Abstract
Matrix-assisted ionization (MAI) is a recently developed ionization technique that produces multiply charged ions on either electrospray ionization (ESI) or matrix-assisted laser desorption/ionization (MALDI) platform without the need of high voltage or laser ablation. In this study, MAI has been coupled to a high resolution accurate mass (HRAM) hybrid instrument, the Orbitrap Elite mass spectrometer, with electron transfer dissociation (ETD) module for fast peptide and intact protein characterization. The softness of MAI process preserves labile post-translational modifications (PTM) and allows fragmentation and localization by ETD. Moreover, MAI on ESI platform allows rapid sample preparation and analysis (~ 1 min/sample) due to the easiness of sample introduction. It significantly improves the throughput compared to ESI direct infusion and MAI on MALDI platform, which usually takes more than 10 min/sample. Intact protein standards, protein mixtures and neural tissue extracts have been characterized using this instrument platform with both full MS and MS/MS (CID, HCD and ETD) analyses. Furthermore, the performances of ESI, MALDI and MAI on both platforms have been tested to provide a systematic comparison among these techniques. With improved ETD performance and PTM analysis capabilities, we anticipate that the HRAM MAI-MS with ETD module will offer greater utilities in large molecule characterization with enhanced speed and coverage. These advancements will enable promising applications in bottom-up and top-down protein analyses.
Keywords: matrix-assisted ionization, post-translational modification, electron transfer dissociation, high resolution accurate mass
Graphical Abstract
Introduction
The development of matrix-assisted laser desorption/ionization (MALDI) [1] and electrospray ionization (ESI) [2] techniques in late 1980s have revolutionized the field of biological mass spectrometry. These soft ionization techniques allow ionizing biomolecules without extensive fragmentation. While MALDI and ESI are extensively used in biomolecule analyses, some limitations of these techniques have been noticed, such as limited fragmentation efficiency for singly charged ions produced by MALDI and difficulty in preserving spatial information during tissue analysis by ESI [3].
Several novel ionization techniques have been developed to overcome the limitations to MALDI and ESI. Matrix-assisted laser desorption electrospray ionization (MALDESI) combines the benefit of MALDI and ESI, which generates ESI-like multiply charged ions on sample surface with matrix [4, 5]. Desorption electrospray ionization (DESI) utilizes electrospray generated charged droplets and solvent ions to ionize analyte from surface [6, 7]. Electrospray-assisted laser desorption ionization (ELDI) ionizes biomolecules from surface with laser desorption and post-ionization electrospray [8–12]. Solid-substrate ESI provides a cost-efficient approach for high throughput analysis using wooden tips or C18 pipette tips [13, 14]. Other ambient surface analysis techniques, such as laser-ablation electrospray ionization (LAESI) [15], liquid extraction surface analysis (LESA) [16] and direct analysis in real time (DART) [17], have also been reported and widely used in biomolecule and pharmaceutical analyses.
Introduced in 2013 by Trimpin and Inutan [18, 19], matrix-assisted ionization (MAI) is considered as a “magic” ionization technique [20] that can produce multiply charged ions on either MALDI or ESI platform (MAI (MALDI) or MAI (ESI)) without laser ablation or high voltage. The only requirements for the ionization process are the inherent vacuum of the instrument and a volatile small molecule matrix, such as 3-nitrobenzonitrile (3-NBN). MAI has several advantages compared to conventional ionization techniques. Compared to MALDI, which predominantly generates singly charged ions, MAI produces multiply charged ions. Compared to ESI, MAI offers higher throughput and less sample carryover or contamination [21]. Compared to laserspray ionization (LSI), which is also a novel ionization technique for generating multiply charged ions [22–24], MAI produces higher charge state ions without the requirement of laser ablation. Therefore, MAI efficiently improves the throughput and expands the mass detection range compared to MALDI and LSI on high resolution accurate mass (HRAM) mass analyzers, such as Orbitrap, which usually has an upper m/z limit of 4,000 or 6,000. In previous studies on MAI process, the influences of source temperature, matrix, solvent, sample introduction route were studied on various instruments. It was also applied to the analyses of various samples, such as small molecules (drugs, pesticides, dyes), peptides, intact proteins as well as tissue extracts and tissue sections, for quantitative and qualitative analyses[18, 19, 24, 25, 3, 26, 27]. Woodall et al. demonstrated MAI-MS analysis of a wide range of molecules with automated sample introduction and studied the relationship between source temperature, sublimation time and ion abundance [21]. A triboluminescence-based mechanism was proposed to explain the production of highly charged ions during the MAI process. The 3-NBN matrix produces strong dinitrogen discharge emission during the fracturing of crystal under vacuum and produced charged particles upon sublimation [19, 28].
Our previous study demonstrated the potential of MAI on analyzing peptides with labile PTMs on the MALDI-LTQ-Orbitrap XL platform to reduce in-source fragmentation [3]. During MALDI-MS acquisition, high intensities of dephosphorylated and deglycosylated peptides were observed with very low intensity of intact peptides. Under MAI condition, high intensities of intact peptides were observed, with only small amount of neutral loss ions. However, when we performed CID and HCD fragmentations on the intact peptide ions, the labile PTM groups were not preserved on fragment ions, suggesting limitations of these slow heating fragmentation techniques (CID and HCD) for more comprehensive structural analysis of peptides with labile PTMs. In contrast, electron capture dissociation (ECD) [29] and electron transfer dissociation (ETD) [30], which produce c and z ions upon the reaction of a multiply protonated peptide with a low energy electron, can potentially preserve the labile PTM groups. ETD has been widely applied to commercially available quadrupole ion trap, linear 2D quadrupole ion trap and hybrid quadrupole time-of-flight (Q-TOF) and LTQ-Orbitrap instruments [31]. The Orbitrap Elite mass spectrometer is a HRAM hybrid instrument with dual-pressure linear ion trap and Orbitrap mass analyzer, which enables superior resolution of up to 240,000 at m/z 400. Moreover, multiple fragmentation techniques, including CID, HCD and ETD, are available on this instrument, which offers versatility of structural elucidation [32].
Herein, we coupled MAI onto the HRAM Orbitrap Elite mass spectrometer with ETD module for high throughput peptide and protein analyses. In this study, we demonstrated utility and evaluated performance of the MAI-HRAM MS system using peptide and protein standards, protein mixture, tissue extract and protein digest for full MS and different types of MS/MS (CID, HCD and ETD). Moreover, the performances of four ionization techniques MAI (ESI), MAI (MALDI), ESI and MALDI were compared. We anticipate that the HRAM MAI-MS with ETD module could be widely applied in large molecule characterization, especially for peptides and proteins with labile PTMs. These advancements, when coupled with off-line separation methods will facilitate both bottom-up and top-down high throughput protein characterization.
Materials and Methods
Materials
All standards and reagents were used without additional purification. Acetonitrile (ACN), methanol (MeOH), ethanol (EtOH), formic acid (FA), acetic acid (AA), urea and ammonium bicarbonate and water were purchased from Fisher Scientific (Pittsburgh, PA). Iodoacetamide (IAA), 3-NBN, α-cyano-4-hydroxycinnamic acid (CHCA), cytochrome C (bovine heart), lysozyme (chicken egg white), alpha-casein (bovine milk), myoglobin (equine heart) and histone (calf thymus) were purchased from Sigma Aldrich (St. Louis, MO). Neuropeptide Y (human, rat) was purchased from American Peptide Company (Sunnyvale, CA, now Bachem, Torrance, CA). Phosphopeptide standard kit was purchased from AnaSpec (Fremont, CA). Glycosylated erythropoietin (EPO) 117–131 was purchased from Protea Biosciences Group, Inc (Morgantown, WV). Dithiothreitol (DTT) and sequencing grade modified trypsin were purchased from Promega (Madison, WI). The Ni-NTA magnetic agarose beads were purchased from QIAGEN (Valencia, CA).
Sample preparation
Peptide and protein stock solutions were prepared by dissolving standards in 50% ACN solution at 10 mg mL−1. Serial dilutions of 1 mg mL−1, 100 μg mL−1, 10 μg mL−1 and 1 μg mL−1 standards were diluted from the stock solution with 50% ACN solution. Samples were stored in −20 °C until analysis.
Animal experiments were conducted following institutional guidelines (University of Wisconsin-Madison IACUC). Perfused brain tissue of female Sprague-Dawley rats were homogenized manually in acidified MeOH solution (MeOH:H2O:AA (v/v/v) 90 : 9: 1) for peptide and protein extractions. The tissue was homogenized for approximately 5 min until no visible tissue chunk could be observed. The homogenized tissue was sonicated for 1 min followed by centrifugation (16,100 rcf, 10 min). The supernatant was transferred to a new micro-centrifuge tube and dried down in a speed vacuum concentrator. The tissue extract was reconstituted in water (with 0.1% FA) and cleaned up with C18 ZipTip (EMD Millipore Corporation, Darmstadt, Germany) according to manufacturer’s instruction. The eluted fraction was dried down and reconstitute in 50% ACN for MS analysis.
MAI matrix, 3-NBN, was prepared as described by Inutan and Trimpin [18, 19]. Briefly, 50 μL ACN was added to 10 mg 3-NBN, the mixture was vortexed until 3-NBN was completely dissolved. A volume of 150 μL 50% ACN was then added to the mixture. The matrix solution was prepared freshly before each experiment. Before MS analysis, 1 μL analyte was mixed with 1 μL matrix solution on Parafilm (Bemis, Neenah, WI). One μL of the mixture was loaded onto a 10 μL gel loading pipette tip and waited until the mixture was crystalized before MS analysis. CHCA matrix, which was used in comparison MALDI experiment, was prepared by dissolving 10 mg of CHCA in 1 mL solution of ACN:EtOH:H2O:FA (v/v/v/v) 84:13:2.997:0.003. The sample and matrix were premixed at a 1:1 ratio and 1 μL mixture was spotted onto MALDI plate.
Protein digestion and IMAC enrichment of phosphopeptides
One mg of alpha-casein standard was dissolved in 400 μL digestion solution (8M urea, 50 mM ammonium bicarbonate) to a final concentration of 2.5 mg/mL. The protein was reduced with 10 mM DTT at room temperature for 1 hr and alkylated with 15 mM IAA in dark at room temperature for 1 hr. Extra DTT was added to the solution to make a final concentration of 20 mM in order to quench the alkylation reaction. Ammonium bicarbonate solution (2.2 mL 50 mM) was added to the sample in order to dilute urea (less than 1 M). Trypsin (20 μg) was reconstituted in 200 μL 50 mM ammonium bicarbonate and added to protein solution. The mixture was incubated in 37 °C water bath for 18 hours and dried down in a speed vacuum concentrator.
The digested peptide was reconstituted in 500 μL 0.1% FA/H2O and desalted using a Sep-Pak C18 cartridge (Waters, Milford, MA) according to manufacturer’s instruction. The samples were dried down in a speed vacuum concentrator and stored in freezer. Prior to immobilized metal affinity chromatography (IMAC) for phosphopeptide enrichment, 500 μL magnetic Ni-NTA magnetic agarose beads suspension was pipetted into a 2 mL tube and used with a magnetic stand to separate beads from solvent. The beads were washed with 1 mL H2O for 3 times and shaken in 1 mL EDTA (pH ~8) solution for 30 min. The beads were then washed with 1 mL H2O for 5 more times and shaken in 1 mL 100 mM FeCl3 for 30 min. The beads were then washed with 1 mL ACN:H2O:TFA (v/v/v) 80:19.85:0.15 for 4 times. The sample was re-suspended in 1 mL ACN:H2O:TFA (v/v/v) 80:19.85:0.15 and quantitatively transferred to the beads. The mixture was shaken for 30 min and the supernatant was saved as phosphopeptide depleted sample. The beads were washed again with 1 mL ACN:H2O:TFA (v/v/v) 80:19.85:0.15 and the supernatant was combined with the phosphopeptide depleted sample. The phosphopeptides were eluted with 100 μL ACN:H2O:NH4OH (v/v/v) 50:49.3:0.7 twice (1 min vortex for each wash) and added to a tube which was pre-loaded with 50 μL of ACN:FA (v/v) 96:4. Both fractions were dried in a speed vacuum concentrator and stored in −80 °C until analysis [33].
Mass spectrometry
The Orbitrap Elite hybrid ion trap-Orbitrap mass spectrometer with NanoSpray Flex ion source (Thermo Scientific, Bremen, Germany) was used for data acquisition. The instrument was controlled with Tune Plus software (Thermo Scientific, Bremen, Germany). Spray voltage was set to be 0 kV and the capillary temperature was set to be 50 °C for all MAI acquisitions. As MAI is a rapid sublimation process, the instrument parameters and file information (name and location) must be prepared before acquisition starts. The gel loading tip with sample/matrix mixture was quickly brought to the inlet of ion transfer tube immediately after acquisition started. The acquisition was stopped after no visible analyte signal could be observed on the real time spectrum. NanoESI data was acquired by direct infusing samples to the mass spectrometer at a rate of 0.3 μL/min. The spray voltage was set to be 1.9 kV and capillary temperature was 250 °C. All data on the Orbitrap Elite was acquired with resolution of 120,000 (m/z 400) with automatic gain control (AGC) target at 1e6 and maximum injection time at 300 ms. The normalized collision energies (NCE) of CID, HCD and the activation time for ETD were optimized for each sample and each charge state, which would be specified in the result. The resulting spectra were processed with XCalibur (Thermo Scientific, Bremen, Germany). The MS/MS spectra were compared with the theoretical mass list generated by MS-Product protein of ProteinProspector (UCSF Mass Spectrometry Facility, San Francisco, CA) and annotated manually.
In the comparison study, the MALDI and MAI (MALDI) spectra were acquired on MALDI-LTQ-Orbitrap XL mass spectrometer (Thermo Scientific, Bremen, Germany), which is also a hybrid ion-trap-Orbitrap mass spectrometer but with MALDI source. The instrument was controlled with Tune Plus software and the data was processed with Xcalibur software. The MALDI-TOF full scan spectrum of histone mixture was acquired on UltrafleXtreme II MALDI TOF/TOF mass spectrometer (Bruker, Billerica, MA) with linear positive mode at m/z 3,000 – 30,000. Laser size was set at “4_large” and laser energy was set to be 80%. Ten thousand laser shots were accumulated. The resulting spectrum was processed with flexAnalysis to perform smooth and baseline subtraction.
Results and discussion
In this study, a HRAM MAI-MS platform was set up on the ESI-LTQ-Orbitrap (Orbitrap Elite) system with ETD module for high throughput top-down and bottom-up analyses of protein standards and mixtures. Detailed comparisons among four ionization techniques: MAI (ESI), MAI (MALDI), ESI and MALDI were performed for better understanding of their properties.
Instrument setup
The ESI-LTQ-Orbitrap (Orbitrap Elite) system was quickly switched to MAI-MS mode by simply removing the emitter tip holder on the Nanospray flex ion source (Figure 1). The capillary temperature was set to be 50 °C and the voltage was set to be 0 kV. If the system was used for ESI experiments before, an extra half-hour was required for the system to cool down. As the relationship between sublimation time, temperature and ion abundance were carefully examined and optimized in the study published by Woodall et al. [21], no further optimization was performed in this study. The capillary temperature of 50 °C was chosen as a good compromise of sensitivity and acquisition time according to Woodall et al., sublimation time was around 20 seconds while ion abundance was at a relatively high level (around 1E6) at 50 °C. A 10 μL pipette with a gel loading pipette tip was used for sample introduction (Figure 1a). The matrix/analyte mixture (0.5 μL to 2 μL), which was crystalized on top of the pipette tip, was attached to the inlet of ion transfer tube for sample introduction (Figure 1 b&c). The matrix/analyte mixture was usually sublimated within 5 min (usually 30 sec) after attaching to the ion transfer tube. Due to the fast sublimation time of the analyte, the instrument was prepared with all parameters entered and data acquisition turned on before sample introduction. The ion signal was monitored instantaneously on the Tune Plus software. The data acquisition was stopped after no ion signal could be observed.
Figure 1. Instrument setup of MAI on Orbitrap-Elite platform with Nanospray flex ion source.
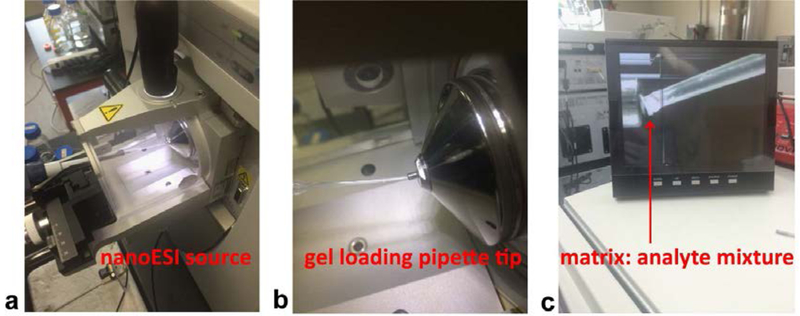
A 10 μL pipette with a gel loading pipette tip was used for sample introduction. The gel loading tip was directly attached to the inlet of ion transfer tube which maximized sample transfer. (a) Picture of the Nanospray flex ion source with pipette inserted for sample introduction. (b) Zoom in image of gel loading pipette tip attached to the inlet of ion transfer tube. (c) Picture of side camera showing the inlet of ion transfer tube.
HRAM MAI-MS full scan and MS/MS analysis of peptide and protein standards and mixtures
HRAM MAI full MS analyses on the Orbitrap Elite platform were performed on both standards and mixtures. Neuropeptide Y, which is a mid-size peptide with molecular weight of 4269.08 Da, was detected with charge states from 3 to 7 (Figure 2a). The lower limit of detection (LLOD), which was determined by the lowest concentration detected with a signal to noise (S/N) ratio above 3, was tested to be 5 nM (5 fmol introduced). Intact protein standards, including cytochrome C (12360.97 Da, Figure 2b), lysozyme (14295.81, Figure 2c) and myoglobin (16952.30, Figure 2d) were also detected on the system with very spread out charge states. The LLODs of cytochrome C, lysozyme and myoglobin were determined to be 20.2 nM (20.2 fmol introduced), 75 pM (75 amol introduced) and 2.95 μM (2.95 pmol introduced) respectively. Myoglobin was the largest molecule detected on this instrument platform, which is consistent with the MAI study on MALDI-LTQ-Orbitrap system [3]. The sensitivity of this system was similar to Hoang’s study [34], which less than 50 amol of angiotensin II was detected on the Orbitrap Exactive system.
Figure 2. HRAM full scan spectra of intact peptide and protein standards: 1.0 μM neuropeptide Y (a), 1.0 μM cytochrome C (b), 1.5 μM lysozyme (c) and 3.0 μM myoglobin (d).
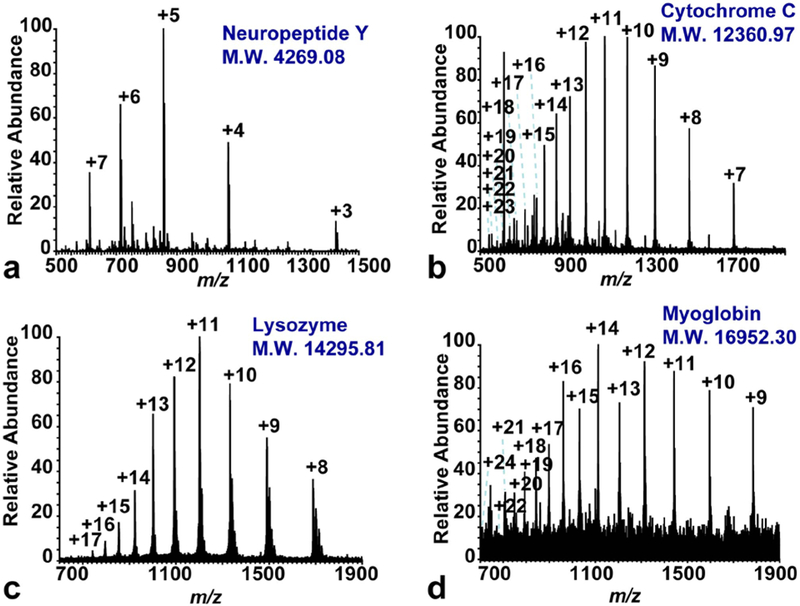
The molecular weight (M.W.) of each standard was shown in the figure. The m/z range of each spectra was chosen based on each sample in order to reveal all detected charge states.
A comparison of MAI (ESI/Orbitrap Elite), MAI (MALDI-LTQ-Orbitrap), ESI (Orbitrap Elite) and MALDI (MALDI-LTQ-Orbitrap) ionization techniques was performed (see Electronic Supplementary Material Fig. S1). The sampling throughputs varied among these ionization methods. The MAI (ESI) underwent a fast sublimation process and could achieve high-throughput analysis with approximately 1 min/sample. The MAI (MALDI) was also a fast sublimation process, but extended time periods were required for inserting and taking out plate as well as pumping down of the MALDI chamber. Therefore, it usually took approximately 10 min/sample on the MAI (MALDI) system as well as on the MALDI system. Direct infusion based ESI analysis also required about 10 min/sample or even longer, as the fluid system needed to be flushed with solvent when switching samples to minimize sample carryover.
Lysozyme standard was used to compare the charge state, noise level and sensitivity of each ionization mode. MAI (ESI) produced the broadest charge state, from 8 to 17 (see Electronic Supplementary Material Fig. S1a); MAI (MALDI) and ESI produced the same charge states, from 8 to 12 (see Electronic Supplementary Material Fig. S1 b&c); no lysozyme ion could be detected by MALDI (see Electronic Supplementary Material Fig. S1d) as the molecular weight of lysozyme exceeded the detection limit of the mass analyzer assuming only singly charged ion was produced. The MAI (MALDI) platform produced very high noise signal; MAI (ESI) produced less noise signal and the ESI had the least. The LLODs of MAI were determined to be 75 pM on the ESI/Orbitrap Elite platform and 175 nM on the MALDI-LTQ-Orbitrap platform. The significant differences of LLOD on these two platforms could be caused by the means of sample introduction and the differences in source pressure. On the ESI/Orbitrap Elite platform, the matrix/analyte mixture was directly attached to the inlet of the MS with no gap, allowing high transmission rate of the analyte into the instrument. Moreover, the matrix/analyte was stored at atmospheric pressure with a very slow sublimation rate before sample introduction, thus there was very little sample loss in the process. In contrast, on the MALDI-LTQ-Orbitrap system, the matrix/analyte was crystalized on the MALDI target plate, which was inserted into an intermediate vacuum chamber. Due to the gap between the MALDI target plate and the ion optics, there might be sample loss in this process. Moreover, when inserting the MALDI plate, the matrix/analyte was exposed to intermediate vacuum environment for a few minutes, which could stimulate the sublimation process before starting data acquisition. Therefore, a significant difference in sensitivity between the MAI (ESI) and MAI (MALDI) platform was observed.
MAI-MS/MS analyses were performed using neuropeptide Y standard. Multiple fragmentation types, including CID, HCD and ETD, were compared and contrasted on the +6 charged neuropeptide precursor ion (Figure 3). The NCEs of CID and HCD as well as the activation time (ms) of ETD were optimized and annotated in the figure. The CID and HCD MS/MS spectra (Figure 3 a&b) revealed incomplete fragmentation. In contrast, the ETD MS/MS spectrum revealed nearly complete sequence coverage (Figure 3c). As annotated on the spectrum and on the sequence, most of the c and z ions were detected except for the ones next to a proline residue which could not be cleaved by ETD. From a mechanistic standpoint, the fragmentation of CID and HCD relies on randomized amide bond protonation, which is difficult to achieve for larger peptides as Arg residues tend to inhibit the random protonation [35]. In contrast, the ETD process does not depend on amide bond protonation, which promotes more uniform fragmentation along the peptide backbone and achieve high sequence coverage [30].Therefore, this experiment showed evidence to suggest that ETD can improve sequence coverage, which made it applicable towards intact protein analysis.
Figure 3. MAI CID (a), HCD (b) and ETD (c) MS/MS spectra for +6 charged neuropeptide Y at m/z 712.86.
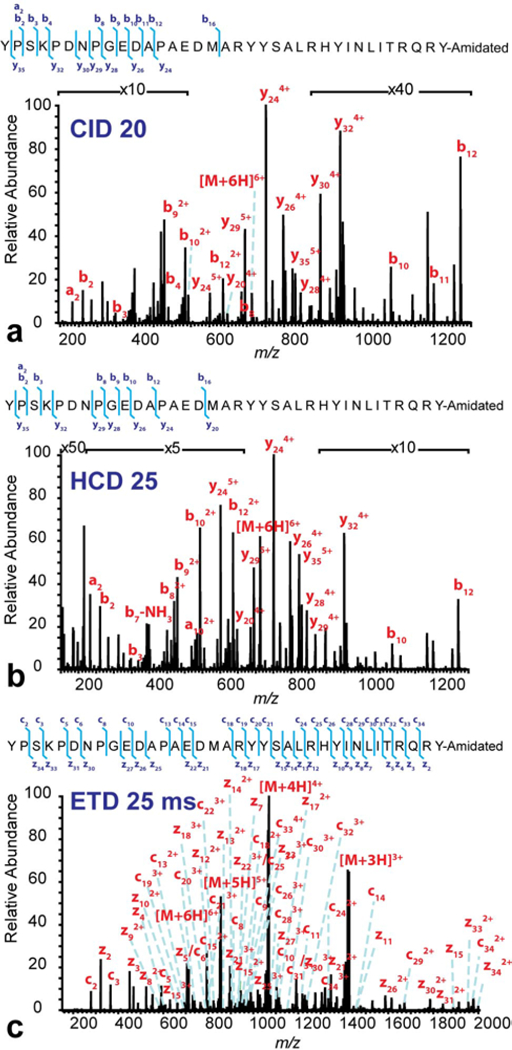
The NCEs for CID and HCD as well as the reaction time for ETD were labeled in the figure. The sequence of neuropeptide Y was shown in each figure with the detected fragment ions labeled.
The HRAM MAI-MS on the Orbitrap Elite system could also be used to analyze protein mixtures (Figure 4), such as histone mixture (Figure 4 a&b) and rat brain protein extract (Figure 4c). A full scan spectrum of histone mixture was acquired on the MALDI-TOF system with m/z 5,000–25,000 to show the relative abundance and mass of each histone molecule. In the MALDI-TOF spectrum, H4 had the highest intensity, followed by H2a and H2b. A very small peak of H3 was detected and no H1 peak could be detected. Multiply charged histone molecules were detected on the MAI (ESI) platform: H4 molecule was detected with charge states from 7 to 15; H2a was detected with charge state from 9 to 13 and H2b was detected with charge states of 9 and 10. No H1 and H3 molecule was detected due to their low abundances. The same analyte was tested on the MAI (MALDI) platform, but no signal was detected (data not shown), presumably due to the lower sensitivity of the MAI (MALDI) platform. Rat brain protein extract was used as a complex sample to further test the MAI (ESI) system. Several abundant proteins were detected in the mixture with various charge states, including thymosin beta-4 (a), ATP synthase-coupling factor (b), ubiquitin (c), acyl-CoA binding protein (d), cytochrome C oxidase subunit 6b (e), mitochondria import inner membrane translocase (f), superoxidase dismutase [Cu-Zn] (g). The proteins were identified by accurate mass matching and a lot of multiply charged peaks in the spectrum were yet to be identified. The protein name, theoretical mass, experimental mass and mass error were listed in Electronic Supplementary Material Table S1. According to a previous study by Ye et al. [36], rat brain proteins detected by MALDI-TOF platform were similar to the ones detected on the MAI (MALDI) platform. Abundant proteins such as thymosin beta-4, acyl-CoA-binding protein, ubiquitin, cytochrome C oxidase subunit 6b and ATP synthase-coupling factor 6 were detected on both platforms.
Figure 4. Full scan spectra of protein mixtures: MALDI-TOF spectrum of histone mixture, 10 mg/mL (a), HRAM MAI-Orbitrap spectrum of histone mixture, 1 mg/mL (b) and HRAM MAI-Orbitrap spectrum of rat brain protein extract.
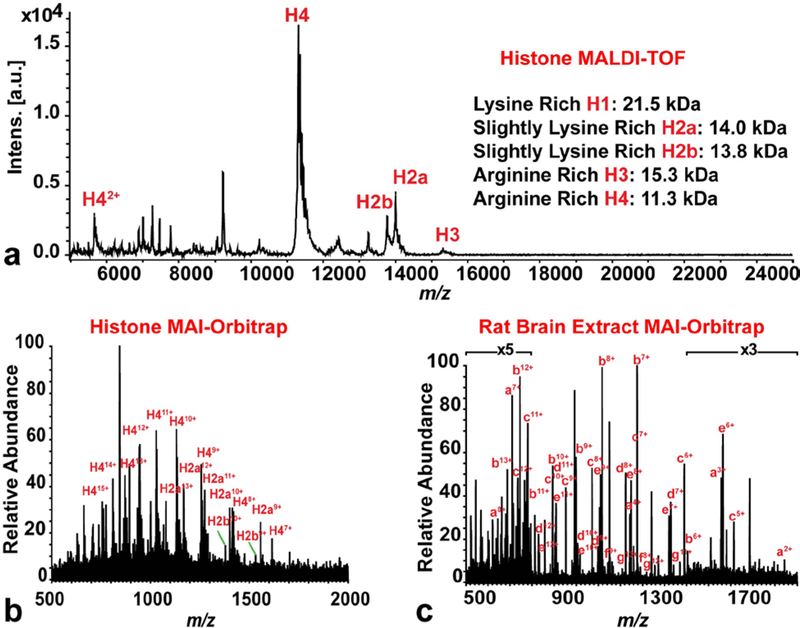
The masses of histone molecules (H1, H2a, H2b, H3 and H4) were listed in the figure. In the spectrum of rat brain extract, single letter code was used to annotate the spectrum. Each letter represents a single protein (identified by accurate mass matching): a: thymosin beta-4, b: ATP synthase-coupling factor, c: ubiquitin, d: Acyl-CoA binding protein, e: cytochrome C oxidase subunit 6b, f: mitochondria import inner membrane translocase, g: superoxidase dismutase [Cu-Zn]. Usually, more than one charge states of one protein were observed in the spectrum.
PTM analysis of glycopeptide and phosphopeptides
The characterization of labile PTMs, such as glycosylation and phosphorylation, presents unique challenges for MS research. The modification groups could be easily cleaved from the peptide backbone during the ionization or fragmentation process and could lead to mis-identification and often become an unsolvable piece of puzzle for peptide mapping. Due to the softness of MAI, we anticipated that it could minimize in-source fragmentation. Our previous study demonstrated MAI’s potential in analyzing labile PTM on the MALDI-LTQ-Orbitrap platform at full MS level: only a small peak corresponding to the doubly charged neutral loss ion was detected. However, the slow heating fragmentation techniques (CID and HCD) prevented us from further localizing the PTM groups on the peptide backbone, as most of them were cleaved first from the peptide backbone before breaking the peptide bonds [3]. Another study published by Marshall et al. demonstrated promising data on sequencing phosphorylated peptide standard using MAI [38].
This puzzle of PTM analysis could be solved on the MAI-MS Orbitrap Elite platform with ETD module. A glycopeptide standard EPO (117–131) and a phosphopeptide standard UOM9 (phosphorylated PKC substrate) were tested on this system (Figure 5, Figure S2 & S3). The HRAM MAI-MS full scan (Figure 5a) revealed most intact EPO peptides, with some singly and doubly charged neutral loss ions. No triply charged neutral loss ion was detected. The triply charged intact ion [M+3H]3+ was selected for MS/MS analysis for structure elucidation and PTM localization. In the CID (Figure 5b) and HCD (Figure 5c) spectra, only a few b and y ions were detected and the GalNAc group was cleaved from most of the detected ions (except for y7 and y8 ions). The information acquired here was not sufficient to localize the GalNAc group. In contrast, all the theoretical c and z ions were detected in the ETD spectrum (Figure 5d) with the GalNAc group preserved, allowing accurate PTM localization. Similar trend was observed for phosphopeptide standard UOM9 (see Electronic Supplementary Material Fig. S2). Peptides with charge states from 2 to 5 were detected in the spectrum with no evident of neutral loss ion (see Electronic Supplementary Material Fig. S2a). Both CID and HCD had incomplete fragmentations with only a few b and y ions detected. In contrast to EPO, most of the phosphate groups were still preserved to the respective b and y ions after CID (see Electronic Supplementary Material Fig. S2b) and HCD (see Electronic Supplementary Material Fig. S2c) fragmentation, as the phosphate group is not as labile as the GalNAc group. In the ETD spectrum, all theoretical c and z ions were detected with phosphate group preserved, allowing accurately localizing the phosphorylation position (see Electronic Supplementary Material Fig. S2d).
Figure 5. MAI full MS (a), CID (b), HCD (c) and ETD (d) spectra of the glycopeptide EPO (117–131).
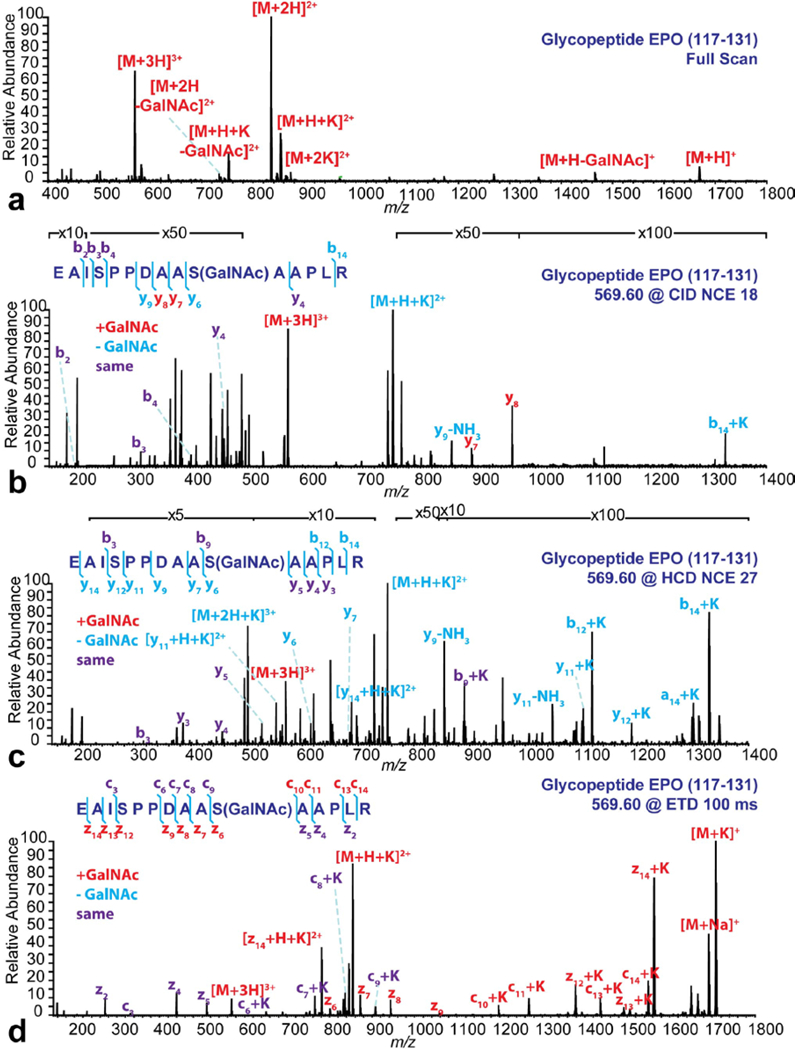
The detected ion species on the full scan were annotated on the spectrum. The MS/MS analyses were all performed on the [M+3H]3+ ions. NCEs or activation time were labeled on the figure. The detected fragments under each condition were annotated on the spectra and listed on the peptide sequence. The red letters represent the fragments with GalNAc group preserved, the blue letters represent the fragments with GalNAc group cleaved and the purple letters represent the fragments before GalNAc group.
A comparison among four ionization modes on analyzing the glycopeptide EPO was illustrated in Figure S3. Neutral loss from singly charged ion was detected in MAI (ESI), MAI (MALDI) and MALDI conditions. The neutral loss peaks became less intense with the charge state increased. The neutral loss peak was no longer detectable for triply charged ion in MAI (ESI) condition and for doubly charged ion in MAI (MALDI) condition. ESI had the best performance in preserving the GalNAc group, as neutral loss peaks could not be detected for any charge state.
To further test the potential of the HRAM MAI-MS system on bottom-up protein analysis, a phosphoprotein standard, alpha-casein, was enzymatically digested and IMAC enriched for phosphopeptide analysis (Figure 6). The phosphopeptide depleted fraction (Figure 6a) and the phosphopeptide enriched fraction (Figure 6b) were analyzed by MAI-MS. The peptides were identified and annotated by accurate mass matching (see Electronic Supplementary Material Table S2). No phosphopeptide was detected in the phosphopeptide depleted fraction, while a lot of phosphopeptides were detected in the phosphopeptide enriched fraction.
Figure 6. HRAM MAI-MS detection of the tryptic digested alpha-casein after IMAC enrichment: (a) phosphopeptide-depleted fraction and (b) phosphopeptide enriched fraction.
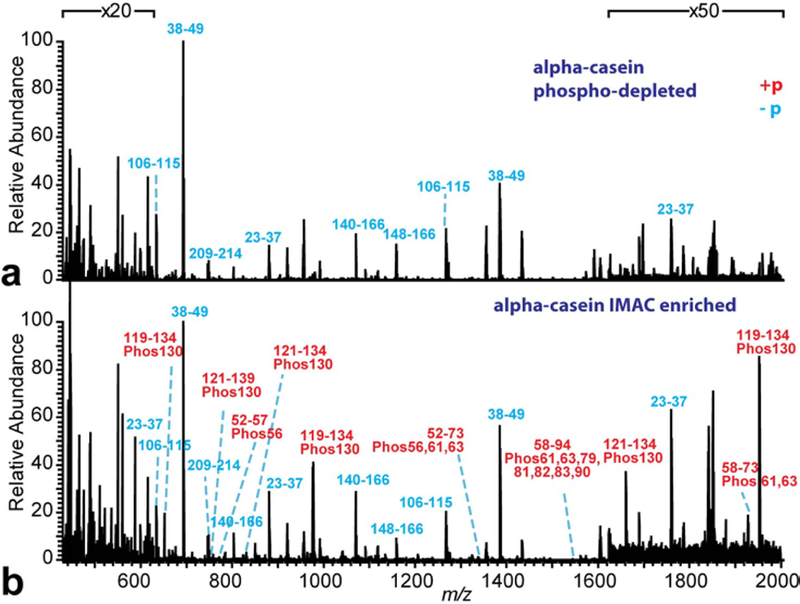
The spectra were annotated by accurate mass matching. The blue letters represent the peptide without phosphorylation and the red letters represent the peptide with phosphorylation.
Challenges
In this study, a HRAM MAI-MS platform was developed on the Orbitrap Elite system with ETD module for high throughput top-down and bottom-up analyses. Even though both full MS and MS/MS were successfully performed on this platform for both standards and complex tissue mixtures, several challenges remained. First of all, the matrix/analyte samples were usually sublimated within 5 min. This limited the researchers from performing long acquisitions, such as data dependent acquisitions or several parallel targeted MS/MS, which required pre-designed instrument methods. Secondly, although the MAI process does not require any high-energy process, such as high voltage and laser ablation, it still could not completely avoid in-source or post-source fragmentation for labile PTMs (Figure 5 and Figure S3). We considered the strong dinitrogen discharge emission during the triboluminescent process could cause the dissociation of labile PTM groups during the MAI process. Thirdly, on-line coupling MAI-MS with separation methods, such as capillary electrophoresis or liquid chromatography, could be challenging. This prevents the platform from performing in depth proteomics analysis from complex mixture. Lastly, in comparison to MAI (MALDI), in situ tissue analysis on the MAI (ESI) platform is challenging. Extending the sublimation time while preserving the performance of the MAI (ESI) platform will be further investigated and explored in the future.
Conclusions
A HRAM MAI-MS method on the Orbitrap Elite platform was developed in this study with improved ETD analysis and labile PTM mapping capability. A volatile matrix, 3-NBN, was premixed with analyte and a gel loading pipette tip was used for sample introduction. The ionization process requires no voltage or laser and could be accomplished in a high throughput fashion (1 min/sample). Intact peptide, protein, protein mixture and tissue extract were analyzed on this system. Various dissociation types, including CID, HCD and ETD, were used for sequence elucidation. Labile PTM analyses of phosphopeptide and glycopeptide were achieved on this platform due to the softness of MAI and the coupling of ETD module. Compared to MAI (MALDI), the MAI (ESI) platform provides high sensitivity, better performance for protein mixture analysis and significantly higher throughput. Compared to direct infusion based ESI analysis on the same instrument, the MAI (ESI) platform has also significantly improved the throughput with reduced sample consumption and elimination of carrying over. Although challenges still exist, this instrument platform provides a promising alternative to the conventional MALDI and ESI instrument platforms and could be beneficial for large molecule characterization, and eventually coupled with off-line separation methods for both bottom-up and top-down proteomics and biopharmaceutics analyses.
Supplementary Material
Acknowledgements
The authors would like to acknowledge Dr. Sarah Trimpin’s laboratory at Wayne State University for the inspiration and Dr. Robert Thorne’s laboratory at University of Wisconsin-Madison for providing rat tissue samples. Dr. Xuefei Zhong, Dr. Xueqin Pang and Dr. Dustin Frost provided helpful discussion and assisted with instrument maintenance for this project. The instrument was purchased through the funding support from NIH S10 RR029531. This work was supported by NIH NIDDK R01DK071801 and NIMH 1R56MH110215. C.L. acknowledges an NIH-supported Chemistry Biology Interface Training Program Predoctoral Fellowship (grant number T32-GM008505) and an NSF Graduate Research Fellowship (DGE-1256259). L.L. acknowledges a Vilas Distinguished Achievement Professorship and a Janis Apinis Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy.
Footnotes
Compliance with Ethical Standards
The authors declare that they have no conflict of interest.
References
- 1.Tanaka K, Waki H, Ido Y, Akita S, Yoshida Y, Yoshida T et al. Protein and polymer analyses up to m/z 100 000 by laser ionization time-of-flight mass spectrometry. Rapid Commun in Mass Spectrom. 1988;2(8):151–3. doi: 10.1002/rcm.1290020802. [DOI] [Google Scholar]
- 2.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246(4926):64–71. [DOI] [PubMed] [Google Scholar]
- 3.Chen B, Lietz CB, OuYang C, Zhong X, Xu M, Li L. Matrix-assisted ionization vacuum for protein detection, fragmentation and PTM analysis on a high resolution linear ion trap-orbitrap platform. Anal Chim Acta. 2016;916:52–9. doi: 10.1016/j.aca.2016.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robichaud G, Barry JA, Muddiman DC. IR-MALDESI mass spectrometry imaging of biological tissue sections using ice as a matrix. J Am Soc Mass Spectrom. 2014;25(3):319–28. doi: 10.1007/s13361-013-0787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sampson JS, Hawkridge AM, Muddiman DC. Generation and detection of multiply-charged peptides and proteins by matrix-assisted laser desorption electrospray ionization (MALDESI) Fourier transform ion cyclotron resonance mass spectrometry. J Am Soc Mass Spectrom. 2006;17(12):1712–6. doi: 10.1016/j.jasms.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Cooks RG, Manicke NE, Dill AL, Ifa DR, Eberlin LS, Costa AB et al. New ionization methods and miniature mass spectrometers for biomedicine: DESI imaging for cancer diagnostics and paper spray ionization for therapeutic drug monitoring. Faraday discussions. 2011;149:247–67; discussion 333–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takats Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306(5695):471–3. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 8.Cho YT, Huang MZ, Wu SY, Hou MF, Li J, Shiea J. Using electrospray laser desorption ionization mass spectrometry to rapidly examine the integrity of proteins stored in various solutions. Anal Bioanal Chem. 2014;406(2):577–86. doi: 10.1007/s00216-013-7491-z. [DOI] [PubMed] [Google Scholar]
- 9.Huang MZ, Hsu HJ, Lee JY, Jeng J, Shiea J. Direct protein detection from biological media through electrospray-assisted laser desorption ionization/mass spectrometry. J Proteome Res. 2006;5(5):1107–16. doi: 10.1021/pr050442f. [DOI] [PubMed] [Google Scholar]
- 10.Peng IX, Ogorzalek Loo RR, Shiea J, Loo JA. Reactive-electrospray-assisted laser desorption/ionization for characterization of peptides and proteins. Anal Chem. 2008;80(18):6995–7003. doi: 10.1021/ac800870c. [DOI] [PubMed] [Google Scholar]
- 11.Peng IX, Shiea J, Ogorzalek Loo RR, Loo JA. Electrospray-assisted laser desorption/ionization and tandem mass spectrometry of peptides and proteins. Rapid Commun Mass Spectrom. 2007;21(16):2541–6. doi: 10.1002/rcm.3154. [DOI] [PubMed] [Google Scholar]
- 12.Shiea J, Huang MZ, Hsu HJ, Lee CY, Yuan CH, Beech I et al. Electrospray-assisted laser desorption/ionization mass spectrometry for direct ambient analysis of solids. Rapid Commun Mass Spectrom. 2005;19(24):3701–4. doi: 10.1002/rcm.2243. [DOI] [PubMed] [Google Scholar]
- 13.Hu B, So PK, Chen H, Yao ZP. Electrospray ionization using wooden tips. Anal Chem. 2011;83(21):8201–7. doi: 10.1021/ac2017713. [DOI] [PubMed] [Google Scholar]
- 14.Yang Y, Deng J, Yao ZP. Pharmaceutical analysis by solid-substrate electrospray ionization mass spectrometry with wooden tips. J Am Soc Mass Spectrom. 2014;25(1):37–47. doi: 10.1007/s13361-013-0748-0. [DOI] [PubMed] [Google Scholar]
- 15.Nemes P, Vertes A. Laser ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry. Anal Chem. 2007;79(21):8098–106. doi: 10.1021/ac071181r. [DOI] [PubMed] [Google Scholar]
- 16.Kertesz V, Weiskittel TM, Vavrek M, Freddo C, Van Berkel GJ. Extraction efficiency and implications for absolute quantitation of propranolol in mouse brain, liver and kidney tissue sections using droplet-based liquid microjunction surface sampling high-performance liquid chromatography/electrospray ionization tandem mass spectrometry. Rapid Commun Mass Spectrom. 2016;30(14):1705–12. doi: 10.1002/rcm.7607. [DOI] [PubMed] [Google Scholar]
- 17.Pierce CY, Barr JR, Cody RB, Massung RF, Woolfitt AR, Moura H et al. Ambient generation of fatty acid methyl ester ions from bacterial whole cells by direct analysis in real time (DART) mass spectrometry. Chem Commun. 2007(8):807–9. doi: 10.1039/b613200f. [DOI] [PubMed] [Google Scholar]
- 18.Trimpin S, Inutan ED. Matrix assisted ionization in vacuum, a sensitive and widely applicable ionization method for mass spectrometry. J Am Soc Mass Spectrom. 2013;24(5):722–32. doi: 10.1007/s13361-012-0571-z. [DOI] [PubMed] [Google Scholar]
- 19.Inutan ED, Trimpin S. Matrix assisted ionization vacuum (MAIV), a new ionization method for biological materials analysis using mass spectrometry. Mol Cell Proteomics. 2013;12(3):792–6. doi: 10.1074/mcp.M112.023663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trimpin S “Magic” Ionization Mass Spectrometry. J Am Soc Mass Spectrom. 2016;27(1):4–21. doi: 10.1007/s13361-015-1253-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woodall DW, Wang B, Inutan ED, Narayan SB, Trimpin S. High-throughput characterization of small and large molecules using only a matrix and the vacuum of a mass spectrometer. Anal Chem. 2015;87(9):4667–74. doi: 10.1021/ac504475x. [DOI] [PubMed] [Google Scholar]
- 22.Chen B, Lietz CB, Li L. In Situ characterization of proteins using laserspray ionization on a high-performance MALDI-LTQ-Orbitrap mass spectrometer. J Am Soc Mass Spectrom. 2014;25(12):2177–80. doi: 10.1007/s13361-014-0986-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inutan ED, Richards AL, Wager-Miller J, Mackie K, McEwen CN, Trimpin S. Laserspray ionization, a new method for protein analysis directly from tissue at atmospheric pressure with ultrahigh mass resolution and electron transfer dissociation. Mol Cell Proteomics. 2011;10(2):M110 000760. doi: 10.1074/mcp.M110.000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trimpin S, Inutan ED, Herath TN, McEwen CN. Laserspray ionization, a new atmospheric pressure MALDI method for producing highly charged gas-phase ions of peptides and proteins directly from solid solutions. Mol Cell Proteomics. 2010;9(2):362–7. doi: 10.1074/mcp.M900527-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Devereaux ZJ, Reynolds CA, Foley CD, Fischer JL, DeLeeuw JL, Wager-Miller J et al. Matrix-Assisted Ionization (MAI) on a Portable Mass Spectrometer: Analysis Directly from Biological and Synthetic Materials. Anal Chem. 2016. doi: 10.1021/acs.analchem.6b00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang B, Tisdale E, Trimpin S, Wilkins CL. Matrix-assisted ionization vacuum for high-resolution Fourier transform ion cyclotron resonance mass spectrometers. Anal Chem. 2014;86(14):6792–6. doi: 10.1021/ac500511g. [DOI] [PubMed] [Google Scholar]
- 27.Chakrabarty S, Pagnotti VS, Inutan ED, Trimpin S, McEwen CN. A new matrix assisted ionization method for the analysis of volatile and nonvolatile compounds by atmospheric probe mass spectrometry. J Am Soc Mass Spectrom. 2013;24(7):1102–7. doi: 10.1007/s13361-013-0634-9. [DOI] [PubMed] [Google Scholar]
- 28.Sweeting LM, Cashel ML, Rosenblatt MM. Triboluminescence Spectra of Organic-Crystals Are Sensitive to Conditions of Acquisition. J Lumin. 1992;52(5–6):281–91. doi:Doi 10.1016/0022-2313(92)90032-5. [DOI] [Google Scholar]
- 29.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J Am Chem Soc. 1998;120(13):3265–6. doi:Doi 10.1021/Ja973478k. [DOI] [Google Scholar]
- 30.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101(26):9528–33. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim MS, Pandey A. Electron transfer dissociation mass spectrometry in proteomics. Proteomics. 2012;12(4–5):530–42. doi: 10.1002/pmic.201100517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michalski A, Damoc E, Lange O, Denisov E, Nolting D, Muller M et al. Ultra high resolution linear ion trap Orbitrap mass spectrometer (Orbitrap Elite) facilitates top down LC MS/MS and versatile peptide fragmentation modes. Mol Cell Proteomics. 2012;11(3):O111 013698. doi: 10.1074/mcp.O111.013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Villen J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nature protocols. 2008;3(10):1630–8. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoang K, Pophristic M, Horan AJ, Johnston MV, McEwen CN. High Sensitivity Analysis of Nanoliter Volumes of Volatile and Nonvolatile Compounds using Matrix Assisted Ionization (MAI) Mass Spectrometry. J Am Soc Mass Spectrom. 2016;27(10):1590–6. doi: 10.1007/s13361-016-1433-x. [DOI] [PubMed] [Google Scholar]
- 35.Dikler S, Kelly JW, Russell DH. Improving mass spectrometric sequencing of arginine-containing peptides by derivatization with acetylacetone. J Mass Spectrom. 1997;32(12):1337–49. doi:10.1002/ [DOI] [PubMed] [Google Scholar]
- 36.Ye H, Mandal R, Catherman A, Thomas PM, Kelleher NL, Ikonomidou C et al. Top-down proteomics with mass spectrometry imaging: a pilot study towards discovery of biomarkers for neurodevelopmental disorders. PloS one. 2014;9(4):e92831. doi: 10.1371/journal.pone.0092831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pierson J, Norris JL, Aerni HR, Svenningsson P, Caprioli RM, Andren PE. Molecular profiling of experimental Parkinson’s disease: direct analysis of peptides and proteins on brain tissue sections by MALDI mass spectrometry. J Proteome Res. 2004;3(2):289–95. [DOI] [PubMed] [Google Scholar]
- 38.Marshall DD, Inutan ED, Wang B, Liu CW, Thawoos S, Wager-Miller J et al. A broad-based study on hyphenating new ionization technologies with MS/MS for PTMs and tissue characterization. Proteomics. 2016;16(11–12):1695–706. doi: 10.1002/pmic.201500530. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


