Abstract
Objectives
To characterize the in vitro replication fitness, viral diversity and phylogeny of dengue viruses (DENV) isolated from Indian patients.
Methods
DENV was isolated from whole blood collected from patients by passaging in cell culture. Passage 3 viruses were used for growth kinetics in C6/36 mosquito cells. Parallel efforts also focused on isolation of DENV RNA from plasma samples of the same patients and processed for next generation sequencing.
Results
We were able to isolate 64 clinical isolates, mostly DENV-2, of which 25 were further used for growth curve analysis in vitro which showed a wide range of replication kinetics. Highest viral titers were of isolates from dengue with warning signs and severe dengue cases. We obtained full genome sequences of 21 DENV isolates. Genome analysis mapped the circulating DENV-2 strains to the Cosmopolitan genotype.
Conclusions
The replication kinetics of isolates from patients with mild or severe infection was not significantly different but the viral titers between the isolates varied by two orders of magnitude suggesting differences in replication fitness among the circulating DENV-2 isolates.
Keywords: Dengue virus, growth curve, phylogenetics, genotype, mutations
Background
Dengue virus is a common arbovirus which causes about 60 million apparent infections every year leading to over 10,000 deaths and the number of dengue cases have doubled every ten years between 1990 and 2013 (Stanaway et al., 2016). Antibody-dependent enhancement of viral infection is a major driver of severe dengue infections but pre-existing antibodies do not always lead to severe infection for all the serotypes (Endy et al., 2004). The antibody response in primary infections, which does provide cross-protection albeit transiently, has also been shown to differ between serotypes suggesting that the genotype and serotype of dengue isolates elicit differential immune responses (Chaudhury et al., 2017, Clapham et al., 2016, VanBlargan et al., 2013). Serotype-specific differences in the efficacy has been reported for the only licensed live-attenuated dengue vaccine constructed on yellow fever virus backbone (Capeding et al., 2014). Some of the reasons for these differences could be the lack of T-cell epitopes of DENV non-structural regions and possible divergence in the sequence of the strains used in the vaccine and circulating serotypes in the endemic countries where the vaccine trials were conducted (Guy et al., 2015, Juraska et al., 2018).
We and others have reported earlier that a significant proportion of primary dengue cases also result in severe dengue disease in endemic countries suggesting that the circulating viral strains and intrinsic host responses play an important role in disease outcomes (Balmaseda et al., 2006, Ngwe Tun et al., 2013, Nunes et al., 2018, Singla et al., 2016). Cohort studies tracking the dengue epidemics over many years have identified viral evolution as a key player that impacts serotype-specific responses. Different clades of DENV-2 led to different disease outcomes in children who had prior immunity to DENV-1 or DENV-3 (OhAinle et al., 2011). Genetic variation in dengue virus sequences is of significance for both epidemiology and vaccine development. It has been well documented by many earlier reports that the replication fitness, infection dynamics and pathogenicity vary among DENV isolates (Balmaseda et al., 2006, Borges et al., 2018, Fontaine et al., 2018, Leitmeyer et al., 1999, Vaughn et al., 2000, Watts et al., 1999). India contributes to about 34% of the global DENV cases and there is limited information on the evolution of DENV strains circulating in India (Bhatt et al., 2013, Dias et al., 2018). Considering the recent progress in dengue vaccine development, it is important to understand the drivers of positive selection and the evolution of dengue virus strains in India to better prepare for vaccine trials (Wichmann et al., 2017). In this study, we isolated dengue virus from patient blood, characterized their growth kinetics in vitro, recovered complete dengue virus genomes from plasma and compared them to create a molecular profile of the DENV strains circulating in India.
Materials and Methods
Study population, isolation of viral RNA from whole blood or plasma and estimation of viremia has been described before and is described in the supplementary information.
Virus isolation from whole blood
Blood samples were diluted 1:10 in minimal essential media (MEM) containing 2% fetal bovine serum (FBS) (Gibco) and added onto cultured monolayer of C6/36 cells for 1 hour with gentle rocking in a 28°C CO2 incubator. After 1 hour, the inoculum was removed and cells were cultured for 7 days in MEM with 2% FBS and antibiotics. Culture supernatant was collected on day 7 and used to infect new batch of C6/36 cells as above. This was repeated three times to achieve passage 3 (P3) isolates. Viral titers in the supernatant was measured by plaque assays on BHK-21 cells as described earlier (Agrawal et al., 2013).
Whole genome sequencing
Specific cDNA synthesis was performed using a primer specific for 3’ untraslated region (UTR) of flaviviruses (5’GGGTCTCCWCTAACCTCTAGTCCT 3’) using the Thermofisher Scientific Maxima H minus Reverse transcriptase. Second strand synthesis was performed using the NEBNext 2nd strand synthesis module. Resulting DNA was purified using magnetics beads. Sequencing library preparation was performed using the Nextra XT Illumina sequencing kit with the 96 barcodes. About 10 pM DNA from pooled libraries was taken for sequencing of the MiSeq platform. Fastq reads were extracted from the run, demultiplexed and used for de novo assembly using SPAdes (ver. 3.12.0) (Nurk et al., 2013).
Reference recruitment to the closest Blastn hit and genome editing was performed in the Geneious software (ver. 11.15) – with a least coverage of 9X and majority rule for calling the consensus. Resulting consensus sequences were annotated by transferring annotations from the reference sequence (RefSeq) and submitted to GenBank. The GenBank accession numbers are provided in the supplementary information.
Molecular Phylogenetic analysis
Multiple sequence alignment was performed to representative sequences from known genotypes (from Virus Pathogen Database and Analysis Resource, ViPR) for which complete genomes are available (accession numbers of the sequences are provided in Supplementary Table T4), using MUSCLE. The phylogenetic trees for whole genome sequences were constructed using Maximum likelihood method in MEGA X software (Kumar et al., 2018). Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach and then selecting the topology with superior log-likelihood value. Bootstrapping with 1000 replicates was used to get improve the statistical significance of each node. Around 91% of the nodes in all trees have bootstrap values > 90%. Phylogenetic tree for the envelope protein of DENV-II was constructed using Neighbour-joining method with 100 bootstraps. FigTree (v1.4.3) was used for graphical presentation of the phylogenetic trees.
Statistical methods
Appropriate statistical tests were performed as indicated in the figure legends and tables. Prism 7 was used for graphical representation of the data.
Dengue cohort
The characteristics and clinical parameters of the 119 patients enrolled into the study is presented (Supplementary Table T1). As expected, most patients with severe dengue disease came to the hospital at later days of fever as compared to mild dengue and dengue with warning signs. Patients with severe dengue had significantly lower platelet counts at the time of enrolment as compared to DI and DW cases. Based on the IgM and IgG ELISA, 56 (47%) patients were classified as either seronegative or primary infections and the remaining 53% were secondary infections.
Isolation of DENV RNA is more efficient from whole blood
Patients were enrolled into the study based on a positive dengue NS-1 test using a point-of-care kit. To further confirm DENV positivity, determine the infecting DENV serotype and to estimate viremia, we isolated RNA from whole blood sampled from patients at the time of enrolment. Furthermore, to compare whether RNA isolation from the plasma preparation of the same samples is equally efficient, we performed RNA isolation using plasma samples from most of these patients. Total RNA isolated from blood or plasma was used for detection and quantitation of viral RNA as described previously (Agrawal et al., 2013). Viral RNA was detected in significantly higher number of samples from whole blood as compared to plasma samples. Overall, 92% of the RNA samples isolated from whole blood had detectable levels of viral RNA whereas the same was 39% with plasma samples (Supplementary Figure S1A). This suggests that the efficiency of RNA isolation is higher with whole blood samples which may be due to the presence of cellular RNA. Although carrier RNA is added as per the manufacturer’s instructions during RNA isolation from plasma, the process is not as optimal to detect viral RNA under our conditions. The DENV genome copy numbers in whole blood between different disease severities was not significantly different (Supplementary Table T1) as reported earlier (Singla et al., 2016). However, viral genome copy numbers were significantly higher in plasma samples as compared to whole blood as the total RNA obtained from plasma is likely to be predominantly of viral origin as compared to whole blood RNA which will be dominated by cellular RNA (Supplementary Figure S1B). 80% of the patients were infected with DENV-2 as determined by serotyping PCR (Supplementary Table T2). About 11% of the samples were either negative or undetermined in serotyping PCR but they were confirmed to be dengue positive by antibody ELISA or viral RNA detection or virus isolation experiments.
DENV isolation from patient samples
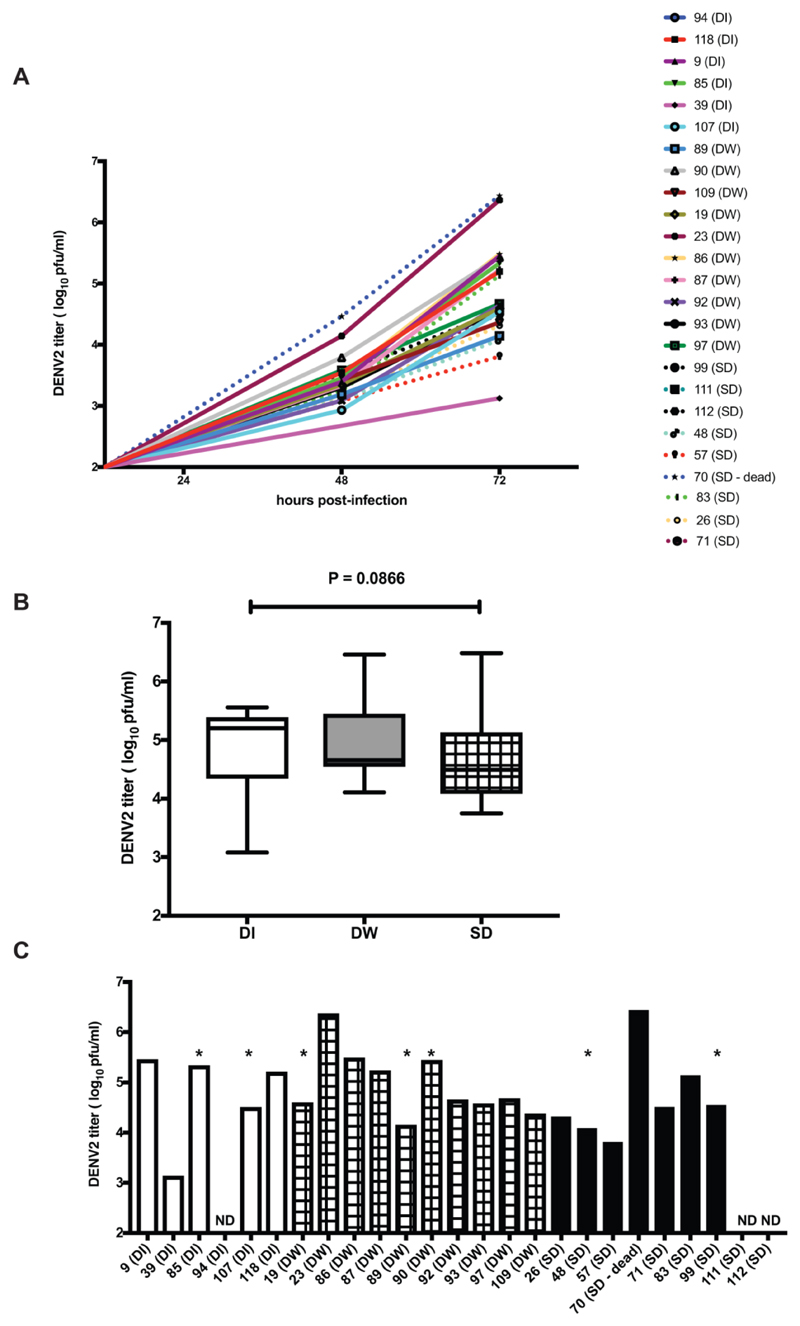
Dengue viruses, like all other RNA viruses, evolve rapidly and changes in the genome regions encoding the T cell and B cell epitopes have an impact on vaccine development and immune response. Therefore, we next invested our efforts in isolation of DENV from patient samples collected in our study. The flow diagram of the process that was followed is shown (Figure 1). We used whole blood for isolation of viruses by three blind passages in C6/36 cells. We obtained 64 isolates with measurable viral titers in P3 supernatants. Segregation of these isolates based on the day of fever (DOF) of the original sample shows that 72.5 % of the isolates from samples collected between DOF 1-3 yielded isolates and 44 % of the DOF 4-6 samples yielded isolates clearly demonstrating that the samples collected between DOF 1-6 would be ideal for virus isolation (Supplementary Figure S2A). The number of virus isolates obtained and the viral titers at P3 was not significantly different between samples obtained from DI, DW or SD patients (Supplementary Figure S2B and S2C). 46 of the isolates were confirmed to be of DENV-2 serotype by RT-PCR and 25 of these isolates had plaque assay titers high enough (≥ 105 – 108 pfu/ml) for growth curve assays in C6/36 cells. We performed a three-day growth kinetics of P3 isolates by infecting C6/36 cells with 0.1 MOI of P3 supernatants. At 24, 48 and 72 h pi, supernatants were collected and viral titers were measured by plaque assay. Of the 25 isolates, we were able to study 22 isolates in C6/36 cells and we observed that the growth rates of isolates were drastically different and the range of viral titers on day 3 post-infection from the clinical isolates was from 103 pfu/ml to 106 pfu/ml (Figure 2A). Highest titers (>106 pfu/ml) were from the sample of a SD patient who had succumbed to the disease and other was from a DW case. Nevertheless, viral titers on day 3 post-infection were not significantly different between viruses isolated from samples with different disease severities (Figure 2B and 2C). Of the three isolates that failed to amplify, one was from a DI patient and two were from SD patients and all the three samples were from within day 3 of fever. These in vitro data suggest that the isolation efficiency and growth kinetics of clinical isolates from DENV patients with different disease severity are not significantly different in cell culture conditions.
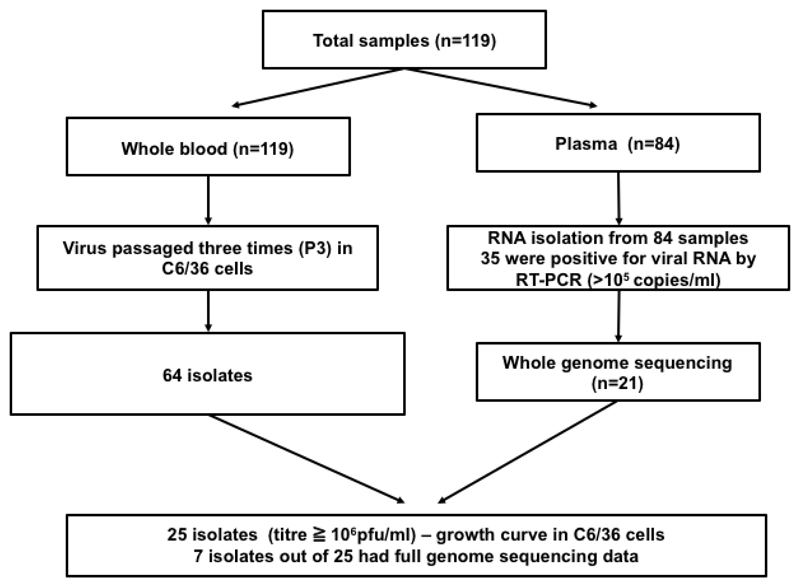
Figure 1.
Work flow of the study. Flowchart of activities depicting virus isolation and growth curve analysis from whole blood/plasma in C6/36 cells and viral RNA isolation from plasma and whole genome sequencing.
Figure 2.
Growth kinetics of patient isolates (passage 3). A) C6/36 cells were infected at a multiplicity of infection (MOI) of 0.1 and culture supernatants were collected on day 1-3. Viral titers were estimated by plaque assays. B) Comparison of day 3 viral titers from the above isolates by segregating into the disease classification of the samples at origin. C) Samples indicating individual viral titers at day 3. ND is not detected. * indicates those isolates for which we obtained whole genome sequences.
Whole genome sequencing of clinical DENV isolates
We next isolated viral RNA from 84 plasma samples and the amount of DENV RNA was determined by quantitative RT-PCR. 35 samples with copy numbers exceeding 105 DENV genome equivalents/ml were processed for whole genome sequencing. A modified RNA sequencing protocol was used to generate the whole genome sequences (see Methods). Using de novo assembly, followed by reference recruitment (mapping) we recovered complete genomes, with greater than 9X coverage across the genome from 21 RNA samples. These included 18 DENV-2, 2 DENV-1 and 1 DENV-3 genomes (Supplementary Table T3). GenBank accession numbers are provided in supplementary information (Supplementary Table T4).
Phylogenetic analysis
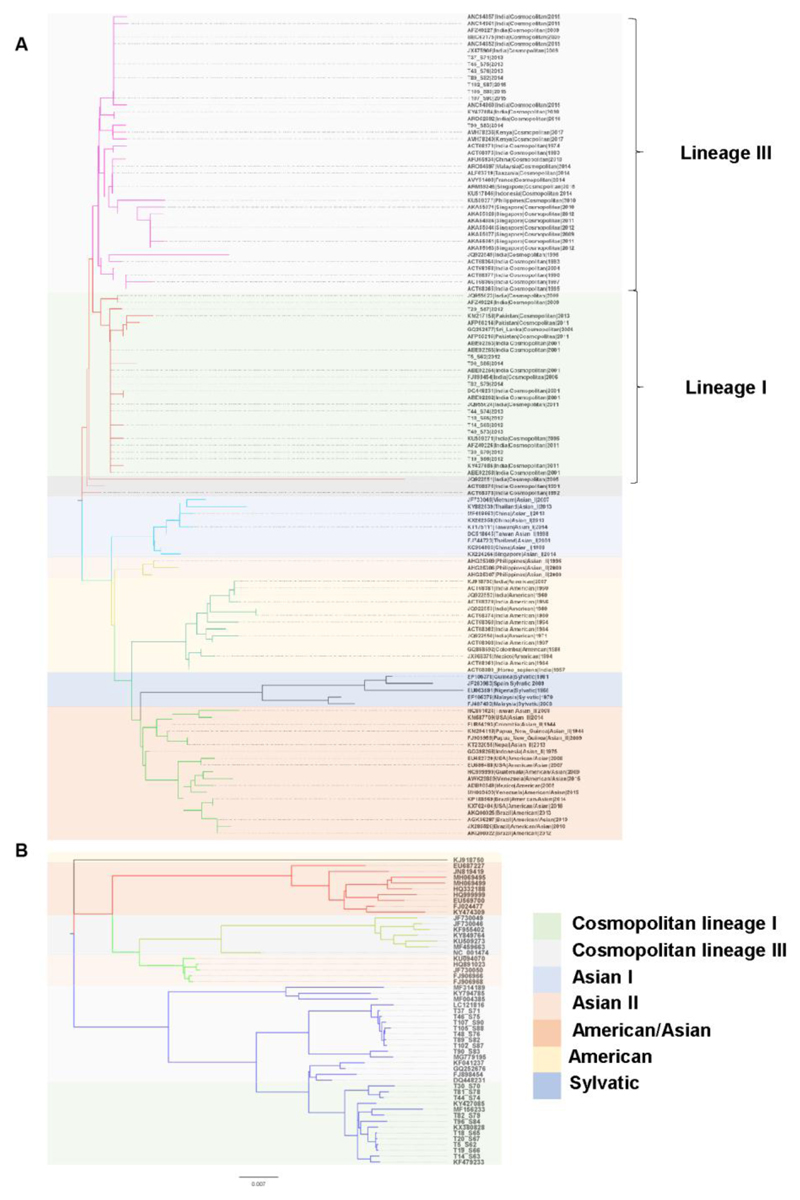
We used MEGA X (Kumar et al. 2018) to reconstruct the evolutionary tree(s) for the 21 near-complete DENV genome sequences. The DENV-3 samples mapped to the genotype III and DENV-1 sample mapped to the genotype V (Supplementary Figure S3A and S3B). Since most of the whole genome sequences recovered belonged to DENV-2 serotype, we focused on further analysis of these samples. The DENV-2 serotype has 6 genotypes that are largely, spatially related, namely American, American-Asian, Cosmopolitan, Asian-I, Asian-II and Sylvatic (Waman et al., 2016). Based on Neighbour-Joining (NJ) tree of the Envelope protein (E), we observed that the Indian DENV strains from pre-2000 were of the American genotype while the recent strains (post-2000) largely seem to be dominated by the Cosmopolitan genotype (Figure 3A). Even within the Cosmopolitan genotype, recent studies have reported prevalence of three lineage clusters in India (Afreen et al., 2016). We also observed the clustering of the DENV-2 genomes with two distinct lineages within the Cosmopolitan genotype. Lineage III sequences were largely prevalent in the second half of the epidemic (2014-15) and lineage I sequences dominated the first half of the epidemic (2012-13) while co-circulating at the same time in the Delhi region. We next built the maximum likelihood phylogenetic tree using the whole genome sequences of the DENV-2 from the GenBank (Figure 3B). DENV-2 genomes from our study largely followed the phylogenetic analysis of the E protein sequences suggesting insignificant recombination in the lineages.
Figure 3.
Phylogenetics analysis of DENV-2 clinical isolates. A) The optimal tree with the sum of branch length = 0.43786479 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances are in the units of the number of amino acid substitutions per site. The analysis involved 121 amino acid sequences of the E protein. There were a total of 473 positions in the final dataset. B) Phylogenetic tree for whole genomes of dengue 2 virus using Maximum Likelihood method based on the General Time Reversible model. The tree with the highest log likelihood (-47856.75) is shown. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 53 nucleotide sequences. There were a total of 10743 positions in the final dataset. Samples from this study are indicated as in Supplementary Table T3.
Sequence diversity from global DENV-2 strains
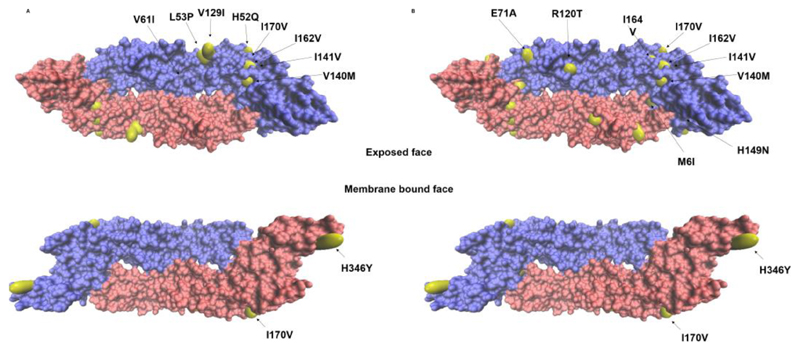
Next, we evaluated the level of diversity between our DENV-2 sequences to the DENV-2 Cosmopolitan genotype. We first generated a consensus DENV-2 Cosmopolitan genome using the 115 whole genomes in the GenBank database (Supplementary Table T5). Synonymous single nucleotide polymorphisms (SNPs) in the polyprotein coding region of virus genome were high (1051 mutations) and uniformly distributed across the whole genome as expected for RNA viruses. The non-synonymous mutations were rare but significant (138 mutations) and spread non-uniformly across the polyprotein (Supplementary Figure S4 and Supplementary Table T6). Most of the changes in the E protein were in the B cell epitopes and two of the changes in NS3 region were part of CD8 T cell epitope (Supplementary Table T6) (Vaughan et al., 2010). We next located the amino acid changes in the E protein from the consensus and the 16881 DENV-2 sequence that has been adapted to generate the DNA infectious clone (Kinney et al., 1997). Almost all of the E protein mutations mapped to the exposed face of the E protein in its dimeric conformation possibly suggesting immunological selection being the driving force (Figure 4A and 4B). For example, a large proportion of the mutations spanned the EDI/II region which is the binding epitope for human monoclonal antibody (hMAb) 3F9 whereas the mutation, H346Y mapped to EDIII binding epitope recognized by hMAbs, 2D22 and 1L12 (Gallichotte et al., 2018). We also compared the untranslated regions (UTR) of the genomes since they are reported to be critical for virus replication (Gebhard et al., 2011, Ng et al., 2017). The largely conserved 5’ UTR among the Cosmopolitan genotype carried a C-U mutation that lies in the RNA polymerase binding SLA loop (Supplementary Figure S5). The 3’ UTR on the other hand displayed several mutations that fell into two categories; one set that was common among all the cosmopolitan genotype and another set of mutations that set the two lineages apart within the genotype (Supplementary Figure S6).
Figure 4.
Nonsynonymous mutations (yellow) mapped on the envelope protein (Protein Data Bank [PDB] accession code 1OAN) mutations considered compared to (A) the consensus generated from 115 cosmopolitan DENV-2 sequences from all over the world, (B) 16881 strain (NC_001474) are shown.
Discussion
Previous reports have compared DENV viral detection using different RNA isolation methods from serum, plasma or whole blood (Anwar et al., 2009, Dettogni and Louro, 2011). Our report confirms previous observations where viral detection was reported to be more efficient with whole blood as compared to plasma (Klungthong et al., 2007). Dengue serotyping using rapid diagnostic tests have also demonstrated the convenience and utility of using whole blood for isolation of viral RNA (Vongsouvath et al., 2016). Another study which used cellular components of whole blood, serum, clot specimens and plasma from blood bank donors who were DENV RNA positive also demonstrated that the detection of viral RNA by RT-PCR was more consistent with cellular components of blood (Añez et al., 2016). We found that the viral RNA isolation was more efficient using whole blood and in samples collected within day 6 of fever. Blood viremia was not different between samples obtained from patients with different disease severities, however, the study has a caveat in that most of the severe dengue patients come to seek clinical care at later stages of infection when the viremia is on the decline. It is possible that high viral load at early stages of infection leads to higher inflammation resulting in severe disease. Therefore, estimation of viremia as a function of time starting from earliest possible time upon infection till hospitalization/recovery would provide a clear picture of the role of viral load in disease progression. About half of the patients enrolled into our study had primary/seronegative status based on IgG and IgM ELISA. 32% of the severe dengue cases were also primary infections (no dengue IgG at the time enrolment) suggesting that in a subset of patients, severe disease was a consequence independent of infection-enhancing antibodies.
There are very few reports on the characterization of clinical isolates from India and it is necessary to determine whether the antibodies generated from previous natural infections or vaccine trials is capable of neutralizing circulating viruses in the country. In addition, sequencing information and corresponding virus neutralization data will also provide clues to emergence of escape variants. Clinical isolates minimally passaged in vitro have been used to assess replication fitness in cell culture models of infection, in immunocompetent and immunocompromised mouse models or rhesus macaques (Borges et al., 2018, Ferreira et al., 2010, Sarathy et al., 2018, Tuiskunen et al., 2011a, Tuiskunen et al., 2011b). It has been demonstrated that isolates from patients with severe or fatal dengue disease demonstrate enhanced replication and induce inflammation at a much higher level (Murgue et al., 1997, Silveira et al., 2011). We show that the low passage clinical DENV-2 isolates do show difference in growth kinetics in cell culture but the differences do not associate with disease severity in patients. Although we have passaged these isolates only three times in cell culture to limit adaptive mutations, we cannot rule out the possibility that the sequence of the virus obtained after third passage may not be identical to the one infecting the patient. Further sequencing of P3 isolates will clarify this issue.
Full genome sequencing data from recent reports suggest that in most parts of South America and Latin America, the circulating DENV-2 strains belong to the American/Asian genotype and that phylogenetically close viruses are circulating between these countries (Lizarazo et al., 2018, Stewart-Ibarra et al., 2018, Williams et al., 2014). Similarly, full genome sequence of a DENV-2 isolate from Myanmar identified Asian genotype which was closely related to the isolates circulating in Vietnam, Thailand and Cambodia (Zeng et al., 2018). Asian genotype of DENV-2 dominated in China prior to 2010 which has been replaced by the cosmopolitan genotype which is similar to the isolates circulating in Singapore (Jiang et al., 2018, Zhao et al., 2012). All the four serotypes are circulating in India although DENV-2 has dominated in the recent years in our study region and also in other parts of the country (Mishra et al., 2015, Mukherjee et al., 2017, Shrivastava et al., 2018, Singla et al., 2016). Phylogenetic analysis of DENV-2 sequences from Pakistan suggests that the Cosmopolitan genotype was introduced from India and Sri Lanka about 30 years ago and a sub-lineage within the Cosmopolitan genotype has emerged in Pakistan and in the Indian sub-continent causing major epidemics (Akram et al., 2015, Khan et al., 2013). This is further confirmed by DENV-2 sequences (complete genome or the structural region) from India starting from 1960 to 2012 which indicate a shift from American genotype to a unique South Asian clade within Cosmopolitan genotype (Dash et al., 2013, Mishra et al., 2015). DENV-2 envelope sequence of isolates from Nepal has shown co-circulation of two DENV-2 genotypes (Asian II and Cosmopolitan IVa and IVb). Among the DENV serotypes, DENV-2 has been the most genetically diverse population. It is the most frequent cause of dengue epidemic worldwide and has also been associated with severe dengue cases (Wei and Li, 2017). We obtained complete genome sequences of 21 DENV-2 isolates from India isolated between 2012 to 2015 which further confirms a positive selection of Cosmopolitan genotype in India. 15 out of the 21 sequences were obtained from seronegative or primary infections and these were isolated at very early days of fever (median DOF 2.5) which we believe is a unique feature of the study and will serve as useful information. DENV-2 sequences from primary and secondary infections in Thailand has shown that the sequences were homogenous in secondary infection while the viral diversity was much more heterogenous in primary infections suggesting that most primary infections could drive generation of quasispecies leading to evolution and selection of dominant isolates during subsequent exposure (Kurosu et al., 2014). We speculate that non-synonymous mutations in the capsid, NS2A and NS5 could be acting as a strong driver of positive selection. In addition, some of the mutations in the E protein mapped to B cell epitopes. We need to further verify if any of these isolates are neutralization escape mutants. Future studies would be designed with focus on capturing the kinetics of viral replication by isolation of viral RNA and viruses from serial samples from both primary and secondary infections to understand the inter-host diversity and evolution of the viral genome with disease progression or recovery.
Supplementary Material
Highlights.
64 dengue virus 2 isolates were amplified by blind passaging
25 clinical isolates of dengue virus 2 from passage 3 were evaluated by growth kinetics in vitro
Clinical isolates exhibited differences in replication fitness
Whole genome sequences of 21 clinical isolates obtained
Most of the changes in the E protein were in the B cell epitopes and two of the changes in NS3 region were part of CD8 T cell epitope
E protein mutations suggest immunological selection as the driving force
Acknowledgements
We thank Amresh Kumar Singh for technical support.
Funding source
This work was supported by funding to GRM, SKK, RL and AC under the “Vaccine Grand Challenge program” of the Department of Biotechnology, Govt. of. India. Sanction No. BT/PR5132/MED/15/85/2012. and from Wellcome trust-DBT India Alliance Intermediate fellowships to GM (IA/S/14/1/501291) and RR (IA/I/13/1/500901). MK received fellowship from the Department of Biotechnology, India. This study was also supported by the Department of Biotechnology, Glue Grant, and NCBS core funds to SK and a grant for dengue sequencing from Mr. Narayana Murthy (Infosys). CP was supported by the Department of Biotechnology Research Associate fellowship, Royal Society-SERB Newton International fellowship and the India Alliance (DBT-Wellcome Trust) Early Career Fellowship. The funders had no role in study design, data collection and interpretation or the decision to submit the work for publication.
Footnotes
Author Contributions
MK, AN, AK, SJ, JS, MS, SM and AP performed experiments and analysed data. AC, SKK, SK, RL contributed reagents and analysed data. RR, CP and GM conceived the study, designed and performed experiments, analyzed data and wrote the manuscript. All authors have reviewed the final version of the manuscript.
Conflict of interest statement
The authors declare that they have no conflict of interest to disclose.
Ethics Statement
The study was approved by the Institutional Ethics committees of all the three participating institutes (Ethics/THSTI/2011/2.1 dated 16 Nov, 2011; AIIMS: IEC/NP-338/2011 dated 17 Nov 2011; ICGEB/IEC/2011/01 dated 12 Nov 2011). Written informed consent for the study was taken from parents/guardians to collect blood samples at the time of admission.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers thatapply to the journal pertain.
References
- Afreen N, Naqvi IH, Broor S, Ahmed A, Kazim SN, Dohare R, et al. Evolutionary Analysis of Dengue Serotype 2 Viruses Using Phylogenetic and Bayesian Methods from New Delhi, India. PLoS Negl Trop Dis. 2016;10(3):e0004511. doi: 10.1371/journal.pntd.0004511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal T, Schu P, Medigeshi GR. Adaptor protein complexes-1 and 3 are involved at distinct stages of flavivirus life-cycle. Sci Rep. 2013;3 doi: 10.1038/srep01813. 1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akram M, Fatima Z, Purdy MA, Sue A, Saleem S, Amin I, et al. Introduction and evolution of dengue virus type 2 in Pakistan: a phylogeographic analysis. Virol J. 2015;12 doi: 10.1186/s12985-015-0371-8. 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Añez G, Heisey DAR, Chancey C, Fares RCG, Espina LM, Souza KPR, et al. Distribution of Dengue Virus Types 1 and 4 in Blood Components from Infected Blood Donors from Puerto Rico. PLOS Neglected Tropical Diseases. 2016;10(2):e0004445. doi: 10.1371/journal.pntd.0004445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar A, Wan G, Chua K-B, August JT, Too H-P. Evaluation of Pre-Analytical Variables in the Quantification of Dengue Virus by Real-Time Polymerase Chain Reaction. The Journal of Molecular Diagnostics. 2009;11(6):537–42. doi: 10.2353/jmoldx.2009.080164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmaseda A, Hammond SN, Perez L, Tellez Y, Saborio SI, Mercado JC, et al. Serotype-specific differences in clinical manifestations of dengue. Am J Trop Med Hyg. 2006;74(3):449–56. [PubMed] [Google Scholar]
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature. 2013;496(7446):504–7. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges MB, Marchevsky RS, Mendes YS, Mendes LG, Duarte AC, Cruz M, et al. Characterization of recent and minimally passaged Brazilian dengue viruses inducing robust infection in rhesus macaques. PLOS ONE. 2018;13(4):e0196311. doi: 10.1371/journal.pone.0196311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capeding MR, Tran NH, Hadinegoro SR, Ismail HI, Chotpitayasunondh T, Chua MN, et al. Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: a phase 3, randomised, observer-masked, placebo-controlled trial. Lancet. 2014;384(9951):1358–65. doi: 10.1016/S0140-6736(14)61060-6. [DOI] [PubMed] [Google Scholar]
- Chaudhury S, Gromowski GD, Ripoll DR, Khavrutskii IV, Desai V, Wallqvist A. Dengue virus antibody database: Systematically linking serotype-specificity with epitope mapping in dengue virus. PLoS Negl Trop Dis. 2017;11(2):e0005395. doi: 10.1371/journal.pntd.0005395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham HE, Rodriguez-Barraquer I, Azman AS, Althouse BM, Salje H, Gibbons RV, et al. Dengue Virus (DENV) Neutralizing Antibody Kinetics in Children After Symptomatic Primary and Postprimary DENV Infection. J Infect Dis. 2016;213(9):1428–35. doi: 10.1093/infdis/jiv759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash PK, Sharma S, Soni M, Agarwal A, Parida M, Rao PVL. Complete genome sequencing and evolutionary analysis of Indian isolates of Dengue virus type 2. Biochemical and Biophysical Research Communications. 2013;436(3):478–85. doi: 10.1016/j.bbrc.2013.05.130. [DOI] [PubMed] [Google Scholar]
- Dettogni RS, Louro ID. Dengue virus RNA purification from human plasma: a comparison of two techniques. Mol Biol Rep. 2011;38(8):4979–83. doi: 10.1007/s11033-010-0642-9. [DOI] [PubMed] [Google Scholar]
- Dias M, Pattabiraman C, Siddappa S, Gowda M, Shet A, Smith D, et al. Complete assembly of a dengue virus type 3 genome from a recent genotype III clade by metagenomic sequencing of serum [version 1; referees: 2 approved] 2018;3(44) doi: 10.12688/wellcomeopenres.14438.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endy TP, Nisalak A, Chunsuttitwat S, Vaughn DW, Green S, Ennis FA, et al. Relationship of preexisting dengue virus (DV) neutralizing antibody levels to viremia and severity of disease in a prospective cohort study of DV infection in Thailand. J Infect Dis. 2004;189(6):990–1000. doi: 10.1086/382280. [DOI] [PubMed] [Google Scholar]
- Ferreira GP, Figueiredo LB, Coelho LF, S PA, Jr, Cecilio AB, Ferreira PC, et al. Dengue virus 3 clinical isolates show different patterns of virulence in experimental mice infection. Microbes Infect. 2010;12(7):546–54. doi: 10.1016/j.micinf.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Fontaine A, Lequime S, Moltini-Conclois I, Jiolle D, Leparc-Goffart I, Reiner RC, Jr, et al. Epidemiological significance of dengue virus genetic variation in mosquito infection dynamics. PLOS Pathogens. 2018;14(7):e1007187. doi: 10.1371/journal.ppat.1007187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallichotte EN, Baric TJ, Yount BL, Jr, Widman DG, Durbin A, Whitehead S, et al. Human dengue virus serotype 2 neutralizing antibodies target two distinct quaternary epitopes. PLoS Pathog. 2018;14(2):e1006934. doi: 10.1371/journal.ppat.1006934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhard LG, Filomatori CV, Gamarnik AV. Functional RNA elements in the dengue virus genome. Viruses. 2011;3(9):1739–56. doi: 10.3390/v3091739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy B, Briand O, Lang J, Saville M, Jackson N. Development of the Sanofi Pasteur tetravalent dengue vaccine: One more step forward. Vaccine. 2015;33(50):7100–11. doi: 10.1016/j.vaccine.2015.09.108. [DOI] [PubMed] [Google Scholar]
- Jiang L, Ma D, Ye C, Li L, Li X, Yang J, et al. Molecular Characterization of Dengue Virus Serotype 2 Cosmospolitan Genotype From 2015 Dengue Outbreak in Yunnan, China. Front Cell Infect Microbiol. 2018;8:219. doi: 10.3389/fcimb.2018.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juraska M, Magaret CA, Shao J, Carpp LN, Fiore-Gartland AJ, Benkeser D, et al. Viral genetic diversity and protective efficacy of a tetravalent dengue vaccine in two phase 3 trials. Proc Natl Acad Sci U S A. 2018 doi: 10.1073/pnas.1714250115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Ellis EM, Tissera HA, Alvi MY, Rahman FF, Masud F, et al. Emergence and Diversification of Dengue 2 Cosmopolitan Genotype in Pakistan, 2011. PLOS ONE. 2013;8(3):e56391. doi: 10.1371/journal.pone.0056391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney RM, Butrapet S, Chang GJ, Tsuchiya KR, Roehrig JT, Bhamarapravati N, et al. Construction of infectious cDNA clones for dengue 2 virus: strain 16681 and its attenuated vaccine derivative, strain PDK-53. Virology. 1997;230(2):300–8. doi: 10.1006/viro.1997.8500. [DOI] [PubMed] [Google Scholar]
- Klungthong C, Gibbons RV, Thaisomboonsuk B, Nisalak A, Kalayanarooj S, Thirawuth V, et al. Dengue virus detection using whole blood for reverse transcriptase PCR and virus isolation. J Clin Microbiol. 2007;45(8):2480–5. doi: 10.1128/JCM.00305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Molecular Biology and Evolution. 2018;35(6):1547–9. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosu T, Chaichana P, Phanthanawiboon S, Khamlert C, Yamashita A, A An, et al. Sequence variation of dengue type 2 virus isolated from clinical cases in Thailand. Jpn J Infect Dis. 2014;67(2):132–4. doi: 10.7883/yoken.67.132. [DOI] [PubMed] [Google Scholar]
- Leitmeyer KC, Vaughn DW, Watts DM, Salas R, Villalobos I, de C, et al. Dengue virus structural differences that correlate with pathogenesis. J Virol. 1999;73(6):4738–47. doi: 10.1128/jvi.73.6.4738-4747.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizarazo E, Couto N, Vincenti-Gonzalez M, Raangs EC, Jaenisch T, Friedrich AW, et al. Complete Coding Sequences of Five Dengue Virus Type 2 Clinical Isolates from Venezuela Obtained through Shotgun Metagenomics. Genome Announc. 2018;6(25) doi: 10.1128/genomeA.00545-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra G, Jain A, Prakash O, Prakash S, Kumar R, Garg RK, et al. Molecular characterization of dengue viruses circulating during 2009–2012 in Uttar Pradesh, India. Journal of Medical Virology. 2015;87(1):68–75. doi: 10.1002/jmv.23981. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Dutta SK, Sengupta S, Tripathi A. Evidence of dengue and chikungunya virus co-infection and circulation of multiple dengue serotypes in a recent Indian outbreak. Eur J Clin Microbiol Infect Dis. 2017;36(11):2273–9. doi: 10.1007/s10096-017-3061-1. [DOI] [PubMed] [Google Scholar]
- Murgue B, Cassar O, Guigon M, Chungue E. Dengue Virus Inhibits Human Hematopoietic Progenitor Growth in vitro. The Journal of Infectious Diseases. 1997;175(6):1497–501. doi: 10.1086/516486. [DOI] [PubMed] [Google Scholar]
- Ng WC, Soto-Acosta R, Bradrick SS, Garcia-Blanco MA, Ooi EE. The 5' and 3' Untranslated Regions of the Flaviviral Genome. Viruses. 2017;9(6) doi: 10.3390/v9060137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngwe Tun MM, Thant KZ, Inoue S, Kurosawa Y, Lwin YY, Lin S, et al. Serological characterization of dengue virus infections observed among dengue hemorrhagic fever/dengue shock syndrome cases in upper Myanmar. J Med Virol. 2013;85(7):1258–66. doi: 10.1002/jmv.23577. [DOI] [PubMed] [Google Scholar]
- Nunes PCG, de Filippis AMB, Lima M, Faria N, de Bruycker-Nogueira F, Santos JB, et al. 30 years of dengue fatal cases in Brazil: a laboratorial-based investigation of 1047 cases. BMC Infect Dis. 2018;18(1):346. doi: 10.1186/s12879-018-3255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol. 2013;20(10):714–37. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OhAinle M, Balmaseda A, Macalalad AR, Tellez Y, Zody MC, Saborio S, et al. Dynamics of dengue disease severity determined by the interplay between viral genetics and serotype-specific immunity. Sci Transl Med. 2011;3(114):114ra28. doi: 10.1126/scitranslmed.3003084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarathy VV, White M, Li L, Kaiser JA, Campbell GA, Milligan GN, et al. Characterization of a murine model of non-lethal, symptomatic dengue virus infection. Scientific Reports. 2018;8(1) doi: 10.1038/s41598-018-22618-w. 4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava S, Tiraki D, Diwan A, Lalwani SK, Modak M, Mishra AC, et al. Co-circulation of all the four dengue virus serotypes and detection of a novel clade of DENV-4 (genotype I) virus in Pune, India during 2016 season. PLoS One. 2018;13(2):e0192672. doi: 10.1371/journal.pone.0192672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira GF, Meyer F, Delfraro A, Mosimann ALP, Coluchi N, Vasquez C, et al. Dengue Virus Type 3 Isolated from a Fatal Case with Visceral Complications Induces Enhanced Proinflammatory Responses and Apoptosis of Human Dendritic Cells. Journal of Virology. 2011;85(11):5374. doi: 10.1128/JVI.01915-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla M, Kar M, Sethi T, Kabra SK, Lodha R, Chandele A, et al. Immune Response to Dengue Virus Infection in Pediatric Patients in New Delhi, India--Association of Viremia, Inflammatory Mediators and Monocytes with Disease Severity. PLoS Negl Trop Dis. 2016;10(3):e0004497. doi: 10.1371/journal.pntd.0004497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanaway JD, Shepard DS, Undurraga EA, Halasa YA, Coffeng LE, Brady OJ, et al. The global burden of dengue: an analysis from the Global Burden of Disease Study 2013. Lancet Infect Dis. 2016;16(6):712–23. doi: 10.1016/S1473-3099(16)00026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Ibarra AM, Ryan SJ, Kenneson A, King CA, Abbott M, Barbachano-Guerrero A, et al. The Burden of Dengue Fever and Chikungunya in Southern Coastal Ecuador: Epidemiology, Clinical Presentation, and Phylogenetics from the First Two Years of a Prospective Study. Am J Trop Med Hyg. 2018;98(5):1444–59. doi: 10.4269/ajtmh.17-0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuiskunen A, Monteil V, Plumet S, Boubis L, Wahlström M, Duong V, et al. Phenotypic and genotypic characterization of dengue virus isolates differentiates dengue fever and dengue hemorrhagic fever from dengue shock syndrome. Archives of Virology. 2011a;156(11):2023. doi: 10.1007/s00705-011-1100-2. [DOI] [PubMed] [Google Scholar]
- Tuiskunen A, Wahlström M, Bergström J, Buchy P, Leparc-Goffart I, Lundkvist Å. Phenotypic characterization of patient dengue virus isolates in BALB/c mice differentiates dengue fever and dengue hemorrhagic fever from dengue shock syndrome. Virology Journal. 2011b;8(1):398. doi: 10.1186/1743-422X-8-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBlargan LA, Mukherjee S, Dowd KA, Durbin AP, Whitehead SS, Pierson TC. The type-specific neutralizing antibody response elicited by a dengue vaccine candidate is focused on two amino acids of the envelope protein. PLoS Pathog. 2013;9(12):e1003761. doi: 10.1371/journal.ppat.1003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan K, Greenbaum J, Blythe M, Peters B, Sette A. Meta-analysis of all immune epitope data in the Flavivirus genus: inventory of current immune epitope data status in the context of virus immunity and immunopathology. Viral Immunol. 2010;23(3):259–84. doi: 10.1089/vim.2010.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn DW, Green S, Kalayanarooj S, Innis BL, Nimmannitya S, Suntayakorn S, et al. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J Infect Dis. 2000;181(1):2–9. doi: 10.1086/315215. [DOI] [PubMed] [Google Scholar]
- Vongsouvath M, Phommasone K, Sengvilaipaseuth O, Kosoltanapiwat N, Chantratita N, Blacksell SD, et al. Using Rapid Diagnostic Tests as a Source of Viral RNA for Dengue Serotyping by RT-PCR - A Novel Epidemiological Tool. PLOS Neglected Tropical Diseases. 2016;10(5):e0004704. doi: 10.1371/journal.pntd.0004704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waman VP, Kolekar P, Ramtirthkar MR, Kale MM, Kulkarni-Kale U. Analysis of genotype diversity and evolution of Dengue virus serotype 2 using complete genomes. PeerJ. 2016;4:e2326. doi: 10.7717/peerj.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts DM, Porter KR, Putvatana P, Vasquez B, Calampa C, Hayes CG, et al. Failure of secondary infection with American genotype dengue 2 to cause dengue haemorrhagic fever. Lancet. 1999;354(9188):1431–4. doi: 10.1016/S0140-6736(99)04015-5. [DOI] [PubMed] [Google Scholar]
- Wei K, Li Y. Global evolutionary history and spatio-temporal dynamics of dengue virus type 2. Sci Rep. 2017;7 doi: 10.1038/srep45505. 45505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann O, Vannice K, Asturias EJ, de Albuquerque Luna EJ, Longini I, Lopez AL, et al. Live-attenuated tetravalent dengue vaccines: The needs and challenges of post-licensure evaluation of vaccine safety and effectiveness. Vaccine. 2017;35(42):5535–42. doi: 10.1016/j.vaccine.2017.08.066. [DOI] [PubMed] [Google Scholar]
- Williams M, Mayer SV, Johnson WL, Chen R, Volkova E, Vilcarromero S, et al. Lineage II of Southeast Asian/American DENV-2 is associated with a severe dengue outbreak in the Peruvian Amazon. Am J Trop Med Hyg. 2014;91(3):611–20. doi: 10.4269/ajtmh.13-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z, Shi J, Guo X, Mo L, Hu N, Sun J, et al. Full-length genome and molecular characterization of dengue virus serotype 2 isolated from an imported patient from Myanmar. Virol J. 2018;15(1):131. doi: 10.1186/s12985-018-1043-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Deng Y-Q, Hong W-X, Yu X-D, Jiang T, Yu M, et al. Complete Genome Sequence of Dengue Virus Serotype 2 Cosmopolitan Genotype Strain in Guangdong, China. Journal of Virology. 2012;86(24) doi: 10.1128/JVI.02562-12. 13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.