibit If current.
Statistical analysis
The reported data are expressed as mean ± standard error of the mean. The statistical significance of the differences between two groups was determined using the Student's t-test. All data were subjected to formal tests for normality. Data not exhibiting a normal distribution were evaluated by nonparametric tests. A value of p < 0.05 was considered to indicate a statistically significant difference.
Results
Identification and characterization of BADSCs by flow cytometric analysis
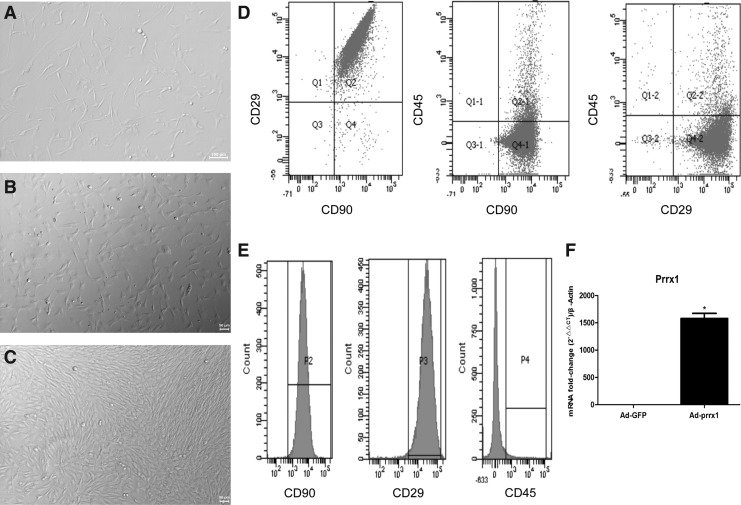
Freshly isolated BADSCs were round and floated on the surface of the 10% FBS medium. Several single, fusiform, or shuttle-shaped cells started to adhere at 24 h (Fig. 1A). After 48 h of culture, many spindle-shaped cells were formed (Fig. 1B). When cell density reached 80–90% of the whole dish after culturing for 5–7 days and tight connections were formed between adjacent cells, BADSCs appeared scrolled-like and oat-like shape (Fig. 1C).
FIG. 1.
Characteristics and identification of BADSCs and expression of Prrx1. (A–C) Isolated BADSCs after 24 h, 48 h, and 7 days under a light microscope (magnification, × 100), respectively; (D–E) identification of BADSCs (n = 3). PE, CD90; APC-A, CD45; (F) mRNA expression of human Prrx1 in the transfected groups (n = 3); *p < 0.05, versus Ad-GFP. Ad, adenovirus; BADSCs, brown adipose-derived stem cells; Prrx1, paired-related homeobox 1; GFP, green fluorescent protein.
Flow cytometric analysis was conducted to identify the phenotypes of the isolated cells. As given in Figure 1D–E, BADSCs highly expressed stromal cell markers CD90 (89.4%) and CD29 (87.2%), but the hematopoietic marker, CD45, was nearly not detected (3.4%). These findings suggested that the isolated cells were pure BADSCs.
Optimal MOI for transfection and expression of the prrx1 gene
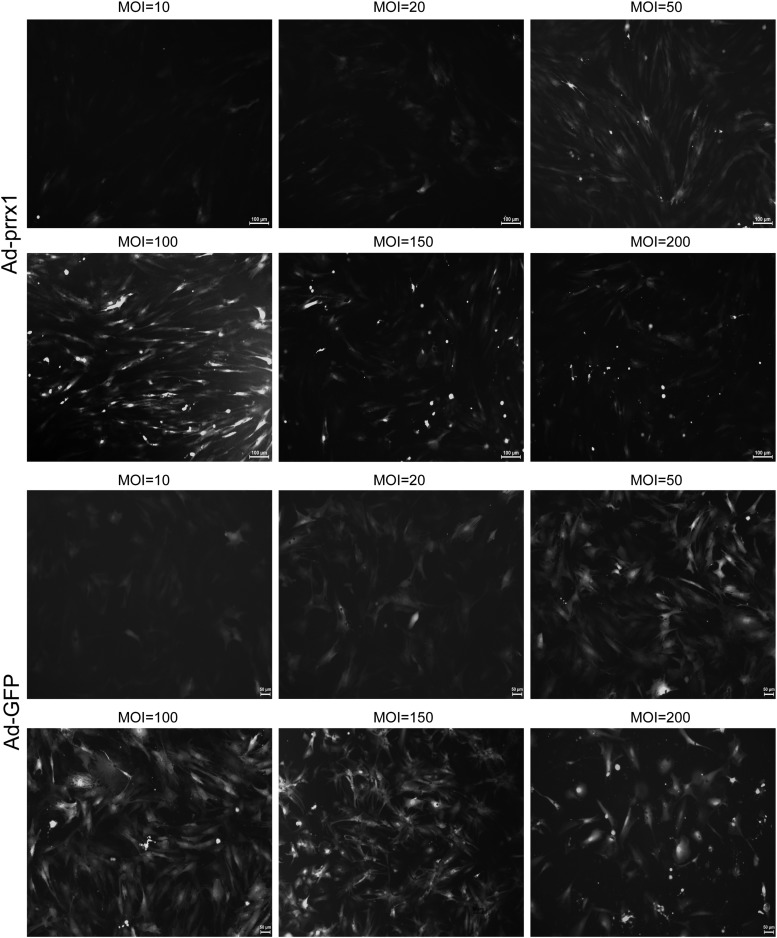
BADSCs of passages 3–6 were digested in culture dishes and then incubated in 24-well plates and 6-well plates. When the cell confluence reached 70–80%, Ad-GFP-prrx1 and Ad-GFP in DMEM/F12 were transfected to BADSCs at different MOI values (MOI = 0, 10, 20, 50, 100, 150, and 200). After a 12-h incubation period, a fresh complete medium was added. The cells were observed under a light and fluorescent microscope (BX51 systems; Olympus Corporation). After successful transfection, a green fluorescence was observed. The results showed that the fluorescence intensity of Ad-prrx1 increased with MOI increment (Fig. 2). The highest MOI that did not reduce the number of cells was chosen as the optimal MOI value. Figure 2 shows that the optimal MOI used in this study for Ad-GFP and Ad-prrx1 transfection were 100. The mRNA level of prrx1 was significantly higher in Ad-prrx1 group than that in Ad-GFP group after 5 days of transfection, suggesting that prrx1 was stably expressed in BADSCs.
FIG. 2.
BADSCs transfected with Ad-GFP and Ad-prrx1, respectively, at different MOI value (magnification, × 100) (n = 3).
Expression of proteins associated with pacemaker-like cells
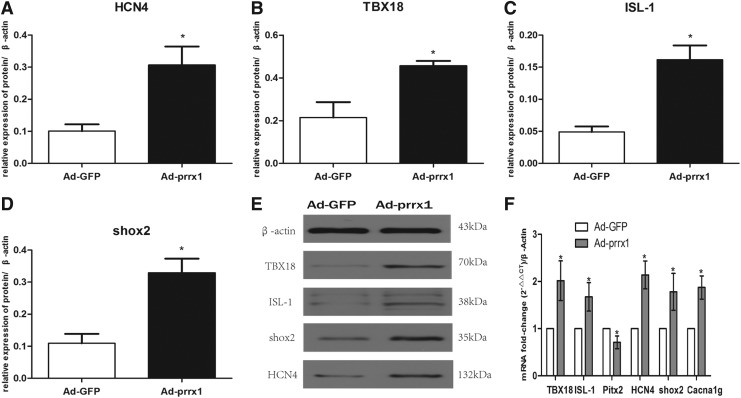
To discover the role of prrx1 in the differentiation of SAN cells, several genes associated with the development of SAN were detected: TBX18, ISL-1, pitx2, and shox2; HCN4, crucial markers of pacemaker cells and the T type calcium channel protein encoded by Cacna1g (representing the alpha 1G subunit, also known as Cav3.1). In the heart, T type Ca2+ channels are expressed in the SAN and conducting cells. As given in Figure 3A–F, the mRNA and protein levels of TBX18, ISL-1, HCN4, shox2, and Cacna1g were higher, but pitx2 was lower, in the Ad-prrx1 group compared with the Ad-GFP group.
FIG. 3.
Expression of related genes by western blot and RT-qPCR analyses after transfected 5 days (n = 3). (A–D) Quantitative assessment of protein levels of HCN4, TBX18, ISL-1, and shox2 by integrated optical density analyses. (E) Protein expression of TBX18, ISL-1, shox2, and HCN4 was examined using western blotting. (F) Gene expression levels of TBX18, ISL-1, Pitx2, HCN4, shox2, and Cacna1g were examined by RT-qPCR analysis. Similar results were obtained in three independent experiments. β-actin was used as the protein control. *p < 0.05 versus Ad-GFP. Cacna1g, calcium channel, voltage-dependent, T type, alpha 1G subunit; HCN4, hyperpolarization-activated cyclic nucleotide-gated potassium channel 4; ISL-1, insulin gene enhancer binding protein 1; Pitx2, paired-like homeodomain transcription factor 2; RT-qPCR, reverse transcription–quantitative polymerase chain reaction; shox2, short stature homeobox 2; TBX18, T-box 18.
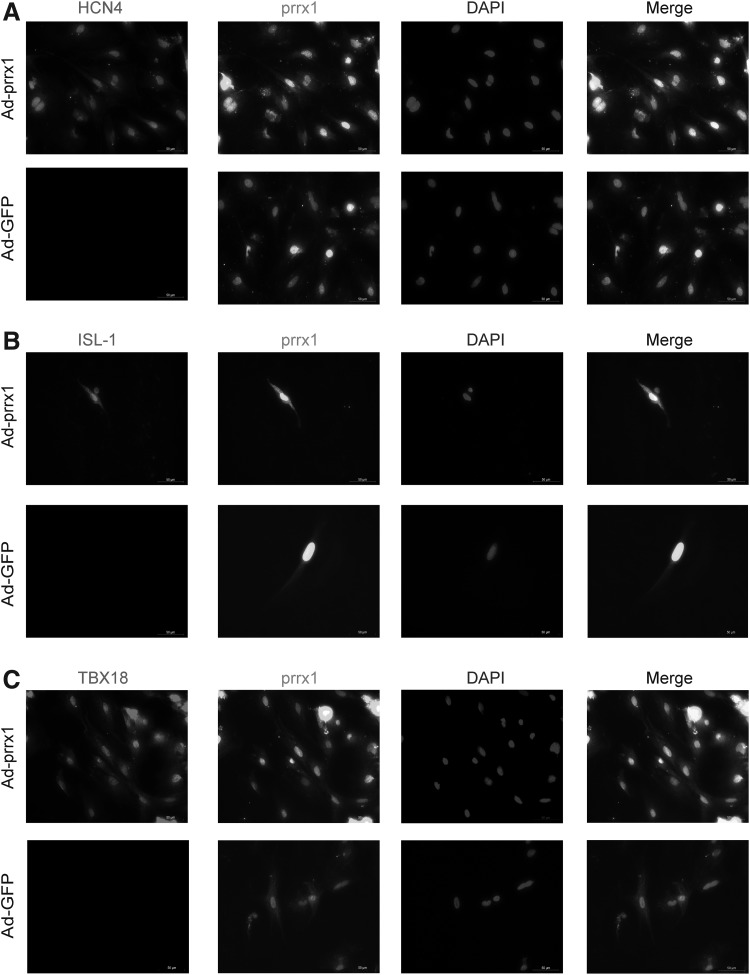
Confirmation of SAN-like cells by immunocytochemical analysis
The immunocytochemical analysis confirmed that Ad-prrx1 transfection enhanced the expression of HCN4, TBX18 and ISL-1. Figure 4A–C shows that prrx1 was coexpressed with HCN4, TBX18, and ISL-1, but not in the Ad-GFP group.
FIG. 4.
SAN-specific proteins examined by immunofluorescence staining in differentiated BADSCs after transfected 5–7 days (magnification, × 400) (n = 6). (A) Clear positive staining for HCN4 was observed in BADSCs transfected with Ad-GFP or Ad-prrx1. (B) Clear positive staining for TBX18 was observed in BADSCs transfected with Ad-GFP or Ad-prrx1. (C) Clear positive staining for ISL-1 was observed in BADSCs transfected with Ad-GFP or Ad-prrx1 scale bar, 50 μm. The nuclei were stained with DAPI, Ad-prrx1 were detected by immunofluorescence. Representative positive staining of HCN4, TBX18, and ISL-1 is shown. DAPI, 4′,6-diamidino-2-phenylindole.
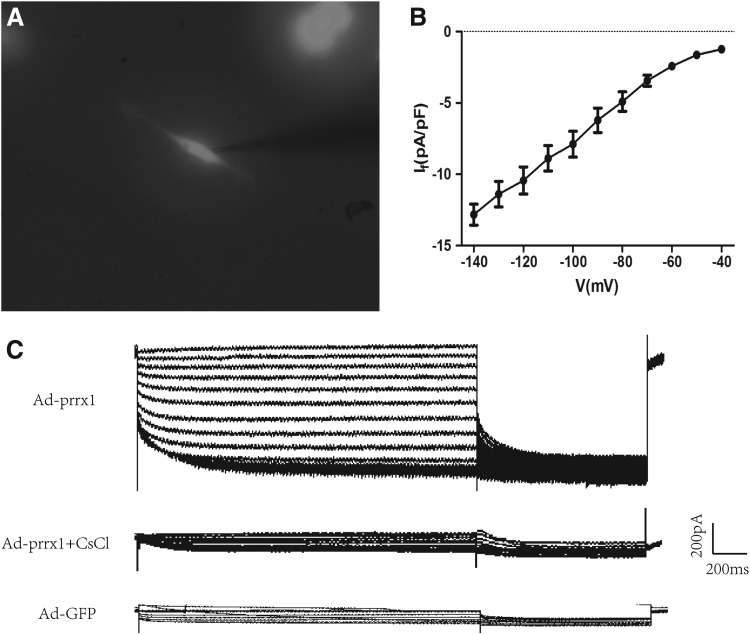
Patch-clamp recording of If current
The green fluorescent cells was used for detecting the intracellular electrical activity by patch-clamp method (Fig. 5A). The If current, which modulates spontaneous phase 4 depolarization, is used as a marker of SAN cells. Ad-prrx1 transfection significantly increased If current, which was inhibited by 4 mM Cscl, but no If current was detected in the Ad-GFP group. In addition, the inward current gradually increased along with the rise in density–voltage increment over time (Fig. 5B–C).
FIG. 5.
Spontaneous electrical activity in differentiated BADSCs. (A) Spindle-shaped cells were used for electrophysiological recordings (magnification, × 400). (B) Current density–voltage relationships of Ad-prrx1 groups (black, n = 8). (C) If current was detected in Ad-prrx1 using the patch clamp technique and was blocked by CsCl (4 mM) (n = 8). If, hyperpolarization-activated inward current.
Discussion
Since the 21st century, numerous studies have investigated the mechanisms of biological pacing. Consequently, several advancements have been made over the decades. Biological pace making is defined as the introduction of specific genes into the myocardium by genetic engineering or the differentiation of stem cells into SAN-like cells in vitro or in vivo. These methods culminate in the induction of pacemaker cells that are similar to the natural pacemaker cells. An ectopic pacing site is constructed to replace the damaged SAN. Patients with a slow heart rate benefit from this technique as it increases the basic heart rate needed to maintain normal activities.
Based on the development of lineage tracing techniques and the establishment of transgenic mice models, several discoveries on the embryonic development of SAN have been made. It was reported that the sinus venosus develops from TBX18+/Nkx2.5−/ISL-1− progenitors around E8 after which some progenitors begin to express ISL-1 (TBX18+/Nkx2.5−/ISL-1+) (Mommersteeg et al., 2010). In the embryo, the sinus venosus is a symmetric structure, whereas the sinus node, which is regulated by the homeobox factor pitx2, only exists in the right horn of the sinus venosus. A previous study reported that two SAN primordia were formed in a pitx2-deficient mice. However, in a normal embryo, pitx2 is expressed in the left sinus node where it negatively regulates SAN gene expression (Wang et al., 2014). In addition, HCN4 is a specific marker used to characterize pacemaker cells. Cardiac-specific HCN4 knockout mice die in utero between E9.5 and E11.5, which matches with the period of SAN development (Stieber et al., 2003; Verkerk and Wilders, 2015).
During embryonic development process, prrx1 and prrx3 (known as shox2) are homologous genes, which influence embryonic skeletal and cardiovascular development. Prrx1 plays an important role in the whole process of embryo formation, especially in the early embryo. During the process of embryo, prrx1 gradually increased its expressions in the outer membrane and involved with the development of the cardiovascular system. Previous studies showed that prrx1 gene mutations impaired the development of the cardiovascular system. For example, prrx1 knockout resulted in arterial catheter dislocation, abnormal aortic arch, the abnormal tilt of right subclavian artery, and other vascular abnormalities. Besides, prrx1 has been found to control vascular smooth muscle cells amplification, which implicated prrx2 in pulmonary vascular disease regulation. Furthermore, in adults, prrx1 mutations were one of the risk factors of atrial fibrillation (Hsu et al., 2018). Many studies have shown that prrx1 mutations increased the susceptibility to atrial fibrillation by reducing the heart's action potential phase (Tucker et al., 2017).
This study found that overexpression of prrx1 gene promoted the expressions of TBX18, ISL-1, and HCN4 and inhibited the levels of pitx2, which induced SAN-like cells in vitro. In addition, the induced cells displayed the electrophysiological functions of SAN. Thus, we reported, for the first time, that prrx1 might be an upstream regulator of SAN genes that can differentiate BADSCs into SAN-like cells.
It is not clear whether prrx1 plays similar roles with its homologous gene shox2 (an important transcription factor of SAN). Further studies are required to explore this concept. Hoffmann et al. (2013) found that ISL-1 is a direct transcriptional target of the homeodomain transcription factor Shox2 that rescues the Shox2-mediated bradycardia. In our study, prrx1 not only increased the expression of ISL-1 but also regulated pitx2 expression that has been reported to inhibit shox2 expression. Besides, prrx1 increased the expression of shox2 in this study. Therefore, we inferred that prrx1 may not only play the same role with homologous gene of shox2 but also locate former site of SAN development.
There are several reasons why we choose BADSCs as the targeted cells in this vivo study. First, BADSCs are easy to obtain and have the general characteristics of stem cells—the ability of self-proliferation. Second, compared with embryonic stem cells, we do not need to take human ethical issues into consideration. Moreover, the advantages of weak immunogenicity and strong multidirectional differentiation of BADSCs make it a classic choice for many stem cell-related studies. Finally, a range of researches demonstrate that BADSCs have their specific advantages compared with white ADSCs (WADSCs). Recent studies have shown that ADSCs can directly differentiate into SAN-like pacemaker cells under the induction of TBX18 transcription factors. Sun et al (2018) compared the efficiency of differentiation of BADSCs and WADSCs into sinus node-like cells induced by TBX18 transduction in terms of morphology and function. They found that there was no significant difference in the transfection rate between the two cells. The ultrastructure of BADSCs was more complicated than that of WADSCs, indicating that BADSCs may have a better structural basis and can differentiate into pacemaker-like cells easier. Furthermore, the expression levels in TBX18-BADSCs were significantly higher than TBX18-WADSCs. In conclusion, TBX18 gene transduction promotes the differentiation of BADSCs and WADSCs into pacemaker-like cardiomyocytes, whereas BADSCs may have higher differentiation capacity than WADSCs.
Our study has the following limitations: (1) we did not perform animal experiments in vivo. (2) prrx1 was only tested in BADSCs and not in cardiomyocytes or iPSCs. This means that our study did not show any results linked with heart rate. Further studies are therefore required to explore the relationship between prrx1 and heart rate. In conclusion, our study demonstrated that prrx1 could induce SAN-like cells and might be an upstream transcription factor of SAN embryonic development.
Conclusion
The study showed that prrx1 can induce the SAN-like cells for the first time and it may be an upstream regulation factor in SAN embryonic development by inducing the expression of TBX18 and ISL-1.
Authors' Contributions
L.Y. and M.X.L. made substantial contributions to the conception and design of the study, performed the experiments and wrote the article. F.Y.W. assisted in the performance of experiments. X.W., Y.H.T., Q.Y.Z., and T.W. participated in research design and coordinated the study. C.X.H. revised the article and gave final approval of the version to be published. All authors read and approved the final article.
Disclosure Statement
No competing financial interests exist.
Funding Information
This study was supported by the Fundamental Research Funds for the Central Universities of China (grant no. 2042015kf0229).
References
- Choudhury M., Black N., Alghamdi A., D'Souza A., Wang R., Yanni J., et al. (2018). TBX18 overexpression enhances pacemaker function in a rat subsidiary atrial pacemaker model of sick sinus syndrome. J Physiol 596, 6141–6155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani E., Goldhaber J.I., and Marban E. (2018). Next-generation pacemakers: from small devices to biological pacemakers. Nat Rev Cardiol 15, 139–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza A., Pearman C.M., Wang Y., Nakao S., Logantha S., Cox C., et al. (2017). Targeting miR-423-5p reverses exercise training-induced HCN4 channel remodeling and sinus bradycardia. Circ Res 121, 1058–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorabi A.M., Hajighasemi S., Khori V., Soleimani M., Rajaei M., Rabbani S., et al. (2019a). Functional biological pacemaker generation by T-box18 protein expression via stem cell and viral delivery approaches in a murine model of complete heart block. Pharmacol Res 141, 443–450 [DOI] [PubMed] [Google Scholar]
- Gorabi A.M., Hajighasemi S., Tafti H.A., Atashi A., Soleimani M., Aghdami N., et al. (2019b). TBX18 transcription factor overexpression in human-induced pluripotent stem cells increases their differentiation into pacemaker-like cells. J Cell Physiol 234, 1534–1546 [DOI] [PubMed] [Google Scholar]
- Hao T., Li J., Yao F., Dong D., Wang Y., Yang B., et al. (2017). Injectable fullerenol/alginate hydrogel for suppression of oxidative stress damage in brown adipose-derived stem cells and cardiac repair. ACS Nano 11, 5474–5488 [DOI] [PubMed] [Google Scholar]
- Hoffmann S., Berger I.M., Glaser A., Bacon C., Li L., Gretz N., et al. (2013). Islet1 is a direct transcriptional target of the homeodomain transcription factor Shox2 and rescues the Shox2-mediated bradycardia. Basic Res Cardiol 108, 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth F.C., Qureshi M.A., Jayaprakash P., Parekh K., Oz M., Dobrzynski H., et al. (2018). The pattern of mRNA expression is changed in sinoatrial node from Goto-Kakizaki type 2 diabetic rat heart. J Diabetes Res 2018, 8454078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J., Gore-Panter S., Tchou G., Castel L., Lovano B., Moravec C.S., et al. (2018). Genetic control of left atrial gene expression yields insights into the genetic susceptibility for atrial fibrillation. Circ Genom Precis Med 11, e002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Xin Y., Zhao Y., and Hu J. (2018). Shox2: the role in differentiation and development of cardiac conduction system. Tohoku J Exp Med 244, 177–186 [DOI] [PubMed] [Google Scholar]
- Kapoor N., Liang W., Marban E., and Cho H.C. (2013). Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat Biotechnol 31, 54–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehat I., Khimovich L., Caspi O., Gepstein A., Shofti R., Arbel G., et al. (2004). Electromechanical integration of cardiomyocytes derived from human embryonic stem cells. Nat Biotechnol 22, 1282–1289 [DOI] [PubMed] [Google Scholar]
- Liang X., Zhang Q., Cattaneo P., Zhuang S., Gong X., Spann N.J., et al. (2015). Transcription factor ISL1 is essential for pacemaker development and function. J Clin Invest 125, 3256–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A.D., Hayrapetyan V., Moreno A.P., and Beyer E.C. (2002). Connexin43 and connexin45 form heteromeric gap junction channels in which individual components determine permeability and regulation. Circ Res 90, 1100–1107 [DOI] [PubMed] [Google Scholar]
- Miake J., Marban E., and Nuss H. B. (2002). Biological pacemaker created by gene transfer. Nature 419, 132–133 [DOI] [PubMed] [Google Scholar]
- Mommersteeg M.T., Dominguez J.N., Wiese C., Norden J., de Gier-de V. C., Burch J.B., et al. (2010). The sinus venosus progenitors separate and diversify from the first and second heart fields early in development. Cardiovasc Res 87, 92–101 [DOI] [PubMed] [Google Scholar]
- Ocana O.H., Coskun H., Minguillon C., Murawala P., Tanaka E.M., Galceran J., et al. (2017). A right-handed signalling pathway drives heart looping in vertebrates. Nature 549, 86–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protze S.I., Liu J., Nussinovitch U., Ohana L., Backx P.H., Gepstein L., et al. (2017). Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker. Nat Biotechnol 35, 56–68 [DOI] [PubMed] [Google Scholar]
- Qu J.H., Plotnikov A.N., Danilo P., Shlapakova I., Cohen I.S., Robinson R.B., et al. (2003). Expression and function of a biological pacemaker in canine heart. Circulation 107, 1106–1109 [DOI] [PubMed] [Google Scholar]
- Scavone A., Capilupo D., Mazzocchi N., Crespi A., Zoia S., Campostrini G., et al. (2013). Embryonic stem cell-derived CD166+ precursors develop into fully functional sinoatrial-like cells. Circ Res 113, 389–398 [DOI] [PubMed] [Google Scholar]
- Stieber J., Herrmann S., Feil S., Loster J., Feil R., Biel M., et al. (2003). The hyperpolarization-activated channel HCN4 is required for the generation of pacemaker action potentials in the embryonic heart. Proc Natl Acad Sci U S A 100, 15235–15240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun A.J., Qiao L., Huang C., Zhang X., Li Y.Q., and Yang X.Q. (2018). Comparison of mouse brown and white adiposederived stem cell differentiation into pacemakerlike cells induced by TBX18 transduction. Mol Med Rep 17, 7055–7064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker N.R., Dolmatova E.V., Lin H., Cooper R.R., Ye J., Hucker W.J., et al. (2017). Diminished PRRX1 expression is associated with increased risk of atrial fibrillation and shortening of the cardiac action potential. Circ Cardiovasc Genet 10, e001902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerk A.O., and Wilders R. (2015). Pacemaker activity of the human sinoatrial node: an update on the effects of mutations in HCN4 on the hyperpolarization-activated current. Int J Mol Sci 16, 3071–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Bai Y., Li N., Ye W., Zhang M., Greene S.B., et al. (2014). Pitx2-microRNA pathway that delimits sinoatrial node development and inhibits predisposition to atrial fibrillation. Proc Natl Acad Sci U S A 111, 9181–9186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Yang M., Yang A., Wang X., Tang Y., Zhao Q., et al. (2019). Insulin gene enhancer binding protein 1 induces adipose tissue-derived stem cells to differentiate into pacemaker-like cells. Int J Mol Med 43, 879–889 [DOI] [PMC free article] [PubMed] [Google Scholar]