Located at the head of the Indo-Burma biodiversity hotspot, the Kunming Institute of Zoology (KIZ), Chinese Academy of Sciences (CAS), serves as China’s main center for research into the diverse animal and ecological resources of southwestern China, Eastern Himalayas, and Southeast Asia. As of October 2019, it has been 60 years since the inception of KIZ. Since 1959, strong roots have been laid down by generations of researchers, allowing KIZ to grow and evolve into a comprehensive research institution renowned for its remarkable achievements in evolutionary mechanisms of animal biodiversity, animal resources protection, and sustainable utilization. It is now recognized as “a major powerhouse in evolutionary biology research in China” and is“establishing itself in the world stage” (Overseas Experts Review Committee, organized by the Bureau of Development Planning, CAS, during international evaluation in 2014).
To celebrate the 60th anniversary of KIZ and the 70th anniversary of CAS, Zoological Research presents this commemorative issue, composed primarily of contributions from KIZ researchers. In addition, it is our great honor to provide here a brief retrospective of the pioneering work undertaken by the earlier scientists at KIZ and recent achievements, which will hopefully serve to motivate and inspire present and future successors.
During its first two decades (1960s–1970s), KIZ carried out various comprehensive field surveys, including the “Joint Survey of Western Yunnan Province”, “Sino-Vietnamese Joint Exploration of Fishery Resources of the Honghe Area”, “Animal Resources Exploration of Southwestern China”, “Field Survey of the Mountain Gaoligong” (which was also recorded as the first scientific documentary film of China “Wild Animals of Yunnan Province”), “Field Survey of the Bilou Snow Mountain”, and “Comprehensive Exploration of the Hengduan Mountain”. These fundamental works formed the roots from which subsequent research have thrived. In these surveys, many animal specimens were collected, and a substantial amount of original data were accumulated, allowing for the advancement of studies in both classical zoological disciplines (e.g., new species identification and vertebrate fauna classification, comparative anatomy, and animal physiology) and thereafter cutting-edge subjects (e.g., de novo gene origin, genome evolution and adaptation, and genomic diversity of animals). Many remarkable breakthroughs were achieved during these years, including the compilation of an extensive vertebrate zoography, establishment of the first captive-bred macaque population in Menglun, Xishuangbanna, and a comprehensive animal field survey in western Sichuan Province, which was reported in KIZ’s first foreign language academic paper published in Nature by Prof. Hung-Shou Pen (彭鸿绶) (Pen, 1962). Sadly, Prof. Pen lost his life during a field survey in August 1981 due to high-altitude pulmonary edema, which was not only a significant loss to KIZ but to the scientific world at large. In 1978, KIZ won the “CAS Award for Major Science and Technology Achievements” for its timely research on the “Biological Effect of Nuclear Weapons”. Furthermore, during many difficult field surveys, KIZ researchers successfully collected abundant specimens, resulting in the considerable expansion of the Kunming Natural History Museum of Zoology, which now holds one of the richest tropical and sub-tropical animal collections in China (with over 800 000 specimens to date).
Over the following two decades (1980s–1990s), KIZ emphasized and strengthened its research directions and increasingly developed its international cooperation. It established key research groups focusing on animal ecology, animal pathology, experimental animals, animal toxinology, and animal cytogenetics. Many significant innovations and achievements were made during this period, with wide international impact. For instance, Shi and coworkers provided strong support for the tandem chromosome fusion theory of Indian muntjac karyotype evolution (Shi et al., 1980), as mentioned by the Nobel laureate Dr. Barbara McClintock in her acceptance speech (McClintock, 1983). Ma et al. (1990) identified a new mammalian species in the genus Muntiacus using morphological analysis and karyotype structure comparison. Professor Yu-Liang Xiong and other KIZ coworkers advanced the treatment of snakebites with the invention of an emergency kit for snake envenomation (Lu et al., 2010). KIZ researchers also published many books based on non-human primate studies, constituting the first monographs in the field at that time.
In 1986, KIZ established the first wild animal cell bank of China under the leadership of the late Prof. Li-Ming Shi, which featured cell lines of endangered species and is currently listed as an affiliate of the Committee on Type Culture Collection of CAS. These rich genetic resources are valuable components of the animal division of the Germplasm Bank of Wild Species, and have received wide attention (Ryder et al., 2000). KIZ researchers also established lymphoblastoid cell lines for ethnic minorities. In 1988, in cooperation with the Primate Center of Yunnan Province, KIZ established the “Joint Laboratory of Primatology”, which progressed into the “China-US Primate Biology Laboratory” in 1996 after collaboration with the University of Wisconsin, USA. The Laboratory of Cellular and Molecular Evolution, established by Prof. Li-Ming Shi in 1990, was among the first laboratories to use molecular techniques to study animal diversity and evolution in China. This laboratory evolved into the “State Key Laboratory of Genetic Resources and Evolution” in 2007 and now serves as one of the core units of KIZ. These platforms and key laboratories have laid a solid foundation for current development and future planning and have strongly influenced the dominant research areas of KIZ.
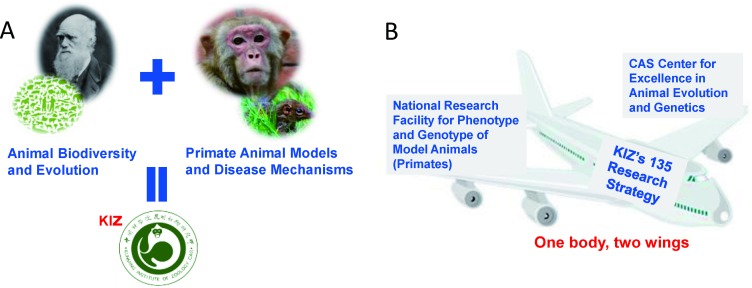
In the new millennium (2000–2019), after being selected as a pioneer institute to undertake the “Knowledge Innovation Project of CAS”, KIZ realigned its research focus, which has proven to be highly successful. With the recent “One-Three-Five” strategic plan, KIZ has further strengthened and established itself as a premier research institute within CAS, concentrating on regional animal biodiversity, evolution, and preservation (Figure 1). At present, KIZ is focused on accomplishing three major breakthroughs: i.e., developing non-human primate models for disease and pharmaceutical research, achieving theoretical discoveries in evolution and genomics, and cultivating genetic resources of domestic animals and their wild counterparts. Based on the “One-Three-Five” strategic plan, KIZ has incorporated its traditional strengths to adapt to new trends in frontier and interdisciplinary sciences. In addition, KIZ has emphasized innovation with profound implications for public education, health, conservation, policy recommendations, and scientifically sound management of diverse biological resources.
Figure 1. Dominant research areas (A) and“one body two wings” strategic planning (B) of the Kunming Institute of Zoology (KIZ), Chinese Academy of Sciences .
Small figure sections were taken from the web or were provided by KIZ.
During the “One-Three-Five” strategic plan, both research output and scientific accomplishments have flourished. For instance, KIZ was granted the State Natural Science Award and State Technological Invention Award of China for various achievements, such as the “Mitochondrial genome diversity and population history of East Asian”, “Mechanism of origin and genetic evolution of newly evolved genes”, “Genome diversity and evolution of Asia populations”, and “ Directional mining technology system of medical functional components from poisonous animals based on organism’s surviving strategy”.
KIZ’s research on non-human primates has greatly advanced the development of animal models of human diseases resistant to existing treatment options (such as HIV/AIDS, Alzheimer’s disease, Parkinson’s disease, and chronic social defeat depression) and has helped to elucidate the underpinnings of these complex diseases. Moreover, the accumulation of knowledge on the reproductive biology of non-human primates has had important implications for human infertility and genetic manipulation of monkeys. For instance, Zheng et al. (2001) created the first serum-free culture medium for in vitro-matured rhesus monkey oocytes, which proved to be crucial for evaluating the effects of in vitro maturation on developmental capacity (Zheng, 2007). Niu et al. (2010) developed the first transgenic monkey in China. Most recently, KIZ researchers and collaborators have attracted world-wide attention for creating transgenic monkeys carrying human MCPH1 gene copies and showing human-like neoteny of brain development, which is the first transgenic monkey model used to study human brain evolution (Shi et al., 2019; Shi & Su, 2019). KIZ has also investigated the use of the Chinese tree shrew as an alternative to non-human primates for animal models of human diseases and has made remarkable progress, e.g., determination of the Chinese tree shrew genome ( Fan et al., 2013, 2019) and creation of the first transgenic Chinese tree shrew (Li et al., 2017). The availability of genetic modification for monkeys and tree shrews will undoubtedly continue to facilitate animal model studies in KIZ. Furthermore, the rich primate resources and studies of KIZ have also attracted global attention (Cyranoski, 2004; Hao, 2007). In recent years, to match the urgent need and rapid development of studies on non-human primate animal models and human disease mechanisms, KIZ commenced construction of the National Research Facility for Phenotype and Genotype of Model Animals, which is one of the “12th Five-Year” Major National Technological Infrastructure Projects. The construction of this mega science facility (MPF: model primate facility) will provide a cutting-edge platform for studying genotype-phenotype relationships using non-human primates and will boost original findings and international collaborations. The construction of this MPF was also featured in Naturenews (Cyranoski, 2016) and will hopefully promote continued research using primate resources.
KIZ has a long tradition for studies on animal toxins and on active peptides and proteins derived from natural biodiversity resources, such as those found in insects, centipedes, and venomous animals, with promising results for the development of novel pharmaceuticals and pain killers (Luo et al., 2018; Xu & Lai, 2015; Zhang, 2015). Relying on these top-tier resources, platforms, and talents, KIZ has been recognized for its achievements in vaccine and medicine development for HIV-1 infection, bacterial infection, and neuropsychiatric disorders. Several new drugs for Alzheimer’s disease, depression, AIDS, vaginosis, and pain relief have been successfully developed by KIZ teams and collaborators. These drugs are currently on the market (e.g., analgesic treatment Ketongning, Tramadol Hydrochloride and Ibuprofen Tablets) or in clinical trial (e.g., antidepressant Orcinocide (phase II of clinical trials) and anti-Alzheimer’s treatment Phenchlobenpyrrone (phase II of clinical trials)).
Yunnan and its surrounding regions boast some of the world’s richest biodiversity, both in regard to wild and domestic animals. These areas provide excellent models for understanding evolution and adaptation to diverse environments and the reasons for high diversity. Research on domestic animals and wild counterparts covers a wide range of scientific questions, from understanding the genetic mechanisms associated with domestication processes to biodiversity monitoring and restoration. KIZ researchers have made remarkable achievements in these fields, e.g., generation of the reference genome sequences of the goat and sheep (Dong et al., 2013; Jiang et al., 2014), comparative genomic analysis of wild versus domesticated species to elucidate the origin and domestication of chickens and dogs (Wang et al. 2014), parallel evolution of dog and human genomes (Wang et al., 2013), and high-plateau adaptation of domestic animals (Wu et al., 2018). A detailed study on genomic selection and adaptation in animals has deepened our understanding of evolutionary genotype-phenotype systems biology (eGPS) (Wang et al., 2019), which is the core idea of the CAS Strategic Priority Research Program “Evolutionary Analysis and Functional Regulation of Animal Complex Traits” chaired by KIZ researchers. Based on eGPS, one would expect that the evolution and genetic basis of complex traits could be elucidated, and we could predict certain phenotypes based on the selected genes/loci or genetic manipulation of these loci (Wang et al., 2019). Because of its outstanding research on animal evolution and genetics, KIZ was approved by CAS to establish the first CAS Center for Excellence in Western China–“CAS Center for Excellence in Animal Evolution and Genetics”. This center is hosted by KIZ, with essential input from other CAS institutes across China, and has the ambitious goal to become one of the top research centers in evolution and genetics.
The protection of wild animal diversity and sustainable utilization of natural resources are key questions that KIZ continues to tackle. For instance, KIZ’s wetland restoration model using indigenous species from Dianchi Lake has proven to be successful at controlling water pollution and protecting the endangered golden-line barbel (Sinocyclocheilus graham), as reported in Science news (Stone, 2008). A new breed of S. graham with very good nutritional components, high growth rate, and disease resistance was fostered during the reproduction and breeding of this endangered species. This serves as a good example for the protection and sustainable use of animal resources. The profound efforts expended by KIZ researchers in protecting rare and endangered species, e.g., Hoolock tianxing (Fan et al., 2017) and green peafowl (Pavo muticus) (Wu et al., 2019), have been instrumental in arousing both public and government attention.
KIZ research into biodiversity has increasingly focused on making sound policy recommendations for protection as well as regional land and water usage. With the introduction of meta-barcoding for biodiversity using cutting-edge sequencing technologies, KIZ researchers put forward the concept of “biodiversity soup” for evaluating biodiversity (Ji et al., 2013; Yu et al., 2012). Similarly, the project “Study on Biodiversity Evolution and Protection of the Himalayas in China” chaired by KIZ has shed light on the biodiversity evolution in these areas, which is critical for implementing effective measures for the sustainable utilization of animal resources. Furthermore, the Cold Code project signifies KIZ’s central role in the development of the International Barcode of Life (iBOL) for amphibians and reptiles (Murphy et al., 2013). Recently, together with Dr. Jonathan Baillie, renowned KIZ researcher Prof. Ya-Ping Zhang discussed the key question regarding how much space we should leave for other forms of life (Baillie & Zhang, 2018), stating that they “encourage governments to set minimum targets of 30% of the oceans and land protected by 2030, with a focus on areas of high biodiversity and/or productivity, and to aim to secure 50% by 2050”. This information will undoubtedly have positive impacts on policy recommendations.
There are many more amazing achievements over the past 60 years that we wish could be shared. It is a credit to the efforts of every colleague that KIZ has evolved from a small working station with only a few dozen staff to an internationally recognized research institute with over 450 colleagues and 400 students. Given its deep roots in the Indo-Burma biodiversity hotspot of southwestern China and the devotion of its staff, we expect KIZ will continue to grow and realize a promising future.

Yong-Gang Yao, Director General
Kunming Institute of Zoology, Chinese Academy of Sciences,Kunming Yunnan 650223, China
Hua Shen, Secretary of CPC Committee and Vice Director
Kunming Institute of Zoology, Chinese Academy of Sciences,Kunming Yunnan 650223, China
Acknowledgments
We thank Ms. Su-Qing Liu, Miss Lin-Xi He, Miss Fang-Fei Rao, Miss Xi Yang, Ms. Jia Tang, and Prof. Bing-Yu Mao for help with the collection of material and draft discussions. We thank Ms. Christine Watts for language editing. We would like to apologize to the wonderful staff at KIZ, including both historic and current colleagues, for not discussing every important achievement here. Unfortunately, given the limits of this editorial, we are unable to mention all the fantastic accomplishments and studies of those who have worked for and with KIZ over the past 60 years.
REFERENCES
- Baillie J, Zhang YP. 2018. Space for nature. Science, 361(6407): 1051. [DOI] [PubMed] [Google Scholar]
- Cyranoski D. 2004. China takes steps to secure pole position in primate research. Nature, 432( 7013): 3. [DOI] [PubMed] [Google Scholar]
- Cyranoski D. 2016. Monkey kingdom. Nature, 532(7599): 300–302. [DOI] [PubMed] [Google Scholar]
- Dong Y, Xie M, Jiang Y, Xiao N, Du X, Zhang W, Tosser-Klopp G, Wang J, Yang S, Liang J, Chen W, Chen J, Zeng P, Hou Y, Bian C, Pan S, Li Y, Liu X, Wang W, Servin B, Sayre B, Zhu B, Sweeney D, Moore R, Nie W, Shen Y, Zhao R, Zhang G, Li J, Faraut T, Womack J, Zhang Y, Kijas J, Cockett N, Xu X, Zhao S, Wang J, Wang W. 2013. Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus) . Nature Biotechnology, 31(2): 135–141. [DOI] [PubMed] [Google Scholar]
- Fan PF, He K, Chen X, Ortiz A, Zhang B, Zhao C, Li YQ, Zhang HB, Kimock C, Wang WZ, Groves C, Turvey ST, Roos C, Helgen KM, Jiang XL. 2017. Description of a new species of Hoolock gibbon (Primates: Hylobatidae) based on integrative taxonomy . American Journal of Primatology, 79(5): e22631. [DOI] [PubMed] [Google Scholar]
- Fan Y, Huang ZY, Cao CC, Chen CS, Chen YX, Fan DD, He J, Hou HL, Hu L, Hu XT, Jiang XT, Lai R, Lang YS, Liang B, Liao SG, Mu D, Ma YY, Niu YY, Sun XQ, Xia JQ, Xiao J, Xiong ZQ, Xu L, Yang L, Zhang Y, Zhao W, Zhao XD, Zheng YT, Zhou JM, Zhu YB, Zhang GJ, Wang J, Yao YG. 2013. Genome of the Chinese tree shrew. Nature Communications, 4: 1426. [DOI] [PubMed] [Google Scholar]
- Fan Y, Ye MS, Zhang JY, Xu L, Yu DD, Gu TL, Yao YL, Chen JQ, Lv LB, Zheng P, Wu DD, Zhang GJ, Yao YG. 2019. Chromosomal level assembly and population sequencing of the Chinese tree shrew genome. Zoological Research, 40( 6): 506–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao X. 2007. Monkey research in China: developing a natural resource. Cell, 129(6): 1033–1036. [DOI] [PubMed] [Google Scholar]
- Ji Y, Ashton L, Pedley SM, Edwards DP, Tang Y, Nakamura A, Kitching R, Dolman PM, Woodcock P, Edwards FA, Larsen TH, Hsu WW, Benedick S, Hamer KC, Wilcove DS, Bruce C, Wang X, Levi T, Lott M, Emerson BC, Yu DW. 2013. Reliable, verifiable and efficient monitoring of biodiversity via metabarcoding. Ecological Letters, 16( 10): 1245–1257. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Xie M, Chen W, Talbot R, Maddox JF, Faraut T, Wu C, Muzny DM, Li Y, Zhang W, Stanton JA, Brauning R, Barris WC, Hourlier T, Aken BL, Searle SMJ, Adelson DL, Bian C, Cam GR, Chen Y, Cheng S, DeSilva U, Dixen K, Dong Y, Fan G, Franklin IR, Fu S, Guan R, Highland MA, Holder ME, Huang G, Ingham AB, Jhangiani SN, Kalra D, Kovar CL, Lee SL, Liu W, Liu X, Lu C, Lv T, Mathew T, McWilliam S, Menzies M, Pan S, Robelin D, Servin B, Townley D, Wang W, Wei B, White SN, Yang X, Ye C, Yue Y, Zeng P, Zhou Q, Hansen JB, Kristensen K, Gibbs RA, Flicek P, Warkup CC, Jones HE, Oddy VH, Nicholas FW, McEwan JC, Kijas J, Wang J, Worley KC, Archibald AL, Cockett N, Xu X, Wang W, Dalrymple BP. 2014. The sheep genome illuminates biology of the rumen and lipid metabolism. Science, 344( 6188):1168–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CH, Yan LZ, Ban WZ, Tu Q, Wu Y, Wang L, Bi R, Ji S, Ma YH, Nie WH, Lv LB, Yao YG, Zhao XD, Zheng P. 2017. Long-term propagation of tree shrew spermatogonial stem cells in culture and successful generation of transgenic offspring. Cell Research, 27(2): 241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QM, Lai R, Zhang Y. 2010. Animal toxins and human disease: from single component to venomics, from biochemical characterization to disease mechanisms, from crude venom utilization to rational drug design. Zoological Research, 31(1): 2–16. [DOI] [PubMed] [Google Scholar]
- Luo L, Li B, Wang S, Wu F, Wang X, Liang P, Ombati R, Chen J, Lu X, Cui J, Lu Q, Zhang L, Zhou M, Tian C, Yang S, Lai R. 2018. Centipedes subdue giant prey by blocking KCNQ channels. Proceedings of the National Academy of Sciences of the United States of America, 115(7): 1646– 1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma SL, Wang YX, Shi LM. 1990. A new species from the Genus Muntiacus in Yunnan, China . Zoological Research, 11(1): 47–53. [Google Scholar]
- McClintock B. 1983. The significance of responses of the genome to challenge. https://www.nobelprize.org/uploads/2018/06/mcclintock-lecture.pdf. [DOI] [PubMed] [Google Scholar]
- Murphy RW, Crawford AJ, Bauer AM, Che J, Donnellan SC, Fritz U, Haddad CFB, Nagy ZT, Poyarkov NA, Vences M, Wang WZ, Zhang YP. 2013. Cold Code: the global initiative to DNA barcode amphibians and nonavian reptiles. Molecular Ecology Resources, 13( 2): 161–167. [Google Scholar]
- Niu Y, Yu Y, Bernat A, Yang S, He X, Guo X, Chen D, Chen Y, Ji S, Si W, Lv Y, Tan T, Wei Q, Wang H, Shi L, Guan J, Zhu X, Afanassieff M, Savatier P, Zhang K, Zhou Q, Ji W. 2010. Transgenic rhesus monkeys produced by gene transfer into early-cleavage-stage embryos using a simian immunodeficiency virus-based vector. Proceedings of the National Academy of Sciences of the United States of America, 107(41):17663– 17667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pen HS.1962. Animals of western Szechuan. Nature, 196(4849): 14–16. [Google Scholar]
- Ryder OA, McLaren A, Brenner S, Zhang YP, Benirschke K. 2000. DNA banks for endangered animal species. Science, 288( 5464): 275–277. [DOI] [PubMed] [Google Scholar]
- Shi L, Luo X, Jiang J, Chen Y, Liu C, Hu T, Li M, Lin Q, Li Y, Huang J, Wang H, Niu Y, Shi Y, Styner M, Wang J, Lu Y, Sun X, Yu H, Ji W, Su B. 2019. Transgenic rhesus monkeys carrying the human MCPH1 gene copies show human-like neoteny of brain development . National Science Review, 6(3): 480– 493, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Su B. 2019. A transgenic monkey model for the study of human brain evolution. Zoological Research, 40( 3): 236–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LM, Ye YY, Duan XS. 1980. Comparative cytogenetic studies on the red muntjac, Chinese muntjac, and their F1 hybrids. Cytogenetics and Cell Genetics, 26 (1): 22–27. [DOI] [PubMed] [Google Scholar]
- Stone R. 2008. From remarkable rescue to restoration of lost habitat. Science, 322( 5899): 184. [DOI] [PubMed] [Google Scholar]
- Wang B, Chen L, Wang W. 2019. Genomic insights into ruminant evolution: from past to future prospects. Zoological Research, 40( 6): 476–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GD, Xie HB, Peng MS, Irwin D, Zhang YP. 2014. Domestication genomics: evidence from animals. Annual Review of Animal Biosciences, 2: 65–84. [DOI] [PubMed] [Google Scholar]
- Wang GD, Zhai W, Yang HC, Fan RX, Cao X, Zhong L, Wang L, Liu F, Wu H, Cheng LG, Poyarkov AD, Poyarkov NA Jr, Tang SS, Zhao WM, Gao Y, Lv XM, Irwin DM, Savolainen P, Wu CI, Zhang YP. 2013. The genomics of selection in dogs and the parallel evolution between dogs and humans. Nature Communications, 4: 1860. [DOI] [PubMed] [Google Scholar]
- Wu DD, Ding XD, Wang S, Wójcik JM, Zhang Y, Tokarska M, Li Y, Wang MS, Faruque O, Nielsen R, Zhang Q, Zhang YP. 2018. Pervasive introgression facilitated domestication and adaptation in the Bosspecies complex . Nature Ecology and Evolution, 2(7): 1139–1145. [DOI] [PubMed] [Google Scholar]
- Wu F, Kong DJ, Shan PF, Wang J, Kungu GN, Lu GY, Yang XJ. 2019. Ongoing green peafowl protection in China. Zoological Research, 40(6): 580–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Lai R. 2015. The chemistry and biological activities of peptides from amphibian skin secretions. Chemical Reviews, 115(4): 1760–846. [DOI] [PubMed] [Google Scholar]
- Yu DW, Ji Y, Emerson BC, Wang X, Ye C, Yang C, Ding Z. 2012. Biodiversity soup: metabarcoding of arthropods for rapid biodiversity assessment and biomonitoring. Methods in Ecology and Evolution, 3(4): 613–623. [Google Scholar]
- Zhang Y. 2015. Why do we study animal toxins?. Zoological Research, 36(4): 183– 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P. 2007. Effects of in vitro maturation of monkey oocytes on their developmental capacity . Animal Reproduction Science, 98(1– 2): 56–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Wang H, Bavister BD, Ji W. 2001. Maturation of rhesus monkey oocytes in chemically defined culture media and their functional assessment by IVF and embryo development. Human Reproduction, 16( 2): 300–305. [DOI] [PubMed] [Google Scholar]