Abstract
The discovery of antibiotics marked a golden age in the revolution of human medicine. However, decades later, bacterial infections remain a global healthcare threat, and a return to the pre-antibiotic era seems inevitable if stringent measures are not adopted to curb the rapid emergence and spread of multidrug resistance and the indiscriminate use of antibiotics. In hospital settings, multidrug resistant (MDR) pathogens, including carbapenem-resistant Pseudomonas aeruginosa, vancomycin-resistant enterococci (VRE), methicillin-resistant Staphylococcus aureus (MRSA), and extended-spectrum β-lactamases (ESBL) bearing Acinetobacter baumannii, Escherichia coli, and Klebsiella pneumoniae are amongst the most problematic due to the paucity of treatment options, increased hospital stay, and exorbitant medical costs. Antimicrobial peptides (AMPs) provide an excellent potential strategy for combating these threats. Compared to empirical antibiotics, they show low tendency to select for resistance, rapid killing action, broad-spectrum activity, and extraordinary clinical efficacy against several MDR strains. Therefore, this review highlights multidrug resistance among nosocomial bacterial pathogens and its implications and reiterates the importance of AMPs as next-generation antibiotics for combating MDR superbugs.
Keywords: Multidrug resistance, Nosocomial infections, Antimicrobial peptide, Antibiotic alternatives
INTRODUCTION
A perturbing prediction by the World Health Organization (WHO) is that by the year 2050, drug-resistant infections, largely exacerbated by the indiscriminate use of antibiotics, will kill 10 million people annually, ignite a financial cataclysm, and impose extreme poverty upon millions of people (de Kraker et al., 2016). The increasing incidence and prevalence of antibiotic resistance among nosocomial bacterial pathogens and the rapid spread of resistance genes in the environment are major global healthcare threats due to the associated increase in morbidity and mortality and huge burden on the economy (Aslam et al., 2018; Zaman et al., 2017).
Since the discovery of penicillin in 1928 by Alexander Fleming, antibiotics have saved countless lives and marked an important medical revolution in plant, animal, and human prophylaxis. Unfortunately, the continued use of antibiotics has been accompanied by the rapid emergence and spread of multidrug resistant (MDR) strains and many medical specialists are warning of an inevitable return to the pre-antibiotic era (Adedeji, 2016). For example, bacterial genomic analysis has revealed the presence of more than 20 000 genes associated with resistance (Liu & Pop, 2009), with a higher number expected over the coming years. Therefore, stringent measures are necessary to curb the spread of resistant strains as they present a major challenge to global public health not only in the war against microbial infections, but also in other clinical applications such as cancer treatment, invasive surgery, and graft transplantation (Gudiol & Carratalà, 2014; Lupei et al., 2010).
Several factors that have been implicated in the current upsurge of antimicrobial resistance (AMR) in hospitals and the community, including the indiscriminate use of antibiotics in human and animal medicine and in agricultural practices involving growth promoters (Silveira et al., 2009; van Boeckel et al., 2015), lack of proper regulation regarding over-the-counter antibiotics, especially in developing countries where they are easily available without proper medical prescription (Ayukekbong et al., 2017), and poor sanitation practices leading to the introduction of unmetabolized antibiotics into the environment through human and animal waste (Davies & Davies, 2010).
Natural selection is an inherent process and key driver of evolution, conferring organisms with traits for increased environmental adaptability and survival (Jesus et al., 2002). Over the years, the widespread use of antibiotics has led to the selection of diverse strains of microorganisms possessing MDR traits or genes (Martínez & Baquero, 2002; Ochoa et al., 2009). Such resistance-conferring properties, through bacterial and genomic associations, can be easily transferred across different ecological niches, e.g., from animals to humans and vice versa, and from hospital settings to the community, where they present an unprecedented crisis (Groisman & Ochman, 1996; Kümmerer, 2004; Phillips et al., 2004).
In bacteria, mechanisms conferring resistance to almost all available classes of antibiotics have been studied extensively and described in detail in previous literature (Blair et al., 2014; Dever & Dermody, 1991; Lin et al., 2015; Lombardi et al., 2019; Morita et al., 2012). These mechanisms include enzymatic degradation of antibiotics, drug-target modification, altered membrane permeability, and enhanced expression of efflux pumps that actively eliminate antibiotics (Alanis, 2005; Laxminarayan & Brown, 2001; Munita & Arias, 2016).
Initially associated with severe nosocomial infections in immunocompromised patients, multidrug resistance has spread to the wider community, resulting in severe infections associated with growing death tolls and huge economic burdens due to increased disability and high medical costs (Jing et al., 2019; Peters et al., 2019). Currently, common problematic MDR bacteria include methicillin-resistant S. aureus (MRSA), vancomycin-resistant MRSA, MDR P. aeruginosa, carbapenem-resistant A. baumannii, E. coli, and K. pneumoniae, vancomycin-resistant enterococci (VRE), and extensively drug-resistant (XDR) Mycobacterium tuberculosis (Levin et al., 1999; Miller et al., 2005). Recent reports have also indicated cases of bacterial strains completely resistant to all available antibiotics. For example, the extensive use of colistin, a drug of last resort for the treatment of MDR pathogens such as P. aeruginosa, A. baumannii,and K. pneumoniae in both human medicine and agriculture, has led to the emergence of a plasmid-mediated MCR-1 gene that encodes for its resistance (Liu et al., 2016; MacNair et al., 2018; Paterson & Harris, 2016).
The increasing incidence of infections resulting from MDR pathogens in clinical settings has intensified the demand for alternative therapies. Antimicrobial peptides (AMPs) with potent antimicrobial activities and diverse mechanisms of action (MOA) are considered important alternatives to solving the issues of multidrug resistance.
IMPLICATIONS OF MULTIDRUG RESISTANCE IN NOSOCOMIAL PATHOGENS
Epidemiological surveillance data worldwide indicate that anomalous use of antibiotics has resulted in the evolution of several human pathogens into MDR strains that are highly tolerant or resistant to antibiotic therapies, thus posing a serious threat to public health (Davies & Davies, 2010; WHO, 2014). Multidrug resistance, especially in nosocomial pathogens, is of great clinical concern due to the increased morbidity and mortality and enhanced virulence and transmissibility (Bhat et al., 2006; Khan et al., 2017). Such pathogens are implicated in severe infections such as ventilator-associated pneumonia (Koenig & Truwit, 2006) as well as bloodstream (Martinez & Wolk, 2016), surgical site (Anderson, 2011), and implant-associated urinary tract infections (Nicolle, 2014). These infections occur often and are severe in immunocompromised patients, although recent evidence also suggests the spread of MDR genes into the general community (Khan et al., 2017).
For gram-negative nosocomial pathogens, especially A. baumannii, P. aeruginosa, K. pneumoniae, S. aureus, and Enterobacter spp.,the emergence of multidrug resistance to several available classes of antibiotics, such as penicillins, aminoglycosides, cephalosporins, and fluoroquinolones, has increased over the years as a result of extensive use, especially in intensive care units (Richard et al., 1994; Struelens, 1998). In many cases, these MDR strains show reduced susceptibility to all available antibiotic therapies and are implicated in serious high mortality rate-nosocomial infections (Hirsch & Tam, 2010; Manchanda et al., 2010). Colistin, an AMP consisting of two polypeptides (polymyxin A and B), is often used as a drug of last resort for MDR strains, notwithstanding its adverse side effects such as nephrotoxicity and neurotoxicity, due to the paucity of treatment options (Falagas et al., 2005; Yamamoto et al., 2018). However, current reports indicate the emergence of MDR strains that exhibit reduced susceptibility to colistin following long-term clinical or laboratory exposure (Jeannot et al., 2017). Resistance to colistin is attributed to the presence of the MCR-1 gene, which is highly transmissible across different bacterial strains and has been isolated from strains on hospital surfaces and in animal and clinical human samples (Yamamoto et al., 2018). These strains also exhibit low susceptibility to almost all other antibiotics, including those designated as “last-resort” such as polymyxins (colistin) and carbapenems, thus raising a new crisis in the war against multidrug resistance (Liu et al., 2016; Rapoport et al., 2016; Yamamoto et al., 2018).
Gram-positive S. aureus is another important nosocomial pathogen and is transmitted through direct contact with contaminated surfaces, clinical waste, open wounds, and clinical staff (Denis, 2017; Price et al., 2017). It is a major cause of skin infections, sepsis, and pneumonia in hospitalized patients, with recent data indicating increased incidences of community-acquired S. aureus infections due to improper disposal of hospital waste, poor hygiene, and the spread of resistant genes to the community (Dotel et al., 2017). The rising incidence of MRSA infections is a growing healthcare threat associated with increased mortality rates in hospitals and the community (Othman et al., 2019). Additionally, most MRSA strains exhibit resistance to all other antibiotics, including the beta-lactams, and recent reports indicate the emergence and spread of strains with reduced susceptibility to glycopeptide compounds such as vancomycin (vancomycin-resistant MRSA), therefore making treatment almost impossible (Bhat et al., 2006; Centers for Disease & Prevention, 1997; Othman et al., 2019).
Over the past few years, enterococci, which are normal flora of the gut and genital tract, have become problematic nosocomial pathogens and a growing clinical predicament. This trend is concomitant with their inherent resistance to several commonly used drugs, including penicillin, ampicillin, cephalosporin, and clindamycin (Gold & Moellering, 1996). In addition to inherently acquired resistance, enterococci can rapidly acquire resistance to virtually any antibiotic either through rapid mutation or the acquisition of foreign genetic material, thus expressing a repertory of resistance mechanisms to antibiotics such as enhanced expression of efflux pumps, modification of drug targets, and enzymatic degradation of drug agents (Linden, 2002). MDR enterococci exhibit high adaptability under antibiotic pressure and can rapidly acquire resistant genes to enhance their survival. One notable example is the rapid transfer of resistant genes associated with the extensive use of vancomycin among several strains of enterococci, especially Enterococcus faecalis and Enterococcus faecium,two important nosocomial pathogens (Arias & Murray, 2012). Resistance to vancomycin presents a major clinical crisis as most of these strains are also resistant to most other antibiotics and the transfer of vancomycin-resistant genes can occur from enterococci strains to even more lethal pathogens such as MRSA, therefore leaving very few or no therapeutic options (Chang et al., 2003; Franchi et al., 1999; Noble et al., 1992).
Bacterial biofilms are of great clinical significance to global healthcare due to their important role in nosocomial and implant-related infections. Most nosocomial pathogens produce biofilms, which complement disease pathogenicity and resistance (Dunne, 2002; Gurung et al., 2013). Biofilm-producing organisms such as A. baumannii and P. aeruginosa exhibit extreme resistance to almost all antibiotics compared with non-biofilm-producing microorganisms. Biofilms form a protective coating around bacterial cells, thus hindering the killing action of antibiotics (Stewart, 2002). Other factors that contribute to resistance in biofilm-producing organisms include the formation of persister cells (dormant and highly resilient to almost all available antibiotics), slow bacterial growth within the biofilm, and adaptability to stressful conditions (Keren et al., 2012; Lewis, 2008).
Multidrug resistance to useful classes of antibiotics has increased gradually among several bacterial nosocomial pathogens. Thus, great efforts have been expended on the discovery of novel antibiotic alternative therapies. Currently, several AMPs with potent efficacy are under clinical trial and present excellent mitigation strategies for the multidrug resistance crisis (de Breij et al., 2018; Koo & Seo, 2019; Stefania et al., 2015). Moreover, compared with traditional antibiotics, AMPs possess many important qualities that make them excellent candidates for therapeutic exploitation. For instance, AMPs are multifunctional with diverse MOA, including membrane disruption, inhibition of DNA and protein synthesis, and impairment of key cellular processes such as metabolism and cell wall synthesis (Kumar et al., 2018; Tzong-Hsien et al., 2016; Wimley, 2010). Their diverse MOA are important as they minimize the tendency of pathogens to select for resistance, as observed in most conventional antibiotics that only act on single targets unless used in combination (Alanis, 2005; Bechinger & Gorr, 2017). Moreover, most AMPs exhibit potent antimicrobial properties against both gram-negative and gram-positive bacteria, fungi, and viruses in the nanomolar and micromolar range (Frecer et al., 2004; Wakabayashi et al., 1996; Yamauchi et al., 1993), rapidly kill pathogens within minutes, and have a low proclivity to select for resistance compared with conventional antibiotics (de Breij et al., 2018; Nagarajan et al., 2019).
AMPs: ORIGIN AND PROSPECTS
AMPs or host defense peptides are a notably class of compounds of the innate immune system with both microbicidal and immunomodulatory activities, providing a first line of defense against pathogenic invasion (Falanga & Galdiero, 2018; Kang et al., 2017; Pasupuleti et al., 2012). AMPs are polypeptides of varying molecular weight and amino acid residues (ranging from five to over 100) and are found in virtually all living organisms, from simple prokaryotes to complex eukaryotes (Figure 1). They play important roles in the direct killing of infectious agents, viruses, bacteria, fungi, and parasites and through the modulation of immune processes such as activation and recruitment of immune cells, wound healing, angiogenesis, and inflammation (Haney & Hancock, 2013; Kumar et al., 2018; Maróti et al., 2011; Oppenheim et al., 2003; Zhao et al., 2013).
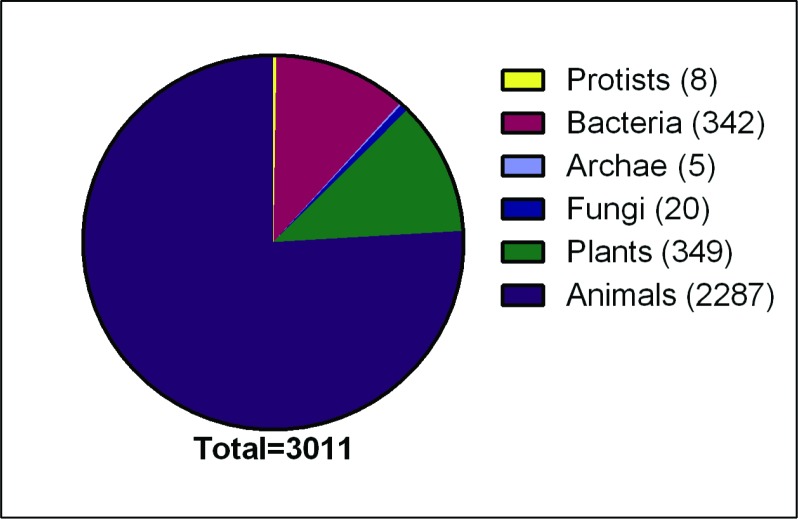
Figure 1. Number of natural and synthetic antimicrobial peptides from different kingdoms (total 3 011) as of July 2019.
Data obtained from antimicrobial peptide database http://aps.unmc.edu/AP/ (Wang et al., 2016).
In most organisms, high concentrations of AMPs are expressed on surfaces that are constantly exposed to pathogens. For example, the mucosal lining and skin of vertebrates display broad-spectrum antimicrobial activities (Brogden et al., 2003; Rinaldi., 2002). In general, AMPs are either produced continuously or up-regulated following exposure to a threat (Ganz, 2003). For example, human skin is continuously protected by an abundance of psoriasin, dermcidin, and lactoferrin, with cathelicidin LL-37 also up-regulated following infection (Harder et al., 2013). In bacteria, AMPs play an important role in enhancing adaptability and survival during antagonistic competition for resources with other bacteria occupying the same ecological niche (Kumar et al., 2018). Several peptides with potential therapeutic activity have been isolated from bacteria and demonstrate potent antimicrobial activities against both gram-negative and gram-positive bacteria and even fungi. These AMPs are known as bacteriocins and play important roles in inhibiting or killing antagonistic bacterial strains with no self-harm, thus conferring a critical survival strategy (Yang et al., 2014). Bacteriocins provide important prospects for the development of alternative antibiotic strategies against different bacterial pathogens and are also useful in the food industry as preservatives (Mills et al., 2011; Silva et al., 2018). Bacteria-derived AMPs include nisin, a 34-amino acid residue peptide and an important food preservative isolated from Lactococcus lactis (Tong et al., 2014),and colistin, isolated from Paenibacillus polymyxa and used extensively in agriculture and human prophylaxis (Poirel et al., 2017). Colistin is an important last-resort peptide drug used against several MDR nosocomial pathogens such as MRSA, MDR P. aeruginosa, and carbapenem-resistant pathogens, although its use is somewhat limited due to its side effects (Falagas et al., 2005; Levin et al., 1999; MacNair et al., 2018).
In vertebrates, AMPs can directly kill microorganisms and play a role in immunomodulation through the activation and recruitment of immune cells during infection (Kumar et al., 2018; Mahlapuu et al., 2016). Several different classes of AMPs, such as cathelicidins and defensins, have been isolated and characterized from immune cells, bodily secretions, and epithelial tissues of amphibians and marine and terrestrial animals (Lu et al., 2008; Wang et al., 2016). For example, several AMPs have been identified from amphibian skins, where they are produced in glands of the dermal skin layer and released following pathogenic exposure, inducing a microbicidal action through membrane disruption (Rollins-Smith et al., 2005; Woodhams et al., 2007). In humans, psoriasin, dermcidin, and lactoferrin are continuously secreted by the sweat glands to form an important barrier against infection, and cathelicidin LL-37 is secreted following microbial introduction (Harder et al., 2013). Nearly 30 cathelicidins have been characterized from domestic and wild mammals (Kościuczuk et al., 2012), and are stored as inactive precursors in neutrophil granules and activated following microbial exposure (Treffers et al., 2005). Apart from cathelicidins, several other classes of antimicrobial peptides have been isolated and identified in vertebrates (Brogden et al., 2003; Zhang, 2006; Zhang et al., 2008). These AMPs have diverse MOA and play multifunctional roles, including modulation of immune responses, prevention of excessive tissue damage, alleviation of inflammation, and destruction of invading pathogens to mitigate infection onset (Haney & Hancock, 2013; Maróti et al., 2011; Oppenheim et al., 2003).
Several vertebrate AMPs show excellent therapeutic potential due to their antibacterial, antiviral, antifungal, and anti-inflammatory properties (Oppenheim et al., 2003; Qi et al., 2019), whereas others show improved clinical efficacy, structural stability, and potent antimicrobial activity against several MDR pathogens following modification (Kumar et al., 2018; Schmidtchen et al., 2014; Zhang et al., 2019).
In contrast to vertebrates, insects and plants lack an adaptive immune system, relying entirely on the innate immune system for defensive purposes, with AMPs playing a pivotal role in the protection against invading pathogens. Several AMPs have been isolated and analyzed from the epithelial cells, hemocytes, and hemolymphs of insects (Brown et al., 2009; Hancock et al., 2006; Philippe & Reto, 2005). Peptides isolated from insects are usually cationic and kill bacteria through permeabilizing bacterial membranes, and further possess potent microbicidal activity either singly or synergistically with traditional antibiotics (Yi et al., 2014). Notable examples of insect-based AMPs include cecropins, defensins, drosocins, and diptericins, which exhibit activities against both gram-negative and gram-positive bacteria as well as fungi (Mylonakis et al., 2016; Rozgonyi et al., 2009; Wu et al., 2018; Zhang & Gallo, 2016).
AMPs also play important defensive roles in plants (which also lack an innate immune system). Several peptides have been isolated and characterized from the leaves, roots, seeds, and tubers of plants (Tam et al., 2015). Generally, they exhibit high proteolytic, thermal, and chemical stability due to the presence of multiple disulfide bonds conserved within their rich cysteine residues (Hammami et al., 2009; Hilchie et al., 2013; Stotz et al., 2009; Tam et al., 2015). Plant AMPs have been extensively studied and discussed (Craik, 2012; Hammami et al., 2009; Nawrot et al., 2014; Stec, 2006). Much like AMPs from vertebrates, plant-based AMPs confer protection against invading pathogens through membrane disruption and pore formation and by targeting key microbial processes such as DNA and protein synthesis (Nawrot et al., 2014). Notable examples of plant AMPs include plant defensins and thionins (Hilchie et al., 2013; Stotz et al., 2009). Plant defensins are small cationic peptides with 45–55 amino acid residues and exhibit potent antibacterial and fungal activities (Gao et al., 2000; Tavares et al., 2008). Thionins are cationic peptides with 45–48 amino acid residues and three or four disulfide bonds (Stec, 2006). They exhibit potent activities against bacteria and fungi and cytotoxicity on animal cells (Evans et al., 1989; Fernandez De Caleya et al., 1972).
Used singly or in combination with traditional therapeutics, AMPs are quite effective at combating infectious agents, including MDR bacteria. They exhibit potent microbicidal activity in the micromolar range, rapid killing action, and low selection of resistance, thus constituting an important strategy for curbing MDR pathogens (Deslouches & Di, 2017; Loeffler et al., 2001; van’t Hof et al., 2001). In contrast to empirical antibiotics that target single or specific bacterial processes, AMPs exhibit multifunctional bacterial killing effects, including disruption of the plasma membrane, and also target microbial intracellular processes, including inhibition of transcription and translation, protein synthesis, and bacterial cell wall formation (Gee et al., 2013; Hurdle et al., 2011; Reddy et al., 2004).
Presently, much effort is being directed toward obtaining novel antimicrobial agents with broad-spectrum activity against pathogenic microorganisms. Naturally occurring AMPs have proven to be invaluable templates for the design and synthesis of synthetic AMPs with increased potency, which are easier to produce and less sensitive to proteolytic degradation (Falanga et al., 2016; Hurdle et al., 2011).
CLASSIFICATION AND STRUCTURE OF AMPs
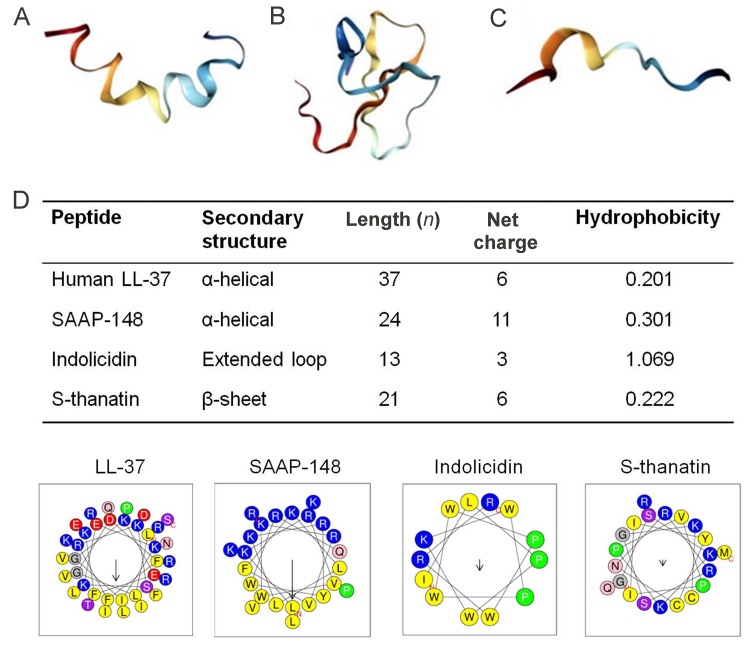
With more than 3 000 sequences reported to date, AMPs are often classified based on their secondary structures and MOA. Based on structure, the three currently established classes include α-helical, β-sheet, and extended-coiled peptides (Figure 2) (Falanga & Galdiero, 2018; Lombardi et al., 2019). The α-helical AMPs are unordered in aqueous solution but once in contact with a biological membrane, they assume an amphipathic α-helical structure (Kumar et al., 2018). Important α-helical AMPs include magainin, a 23-amino acid residue peptide isolated from the skin of the African clawed frog(Xenopus laevis),which exhibits highly potent tumoricidal and broad-spectrum antimicrobial activities against bacteria, fungi, and protozoa due to membrane disruption (Matsuzaki, 1999; Zerweck et al., 2017). The cathelicidin family of AMPs are also important members of this group and are found widely in nature in all vertebrates (Kościuczuk et al., 2012). For example, LL-37, a prominent cathelicidin in humans, exhibits broad-spectrum microbicidal activities against gram-positive and gram-negative bacteria by plasma-membrane disruption (Björstad et al., 2009). LL-37 has been extensively studied and used as a template for the design of several synthetic AMPs, many of which are currently under clinical trial (de Breij et al., 2018; Dürr et al., 2006; Kościuczuk et al., 2012). Other α-helical AMPs include aurein from the granular dorsal glands of the frog Litoria aurea (Rai & Qian, 2017), melittin from honey-bee venom, and cecropin derived from the hemolymph of Hyalophora cecropia (Yi et al., 2014).
Figure 2. Structural diversity and helical wheel projections of representative AMPs.
A: α-helical-magainin (PDB ID 2LSA). B: β-sheet-chicken ovo-defensin (PDB ID 2MJK). C: Extended coil-tritrpticin. Images were created with Protein Data Bank (PDP) (Bioinformaticsdoi:10.1093/bioinformatics/bty419) (Rose et al., 2018) and visualized with Jmol software. D: Helical wheel projections of four representative peptides showing physical properties canonical to all AMPs, including distribution of amino acid residues, net charge, and hydrophobicity established to correlate with antimicrobial activity, selectivity, and cytotoxicity. Positively charged residues (polar) are represented as blue circles and hydrophobic (nonpolar) residues are yellow circles. Wheels projections, net charge, and hydrophobicity of AMPs were generated with HeliQuest webserver (http://heliquest.ipmc. cnrs.fr/).
The β-sheet peptides, which constitute the largest group of AMPs, are produced in many organisms such as marine invertebrates, amphibians, plants, and animals (Haney & Hancock, 2013; Maróti et al., 2011; Oppenheim et al., 2003; Wang et al., 2016) and show both antibacterial and antifungal properties (Harwig et al., 1996). In contrast to α-helical peptides, these peptides are ordered in aqueous solution and contain conserved cysteine residues that form disulfide bonds that enhance structural stability and minimize proteolytic degradation (Tzong-Hsien et al., 2016). They exhibit boundless therapeutic prospects, e.g., as antifungal, antibacterial, antiviral, and anti-inflammatory agents, and primarily kill bacteria through plasma-membrane disruption (Kumar et al., 2018; Panteleev et al., 2015). Prominent members of this family include defensins, protegrins, and tachyplesins. Defensins are found in plants, invertebrates, and vertebrates and exhibit activity against both gram-positive and gram-negative bacteria, fungi, and viruses (Panteleev et al., 2015; Yamaguchi & Ouchi, 2012). Tachyplesins, a class of peptides isolated from hemocytes of the horseshoe crab, exhibit strong microbicidal activity, although their use is hampered by their potential toxicity to mammalian cells (Edwards et al., 2017; Liu et al., 2018; Miyata et al., 1989).
The third class of AMPs include those with an extended-coil structure. These AMPs lack secondary structures present in other AMPs such as α helices and β sheets and mostly consist of specific amino acid residues including arginine (Arg), proline, or tryptophan (Mahlapuu et al., 2016). They exhibit broad-spectrum activity against gram-negative bacteria via membrane disruption and targeting internal processes and also possess antitumor activity (Falla et al., 1996). Important extended-coil AMPs include indolicidin, a 13-amino acid peptide produced by bovine leukocytes that shows potent antimicrobial activity (Falla et al., 1996), as well as tritrpticin (Yang et al., 2002) and human salivary histatin (Du et al., 2017).
MECHANISM OF ACTION (MOA) OF AMPs
For AMPs, there is strong evidence suggesting a close correlation between their MOA and physical features or primary structure, which are characterized by a net positive charge and hydrophobicity (Figure 2) (Dathe & Wieprecht, 1999; Matsuzaki, 1999). The cationic charge influenced by amino acid residues such as Arg and lysine (Lys) enhances selectivity for negatively charged bacterial plasma membranes while exhibiting low electrostatic interactions with the uncharged outer layer of the eukaryotic cell membrane (Guilhelmelli et al., 2013). Current efforts at improving the selectivity for and electrostatic interaction of AMPs with bacterial membranes involve the substitution of amino acids with positively charged amino acid residues like Arg and Lys (Arias et al., 2018; Yeaman & Yount, 2003). For example, Jin et al. (2016) reported that substitution of amino acids improved antimicrobial efficacy, increased stability, and reduced the hemolytic activity of the designed peptide ZY13 compared to its precursor peptide cathelicidin-BF15.
Previous studies have also indicated a strong correlation between the cationic charge of an AMP and its antimicrobial activity, with data showing improved antimicrobial activity of several AMPs against both bacterial and fungal pathogens paralleled with an increase in cationic charge (Gagnon et al., 2017; Hong et al., 2001; Lyu et al., 2016); however, increased hemolytic activity has been observed for several AMPs following an increase in net charge (Chen et al., 2005; Jiang et al., 2008). Secondly, hydrophobicity is an important parameter of all AMPs and is attributed to a substantial proportion (almost 50%) of hydrophobic residues in peptide sequences, such as leucine, valine, isoleucine, alanine, methionine, tyrosine, tryptophan, and phenylalanine. Hydrophobicity is crucial for peptide selectivity to biological membranes and increased hydrophobicity is correlated with increased hemolytic activity as highly hydrophobic AMPs have a stronger ability to penetrate the hydrophobic core of erythrocyte membranes (Chen et al., 2007; Tachi et al., 2002).
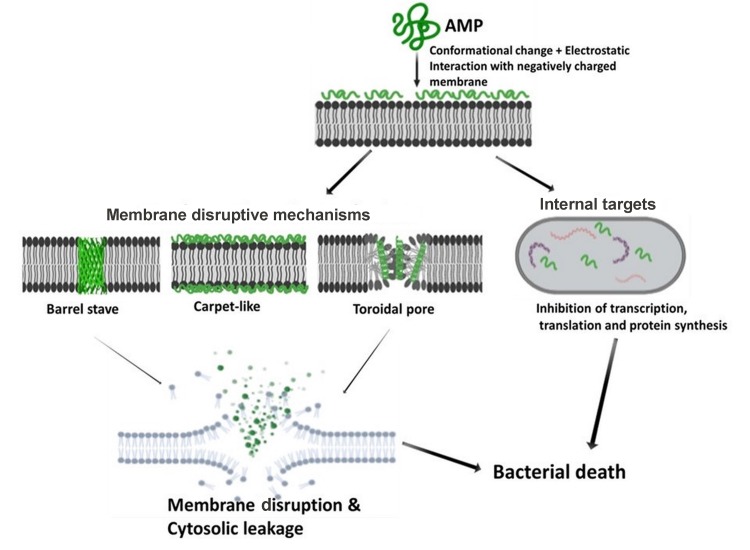
Understanding the MOA of AMPs is paramount for unlocking their full potential as next-generation antibiotics. Extensive research has been conducted, and is still ongoing, to unveil the MOA of AMPs (Guilhelmelli et al., 2013; Hancock & Lehrer, 1998; Ulm et al., 2012). Based on their MOA, AMPs can be classified into those that kill through membrane disruptive mechanisms and non-membrane disruptive mechanisms, as illustrated in Figure 3. For the membrane disruptive killing action, AMPs produce microbicidal activity by targeting and disrupting the bacterial plasma-membrane structure, mostly through permeabilization, thus resulting in leakage of intracellular content (Huang et al., 2010). An important aspect to consider for this MOA is the concentration threshold of peptide molecules on the bacterial membrane surface for effective permeabilization via interaction of positively charged AMP residues with negatively charged moieties on the bacterial membrane surface (Chen et al., 2007; Shai, 2002). To drive this membrane disruptive MOA, three models have been postulated, i.e., barrel-stave, carpet-like, and toroidal pore models. For all models, the MOA begins with the accumulation and organization of AMP molecules parallel to the membrane surface, followed by electrostatic interaction between the AMPs’ cationic charged residues and negatively charged phospholipids on the membrane surface (Falanga et al., 2016).
Figure 3. Schematic of membrane disruptive and non-membrane disruptive bacterial killing mechanisms of AMPs.
Illustration created with BIORENDER.COM.
Typical for α-helical AMPs, the barrel-stave model constitutes the formation of hydrophilic pores on the hydrophobic core of the bacteria membrane structure, resulting in membrane disruption and leakage of extracellular content (Kumar et al., 2018). For the carpet-like mechanism, peptide molecules accumulate parallel to the membrane surface in a carpet-like fashion, followed by penetration into the membrane and disruption of the lipid bilayer (Brogden, 2005; Falanga et al., 2016; Rodríguez-Vázquez et al., 2014). In contrast, toroidal pore models, e.g., magainin peptide, cause membrane disruption by perpendicular insertion into the lipid bilayer (Shai, 2002; Wimley, 2010).
Direct killing through non-membrane disruptive mechanisms involves targeting microbial processes or physiological functions other than the cell membrane, utilizing similar killing mechanisms as those by conventional antibiotics such as inhibition of cell wall, protein, and DNA synthesis and reduced enzymatic activity (Hancock & Rozek, 2002; Hancock & Sahl, 2006). Firstly, the AMP interacts with the plasma membrane before penetrating with or without membrane permeabilization (e.g., activity of buforin II on E. coli) and accumulating intracellularly, where it targets and acts on key processes such as transcription and translation, protein synthesis, enzymatic activity, and microbial death (Brogden, 2005; Park et al., 1998). For example, human α-defensin 1, human α-defensin 5, human β-defensin 4, and indolicidin all have intracellular targets (Falla et al., 1996; Lehrer et al., 1989; Sharma & Nagaraj, 2015).
In addition to direct microbial killing, AMPs are required for other immunomodulatory functions. They are secreted by a wide range of immune cells, including phagocytes, neutrophils, and macrophages, and are involved in the mitigation of infection such as controlled secretion of proinflammatory cytokines to prevent cytokine storm and tissue damage, recruitment and activation of immune cells, promotion of angiogenesis, and suppression of excessive reactive oxygen species release (Hancock et al., 2012; Hilchie et al., 2013; Nicole et al., 2012; Nijnik & Hancock, 2009). For example, human cathelicidin LL-37 exhibits immunomodulation properties such as chemoattraction and activation of various immune cells such as monocytes, neutrophils, and mast cells by using formyl peptide receptor-like 1 (De et al., 2000; Nijnik & Hancock, 2009). A thorough understanding of the antimicrobial and immunomodulatory properties of AMPs provides excellent opportunities for the discovery and design of novel therapies for bacterial infection as well as inflammatory and cardiovascular diseases such as atherosclerosis and thrombosis (Bei et al., 2019; Zhang et al., 2015).
ACTIVITY OF AMPs AGAINST MDR NOSOCOMIAL BACTERIAL PATHOGENS
The rapid spread of antibiotic resistance presents a daunting clinical challenge due to the recalcitrance of pathogens to traditional antibiotics, especially among nosocomial pathogens. In both developing and developed countries, hospital-acquired infections, commonly referred to as nosocomial infections, are a growing concern often associated with prolonged hospital stay, increased mortality rates, and huge economic burden. Therefore, the discovery of novel alternative therapies is paramount. Several AMPs have been found to show potent microbicidal activity in the micromolar range against several bacteria strains, with a low likelihood for selection of resistance, therefore creating hope in the war against MDR pathogens (Deslouches & Di, 2017; Loeffler et al., 2001; van’t Hof et al., 2001).
Natural and synthetic peptides have proven to be effective in combating several pathogens, including MDR gram-positive and gram-negative bacteria (see Table 1), with continued effort aimed at designing peptides with improved therapeutic potential and reduced side effects (Chou et al., 2008; Liu et al., 2015). Such strategies have focused on improving efficacy, structural stability, and protection from proteolytic degradation, and include chemical modifications such as acetylation, enrichment with non-natural D-amino acids, substitution of amino acid residues, and use of delivery systems (Nordström & Malmsten, 2017; Zhang et al., 2019; Zhao et al., 2016).
Table 1.
Select antimicrobial peptides with potent activity against MDR pathogens
| Peptide | Sequence | Development phase | Description |
|---|---|---|---|
| Human LL-37 | LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES | From human leucocytes, kills bacteria through pore formation and possesses immunomodulation activities. | |
| SAAP-148 (de Breij et al., 2018) | LKRVWKRVFKLLKRYWRQLKKPVR | Preclinical | LL-37 derivative, shows potent bactericidal activity through membrane permeabilization and wound healing activity. |
| Cathelicidin-BF (Wang et al., 2008) | KFFRKLKKSVKKRAKEFFKKPRVIGVSIPF | Lysine-phenylalanine-rich peptide from snake venom. Shows potent efficacy against fungal and bacterial strains including MDR pathogens. Low hemolytic activity. | |
| D-OH-CATH30 (Zhao et al., 2018) | KFFKKLKNSVKKRAKKFFKKPRVIGVSIPF | Preclinical | Cathelicidin from snake venom, rapidly kills MDR gram-positive and gram-negative pathogens, with low hemolytic activity and in vivo toxicity. |
| Indolicidin (Falla et al., 1996) | ILPWKWPWWPWRR | III | Isolated from bovine leucocytes, shows potent bactericidal activity through pore formation. |
| Omiganan (Melo & Castanho, 2007) | ILRWPWWPWRRK-NH2 | III | Indolicidin derivative, broad-spectrum antimicrobial activity, therapeutic agent against acne and catheter related infections. |
| Ci-MAM-A24 (Fedders et al., 2010) | WRSLGRTLLRLSHALKPLARRSGW-NH2 | Preclinical | Isolated from Ciona intestinalis, shows potent bactericidal activity against MRSA, VRE, and MDR P. aeruginosa through pore formation. |
| Pexiganan (Ge et al., 1999) | GIGKFLKKAKKFGKAFVKILKK-NH2 | III | Magainin analog, phase III clinical trials for treatment of bacterial infections and diabetic foot ulcers. Potent antimicrobial activity. |
| S-thanatin (Wu et al., 2011) | GSKKPVPIIYCNRRSGKCQRM | Preclinical | Thanatin derivative, shows improved antimicrobial activity with reduced hemolytic activity. Potently inhibits gram-negative growth in vitro and in vivo and alleviates sepsis in mice. |
| AA139 (van der Weide et al., 2019) | GFCWYVCARRNGARVCYRRCN | Preclinical | Analog of arenicin-3 with β-hairpin structure, exhibits potent microbicidal activity against MDR gram-negative pathogens, and excellent candidate for in vivo application. |
| SET-M33 (Van De Weide et al., 2019) | KKIRVRLSA)4K2KβΑ-ΟΗ | Preclinical | Synthetic tetra-branched peptide with potent microbicidal activity against MDR bacteria and in vivo therapeutic potential. Currently under development for treatment of sepsis and lung infections (Brunetti et al., 2016a). |
| EC-hepcidin3 | APAKCTPYCYPTHDGVFCGVRCDFQ | Preclinical | Cysteine-rich peptide cloned from marine fish. Potent microbicidal activity against S. aureus and Pseudomonas spp. |
| Tachyplesin-1 (Ohta et al., 1992) | KWCFRVCYRG ICYRRCR | II | Cationic β-hairpin peptide from horseshoe crab. Potent microbicidal activity against gram-negative and gram-positive bacteria. Use limited by high cytotoxicity. |
Cathelicidin AMPs, which are found in humans and other animals, are excellent candidates for therapeutic agents. They display multifunctional roles, including potent broad-spectrum antimicrobial activity against infectious agents such as fungi, viruses, and bacteria, and can trigger specific immune responses such as activation and recruitment of immune cells (Gennaro et al., 1989; Kościuczuk et al., 2012). The human cathelicidin LL-37, expressed in various tissues and circulating cells, is the most extensively studied peptide and exhibits potent bactericidal activity against bacterial strains and fungi (Björstad et al., 2009). Furthermore, it has been used as a template for the design of several synthetic peptides with improved antibiotic activity against nosocomial pathogens, biofilms, and persister cells (Dürr et al., 2006; Xhindoli et al., 2016). A notable LL-37 derivative (SAAP-148) designed by de Breij et al. (2018) through amino acid substitution of the C-terminal chain of LL-37 shows potent microbicidal activity (minimum inhibitory concentration (MIC) 0.4 to 12.8 µm) against several ESKAPE pathogens (e.g., E. faecium, S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa, and Enterobacter species) without selection of resistance. This derivative can also eliminate biofilms and persister cells of S. aureus, A. baumannii and P. aeruginosa in the micromolar range.
Cathelicidins make up the bulk of naturally occurring AMPs in snake venom. Several snake venom-based AMPs have been established, which potently kill both fungal and bacterial pathogens (Jin et al., 2016; Wang et al., 2011; Zhang, 2015). Zhao et al. (2018)identified a novel 34-amino acid residue cathelicidin (OH-CATH) from the king cobra, with its analog D-OH-CATH30 found to exhibit potent microbicidal activity (MIC 1.56 to 12.5 µg/mL) against several gram-negative and gram-positive bacteria, including MDR A. baumannii, MDR P. aeruginosa, MRSA, and E. coli, compared to nine commonly used antibiotics. Remarkably, bacteria-killing kinetics revealed that D-OH-CATH30 rapidly killed E. coli within 6 min and exhibited low hemolytic activity on red blood cells at high concentration. Furthermore, in vivo toxicity testing using mice revealed a high lethal dose, with no deaths observed at high concentrations of 160 mg/kg (Li et al., 2012; Zhao et al., 2018), thus highlighting their potential as excellent therapeutic candidates.
Cathelicidin-BF is a 30-amino acid residue cathelicidin isolated from the venom of Bungarus fasciatus and possesses potent antimicrobial activity, even against MDR clinical isolates (Wang et al. 2008). Its designed and shortened 15-amino acid residue peptide cathelicidin-BF15 also shows strong efficacy against fungal species such as Candida albicans (Jin et al., 2016; Wang et al., 2008) and against several bacterial strains including MDR clinical isolates of E. coli, P. aeruginosa,and S. aureus through membrane permeabilization (Wang et al., 2008), with low hemolytic activity on red blood cells and therapeutic potential against acne vulgaris (Wang et al., 2011).
Several AMPs with potent antimicrobial properties have also been isolated from amphibians such as salamanders, frogs, and toads (Patocka et al., 2018; Liu et al., 2011; Xiao et al., 2011). Such peptides are viable targets for the development of therapeutic agents. For example, Qi et al., (2019)identified two novel cathelicidin peptides, OL-CATH-1 and -2, from the frog Odorrana livida, which both show potent antimicrobial and anti-inflammatory activities. Wang et al. (2013)isolated five novel AMPs from Limnonectes kuhlii frog skin secretions, which exhibited potent antimicrobial activity against several gram-negative and gram-positive bacterial strains, but with low hemolytic activity on mammalian cells.
Other extensively studied cathelicidins include human-β defensin 3 (hBD-3), sheep myeloid peptide (AMP-29), rat cathelin-related antimicrobial peptide (rCRAMP), and bovine myeloid antimicrobial peptide 27 (BMAP-27), which demonstrate potent microbicidal activity against pathogenic strains such as E. coli, P. aeruginosa, MRSA, and A. baumannii, inhibition of biofilm formation, and immunomodulation activity (Dhople et al., 2006; Giacometti et al., 2004; Guo et al., 2018; Kościuczuk et al., 2012; Kurosaka et al., 2005).
Besides cathelicidins, other AMPs with diverse structural scaffolds and properties have been reported. Indolicidin, a natural cationic peptide from bovine neutrophils, also exhibits potent bactericidal activity through membrane permeabilization against gram-negative and gram-positive nosocomial pathogens such as E. coli, P. aeruginosa, and S. aureus (Falla et al., 1996). Several synthesized derivates of indolicidin have shown improved antimicrobial activities against a wide panel of MDR nosocomial pathogens with low MIC values, e.g., RN7-IN10 and RN7-IN9 (Jindal et al., 2015). Omiganan, a novel peptide currently under phase III clinical trials as a therapeutic agent for bacterial-caused acne and catheter-associated bloodstream infection (Melo & Castanho, 2007), also exhibits potent broad-spectrum antimicrobial activity (Sader et al., 2004).
Also currently under preclinical trial, Ci-MAM-A24, a synthetic peptide derivative of a peptide precursor isolated from Ciona intestinalis, exhibits potent bactericidal activity through membrane permeabilization at low concentration (MIC<10 µg/mL) against MDR nosocomial pathogens, including MRSA, VRE, MDR P. aeruginosa, ESBL-producing E. coli (Fedders et al., 2010). Thus, it is an important candidate as a therapeutic agent against MDR nosocomial pathogens. The pexiganan peptide, a synthetic 22-amino acid residue analog of magainin isolated from the African clawed frog Xenopus laevis (Ge et al., 1999; Zasloff, 1987),also exhibits potent broad-spectrum bactericidal activity against several pathogens at low concentration. It is currently in phase III clinical trials as an agent for diabetic foot ulcers caused by bacterial infections and can rapidly kill pathogens such as P. aeruginosa and several other gram-positive and gram-negative bacteria, with a low tendency to induce resistance selection (Ge et al., 1999; Mahlapuu et al., 2016). However, several reports demonstrate selection of resistance to pexiganan following long-term laboratory exposure of pathogens (Habets & Brockhurst, 2012; Perron et al., 2006).
Thanatin is another potential therapeutic candidate for MDR pathogens. It is a 21-amino acid residue peptide and its analog, S-thanatin, exhibits low hemolytic activity and potent antimicrobial activity against several strains of gram-negative and gram-positive bacteria (Wu et al., 2011). Furthermore, with improved therapeutic activity, S-thanatin shows potent activity against nosocomial pathogens K. pneumoniae and E. coli at very low concentrations, and can alleviate sepsis in mice models, thus making it an excellent therapeutic candidate (Ding et al., 2009; Wu et al., 2013).
Colistin is an important AMP used as a last-resort drug for the treatment of MDR pathogen infections. Recent reports of selection of resistance to colistin thus present a concerning global healthcare predicament in the fight against MDR bacterial infections (Falagas et al., 2005; MacNair et al., 2018; Yamamoto et al., 2018). Remarkably, two novel AMPs (AA139 and SET-M33) with similar MOA as colistin are currently in development and have shown excellent therapeutic potential both in vitro against several MDR gram-pathogens and in-vivo in animal disease models (van der Weide et al., 2019). AA139 is a 21-amino acid amphipathic peptide with reduced cytotoxic and hemolytic activity. It was designed from arenicin-3, a peptide with potent microbicidal activity against several MDR bacterial strains and isolated from the marine lugworm Arenicola marina (Sierra et al., 2017; Wang et al., 2018). SET-M33 is a synthetic AMP exhibiting potent microbicidal activity against gram-negative bacteria through membrane disruption and an excellent candidate for in vivo application, as evidenced from animal models of infectious diseases having both anti-inflammatory and immunomodulatory activities (Brunetti et al., 2016b; Falciani et al., 2012; van der Weide et al., 2017).
Several AMPs with potent activity against gram-positive and gram-negative MDR and showing excellent prospects as therapeutic agents have also been isolated and characterized from marine animals (Destoumieux-Garzón et al., 2016; Tincu & Taylor., 2004). For example, mytimacin-AF, an 80-cysteine-rich amino acid residue isolated from marine mollusks, shows potent activity against both gram-positive and gram-negative bacteria, especially against nosocomial pathogens S. aureus and K. pneumoniae (MIC<2 μg/mL) (Zhong et al., 2013). EC-hepcidin3, cloned from marine fish Epinephelus coioides,is a novel cysteine-rich AMP with rapid and potent microbicidal activity against S. aureus and Pseudomonasstutzeri (MIC<3 μmol/L) (Qu et al., 2013). Tachyplesins, a class of peptides isolated from horseshoe crab hemocytes, are cationic β-hairpin structured peptides with potent antimicrobial activities against MDR gram-negative and gram-positive bacteria at low concentrations (Falanga et al., 2016; Liu et al., 2018; Nakamura et al., 1988), although with increasing hemolytic and cytotoxic activity at higher levels (Edwards et al., 2017; Liu et al., 2018; Ohta et al., 1992).
Continued research on AMPs has yielded a new class of highly potent peptides called “Selectively Targeted AMPs” (STAMPs), which demonstrate increased sensitivity to target pathogens without harming normal flora and increased bacterial killing potency and kinetics (Chung & Khanum, 2017). Such AMPs have been designed to target pathogens through specific determinants followed by selective killing (Sarma et al., 2018). Notable STAMPs include synthetic peptide M8(KH)-20, a multi-headed peptide specifically targeting and potently killing P. aeruginosa and Streptococcus mutans in vitro with very little effect on other pathogens (He et al., 2009). Oritavancin, another synthetic STAMP designed from the naturally occurring glycopeptide chloroeremoycin and currently in clinical development, is reported to be highly selective and potently kills MDR pathogens such as methicillin-resistant S. aureus and vancomycin-resistant S. aureus (VRSA)more rapidly than vancomycin through membrane permeabilization and inhibition of cell wall synthesis (Allen & Nicas, 2003; Chung & Khanum, 2017).
CHALLENGES FACING EXPLOITATION OF AMPs AS THERAPEUTIC AGENTS AND SOLUTION STRATEGIES
To be considered for approval as a therapeutic agent, effective AMPs need to show broad-spectrum activity, high selectivity for bacterial pathogens, and low cytotoxicity on mammalian cells (Dathe & Wieprecht, 1999). Currently, various AMPs are under clinical trial; however, several factors curtail their full utilization and approval as antibiotic alternatives (Falanga et al., 2016; Lombardi et al., 2019; Wang et al., 2016). The main factor limiting the systemic application of AMPs is their sensitivity to proteolytic digestion. For instance, host proteolytic enzymes in intestinal mucosa, gastrointestinal tract, and blood plasma can readily degrade antimicrobial peptides, which negatively impacts their in vivo stability and pharmacokinetics (Moncla et al., 2011; Starr & Wimley, 2017). Therefore, as a result, many AMPs are limited to topical application rather than oral or intravenous administration. Moreover, studies have demonstrated that, as a defensive mechanism in the presence of AMPs, certain bacteria will up-regulate the secretion of proteolytic enzymes and metalloproteases (MMPs) that can partially or completely degrade AMPs (Lai et al., 2007; Sieprawska-Lupa et al., 2004).
Toxicity and efficacy of AMPs is another major drawback of their use. Several AMPs have adverse side effects in vivo, which limits their use to topical application only (McPhee & Hancock, 2005). Moreover, in addition to the low correlation between in vitro- and in vivo-based results in some cases, physiological conditions such as high salt concentration and serum components can negatively affect the antimicrobial activity of many AMPs (Cantisani et al., 2014; de Breij et al., 2018; Han et al., 2016). For most AMPs, antimicrobial activity is dependent on the electrostatic interactions between positively charged peptides and negatively charged plasma membranes (Guilhelmelli et al., 2013). Such interactions can be affected by elevated salt levels or binding of AMPs to serum components (e.g., lipoproteins) (Li et al., 2017). Therefore, efficacy screening of AMPs under different physiological conditions and in the presence of host components is necessary to affirm their activity.
Compared to conventional antibiotics, AMPs are also expensive to produce. Therefore, current efforts are being directed at designing shorter peptides with reduced side effects and improved stability under different physiological conditions (Kim et al., 2013; Zhao et al., 2015). For example, Li et al. (2017)pioneered an important strategy for designing AMPs with trypsin inhibitor activity based on a peptide isolated from the frog Odorrana grahami.
Although AMPs have a low likelihood to select for resistance, reports already exist detailing the development of resistance against some AMPs (Andersson et al., 2016; Omardien et al., 2016); for example, the development of resistance to colistin by A. baumannii following long-term clinical application (Jeannot et al., 2017; Liu et al., 2016). Colistin is a last-resort drug used for the treatment of MDR nosocomial pathogens, and thus resistance to colistin is an important clinical issue (Liu et al., 2016; MacNair et al., 2018; Paterson & Harris., 2016). Several mechanisms have been reported to be responsible for resistance to AMPs, including expression of efflux pumps, surface charge modification to impede membrane-peptide electrostatic interactions, and increased secretion of proteolytic enzymes (Andersson et al., 2016; Bechinger & Gorr, 2017; Morita et al., 2012). For example, S. aureus can alter surface charge by adding basic amino groups from D-alanine from the cytoplasm to teichoic acid on the membrane surface (Peschel et al., 1999). Resistance to AMPs also raises other critical issues such as the potential development of cross-resistance to antibiotic regimens and host AMPs (Conlon, 2015; Kubicek-Sutherland et al., 2017), which could create unprecedented detrimental consequences.
Presently, studies are focusing on developing strategies to improve the efficacy of AMPs in vivo, enhance selectivity for microbial cells while reducing cytotoxicity, increase stability, and minimize proteolytic degradation. Chemical modifications such as the addition of D-amino acids, cyclization, or acetylation are important strategies used to improve the stability of AMPs and reduce sensitivity to proteolytic degradation (Gao et al., 2018; Zhao et al., 2016). However, additional modifications add to production costs. To solve this, efforts are currently being directed at the design and synthesis of shorter peptides with increased potency and stability and reduced toxicity (Jin et al., 2016; Kim et al., 2013).
The use of delivery systems is another important strategy for improving the stability and efficacy of AMPs (Nordström & Malmsten, 2017). Of particular interest is the use of nanocarriers, which are designed and covalently attached to AMPs to prevent self-aggregation, improve chemical stability, and release profiles of AMPs to target sites. Importantly, such nanocarriers are synthesized from materials that are easily biodegradable, including lipids (such as triglycerides and cholesterol) and polymers such as gels, cellulose, and fiber (D’angelo et al., 2015; Mahlapuu et al., 2016; Yüksel et al., 2016).
FUTURE PROSPECTS AND CONCLUSIONS
In this review, we reiterated the importance of AMPs as potential next-generation antibiotics to mitigate a wide array of microbial infections, including those caused by MDR strains. Natural and synthetic AMPs are unique and exhibit boundless therapeutic potential. Remarkably, AMPs display diverse MOA, with a low tendency to select for resistance, rapid killing ability, and multifunctional activities, thus conferring great advantages over empirical antibiotics. Extensive research on the origin, structure, and biological properties has improved our understanding and use of AMPs. Of particular importance is the potential of these peptides to act as templates and precursor molecules for the development of novel anticancer and antimicrobial agents for bacterial, fungal, and viral infections where most available therapies have been rendered ineffective due to the rapid emergence and spread of resistance. Furthermore, these studies allow for the generation of models mimicking the interaction of these peptides with biological membranes, a key element in their microbicidal activity. Such insight is important in the design and synthesis of AMPs with increased potency against MDR strains and improved chemical stability.
Presently, substantial effort is being dedicated to the design and synthesis of shorter peptides with improved therapeutic activity and reduced cytotoxicity. Such efforts are bearing fruit, as evidenced in the presence of short synthetic AMPs with potent and rapid microbicidal activity against several MDR pathogens, even those showing resistance to last-resort drugs, thus presenting hope in the war against multidrug resistance.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
J.M., X.H., R.L., and Y.Z.Z. conceived the review and prepared the manuscript. All authors contributed to the discussions. All authors read and approved the final version of the manuscript.
Funding Statement
This work was supported by the National Natural Science Foundation of China (21761142002 and 31801975), Chinese Academy of Sciences (XDB31000000, SAJC201606, KFZD-SW-219-2, KFJ-BRP-008, and KGFZD-135-17-011), and Yunnan Province Grant (2015HA023)
REFERENCES
- Adedeji WA. 2016. The treasure called antibiotics. Annals of Ibadan Postgraduate Medicine, 14(2): 56–57. [PMC free article] [PubMed] [Google Scholar]
- Alanis AJ. 2005. Resistance to antibiotics: Are we in the post-antibiotic era?. Archives of Medical Research, 36(6): 697–705. [DOI] [PubMed] [Google Scholar]
- Allen NE, Nicas TI. 2003. Mechanism of action of oritavancin and related glycopeptide antibiotics. FEMS Microbiology Reviews, 26(5): 511–532. [DOI] [PubMed] [Google Scholar]
- Anderson DJ. 2011. Surgical site Infections. Infectious Disease Clinics of North America, 25(1): 135–153. [DOI] [PubMed] [Google Scholar]
- Andersson DI, Hughes D, Kubicek-Sutherland JZ. 2016. Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resistance Updates, 26: 43–57. [DOI] [PubMed] [Google Scholar]
- Arias CA, Murray BE. 2012. The rise of the enterococcus: beyond vancomycin resistance. Nature Reviews Microbiology, 10(4): 266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias M, Piga KB, Hyndman ME, Vogel HJ. 2018. Improving the activity of trp-rich antimicrobial peptides by arg/lys substitutions and changing the length of cationic residues. Biomolecules, 8(2): 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam B, Wang W, Arshad MI, Khurshid M, Muzammil S, Rasool MH, Nisar MA, Alvi RF, Aslam MA, Qamar MU, Salamat MKF, Baloch Z. 2018. Antibiotic resistance: a rundown of a global crisis. Infection and Drug Resistance, 11: 1645–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayukekbong JA, Ntemgwa M, Atabe AN . 2017. The threat of antimicrobial resistance in developing countries: causes and control strategies. Antimicrobial Resistance & Infection Control, 6(1): 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechinger B, Gorr SU . 2017. Antimicrobial peptides: mechanisms of action and resistance. Journal of Dental Research, 96(3): 254–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bei YH, Pan LL, Zhou QL, Zhao CM, Xie Y, Wu CF, Meng XM, Gu HY, Xu JH, Zhou L, Sluijter JPG, Das S, Agerberth B, Sun J, Xiao JJ. 2019. Cathelicidin-related antimicrobial peptide protects against myocardial ischemia/reperfusion injury. BMC Medicine, 17(1): 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat G, Kamath S, Hussain A. 2006. Nosocomial methicillin--resistant Staphylococcus aureus with reduced susceptibility to vancomycin. Indian Journal of Pathology & Microbiology, 49(2): 311–312. [PubMed] [Google Scholar]
- Å Björstad, Askarieh G, Brown KL, Christenson K, Forsman H, Önnheim K, Li H-N, Teneberg S, Maier O, Hoekstra D, Dahlgren C, Davidson DJ, Bylund J. 2009. The host defense peptide LL-37 selectively permeabilizes apoptotic leukocytes. Antimicrobial Agents and Chemotherapy, 53(3): 1027–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJV. 2014. Molecular mechanisms of antibiotic resistance. Nature Reviews Microbiology, 13: 42–51. [DOI] [PubMed] [Google Scholar]
- Brogden KA. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria?. Nature Reviews Microbiology, 3(3): 238–250. [DOI] [PubMed] [Google Scholar]
- Brogden KA, Ackermann M, Mccray Jr PB, Tack BF. 2003. Antimicrobial peptides in animals and their role in host defences. International Journal of Antimicrobial Agents, 22(5): 465–478. [DOI] [PubMed] [Google Scholar]
- Brown SE, Howard A, Kasprzak AB, Gordon KH, East PD. 2009. A peptidomics study reveals the impressive antimicrobial peptide arsenal of the wax moth Galleria mellonella . Insect Biochemistry and Molecular Biology, 39(11): 792–800. [DOI] [PubMed] [Google Scholar]
- Brunetti J, Falciani C, Roscia G, Pollini S, Bindi S, Scali S, Arrieta UC, Gómez-Vallejo V, Quercini L, Ibba E, Prato M, Rossolini GM, Llop J, Bracci L, Pini A. 2016a. In vitro and in vivo efficacy, toxicity, bio-distribution and resistance selection of a novel antibacterial drug candidate. Scientific Reports, 6: 26077–26077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti J, Roscia G, Lampronti I, Gambari R, Quercini L, Falciani C, Bracci L, Pini A. 2016b. Immunomodulatory and anti-inflammatory activity in vitro and in vivo of a novel antimicrobial candidate. The Journal of Biological Chemistry, 291(49): 25742–25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantisani M, Finamore E, Mignogna E, Falanga A, Nicoletti GF, Pedone C, Morelli G, Leone M, Galdiero M, Galdiero S. 2014. Structural insights into and activity analysis of the antimicrobial peptide myxinidin. Antimicrobial Agents and Chemotherapy, 58(9): 5280–5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention 1997. Update: staphylococcus aureus with reduced susceptibility to vancomycin--united states, 1997. mmwr. Morbidity and Mortality Weekly Report, 46(35): 813–815. [PubMed] [Google Scholar]
- Chang S, Sievert DM, Hageman JC, Boulton ML, Tenover FC, Downes FP, Shah S, Rudrik JT, Pupp GR, Brown WJ, Cardo D, Fridkin SK. 2003. Infection with vancomycin-resistant Staphylococcus aureus containing the vana resistance gene. New England Journal of Medicine, 348(14): 1342–1347. [DOI] [PubMed] [Google Scholar]
- Chen YX, Guarnieri MT, Vasil AI, Vasil ML, Mant CT, Hodges RS. 2007. Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides. Antimicrobial Agents and Chemotherapy, 51(4): 1398–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YX, Mant CT, Farmer SW, Hancock REW, Vasil ML, Hodges RS. 2005. Rational design of alpha-helical antimicrobial peptides with enhanced activities and specificity/therapeutic index. The Journal of Biological Chemistry, 280(13): 12316–12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HT, Kuo TY, Chiang JC, Pei MJ, Yang WT, Yu HC, Lin SB, Chen WJ. 2008. Design and synthesis of cationic antimicrobial peptides with improved activity and selectivity against Vibrio spp. International Journal of Antimicrobial Agents, 32(2): 130–138. [DOI] [PubMed] [Google Scholar]
- Chung PY, Khanum R. 2017. Antimicrobial peptides as potential anti-biofilm agents against multidrug-resistant bacteria. Journal of Microbiology, Immunology and Infection, 50(4): 405–410. [DOI] [PubMed] [Google Scholar]
- Conlon JM. 2015. Host-defense peptides of the skin with therapeutic potential: from hagfish to human. Peptides, 67: 29–38. [DOI] [PubMed] [Google Scholar]
- Craik DJ. 2012. Host-defense activities of cyclotides. Toxins (Basel), 4(2): 139–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’angelo I, Casciaro B, Miro A, Quaglia F, Mangoni ML, Ungaro F. 2015. Overcoming barriers in Pseudomonas aeruginosa lung infections: engineered nanoparticles for local delivery of a cationic antimicrobial peptide. Colloids and Surfaces B: Biointerfaces, 135: 717–725. [DOI] [PubMed] [Google Scholar]
- Dathe M, Wieprecht T. 1999. Structural features of helical antimicrobial peptides: their potential to modulate activity on model membranes and biological cells. Biochimica et Biophysica Acta (BBA) - Biomembranes, 1462(1-2): 71–87. [DOI] [PubMed] [Google Scholar]
- Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiology and Molecular Biology Reviews, 74(3): 417–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Breij A, Riool M, Cordfunke RA, Malanovic N, De Boer L, Koning RI, Ravensbergen E, Franken M, van der Heijde T, Boekema BK, Kwakman PHS, Kamp N, El Ghalbzouri A, Lohner K, SaJ Zaat, Drijfhout JW, Nibbering PH. 2018. The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Science Translational Medicine, 10(423): eaan 4044. [DOI] [PubMed] [Google Scholar]
- de Kraker MEA, Stewardson AJ, Harbarth S. 2016. Will 10 million people die a year due to antimicrobial resistance by 2050?. PLoS Medicine, 13(11): e1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Y, Chen Q, Schmidt AP, Anderson GM, Wang JM, Wooters J, Oppenheim JJ, Chertov O. 2000. LL-37, the neutrophil granule-and epithelial cell-derived cathelicidin, utilizes formyl peptide receptor-like 1 (FPRL1) as a receptor to chemoattract human peripheral blood neutrophils, monocytes, and T cells. The Journal of Experimental Medicine, 192(7): 1069–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis O. 2017. Route of transmission of Staphylococcus aureus . The Lancet Infectious Diseases, 17(2): 124–125. [DOI] [PubMed] [Google Scholar]
- Deslouches B, Di YP. 2017. Antimicrobial peptides: a potential therapeutic option for surgical site infections. Clinics in Surgery, 2: 1740. [PMC free article] [PubMed] [Google Scholar]
- Destoumieux-Garzón D, Rosa RD, Schmitt P, Barreto C, Vidal-Dupiol J, Mitta G, Gueguen Y, Bachère E. 2016. Antimicrobial peptides in marine invertebrate health and disease. Philosophical Transactions of the Royal Society B: Biological Sciences, 371(1695): 20150300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever LA, Dermody TS. 1991. Mechanisms of bacterial resistance to antibiotics. JAMA Internal Medicine, 151(5): 886–895. [PubMed] [Google Scholar]
- Dhople V, Krukemeyer A, Ramamoorthy A. 2006. The human beta-defensin-3, an antibacterial peptide with multiple biological functions. Biochimica et Biophysica Acta (BBA) - Biomembranes, 1758(9): 1499–1512. [DOI] [PubMed] [Google Scholar]
- Ding JX, Wu GQ, Zhao R, Li LX, Shen ZL, Xi T. 2009. Activity of S-thanatin thanatin analog against multidrug-resistant gram-negative organisms in vitro and the synergy with 7 kinds of antibiotics. Pharmaceutical biotechnology, 16(2): 158–161. [Google Scholar]
- Dotel R, O'sullivan M, Gilbert G. 2017. Staphylococcus aureus in critical care. The Lancet Infectious Diseases, 17(6): 579–580. [DOI] [PubMed] [Google Scholar]
- Du H, Puri S, Mccall A, Norris HL, Russo T, Edgerton M. 2017. Human salivary protein histatin 5 has potent bactericidal activity against eskape pathogens. Frontiers in Cellular and Infection Microbiology, 7: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunne WM Jr. 2002. Bacterial adhesion: seen any good biofilms lately?. Clinical Microbiology Reviews, 15(2): 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürr UHN, Sudheendra US, Ramamoorthy A. 2006. LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochimica et Biophysica Acta (BBA) - Biomembranes, 1758(9): 1408-1425. [DOI] [PubMed] [Google Scholar]
- Edwards IA, Elliott AG, Kavanagh AM, Blaskovich MAT, Cooper MA . 2017. Structure-activity and-toxicity relationships of the antimicrobial peptide tachyplesin-1. ACS Infectious Diseases, 3(12): 917–926. [DOI] [PubMed] [Google Scholar]
- Evans J, Wang YD, Shaw KP, Vernon LP. 1989. Cellular responses to Pyrularia thionin are mediated by Ca2+ influx and phospholipase A2 activation and are inhibited by thionin tyrosine iodination. Proceedings of the National Academy of Sciences of the United States of America, 86(15): 5849–5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falagas ME, Kasiakou SK, Saravolatz LD. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clinical Infectious Diseases, 40(9): 1333–1341. [DOI] [PubMed] [Google Scholar]
- Falanga A, Galdiero S. 2018. Emerging therapeutic agents on the basis of naturally occurring antimicrobial peptides. The Royal Society of Chemistry, 42: 190–227. [Google Scholar]
- Falanga A, Lombardi L, Franci G, Vitiello M, Iovene MR, Morelli G, Galdiero M, Galdiero S. 2016. Marine antimicrobial peptides: nature provides templates for the design of novel compounds against pathogenic bacteria. International Journal of Molecular Sciences, 17(5): 785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falciani C, Lozzi L, Pollini S, Luca V, Carnicelli V, Brunetti J, Lelli B, Bindi S, Scali S, Di Giulio A, Rossolini GM, Mangoni ML, Bracci L, Pini A. 2012. Isomerization of an antimicrobial peptide broadens antimicrobial spectrum to gram-positive bacterial pathogens. PLoS One, 7(10): e46259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falla TJ, Karunaratne DN, Hancock REW. 1996. Mode of action of the antimicrobial peptide indolicidin. The Journal of Biological Chemistry, 271(32): 19298–19303. [DOI] [PubMed] [Google Scholar]
- Fedders H, Podschun R, Leippe M. 2010. The antimicrobial peptide Ci-MAM-A24 is highly active against multidrug-resistant and anaerobic bacteria pathogenic for humans. International Journal of Antimicrobial Agents, 36(3): 264–266. [DOI] [PubMed] [Google Scholar]
- Fernandez De Caleya R, Gonzalez-Pascual B, García-Olmedo F, Carbonero P. 1972. Susceptibility of phytopathogenic bacteria to wheat purothionins in vitro. Applied Microbiology, 23(5): 998–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi D, Climo MW, Wong AH, Edmond MB, Wenzel RP. 1999. Seeking vancomycin resistant Staphylococcus aureus among patients with vancomycin-resistant enterococci. Clinical Infectious Diseases, 29(6): 1566–1568. [DOI] [PubMed] [Google Scholar]
- Frecer V, Ho B, Ding JL. 2004. De novo design of potent antimicrobial peptides. Antimicrobial Agents and Chemotherapy, 48(9): 3349–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon MC, Strandberg E, Grau-Campistany A, Wadhwani P, Reichert J, Bürck J, Rabanal F, Auger M, Paquin JF, Ulrich AS. 2017. Influence of the length and charge on the activity of α-helical amphipathic antimicrobial peptides. Biochemistry, 56(11): 1680–1695. [DOI] [PubMed] [Google Scholar]
- Ganz T. 2003. The role of antimicrobial peptides in innate immunity1. Integrative and Comparative Biology, 43(2): 300–304. [DOI] [PubMed] [Google Scholar]
- Gao AG, Hakimi SM, Mittanck CA, Wu Y, Woerner BM, Stark DM, Shah DM, Liang J, Rommens CMT. 2000. Fungal pathogen protection in potato by expression of a plant defensin peptide. Nature Biotechnology, 18(12): 1307–1310. [DOI] [PubMed] [Google Scholar]
- Gao YD, Fang HT, Fang L, Liu DW, Liu JS, Su MH, Fang Z, Ren WZ, Jiao HP. 2018. The modification and design of antimicrobial peptide. Current Pharmaceutical Design, 24(8): 904–910. [DOI] [PubMed] [Google Scholar]
- Ge Y, Macdonald DL, Holroyd KJ, Thornsberry C, Wexler H, Zasloff M. 1999. In vitro antibacterial properties of pexiganan, an analog of magainin. Antimicrobial Agents and Chemotherapy, 43(4): 782–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee ML, Burton M, Grevis-James A, Hossain MA, Mcarthur S, Palombo EA, Wade JD, Clayton AHA. 2013. Imaging the action of antimicrobial peptides on living bacterial cells. Scientific Reports, 3: 1557–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennaro R, Skerlavaj B, Romeo D. 1989. Purification, composition, and activity of two bactenecins, antibacterial peptides of bovine neutrophils. Infection and Immunity, 57(10): 3142–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacometti A, Cirioni O, Ghiselli R, Mocchegiani F, D'amato G, Circo R, Orlando F, Skerlavaj B, Silvestri C, Saba V, Zanetti M, Scalise G. 2004. Cathelicidin peptide sheep myeloid antimicrobial peptide-29 prevents endotoxin-induced mortality in rat models of septic shock. American Journal of Respiratory and Critical Care Medicine, 169(2): 187–194. [DOI] [PubMed] [Google Scholar]
- Gold HS, Moellering RC. 1996. Antimicrobial-drug resistance. New England Journal of Medicine, 335(19): 1445–1453. [DOI] [PubMed] [Google Scholar]
- Groisman EA, Ochman H. 1996. Pathogenicity islands: bacterial evolution in quantum leaps. Cell, 87(5): 791–794. [DOI] [PubMed] [Google Scholar]
- Gudiol C, Carratalà J. 2014. Antibiotic resistance in cancer patients. Expert Review of Anti-infective Therapy, 12(8): 1003–1016. [DOI] [PubMed] [Google Scholar]
- Guilhelmelli F, Vilela N, Albuquerque P, Derengowski LDS, Silva-Pereira I, Kyaw CM. 2013. Antibiotic development challenges: the various mechanisms of action of antimicrobial peptides and of bacterial resistance. Frontiers in Microbiology, 4: 353–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YJ, Xun M, Han J. 2018. A bovine myeloid antimicrobial peptide (BMAP-28) and its analogs kill pan-drug-resistant Acinetobacter baumannii by interacting with outer membrane protein A (OmpA). Medicine (Baltimore), 97(42): e12832–e12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung J, Khyriem AB, Banik A, Lyngdoh WV, Choudhury B, Bhattacharyya P. 2013. Association of biofilm production with multidrug resistance among clinical isolates of Acinetobacter baumannii and Pseudomonas aeruginosa from intensive care unit. Indian Journal of Critical Care Medicine, 17(4): 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MGJL Habets, Brockhurst MA . 2012. Therapeutic antimicrobial peptides may compromise natural immunity. Biology Letters, 8(3): 416–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammami R, Ben Hamida J, Vergoten G, Fliss I. 2009. Phytamp: a database dedicated to antimicrobial plant peptides. Nucleic Acids Research, 37 (Database issue): D963–D968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han HM, Gopal R, Park Y. 2016. Design and membrane-disruption mechanism of charge-enriched AMPs exhibiting cell selectivity, high-salt resistance, and anti-biofilm properties. Amino Acids, 48(2): 505–522. [DOI] [PubMed] [Google Scholar]
- Hancock REW, Brown KL, Mookherjee N. 2006. Host defence peptides from invertebrates – emerging antimicrobial strategies. Immunobiology, 211(4): 315–322. [DOI] [PubMed] [Google Scholar]
- Hancock REW, Lehrer R. 1998. Cationic peptides: a new source of antibiotics. Trends in Biotechnology, 16(2): 82–88. [DOI] [PubMed] [Google Scholar]
- Hancock REW, Nijnik A, Philpott DJ. 2012. Modulating immunity as a therapy for bacterial infections. Nature Reviews Microbiology, 10: 243–254. [DOI] [PubMed] [Google Scholar]
- Hancock REW, Rozek A. 2002. Role of membranes in the activities of antimicrobial cationic peptides. FEMS Microbiology Letters, 206(2): 143–149. [DOI] [PubMed] [Google Scholar]
- Hancock REW, Sahl HG. 2006. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nature Biotechnology, 24: 1551–1557. [DOI] [PubMed] [Google Scholar]
- Haney EF, Hancock REW. 2013. Peptide design for antimicrobial and immunomodulatory applications. Peptide Science, 100(6): 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder J, Schröder JM, Gläser R. 2013. The skin surface as antimicrobial barrier: present concepts and future outlooks. Experimental Dermatology, 22(1): 1–5. [DOI] [PubMed] [Google Scholar]
- Harwig SSL, Waring A, Yang HJ, Cho Y, Tan L, Lehrer RI. 1996. Intramolecular disulfide bonds enhance the antimicrobial and lytic activities of protegrins at physiological sodium chloride concentrations. European Journal of Biochemistry, 240(2): 352–357. [DOI] [PubMed] [Google Scholar]
- He J, Anderson MH, Shi WY, Eckert R. 2009. Design and activity of a 'dual-targeted' antimicrobial peptide. International Journal of Antimicrobial Agents, 33(6): 532–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilchie AL, Wuerth K, Hancock REW. 2013. Immune modulation by multifaceted cationic host defense (antimicrobial) peptides. Nature Chemical Biology, 9: 761–768. [DOI] [PubMed] [Google Scholar]
- Hirsch EB, Tam VH. 2010. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Review Pharmacoeconomics & Outcomes Research, 10(4): 441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SY, Park TG, Lee KH. 2001. The effect of charge increase on the specificity and activity of a short antimicrobial peptide. Peptides, 22(10): 1669–1674. [DOI] [PubMed] [Google Scholar]
- Huang YB, Huang JF, Chen YX. 2010. Alpha-helical cationic antimicrobial peptides: relationships of structure and function. Protein & Cell, 1(2): 143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurdle JG, O'neill AJ, Chopra I, Lee RE. 2011. Targeting bacterial membrane function: an underexploited mechanism for treating persistent infections. Nature Reviews Microbiolology, 9(1): 62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeannot K, Bolard A, Plésiat P. 2017. Resistance to polymyxins in gram-negative organisms. International Journal of Antimicrobial Agents, 49(5): 526–535. [DOI] [PubMed] [Google Scholar]
- Jesus B, Antonio O, Jose-Maria GG. 2002. Mutation and evolution of antibiotic resistance: antibiotics as promoters of antibiotic resistance?. Current Drug Targets, 3(4): 345–349. [DOI] [PubMed] [Google Scholar]
- Jiang ZQ, Vasil AI, Hale JD, Hancock REW, Vasil ML, Hodges RS. 2008. Effects of net charge and the number of positively charged residues on the biological activity of amphipathic alpha-helical cationic antimicrobial peptides. Biopolymers, 90(3): 369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Bai XW, Luan N, Yao HM, Zhang ZY, Liu WH, Chen Y, Yan XW, Rong MQ, Lai R, Lu QM. 2016. A designed tryptophan-and lysine/arginine-rich antimicrobial peptide with therapeutic potential for clinical antibiotic-resistant Candida albicans vaginitis. Journal of Medicinal Chemistry, 59(5): 1791–1799. [DOI] [PubMed] [Google Scholar]
- Jindal HM, Le CF, Mohd Yusof MY, Velayuthan RD, Lee VS, Zain SM, Isa DM, Sekaran SD. 2015. Antimicrobial activity of novel synthetic peptides derived from indolicidin and ranalexin against Streptococcus pneumoniae . PLoS One, 10(6): e0128532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y, Yue XC, Yang S, Li S. 2019. Association of aspirin resistance with increased mortality in ischemic stroke. The Journal of Nutrition, Health & Aging, 23(3): 266–270. [DOI] [PubMed] [Google Scholar]
- Kang HK, Kim C, Seo CH, Park Y. 2017. The therapeutic applications of antimicrobial peptides (AMPs): a patent review. Journal of Microbiology, 55(1): 1–12. [DOI] [PubMed] [Google Scholar]
- Keren I, Mulcahy LR, Lewis K. 2012. Persister eradication: lessons from the world of natural products. Methods in Enzymology, 517: 387–406. [DOI] [PubMed] [Google Scholar]
- Khan HA, Baig FK, Mehboob R. 2017. Nosocomial infections: epidemiology, prevention, control and surveillance. Asian Pacific Journal of Tropical Biomedicine, 7(5): 478–482. [Google Scholar]
- Kim H, Jang JH, Kim SC, Cho JH. 2013. De novo generation of short antimicrobial peptides with enhanced stability and cell specificity. Journal of Antimicrobial Chemotherapy, 69(1): 121–132. [DOI] [PubMed] [Google Scholar]
- Koenig SM, Truwit JD. 2006. Ventilator-associated pneumonia: diagnosis, treatment, and prevention. Clinical Microbiogy Reviews, 19(4): 637–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo HB, Seo J. 2019. Antimicrobial peptides under clinical investigation. Peptide Science, doi: 10.1002/pep2.24122. [DOI] [Google Scholar]
- Kościuczuk EM, Lisowski P, Jarczak J, Strzałkowska N, Jóźwik A, Horbańczuk J, Krzyżewski J, Zwierzchowski L, Bagnicka E. 2012. Cathelicidins: family of antimicrobial peptides. A review. Molecular Biolology Reports, 39(12): 10957–10970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek-Sutherland JZ, Lofton H, Vestergaard M, Hjort K, Ingmer H, Andersson DI . 2017. Antimicrobial peptide exposure selects for Staphylococcus aureus resistance to human defence peptides. The Journal of Antimicrobial Chemotherapy, 72(1): 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Kizhakkedathu JN, Straus SK. 2018. Antimicrobial peptides: diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules, 8(1): 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kümmerer K. 2004. Resistance in the environment. Journal of Antimicrobial Chemotherapy, 54(2): 311–320. [DOI] [PubMed] [Google Scholar]
- Kurosaka K, Chen Q, Yarovinsky F, Oppenheim JJ, Yang D. 2005. Mouse cathelin-related antimicrobial peptide chemoattracts leukocytes using formyl peptide receptor-like 1/mouse formyl peptide receptor-like 2 as the receptor and acts as an immune adjuvant. The Journal of Immunology, 174(10): 6257–6265. [DOI] [PubMed] [Google Scholar]
- Lai YP, Villaruz AE, Li M, Cha DJ, Sturdevant DE, Otto M. 2007. The human anionic antimicrobial peptide dermcidin induces proteolytic defence mechanisms in staphylococci. Molecular Microbiology, 63(2): 497–506. [DOI] [PubMed] [Google Scholar]
- Laxminarayan R, Brown GM. 2001. Economics of antibiotic resistance: a theory of optimal use. Journal of Environmental Economics and Management, 42(2): 183–206. [Google Scholar]
- Lehrer RI, Barton A, Daher KA, Harwig SS, Ganz T, Selsted ME. 1989. Interaction of human defensins with Escherichia coli . mechanism of bactericidal activity. The Journal of Clinical Investigation, 84(2): 553–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin AS, Barone AA, Penço J, Santos MV, Marinho IS, Arruda EAG, Manrique EI, Costa SF. 1999. Intravenous colistin as therapy for nosocomial infections caused by multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii . Clinical Infectious Diseases, 28(5): 1008–1011. [DOI] [PubMed] [Google Scholar]
- Lewis K. 2008. Multidrug tolerance of biofilms and persister cells. In: Romeo T. Bacterial Biofilms. Berlin, Heidelberg: Springer Berlin Heidelberg, 107–131. [DOI] [PubMed] [Google Scholar]
- Li JG, Koh JJ, Liu SP, Lakshminarayanan R, Verma CS, Beuerman RW. 2017. Membrane active antimicrobial peptides: translating mechanistic insights to design. Frontiers in Neuroscience, 11: 73–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SA, Lee WH, Zhang Y. 2012. Efficacy of OH-CATH30 and its analogs against drug-resistant bacteria in vitro and in mouse models. Antimicrobial Agents and Chemotherapy, 56(6): 3309–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Nishino K, Roberts MC, Tolmasky M, Aminov RI, Zhang LX. 2015. Mechanisms of antibiotic resistance. Frontiers in Microbiology, 6: 34. doi: 10.3389/fmicb.2015.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden PK. 2002. Treatment options for vancomycin-resistant enterococcal infections. Drugs, 62(3): 425–441. [DOI] [PubMed] [Google Scholar]
- Liu B, Pop M. 2009. ARDB--antibiotic resistance genes database. Nucleic Acids Research, 37 (Database issue): D443–D447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CB, Qi JL, Shan B, Ma YB. 2018. Tachyplesin causes membrane instability that kills multidrug-resistant bacteria by inhibiting the 3-ketoacyl carrier protein reductase fabG. Frontiers in Microbiology, 9: 825. doi: 10.3389/fmicb.2018.00825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CB, Shan B, Bai HM, Tang J, Yan LZ, Ma YB. 2015. Hydrophilic/hydrophobic characters of antimicrobial peptides derived from animals and their effects on multidrug resistant clinical isolates. Zoological Research, 36(1): 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Liu H, Ma YF, Wu J, Yang HL, Ye HH, Lai R. 2011. There are abundant antimicrobial peptides in brains of two kinds of Bombina toads. Journal of Proteome Research, 10(4): 1806–1815. [DOI] [PubMed] [Google Scholar]
- Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang XH, Yu LF, Gu DX, Ren HW, Chen XJ, Lv LC, He DD, Zhou HW, Liang Z, Liu J-H, Shen JZ. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. The Lancet Infectious Diseases, 16(2): 161–168. [DOI] [PubMed] [Google Scholar]
- Loeffler JM, Nelson D, Fischetti VA. 2001. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science, 294(5549): 2170–2172. [DOI] [PubMed] [Google Scholar]
- Lombardi L, Falanga A, Del Genio V, Galdiero S. 2019. A new hope: self-assembling peptides with antimicrobial activity. Pharmaceutics, 11(4): 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Ma YF, Wang X, Liang JG, Zhang CX, Zhang KY, Lin GQ, Lai R. 2008. The first antimicrobial peptide from sea amphibian. Molecular Immunology, 45(3): 678–681. [DOI] [PubMed] [Google Scholar]
- Lupei MI, Mann HJ, Beilman GJ, Oancea C, Chipman JG. 2010. Inadequate antibiotic therapy in solid organ transplant recipients is associated with a higher mortality rate. Surgical Infections, 11(1): 33–39. [DOI] [PubMed] [Google Scholar]
- Lyu YF, Yang Y, Lyu X, Dong N, Shan A. 2016. Antimicrobial activity, improved cell selectivity and mode of action of short PMAP-36-derived peptides against bacteria and Candida. Scientific Reports, 6: 27258–27258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnair CR, Stokes JM, Carfrae LA, Fiebig-Comyn AA, Coombes BK, Mulvey MR, Brown ED. 2018. Overcoming mcr-1 mediated colistin resistance with colistin in combination with other antibiotics. Nature Communications, 9(1): 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlapuu M, Håkansson J, Ringstad L, Björn C. 2016a. Antimicrobial peptides: an emerging category of therapeutic agents. Frontiers in Cellular and Infection Microbiology, 6: 194–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchanda V, Sanchaita S, Singh NP. 2010. Multidrug resistant acinetobacter. Journal of Global Infectious Diseases, 2(3): 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maróti G, Kereszt A, É Kondorosi, Mergaert P. 2011. Natural roles of antimicrobial peptides in microbes, plants and animals. Research in Microbiology, 162(4): 363–374. [DOI] [PubMed] [Google Scholar]
- Martínez JL, Baquero F. 2002. Interactions among strategies associated with bacterial infection: pathogenicity, epidemicity, and antibiotic resistance. Clinical Microbiology Reviews, 15(4): 647–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez RM, Wolk DM. 2016. Bloodstream infections. In: Hayden R, Wolk D, Carroll K, Tang YW. Diagnostic Microbiology of the Immunocompromised Host, 2nd edn. Washington DC: ASM Press. [Google Scholar]
- Matsuzaki K. 1999. Why and how are peptide–lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochimica et Biophysica Acta (BBA) - Biomembranes, 1462(1-2): 1–10. [DOI] [PubMed] [Google Scholar]
- Mcphee JB, Hancock REW. 2005. Function and therapeutic potential of host defence peptides. Journal of Peptide Science, 11(11): 677–687. [DOI] [PubMed] [Google Scholar]
- Melo MN, MARB Castanho. 2007. Omiganan interaction with bacterial membranes and cell wall models. Assigning a biological role to saturation. Biochimica et Biophysica Acta (BBA) - Biomembranes, 1768(5): 1277–1290. [DOI] [PubMed] [Google Scholar]
- Miller LG, Perdreau-Remington F, Rieg G, Mehdi S, Perlroth J, Bayer AS, Tang AW, Phung TO, Spellberg B. 2005. Necrotizing fasciitis caused by community-associated methicillin-resistant Staphylococcus aureus in Los Angeles. New England Journal of Medicine, 352(14): 1445–1453. [DOI] [PubMed] [Google Scholar]
- Mills S, Serrano LM, Griffin C, O'connor PM, Schaad G, Bruining C, Hill C, Ross RP, Meijer WC. 2011. Inhibitory activity of Lactobacillus plantarum LMG P-26358 against Listeria innocua when used as an adjunct starter in the manufacture of cheese. Microbial Cell Factories, 10 (Suppl 1): S7–S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Tokunaga F, Yoneya T, Yoshikawa K, Iwanaga S, Niwa M, Takao T, Shimonishi Y. 1989. Antimicrobial peptides, isolated from horseshoe crab hemocytes, tachyplesin II, and polyphemusins I and II: chemical structures and biological activity. The Journal of Biochemistry, 106(4): 663–668. [DOI] [PubMed] [Google Scholar]
- Moncla BJ, Pryke K, Rohan LC, Graebing PW. 2011. Degradation of naturally occurring and engineered antimicrobial peptides by proteases. Advances in Bioscience and Biotechnology, 2(6): 404–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y, Tomida J, Kawamura Y. 2012. Mexxy multidrug efflux system of Pseudomonas aeruginosa . Frontiers in Microbiology, 3: 408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munita JM, Arias CA. 2016. Mechanisms of antibiotic resistance. Microbiology Spectrum, 4(2), doi: 10.1128/microbiolspec.VMBF-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylonakis E, Podsiadlowski L, Muhammed M, Vilcinskas A. 2016. Diversity, evolution and medical applications of insect antimicrobial peptides. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 371(1695): 20150290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan D, Roy N, Kulkarni O, Nanajkar N, Datey A, Ravichandran S, Thakur C, T S, Aprameya IV, Sarma SP, Chakravortty D, Chandra N. 2019. Ω76: A designed antimicrobial peptide to combat carbapenem-and tigecycline-resistant Acinetobacter baumannii . Science Advances, 5(7): eaax1946–eaax1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Furunaka H, Miyata T, Tokunaga F, Muta T, Iwanaga S, Niwa M, Takao T, Shimonishi Y. 1988. Tachyplesin, a class of antimicrobial peptide from the hemocytes of the horseshoe crab (Tachypleus tridentatus). Isolation and chemical structure. Journal of Biological Chemistry, 263(32): 16709–16713. [PubMed] [Google Scholar]
- Nawrot R, Barylski J, Nowicki G, Broniarczyk J, Buchwald W, Goździcka-Józefiak A. 2014. Plant antimicrobial peptides. Folia Microbiologica, 59(3): 181–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicole JA, Amy TYY, Olga MP, Robert EWH. 2012. Therapeutic potential of host defense peptides in antibiotic-resistant infections. Current Pharmaceutical Design, 18(6): 807–819. [DOI] [PubMed] [Google Scholar]
- Nicolle LE 2014. Catheter associated urinary tract infections. Antimicrobial Resistance and Infection Control, 3: 23–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijnik A, Hancock REW. 2009. Host defence peptides: antimicrobial and immunomodulatory activity and potential applications for tackling antibiotic-resistant infections. Emerging Health Threats Journal, 2: e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble WC, Virani Z, Cree RGA. 1992. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus . FEMS Microbiology Letters, 93(2): 195–198. [DOI] [PubMed] [Google Scholar]
- Nordström R, Malmsten M. 2017. Delivery systems for antimicrobial peptides. Advances in Colloid and Interface Science, 242: 17–34. [DOI] [PubMed] [Google Scholar]
- Ochoa TJ, Ruiz J, Molina M, Del Valle LJ, Vargas M, Gil AI, Ecker L, Barletta F, Hall E, Cleary TG, Lanata CF. 2009. High frequency of antimicrobial drug resistance of diarrheagenic Escherichia coli in infants in peru. The American Journal of Tropical Medicine and Hygiene, 81(2): 296–301. [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Ito H, Masuda K, Tanaka S, Arakawa Y, Wacharotayankun R, Kato N. 1992. Mechanisms of antibacterial action of tachyplesins and polyphemusins, a group of antimicrobial peptides isolated from horseshoe crab hemocytes. Antimicrobial Agents and Chemotherapy, 36(7): 1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omardien S, Brul S, Zaat SAJ. 2016. Antimicrobial activity of cationic antimicrobial peptides against gram-positives: current progress made in understanding the mode of action and the response of bacteria. Frontiers in Cell and Developmental Biology, 4: 111–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheim JJ, Biragyn A, Kwak LW, Yang D. 2003. Roles of antimicrobial peptides such as defensins in innate and adaptive immunity. Annals of the Rheumatic Diseases, 62(suppl 2): ii17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Othman HB, Halim RMA, Gomaa FAM, Amer MZ. 2019. Vancomycin MIC distribution among methicillin-resistant Staphylococcus aureus . Is reduced vancomycin susceptibility related to MIC creep?. Open Access Macedonian Journal of Medical Sciences, 7(1): 12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panteleev PV, Bolosov IA, Balandin SV, Ovchinnikova TV. 2015. Structure and biological functions of β-hairpin antimicrobial peptides. Acta Naturae, 7(1): 37–47. [PMC free article] [PubMed] [Google Scholar]
- Park CB, Kim HS, Kim SC. 1998. Mechanism of action of the antimicrobial peptide buforin II: buforin ii kills microorganisms by penetrating the cell membrane and inhibiting cellular functions. Biochemical and Biophysical Research Communications, 244(1): 253–257. [DOI] [PubMed] [Google Scholar]
- Pasupuleti M, Schmidtchen A, Malmsten M. 2012. Antimicrobial peptides: key components of the innate immune system. Critical Reviews in Biotechnology, 32(2): 143–171. [DOI] [PubMed] [Google Scholar]
- Paterson DL, Harris PNA. 2016. Colistin resistance: a major breach in our last line of defence. The Lancet Infectious Diseases, 16(2): 132–133. [DOI] [PubMed] [Google Scholar]
- Patocka J, Nepovimova E, Klimova B, Wu QH, Kuca K. 2018. Antimicrobial peptides: amphibian host defense peptides. Current Medicinal Chemistry, 25: 1–21. [DOI] [PubMed] [Google Scholar]
- Perron GG, Zasloff M, Bell G. 2006. Experimental evolution of resistance to an antimicrobial peptide. Proceedings of the Royal Society B: Biological sciences, 273(1583): 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Götz F. 1999. Inactivation of the dlt operon Instaphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. Journal of Biological Chemistry, 274(13): 8405–8410. [DOI] [PubMed] [Google Scholar]
- Peters L, Olson L, Khu DTK, Linnros S, Le NK, Hanberger H, Hoang NTB, Tran DM, Larsson M. 2019. Multiple antibiotic resistance as a risk factor for mortality and prolonged hospital stay: a cohort study among neonatal intensive care patients with hospital-acquired infections caused by gram-negative bacteria in Vietnam. PLoS One, 14(5): e0215666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe B, Reto S. 2005. Insect antimicrobial peptides: structures, properties and gene regulation. Protein & Peptide Letters, 12(1): 3–11. [DOI] [PubMed] [Google Scholar]
- Phillips I, Casewell M, Cox T, De Groot B, Friis C, Jones R, Nightingale C, Preston R, Waddell J. 2004. Does the use of antibiotics in food animals pose a risk to human health? A critical review of published data. Journal of Antimicrobial Chemotherapy, 53(1): 28–52. [DOI] [PubMed] [Google Scholar]
- Poirel L, Jayol A, Nordmann P. 2017. Polymyxins: Antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clinical Microbiology Reviews, 30(2): 557–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JR, Cole K, Bexley A, Kostiou V, Eyre DW, Golubchik T, Wilson DJ, Crook DW, Walker AS, Peto TEA, Llewelyn MJ, Paul J. 2017. Transmission of Staphylococcus aureus between health-care workers, the environment, and patients in an intensive care unit: a longitudinal cohort study based on whole-genome sequencing. The Lancet Infectious Diseases, 17(2): 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi RH, Chen Y, Guo ZL, Zhang F, Fang Z, Huang K, Yu HN, Wang YP. 2019. Identification and characterization of two novel cathelicidins from the frog Odorrana livida . Zoological Research, 40(2): 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu HD, Chen B, Peng H, Wang KJ. 2013. Molecular cloning, recombinant expression, and antimicrobial activity of EC-hepcidin3, a new four-cysteine hepcidin isoform from Epinephelus coioides . Bioscience, Biotechnology, and Biochemistry, 77(1): 103–110. [DOI] [PubMed] [Google Scholar]
- Rai DK, Qian S. 2017. Interaction of the antimicrobial peptide aurein 1.2 and charged lipid bilayer. Scientific Reports, 7(1): 3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport M, Faccone D, Pasteran F, Ceriana P, Albornoz E, Petroni A, Corso A. 2016. First description of mcr-1-mediated colistin resistance in human infections caused by Escherichia coli in Latin America. Antimicrobial Agents and Chemotherapy, 60(7): 4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KVR, Yedery RD, Aranha C. 2004. Antimicrobial peptides: premises and promises. International Journal of Antimicrobial Agents, 24(6): 536–547. [DOI] [PubMed] [Google Scholar]
- Richard P, Floch RL, Chamoux C, Pannier M, Espaze E, Richt H. 1994. Pseudomonas aeruginosa outbreak in a burn unit: role of antimicrobials in the emergence of multiply resistant strains. The Journal of Infectious Diseases, 170(2): 377–383. [DOI] [PubMed] [Google Scholar]
- Rinaldi AC. 2002. Antimicrobial peptides from amphibian skin: an expanding scenario: commentary. Current Opinion in Chemical Biology, 6(6): 799–804. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Vázquez N, Lionel Ozores H, Guerra A, González-Freire E, Fuertes A, Panciera M, Priegue JM,Outeiral J, Montenegro J, Garcia-Fandino R, Amorín M, Granja JR. 2014. Membrane-targeted self-assembling cyclic peptide nanotubes. Current Topics in Medicinal Chemistry, 14(23): 2647–2461. [DOI] [PubMed] [Google Scholar]
- Rollins-Smith LA, Reinert LK, O'leary CJ, Houston LE, Woodhams DC. 2005. Antimicrobial peptide defenses in amphibian skin. Integrative and Comparative Biology, 45(1): 137–142. [DOI] [PubMed] [Google Scholar]
- Rose AS, Bradley AR, Valasatava Y, Duarte JM, Prlić A, Rose PW. 2018. NGL viewer: web-based molecular graphics for large complexes. Bioinformatics, 34(21): 3755–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozgonyi F, Szabo D, Kocsis B, Ostorhazi E, Abbadessa G, Cassone M, Wade JD, L . Otvos J. 2009. The antibacterial effect of a proline-rich antibacterial peptide A3-APO. Current Medicinal Chemistry, 16(30): 3996–4002. [DOI] [PubMed] [Google Scholar]
- Sader HS, Fedler KA, Rennie RP, Stevens S, Jones RN. 2004. Omiganan pentahydrochloride (MBI 226), a topical 12-amino-acid cationic peptide: spectrum of antimicrobial activity and measurements of bactericidal activity. Antimicrobial Agents and Chemotherapy, 48(8): 3112–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma P, Mahendiratta S, Prakash A, Medhi B. 2018. Specifically targeted antimicrobial peptides: a new and promising avenue in selective antimicrobial therapy. Indian Journal of Pharmacology, 50(1): 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidtchen A, Pasupuleti M, Malmsten M. 2014. Effect of hydrophobic modifications in antimicrobial peptides. Advances in Colloid and Interface Science, 205: 265–274. [DOI] [PubMed] [Google Scholar]
- Shai Y. 2002. Mode of action of membrane active antimicrobial peptides. Peptide Science, 66(4): 236–248. [DOI] [PubMed] [Google Scholar]
- Sharma H, Nagaraj R. 2015. Human β-defensin 4 with non-native disulfide bridges exhibit antimicrobial activity. PLoS One, 10(3): e0119525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieprawska-Lupa M, Mydel P, Krawczyk K, Wójcik K, Puklo M, Lupa B, Suder P, Silberring J, Reed M, Pohl J, Shafer W, Mcaleese F, Foster T, Travis J, Potempa J. 2004. Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases. Antimicrobial Agents and Chemotherapy, 48(12): 4673–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra JM, Fusté E, Rabanal F, Vinuesa T, Viñas M. 2017. An overview of antimicrobial peptides and the latest advances in their development. Expert Opinion on Biological Therapy, 17(6): 663–676. [DOI] [PubMed] [Google Scholar]
- Silva CCG, Silva SPM, Ribeiro S. 2018. Application of bacteriocins and protective cultures in dairy food preservation. Frontiers in Microbiology, 9: 594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira E, Freitas AR, Peixe L, Novais C. 2009. Environmental spread of antibiotic molecules, antibiotic resistant bacteria and genes: jigsaw pieces of a public health problem. Revista da Faculdade de Ciencias da Sande, 6: 244–253. [Google Scholar]
- Starr CG, Wimley WC. 2017. Antimicrobial peptides are degraded by the cytosolic proteases of human erythrocytes. Biochimica et Biophysica Acta - Biomembranes, 1859(12): 2319–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stec B. 2006. Plant thionins – the structural perspective. Cellular and Molecular Life Sciences CMLS, 63(12): 1370–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefania G, Annarita F, Rita B, Paolo G, Giancarlo M, Massimiliano G. 2015. Antimicrobial peptides as an opportunity against bacterial diseases. Current Medicinal Chemistry, 22(14): 1665–1677. [DOI] [PubMed] [Google Scholar]
- Stewart PS. 2002. Mechanisms of antibiotic resistance in bacterial biofilms. International Journal of Medical Microbiology, 292(2): 107–113. [DOI] [PubMed] [Google Scholar]
- Stotz HU, Thomson JG, Wang Y. 2009. Plant defensins: defense, development and application. Plant Signaling & Behavior, 4(11): 1010–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struelens MJ. 1998. The epidemiology of antimicrobial resistance in hospital acquired infections: problems and possible solutions. British Medical Journal, 317(7159): 652–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachi T, Epand RF, Epand RM, Matsuzaki K. 2002. Position-dependent hydrophobicity of the antimicrobial magainin peptide affects the mode of peptide-lipid interactions and selective toxicity. Biochemistry, 41(34): 10723–10731. [DOI] [PubMed] [Google Scholar]
- Tam JP, Wang SJ, Wong KH, Tan WL. 2015. Antimicrobial peptides from plants. Pharmaceuticals (Basel), 8(4): 711–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares LS, Santos MDO, Viccini LF, Moreira JS, Miller RNG, Franco OL. 2008. Biotechnological potential of antimicrobial peptides from flowers. Peptides, 29(10): 1842–1851. [DOI] [PubMed] [Google Scholar]
- Tincu JA, Taylor SW. 2004. Antimicrobial peptides from marine invertebrates. Antimicrobial Agents and Chemotherapy, 48(10): 3645–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong ZC, Ni LX, Ling JQ. 2014. Antibacterial peptide nisin: a potential role in the inhibition of oral pathogenic bacteria. Peptides, 60: 32–40. [DOI] [PubMed] [Google Scholar]
- Treffers C, Chen L, Anderson RC, Yu PL. 2005. Isolation and characterisation of antimicrobial peptides from deer neutrophils. International Journal of Antimicrobial Agents, 26(2): 165–169. [DOI] [PubMed] [Google Scholar]
- Tzong-Hsien L, Kristopher NH, Marie-Isabel A. 2016. Antimicrobial peptide structure and mechanism of action: a focus on the role of membrane structure. Current Topics in Medicinal Chemistry, 16(1): 25–39. [DOI] [PubMed] [Google Scholar]
- Ulm H, Wilmes M, Shai Y, Sahl HG. 2012. Antimicrobial host defensins- specific antibiotic activities and innate defense modulation. Frontiers in Immunology, 3: 249–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boeckel TP, Brower C, Gilbert M, Grenfell BT, Levin SA, Robinson TP, Teillant A, Laxminarayan R. 2015. Global trends in antimicrobial use in food animals. Proceedings of the National Academy of Sciences of the United States of America, 112(18): 5649–5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weide H, Brunetti J, Pini A, Bracci L, Ambrosini C, Lupetti P, Paccagnini E, Gentile M, Bernini A, Niccolai N, Jongh DV, IAJM Bakker-Woudenberg, Goessens WHF, Hays JP, Falciani CS. 2017. Investigations into the killing activity of an antimicrobial peptide active against extensively antibiotic-resistant K . pneumoniae and P. aeruginosa. Biochimica et Biophysica Acta (BBA) - Biomembranes, 1859(10): 1796–1804. [DOI] [PubMed] [Google Scholar]
- van der Weide H, Vermeulen-De Jongh DMC, van der Meijden A, Boers SA, Kreft D, Ten Kate MT, Falciani C, Pini A, Strandh M, IAJM Bakker-Woudenberg, Hays JP, Goessens WHF. 2019. Antimicrobial activity of two novel antimicrobial peptides AA139 and SET-M33 against clinically and genotypically diverse Klebsiella pneumoniae isolates with differing antibiotic resistance profiles. International Journal of Antimicrobial Agents, 54(2): 159–166. [DOI] [PubMed] [Google Scholar]
- van't Hof W, Veerman ECI, Helmerhorst EJ, Amerongen AVN. 2001. Antimicrobial peptides: Properties and applicability. Biological Chemistry, 382(4): 597–619. [DOI] [PubMed] [Google Scholar]
- Wakabayashi H, Hiratani T, Uchida K, Yamaguchi H. 1996. Antifungal spectrum and fungicidal mechanism of an N-terminal peptide of bovine lactoferrin. Journal of Infection and Chemotherapy, 1(3): 185–189. [DOI] [PubMed] [Google Scholar]
- Wang GS, Li X, Wang Z. 2016. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Research, 44(D1): D1087–D1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GX, Wang Y, Ma DY, Liu H, Li JX, Zhang KY, Yang XL, Lai R, Liu JZ. 2013. Five novel antimicrobial peptides from the kuhl’s wart frog skin secretions, Limnonectes kuhlii . Molecular Biology Reports, 40(2): 1097–1102. [DOI] [PubMed] [Google Scholar]
- Wang YP, Hong J, Liu XH, Yang HL, Liu R, Wu J, Wang A, Lin DH, Lai R. 2008. Snake cathelicidin from Bungarus fasciatus is a potent peptide antibiotics. PLoS One, 3(9): e3217–e3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YP, Zhang ZY, Chen LL, Guang HJ, Li Z, Yang HL, Li JX, You DW, Yu HN, Lai R. 2011. Cathelicidin-BF, a snake cathelicidin-derived antimicrobial peptide, could be an excellent therapeutic agent for acne vulgaris. PLoS One, 6(7): e22120–e22120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZL, Wang XM, Wang JH. 2018. Recent Advances in antibacterial and antiendotoxic peptides or proteins from marine resources. Marine Drugs, 16(2): 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO 2014. Antimicrobial resistance: Global report on surveillance. https://www.who.int/drugresistance/documents/AMR_report_Web_slide_set.pdf. [Google Scholar]
- Wimley WC. 2010. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chemical Biology, 5(10): 905–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhams DC, Ardipradja K, Alford RA, Marantelli G, Reinert LK, Rollins-Smith LA. 2007. Resistance to chytridiomycosis varies among amphibian species and is correlated with skin peptide defenses. Animal Conservation, 10(4): 409–417. [Google Scholar]
- Wu GQ, Li XF, Fan XB, Wu HB, Wang SL, Shen ZL, Xi T. 2011. The activity of antimicrobial peptide S-thanatin is independent on multidrug-resistant spectrum of bacteria. Peptides, 32(6): 1139–1145. [DOI] [PubMed] [Google Scholar]
- Wu GQ, Wu PP, Xue XL, Yan XJ, Liu S, Zhang C, Shen ZL, Xi T. 2013. Application of S-thanatin, an antimicrobial peptide derived from thanatin, in mouse model of Klebsiella pneumoniae infection. Peptides, 45: 73–77. [DOI] [PubMed] [Google Scholar]
- Wu QH, Patočka J, Kuča K. 2018. Insect antimicrobial peptides, a mini review. Toxins (Basel), 10(11): 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xhindoli D, Pacor S, Benincasa M, Scocchi M, Gennaro R, Tossi A. 2016. The human cathelicidin LL-37 — a pore-forming antibacterial peptide and host-cell modulator. Biochimica et Biophysica Acta (BBA) - Biomembranes, 1858(3): 546-–566. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Liu CB, Lai R. 2011. Antimicrobial peptides from amphibians. BioMolecular Concepts, 2(1-2): 27–38. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Ouchi Y. 2012. Antimicrobial peptide defensin: identification of novel isoforms and the characterization of their physiological roles and their significance in the pathogenesis of diseases. Proceedings of the Japan Academy. Series B, Physical and Biological Sciences, 88(4): 152–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Kawahara R, Fujiya Y, Sasaki T, Hirai I, Khong DT, Nguyen TN, Nguyen BX. 2018. Wide dissemination of colistin-resistant Escherichia coli with the mobile resistance gene mcr in healthy residents in Vietnam. Journal of Antimicrobial Chemotherapy, 74(2): 523–524. [DOI] [PubMed] [Google Scholar]
- Yamauchi K, Tomita M, Giehl TJ, Ellison RT, 3rd . 1993. Antibacterial activity of lactoferrin and a pepsin-derived lactoferrin peptide fragment. Infection and Immunity, 61(2): 719–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SC, Lin CH, Sung CT, Fang JY. 2014. Antibacterial activities of bacteriocins: application in foods and pharmaceuticals. Frontiers in Microbiology, 5: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ST, Yub Shin S, Kim YC, Kim Y, Hahm KS, Kim JI. 2002. Conformation-dependent antibiotic activity of tritrpticin, a cathelicidin-derived antimicrobial peptide. Biochemical and Biophysical Research Communications, 296(5): 1044–1050. [DOI] [PubMed] [Google Scholar]
- Yeaman MR, Yount NY. 2003. Mechanisms of antimicrobial peptide action and resistance. Pharmacological Reviews, 55(1): 27–55. [DOI] [PubMed] [Google Scholar]
- Yi HY, Chowdhury M, Huang YD, Yu XQ. 2014. Insect antimicrobial peptides and their applications. Applied Microbiology and Biotechnology, 98(13): 5807–5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yüksel E, Karakeçili A, Demirtaş TT, Gümüşderelioğlu M. 2016. Preparation of bioactive and antimicrobial PLGA membranes by magainin II/EGF functionalization. International Journal of Biological Macromolecules, 86: 162–168. [DOI] [PubMed] [Google Scholar]
- Zaman SB, Hussain MA, Nye R, Mehta V, Mamun KT, Hossain N. 2017. A review on antibiotic resistance: alarm bells are ringing. Cureus, 9(6): e1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasloff M. 1987. Magainins, a class of antimicrobial peptides from xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proceedings of the National Academy of Sciences of the United States of America, 84(15): 5449–5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerweck J, Strandberg E, Kukharenko O, Reichert J, Bürck J, Wadhwani P, Ulrich AS. 2017. Molecular mechanism of synergy between the antimicrobial peptides PGLa and magainin 2. Scientific Reports, 7(1): 13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Guo ZL, Chen Y, Li L, Yu HN, Wang YP. 2019. Effects of C-terminal amidation and heptapeptide ring on the biological activities and advanced structure of amurin-9KY, a novel antimicrobial peptide identified from the brown frog, Rana kunyuensis . Zoological Research, 40(3): 198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LJ, Gallo RL. 2016. Antimicrobial peptides. Current Biology, 26(1): R14–R19. [DOI] [PubMed] [Google Scholar]
- Zhang Y. 2006. Amphibian skin secretions and bio-adaptive significance —implications from Bombina maxima skin secretion proteome. Zoological Research, 27(1): 101–112. [Google Scholar]
- Zhang Y. 2015. Why do we study animal toxins?. Zoological Research, 36(4): 183–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YX, Zou AH, Manchu RG,Zhou YC,Wang SF. 2008. Purification and antimicrobial activity of antimicrobial protein from brown-spotted grouper, Epinephelus fario . Zoological Research, 29(6): 627–632. [Google Scholar]
- Zhang ZY, Meng P, Han Y, Shen CB, Li BW, Hakim MA, Zhang XG, Lu QM, Rong MQ, Lai R. 2015. Mitochondrial DNA-LL-37 complex promotes atherosclerosis by escaping from autophagic recognition. Immunity, 43(6): 1137–1147. [DOI] [PubMed] [Google Scholar]
- Zhao CX, Dwyer MD, Yu AL, Wu Y, Fang S, Middelberg APJ. 2015. A simple and low-cost platform technology for producing pexiganan antimicrobial peptide in E . coli. Biotechnology and Bioengineering, 112(5): 957–964. [DOI] [PubMed] [Google Scholar]
- Zhao F, Lan XQ, Du Y, Chen PY, Zhao J, Zhao F, Lee WH, Zhang Y. 2018. King cobra peptide OH-CATH30 as a potential candidate drug through clinic drug-resistant isolates. Zoological Research, 39(2): 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XW, Wu HY, Lu HR, Li GD, Huang QS. 2013. LAMP: A database linking antimicrobial peptides. PLoS One, 8(6): e66557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YY, Zhang M, Qiu S, Wang JY, Peng JX, Zhao P, Zhu RR, Wang HL, Li Y, Wang KR, Yan WJ, Wang R. 2016. Antimicrobial activity and stability of the D-amino acid substituted derivatives of antimicrobial peptide polybia-MPI. AMB Express, 6(1): 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Wang WH, Yang XM, Yan XW, Liu R. 2013. A novel cysteine-rich antimicrobial peptide from the mucus of the snail of Achatina fulica . Peptides, 39: 1–5. [DOI] [PubMed] [Google Scholar]