Abstract
It has been 20 years since West Nile virus first emerged in the Americas, and since then, little progress has been made to control outbreaks caused by this virus. After its first detection in New York in 1999, West Nile virus quickly spread across the continent, causing an epidemic of human disease and massive bird die-offs. Now the virus has become endemic to the United States, where an estimated 7 million human infections have occurred, making it the leading mosquito-borne virus infection and the most common cause of viral encephalitis in the country. To bring new attention to one of the most important mosquito-borne viruses in the Americas, we provide an interactive review using Nextstrain: a visualization tool for real-time tracking of pathogen evolution (nextstrain.org/WNV/NA). Nextstrain utilizes a growing database of more than 2,000 West Nile virus genomes and harnesses the power of phylogenetics for students, educators, public health workers, and researchers to visualize key aspects of virus spread and evolution. Using Nextstrain, we use virus genomics to investigate the emergence of West Nile virus in the U S, followed by its rapid spread, evolution in a new environment, establishment of endemic transmission, and subsequent international spread. For each figure, we include a link to Nextstrain to allow the readers to directly interact with and explore the underlying data in new ways. We also provide a brief online narrative that parallels this review to further explain the data and highlight key epidemiological and evolutionary features (nextstrain.org/narratives/twenty-years-of-WNV). Mirroring the dynamic nature of outbreaks, the Nextstrain links provided within this paper are constantly updated as new West Nile virus genomes are shared publicly, helping to stay current with the research. Overall, our review showcases how genomics can track West Nile virus spread and evolution, as well as potentially uncover novel targeted control measures to help alleviate its public health burden.
Background
West Nile virus (WNV; family Flaviviridae; genus Flavivirus) is globally distributed and maintained by a complex transmission cycle involving multiple species of mosquitoes and birds [1–3]. Since its emergence in the Americas in 1999, the virus has resulted in over 48,000 reported cases, 24,000 reported neuroinvasive cases, over 2,300 deaths [4], and an estimated 7 million total human infections in the continental US [5]. At present, WNV is considered one of the most important zoonotic diseases of concern to the US population [6]. WNV is also a significant animal pathogen, having caused over 28,000 reported equine cases [7] and mortality in over 300 bird species [2], resulting in massive population declines reported among at least 23 bird species [8,9]. This includes a reported 45% decline in the American Crow (Corvus brachyrhynchos) population following the introduction of WNV [9]. The impact of WNV has not been limited to the US, as more than 5,000 human infections have been reported in Canada [10], and the virus is recognized as an emerging threat across the Americas [11–13].
In the two decades since WNV became established in the US, limited progress has been made in controlling transmission of the virus. Thus, renewed investment in understanding the fundamental aspects of endemic WNV transmission and innovative research directions are needed to mitigate the public health burden of WNV for the next 20 years. For example, advances in virus sequencing and phylogenetic analysis (comparing genetic variation over space and time) have transformed our ability to map the spread and evolution of viruses [14,15]. Such “genomic epidemiology” approaches, for example, were recently deployed to reconstruct the emergence of Zika virus in the Americas [16–18] and revealed the sources causing the yellow fever outbreaks in Brazil [19,20]. For WNV, virus genomics played a central role in uncovering how the virus emerged and became endemic in a new region after its introduction in the US (e.g., [21–27]), and more detailed studies may identify transmission networks and source areas of potential reemergence to be targeted during future interventions [28,29]. While powerful, these phylogenetic tools require technical expertise and computing resources that make them out of reach for some groups. To fill this need, Nextstrain (nextstrain.org) [30] was created to provide everyone the ability to explore pathogen spread and evolution using genomic epidemiology. In this review, we use Nextstrain to uncover key epidemiological and evolutionary processes during the emergence and establishment of WNV in the Americas (nextstrain.org/WNV/NA) and discuss collaborative opportunities to advance this work for future outbreak control.
Visualizing spread and evolution using Nextstrain
Nextstrain (nextstrain.org) is an open-source initiative to use pathogen genome data to provide a real-time interactive view of the spread and evolution of significant human pathogens [30]. The concept was born from a large collaborative project to track influenza virus evolution using rapid virus sequencing and data sharing [31]. This aided the seasonal influenza vaccine selection process by identifying emerging virus strains that may have evolved resistance to previous vaccines [32,33]. Moreover, recent epidemics of Zika and Ebola have highlighted the need for not only real-time virus genome sequencing but also rapid dissemination of epidemiologically relevant results [34]. Thus, Nextstrain was created to provide a publicly accessible and up-to-date overview of pathogen spread and evolution during outbreaks, facilitated through collaborations with subject matter experts. Alongside platforms such as Virological (virological.org), Nextstrain has become an important venue to quickly disseminate prepublication results, which is especially critical when the information can lead to public health interventions or improved public understanding [34].
Nextstrain is powered by a collection of open-source bioinformatic tools to curate, analyze, and visualize available virus genomes [30]. These datasets are used to reconstruct the geographic structure and estimated virus spread using incorporated phylogenetic tools [14,15]. To reduce runtimes, Nextstrain leverages TreeTime for the maximum likelihood phylogenetic analysis [35], through which the entire WNV dataset currently presented on Nextstrain (n = 2,267 genomes as of June 2019) can be analyzed in under an hour on a modern computer.
We created a WNV Nextstrain resource in 2018 to convey the history of WNV in the Americas and promote new research directions (nextstrain.org/WNV/NA; S1 Fig). All of the figures herein were created using Nextstrain and contain live display links in each legend to allow the reader to explore the data. The analysis presented on Nextstrain can be recreated using our build pipeline found at github.com/grubaughlab/WNV-nextstrain, which can be used to analyze any WNV dataset offline. Additionally, we designed a new narrative function in Nextstrain to accompany this review—accessible at nextstrain.org/narratives/twenty-years-of-WNV—which walks users through the epidemiological findings discussed here in a more condensed and interactive format. Mirroring the dynamic nature of such research, Nextstrain is constantly updated as new WNV genomes are shared publicly. Thus, the live display links will always contain the most up-to-date information, helping the readers stay current with the research.
Emergence of an exotic virus
During the summer of 1999, New York City experienced a remarkably high mortality among crows and several exotic captive birds, including Chilean flamingos (Phoenicopterus chilensis) in the Bronx Zoo [36]. Meanwhile, health professionals in Queens observed an unusual peak of unexplained human encephalitis cases, but the connection was not initially made with the increased bird mortality [37]. Early serological investigations pointed to Saint Louis encephalitis virus (SLEV), a mosquito-borne flavivirus endemic to the Americas, as the causative agent of the human encephalitis cases [38]. Electron microscopy investigation of viruses isolated from bird tissues also revealed flavivirus-like particles [21]. However, SLEV is usually not associated with mortality in birds, triggering further investigation of a possible link between the virus causing encephalitis in birds and humans [36]. Sequencing of viruses from bird and human tissues ultimately revealed that the 1999 outbreak was not caused by SLEV but, rather, a virus that had never before been observed in the Americas—WNV [21,36,39].
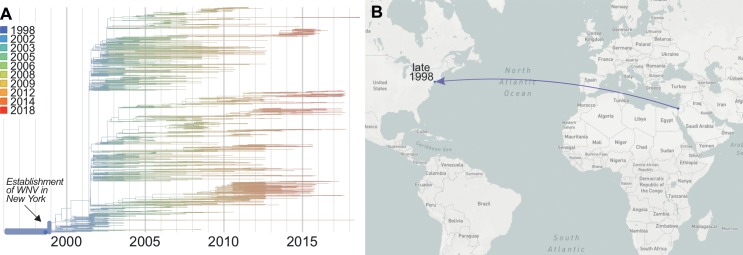
WNV was first isolated in Uganda in 1937 [40] and has since become endemic to many parts of Africa, Europe, Asia, Australia, and the Middle East [41]. Despite its global distribution, WNV was not detected in the Americas before the outbreak in 1999, raising two important questions: (1) where did the virus originate from and (2) when was it first introduced? Phylogenetic comparisons of the virus sequences isolated from New York in 1999 to WNV sequences from around the world revealed that it was most closely related to a WNV isolate from the brain of a dead goose found in Israel in 1998 [21,39], suggesting a potential origin from the Middle East [42] (Fig 1). Further genetic studies revealed that WNV was likely introduced in 1998 [24,43] (Fig 1). Thus, WNV may have been circulating in the US for a year before the first outbreak was detected during the summer of 1999.
Fig 1. Emergence of WNV in New York.
Though WNV was first detected in New York in 1999, (A) it likely became established in 1998 (confidence interval = October to December 1998) (B) by an introduction possibly from the Middle East, although the exact location cannot be inferred. Data from specific times can be visualized on Nextstrain using the “Date Range” function. A live display can be found at nextstrain.org/WNV/NA?c=num_date&d=tree,map&dmax=1998-12-01&p=grid.
A still unsettled question is how the virus was brought into the US [44]. The primary mechanism by which dengue and Zika viruses spread around the world is the long-distance travel of infected humans (e.g., [16,45]). However, unlike these viruses, WNV is maintained in a transmission cycle between birds and mosquitoes, in which humans are considered “dead-end” hosts (i.e., do not contribute to the cycle). Therefore, mobility of infected birds or mosquitoes, rather than infected humans, is the more plausible mechanism for the introduction of WNV from the Middle East into the US. While migratory birds might be important dispersers of WNV worldwide [46–48], flyways between the Middle East and North America are not common [3]. A more likely scenario is commercial or unintentional human transportation of birds and/or mosquitoes. Mosquitoes are notorious hitchhikers on airplanes [39,40], making an introduction via one of the two international airports located within the area of the 1999 New York City WNV outbreak a plausible hypothesis. Although it is impossible to determine exactly how WNV was introduced, more detailed future investigations into the patterns of WNV introductions worldwide may reveal new insights.
Conquering a continent
Following the initial outbreak in New York in 1999 [37], surveillance of mosquitoes and birds showed WNV spread along the eastern seaboard reaching Florida by 2001 [4] and west to the Rocky Mountains and Pacific Northwest (Washington state) by 2002 [4,49]. Finally, in 2003, WNV was detected in Southern California [4,50], marking its successful establishment across the continental US.
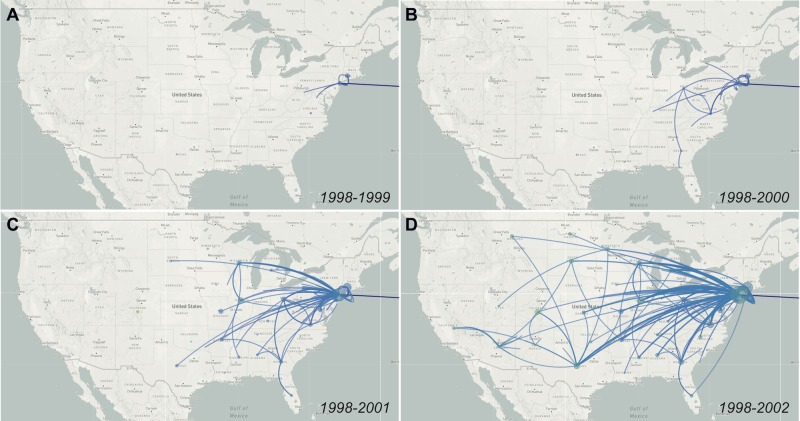
The virus genomic data reveal that WNV spread even faster across the US than the surveillance data show (Fig 2). When combining spatial and phylogenetic data, it is estimated that the WNV “wave” moved from the East Coast to the West Coast at an average dispersal velocity of approximately 1,000 km/year during the first few years (1999–2003) [26]. Moreover, by the time WNV was first detected in New York, the virus had already spread to neighboring states [26] (Fig 2A), before reaching parts of the Midwest and Southeast by 2000 (Fig 2B). These data also suggest that WNV was established in Texas by 2001 [26] (Fig 2C) and California by 2002 [23] (Fig 2D), a year before it was detected by local surveillance systems [4,50]. The rapid geographical expansion of WNV between 2001 and 2002 is consistent with a large increase in virus genetic diversity (i.e., a “polytomy,” as a result of many new transmission chains being introduced; Fig 1A) [24] and a significant jump in human cases (66 human cases in 2001 to 4,156 in 2002) [4].
Fig 2. Genomics reveals rapid spread of WNV across the continent.
By reconstructing the ancestral traits using TreeTime, Nextstrain infers state of all internal nodes of the phylogenetic tree to map the pattern of WNV spread. The phylogenetic data show that WNV spread to new regions occurred about a year before it was detected by local surveillance. Data from specific times can be visualized on Nextstrain using the “Date Range” function. Live displays can be found at (A) nextstrain.org/WNV/NA?c=num_date&d=map&dmax=1999-12-31&f_country=USA, (B) nextstrain.org/WNV/NA?c=num_date&d=map&dmax=2000-12-31&f_country=USA, (C) nextstrain.org/WNV/NA?c=num_date&d=map&dmax=2001-12-31&f_country=USA, (D) nextstrain.org/WNV/NA?c=num_date&d=map&dmax=2002-12-31&f_country=USA.
During the initial rapid spread of WNV in the US, the lack of geographic structure (i.e., “bush-like’ tree topology; S2 Fig) detected from the numerous national [24,26,51–54] and regional [51,55–58] phylogenetic studies suggests that WNV encountered a highly conducive environment with few barriers after it was introduced. This is likely due in part to the large diversity of hosts and vectors that WNV can utilize for transmission [1–3]. Among these, highly abundant passerine birds (e.g., thrushes and sparrows) [59] and Culex mosquitoes (e.g., C. tarsalis and C. pipiens) [60–62] are the most important for maintaining WNV transmission. Therefore, it is likely that a large population of susceptible hosts and vectors already present in the Americas helped WNV to quickly conquer a continent despite the varied landscapes that it needed to traverse.
While the typical home ranges of residential birds [23,63] and mosquitoes [64] may account for a large portion of WNV dispersal, the rapid rates of spread, nonuniform diffusion pattern, and lack of geographical structure are best explained by frequent mixing of local and nonlocal viruses [26]. Laboratory [65], field [66,67], and phylogenetic studies [24,27] support the hypothesis that the introduction of nonlocal viruses was facilitated by the movement of WNV-infected migratory birds. Indeed, the early spread of WNV along the eastern seaboard aligns with a major bird migration flyway; and while the routes primarily run north and south, the elliptical migration patterns of passerines may account for the East to West WNV expansion [27,48,68]. However, it is also possible that other mechanisms may account for the rapid spread and frequent virus mixing. For example, the impact of human behavior on WNV spread [63], specifically through the unintentional transport of WNV-infected birds or mosquitoes via the dense trucking industry in North America, should be examined more thoroughly. Better understanding of how WNV spreads long distances may be critical to inform future mitigation strategies.
Aided by evolution?
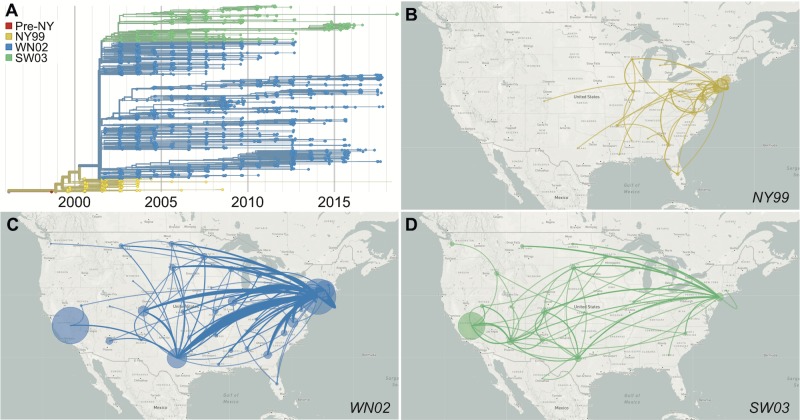
While WNV continues to diversify, the direct role that evolution played during the emergence of WNV in the Americas is currently difficult to discern. Evolution of WNV and other mosquito-borne viruses is complicated due to their requirement to maintain fitness in alternating and quite disparate mosquito vectors and vertebrate hosts [69–73]. As a result, the WNV evolutionary rate (approximately 4 × 10 −4 substitutions/site/year; S3 Fig) is slower than many other single-host RNA viruses [74]; and little evidence for positive selection in the virus population has been detected since the emergence of WNV in the Americas [24]. Yet, the displacement of initial WNV genotype (termed NY99) by locally derived genotypes (WN02 and SW03) [25,51,55,75,76] suggests that the virus may have undergone adaptive evolution in the US (Fig 3).
Fig 3. Displacement of the introduced WNV by locally derived genotypes.
(A) The WNV phylogenetic data support three main genotypes in the US: NY99, the introduced genotype; WN02, defined by the amino acid substitution E-V159A; and SW03, defined by the amino acid substitutions NS4A-A85T and NS5-K314R. The phylogeographic data from the US reveal that (B) NY99 rarely migrated past the Mississippi River, while (C) WN02 and (D) SW03 emerged in the East and spread all the way to the West Coast. WN02 and SW03 continue to cocirculate across the continent. Circle size is relative to the number of genomes that remained local. Data from specific genotypes can be visualized on Nextstrain using the “Color By: Strain” and “Filter by Lineage” functions. Live displays can be found at (A) nextstrain.org/WNV/NA?c=lineage&d=tree&f_country=USA, (B) nextstrain.org/WNV/NA?c=lineage&d=map&f_country=USA&f_lineage=NY99, (C) nextstrain.org/WNV/NA?c=lineage&d=map&f_country=USA&f_lineage=WN02, (D) nextstrain.org/WNV/NA?c=lineage&d=map&f_country=USA&f_lineage=SW03.
The defining feature of the NY99 genotype displacement is a single amino acid substitution in the WNV envelope protein (E-V159A) that emerged around 2001 [25,51,76] (Fig 3A). By 2003, the WN02 genotype containing the E-V159A substitution became dominant (Fig 3A), corresponding to the westward spread of the virus (Fig 3B and 3C). Experimental studies have shown that compared to NY99 strains, WN02 strains may be more efficiently transmitted by C. pipiens and C. tarsalis mosquitoes [76–78] and produce higher viremias during infection of house sparrows (Passer domesticus) [79]. Other studies, however, have not found differences in mosquito transmission rates between the genotypes [80–82] and evidence for selection of this allele was not found during phylogenetic [24] or experimental evolution studies [83,84]. Thus, the events leading to the extinction of NY99, either by stochastic processes or through an unknown selective disadvantage, remain unknown.
Soon after the emergence of WN02 in 2001, a southwest genotype (SW03), defined by two amino acid substitutions NS4A-A85T and NS5-K314R, arose from within this clade [24,85] (Fig 3A). Both of these substitutions have occurred independently multiple times (S4 Fig) and appear to be under positive selection [85,86]. WN02 and SW03 continue to coexist within the US (Fig 3) and even cocirculate within the same locations, years, and mosquito vectors (e.g., Arizona [22], California [23], and New York [87]; S5 Fig), suggesting that there are not any major barriers segregating these genotypes. However, there is some evidence that SW03 has a more rapid rate of spread than WN02 in California [23]. This could suggest that NS4A-A85T and/or NS5-K314R are the result of recent adaptations; however, the direct fitness of either allele has yet to be experimentally evaluated. Altogether, the evidence that supports the hypothesis that rapid evolution of WNV facilitated its westwardly spread is limited, and additional studies are required to tease apart if locally derived genotypes, such as WN02 and SW03, are specifically adapted to the environment in the Americas.
Becoming an entrenched virus
Following its emergence, WNV quickly established endemicity and became one of the most important mosquito-borne viruses in North America [4,6]. Since 2002, between 662 (in 2009) and 9,438 (in 2003) human WNV cases have been reported every year in the US [4]. Evidence for endemicity from the genomic data is clear in New York, with some local transmission chains (i.e., branches on the phylogenetic tree) persisting for at least 10 years (S6 Fig) [87]. Likely due to the constant repopulation of susceptible mosquito vectors and avian hosts, the data indicate that WNV poses as an annual public health threat with no signs of remission.
Though WNV transmission is consistently detected throughout the continental US, its burden is not uniform, with a large variation in year-to-year human cases and the highest incidence rates often occurring in the central great plains (e.g., Wyoming, South Dakota, and North Dakota) [4]. Dynamic extrinsic factors, such as rainfall and temperature, that influence mosquito and bird populations can be predictive of WNV intensity [88–94], yet the contributions of these extrinsic factors vary across the US due to differences in regional ecology [3,95,96]. This is supported by the nonuniform abundance of C. tarsalis, C. pipiens, and C. quinquefasciatus across the US [97,98], where the presence or absence of these primary mosquito vectors in a given location is mediated by many factors including human land use, elevation, and winter temperatures. Together, these ecological differences create unique transmission networks at the local and regional levels that can significantly influence the likelihood of human disease risks [96–100]. These ecological barriers may further influence mosquito genetic diversity [64,101], causing potential differences in their efficiency for WNV transmission [95,102]. Combined with varied avian ecology and migratory bird flyways across the US [3,96], these data suggest that WNV is maintained in regional transmission networks with unique selective pressures that influence the stability and emergence of new WNV strains [87].
The slowed dispersal velocity and formation of geographical segregation [22–24,26,87] (S2 Fig, S6 Fig) after the initial rapid spread stage (Fig 2) reinforce the hypothesis that WNV is now primarily maintained at local or regional levels with less long-distance mixing. However, it is unclear if dynamic clustering of the virus across the US will lead to the emergence of new adaptive genotypes and if locally adapted viruses are more likely to cause outbreaks. Understanding such fundamental questions of the epidemiology and biology of WNV strains between and within states may be important for effectively combating this entrenched virus.
An international concern
The spread of WNV north to Canada and as far south as Argentina highlights that the growing burden of WNV is not limited to the US. The first human WNV case in Canada was reported in Ontario in 2002, and it has now become endemic in several southern Canadian provinces [10,103]. Surveillance between 2001 and 2004 showed that WNV had spread to Central America and the Caribbean [12,13,104–107]. Since then, evidence of WNV transmission has been found across South America, including Colombia, Venezuela, Brazil, and Argentina [13,108–111]. Detection of WNV antibodies in migratory birds suggests that this route may be an important factor for WNV dissemination throughout the Americas [106]; but as described above, it is still unclear if other mechanisms may influence the spread of WNV to Central and South America.
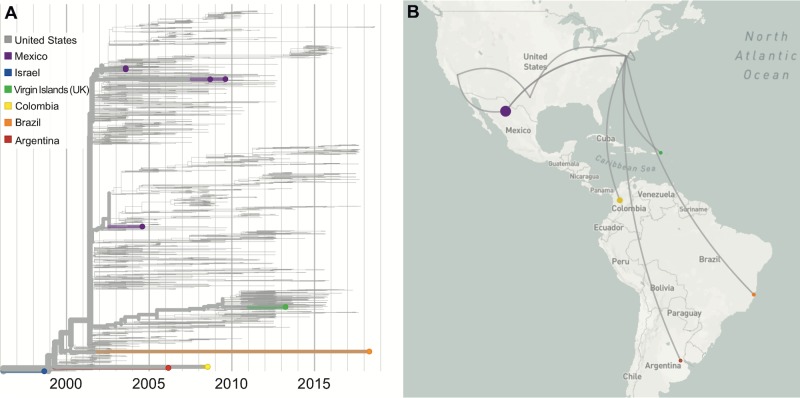
Although many partial WNV genomes (primarily envelope coding sequences) were generated from the Americas outside of the US (e.g., [52,112]), complete or near-complete virus genomes are necessary to accurately reconstruct spread and outbreak dynamics [113]. To this end, there are currently only 11 WNV genomes available from Mexico [114,115], seven from the rest of Latin America and the Caribbean [116–119], and none from Canada. These limited data do reveal that the southwards spread into Latin America and the Caribbean began during the early stages of the outbreak in the US and occurred independently multiple times [115–120] (Fig 4). In Mexico, multiple introductions of the WN02 and SW03 genotypes suggest that viruses may be commonly moving across the US–Mexico border [114,115]. Moreover, WNV genetic data from the British Virgin Islands [119] and Puerto Rico (partial sequences [121]) reveal that at least the WN02 genotype was introduced into the Caribbean. Interestingly, the NY99 genotype that was displaced in the US (Fig 3) was detected in Colombia, Argentina, and Brazil, likely via separate introductions, and may still be in circulation in South America [116–118]. However, the paucity of complete WNV genomes makes it difficult to understand the patterns of spread and diversity outside of the US.
Fig 4. International spread of WNV following its emergence in the US.
Following the emergence of WNV in the US, the virus was detected throughout the Americas, from Canada to Argentina. (A) The colored branches on the tree correspond to (B) the lines on the map depicting spread. However, the limited number of available near-complete WNV genomes from each location and the long branches make it difficult to accurately map the patterns of spread and timing of introductions (e.g., the long orange branch on the tree may indicate that the virus may have traveled to many locations before being introduced in Brazil sometime between 2002 and 2018). Data from specific countries can be visualized on Nextstrain using the “Color By: Country” and “Filter by Country” functions. A live display can be found at nextstrain.org/WNV/NA?c=country&f_country=Argentina,Brazil,British-Virgin-Islands,Colombia,Mexico&p=grid.
An important question is why large human WNV outbreaks have not been detected in Latin American countries [12,122]. The tropical and subtropical regions of the Americas have suitable conditions for the establishment of WNV, including warm temperatures, diverse avian populations, and a variety of Culex mosquitoes [11,122]. Moreover, SLEV, a similar flavivirus maintained by Culex vectors and avian hosts is endemic throughout the Americas [123]. Considering that the “old” NY99 genotype virus was recently detected in Brazil [118] and serological evidence of WNV is prevalent in resident birds from Latin America [13,104–106], it is likely that WNV is endemic throughout the Americas. Perhaps the presence of other mosquito-borne viruses has thus far masked human cases (e.g., misdiagnosed as dengue) or may convey some level of cross-protection [12,122]. Alternatively, as WNV may have initially outcompeted SLEV in the US [124], the presence of competitive SLEV genotypes in Central and South America may have at least temporarily slowed WNV transmission in these regions. Given these uncertainties, initiatives to introduce human and equine serosurveys and dead bird and mosquito surveillance for WNV in regions throughout the Americas are paramount for determining its true burden.
Future role of genomics
Given how much WNV has thrived in the US, significant national investment will be required to control future outbreaks. Among this investment should include sustained and collaborative efforts to fill in the many gaps in WNV genomic sampling throughout the country, especially from the last 10 years (Fig 5), to better understand endemic WNV transmission using virus genomics approaches [14]. Fortunately, the availability of scalable, easy, and relatively inexpensive sequencing tools (e.g., [125,126]) make these efforts more achievable, and recent work by Swetnam and colleagues [27], Hepp and colleagues [22], Bialosuknia and colleagues [87], and the WestNile 4K Project (westnile4k.org/) provides a template for future public health and academic lab partnerships. Platforms such as Nextstrain (nextstrain.org/WNV/NA) and Virological (virological.org) allow these new data to be continually communicated to the public.
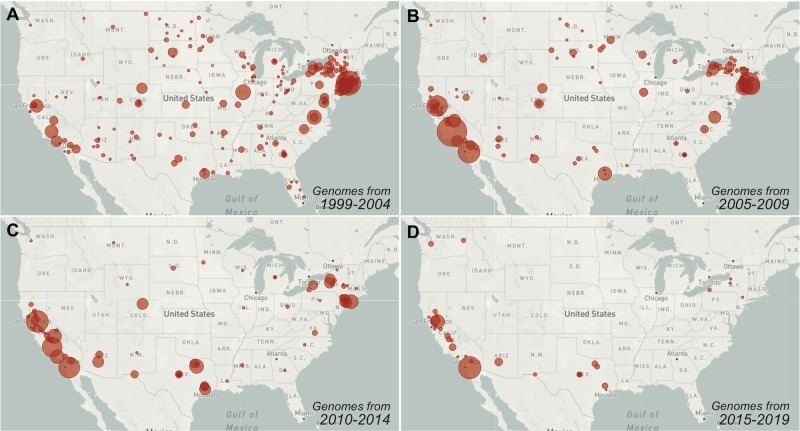
Fig 5. Gaps in recent WNV sampling hinder the usefulness of virus genomic analyses.
Following the initial interest in sequencing WNV, the number of available genomes for research has continued to decrease. The circle sizes represent the relative number of near-complete WNV genomes available from each collection location (county) per 5-year period. There are currently (A) 728 available WNV genomes from 37 states from 1999 to 2004, (B) 789 WNV genomes from 22 from 2005 to 2009, (C) 541 WNV genomes from 22 states from 2010 to 2014, and (D) 190 WNV genomes from 5 states from 2015 to 2019 (all counts are as of June 2019). Data from specific times can be visualized on Nextstrain using the “Date Range” function. Live displays of the data can be found at (A) nextstrain.org/WNV/NA?c=num_date&d=map&dmax=2004-12-31&f_country=USA&r=division, (B) nextstrain.org/WNV/NA?c=num_date&d=map&dmax=2009-12-31&dmin=2005-01-01&f_country=USA&r=division, (C) nextstrain.org/WNV/NA?c=num_date&d=map&dmax=2014-12-31&dmin=2010-01-01&f_country=USA&r=division, (D) nextstrain.org/WNV/NA?c=num_date&d=map&dmax=2019-12-31&dmin=2015-01-01&f_country=USA&r=division.
Generating a well-distributed temporal and spatial WNV genomic dataset can be used to determine which strains are causing outbreaks and, importantly, where those strains came from and when they became locally established. This work will help to identify if WNV outbreaks in Ames, Iowa, for example, are connected to outbreaks as far away as Des Moines, Iowa (approximately 60 km); Omaha, Nebraska (approximately 270 km); or even Chicago, Illinois (approximately 560 km). Systematically searching for connections among WNV outbreaks around the country may reveal endemic “transmission networks,” which could be exploited for control and forecasting purposes. WNV control is primarily based on local interventions to temporarily reduce the populations of mosquito vectors [127], but the long-term effects are likely limited by recolonization of mosquitoes and reintroductions of viruses once the pressure is removed. Thus, a highly coordinated effort among several health departments and mosquito abatement districts to synchronize vector control within a transmission network may reduce reintroductions and limit WNV transmission beyond a single season. In addition, genomic epidemiology approaches with large-scale efforts to collect and share mosquito abundance and infection rate data [128] may help to identify potential patterns of virus outbreaks (e.g., as used to identify factors contributing to the Ebola epidemic [29,129]). This, in turn, could be used to maximize vector control efforts by strategically focusing resources at a precise time and location to limit potential outbreaks [29]. After 20 years of WNV in the Americas, it is time to recognize that alleviating the burden of WNV for the next generation will likely depend on significant investment, new approaches, and large collaborations.
Supporting information
A depiction of the main interactive interface on Nextstrain to visualize the virus (A) phylogeny (time-resolved phylogenetic tree created with TreeTime, states listed by two-letter abbreviations, countries listed by three-letter abbreviations), (B) spread (inferred patterns of spread based on the tree, circle size is relative to the number of genomes that remained local), and (C) genetic diversity (amino acid diversity plotted as entropy and positioned by codon sequence). In the WNV genome, the protein abbreviations are as follows: C = capsid, prM = premembrane, E = envelope, NS1-5 = nonstructural protein 1–5. A live display, which can be used to select and zoom in on various features, can be found at nextstrain.org/WNV/NA?p=grid. WNV, West Nile virus.
(PDF)
An unrooted maximum likelihood WNV phylogenetic tree shows that there is little geographic structure (i.e., bush-like topology) during the early spread phase, as best demonstrated by the Texas (gold) and New York (purple) genomes often clustering together. This suggests that environment was highly conducive for transmission and that the virus could freely spread. The minimal mixing between California and New York genomes, however, suggests that geographic structure is starting to form as the virus transitioned into its endemic phase. Additional WNV sequencing throughout the US from the last 10 years will help to better understand if structure is starting to form and at what scale. States are listed by two-letter abbreviations and countries are listed by three-letter abbreviations. This “unrooted” tree view can be visualized on Nextstrain by toggling between the “Tree Options: Layout”. A live display can be found at nextstrain.org/WNV/NA?l=unrooted&m=div&p=full. WNV, West Nile virus.
(PDF)
A root-to-tip plot showing the divergence (substitutions per site) of sequenced WNV genomes (tips) from the inferred ancestral sequence (root) by the collection dates (shown in years) is used to estimate the evolutionary rate at approximately 4×10−4 substitutions/site/year. The tips are colored by increasing time. This “clock” view can be visualized on Nextstrain by toggling between the “Tree Options: Layout.” A live display can be found at nextstrain.org/WNV/NA?c=num_date&d=tree&l=clock. WNV, West Nile virus.
(PDF)
WNV genotype SW03 is defined by two amino acid substitutions, (A) nonstructural protein 4A (NS4A) A85T and (B) NS5-K314R. Branches (inferred ancestral genome) and tips (sequenced genome) are colored by amino acid at position (A) NS4A site 85 (nucleotide position 6721) and (B) NS5 site 314 (nucleotide position 8621). Both (A) NS4A-A85T and (B) NS5-K314R help to form a well-supported clade (yellow branches at the top) but also occur independently throughout the tree (yellow scattered throughout the lower half). All other alleles (nucleotide and amino acid changes) can be visualized using Nextstrain by entering the loci using the “Color By: genotype” function or by selecting a loci on the “Diversity” plot (i.e., S1C Fig). Live displays can be found at (A) nextstrain.org/WNV/NA?c=gt-NS4A_85 and (B) nextstrain.org/WNV/NA?c=gt-NS5_314. WNV, West Nile virus.
(PDF)
Since the emergence of WN02 (blue) and SW03 (green) in 2001, the genotypes continue to cocirculate in locations throughout the US For example, both genotypes were detected during the same years and vector species (A) in Maricopa County, Arizona [22] (B), throughout California [23], and (C) throughout New York [87]. Data from other studies can be visualized on Nextstrain by using the “Filter by Authors” function. Live displays can be found at (A) https://nextstrain.org/WNV/NA?c=lineage&f_authors=Hepp%20et%20al, (B) https://nextstrain.org/WNV/NA?c=lineage&f_authors=Duggal%20et%20al, and (C) https://nextstrain.org/WNV/NA?c=lineage&f_authors=Shabman%20et%20al. WNV, West Nile virus.
(PDF)
WNV is likely establishing persistent local transmission networks throughout the US, but this can be easily demonstrated from the 570 genomes available from New York (most generated by [87]). Multiple co-occurring transmission chains (branches) exist derived from either local evolution or separate (re-)introductions, but several persist locally for 5 or more years indicating that those viruses became established. Data from other states can be visualized on Nextstrain by using the “Filter by State” function. A live display can be found at nextstrain.org/WNV/NA?f_state=NY&d=tree. WNV, West Nile virus.
(PDF)
Acknowledgments
This work is possible due to the groups who have made their data openly available for analysis—with a special thanks to Alex Ciota, Nikos Gurfield, Saran Grewal, Chris Barker, Ying Fang, and Amy Salamone for making their data available ahead of publication. We also thank Simon Dellicour, Sebastian Lequime, Bram Vrancken, Philippe Lemey, Karthik Gangavarapu, Nate Matteson, Sharada Saraf, Glenn Oliveira, Refugio Robles, Zhe Zheng, Sophie Taylor, and Philip Jack for curating the original dataset, generating new data, and helping with the Nextstrain updates.
Funding Statement
DMS acknowledges funding support from the Pacific Southwest Regional Center of Excellence for Vector-Borne Diseases funded by the U.S. Centers for Disease Control and Prevention (CDC, Cooperative Agreement 1U01CK000516). CBFV is supported by NWO Rubicon 019.181EN.004. KGA is a Pew Biomedical Scholar, and is supported by NIH NCATS CTSA UL1TR002550, NIAID contract HHSN272201400048C, NIAID R21AI137690, NIAID U19AI135995, and The Ray Thomas Foundation. RCS and RET are supported in part by the Epidemiology and Laboratory Capacity for Infectious Diseases (ELC) Program Components Contract 5887EL11 through the Iowa Department of Public Health and the Midwest Center of Excellence for Vector-Borne Diseases by Cooperative Agreement U01CK000505, funded by the CDC. TB is a Pew Biomedical Scholar. TB and JH are supported by NIH R35 GM119774-01. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the CDC or the Department of Health and Human Services. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kramer LD, Styer LM, Ebel GD. A global perspective on the epidemiology of West Nile virus. Annu Rev Entomol. 2008;53: 61–81. 10.1146/annurev.ento.53.103106.093258 [DOI] [PubMed] [Google Scholar]
- 2.Komar N. West Nile virus: epidemiology and ecology in North America. Adv Virus Res. 2003;61: 185–234. [DOI] [PubMed] [Google Scholar]
- 3.Reisen WK. Ecology of West Nile virus in North America. Viruses. 2013;5: 2079–2105. 10.3390/v5092079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CDC. West Nile Virus Final Cumulative Maps and Data [Internet]. 10 Dec 2018. Available from: https://www.cdc.gov/westnile/statsmaps/finalmapsdata/index.html. [cited 2019 May 5].
- 5.Ronca SE, Murray KO, Nolan MS. Cumulative incidence of West Nile virus infection, continental United States, 1999–2016. Emerg Infect Dis. 2019;25: 325 10.3201/eid2502.180765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.CDC. 8 Zoonotic Diseases Shared Between Animals and People of Most Concern in the U.S [Internet]. 6 May 2019. Available from: https://www.cdc.gov/media/releases/2019/s0506-zoonotic-diseases-shared.html. [cited 2019 May 13].
- 7.Aphis U. West Nile Virus Maps- States with Equine Cases [Internet]. Available from: https://www.aphis.usda.gov/aphis/ourfocus/animalhealth/animal-disease-information/horse-disease-information/wnv/west-nile-virus. [cited 2019 Jan 25].
- 8.George TL, Harrigan RJ, LaManna JA, DeSante DF, Saracco JF, Smith TB. Persistent impacts of West Nile virus on North American bird populations. Proc Natl Acad Sci U S A. 2015;112: 14290–14294. 10.1073/pnas.1507747112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LaDeau SL, Kilpatrick AM, Marra PP. West Nile virus emergence and large-scale declines of North American bird populations. Nature. 2007;447: 710–713. 10.1038/nature05829 [DOI] [PubMed] [Google Scholar]
- 10.Public Health Agency of Canada. Surveillance of West Nile virus [Internet]. 26 Jun 2015. Available from: https://www.canada.ca/en/public-health/services/diseases/west-nile-virus/surveillance-west-nile-virus.html. [cited 2019 May 26].
- 11.Castro-Jorge LA de, Siconelli MJL, Ribeiro BDS, Moraes FM de, Moraes JB de, Agostinho MR, et al. West Nile virus infections are here! Are we prepared to face another flavivirus epidemic? Rev Soc Bras Med Trop. 2019;52: e20190089 10.1590/0037-8682-0089-2018 [DOI] [PubMed] [Google Scholar]
- 12.Elizondo-Quiroga D, Elizondo-Quiroga A. West nile virus and its theories, a big puzzle in Mexico and Latin America. J Glob Infect Dis. 2013;5: 168–175. 10.4103/0974-777X.122014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Who P. West nile virus Epidemiological alerts and updates [Internet]. Available from: https://www.paho.org/hq/index.php?option=com_topics&view=rdmore&cid=2195&Itemid=40782&lang=en. [cited 2019 May 26].
- 14.Grubaugh ND, Ladner JT, Lemey P, Pybus OG, Rambaut A, Holmes EC, et al. Tracking virus outbreaks in the twenty-first century. Nat Microbiol. 2019;4: 10–19. 10.1038/s41564-018-0296-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Volz EM, Koelle K, Bedford T. Viral phylodynamics. PLoS Comput Biol. 2013;9: e1002947 10.1371/journal.pcbi.1002947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grubaugh ND, Ladner JT, Kraemer MUG, Dudas G, Tan AL, Gangavarapu K, et al. Genomic epidemiology reveals multiple introductions of Zika virus into the United States. Nature. 2017;90: 4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metsky HC, Matranga CB, Wohl S, Schaffner SF, Freije CA, Winnicki SM, et al. Zika virus evolution and spread in the Americas. Nature. 2017;66: 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faria NR, Quick J, Claro IM, Thézé J, de Jesus JG, Giovanetti M, et al. Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature. 2017; 10.1038/nature22401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faria NR, Kraemer MUG, Hill SC, Goes de Jesus J, Aguiar RS, Iani FCM, et al. Genomic and epidemiological monitoring of yellow fever virus transmission potential. Science. 2018; 10.1126/science.aat7115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moreira-Soto A, Torres MC, Lima de Mendonça MC, Mares-Guia MA, Dos Santos Rodrigues CD, Fabri AA, et al. Evidence for multiple sylvatic transmission cycles during the 2016–2017 yellow fever virus outbreak, Brazil. Clin Microbiol Infect. 2018;24: 1019.e1–1019.e4. [DOI] [PubMed] [Google Scholar]
- 21.Lanciotti RS, Roehrig JT, Deubel V, Smith J, Parker M, Steele K, et al. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science. 1999;286: 2333–2337. 10.1126/science.286.5448.2333 [DOI] [PubMed] [Google Scholar]
- 22.Hepp CM, Cocking JH, Valentine M, Young SJ, Damian D, Samuels-Crow KE, et al. Phylogenetic analysis of West Nile Virus in Maricopa county, Arizona: Evidence for dynamic behavior of strains in two major lineages in the American Southwest. PLoS ONE. 2018;13: e0205801 10.1371/journal.pone.0205801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duggal NK, Reisen WK, Fang Y, Newman RM, Yang X, Ebel GD, et al. Genotype-specific variation in West Nile virus dispersal in California. Virology. 2015;485: 79–85. 10.1016/j.virol.2015.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Giallonardo F, Geoghegan JL, Docherty DE, McLean RG, Zody MC, Qu J, et al. Fluid spatial dynamics of West Nile virus in the United States: Rapid spread in a permissive host environment. J Virol. 2016;90: 862–872. 10.1128/JVI.02305-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis CT, Ebel GD, Lanciotti RS, Brault AC, Guzman H, Siirin M, et al. Phylogenetic analysis of North American West Nile virus isolates, 2001–2004: Evidence for the emergence of a dominant genotype. Virology. 2005;342: 252–265. 10.1016/j.virol.2005.07.022 [DOI] [PubMed] [Google Scholar]
- 26.Pybus OG, Suchard MA, Lemey P, Bernardin FJ, Rambaut A, Crawford FW, et al. Unifying the spatial epidemiology and molecular evolution of emerging epidemics. Proc Natl Acad Sci U S A. 2012;109: 15066–15071. 10.1073/pnas.1206598109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swetnam D, Widen SG, Wood TG, Reyna M, Wilkerson L, Debboun M, et al. Terrestrial bird migration and West Nile virus circulation, United States. Emerg Infect Dis. 2018;24: 2184–2194. 10.3201/eid2412.180382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ladner JT, Grubaugh ND, Pybus OG, Andersen KG. Precision epidemiology for infectious disease control. Nat Med. 2019;25: 206–211. 10.1038/s41591-019-0345-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dellicour S, Baele G, Dudas G, Faria NR, Pybus OG, Suchard MA, et al. Phylodynamic assessment of intervention strategies for the West African Ebola virus outbreak. Nat Commun. 2018;9: 2222 10.1038/s41467-018-03763-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, et al. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics. 2018;34: 4121–4123. 10.1093/bioinformatics/bty407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.GISAID. Global Initiative on Sharing All Influenza Data [Internet]. Available from: https://www.gisaid.org/. [cited 2019 May 18].
- 32.Neher RA, Bedford T. Nextflu: Real-time tracking of seasonal influenza virus evolution in humans. Bioinformatics. 2015;31: 3546–3548. 10.1093/bioinformatics/btv381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neher RA, Bedford T, Daniels RS, Russell CA, Shraiman BI. Prediction, dynamics, and visualization of antigenic phenotypes of seasonal influenza viruses. Proc Natl Acad Sci U S A. 2016;113: E1701–9. 10.1073/pnas.1525578113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gardy JL, Loman NJ. Towards a genomics-informed, real-time, global pathogen surveillance system. Nat Rev Genet. 2018;19: 9–20. 10.1038/nrg.2017.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sagulenko P, Puller V, Neher RA. TreeTime: Maximum-likelihood phylodynamic analysis. Virus Evol. 2018;4: vex042 10.1093/ve/vex042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Briese T, Jia XY, Huang C, Grady LJ, Lipkin WI. Identification of a Kunjin/West Nile-like flavivirus in brains of patients with New York encephalitis. Lancet. 1999;354: 1261–1262. 10.1016/s0140-6736(99)04576-6 [DOI] [PubMed] [Google Scholar]
- 37.CDC. Outbreak of West Nile-like viral encephalitis—New York, 1999. MMWR Morb Mortal Wkly Rep. 1999;48: 845–849. [PubMed] [Google Scholar]
- 38.Exotic diseases close to home. Lancet. 1999;354: 1221 [PubMed] [Google Scholar]
- 39.Jia XY, Briese T, Jordan I, Rambaut A, Chi HC, Mackenzie JS, et al. Genetic analysis of West Nile New York 1999 encephalitis virus. Lancet. 1999;354: 1971–1972. 10.1016/s0140-6736(99)05384-2 [DOI] [PubMed] [Google Scholar]
- 40.Smithburn KC, Hughes TP, Burke AW, Paul JH. A neurotropic virus isolated from the blood of a native of Uganda. Am J Trop Med Hyg. 1940;s1-20: 471–492. [Google Scholar]
- 41.Mackenzie JS, Gubler DJ, Petersen LR. Emerging flaviviruses: The spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat Med. 2004;10: S98–109. 10.1038/nm1144 [DOI] [PubMed] [Google Scholar]
- 42.May FJ, Davis CT, Tesh RB, Barrett ADT. Phylogeography of West Nile virus: From the cradle of evolution in Africa to Eurasia, Australia, and the Americas. J Virol. 2011;85: 2964–2974. 10.1128/JVI.01963-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zehender G, Ebranati E, Bernini F, Lo Presti A, Rezza G, Delogu M, et al. Phylogeography and epidemiological history of West Nile virus genotype 1a in Europe and the Mediterranean basin. Infection, Genetics and Evolution. 2011;11: 646–653. 10.1016/j.meegid.2011.02.003 [DOI] [PubMed] [Google Scholar]
- 44.Johnston BL, Conly JM. West Nile virus—where did it come from and where might it go? Canadian Journal of Infectious Diseases. 2000;11: 175–178. 10.1155/2000/856598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian H, Sun Z, Faria NR, Yang J, Cazelles B, Huang S, et al. Increasing airline travel may facilitate co-circulation of multiple dengue virus serotypes in Asia. PLoS Negl Trop Dis. 2017;11: e0005694 10.1371/journal.pntd.0005694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Viana DS, Santamaría L, Figuerola J. Migratory birds as global dispersal vectors. Trends Ecol Evol. 2016;31: 763–775. 10.1016/j.tree.2016.07.005 [DOI] [PubMed] [Google Scholar]
- 47.Hernández-Triana LM, Jeffries CL, Mansfield KL, Carnell G, Fooks AR, Johnson N. Emergence of West Nile virus lineage 2 in Europe: A review on the introduction and spread of a mosquito-borne disease. Front Public Health. 2014;2: 271 10.3389/fpubh.2014.00271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reed KD, Meece JK, Henkel JS, Shukla SK. Birds, migration and emerging zoonoses: West Nile virus, Lyme disease, influenza A and enteropathogens. Clin Med Res. 2003;1: 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.CDC. West Nile Virus Activity—United States, September 26-October 2, 2002, and Investigations of West Nile Virus Infections in Recipients of Blood Transfusion and Organ Transplantation [Internet]. 23 Oct 2002. Available from: https://www.cdc.gov/mmwr/preview/mmwrhtml/mm5139a5.htm. [cited 2019 May 5]. [PubMed]
- 50.Reisen W, Lothrop H, Chiles R, Madon M, Cossen C, Woods L, et al. West Nile virus in California. Emerg Infect Dis. 2004;10: 1369–1378. 10.3201/eid1008.040077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beasley DWC, Davis CT, Guzman H, Vanlandingham DL, Travassos da Rosa APA, Parsons RE, et al. Limited evolution of West Nile virus has occurred during its southwesterly spread in the United States. Virology. 2003;309: 190–195. 10.1016/s0042-6822(03)00150-8 [DOI] [PubMed] [Google Scholar]
- 52.Herring BL, Bernardin F, Caglioti S, Stramer S, Tobler L, Andrews W, et al. Phylogenetic analysis of WNV in North American blood donors during the 2003–2004 epidemic seasons. Virology. 2007;363: 220–228. 10.1016/j.virol.2007.01.019 [DOI] [PubMed] [Google Scholar]
- 53.Grinev A, Chancey C, Volkova E, Añez G, Heisey DAR, Winkelman V, et al. Genetic variability of West Nile virus in US blood donors from the 2012 epidemic season. PLoS Negl Trop Dis. 2016;10: e0004717 10.1371/journal.pntd.0004717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anez G, Grinev A, Chancey C, Ball C, Akolkar N, Land KJ, et al. Evolutionary dynamics of West Nile virus in the United States, 1999–2011: Phylogeny, selection pressure and evolutionary time-scale analysis. PLoS Negl Trop Dis. 2013;7: e2245 10.1371/journal.pntd.0002245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis CT, Beasley DWC, Guzman H, Raj R, D’Anton M, Novak RJ, et al. Genetic variation among temporally and geographically distinct West Nile virus isolates, United States, 2001, 2002. Emerg Infect Dis. 2003;9: 1423–1429. 10.3201/eid0911.030301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bertolotti L, Kitron UD, Walker ED, Ruiz MO, Brawn JD, Loss SR, et al. Fine-scale genetic variation and evolution of West Nile Virus in a transmission “hot spot” in suburban Chicago, USA. Virology. 2008;374: 381–389. 10.1016/j.virol.2007.12.040 [DOI] [PubMed] [Google Scholar]
- 57.Amore G, Bertolotti L, Hamer GL, Kitron UD, Walker ED, Ruiz MO, et al. Multi-year evolutionary dynamics of West Nile virus in suburban Chicago, USA, 2005–2007. Philos Trans R Soc Lond B Biol Sci. 2010;365: 1871–1878. 10.1098/rstb.2010.0054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bertolotti L, Kitron U, Goldberg TL. Diversity and evolution of West Nile virus in Illinois and the United States, 2002–2005. Virology. 2007;360: 143–149. 10.1016/j.virol.2006.10.030 [DOI] [PubMed] [Google Scholar]
- 59.Komar N, Langevin S, Hinten S, Nemeth N, Edwards E, Hettler D, et al. Experimental infection of North American birds with the New York 1999 strain of West Nile virus. Emerg Infect Dis. 2003;9: 311–322. 10.3201/eid0903.020628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Turell MJ, O’Guinn M, Oliver J. Potential for New York mosquitoes to transmit West Nile virus. Am J Trop Med Hyg. 2000;62: 413–414. 10.4269/ajtmh.2000.62.413 [DOI] [PubMed] [Google Scholar]
- 61.Goddard LB, Roth AE, Reisen WK, Scott TW. Vector competence of California mosquitoes for West Nile virus. Emerg Infect Dis. 2002;8: 1385–1391. 10.3201/eid0812.020536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sardelis MR, Turell MJ, Dohm DJ, O’Guinn ML. Vector competence of selected North American Culex and Coquillettidia mosquitoes for West Nile virus. Emerg Infect Dis. 2001;7: 1018–1022. 10.3201/eid0706.010617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rappole JH, Compton BW, Leimgruber P, Robertson J, King DI, Renner SC. Modeling movement of West Nile virus in the western hemisphere. Vector Borne Zoonotic Dis. 2006;6: 128–139. 10.1089/vbz.2006.6.128 [DOI] [PubMed] [Google Scholar]
- 64.Venkatesan M, Rasgon JL. Population genetic data suggest a role for mosquito-mediated dispersal of West Nile virus across the western United States. Mol Ecol. 2010;19: 1573–1584. 10.1111/j.1365-294X.2010.04577.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Owen J, Moore F, Panella N, Edwards E, Bru R, Hughes M, et al. Migrating birds as dispersal vehicles for West Nile virus. Ecohealth. 2006;3: 79. [Google Scholar]
- 66.Dusek RJ, McLean RG, Kramer LD, Ubico SR, Dupuis AP 2nd, Ebel GD, et al. Prevalence of West Nile virus in migratory birds during spring and fall migration. Am J Trop Med Hyg. 2009;81: 1151–1158. 10.4269/ajtmh.2009.09-0106 [DOI] [PubMed] [Google Scholar]
- 67.Reisen WK, Wheeler SS, Garcia S, Fang Y. Migratory birds and the dispersal of arboviruses in California. Am J Trop Med Hyg. 2010;83: 808–815. 10.4269/ajtmh.2010.10-0200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rappole JH, Derrickson SR, Hubálek Z. Migratory birds and spread of West Nile virus in the western hemisphere. Emerg Infect Dis. 2000;6: 319–328. 10.3201/eid0604.000401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weaver SC, Rico-Hesse R, Scott TW. Genetic diversity and slow rates of evolution in New World alphaviruses. Current Topics in Microbiology and Immunology. 1992; 99–117. 10.1007/978-3-642-77011-1_7 [DOI] [PubMed] [Google Scholar]
- 70.Weaver SC, Brault AC, Kang W, Holland JJ. Genetic and fitness changes accompanying adaptation of an arbovirus to vertebrate and invertebrate cells. J Virol. 1999;73: 4316–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ciota AT, Lovelace AO, Ngo KA, Le AN, Maffei JG, Franke MA, et al. Cell-specific adaptation of two flaviviruses following serial passage in mosquito cell culture. Virology. 2007;357: 165–174. 10.1016/j.virol.2006.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grubaugh ND, Fauver JR, Ruckert C, Weger-Lucarelli J, Garcia-Luna S, Murrieta RA, et al. Mosquitoes transmit unique West Nile virus populations during each feeding episode. Cell Rep. 2017;19: 709–718. 10.1016/j.celrep.2017.03.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deardorff ER, Fitzpatrick KA, Jerzak GVS, Shi P-Y, Kramer LD, Ebel GD. West Nile virus experimental evolution in vivo and the trade-off hypothesis. PLoS Pathog. 2011;7: e1002335 10.1371/journal.ppat.1002335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jenkins GM, Rambaut A, Pybus OG, Holmes EC. Rates of molecular evolution in RNA viruses: A quantitative phylogenetic analysis. J Mol Evol. 2002;54: 156–165. 10.1007/s00239-001-0064-3 [DOI] [PubMed] [Google Scholar]
- 75.Granwehr BP, Li L, Davis CT, Beasley DWC, Barrett ADT. Characterization of a West Nile virus isolate from a human on the Gulf Coast of Texas. J Clin Microbiol. 2004;42: 5375–5377. 10.1128/JCM.42.11.5375-5377.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ebel GD, Carricaburu J, Young D, Bernard KA, Kramer LD. Genetic and phenotypic variation of West Nile virus in New York, 2000–2003. Am J Trop Med Hyg. 2004;71: 493–500. [PubMed] [Google Scholar]
- 77.Moudy RM, Meola MA, Morin L-LL, Ebel GD, Kramer LD. A newly emergent genotype of West Nile virus is transmitted earlier and more efficiently by Culex mosquitoes. Am J Trop Med Hyg. 2007;77: 365–370. [PubMed] [Google Scholar]
- 78.Kilpatrick AM, Meola MA, Moudy RM, Kramer LD. Temperature, viral genetics, and the transmission of West Nile virus by Culex pipiens mosquitoes. PLoS Pathog. 2008;4: e1000092 10.1371/journal.ppat.1000092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Duggal NK, Bosco-Lauth A, Bowen RA, Wheeler SS, Reisen WK, Felix TA, et al. Evidence for co-evolution of West Nile virus and house sparrows in North America. PLoS Negl Trop Dis. 2014;8: e3262 10.1371/journal.pntd.0003262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Anderson JF, Main AJ, Cheng G, Ferrandino FJ, Fikrig E. Horizontal and vertical transmission of West Nile virus genotype NY99 by Culex salinarius and genotypes NY99 and WN02 by Culex tarsalis. Am J Trop Med Hyg. 2012;86: 134–139. 10.4269/ajtmh.2012.11-0473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vanlandingham DL, McGee CE, Klingler KA, Galbraith SE, Barrett ADT, Higgs S. Short report: Comparison of oral infectious dose of West Nile virus isolates representing three distinct genotypes in Culex quinquefasciatus. Am J Trop Med Hyg. 2008;79: 951–954. [PMC free article] [PubMed] [Google Scholar]
- 82.Danforth ME, Reisen WK, Barker CM. Extrinsic incubation rate is not accelerated in recent California strains of West Nile virus in Culex tarsalis (Diptera: Culicidae). J Med Entomol. 2015;52: 1083–1089. 10.1093/jme/tjv082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grubaugh ND, Smith DR, Brackney DE, Bosco-Lauth AM, Fauver JR, Campbell CL, et al. Experimental evolution of an RNA virus in wild birds: Evidence for host-dependent impacts on population structure and competitive fitness. PLoS Pathog. 2015;11: e1004874 10.1371/journal.ppat.1004874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Grubaugh ND, Weger-Lucarelli J, Murrieta RA, Fauver JR, Garcia-Luna SM, Prasad AN, et al. Genetic drift during systemic arbovirus infection of mosquito vectors leads to decreased relative fitness during host switching. Cell Host Microbe. 2016;19: 481–492. 10.1016/j.chom.2016.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McMullen AR, May FJ, Li L, Guzman H, Bueno R Jr, Dennett JA, et al. Evolution of new genotype of West Nile virus in North America. Emerg Infect Dis. 2011;17: 785–793. 10.3201/eid1705.101707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Añez G, Grinev A, Chancey C, Ball C, Akolkar N, Land KJ, et al. Evolutionary dynamics of West Nile virus in the United States, 1999–2011: Phylogeny, selection pressure and evolutionary time-scale analysis. PLoS Negl Trop Dis. 2013;7: e2245 10.1371/journal.pntd.0002245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bialosuknia SM, Tan Y, Zink SD, Koetzner CA, Maffei JG, Halpin RA, et al. Evolutionary dynamics and molecular epidemiology of West Nile virus in New York State: 1999–2015. Virus Evol. 2019;5: vez020 10.1093/ve/vez020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Paull SH, Horton DE, Ashfaq M, Rastogi D, Kramer LD, Diffenbaugh NS, et al. Drought and immunity determine the intensity of West Nile virus epidemics and climate change impacts. Proceedings of the Royal Society B. 2017; 10.1098/rspb.2016.2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Landesman WJ, Allan BF, Langerhans RB, Knight TM, Chase JM. Inter-annual associations between precipitation and human incidence of West Nile virus in the United States. Vector Borne Zoonotic Dis. 2007;7 10.1089/vbz.2006.0590 [DOI] [PubMed] [Google Scholar]
- 90.Shaman J, Day JF, Stieglitz M. Drought-induced amplification and epidemic transmission of West Nile virus in southern Florida. J Med Entomol. 2005;42: 134–141. 10.1093/jmedent/42.2.134 [DOI] [PubMed] [Google Scholar]
- 91.Wimberly MC, Lamsal A, Giacomo P, Chuang TW. Regional variation of climatic influences on West Nile virus outbreaks in the United States. Am J Trop Med Hyg. 2014;91: 677–684. 10.4269/ajtmh.14-0239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ruiz MO, Chaves LF, Hamer GL, Sun T, Brown WM, Walker ED, et al. Local impact of temperature and precipitation on West Nile virus infection in Culex species mosquitoes in northeast Illinois, USA. Parasit Vectors. 2010;3: 19 10.1186/1756-3305-3-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Davis JK, Vincent G, Hildreth MB, Kightlinger L, Carlson C, Wimberly MC. Integrating environmental monitoring and mosquito surveillance to predict vector-borne disease: Prospective forecasts of a West Nile virus outbreak. PLoS Curr. 2017; 10.1371/currents.outbreaks.90e80717c4e67e1a830f17feeaaf85de [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chuang TW, Wimberly MC. Remote sensing of climatic anomalies and West Nile virus incidence in the northern great plains of the United States. PLoS ONE. 2012;7: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ciota AT, Kramer LD. Vector-virus interactions and transmission dynamics of West Nile virus. Viruses. 2013;5: 3021–3047. 10.3390/v5123021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kilpatrick AM. Globalization, land use, and the invasion of West Nile virus. Science. 2011;334: 323–327. 10.1126/science.1201010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dunphy BM, Kovach KB, Gehrke EJ, Field EN, Rowley WA, Bartholomay LC, et al. Long-term surveillance defines spatial and temporal patterns implicating Culex tarsalis as the primary vector of West Nile virus. Sci Rep. 2019;9: 6637 10.1038/s41598-019-43246-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Andreadis TG. The contribution of Culex pipiens complex mosquitoes to transmission and persistence of West Nile virus in North America. Journal of American Mosquito Control Association. 2012;28: 137–151. [DOI] [PubMed] [Google Scholar]
- 99.Gibbs SEJ, Wimberly MC, Madden M, Masour J, Yabsley MJ, Stallknecht DE. Factors affecting the geographic distribution of West Nile virus in Georgia, USA: 2002–2004. Vector Borne Zoonotic Dis. 2006;6: 73–82. 10.1089/vbz.2006.6.73 [DOI] [PubMed] [Google Scholar]
- 100.Bowden SE, Magori K, Drake JM. Regional differences in the association between land cover and West Nile virus disease incidence in humans in the United States. Am J Trop Med Hyg. 2011;84: 234–238. 10.4269/ajtmh.2011.10-0134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kothera L, Godsey MS, Doyle MS, Savage HM. Characterization of Culex pipiens complex (Diptera: Culicidae) populations in Colorado, USA using microsatellites. PLoS ONE. 2012;7: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ciota AT, Chin PA, Kramer LD. The effect of hybridization of Culex pipiens complex mosquitoes on transmission of West Nile virus. Parasit Vectors. 2013;6: 305 10.1186/1756-3305-6-305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Giordano BV, Kaur S, Hunter FF. West Nile virus in Ontario, Canada: A twelve-year analysis of human case prevalence, mosquito surveillance, and climate data. PLoS ONE. 2017;12: e0183568 10.1371/journal.pone.0183568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dupuis AP 2nd, Marra PP, Kramer LD. Serologic evidence of West Nile virus transmission, Jamaica, West Indies. Emerg Infect Dis. 2003;9: 860–863. 10.3201/eid0907.030249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Estrada-Franco JG, Navarro-Lopez R, Beasley DW, Coffey L, Carrara AS, da Rosa A T, et al. West Nile virus in Mexico: Evidence of widespread circulation since July 2002. Emerging Infectious Diseases. 2003;9 10.3201/eid0912.030564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Komar N, Clark GG. West Nile virus activity in Latin America and the Caribbean. Revista Panamericana de Salud Pública. 2006;19: 112–117. 10.1590/s1020-49892006000200006 [DOI] [PubMed] [Google Scholar]
- 107.Elizondo-Quiroga D, Davis CT, Fernandez-Salas I, Escobar-Lopez R, Velasco Olmos D, Soto Gastalum LC, et al. West Nile Virus isolation in human and mosquitoes, Mexico. Emerg Infect Dis. 2005;11: 1449–1452. 10.3201/eid1109.050121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mattar S, Edwards E, Laguado J, González M, Alvarez J, Komar N. West Nile virus antibodies in Colombian horses. Emerg Infect Dis. 2005;11: 1497–1498. 10.3201/eid1109.050426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Morales MA, Barrandeguy M, Fabbri C, Garcia JB, Vissani A, Trono K, et al. West Nile virus isolation from equines in Argentina, 2006. Emerg Infect Dis. 2006;12: 1559–1561. 10.3201/eid1210.060852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bosch I, Herrera F, Navarro J-C, Lentino M, Dupuis A, Maffei J, et al. West Nile virus, Venezuela. Emerg Infect Dis. 2007;13: 651–653. 10.3201/eid1304.061383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pauvolid-Corrêa A, Morales MA, Levis S, Figueiredo LTM, Couto-Lima D, Campos Z, et al. Neutralising antibodies for West Nile virus in horses from Brazilian Pantanal. Mem Inst Oswaldo Cruz. 2011;106: 467–474. 10.1590/s0074-02762011000400014 [DOI] [PubMed] [Google Scholar]
- 112.Ulloa A, Ferguson HH, Méndez-Sánchez JD, Danis-Lozano R, Casas-Martínez M, Bond JG, et al. West Nile virus activity in mosquitoes and domestic animals in Chiapas, México. Vector Borne Zoonotic Dis. 2009;9: 555–560. 10.1089/vbz.2008.0087 [DOI] [PubMed] [Google Scholar]
- 113.Dudas G, Bedford T. The ability of single genes vs full genomes to resolve time and space in outbreak analysis [Internet]. bioRxiv. 2019. p. 582957 10.1101/582957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mann BR, McMullen AR, Guzman H, Tesh RB, Barrett ADT. Dynamic transmission of West Nile virus across the United States-Mexican border. Virology. 2013;436: 75–80. 10.1016/j.virol.2012.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Deardorff E, Estrada-Franco J, Brault AC, Navarro-Lopez R, Campomanes-Cortes A, Paz-Ramirez P, et al. Introductions of West Nile virus strains to Mexico. Emerg Infect Dis. 2006;12: 314–318. 10.3201/eid1202.050871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Osorio JE, Ciuoderis KA, Lopera JG, Piedrahita LD, Murphy D, Levasseur J, et al. Characterization of West Nile viruses isolated from captive American flamingoes (Phoenicopterus ruber) in Medellin, Colombia. Am J Trop Med Hyg. 2012;87: 565–572. 10.4269/ajtmh.2012.11-0655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fabbri CM, García JB, Morales MA, Enría DA, Levis S, Lanciotti RS. Complete genome sequences and phylogenetic analysis of two West Nile virus strains isolated from equines in Argentina in 2006 could indicate an early introduction of the virus in the Southern Cone. Vector Borne Zoonotic Dis. 2014;14: 794–800. 10.1089/vbz.2014.1588 [DOI] [PubMed] [Google Scholar]
- 118.Martins LC, Silva EVP da, Casseb LMN, Silva SP da, Cruz ACR, Pantoja JA de S, et al. First isolation of West Nile virus in Brazil. Mem Inst Oswaldo Cruz. 2019;114: e180332 10.1590/0074-02760180332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Anthony SJ, Garner MM, Palminteri L, Navarrete-Macias I, Sanchez-Leon MD, Briese T, et al. West Nile virus in the British Virgin Islands. Ecohealth. 2014;11: 255–257. 10.1007/s10393-014-0910-6 [DOI] [PubMed] [Google Scholar]
- 120.Mann BR, McMullen AR, Swetnam DM, Barrett ADT. Molecular epidemiology and evolution of West Nile virus in North America. Int J Environ Res Public Health. 2013;10: 5111–5129. 10.3390/ijerph10105111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Barrera R, Hunsperger E, Muñoz-Jordán JL, Amador M, Diaz A, Smith J, et al. First isolation of West Nile virus in the Caribbean. Am J Trop Med Hyg. 2008;78: 666–668. [PubMed] [Google Scholar]
- 122.Gubler DJ. The continuing spread of West Nile virus in the western hemisphere. Clin Infect Dis. 2007;45: 1039–1046. 10.1086/521911 [DOI] [PubMed] [Google Scholar]
- 123.Kopp A, Gillespie TR, Hobelsberger D, Estrada A, Harper JM, Miller RA, et al. Provenance and geographic spread of St. Louis encephalitis virus. MBio. 2013;4: e00322–13. 10.1128/mBio.00322-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Diaz A, Coffey LL, Burkett-Cadena N, Day JF. Reemergence of St. Louis Encephalitis Virus in the Americas. Emerg Infect Dis. 2018;24 10.3201/eid2412.180372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Quick J, Grubaugh ND, Pullan ST, Claro IM, Smith AD, Gangavarapu K, et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nature Protocols. 2017;12: 1261–1276. 10.1038/nprot.2017.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Metsky HC, Siddle KJ, Gladden-Young A, Qu J, Yang DK, Brehio P, et al. Capturing sequence diversity in metagenomes with comprehensive and scalable probe design. Nat Biotechnol. 2019;37: 160–168. 10.1038/s41587-018-0006-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gubler DJ, Campbell GL, Nasci R, Komar N, Petersen L, Roehrig JT. West Nile virus in the United States: Guidelines for detection, prevention, and control. Viral Immunology. 2000;13: 469–475. 10.1089/vim.2000.13.469 [DOI] [PubMed] [Google Scholar]
- 128.Rund SSC, Martinez ME. Rescuing hidden ecological data to tackle emerging mosquito-borne diseases. Available from: https://www.biorxiv.org/content/early/2017/08/09/096875.abstract. 2017; 096875. [cited 2019 June 11]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dudas G, Carvalho LM, Bedford T, Tatem AJ, Baele G, Faria NR, et al. Virus genomes reveal factors that spread and sustained the Ebola epidemic. Nature. 2017;544: 309–315. 10.1038/nature22040 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A depiction of the main interactive interface on Nextstrain to visualize the virus (A) phylogeny (time-resolved phylogenetic tree created with TreeTime, states listed by two-letter abbreviations, countries listed by three-letter abbreviations), (B) spread (inferred patterns of spread based on the tree, circle size is relative to the number of genomes that remained local), and (C) genetic diversity (amino acid diversity plotted as entropy and positioned by codon sequence). In the WNV genome, the protein abbreviations are as follows: C = capsid, prM = premembrane, E = envelope, NS1-5 = nonstructural protein 1–5. A live display, which can be used to select and zoom in on various features, can be found at nextstrain.org/WNV/NA?p=grid. WNV, West Nile virus.
(PDF)
An unrooted maximum likelihood WNV phylogenetic tree shows that there is little geographic structure (i.e., bush-like topology) during the early spread phase, as best demonstrated by the Texas (gold) and New York (purple) genomes often clustering together. This suggests that environment was highly conducive for transmission and that the virus could freely spread. The minimal mixing between California and New York genomes, however, suggests that geographic structure is starting to form as the virus transitioned into its endemic phase. Additional WNV sequencing throughout the US from the last 10 years will help to better understand if structure is starting to form and at what scale. States are listed by two-letter abbreviations and countries are listed by three-letter abbreviations. This “unrooted” tree view can be visualized on Nextstrain by toggling between the “Tree Options: Layout”. A live display can be found at nextstrain.org/WNV/NA?l=unrooted&m=div&p=full. WNV, West Nile virus.
(PDF)
A root-to-tip plot showing the divergence (substitutions per site) of sequenced WNV genomes (tips) from the inferred ancestral sequence (root) by the collection dates (shown in years) is used to estimate the evolutionary rate at approximately 4×10−4 substitutions/site/year. The tips are colored by increasing time. This “clock” view can be visualized on Nextstrain by toggling between the “Tree Options: Layout.” A live display can be found at nextstrain.org/WNV/NA?c=num_date&d=tree&l=clock. WNV, West Nile virus.
(PDF)
WNV genotype SW03 is defined by two amino acid substitutions, (A) nonstructural protein 4A (NS4A) A85T and (B) NS5-K314R. Branches (inferred ancestral genome) and tips (sequenced genome) are colored by amino acid at position (A) NS4A site 85 (nucleotide position 6721) and (B) NS5 site 314 (nucleotide position 8621). Both (A) NS4A-A85T and (B) NS5-K314R help to form a well-supported clade (yellow branches at the top) but also occur independently throughout the tree (yellow scattered throughout the lower half). All other alleles (nucleotide and amino acid changes) can be visualized using Nextstrain by entering the loci using the “Color By: genotype” function or by selecting a loci on the “Diversity” plot (i.e., S1C Fig). Live displays can be found at (A) nextstrain.org/WNV/NA?c=gt-NS4A_85 and (B) nextstrain.org/WNV/NA?c=gt-NS5_314. WNV, West Nile virus.
(PDF)
Since the emergence of WN02 (blue) and SW03 (green) in 2001, the genotypes continue to cocirculate in locations throughout the US For example, both genotypes were detected during the same years and vector species (A) in Maricopa County, Arizona [22] (B), throughout California [23], and (C) throughout New York [87]. Data from other studies can be visualized on Nextstrain by using the “Filter by Authors” function. Live displays can be found at (A) https://nextstrain.org/WNV/NA?c=lineage&f_authors=Hepp%20et%20al, (B) https://nextstrain.org/WNV/NA?c=lineage&f_authors=Duggal%20et%20al, and (C) https://nextstrain.org/WNV/NA?c=lineage&f_authors=Shabman%20et%20al. WNV, West Nile virus.
(PDF)
WNV is likely establishing persistent local transmission networks throughout the US, but this can be easily demonstrated from the 570 genomes available from New York (most generated by [87]). Multiple co-occurring transmission chains (branches) exist derived from either local evolution or separate (re-)introductions, but several persist locally for 5 or more years indicating that those viruses became established. Data from other states can be visualized on Nextstrain by using the “Filter by State” function. A live display can be found at nextstrain.org/WNV/NA?f_state=NY&d=tree. WNV, West Nile virus.
(PDF)