Abstract
Heterochromatin is a key architectural feature of eukaryotic chromosomes, which endows particular genomic domains with specific functional properties. The capacity of heterochromatin to restrain the activity of mobile elements, isolate DNA repair in repetitive regions and ensure accurate chromosome segregation is crucial for maintaining genomic stability. Nucleosomes at heterochromatin regions display histone post-translational modifications that contribute to developmental regulation by restricting lineage-specific gene expression. The mechanisms of heterochromatin establishment and of heterochromatin maintenance are separable and involve the ability of sequence-specific factors bound to nascent transcripts to recruit chromatin-modifying enzymes. Heterochromatin can spread along the chromatin from nucleation sites. The propensity of heterochromatin to promote its own spreading and inheritance is counteracted by inhibitory factors. Because of its importance for chromosome function, heterochromatin has key roles in the pathogenesis of various human diseases. In this Review, we discuss conserved principles of heterochromatin formation and function using selected examples from studies of a range of eukaryotes, from yeast to human, with an emphasis on insights obtained from unicellular model organisms.
Heterochromatin is a fundamental architectural feature of eukaryotic chromosomes that endows particular genomic regions with specific functional properties. The term ‘heterochromatin’ was coined based on the differential staining of chromosomal regions but now generally refers to molecular subtypes of transcriptionally repressed chromatin domains that extend beyond a single gene or regulatory element (BOX 1). Different varieties of heterochromatin are distinguished by their combination of histone post-translational modifications (PTMs). These affect the recruitment of proteins to, and the folding of, chromatin. Sequences embedded in heterochromatin often contain repetitive elements, such as satellite repeats and transposable elements. A crucial function of heterochromatin, which is generally more compact than euchromatin, is to prevent such selfish sequences from producing genetic instability. Additional heterochromatin roles include asserting cell-type-specific transcription and centromere function.
Box 1 |. Heterochromatin history.
The term heterochromatin was first coined in 1928 by Emil Heitz, who developed chromatin staining methods and found that chromosomes are composed of regions that are not stained after telophase (euchromatin) and regions that stained throughout the cell cycle (heterochromatin). He noted that staining patterns are chromosome-specific and later suggested that genes are found in euchromatin, whereas heterochromatin is genetically inert. He also noted that heterochromatin is often associated with sex chromosomes (reviewed in REF. 228). Finally, Heitz recognized that some regions are stained only in certain cells; these were later termed facultative heterochromatin to distinguish them from constitutively stained regions, which were dubbed ‘constitutive heterochromatin’.
In the early 1930s, Hermann Muller isolated the experimentally induced white mottled mutations in Drosophila melanogaster, which exhibited a mosaic or variegated pattern of red (wild-type) or white (mutant) eye facets as a result of chromosome rearrangements that displaced the white gene from its original position229. In 1936, the examination of D. melanogaster polytene chromosomes revealed that variegating mutants were freguently associated with rearrangement breakpoints in heterochromatic regions230. Thus, the vague cytological entity ‘heterochromatin’ became intertwined with a phenomenon that was termed ‘position-effect variegation’, which refers to phenotypic variegation due to variable inactivation of a gene triggered by its placement in or near heterochromatin. Extra copies of heterochromatic chromosomes were found to alleviate position-effect variegation, perhaps because they compete for binding by limiting factors231,232. Later, mutations were isolated in single genes that increased or decreased the variegated eye colour phenotype233–235.
In the 1960s, reassociation kinetics of sheared denatured DNA revealed that a substantial fraction of eukaryotic genomes is repetitive236. These rapidly annealing fractions were found to correspond to genomic sequences that exhibited distinct buoyant density on CsCl gradients because of their skewed base composition relative to the rest of the genome237–238. Because these sequences formed an ancillary peak in the density profile, they were termed ‘satellites’. As satellite peaks form with both sheared, low-molecular-weight DNA and with high-molecular-weight DNA, it was concluded that the constituent repeats are organized in long, tandem arrays239–241. Because of its repetitive nature, satellite DNA was the first eukaryotic DNA to be sequenced by early methods242,243.
The use of purified satellite DNA as labelled probes for in situ hybridization to metaphase chromosomes revealed that these satellites are located in the pericentromeric heterochromatin regions of metaphase chromosomes244,245 and colocalize with dense chromatin at the nuclear periphery in interphase cells246. Thus, it became apparent that large blocks of constitutive pericentromeric heterochromatin contain arrays of repetitive sequences and that artificial juxtaposition of genes with such regions by a chromosomal rearrangement led to their inactivation.
The above findings, coupled with the inability to detect satellite-complementary RNA, suggested that heterochromatin is transcriptionally inactive247. Moreover, the differential centrifugal sedimentation of chromatin containing satellite DNA was consistent with heterochromatin being more compact248. In addition, satellite DNA replicated late during S phase249 and under-replicated in polytene nuclei250, suggesting that heterochromatin also affects DNA replication.
Histone PTMs, particularly on lysine residues of the unstructured histone tails that protrude from nucleosomes, are often referred to as ‘epigenetic marks’ because they can confer gene expression properties that are not strictly dependent on DNA sequence (BOX 2). Histone PTMs regulate the propensity of the underlying DNA to participate in the processes of transcription, replication, repair and recombination. Specific PTMs control binding of particular proteins to nucleosomes via specific protein domains known as ‘reader’ domains or modules (FIG. 1). Reader domains can be joined in the same protein with domains that modify chromatin, or reader proteins can be part of complexes that contain or recruit chromatin-modifying enzymes, which modify histones by the addition of PTMs (‘writers’) or the removal of PTMs (‘erasers’). Other enzymes recruited by histone PTMs are chromatin remodellers, which alter contacts between the histone octamer core and DNA to accomplish a variety of tasks1.
Box 2 |. The use of the terms ‘epigenetic’ and ‘epigenetics’.
Conrad Waddington originally coined the term ‘epigenetics’ to refer to the mechanisms of acguisition of stable cell fates during development, but subseguently this definition was repeatedly modified (reviewed in REF. 251). Robin Holliday defined epigenetics as the inheritance of changes in gene expression patterns and, more generally, the inheritance of any change in gene function that does not involve a change in DNA sequence. Arthur Riggs defined epigenetics as the study of mitotically and/or meiotically heritable changes in gene function that cannot be explained by changes in DNA sequence. Mark Ptashne defined the phrase ‘epigenetic change’ as a heritable change in the expression of a gene that does not involve a change in its sequence and persists in the absence of the initiating signal. Conversely, Adrian Bird questioned whether heritability should be a compulsory component of a modern definition of epigenetics because it does not specify how many generations of inheritance might be required to satisfy the definition. Instead, Bird suggested as an all-encompassing definition of epigenetics “the structural adaptation of chromosomal regions so as to register, signal or perpetuate altered activity states” (REF. 252). This chromosome-based definition excludes any number of other feedback mechanisms that can mediate heritable change without a change in DNA sequence, such as post-transcriptional positive feedback loops that occur in Drosophila melanogaster sex determination and in prions.
Despite these foundational differences in definition, the use of the noun ‘epigenetics’ and the adjective ‘epigenetic’ has been essentially redefined by many to refer to chemical modifications of histones and DNA because, in some cases, these are required for or contribute to a heritable change in gene expression. The adjective ‘epigenetic’ has thus been used in the context of phrases such as ‘epigenetic mark’ or ‘epigenetic modification’ in a manner synonymous with chemical modification of nucleic acids or associated proteins, or more generally as synonymous with ‘chromatin modification’. The ensemble of such modifications has been referred to as the ‘epigenome’. Such extensions, although entrenched, may be misconstrued or imply an untruth (depending on the definition being applied)—namely, that any chemical modification of a nucleic acid or associated protein mediates a heritable change in the expression of a gene.
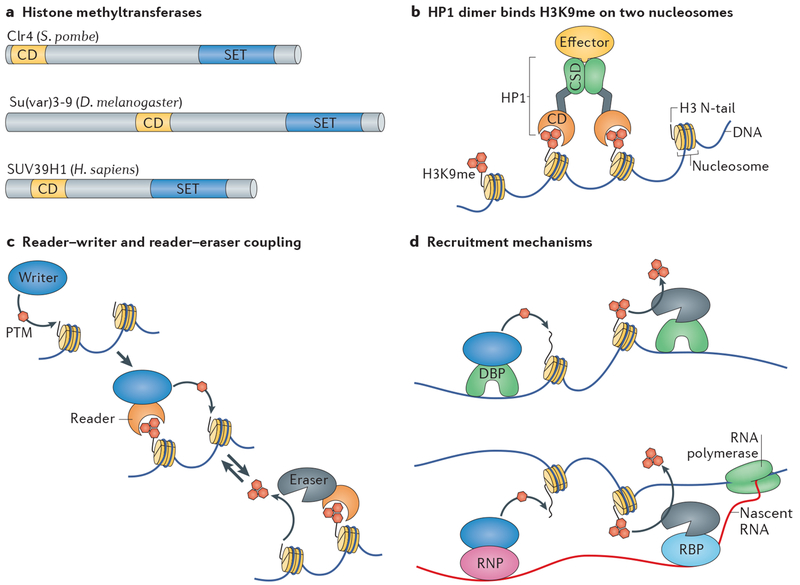
Figure 1 |. Core heterochromatin components and mechanisms.
a | The protein domain organization of the histone–lysine N-methyltransferases (KMTs) cryptic loci regulator 4 (Clr4) of Schizosaccharomyces pombe, suppressor of variegation 3-9 (Su(var)3-9) of Drosophila melanogaster and SUV39H1 (SU(VAR)3-9 homologue 1) of Homo sapiens. The SET (Su(var)3-9, enhancer of zeste and trithorax) domain isthe KMT catalytic domain and uses S-adenosyl-methionine as a methyl donor to methylate histone H3 lysine 9. The chromodomain (CD) specifically recognizes methylated histone H3 lysine 9 (H3K9me). b | Depiction of a heterochromatin protein 1 (HP1) dimer bound to nucleosomes modified with H3K9me (red hexagons). The chromodomain and the chromoshadow domain (CSD), which is a dimerization domain, of HP1 are shown. The platform produced by the CSD dimer enables binding of effector proteins. For simplicity, only one of the two H3 tails that protrude from the octamer core is shown on each nucleosome. c | Heterochromatin assembly and disassembly by reader-modifier coupling. Different ‘writer’ enzymes catalyse the addition of a post-translational modification (PTM) to a histone within a nucleosome, whereas ‘eraser’ enzymes catalyse the removal of PTMs. ‘Reader’ proteins or protein domains recognize and bind PTMs and are often coupled with writer or eraser proteins or protein domains in the same protein, protein complex or via reversible protein-protein interactions, d | Recruitment mechanisms. DNA-binding proteins (DBPs) can recruit writers or erasers to chromatin (top). Alternatively, a nascent transcript associated with the RNA polymerase can harbour recognition signals for a sequence-specific and/or structure-specific ribonucleoprotein (RNP) or RNA binding protein (RBP) (bottom). The latter include the Argonaute family proteins (not shown), which recognize and bind RNA by incorporating cognate small RNAs such as siRNAs or Piwi-associated RNAs (reviewed in REF. 253). In turn the RNP or RBP can recruit writers or erasers that modify chromatin.
The best-studied types of heterochromatin are marked by the addition of one (me1), two (me2) or three (me3) methyl groups to histone H3 lysine 9 (H3K9me) or lysine 27 (H3K27me). In this Review, we focus mostly on H3K9 methylation-dependent heterochromatin, which forms the major blocks of heterochromatin in cells and represents the defining molecular feature of constitutive heterochromatin in many eukaryotes. We also touch on other paradigms, namely, the silent information regulator (SIR) and Polycomb silencing systems, to highlight specific concepts. Histone H3K9 methylation on ε-amine groups of lysine residues is catalysed by suppressor of variegation 3-9 (Su(var)3-9), enhancer of zeste and trithorax (SET) domains of the histone–lysine N-methyltransferase (KMT) orthologues of the Drosophila melanogaster Su(var)3-9 proteins (SUV39H1 and SUV39H2 in mammals and abbreviated here as Suv39 when referring to both fruit fly and mammalian proteins) and Schizosaccharomyces pombe cryptic loci regulator 4 (Clr4)2–4 (FIG. 1a). The H3K9me readers, heterochromatin protein la (HP1a) and HP1b in D. melanogaster (FIG. 1b) and their S. pombe orthologues Swi6 and chromo domain-containing protein 2 (Chp2), selectively bind methylated H3K9 through their chromodomains2–13. Additional factors and epigenetic modifications contribute to the formation and maintenance of heterochromatin; for example, the gene-repressive histone PTM H3K9me can be coupled with the repressive DNA 5-methylcytosine (5meC) in some systems14–17.
In this Review, we discuss the key principles of heterochromatin formation and function. We illustrate these with examples taken mainly from unicellular yeasts, but include selected studies from a variety of model organisms. It is not our intention to be comprehensive; therefore, we have included only limited discussion of system-specific details and caveats. We focus on studies and experimental approaches that illuminate how histone modifications recruit heterochromatin factors, the role of RNA as a recruiting platform, the differences between heterochromatin establishment and maintenance, the processes of heterochromatin spreading and inheritance and the contributions of heterochromatin to genome stability, development and disease.
One: coupling of readers and modifiers
The SIR proteins of the budding yeast Saccharomyces cerevisiae (reviewed in REFS 18,19) comprise the first system in which the molecular mechanism of chromatin silencing was molecularly defined, although the silenced chromatin is distinct from canonical, H3K9me-dependent heterochromatin. In the SIR system, silencer elements are recognized by sequence-specific DNA-binding proteins that then recruit four proteins: Sir1, Sir2, Sir3 and Sir4. Sir2 is a NAD+-dependent histone deacetylase (HDAC) that acts on acetylated histone H4 lysine 16, thereby enabling the Bromo-associated homology domain of Sir3 (a component of the Sir3–Sir4 complex) to bind nucleosomes20. Deacetylation by Sir2 thus promotes Sir3 binding, allowing further cycles of Sir protein recruitment to form silent domains. The SIR system illustrates the principle of reader–modifier coupling (FIG. 1c), in this case between the Sir3 reader and the Sir2 eraser. It also illustrates the principle of gene-silencing initiation by the recruitment of sequence-specific DNA-binding proteins (FIG. 1d). Although paradigmatic, the SIR system is restricted to S. cerevisiae and its relatives and thus evolved quite recently during evolution21.
Reader–modifier coupling is also a key feature of the more canonical H3K9me-dependent heterochromatin5,6,22–24. Both fruitfly and mammalian Suv39 and S. pombe Clr4 H3K9 methyltransferases have a similar organization with an N-terminal chromodomain and C-terminal SET domain (FIG. 1a), thereby coupling writer and reader modules in the same protein. Methylation of H3K9 by the SET domain enables recruitment of Suv39 or Clr4 through their respective chromodomains. HP1 proteins contain not only a chromodomain reader module but also a more C-terminal chromoshadow domain (CSD; FIG. 1b). CSD dimerization forms a binding platform for other effector proteins25,26. Thus, the reading of H3K9me by HP1 proteins provides another route to reader–modifier coupling through CSD dimerization. For example, in S. pombe, recruitment of HDAC (eraser) complexes such as SHREC (Snf2/Hdac-containing repressor complex and Clr6 complex) by Swi6 and Chp2 removes acetylation, thereby allowing H3K9 methylation27,28. The recruitment of SHREC, which harbours the Mit1 (Mi2-like protein interacting with Clr3) chromatin remodeller subunit, also has a role in the elimination of nucleosome-free regions — nucleosome absence is a hallmark of heterochromatin in S. pombe29,30. As part of protein dimers, the reader domains of HP1 are also coupled as pairs with ensuing functions: two dimers of Swi6 bind a single H3K9me-modified nucleosome and provide ‘sticky ends’ that enable Swi6 to bridge two nucleosomes31. In some systems, H3K9 readers can be coupled with DNA modification. In mammals and plants, 5meC DNA methyltransferases can be recruited with H3K9 methyl-transferases so that the two reciprocally bolster each other to ensure that the DNA is rendered inaccessible15–17.
Reader–modifier coupling is also a feature of the more dynamic silencing complexes recruited by H3K27me. Methylation of H3K27 by the Polycomb repressive complex 2 (PRC2) catalytic subunit enhancer of zeste (E(z)) in D. melanogaster or enhancer of zeste homologue 2 (EZH2) in mammals promotes binding of the Polycomb subunit of PRC1 to chromatin through its chromodomain32–36. In addition, the PRC2 subunits extra sex combs (ESC) in D. melanogaster and embryonic ectoderm development (EED) in mammals recognize the H3K27me mark and allosterically activate E(z) or EZH2, respectively37.
Two: noncoding RNAs recruit modifiers
Noncoding RNAs (ncRNAs) that may be processed into small RNAs are transcribed from heterochromatin16,38. This may seem surprising because heterochromatin induces transcription silencing. Nonetheless, a low level of transcription occurs in heterochromatin, and this is important for heterochromatin formation in several organisms. Heterochromatin transcription can be cell-cycle regulated, occurring only during DNA replication, when heterochromatin becomes accessible39–41. One function for this transcription appears to be recruitment of silencing factors (FIG. 1d) through association with nascent transcripts, for example, in S. pombe42–50. The ncRNAs also provide a substrate for the generation of small RNAs, which promote the recruitment of silencing factors through base pairing, likely with nascent transcripts.
In S. pombe, RNA polymerase II (Pol II) transcribes heterochromatin repeats. The Argonaute protein (Ago1) binds single-stranded siRNAs and uses them to target homologous nascent repeat transcripts that emerge from chromatin-associated Pol II and thus to recruit silencing factors47,51,52. Ago1 is part of the three-subunit complex RNA-induced transcriptional silencing (RITS)46, which associates with both the RNA-dependent RNA polymerase complex (RDRC)43 and the histone Clr4 complex (CLRC)45,53–55. RDRC uses primary transcripts as templates for synthesis of double-stranded RNA (dsRNA), which is subsequently processed into siRNAs, thereby increasing siRNA production43–56. In a positive feedback loop, CLRC is required for all H3K9 methylation in S. pombe, and H3K9me promotes efficient siRNA production. Such feedback is, in part, mediated through the recruitment of RITS to H3K9-methylated chromatin through its chromodomain protein Chp1 (REFS 24,57,58). Two bridging factors connect the different heterochromatin-promoting complexes: Stc1 (siRNA to chromatin) recruits CLRC through RITS44, whereas Ers1 (essential for RNA silencing) couples RDRC, RITS and Swi6 (REFS 59–61).
In plants, a similar feedback loop promotes H3K9 methylation. Most details come from studies in Arabidopsis thaliana where, similarly to S. pombe, nascent transcripts provide the platform for the recruitment of ARGONAUTE–siRNA complexes. Pol V, which is a specialized plant Pol II paralogue, produces transcripts that are targeted by siRNA-guided AGO4 (REF. 62). AGO4 recruits the de novo DNA methyltransferase DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2)63, which in turn recruits the KMTs and H3K9 methyltransferases SUVH4, SUVH6 and SUVH9 through the DDR complex (DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1 (DRD1), DEFECTIVE IN MERISTEM SILENCING 3 (DMS3) and RNA-DIRECTED DNA METHYLATION 1 (RDM1))64. Another Pol II paralogue, Pol IV, produces dsRNA from template transcripts in association with RNA-dependent RNA polymerase. The processing of the resulting dsRNA by Dicer generates siRNAs that are loaded into AGO4 and targeted at the complementary, Pol V-generated transcripts65–67. Pol IV is commonly recruited by an H3K9me reader, SAWADEE HOMEODOMAIN HOMOLOGUE 1 (SHH1)68,69. Thus, as in S. pombe, nascent transcripts have two functions in A. thaliana: transcripts produced by Pol V recruit chromatin-modifying enzymes through base pairing with siRNAs, which in turn are produced from homologous, Pol IV-generated transcripts.
Another function of nascent heterochromatin transcripts is to recruit silencing-promoting proteins without the intermediary of small RNAs. S. pombe possesses an RNAi-independent pathway that promotes H3K9 methylation and functions to maintain pericentromeric heterochromatin70. One component of this pathway is seven binding 1 (Seb1), which contains an RNA recognition motif that recognizes GUA trinucleotides in nascent transcripts and a Pol II C-terminal-domain interaction domain71,72. Seb1 acts upstream of the SHREC complex73, which participates in an RNAi-independent silencing pathway74. The Seb1–SHREC pathway is partially redundant with RNAi because H3K9 methylation is eliminated only in mutants where both pathways are inactivated74. As GUA trinucleotides occur frequently, how Seb1 promotes H3K9 methylation selectively at pericentromeric regions is not fully understood, but the selectivity could be facilitated by the relative depletion of GUA sequences from S. pombe protein-coding genes72. Furthermore, recent findings show that Suv39 KMTs are stabilized on heterochromatin by their nonspecific affinity for nascent RNA produced from mammalian centromere repeat arrays75–77.
Similar transcript-driven processes mediate X chromosome inactivation in female mammals, a process that produces a condensed, silenced X chromosome marked by H3K27me3. Although the inactive X is not considered constitutive heterochromatin, this form of silent chromatin serves to illustrate related important principles. The A-repeat region of the long noncoding RNA X-inactive specific transcript (XIST) recruits SPEN (split ends; also known as SMART/HDAC1-associated repressor protein and Msx2-interacting protein), a protein that contains RNA recognition motifs78–81 and that in turn recruits HDAC3 through the adaptor protein SMRT (silencing mediator of retinoid and thyroid hormone receptor; also known as NCOR2)31,82,83. Ensuing histone deacetylation probably triggers the recruitment of at least two redundant silencing machineries: one comprising PRC2 and PRC1, and the other remains to be identified (reviewed in REF. 83). As with Seb1, it is unclear whether SPEN alone has sufficient specificity to target XIST and the X chromosome for inactivation.
Three: establishment versus maintenance
Some signals and factors required to initiate de novo assembly of heterochromatin (that is, to convert euchromatin into heterochromatin) differ from those required for heterochromatin maintenance. This distinction between establishment and maintenance is crucial for understanding how heterochromatin formation occurs.
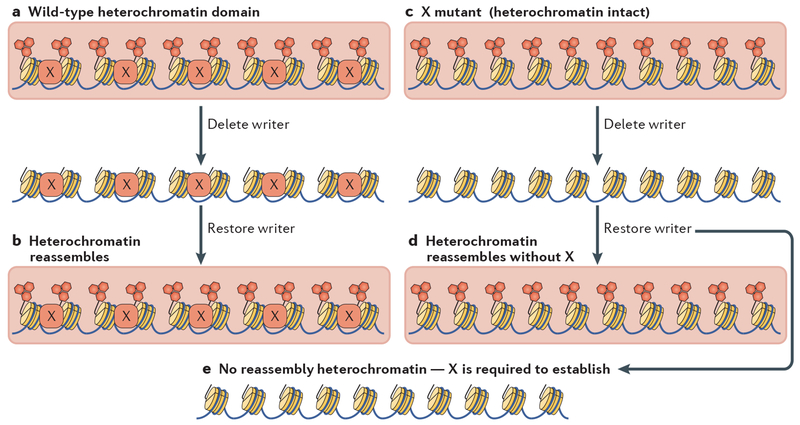
Testing whether a nonessential factor is required to establish heterochromatin (an establishment factor) is performed in S. pombe as follows (FIG. 2). Heterochromatin is first erased by removing the gene encoding a key chromatin modifier (for example, a KMT or HDAC). The re-introduction of that gene into otherwise wild-type cells allows heterochromatin re-establishment; however, cells lacking a heterochromatin establishment factor are unable to assemble heterochromatin24. Another approach compares the outcome of introducing naive DNA that can serve as a heterochromatin template (for example, centromere repeats) into wild-type versus mutant cells24,84,85. A third way is to erase heterochromatin by exposure to inhibitors (for example, the HDAC inhibitor Trichostatin A) and determine whether mutant cells recover heterochromatin after the removal of the inhibitor86,87. Such assays revealed that in S. pombe RNAi has an essential role in establishing heterochromatin. For instance, in the absence of RNAi factors, no H3K9me can be established at centromere repeats or related sequences when the KMT Clr4 is re-introduced into cells lacking Clr4. Likewise, H3K9me is established on repeats transformed into wild-type cells but not cells lacking RNAi. This stands in contrast to the partially redundant role of RNAi with Seb1 or with the HDACs Clr3 or Sir2 in the maintenance of H3K9me at peri-centromeric regions, in which double mutants of RNAi and Seb1 or RNAi and a HDAC are required to eliminate H3K9me2 (REFS 74,85).
Figure 2 |. Determining whether a factor is required for the establishment, but not maintenance, of heterochromatin.
Identifying a factor that is required to maintain repressive heterochromatin is straightforward because deletion of the gene encoding that factor will disrupt heterochromatin formation and associated phenotypes such as gene silencing. Determining whether a factor has a role in heterochromatin establishment requires additional experiments. a | The gene for an endogenous pivotal writer is inactivated, resulting in the loss of a heterochromatin domain (large red rectangles) such as that mediated by histone H3 lysine 9 methylation (red hexagons) in these cells. A heterochromatin-associated factor (protein, RNA or post-translational modification) is marked with ‘X’. b | Restoration of the writer to these cells allows re-establishment of a full heterochromatin domain, indicating that all factors required for heterochromatin nucleation, spreading and maintenance are present, including factor X. c | Cells lacking the heterochromatin-associated factor X are similarly tested. Note that X may be required for heterochromatin establishment but not strictly required for maintenance. d | The full assembly of a silent heterochromatin domain upon restoration of the writer indicates that X is not required for nucleating heterochromatin formation. e | The inability to re-establish a full heterochromatin domain indicates that X is required to trigger heterochromatin assembly but is not required for its maintenance. RNAi in Schizosaccharomyces pombe and the long noncoding RNA X-inactive specific transcript in mammals are examples of such heterochromatin establishment factors.
Establishment of heterochromatin on S. pombe centromeric outer repeats requires RNAi, but it remains unclear how the initiating source of dsRNA is generated. Possibilities include dsRNA produced by convergent, overlapping transcripts38, RNA secondary structures88 and degradation products89. Another possibility is that the RDRC synthesizes the initiating dsRNA from centromere repeat transcripts43,56, as in plants (see above). In the latter case, specific features must exist that distinguish repeat-element transcripts from mRNAs to ensure the specific recruitment of RDRC to the former.
In S. pombe, dsRNA induced by expression of an artificial, hairpin-encoding DNA is sufficient to generate synthetic siRNAs and direct H3K9me–heterochromatin formation in cis at the locus producing the dsRNA90. Here, no inherent special features are required to trigger heterochromatin formation once dsRNA is synthesized. Surprisingly, siRNAs produced from such artificial dsRNAs only weakly induce heterochromatin assembly in trans at transcribed homologous loci in euchromatin91. Such synthetic siRNAs trigger more efficient H3K9me–heterochromatin formation in trans in cells harbouring mutations in the Pol II-associated polymerase associated factor complex92–94. Defective canonical polyadenylation signals at the transcribed target locus also enhance silencing95. Thus, nascent transcripts that are held at native heterochromatin loci owing to inefficient transcription elongation and/or termination could bolster RNAi-mediated H3K9 methylation.
RNAi-independent mechanisms of heterochromatin establishment also exist in S. pombe as RNAi is not required for establishment of heterochromatin adjacent to telomeres. Clr4 is recruited to telomere repeats through the telomere-binding protein complex shelterin96. However, RNAi contributes to subtelomeric silencing in S. pombe at centromere-related, telomere-adjacent repeats97,98.
Heterochromatin establishment-specific factors that function through H3K9 methylation have also been identified in the Caenorhabditis elegans germ line, where small Piwi-associated RNAs (piRNAs) trigger an siRNA–H3K9 methylation feedback loop, much like those in S. pombe and plants99. However, piRNAs are dispensable for the maintenance of the feedback loop. This was revealed through genetic crosses that removed the two Piwi-related genes, prg-1 and prg-2, after triggering heterochromatin formation100. Piwi also has a role in the establishment of HP1a-bound heterochromatin during D. melanogaster development101.
In A. thaliana, where DNA methylation and H3K9 methylation are connected, most loci controlled by RNAi can re-establish silencing following transient disruption of DNA methylation102. However, at a small subset of these loci, DNA methylation cannot be rescued by the re-introduction of the maintenance DNA (CYTOSINE 5)-METHYTRANSFERASE (MET1) to MET1 mutants102. This suggests that once DNA methylation has been erased from these particular loci, they lack the cues required for its re-establishment.
Finally, during X chromosome inactivation in murine epiblasts, the XIST ncRNA is required to establish silencing on one of the X chromosomes (see above). However, removal of XIST later in development has no effect on the maintenance of silencing103,104. Furthermore, analyses in embryonic stem cell models showed that although SPEN and other factors are required to establish XIST-mediated gene silencing, once DNA methylation has been installed on the inactive X, the silent state is inherited in the absence of SPEN or other initiating factors78–81.
Four: heterochromatin can spread
Once nucleated at a particular location, the biochemical properties of heterochromatin components enable the expansion of the domain in a manner that is largely independent of DNA sequence. The classic example of this is position-effect variegation in D. melanogaster, where specific chromosome rearrangements can juxtapose heterochromatin with euchromatin; in such cases, heterochromatin spreads over large distances into euchromatin (reviewed in REF. 105). In D. melanogaster, the presence of additional heterochromatin elsewhere titrates limiting factors away from, and consequently weakens, heterochromatin, thereby alleviating repression at different loci106–108. Thus, spreading requires a surplus of unassembled heterochromatin components and can be driven by their overexpression109–111.
Heterochromatin spreading requires reader–writer coupling. Nucleosomes bearing H3K9me are bound by the chromodomains of H3K9me writers such as Suv39 and Clr4, and mutations in the Clr4 chromodomain impede spreading in S. pombe45,112. However, spreading also requires the HP1-dependent recruitment of HDACs28,43,50,113. Thus, interconnections among reader, writer and eraser modules form positive feedback loops that extend heterochromatin domains.
Single-cell reporter analysis in S. pombe showed that nucleation of heterochromatin at the mating-type locus can take several cell divisions and that the expansion of the domain to its full size requires even longer time114. This indicates that feedback mechanisms act both locally, on adjacent nucleosomes, and more broadly over greater distances to mediate this two-step process114. Thus, the spreading of silent chromatin does not necessarily occur in a linear fashion; random collisions between a heterochromatin domain and chromatin that is spatially located nearby may allow the key modification to be deposited discontinuously through ‘hops’ that decline in frequency with distance from the nucleation site or domain. Subsequently, gaps between the original domain and the new heterochromatin patch could be filled by a pincer-like movement, although exceptions to this scenario have been observed in D. melanogaster108. Modelling of available data suggests that reader–writer-driven feedback coupled with collisions between modified and unmodified sites optimally describes the dynamics of heterochromatin domains115.
Heterochromatin-spreading models may be influenced by recent findings of a role for HP1-induced liquid–liquid phase separation in heterochromatin assembly116,117. Purified D. melanogaster HP1a can form proteinaceous liquid droplets that undergo liquid–liquid de-mixing in vitro in particular conditions117. In D. melanogaster and mammalian cells, heterochromatin domains display properties of phase-separated liquids117. In vitro de-mixing has also been reported for the human HP1α protein. Phosphorylated HP1α de-mixes more efficiently than unphosphorylated HP1α, suggesting potential for regulation in vivo116. Indeed, a mutant that cannot be phosphorylated forms smaller heterochromatic foci when introduced into cells. Nucleosomes and DNA preferentially partition into these phosphor-HP1α droplets in vitro, suggesting that the HP1α ‘solvent’ controls entry of molecules into heterochromatin116. We anticipate that future work will reveal further the functions of phase separation in heterochromatin assembly, spreading and/or function.
Mammalian X chromosome inactivation is initiated by XIST expression from the X inactivation centre. XIST spreads discontinuously along the X chromosome and may first affect noncontiguous chromosomal regions that contact the X inactivation centre in three-dimensional space118,119. XIST spreading, accompanied by gene silencing, is not limited to X chromosomes. Specifically, rearrangements that fuse autosomes to an inactive X result in spreading of silencing into the autosome, albeit with limited efficiency120–122. Likewise, ectopic expression of XIST from autosomes results in reduced gene expression over large adjacent domains83,103,123–127.
Five: spreading is restrained
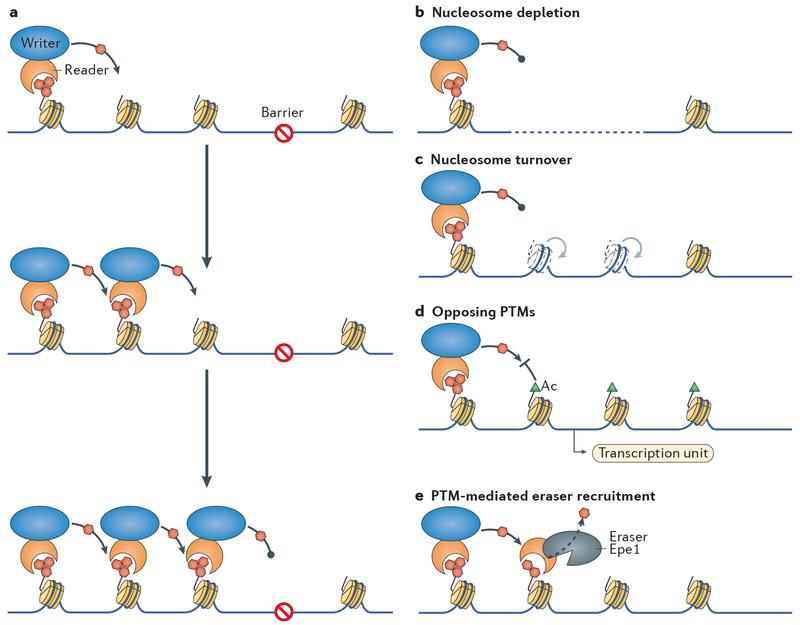
Because heterochromatin can spread, mechanisms to restrict its expansion are necessary to avoid erroneous and potentially deleterious gene silencing (FIG. 3). Mechanisms to create such barriers and interrupt lateral heterochromatin spreading include the following: generation of nucleosome-depleted regions by binding of proteins such as transcription factors; processes that promote nucleosome turnover; recruitment of antisilencing factors by ongoing transcription and associated regulatory elements; recruitment of readers with antisilencing activity; and restricting silencing factors to their sites of prior action.
Figure 3 |. The regulation of heterochromatin spreading.
a | A model for the expansion of a heterochromatin domain, in which a ‘reader’ is associated with a ‘writer’, thereby enhancing the formation of repressive histone post-translational modifications(PTMs; red hexagons) in adjacent nucleosomes. Iterative cycles result in the formation of extensive heterochromatin domains. The barrier represents a series of mechanisms that restrict such spreading, which are shown in parts b–e. b | Sequencesthat are bound by factors that disfavour nucleosome assembly create extensive gaps (dashed line) that prevent heterochromatin from spreading. c | Factors that promote nucleosome turnover through disassembly and reassembly and/or through cycles of histone exchange (light nucleosomes and arrows) effectively block heterochromatin domain expansion. d | Adjacently expressed transcription units mediate the addition of active PTMs (green triangles) to histones, which prevent the intrusion of repressive PTMs and heterochromatin. e | Erasers such as the Schizosaccharomyces pombe demethylase enhancer of position effect 1 (Epe1) are recruited by readers of repressive PTMs at the edge of heterochromatin and prevent heterochromatin expansion. Ac, acetylation.
tRNA genes are a class of heterochromatin-spreading barrier conserved from yeast to man128–130. Binding sites for the Pol III-associated transcription factor complex TFIIIC appear to be crucial for the barrier function as clusters of these sites alone, independent of tRNA genes, function as heterochromatin barriers. One example derives from the boundaries of the silent mating-type region in S. pombe131. These regions have large nucleosome-free regions, which may prevent heterochromatin spreading by forming a ‘gap’ in chromatin over which some reader–writer machineries cannot cross29 (FIG. 3b). tRNA genes, such as the TFIIIC sites at the mating-type locus, are themselves accessible and essentially free of nucleosomes132–134. Turnover of nucleosomes assembled in heterochromatin is low92,135, and factors such as the polymerase associated factor complex, which promote their turnover, are required for barrier function92,94,135 (FIG. 3c). Myriad other types of boundary elements and factors have been described suggesting that there are many mechanisms for interrupting heterochromatin assembly and spreading.
Euchromatin is marked by a variety of chromatin modifications that antagonize heterochromatin assembly. These include the histone variant H2A.Z, which is deposited in response to the formation of nucleosome-free regions at the first nucleosome (+1) downstream of transcription initiation sites136–138, and histone PTMs triggered by active transcription (for example, methylation at H3K4, H3K36 and H3K79 and general histone acetylation). In S. cerevisiae, such PTMs have an antisilencing role at SIR-dependent heterochromatin (see above and FIG. 3d)139–144. Thus, competition between the two opposing mechanisms of heterochromatin spreading and transcription likely explains the classic bistability of heterochromatic (repressed) versus euchromatic (expressed) states implied by the phenomenon of position-effect variegation described above.
Heterochromatin can itself recruit the inhibitors that limit its own spreading through reader–eraser coupling (FIG. 3e). An example in S. pombe is the Epe1 (enhancement of position effect 1; also known as Jmjc domain chromatin-associated protein) protein, which is a putative H3K9 demethylase that is recruited by the reader Swi6 (REFS 145–148). Epe1 is degraded through the action of a ubiquitin ligase, the activity of which is limited to the interior of heterochromatin domains and absent from their edges, thereby providing a mechanism by which heterochromatin can recruit an antisilencing factor in a restricted manner149. Epe1 acts in parallel with boundary elements, as loss of both Epe1 and TFIIIC sites that flank the mating-type locus result in extensive heterochromatin spreading and slow cell growth150. Likewise, cells lacking both Epe1 and the globally acting histone acetyltransferase Mst2 display widespread ectopic heterochromatin assembly and slow growth, again emphasizing the importance of redundancy of antisilencing mechanisms151. Ectopic heterochromatin formation in such double mutants suggests that the processes that trigger heterochromatin assembly at the primary genomic locations can act globally, albeit normally less effectively. The detection of low levels of H3K9me at several euchromatic loci in wild-type cells, in specific conditions or in mutants, may be a manifestation of pathways that are important for gene regulation in response to various cues92,151–155.
Tethering silencing machinery to its sites of prior action provides another mechanism to restrict heterochromatin to particular loci. Numerous chromatin-modifying complexes have domains that recognize the products of their enzymatic activity. In the budding yeast Cryptococcus neoformans, the H3K27-specific methyl-transferase PRC2 contains a subunit with a chromodomain, Ccc1 (chromodomain and coiled coil 1), which recognizes the H3K27me modification. In this yeast, H3K27me3 is selectively generated over sub-telomeric regions156. Mutations that prevent Ccc1 from recognizing H3K27me3 cause ectopic H3K27 methylation at centromeres. This ectopic methylation requires prior H3K9 methylation at centromeres, indicating that tethering of PRC2 to its sites of prior action (subtelomeres) through reader–writer coupling suppresses a latent attraction of PRC2 to H3K9-methylated domains, perhaps through the methyl-lysine binding activity of Eed.
Six: heterochromatin can be inherited
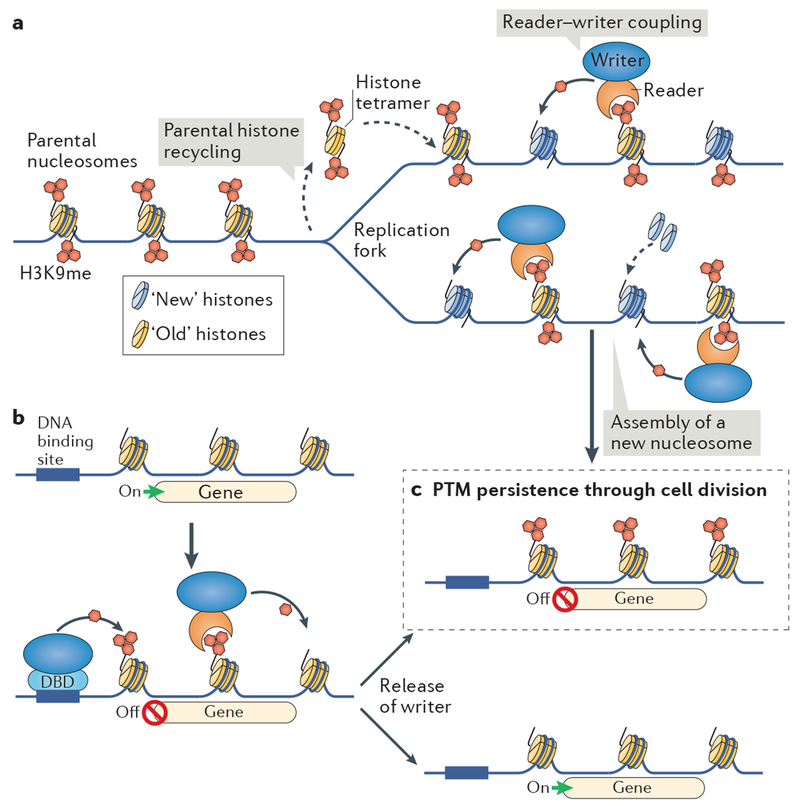
During DNA replication, the H3–H4 tetramers of old, parental nucleosomes are randomly distributed to sister chromatids during their synthesis (reviewed in REF. 157). Nucleosome occupancy on the newly synthesized DNA molecules is fully restored with new nucleosomes that are assembled from free histones. The recruitment of KMTs by the H3K9me modification that they catalyse (reader–writer coupling) may allow the modification of such newly assembled neighbouring nucleosomes and suggests that heterochromatin self-propagates in a manner that is independent of the underlying DNA sequence (FIG. 4a). This process would enable the preservation of silent chromatin through DNA replication into progeny cells. Such inheritance is termed ‘cis inheritance of a chromatin state’.
Figure 4 |. Reader–writer coupling allows the maintenance of repressive chromatin modifications through DNA replication and their transmission through cell division.
a | The maintenance of repressive histone post-translational modifications (PTMs) through DNA replication by reader–writer coupling. During replication, H3–H4 tetramers from pre-existing parental ‘old’ nucleosomes are randomly recycled to either of the two newly synthesized DNA molecules. Conseguently, the number of H3 histones bearing a PTM, such as methylation of H3 Lys 9 (H3K9me), on the two new DNA molecules will be reduced by half compared with the parental DNA. Reader–writer coupling should enable propagation of the PTM from old nucleosomes that retained the PTM to newly assembled nucleosomes, thereby replenishing PTM levels and reinstating the full chromatin domain on both sister chromatids, ultimately allowing its transmission to progeny cells, b | A writer module such as the SET (Su(var)3-9, enhancer of zeste and trithorax) domain of an H3K9 methyltransferase, can be artificially recruited to DNA by its fusion to a DNA-binding domain (DBD) whose binding site is inserted at a neutral genomic location. This generates a region with a specific, newly catalysed chromatin PTM such as H3K9me, which can recruit additional reader–writers that can spread the PTM over a nearby reporter gene, thereby silencing its expression. Release of the artificial writer from DNA by inhibition of its DBD enables assessment of the persistence and heritability of this heterochromatin. c | If heterochromatin and gene silencing persist through cell division (by the mechanism shown in part a), then the modification, in this case H3K9me, must be capable of mediating a heritable epigenetic change (BOX 2).
Epigenetic inheritance is well known to be mediated by DNA methylation in some organisms, where the maintenance DNA (cytosine-5)-methyltransferase 1 is associated with the replisome, recognizes 5meC in CG dinucleotides and adds a methyl group to cytosine in the CG of the complementary strand (reviewed in REF. 158). In the filamentous fungus Neurospora crassa, H3K9 methylation and 5meC can reinforce each other; H3K9me nucleosomes can recruit the DIM2 (defective in methylation 2) DNA methyltransferase through HP1, and DNA methylation recruits the H3K9 methyltransferase DIM5 (REFS 14,159). Connections between H3K9 methylation and DNA methylation are also well established in other organisms (reviewed in REF. 160). Because 5meC on CG dinucleotides is heritable through DNA replication, its influence on H3K9 methylation could mask the cis inheritance of chromatin states mediated by H3K9me read–write systems themselves.
Thus, a strong test of H3K9me-dependent heterochromatin heritability would be enabled by a system lacking DNA methylation. In fission yeast, DNA methylation is undetectable, and stable cis inheritance of heterochromatin occurs at the silent mating-type locus87,161. Domains of synthetic heterochromatin formed when the SET domain of the KMT Clr4 was fused to a DNA-binding domain and through it recruited to cognate binding sites placed at loci in euchromatin, resulting in gene silencing162. A DNA-binding domain controlled by a small molecule allowed conditional use of such an artificial heterochromatin nucleator to test whether endogenous wild-type Clr4, along with other effector proteins, could maintain heterochromatin and gene silencing through cell division163,164 (FIG. 4b). Release of the tethered Clr4 resulted in rapid loss of H3K9me, even when the cell cycle was arrested, suggesting that rather than being passively diluted through rounds of replication, H3K9me is actively removed. The histone demethylase Epe1 was found to be responsible for the rapid removal of this ectopic H3K9 methylation. Cells lacking Epe1 can transmit H3K9me at the manipulated locus into progeny through multiple cell divisions and even through meiosis (FIG. 4c). Thus, H3K9 methylation can be heritable and affect phenotype. Nonetheless, even in the absence of Epe1, such engineered H3K9-methylated heterochromatin and associated gene silencing eventually dissipates, presumably because of imperfect maintenance during replication and/or transcription-coupled loss of H3K9me nucleosomes.
Analogous transient targeting experiments in mammalian cells suggest that H3K9me-mediated repression is reversible, whereas DNA methylation allows the silent state to persist for many cell divisions165,166. Thus, mammalian cells also appear to restrict the heritability of H3K9me-mediated repression after the initial recruiting mechanism is disabled. By contrast, H3K9me-dependent heterochromatin formed by tethering HP1 persisted for many cell divisions following HP1 release from an engineered murine locus, although a potential role for DNA methylation in its maintenance at this locus seems difficult to rule out167.
There is now strong evidence that silencing by the Polycomb proteins can mediate the cis inheritance of a chromatin state168. Interestingly, in D. melanogaster, specific sequences that mediate PRC recruitment are required for this inheritance, suggesting again that the heritability of silent chromatin is tightly regulated169,170. Thus, Polycomb-mediated silencing exhibits similarity to heterochromatin assembly at the S. pombe mating-type locus, which also involves sequence-specific binding proteins161,171,172. In the latter case, heterochromatin maintenance is further promoted by chromatin remodelling enzymes that curb nucleosome turnover, limit euchromatin assembly and affect the spatial positioning of chromatin in the nucleus173–175.
Seven: defending the genome
Repetitive sequences are a threat to genome stability and organism viability. Mechanisms of genome destabilization by DNA repeats include mutations produced by the integration or excision of transposable elements and recombination between repeats. Heterochromatin has a pivotal role in suppressing these deleterious events through diverse mechanisms.
Studies in plants have revealed increased transposon copy numbers in mutants defective in the RNA-dependent DNA methylation (RdDM) silencing pathway described above176. Surprisingly, the copy number of only a single copia-type retrotransposon, EVD (evadé), increases when this pathway is mutated. Additional analyses confirm this observation177, which has several potential implications. First, it suggests that it is the latent activity of this single transposon that drives maintenance of RdDM in A. thaliana. Theoretical work shows that a single transposable element can spread through a sexually reproducing population despite a negative impact on fitness178. Second, the lack of effect of RdDM pathway loss on the copy number of other transposons, despite an increase in their transcript levels, suggests that these other elements are not active or that other mechanisms limit their transposition. Nevertheless, their silencing could be important for genetic stability, as discussed below.
In C. elegans, loss of Piwi proteins, which control a nuclear RNAi pathway that is coupled to H3K9me, has been shown to increase the transposition of the Tc3 family of transposable elements100,179. Worms lacking the H3K9 methyltransferases met-2 and set-25 show widespread upregulation of Tc3 transcripts in both germline and somatic tissues. Strikingly, this resulted in the formation of R-loops, replication stress and increased mutation frequency within repetitive elements179. Thus, transcribed transposons can be mutagenic even without undergoing transposition per se.
A less-appreciated characteristic of heterochromatin is that it can control transposon activity by promoting the biogenesis of specialized small RNAs, rather than by transcriptional silencing. In the D. melanogaster female gonad, mutations in the HP1 paralogue Rhino result in defective piRNA biogenesis from clustered elements180. This is highly reminiscent of the role of H3K9 methylation in siRNA biogenesis in S. pombe. These genetic clusters are heterochromatin islands that produce transposon-homologous piRNAs181,182, which act transcriptionally and post-transcriptionally to silence transposable elements181. Insertion of a transposon into a piRNA cluster in female gonads activates a mechanism that monitors and silences the transposon. The piRNA system also operates in mammalian testes to silence transposons through DNA methylation183–185.
An important mechanism of genome defence is the suppression of chromosomal rearrangements in repetitive elements following DNA damage. Homologous recombination between repeats such as dispersed transposable elements can result in chromosomal deletions, inversions and translocations. Intrachromosomal homologous recombination within repeat arrays often results in array expansion and contraction that may cause little harm (an exception being recombination within ribosomal DNA repeats186). By contrast, homologous recombination between repeats on nonhomologous chromosomes can cause translocations and result in the formation of dicentric and acentric chromosomes. Studies in D. melanogaster and mammalian cells demonstrated that breaks within heterochromatin are sequestered to the periphery of heterochromatin compartments187–189. This is thought to favour repair by homologous recombination within cognate heterochromatin repeats and thereby prevent illegitimate recombination with similar repeats on nonhomologous chromosomes190,191.
Eight: influencing centromere function
Centromeres are the chromosomal loci where kinetochores assemble. Most eukaryotic centromeres are composed of repetitive arrays of DNA; the majority of these repeats are embedded in H3K9me-dependent heterochromatin and their DNA is heavily methylated in mammalian somatic cells. However, patches of repeats assemble unusual nucleosomes, in which histone H3 is replaced by its variant centromere protein A (CENP-A). These centromere-specific nucleosomes form the physical foundation for the kinetochore (reviewed in REF. 192). Heterochromatin has two important roles in centromere and kinetochore function.
First, heterochromatin influences the assembly of CENP-A into nucleosomes. In S. pombe, CENP-A-containing chromatin and functional kinetochores cannot be established on centromere DNA lacking flanking pericentromeric heterochromatin. Heterochromatin provides a crucial, but unknown, function to ensure CENP-A assembly into adjacent chromatin. Heterochromatin-directed histone modifications and/or nuclear–periphery association may promote CENP-A incorporation. Heterochromatin may also act to limit the size of the CENP-A–kinetochore domain130,193. Conversely, inadvertent or forced heterochromatin formation in fission yeast194 or mammalian cell centromeres prevents CENP-A and kinetochore assembly195,196.
A second role for heterochromatin at centromeres involves sister-chromatin cohesion, which is mediated by cohesin197. In most metazoans, at metaphase sister chromatids remain associated through cohesion only at their centromeres. This is because centromeric cohesin, which embraces both sister chromatids, is protected from degradation until anaphase. In S. pombe, centromeric heterochromatin is required to mediate tight physical sister-centromere cohesion by trapping high levels of centromeric cohesin. This occurs through physical association of the cohesin complex with Swi6HP1 (REFS 198,199). In cells lacking heterochromatin, single kinetochores are disorganized and display aberrant attachment to spindles, and sister centromeres prematurely dissociate, leading to chromosome loss and gain200–202. This explains the elevated frequency of chromosome loss in S. pombe cells with defective heterochromatin8,202. Sister-centromere cohesion may also be weaker in human cells with reduced levels of centromeric H3K9me heterochromatin203.
Nine: controlling cell differentiation
Gene silencing by heterochromatin provides the capacity to control cell-type specification. An example in S. pombe is the silencing of two gene cassettes, mat2P and mat3M, that encode transcription factors that programme the cell mating type, either Plus (P) or Minus (M). The heterochromatin domain that silences these cassettes is called the mat2–mat3 region. Lineage-regulated recombination places copies of these transcription-factor-encoding genes into the expression site (mat1), thereby producing a switch in mating type, P to M or M to P204,205. In addition to the silencing of mat2 and mat3, H3K9me-heterochromatin has a role in regulating the directionality of this recombination and therefore the pattern of mating-type switching so that P-to-P and M-to-M are disfavoured205.
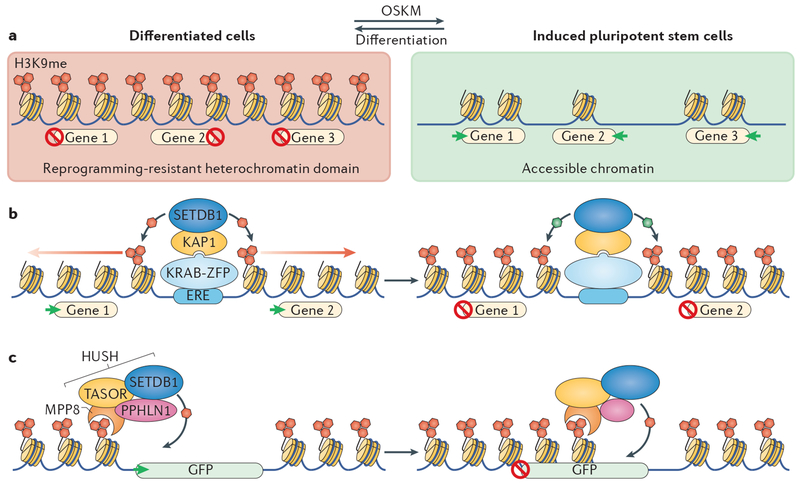
In mammals, megabase-sized islands of H3K9me-dependent heterochromatin are formed in a cell-type-specific manner206. One function of these islands is to form a barrier to transcription-factor-mediated cell-type reprogramming; hence, they are termed ‘differentially bound’ or ‘reprogramming-resistant regions’ (FIG. 5a). This type of heterochromatin is important for preserving the cell-type identity of differentiated cells, as depletion of proteins involved in maintenance of this heterochromatin — chromatin assembly factor 1 (CAF1), SET domain bifurcated 1 (SETDB1), KRAB-associated protein 1 (KAP1; also known as transcription intermediary factor 1β and TRIM28) — enables efficient reprogramming of differentiated cells to induced pluripotent stem cells206–210 or of somatic nuclei transferred into oocytes211. The determinants required to establish these large heterochromatin islands in cis remain unknown, but heterochromatin nucleation may be linked to mechanisms that silence endogenous retroelements (EREs), including endogenous retroviruses (ERVs), and neighbouring genes in somatic cells (FIG. 5b). A family of Krüppel-associated box zinc-finger (KRAB-ZFP) proteins recruits the H3K9 methyltransferase SETDB1 to EREs through the adaptor KAP1, where they elicit the formation of repressive heterochromatin212–215. Thus, ancient transposable elements appear to have been co-opted for the regulation of adjacent chromatin and genes.
Figure 5 |. Heterochromatin functions in mammalian cells.
a | The forced expression of four transcription factors (OCT4, SRY-box 2, Krüppel-like factor 4 and MYC, collectively known as OSKM) induces dedifferentiation of somatic cells into induced pluripotent stem cells. Such cell-type reprogramming is inefficient because large heterochromatin domains (depicted in the large red rectangle) present a barrier to the activation of key genes that are reguired for pluripotency. Reprogramming efficiency can be increased by depletion of proteins that are reguired for heterochromatin maintenance, thereby allowing activation (large green rectangle) of reprogramming pathways. b | In mammalian cells, histone H3 Lys 9 methylation (H3K9me)-dependent heterochromatin formation can be nucleated by transposable elements such as endogenous retroelements (EREs). EREs are bound by members of the large family of Krüppel-associated box zinc-finger proteins (KRAB-ZFPs), which recruit the H3K9me writer methyltransferase SET domain bifurcated 1 (SETDB1) through the adaptor protein KRAB-associated protein 1 (KAP1). This in turn allows the recruitment of H3K9me readers (such as heterochromatin protein 1) and writers to expand the heterochromatin domain. Heterochromatin spreading can silence adjacent genes, suggesting that remnants of transposable elements have been co-opted for defining and regulating heterochromatin domain formation. c | Retroviral GFP reporter constructs can be silenced by heterochromatin spreading mediated by the human silencing hub (HUSH) complex, which comprises the proteins M-phase phosphoprotein 8 (MPP8), periphilin 1 (PPHLN1), transgene activation suppressor protein (TASOR) and SETDB1. MPP8 binds flanking H3K9me and recruits SETDB1 through the adaptor protein TASOR. This silencing mechanism may be used to render pathogenic viruses latent. HUSH might also promote the formation of heterochromatin islands by mediating spreading from dispersed repeats, transposable elements or EREs.
Ten: medical relevance
Heterochromatin function is involved in various aspects of human health. We focus here on a handful of examples that illustrate and extend some of the principles introduced above.
Viral dormancy.
Heterochromatin protects genomes from pathogenic viruses. A fraction of genomic integrations of HIV-1 can occur in heterochromatin regions216. Retroviral reporters in lymphocyte cell lines are subject to silencing by H3K9me-dependent heterochromatin through the human silencing hub (HUSH) complex, which includes the proteins M-phase phosphoprotein 8 (MPP8), periphilin 1 (PPHLN1), TASOR (transgene activation suppressor protein; also known as FAM208A) and SETDB1. HUSH spreads across the viral genome from neighbouring heterochromatin217 (FIG. 5c). Although speculative, silencing of integrated HIV-1 viruses may allow dormant viral genomes to persist in T cells long after therapeutic clearance of circulating virus is achieved. Sporadic reactivation of these proviruses could allow the later reappearance of viruses. Interestingly, a distinct chromodomain protein, MPP8, and not HP1, binds HUSH-installed H3K9me3. Other human viruses may also be rendered dormant by HUSH-mediated heterochromatin spreading216. HUSH-directed silencing is distinct from that mediated by KRAB-ZFPs, which target heterochromatin formation to ERVs and EREs (see above)212–216,218.
Obesity.
The increasing frequency of obesity and its associated health risks in humans have a heritable component. Intriguingly, KAP1 haploinsufficiency in mice results in stochastic production of either normal or obese offspring from genetically identical parents. Analyses of human lean and obese cohorts indicate that KAP1 expression levels correlate with expression patterns of key obesity-associated genes and with body mass index219.
Premature ageing.
The progeroid (premature ageing) Werner syndrome is caused by mutations in the WRN gene, which encodes a helicase. WRN-null human mesenchymal stem cells display disrupted heterochromatin with loss of H3K9me3 from heterochromatin islands220. The WRN protein is targeted to centromeric repeats and associates with the H3K9 methyltransferase SUV39H1 and HP1α. This WRN complex may stabilize repeat arrays within heterochromatin, thereby preventing DNA damage. Comparison of primary human mesenchymal stem cells from young and old individuals revealed reduced levels of WRN protein and heterochromatin loss in the cells of old individuals. This implies that WRN protects heterochromatin and thereby prevents the irreversible genome instability associated with ageing. Alternatively, DNA damage associated with defective WRN might induce the loss of heterochromatin.
Metabolism.
DNA and histone methyltransferases and demethylases require metabolites for their function (reviewed in REFS 221,222). S-Adenosyl-methionine is the methyl donor used by nucleic acid and histone methyltransferases. Many demethylases require α-ketoglutarate, which is a metabolic intermediate of the Krebs cycle, as a co-substrate, whereas other demethylases utilize flavin adenine dinucleotide. Acetyl-CoA is the acetyl donor used by histone acetyltransferases, and the sirtuin family of histone deacetylases requires NAD+ as a cofactor. Consequently, nutritional changes or mutations that affect levels of metabolites can cause the accumulation of inhibitors of writers and erasers, which can alter chromatin.
Mutations in the genes encoding the Krebs cycle enzymes isocitrate dehydrogenase, fumarate hydratase and succinate dehydrogenase cause the accumulation of their substrates 2-hydroxyglutarate, fumarate and succinate, respectively. These metabolites are competitive inhibitors of α-ketoglutarate-dependent histone and DNA demethylases223,224. Consequently, such mutations promote tumorigenesis. Accumulation of 2-hydroxyglutarate results in elevated H3K9me levels and blocks cellular differentiation223; conversely, provision of α-ketoglutarate to embryonic stem cells reduces histone and DNA methylation and promotes pluripotency whereas succinate has the opposite effect. Histone methylation in embryonic stem cells is sensitive to glutamate and thus to fluctuations in α-ketoglutarate levels225. Poor nutrient availability is a feature of many solid tumours, the interiors of which are deprived of glutamine and hence of α-ketoglutarate, leading to elevated histone methylation and cellular dedifferentiation within such tumours226. In S. cerevisiae, equivalent mutations to those that cause 2-hydroxyglutarate accumulation were found to enhance SIR-mediated silencing by inhibiting H3K36 methyltransferases227.
Concluding remarks
We have discussed fundamental principles that have emerged from the study of heterochromatin in a broad range of organisms. Among many unanswered questions in the field, several stand out. What are the signals that initially trigger heterochromatin at specific sites? What determines the heritability or lack of herit ability of heterochromatin? What is the role of liquid-liquid phase separation in maintaining heterochromatin integrity? What enables transcription at heterochromatin regions? How is heterochromatin regulated during stress and development? Addressing these outstanding questions will require new model organisms and technologies as well as ingenious experimental strategies.
Acknowledgements
R.C.A. is a Wellcome Principal Research Fellow; his research is supported by the UK Wellcome Trust (200885) and core funding of the UK Wellcome Centre for Cell Biology (203149). Research in the laboratory of H.D.M. is supported by grants from the US National Institutes of Health. H.D.M. is a Chan-Zuckerberg BioHub investigator. The authors apologize to colleagues whose work could not be cited because of length restrictions. The authors dedicate this piece to the memory of A. Klar, whose pioneering studies of cell-type specification and gene silencing in Saccharomyces cerevisiae and Schizosaccharomyces pombe paved the way for many advances.
Glossary
- Post-translational modifications (PTMs)
Chemical groups (such as methyl or acetyl) on amino acid side chains that are enzymatically added by ‘writer’, removed by ‘eraser’ and recognized by ‘reader’ protein modules.
- Satellite repeats
Short repetitive sequences that exhibit a distinct satellite peak on buoyant density gradients ‘owing ho their skewed base composition.
- Constitutive heterochromatin
In most eukaryotes, heterochromatin that is consistently termed throughout the cell cycle and in many cell types, for example, centrome-reassociated heterochromatin.
- Facultative heterochromatin
Locus-specific and cell-type-specific heterochromatin, for example, the inactive X chromosome in mammals.
- Chromoshadow domain (CSD)
Dimerization domain in heterochromatin protein 1 -related proteins that forms a peptide-binding groove at the dimer interface that can recruit additional heterochromatin proteins.
- Argonaute
Proteins with PAZ and Piwi domains that are loaded with small RNAs, which target them and their associated proteins to long RNAs that bear homology to the small RNA.
- Pericentromeric heterochromatin
Large blocks of heterochromatin formed on the tandem repeats that surround the centromere–kinetochore region.
- X chromosome inactivation
Mechanism of dosage compensation in female mammals in which one of the two X chromosomes is inactivated by the formation of facultative heterochromatin.
- X-inactive specific transcript (XIST)
Long noncoding RNA that designates the X chromosome from which it is expressed for X chromosome inactivation.
- Piwi-associated RNAs (piRNAs)
Small RNAs associated with Piwi members of the Argonaute protein superfamily, which promotes repression of transposable elements in animal gonads.
- R-loops
Nascent RNA that remains associated with its DNA template through hybridization, thereby dislodging the opposite, nontemplate DNA strand.
- Heterochromatin islands
Extensive domains of heterochromatin on chromosome arms, which are distinct from the main centromeric and telomeric heterochromatin domains.
- Reprogramming-resistant regions
Large lineage-specific chromosomal regions that are assembled into heterochromatin and thus resist binding by reprogramming factors.
- Endogenous retroelements
Mobile elements that replicate through reverse transcription followed by genomic integration. The term also includes degenerate, immobile elements.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Zhou CY, Johnson S Gamarra NI & Narlikar GJ Mechanisms of ATP-dependent chromatin remodeling motots. Annu. Rev. Biophys 45, 153–181 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rea S et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Tschiersch B et al. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3–9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 13, 3822 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hissenberg JC et al. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 87, 9923–9927 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lachner M, O’Carroll D, Rea S, Mechtler K &Jenuwein T Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Bannister AJ et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Lorentz A, Ostermann K, Fleck O & Schmidt H Switching gene swi6, involved in repression of silent mating-type loci in fission yeast, encodes a homologue of chromatin-associated proteins from Drosophila and mammals. Gene 143, 139–143 (1994). [DOI] [PubMed] [Google Scholar]
- 8.Allshire RC, Nimmo HR, Ekwall K, Javerzat JP & Cranston G Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9, 218–233 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Thon G & Verhein-Hansen J Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics 155, 551–568 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klar AJ & Bonaduce MJ swi6, a gene required for mating-type switching, prohibits meiotic recombination in the mat2-mat3 “cold spot” of fission yeast. Genetics 129, 1033–1042 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lorentz A, Heim L & Schmidt H The switching gene swi6 affects recombination and gene expression in the mating-type region of Schizosaccharomyces pombe. Mol. Gen. Genet 233, 436–442 (1992). [DOI] [PubMed] [Google Scholar]
- 12.Ekwall K & Ruusala T Mutations in rik1, clr2, clr3 and clr4 genes asymmetrically derepress the silent mating-type loci in fission yeast. Genetics 136, 53–64 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thon G, Cohen A & Klar AJ Three additional linkage groups that repress transcription and meiotic recombination in the mating-type region of Schizosaccharomyces pombe. Genetics 138, 29–38 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamaru H & Selker EU A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414, 277–283 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Jackson JP, Lindroth AM, Cao X & Jacobsen SE Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416, 556–560 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Lehnertz B et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol 13, 1192–1200 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Hashimshony T, Zhang J, Keshet I, Bustin M & Cedar H The role of DNA methylation in setting up chromatin structure during development. Nat. Genet 34, 187–192 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Kueng S, Oppikofer M & Gasser SM SIR proteins and the assembly of silent chromatin in budding yeast. Ann. Rev. Genet 47, 275–306 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Rusche LN, Kirchmaier AL & Rine J The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Ann. Rev. Biochem 72, 481–516 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Armache KJ, Garlick JD, Canzio D, Narlikar GJ & Kingston RE Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 A resolution. Science 334, 977–982 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanson SJ & Wolfe KH An evolutionary perspective on yeast mating-type switching. Genetics 206, 9–32 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ekwall K et al. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J. Cell Sci 109, 2637–2648 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Maison C et al. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet 30, 329–334 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Sadaie M, Iida T, Urano T & Nakayama J A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J. 23, 3825–3835 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brasher SV et al. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J 19, 1587–1597 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cowieson NP, Partridge JF, Allshire RC & McLaughlin PJ Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr. Biol 10, 517–525 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Motamedi MR et al. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol. Cell 32, 778–790 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischer T et al. Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc. Natl Acad. Sci. USA 106, 8998–9003 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia JR, Dumesic PA, Hartley PD, El-Samad H & Madhani HD Combinatorial, site-specific requirement for heterochromatic silencing factors in the elimination of nucleosome-free regions. Genes Dev. 24, 1758–1771 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Creamer KM et al. The Mi-2 homolog Mit1 actively positions nucleosomes within heterochromatin to suppress transcription. Mol. Cell. Biol 34, 2046–2061 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Canzio D et al. A conformational switch in HP1 releases auto-inhibition to drive heterochromatin assembly. Nature 496, 377–381 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao R et al. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science 298, 1039–1043 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Muller J et al. Histone methyltransferase activity of a Drosophila polycomb group repressor complex. Cell 111, 197–208 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Czermin B et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111, 185–196 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P & Reinberg D Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 16, 2893–2905 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fischle W et al. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev 17, 1870–1881 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Margueron R et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–767 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Volpe TA et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Kloc A, Zaratiegui M, Nora E & Martienssen R RNA interference guides histone modification during the S phase of chromosomal replication. Curr. Biol 18, 490–495 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen ES et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 451, 734–737 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Lu J & Gilbert DM Proliferation-dependent and cell cycle regulated transcription of mouse pericentric heterochromatin. J. Cell Biol 179, 411–421 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reinhart B & Bartel DP Small RNAs correspond to centromere heterochromatic repeats. Science 13, 1831 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Motamedi MR et al. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119, 789–802 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Bayne EH et al. Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell 140, 666–677 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang K, Mosch K, Fischle W & Grewal SI Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat. Struct. Mol. Biol 15, 381–388 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Verdel A et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303, 672–676 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buhler M, Verdel A & Moazed D Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell 125, 873–886 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Gerace EL, Halic M & Moazed D The methyltransferase activity of Clr4Suv39h triggers RNAi independently of histone H3K9 methylation. Mol. Cell 39, 360–372 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jain R, Iglesias N & Moazed D Distinct functions of argonaute slicer in siRNA maturation and heterochromatin formation. Mol. Cell 63, 191–205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Colmenares SU, Buker SM, Buhler M, Dlakic M & Moazed D Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol. Cell 27, 449–461 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Kato H et al. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science 309, 467–469 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Djupedal I et al. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev 19, 2301–2306 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hong EJ, Villen J, Gerace EL, Gygi SP & Moazed D A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol. 2, 106–111 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Horn PJ, Bastie JN & Peterson CLA Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev 19, 1705–1714 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jia S, Kobayashi R & Grewal SI Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat. Cell Biol 7, 1007–1013 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Sugiyama T, Cam H, Verdel A, Moazed D & Grewal SI RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc. Natl Acad. Sci. USA 102, 152–157 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Partridge JF, Borgstrøm B & Allshire R Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev 14, 783–791 (2000). [PMC free article] [PubMed] [Google Scholar]
- 58.Cam HP et al. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet 37, 809–819 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Rougemaille M et al. Ers1 links HP1 to RNAi. Proc. Natl Acad. Sci. USA 109, 11258–11263 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayashi A et al. Heterochromatin protein 1 homologue Swi6 acts in concert with Ers1 to regulate RNAi-directed heterochromatin assembly. Proc. Natl Acad. Sci. USA 109, 6159–6164 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rougemaille M, Shankar S, Braun S, Rowley M & Madhani HD Ers1, a rapidly diverging protein essential for RNA interference-dependent heterochromatic silencing in Schizosaccharomyces pombe. J. Biol. Chem 283, 25770–25773 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wierzbicki AT, Ream TS, Haag JR & Pikaard CS RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet 41, 630–634 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhong X et al. Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell 157, 1050–1060 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu ZW et al. The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci. PLoS Genet. 10, e1003948 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Herr AJ, Jensen MB, Dalmay T & Baulcombe DC RNA polymerase IV directs silencing of endogenous DNA. Science 308, 118–120 (2005). [DOI] [PubMed] [Google Scholar]
- 66.Blevins T et al. Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. eLife 4, e09591 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhai J et al. A one precursor one siRNA model for Pol IV-dependent siRNA biogenesis. Cell 163, 445–455 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang H et al. DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proc. Natl Acad. Sci. USA 110, 8290–8295 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; References 67 and 68 show that short double-stranded RNA precursors generated by Pol IV and RNA-dependent polymerase 2 produce a single siRNA duplex through the activity of the Dicer Dcl3.
- 69.Law JA .et al. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 498, 385–389 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reyes-Turcu FE, Zhang K, Zofall M, Chen E & Grewal SI Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nat. Struct. Mol. Biol 18, 1132–1138 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wittmann S et al. The conserved protein Seb1 drives transcription termination by binding RNA polymerase II and nascent RNA. Nat. Commun 8, 14861 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lemay JF et al. The Nrd1 -like protein Seb1 coordinates cotranscriptional 3’ end processing and polyadenylation site selection. Genes Dev. 30, 1558–1572 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sugiyama T et al. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 128, 491–504 (2007). [DOI] [PubMed] [Google Scholar]
- 74.Marina DB, Shankar S, Natarajan P, Finn KJ & Madhani HD A conserved ncRNA-binding protein recruits silencing factors to heterochromatin through an RNAi-independent mechanism. Genes Dev. 27, 1851–1856(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article shows that the conserved RNA-binding protein Seb1, an orthologue of S. cerevisiae Nrd1, is an essential mediator of RNAi-independent heterochromatin assembly at fission yeast pericentromeric repeats.
- 75.Shirai A et al. Impact of nucleic acid and methylated H3K9 binding activities of Suv39h1 on its heterochromatin assembly. eLife 6, e25317 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Johnson WL et al. RNA-dependent stabilization of SUV39H1 at constitutive heterochromatin. eLife 6, e25299 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Velazquez Camacho O et al. Major satellite repeat RNA stabilize heterochromatin retention of Suv39h enzymes by RNA-nucleosome association and RNA: DNA hybrid formation. eLife 6, e25293 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; References 75–77 show that the nucleic acid binding activities of the chromodomain of SUV39 enzymes stabilize their association with heterochromatin.
- 78.Chu C et al. Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Minajigi A et al. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 349, aab2276 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Monfort A et al. Identification of Spen as a crucial factor for Xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep. 12, 554–561 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moindrot B et al. A Pooled shRNA screen identifies Rbm 15, Spen, and Wtap as factors required for Xist RNA-mediated silencing. Cell Rep. 12, 562–572 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McHugh CA et al. The Xist IncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; References 80–82 identify the RNA binding protein SPEN (SHARP) as crucial for the establishment of X inactivation in mice.
- 83.Mira-Bontenbal H & Gribnau J New Xist-interacting proteins in X-chromosome inactivation. Curr. Biol 26, R338–R342 (2016). [DOI] [PubMed] [Google Scholar]
- 84.Folco HD, Pidoux AL, Urano T & Allshire RC Heterochromatin and RNAi are required to establish CHNP-A chromatin at centromeres. Science 319, 94–97 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Buscaino A et al. Distinct roles for Sir2 and RNAi in centromeric heterochromatin nucleation, spreading and maintenance. EMBO J 32, 1250–1264 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ekwall K, Olsson T, Turner BM, Cranston G & Allshire RC Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell 91, 1021–1032 (1997). [DOI] [PubMed] [Google Scholar]
- 87.Hall IM et al. Establishment and maintenance of a heterochromatin domain. Science 297, 2232–2237 (2002). [DOI] [PubMed] [Google Scholar]
- 88.Djupedal I et al. Analysis of small RNA in fission yeast; centromeric siRNAs are potentially generated through a structured RNA. EMBO J. 28, 3832–3844 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Halic M & Moazed D Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell 140, 504–516(2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Simmer F et al. Hairpin RNA induces secondary small interfering RNA synthesis and silencing in trans in fission yeast. EMBO Rep. 11, 112–118 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Iida T, Nakayama J & Moazed D siRNA-mediated heterochromatin establishment requires HP1 and is associated with antisense transcription. Mol. Cell 31, 178–189 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sadeghi L,Prasad P, Ekwall K, Cohen A & Svensson JP The Paf1 complex factors Leo1 and Paf1 promote local histone turnover to modulate chromatin states in fission yeast. EMBO Rep. 16, 1673–1687 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kowalik KM et al. The Paf1 complex represses small-RNA-mediated epigenetic gene silencing. Nature 520, 248–252 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Verrier L et al. Global regulation of heterochromatin spreading by Leo1. Open Biol. 5, 150045 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; References 92–94 show that the Paf1 complex, which is a Pol II elongation factor, inhibits heterochromatin assembly.
- 95.Yu R, Jih G, Iglesias N & Moazed D Determinants of heterochromatic siRNA biogenesis and function. Mol. Cell 53, 262–276 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang J et al. The proper connection between shelterin components is required for telomeric heterochromatin assembly. Genes Dev 30, 827–839 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kanoh J, Sadaie M, Urano T & Ishikawa F Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr. Biol 15, 1808–1819 (2005). [DOI] [PubMed] [Google Scholar]
- 98.Hansen KR, Ibarra PT & Thon G Evolutionary-conserved telomere-linked helicase genes of fission yeast are repressed by silencing factors, RNAi components and the telomere-binding protein Taz1. Nucleic Acids Res. 34, 78–88 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Weick EM & Miska EA piRNAs: from biogenesis to function. Development 141, 3458–3471 (2014). [DOI] [PubMed] [Google Scholar]
- 100.Das PP et al. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol. Cell 31, 79–90 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gu T & Elgin SC Maternal depletion of Piwi, a component of the RNAi system, impacts heterochromatin formation in Drosophila. PLoS Genet 9, e1003780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Blevins T et al. A two-step process for epigenetic inheritance in Arabidopsis. Mol. Cell 54, 30–42 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wutz A & Jaenisch R A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol. Cell 5, 695–705 (2000). [DOI] [PubMed] [Google Scholar]
- 104.Csankovszki G, Nagy A & Jaenisch R Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J. Cell Biol 153, 773–784 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Elgin SC & Reuter G Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb. Perspect. Biol 5, a017780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Spofford JB Parental control of position-effect variegation: I. Parental heterochromatin and expression of the white locus in compound-X Drosophila melanogaster. Proc. Natl Acad. Sci. USA 45, 1003–1007 (1959). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tartof KD, Hobbs C & Jones M A structural basis for variegating position effects. Cell 37, 869–878 (1984). [DOI] [PubMed] [Google Scholar]
- 108.Talbert PB & Henikoff S A reexamination of spreading of position-effect variegation in the white-roughest region of Drosophila melanogaster. Genetics 154, 259–272 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hecht A, Strahl-Bolsinger S & Grunstein M Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 383, 92–96 (1996). [DOI] [PubMed] [Google Scholar]
- 110.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM & Grunstein M Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80, 583–592 (1995). [DOI] [PubMed] [Google Scholar]
- 111.Renauld H et al. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 7, 1133–1145 (1993). [DOI] [PubMed] [Google Scholar]
- 112.Al-Sady B, Madhani HD & Narlikar GJ Division of labor between the chromodomains of HP1 and Suv39 methylase enables coordination of heterochromatin spread. Mol. Cell 51, 80–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yamada T, Fischle W, Sugiyama T, Allis CD & Grewal SI The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol. Cell 20, 173–185 (2005). [DOI] [PubMed] [Google Scholar]
- 114.Obersriebnig MJ, Pallesen EM, Sneppen K, Trusina A & Thon G Nucleation and spreading of a heterochromatic domain in fission yeast. Nat. Commun 7, 11518 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Erdel F & Greene EC Generalized nucleation and looping model for epigenetic memory of histone modifications. Proc. Natl Acad. Sci. USA 113, E4180–E4189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Larson AG et al. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature SAT, 236–240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; The paper demonstrates that human HP1α can undergo phase separation in vitro upon phosphorylation and that this correlates with its ability to form foci in vivo.
- 117.Strom AR et al. Phase separation drives heterochromatin domain formation. Nature 547, 241–245 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; The article shows that D. melanogaster HP1 a can undergo phase separation in vitro and that heterochromatic foci in fly and human cells have the properties of liquid droplets.
- 118.Simon MD et al. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature 504, 465–469 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; High-resolution RNA mapping identifies the phases of XIST assembly during X inactivation, revealing initial binding to gene-rich regions before appearance in gene-poor regions.
- 119.Engreitz JM et al. The Xist IncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341, 1237973 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; The article examines X inactivation in the context of long-range chromatin interactions and concludes that XIST first spreads from the X chromosome inactivation centre to regions in close 3D proximity to the initiating locus.
- 120.White WM, Willard HF, Van Dyke DL & Wolff DJ The spreading of X inactivation into autosomal material of an x;autosome translocation: evidence for a difference between autosomal and X-chromosomal DNA. Am. J. Hum. Genet 63, 20–28 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Popova BC, Tada T, Takagi N, Brockdorff N & Nesterova TB Attenuated spread of X-inactivation in an X;autosome translocation. Proc. Natl Acad. Sci. USA 103, 7706–7711 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bala Tannan N et al. DNA methylation profiling in X;autosome translocations supports a role for L1 repeats in the spread of X chromosome inactivation. Hum. Mol. Genet 23, 1224–1236(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wutz A, Rasmussen TP & Jaenisch R Chromosomal silencing & localization are mediated by different domains of Xist RNA. Nat. Genet 30, 167–174 (2002). [DOI] [PubMed] [Google Scholar]
- 124.Lee JT & Jaenisch R Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature 386, 275–279 (1997). [DOI] [PubMed] [Google Scholar]
- 125.Herzing LB, Romer JT, Horn JM & Ashworth A Xist has properties of the X-chromosome inactivation centre. Nature 386, 272–275 (1997). [DOI] [PubMed] [Google Scholar]
- 126.da Rocha ST & Heard E Novel players in X inactivation: insights into Xist-mediated gene silencing and chromosome conformation. Nat. Struct. Mol. Biol 24, 197–204 (2017). [DOI] [PubMed] [Google Scholar]
- 127.Jiang J et al. Translating dosage compensation to trisomy 21. Nature 500, 296–300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Raab JR, et al. Human tRNA genes function as chromatin insulators. EMBO J 31, 330–350 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Donze D, Adams CR, Rine J & Kamakaka RT The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 13, 698–708 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Scott KC, Merrett SL & Willard HF A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr. Biol 16, 119–129 (2006). [DOI] [PubMed] [Google Scholar]
- 131.Noma K, Cam HP, Maraia RJ & Grewal SI A role for TFIIIC transcription factor complex in genome organization. Cell 125, 859–872 (2006). [DOI] [PubMed] [Google Scholar]
- 132.Coveney J & Woodland HR The DNase I sensitivity of Xenopus laevis genes transcribed by RNA polymerase III. Nature 298, 578–580 (1982). [DOI] [PubMed] [Google Scholar]
- 133.DeLotto R & Schedl P Internal promoter elements of transfer RNA genes are preferentially exposed in chromatin. J. Mol. Biol 179, 607–628 (1984). [DOI] [PubMed] [Google Scholar]
- 134.Takahashi K et al. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol. Biol. Cell 3, 819–835 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Aygun O, Mehta S & Grewal SI HDAC-mediated suppression of histone turnover promotes epigenetic stability of heterochromatin. Nat. Struct. Mol. Biol 20, 547–554 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hartley PD & Madhani HD Mechanisms that specify promoter nucleosome location and identity. Cell 137, 445–458 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Raisner RM et al. Histone variant H2A.Z marks the 5’ ends of both active and inactive genes in euchromatin. Cell 123, 233–248 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Meneghini MD, Wu M & Madhani HD Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112, 725–736 (2003). [DOI] [PubMed] [Google Scholar]
- 139.Venkatasubrahmanyam S, Hwang WW, Meneghini MD, Tong AH & Madhani HD Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Setl and H2A.Z. Proc. Natl Acad. Sci. USA 104, 16609–16614 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tompa R & Madhani HD Histone H3 lysine 36 methylation antagonizes silencing in Saccharomyces cerevisiae independently of the Rpd3S histone deacetylase complex. Genetics 175, 585–593 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Santos-Rosa H, Bannister AJ, Dehe PM, Geli V & Kouzarides I Methylation of H3 lysine 4 at euchromatin promotes Sir3p association with heterochromatin. J. Biol. Chem 279, 47506–47512 (2004). [DOI] [PubMed] [Google Scholar]
- 142.van Leeuwen R, Gafken PR & Gottschling DH Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109, 745–756 (2002). [DOI] [PubMed] [Google Scholar]
- 143.Verzijlbergen KR, Faber AW, Stulemeijer IJ & van Leeuwen F Multiple histone modifications in euchromatin promote heterochromatin formation by redundant mechanisms in Saccharomyces cerevisiae. BMC Mol. Biol 10, 76 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li X et al. Chromatin boundaries require functional collaboration between the hSET1 and NURF complexes. Blood 118, 1386–1394 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ayoub N et al. A novel jmjC domain protein modulates heterochromatization in fission yeast. Mol. Cell. Biol 23, 4356–4370 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Trewick SC, McLaughlin PJ & Allshire RC Methylation: lost in hydroxylation? EMBO Rep. 6, 315–320 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zofall M & Grewal SI Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol. Cell 22, 681–692 (2006). [DOI] [PubMed] [Google Scholar]
- 148.Trewick SC, Minc E, Antonelli R, Urano T & Allshire RC The JmjC domain protein Epe1 prevents unregulated assembly and disassembly of heterochromatin. EMBO J. 26, 4670–4682 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Braun S et al. The Cul4-DdbCdt2 ubiquitin ligase inhibits invasion of a boundary-associated antisilencing factor into heterochromatin. Cell 144, 41–54 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Garcia JP, Al-Sady B & Madhani HD Intrinsic toxicity of unchecked heterochromatin spread is suppressed by redundant chromatin boundary functions in Schizosacchromyces pombe. G3 5, 1453–1461 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang J, Reddy BD & Jia S Rapid epigenetic adaptation to uncontrolled heterochromatin spreading, eLife 4, e06179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; Increased H3K9me-dependent heterochromatin formation in fission yeast cells lacking factors that counteract its assembly can be suppressed by spontaneous heterochromatin-mediated silencing of genes encoding components of the CIr4 histone H3K9 methyltransferase complex.
- 152.Lee NN et al. Mtr4-like protein coordinates nuclear RNA processing for heterochromatin assembly and for telomere maintenance. Cell 155, 1061–1074 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zofall M et al. RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science 335, 96–100 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yamanaka S et al. RNAi triggered by specialized machinery silences developmental genes and retrotransposons. Nature 493, 557–560 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Joh RI et al. Survival in quiescence requires the euchromatic deployment of Clr4/SUV39H by Argonaute-associated small RNAs. Mol. Cell 64, 1088–1101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; References 152–155 report the presence of regulated heterochromatin islands outside of the main heterochromatin domains at centromeres, telomeres and the mating-type loci in fission yeast.
- 156.Dumesic PA et al. Product binding enforces the genomic specificity of a yeast polycomb repressive complex. Cell 160, 204–218 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; Reports the first yeast Polycomb system in C. neoformans and reveals that tethering of its PRC2-Iike complex to sites of previous action at subtelomeric regions prevents it from erroneously modifying H3K9me-marked centromeric heterochromatin.
- 157.Alabert C & Groth A Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell. Biol 13, 153–167 (2012). [DOI] [PubMed] [Google Scholar]
- 158.Jones PA & Liang G Rethinking how DNA methylation patterns are maintained. Nat. Rev. Genet 10, 805–811 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Freitag M, Hickey PC, Khlafallah TK, Read ND & Selker EU HP1 is essential for DNA methylation in Neurospora. Mol. Cell 13, 427–434 (2004). [DOI] [PubMed] [Google Scholar]
- 160.Du J, Johnson LM, Jacobsen SE & Patel DJ DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell. Biol 16, 519–532 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang X & Moazed D DNA sequence-dependent epigenetic inheritance of gene silencing and histone H3K9 methylation. Science 356, 88–91 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kagansky A et al. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science 324, 1716–1719 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Audergon PN et al. Epigenetics. Restricted epigenetic inheritance of H3K9 methylation. Science 348, 132–135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ragunathan K, Jih G & Moazed D Epigenetic inheritance uncoupled from sequence-specific recruitment. Science 348, 1258699 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; References 163 and 164 demonstrate that transient tethering of the CIr4 H3K9 methyltransferase to a genomic site in fission yeast results in the formation of heritable heterochromatin and gene silencing, provided that a histone demethylase, Epe1, is removed from cells.
- 165.Bintu L et al. Dynamics of epigenetic regulation at the single-cell level. Science 351, 720–724 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Amabile A et al. Inheritable silencing of endogenous genes by hit-and-run targeted epigenetic editing. Cell 167, 219–232.e14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hathaway NA et al. Dynamics and memory of heterochromatin in living cells. Cell 149, 1447–1460 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Berry S, Hartley M, Olsson TS, Dean C & Howard M Local chromatin environment of a Polycomb target gene instructs its own epigenetic inheritance, eLife 4, e07205 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Laprell P, Finkl K & Muller J Propagation of Polycomb-repressed chromatin requires sequence-specific recruitment to DNA. Science 356, 85–88 (2017). [DOI] [PubMed] [Google Scholar]
- 170.Coleman RT & Struhl G Causal role for inheritance of H3K27me3 in maintaining the OFF state of a Drosophila HOX gene. Science 10.1126/science.aai8236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; References 169 and 170 show that binding sites for DNA-binding proteins are required for the heritability of H 3K27me-marked silent domains in D. melanogaster.
- 171.Grewal SI & Klar AJ Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis. Cell 86, 95–101 (1996). [DOI] [PubMed] [Google Scholar]
- 172.Jia S, Noma K & Grewal SI RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304, 1971–1976 (2004). [DOI] [PubMed] [Google Scholar]
- 173.Taneja N et al. SNF2 family protein Fft3 suppresses nucleosome turnover to promote epigenetic inheritance and proper replication. Mol. Cell 66, 50–62.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Steglich B et al. The Fun30 chromatin remodeler Fft3 controls nuclear organization and chromatin structure of insulators and subtelomeres in fission yeast. PLoS Genet. 11, el005101 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Stralfors A, Walfridsson J, Bhuiyan H & Ekwall K The FUN30 chromatin remodeler, Fft3, protects centromeric and subtelomeric domains from euchromatin formation. PLOS Genet. 7, el001334 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mari-Ordonez A et al. Reconstructing de novo silencing of an active plant retrotransposon. Nat. Genet 45, 1029–1039 (2013). [DOI] [PubMed] [Google Scholar]
- 177.Lanciano S et al. Sequencing the extrachromosomal circular mobilome reveals retrotransposon activity in plants. PLOS Genet. 13, e1006630 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hickey DA Selfish DNA: a sexually-transmitted nuclear parasite. Genetics 101, 519–531 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zeller P et al. Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nat. Genet 48, 1385–1395 (2016). [DOI] [PubMed] [Google Scholar]; This article reveals that C. elegans mutants lacking H3K9 methylation have increased levels of R-Ioops and consequent genetic instability.
- 180.Klattenhoff C et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138, 1137–1149 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sienski G, Donertas D & Brennecke J Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151, 964–980 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Brennecke J et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103 (2007). [DOI] [PubMed] [Google Scholar]
- 183.Aravin AA et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31, 785–799 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kojima-Kita K et al. MIWI2 as an effector of DNA methylation and gene silencing in embryonic male germ cells. Cell Rep. 16, 2819–2828 (2016). [DOI] [PubMed] [Google Scholar]
- 185.Kuramochi-Miyagawa S et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 22, 908–917 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Peng JC & Karpen GH H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat. Cell Biol 9, 25–35 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Chiolo I et al. Double-strand breaks in heterochromatin move outside of a dynamic HP1 a domain to complete recombinational repair. Cell 144, 732–744 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Jakob B et al. DNA double-strand breaks in heterochromatin elicit fast repair protein recruitment, histone H2AX phosphorylation and relocation to euchromatin. Nucleic Acids Res. 39, 6489–6499 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ryu T et al. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat. Cell Biol 17, 1401–1411 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Tsouroula K et al. Temporal and spatial uncoupling of DNA double strand break repair pathways within mammalian heterochromatin. Mol. Cell 63, 293–305 (2016). [DOI] [PubMed] [Google Scholar]
- 191.Janssen A et al. A single double-strand break system reveals repair dynamics and mechanisms in heterochromatin and euchromatin. Genes Dev. 30, 1645–1657 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.McKinley KL & Cheeseman IM The molecular basis for centromere identity and function. Nat. Rev Mol. Cell. Biol 17, 16–29 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sullivan LL, Maloney KA, Towers AJ, Gregory SG & Sullivan BA Human centromere repositioning within euchromatin after partial chromosome deletion. Chromosome Res. 24, 451–466 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sato H, Masuda P, Takayama Y, Takahashi K & Saitoh S Epigenetic inactivation and subsequent heterochromatinization of a centromere stabilize dicentric chromosomes. Cun: Biol 22, 658–667 (2012). [DOI] [PubMed] [Google Scholar]
- 195.Nakano M et al. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev. Cell 14, 507–522 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Cardinale S et al. Hierarchical inactivation of a synthetic human kinetochore by a chromatin modifier. Mol. Biol. Cell 20, 4194–4204 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Uhlmann P SMC complexes: from DNA to chromosomes. Nat. Rev. Mol. Cell. Biol 17, 399–412 (2016). [DOI] [PubMed] [Google Scholar]
- 198.Bernard P et al. Requirement of heterochromatin for cohesion at centromeres. Science 294, 2539–2542 (2001). [DOI] [PubMed] [Google Scholar]
- 199.Nonaka N et al. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol 4, 89–93 (2002). [DOI] [PubMed] [Google Scholar]
- 200.Pidoux AL, Uzawa S, Perry PE, Cande WZ & Allshire RC Live analysis of lagging chromosomes during anaphase and their effect on spindle elongation rate in fission yeast. J. Cell Sci 113, 4177–4191 (2000). [DOI] [PubMed] [Google Scholar]
- 201.Gregan J et al. The kinetochore proteins Pcsl and Mde4 and heterochromatin are required to prevent merotelic orientation. Curr. Biol 17, 1190–1200 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ekwall K et al. The chromodomain protein Swi6: a key component at fission yeast centromeres. Science 269, 1429–1431 (1995). [DOI] [PubMed] [Google Scholar]
- 203.Tanno Y et al. The inner centromere-shugoshin network prevents chromosomal instability. Science 349, 1237–1240 (2015). [DOI] [PubMed] [Google Scholar]
- 204.Klar AJ, Ishikawa K & Moore SA Unique DNA recombination mechanism of the mating/cell-type switching of fission yeasts: a review. Microbiol. Spectr 2, 10.1128/microbiolspec.MDNA3-0003-2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Jia S, Yamada T & Grewal SI Heterochromatin regulates cell type-specific long-range chromatin interactions essential for directed recombination. Cell 119, 469–480 (2004). [DOI] [PubMed] [Google Scholar]
- 206.Soufi A, Donahue G & Zaret KS Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151, 994–1004 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows that megabase-sized heterochromatin domains in somatic cells impede the binding of pluripotency transcription factors to their targets.
- 207.Becker JS, Nicetto D & Zaret KS H3K9me3-dependent heterochromatin: barrier to cell fate changes. Trends Genet. 32, 29–41 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Chen J et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat. Genet 45, 34–42 (2013). [DOI] [PubMed] [Google Scholar]
- 209.Sridharan R et al. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1 γ in reprogramming to pluripotency. Nat. Cell Biol 15, 872–882 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Cheloufi S et al. The histone chaperone CAF-1 safeguards somatic cell identity. Nature 528, 218–224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Matoba S et al. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell 159, 884–895 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Castro-Diaz N et al. Evolutionally dynamic LI regulation in embryonic stem cells. Genes Dev. 28, 1397–1409 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Jacobs FMJ et al. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 516, 242–245 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wolf D & Goff SP Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458, 1201–1204 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Imbeault M, Helleboid P-Y & Trono D KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature 543, 550–554 (2017). [DOI] [PubMed] [Google Scholar]; This paper shows that transposable elements in tetrapods are recognized and silenced by KRAB-ZFP proteins that coevolve with the elements in an ongoing arms race.
- 216.Timms RL, Tchasovnikarova IA & Lehner PJ Position-effect variegation revisited: HUSHing up heterochromatin in human cells. Bioessays 38, 333–343 (2016). [DOI] [PubMed] [Google Scholar]
- 217.Tchasovnikarova IA et al. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science 348, 1481–1485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; A genetic screen in haploid cells for factors that silence an integrated retroviral reporter identifies a protein complex containing the H3K9 methyltransferase SETDB1 and the H32K9me-binding MPP8 chromodomain protein.
- 218.Wolf G et al. The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes Dev. 29, 538–554 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Dalgaard K et al. Trim28 haploinsufficiency triggers bi-stable epigenetic obesity. Cell 164, 353–364 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; Haploinsufficiency of a component of the KRAB-KAP1 H3K9me-recruiting complex is shown to result in epigenetic variability in obesity in mice.
- 220.Zhang W et al. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science 348, 1160–1163 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Janke R, Dodson AE & Rine J Metabolism and epigenetics. Annu. Rev. Cell Dev Biol 31, 473–496 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Sharma U & Rando OJ Metabolic inputs into the epigenome. Cell Metab. 25, 544–558 (2017). [DOI] [PubMed] [Google Scholar]
- 223.Lu C et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483, 474–478 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Xiao M et al. Inhibition of a-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev 26, 1326–1338 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Carey BW, Finley LWS, Cross JF, Allis CD & Thompson CB Intracellular a-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518, 413–416 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Pan M et al. Regional glutamine deficiency in tumours promotes dedifferentiation through inhibition of histone demethylation. Nat. Cell Biol 18, 1090–1101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; References 223–226 show how imbalances in metabolite levels generated by the tricarboxylic acid cycle because of mutation, oversupply or undersupply of nutrients result in altered cell fate and tumorigenesis.
- 227.Janke R, Iavarone AT & Rine J Oncometabolite D-2-hydroxyglutarate enhances gene silencing through inhibition of specific H3K36 histone demethylases. eLife 6, e22451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Passarge E Emil Heitz and the concept of heterochromatin: longitudinal chromosome differentiation was recognized fifty yearn ago. Am. J. Hum. Genet 31, 106 (1979). [PMC free article] [PubMed] [Google Scholar]
- 229.Muller HJ Types of visible variations induced by X-rays in Drosophila. J. Genet. 22, 299–334 (1930). [Google Scholar]
- 230.Schultz J Variegation in Drosophila and the inert chromosome regions. Proc. Natl Acad. Sci. USA 22, 27–33 (1936). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Gowen J & Gay E Chromosome constitution and behavior in eversporting and mottling in Drosophila melanogaster. Genetics 19, 189 (1934). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Dimitri P & Pisano C Position effect variegation in Drosophila melanogaster. relationship between suppression effect and the amount of Y chromosome. Genetics 122, 793–800 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Spradling AC & Karpen GH Sixty years of mystery. Genetics 126, 779–784 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Henikoff S Position-effect variegation after 60 years. Trends Genet. 6, 422–426 (1990). [DOI] [PubMed] [Google Scholar]
- 235.Reute G & Spierer P Position effect variegation and chromatin proteins. BioEssays 14, 605–612 (1992). [DOI] [PubMed] [Google Scholar]
- 236.Britten RJ & Kohne DE Repeated Sequences in DNA. Science 161, 529–540 (1968). [DOI] [PubMed] [Google Scholar]
- 237.Kit S Equilibrium sedimentation in density gradients of DNA preparations from animal tissues. J. Mol. Biol 3, 711–716(1961). [DOI] [PubMed] [Google Scholar]
- 238.Yasmineh WG & Yunis JJ Localization of mouse satellite DNA in constitutive heterochromatin. Exp. Cell Res 59, 69–75 (1970). [DOI] [PubMed] [Google Scholar]
- 239.Flamm WG, Bond HE, Burr HE & Bond SB Satellite DNA isolated from mouse liver; some physical and metabolic properties. Biochim. Biophys. Acta 123, 652–654 (1966). [DOI] [PubMed] [Google Scholar]
- 240.Flamm WG, McCallum M & Walker PM The isolation of complementary strands from a mouse DNA fraction. Proc. Natl Acad. Sci. USA 57, 1729–1734 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Filipski J, Thiery J-P & Bernard! G An analysis of the bovine genome by Cs2SO4—Ag+ density gradient centrifugation. J. Mol. Biol 80, 177–197 (1973). [DOI] [PubMed] [Google Scholar]
- 242.Southern EM Base sequence and evolution of guinea-pig α-satellite DNA. Nature 227, 794–798 (1970). [DOI] [PubMed] [Google Scholar]
- 243.Fry K et al. Nucleotide sequence of HS-β satellite DNA from kangaroo rat Dipodomys ordii. Proc. Natl Acad. Sci. USA 70, 2642 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Jones KW Chromosomal and nuclear location of mouse satellite DNA in individual cells. Nature 225, 912–915 (1970). [DOI] [PubMed] [Google Scholar]
- 245.Pardue ML & Gall JG Chromosomal localization of mouse satellite DNA. Science 168, 1356–1358 (1970). [DOI] [PubMed] [Google Scholar]
- 246.Rae PMM & Franke WW The interphase distribution of satellite DNA-containing heterochromatin in mouse nuclei. Chromosoma 39, 443–456(1972). [DOI] [PubMed] [Google Scholar]
- 247.Flamm WG, Walker PM & McCallum M Some properties of the single strands isolated from the DNA of the nuclear satellite of the mouse (Mus musculus). J. Mol. Biol 40, 423–443 (1969). [DOI] [PubMed] [Google Scholar]
- 248.Yunis JJ & Yasmineh WG Satellite DNA in constitutive heterochromatin of the guinea pig. Science 168, 263–265 (1970). [DOI] [PubMed] [Google Scholar]
- 249.Lima-de-Faria A & Jaworska H Late DNA Synthesis in heterochromatin. Nature 217, 138–142 (1968). [DOI] [PubMed] [Google Scholar]
- 250.Gall J, Cohen E & Polan M Repetitive DNA sequences in Drosophila. Chromosoma 33, 319–344 (1971). [DOI] [PubMed] [Google Scholar]
- 251.Deans C & Maggert KA What do you mean, “epigenetic”? Genetics 199, 887–896 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Bird A Perceptions of epigenetics. Nature 447, 396–398(2007). [DOI] [PubMed] [Google Scholar]
- 253.Castel SE & Martienssen RA RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev Mol. Cell. Biol 14, 100–112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]