Abstract
Common risk factors for psychiatric and other brain disorders likely converge on biological pathways influencing the development and maintenance of brain structure and function across life. Using structural magnetic resonance imaging data from 45,615 individuals aged 3 to 96 years, we demonstrate distinct patterns of apparent brain aging in several brain disorders and reveal genetic pleiotropy between apparent brain aging in healthy individuals and common brain disorders.
Keywords: Brain age gap, brain disorders, genetic architecture, pleiotropy
Psychiatric disorders and other brain disorders are among the main contributors to morbidity and disability around the world1. The disease mechanisms are complex, spanning a wide range of genetic and environmental contributing factors2. The inter-individual variability is large, but on a group-level, patients with common brain disorders perform worse on cognitive tests, are less likely to excel professionally, and engage in adverse health behaviours more frequently3. It is unclear to what extend these characteristics are a cause, consequence or confounder of disease.
Dynamic processes influencing the rate of brain maturation and change throughout the lifespan play a critical role, as reflected in the wide range of disease onset times from early childhood to old age4. This suggests that the age at which individual trajectories diverge from the norm reflects key characteristics of the underlying pathophysiology. Whereas autism spectrum disorder (ASD) and attention-deficit/hyperactivity disorder (ADHD) emerge in childhood5, schizophrenia (SZ) and bipolar (BD) spectrum disorders likely develop during late childhood and adolescence, before the characteristic outbreak of severe symptoms in early adulthood6. Likewise, multiple sclerosis (MS) most often manifests in early adulthood but the disease process likely starts much earlier7. First episodes in major depressive disorder (MDD) can appear at any stage from adolescence to old age5, whereas mild cognitive impairment (MCI) and dementia (DEM) primarily emerge during senescence8. Beyond such differential temporal evolution across the lifespan, age-related deviations from the norm may also differ between disorders in terms of anatomical location, direction, change rate and magnitude, all of which add complexity to the interpretation of observed effects.
Machine learning techniques enable robust estimation of the biological age of the brain using information provided by magnetic resonance imaging (MRI)9,10, assessing the similarity of a given brain scan with scans of a range of individuals to estimate the age of the tissue from a normative lifespan trajectory. Initial evidence suggested that the deviation between brain age and chronological age – termed the brain age gap - is a promising marker of brain health11, but several issues remain to be addressed. First, while advantageous for narrowing the complexity, reducing a rich set of brain imaging features into a single estimate of brain age inevitably compromises spatial specificity, thereby neglecting disorder-specific patterns. Second, most studies so far have been rather small-scale, performed within a limited age range and focusing on a single disorder, which left them unable to uncover clinical specificity and lifespan dynamics. Third, the genetic underpinnings of brain age gap are not understood, and it is unknown to what degree they overlap with the genetic architecture of major clinical traits. To address these critical knowledge gaps, large imaging genetics samples covering a range of prevalent brain disorders are necessary.
Here, we employed a centralized and harmonized processing protocol including automated surface-based morphometry and subcortical segmentation using Freesurfer on raw structural MRI data from 45,615 individuals aged 3 to 96 years that passed quality control (Suppl. Fig. 1). The sample included data from healthy controls (HC; n = 39,827; 3-95 years) and 5,788 individuals with various brain disorders. We included data from individuals with ASD (n = 925; 5-64 years), ADHD (n = 725; 7-62 years), prodromal SZ or at risk mental state (SZRISK; n = 94; 16-42 years), SZ (n = 1110; 18-66 years), a heterogeneous group with mixed diagnoses in the psychosis spectrum (PSYMIX; n = 300; 18-69 years), BD (n = 459; 18-66 years), MS (n = 254; 19-68 years), MDD (n = 208; 18-71 years), MCI (n = 974; 38-91 years), and DEM (including Alzheimer’s disease; n = 739; 53-96 years). Suppl. Tables 1-3 provide details on the sample’s characteristics and scanning protocols.
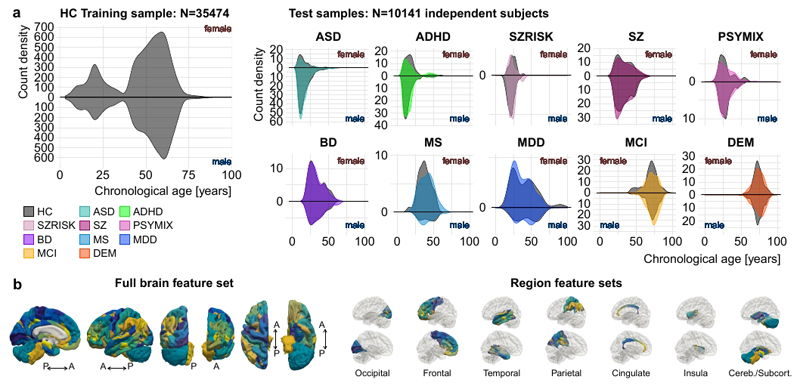
We used machine learning to estimate individual brain age based on structural brain imaging features. First, we grouped all subjects into different samples. For each of the ten clinical groups, we identified a group of healthy individuals of equal size, matched on age, sex and scanning site from a pool of 4353 healthy control subjects. All remaining individuals were joined into one independent sample comprising healthy individuals only. The latter constituted a training sample, used to train and tune the machine learning models for age prediction (n = 35,474 aged 3-89 years; 18,990 females), whereas the ten clinical samples were used as independent test samples. Figure 1a illustrates the respective age distributions per sex and diagnosis.
Figure 1. Sample distributions and imaging features used for brain age prediction.
a, Age distributions of the training (left) and the ten test samples (right) per sex and diagnosis. The grey shades behind each clinical group reflect its age-, sex- and site-matched control group. b, Cortical features from the Human Connectome Project (HCP) atlas as well as cerebellar/subcortical features used for brain age prediction. Colours were assigned randomly to each feature. All features were used in the full brain feature set (left), whereas only those from specific regions (occipital, frontal, temporal, parietal, cingulate, insula, cerebellar/subcortical) were included in the regional feature set (right). For illustration purpose, the left hemisphere is shown.
The large sample size and wide age-span of the training sample allowed us to model male and female brain age separately, thereby accounting for potential sexual dimorphisms in brain structural lifespan trajectories12. For each sex, we built a machine learning model based on gradient tree boosting to predict the age of the brain from a set of thickness, area and volume features extracted using a multi-modal parcellation of the cerebral cortex as well as a set of cerebellar/subcortical volume features (1,118 features in total, Fig. 1b). Five-fold cross-validations revealed high correlations between chronological age and predicted brain age (r=.93 and r=.94 for the female and male model, respectively; Suppl. Fig. 2). Suppl. Fig. 3-6 provide further validation of the prediction approach and Suppl. Table 4 provides details on sex differences in the prediction models. Next, we applied the models to predict age for each individual in the ten independent test samples (predicting brain age using the female model in females and the male model in males) and tested for effects of diagnosis on the brain age gap using linear models. We used mega-analysis (across-site analysis) as the main statistical framework and provide results from a meta-analysis framework in the supplement. We included age, age2, sex, scanning site and a proxy of image quality (Euler number) in all statistical models testing for group differences and clinical associations. To further minimize confounding effects of data quality, we repeated the main analyses using a more stringent quality control and exclusion procedure.
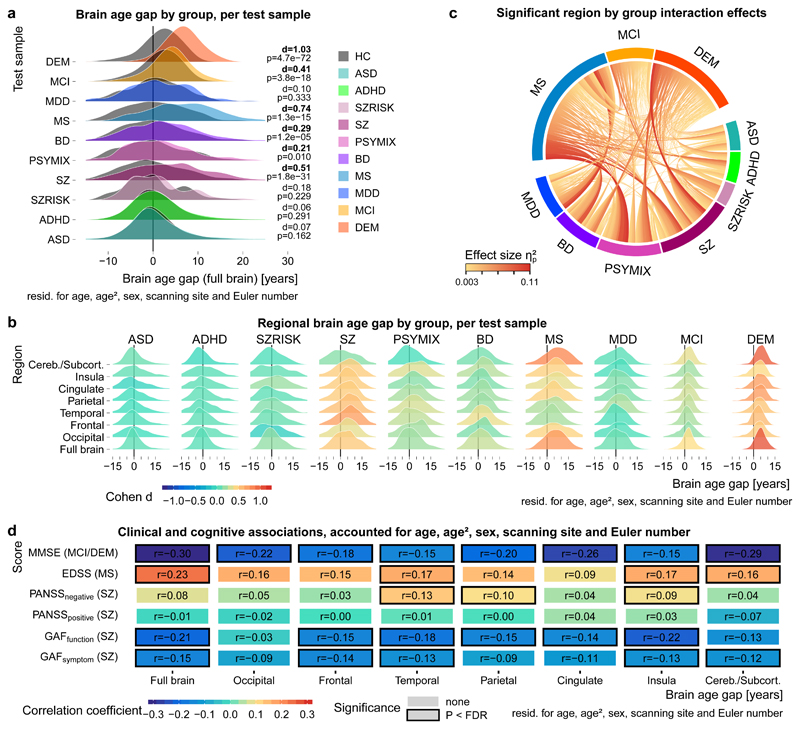
Figure 2a illustrates that the estimated brain age gap was increased in several brain disorders. Strongest effects were observed in SZ (Cohen’s d = 0.51), MS (d = 0.74), MCI (d = 0.41) and DEM (d = 1.03). PSYMIX (d = 0.21) and BD (d = 0.29) showed small effects of increased brain age gap, whereas other groups showed negligible effects (d<0.2). The meta-analysis converged on the same findings (Suppl. Fig. 7) and the results replicated regardless of the quality control exclusion criterion applied (Suppl. Fig. 8). The brain age gap in all clinical groups was positive on average and there were no signs of a negative brain age gap (developmental delay) in children with ASD or ADHD, and no significant group by age interaction effect (Suppl. Table 5).
Figure 2. Apparent brain aging is common in several brain disorders and sensitive to clinical and cognitive measures.
a, The gap between chronological age and brain age was increased in several disorders. The grey shades behind each clinical group reflect its age-, sex- and site-matched controls. The test samples comprised n=925 ASD / n=925 HC, n=725 ADHD / n=725 HC, n=94 SZRISK / n=94 HC, n=1110 SZ / n=1110 HC, n=300 PSYMIX / n=300 HC, n=459 BD / n=459 HC, n=254 MS / n=254 HC, n=208 MDD / n=208 HC, n=974 MCI / n=974 HC, n=739 DEM / n=739 HC; in total n=10,141 independent subjects. Cohen’s d effect sizes (pooled standard deviation units) and two-sided P-values are provided. b, Several disorders showed specific patterns in regional brain age gaps. Colours indicate Cohen’s d effect sizes for group comparisons. Sample size as specified in panel a. Corresponding correlation matrix of the effect sizes is depicted in Suppl. Fig. 9. c, Effect sizes of significant region by group interactions from repeated measures ANOVAs run for each combination of regions and groups (1260 tests in total). Sample size as specified in panel a yet excluding HC; n=5788 independent subjects. Only significant (p<FDR; Benjamini-Hochberg) effects are shown. Suppl. Fig. 10 depicts effect sizes for all 1260 tests. d, Correlation coefficients for linear associations between brain age gaps and cognitive and clinical scores. Sample size comprised n=389 SZ for GAFsymptom, n=269 SZ for GAFfunction, n=646 SZ for PANSSpositive, n=626 SZ for PANSSnegative, n=195 MS for EDSS, n=907 MCI and n=686 DEM for MMSE. Associations were computed using linear models accounting for age, age2, sex, scanning site and Euler number, and the resulting t-statistics were transformed to r. Significant (P<FDR; Benjamini-Hochberg; two-sided) associations are marked with a black box. Corresponding scatter plots are depicted in Suppl. Fig 11.
We assessed specificity of the spatial brain age gap patterns across clinical groups. We trained age prediction models using only occipital, frontal, temporal, parietal, cingulate, insula, or cerebellar/subcortical features (Fig. 1b). Cross-validation confirmed the predictive performance of all regional models (Suppl. Fig. 2) which were used to predict regional brain age in the ten independent test sets. Regional brain age gaps largely corresponded to the full brain level, with some notable differential spatial patterns (Fig. 2b). For example, increased cerebellar/subcortical age gap was most prominent in DEM (d = 0.99) and MS (d = 0.81) but was not present in SZ (d = 0.16). The largest effect in SZ was observed in the frontal lobe (d = 0.70). A brain age gap in the temporal lobe was observed in MDD (d = 0.24), whereas there was no evidence (d<0.2) for a brain age gap in ASD, ADHD or SZRISK in any of the regions. To explore regional differences in brain age patterns, we tested for group by region interactions on each pairwise combination of clinical groups and pairwise combination of regional brain age gaps (1260 tests). Figure 2c illustrates the significant effect sizes, indicating that the rate at which different regions age in relation to each other oftentimes showed opposite patterns between disorders typically considered neurodevelopmental (e.g. SZ) and neurodegenerative (e.g. MS/DEM), respectively.
With converging evidence demonstrating largest brain age gaps in SZ, MS, MCI and DEM, we explored the functional relevance of the regional brain age gaps for these groups by testing for associations with clinical and cognitive data. Clinical data available from individuals with SZ included symptom (n = 389) and function (n = 269) scores of the Global Assessment of Functioning scale (GAF) as well as positive (n = 646) and negative (n = 626) scores of the Positive and Negative Syndrome Scale (PANSS). For MS, we assessed associations with scores from the Expanded Disability Status Scale (EDSS, n = 195). In the dementia spectrum, we assessed associations with Mini Mental State Examination scores (MMSE, n = 907 MCI, n = 686 DEM). Figure 2d depicts association strengths accounted for age, age2, sex, scanning site and Euler number and Suppl. Fig. 11 provides corresponding scatter plots. In SZ, larger brain age gaps were associated with lower functioning, for example full brain age gap with GAF symptom (r = -0.15, P = .003) and insula brain age gap with GAF function (r = -0.22, P = 3 x 10-4), and with more negative symptoms, for example temporal brain age gap with PANSS negative (r = 0.13, P = .001). In MS, larger full brain age gap was associated with higher disability (r = 0.23, P = .001). Finally, lower cognitive functioning was associated with larger brain age gaps in MCI/DEM, with strongest effects for full brain (r = -0.30, P = 7 x 10-33) and cerebellar/subcortical (r = -0.29, P = 2 x 10-30) brain age gaps.
Given the substantial genetic contributions to most brain disorders, our results incite the question to what degree brain age patterns are genetically influenced and if the implicated polymorphisms overlap with the polygenic architectures of the disorders. We used single nucleotide polymorphism (SNP) data from the 20,170 adult healthy individuals with European ancestry available in UK Biobank. We estimated full and regional brain age for these individuals using 5-fold cross-validation in models trained on all healthy controls (n = 39,827 aged 3-95 years; 20,868 females, models trained per sex).
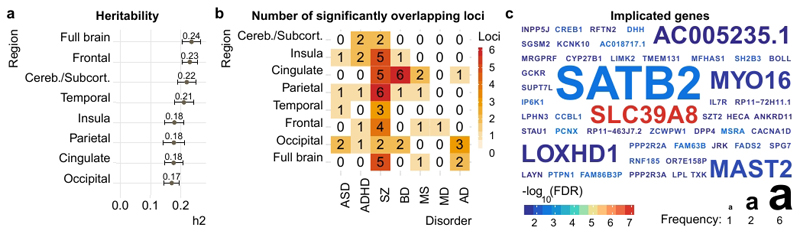
First, we performed one genome-wide association study (GWAS) per brain age gap using PLINK, including the first ten population components from multidimensional scaling, age, age2, sex, scanning site and Euler number as covariates. Next, we assessed heritability using LD score regression on the resulting summary statistics. In line with earlier results from twin studies13, our SNP-based analysis revealed significant heritability (Fig. 3a), with common SNPs explaining 24% of the variance in brain age gap across all individuals (full brain, h2SNP = 0.24, SE = 0.03) and 17-23% of the variance in regional brain age gaps (all SE < 0.03).
Figure 3. The brain age gaps are heritable, and the genetic underpinnings overlap with those observed for several disorders.
Genetic analyses were performed using data from n=20,170 healthy adult individuals with European ancestry a, Heritability (h2) estimated using LD Score regression. Error bars reflect standard error. b, Significantly (P<FDR) overlapping loci between brain age gaps and disorders, identified using conjunctional FDR. c, Corresponding to panel b, the overlapping genes across all disorders, coloured by significance and sized by frequency of detection.
Next, we assessed the overlap between the genetic underpinnings of brain age gap and common brain disorders. We gathered GWAS summary statistics for ASD, ADHD, SZ, BD, MS, major depression (MD), and Alzheimer’s disease (AD) (see online methods). First, using LD score regression, we assessed the genetic correlation between these summary statistics and those from brain age gaps. Correlations were overall weak (Suppl. Fig. 12), with only one surviving FDR correction for the number of tests (cingulate brain age gap with ADHD). Lack of genetic correlation does not preclude genetic dependence as traits may have mixed effect directions across shared genetic variants14. Thus, we next used conjunctional FDR analyses to identify SNPs that are significantly associated with both brain age gap and disorders. We found significant independent loci showing pleiotropy between brain age gaps and all included disorders (Figure 3b). Most loci were identified for SZ (2 occipital, 4 frontal, 3 temporal, 6 parietal, 5 cingulate, 5 insula, 2 cerebellar/subcortical; 161 SNPs in total). Further, 5 independent loci for ASD (76 SNPs), 6 for ADHD (80 SNPs), 10 for BD (94 SNPs), 5 for MS (22 SNPs), 1 for MD (14 SNPs), and 6 for AD (15 SNPs). Suppl. Table 6 provides details. Figure 3c depicts the identified genes coloured by significance and sized by frequency. An intronic variant in protein coding gene SATB2 at chromosome 2q33.1 was most frequently associated with brain age gaps and SZ. A missense variant in protein coding gene SLC39A8 was associated with subcortical brain age gap and SZ and showed the strongest effect in all tested associations (P = 9 x 10-8).
Taken together, our results provide strong evidence that several common brain disorders are associated with an apparent aging of the brain, with effects observed at the full brain or regional level in SZ, PSYMIX, BD, MS, MDD, MCI and DEM; but not in ASD, ADHD or SZRISK. Importantly, our approach revealed differential neuroanatomical distribution of brain age gaps between several disorders. Associations with clinical and cognitive data in patients supported the functional relevance of the brain age gaps and genetic analyses in healthy individuals provided evidence that the brain age gaps are heritable, with overlapping genes between brain age gaps in healthy adults and common brain disorders.
Our approach of estimating regional brain age was useful to reveal differential spatial patterns between disorders. Whereas the implicated regions in the spatial brain age profiles of the disorders largely corresponded with previously reported structural abnormalities (e.g. frontal in SZ15 and substantial subcortical volume loss in AD16), our regional brain age approach preserved the well-established benefit of down-sampling a large number of brain imaging features into a condensed and interpretable score without a total loss of spatial sensitivity. As such, the analysis revealed substantial differences in spatial aging profiles between disorders typically regarded as neurodegenerative (MS, MCI, DEM) and neurodevelopmental, in particular SZ and PSYMIX. For example, whereas these disorders were all associated with increased brain age gap on the full brain level, regional analysis revealed interactions between the frontal brain age patterns observed in SZ and the cerebellar/subcortical patterns observed in MS and DEM, supporting spatial differences in apparent brain age. Moreover, significant associations with clinical and cognitive data, in particular with scores of the GAF and PANSS in SZ, with the EDSS in MS and with MMSE in the dementia spectrum demonstrated functional relevance of brain age gap beyond group differences. By gauging the dynamic associations between changes in brain age and clinical and cognitive function, future longitudinal studies may prove instrumental to dissect the large individual differences among patients with brain disorders, even within the same diagnostic category17. Furthermore, incorporating additional imaging modalities, voxel-level data or different segmentations at various levels of resolution will allow for estimation of tissue-specific brain age gaps or different regional gaps in future studies. Such approaches will also be useful to further investigate the apparent lack of brain age gap differences in ASD and ADHD. In contrast to research from other imaging phenotypes18,19, we did not observe case-control differences in brain age gaps for ASD or ADHD, nor group by age interactions (developmental delays might be reflected in a negative brain age gap in children). Brain age gaps based on different imaging modalities may capture different aspects of pathophysiology and will therefore yield an important contribution in future research.
Conceptually, brain age gaps reflect a prediction error from a machine learning model and can therefore be attributed to both noise (lack of model accuracy, insufficient data quality) and physiology (deviations from normal aging trajectories). The large training sample and accurate model performance, replication of results at different data quality criterions, as well as our approach of comparing brain age gaps of cases to a group of age-, sex- and scanner-matched controls allowed us to reduce the impact of noise and to attribute variation in brain age gaps as likely related to biologically relevant differences. The physiological underpinnings of the brain age gaps are likely diverse, much like the polygenic nature of brain disorders and their profoundly heterogeneous symptomatology. They may reflect differences in disease severity, effects of comorbid disorders, substance use or other adverse lifestyle factors. Genetic analysis offers one way of exploring factors that influence phenotypic variation toward an improved understanding of the multi-faceted sources of lifespan trajectories in the brain. Here, we provided evidence that full and regional brain age gaps represent genetically influenced traits, and illustrated that the genetic variants associated with brain age gaps in healthy individuals partly overlap with those observed in ASD, ADHD, SZ, BD, MS, MD and AD. In line with accumulating evidence that common brain disorders are highly polygenic and partly overlapping20 these results suggest shared molecular genetic mechanisms between brain age gaps and brain disorders. Statistical associations do not necessarily signify causation, and functional interpretations of the identified genes should be made with caution. Larger imaging genetics samples, in particular those including individuals with common brain disorders, may in the future allow the investigation of specificity of the implicated genes, and integrating a wider span of imaging modalities may increase both sensitivity and specificity.
In conclusion, we have established that the brain age gap is increased in several common brain disorders, sensitive to clinical and cognitive phenotypes and genetically influenced. Our results emphasize the potential of advanced lifespan modelling in the clinical neurosciences, highlighting the benefit of big data resources that cover a wide age span and conditions. Delineating dynamic lifespan trajectories within and across individuals will be essential to disentangle the pathophysiological complexity of brain disorders.
Online methods
Additional information is available in the Life Sciences Reporting Summary.
Samples
We have included data collected through collaborations, data sharing platforms, consortia as well as available in-house cohorts. No statistical methods were used to pre-determine sample sizes. We included as much data as we could gather (brain scans from N=45,615 individuals) and sample size of individual clinical groups is thus based on data availability. Suppl. Tables 1 - 3 provide detailed information on the individual cohorts. All included cohorts have been published on, and we refer to a list of publications that can be consulted for a more detailed overview of cohort characteristics. Data collection in each cohort was performed with participants’ written informed consent and with approval by the respective local Institutional Review Boards.
Image pre-processing and quality control
Raw T1 data for all study participants were stored and analysed locally at University of Oslo, following a harmonized analysis protocol applied to each individual subject data (Suppl. Fig. 1). We performed automated surface-based morphometry and subcortical segmentation using Freesurfer 5.321. We deployed an automated quality control protocol executed within each of the contributing cohorts that excluded potential outliers based on the Euler number22 of the respective Freesurfer segmentations. Euler number captures the topological complexity of the uncorrected Freesurfer surfaces and thus comprises a proxy of data quality22. In brief, for each scanning site we regressed age, age2 and sex from the Euler number of the left and right hemispheres and identified scans that deceeded 3 standard deviations (SD) on either of the residualized Euler numbers. Suppl. Fig. 13 provides a validation of the approach against manual quality control. Data from a total of 977 individuals was excluded in this step, yielding 45,615 subjects for the main analysis. To further minimize confounding effects of data quality23, we performed supplementary analyses using a subset of data, where a more stringent threshold was used for exclusion (1 SD on Euler numbers). Thus, supplemental analysis provides a sanity check with those subjects excluded (sample size: n = 40,301).
Brain age prediction
We utilized a recent multi-modal cortical parcellation scheme24 to extract cortical thickness, area and volume for 180 regions of interest (ROI) per hemisphere. In addition, we extracted the classic set of cerebellar/subcortical and cortical summary statistics21. This yielded a total set of 1118 structural brain imaging features (360/360/360/38 for cortical thickness/area/volume as well as cerebellar/subcortical and cortical summary statistics, respectively).
We used machine learning on this feature set to predict the age of each individual’s brain. First, we split the available data into a training sample and ten independent test samples (Fig. 1a). The test samples in total comprised 5788 individuals with brain disorders and 4353 healthy controls. For each of the ten clinical groups, we selected a set of healthy controls from the pool of 4353 individuals, matched for age, sex and scanning site using propensity score matching25. Thus, data from some healthy individuals acted as control data in several test samples, yet each test sample had the same number of patients and controls and all subjects in the test samples were independent of the subjects in the training sample. The remaining datasets (45,615 − (5788+4353) = 35,474) went into the training set. For each sex, we trained machine learning models based on gradient tree boosting26 utilizing the xgboost package in R27, chosen due to its resource efficiency and demonstrated superior performance in previous machine learning competitions26, to predict the age of the brain using data available in the training set. First, model parameters were tuned using a 5-fold cross-validation of the training data. This step identified the optimal number of model training iterations by assessing the prediction error for 1500 rounds and implementing an early stopping if the performance did not improve for 20 rounds. Based on previous experience, the learning rate was pre-set to eta=0.01 and all other parameters were set to default27 for linear xgboost tree models. After determining the optimal number of training iterations, the full set of training data was used to train the final models with the adjusted nrounds parameter. These models were used to predict brain age in the test samples, and the brain age gap (deviation between brain and chronological age) was computed. In line with a recent recommendation28, all statistical analyses on the brain age gap accounted for age, age2, sex, scanning site and Euler number. In addition, to assess overall model performance, prediction models were cross-validated within the training set using a 5-fold cross validation, each fold implementing the above described training procedure and testing on the hold-out part of the training set. Brain age predictions on the level of individual brain regions followed the same procedures as those described for the full brain level, except that the feature set was reduced to cover only those features that overlapped more than 50% with a given lobe. Regions were defined following the Freesurfer lobesStrict segmentation as occipital, frontal, temporal, parietal, cingulate and insula. In addition, given the limited number of cerebellar features available in the Freesurfer summary statistics, cerebellar and subcortical features were grouped into a cerebellar/subcortical region (Fig. 1b). For additional validation, we compared our xgboost approach against two other approaches (Suppl. Fig. 3). One approach implemented a different machine learning algorithm on the same set of features (slm from the care package29), whereas the other approach made use of a fully independent processing pipeline, feature set and algorithm (github.com/james-cole/brainageR13,30). Furthermore, we assessed the impact of sample size on model performance by creating random subsets of data with sample sizes of 100, 500, 1000, 2000, 5000, 10,000, and 20,000 individuals (40 random subsets per sample size). For each subset and sample size we assessed model performance using cross-validation (Suppl. Fig. 5).
The genetic analysis was performed in UK Biobank data, which was part of the training set in the main analysis. We thus trained different brain age models for the genetic analysis. We selected all healthy subjects and estimated their brain age using a 5-fold cross-validation approach, like the one performed when validating performance of the training set. The resulting unbiased estimates of brain age gaps for all UK Biobank individuals with genetic data available went into the genome-wide association analysis, LD score regression and conjunctional FDR.
Main statistical analysis framework
We performed both mega- (across cohorts) and meta- (within cohort) analyses. To estimate group effects on a given measure in a mega-analysis framework, we computed the effect of diagnosis in relation to the healthy controls for each of the ten test samples in a linear model accounting for age, age2, sex, scanning site and Euler number. Cohen’s d effect sizes were estimated based on contrast t-statistics31 following Formula 1:
| (1) |
For the meta-analysis, similar models were computed within cohorts. In addition to estimating Cohen’s d (Formula 1), we estimated the variance of d following Formula 2.
| (2) |
Cumulative effects across cohorts were then estimated using a variance-weighted random-effects model as implemented in the metafor package in R32.
Data distributions were assumed to be normal, but this was not formally tested. Data collection and analysis were not performed blind to the conditions of the experiments.
Assessment of regional specificity
In Suppl. Fig 9, we performed clustering of effect sizes from Figure 2b using heatmap.2 from the gplots package33 in R. A Spearman correlation matrix was computed based on the case-control effect sizes obtained from each test sample and region and hierarchical clustering was performed using the default settings. To further explore regional specificity, we performed an analysis that involved only the clinical groups. We regressed age, age2, sex, scanning site and Euler number from the brain age gaps in each test sample. Next, we joined data from each pair of clinical groups and each pair of regions for repeated measures analysis of variance and estimated the effect sizes of region x group interactions (1260 ANOVAs in total). The significant interaction effects were visualized in Figure 2c using the circlize package34 in R.
Genetic analyses
We restricted all genetic analyses to individuals from the UK Biobank with European ancestry, as determined by the UK Biobank study team35. We applied standard quality control procedures to the UK Biobank v3 imputed genetic data. In brief, we removed SNPs with an imputation quality score below 0.5, with a minor allele frequency less than .05, missing in more than 5% of individuals, and failing the Hardy Weinberg equilibrium tests at a p<1x10-6, yielding SNP data from 20,170 adult healthy individuals. We performed a genome-wide association analysis using PLINK v1.936, accounting the analysis for 10 genetic principal components, age, age2, sex, scanning site and Euler number. We used LD Score regression37 to estimate narrow sense heritability.
Furthermore, we used cross-trait LD Score regression37,38 to calculate genetic correlations, and conjunctional FDR analyses39,40 to assess genetic overlap between two complex traits. We gathered genome-wide association analysis (GWAS) summary statistics for ASD41, ADHD42, SZ43, BD44, MS45, MD46, and AD47; and assessed genetic overlap with brain age gap genetics. The MHC region was excluded from all analysis. Conjunctional FDR was run for each pair of full brain / regional brain age gap and group, using conjunctional FDR threshold of 0.05. SNPs were annotated using the Ensembl Variant Effect Predictor48.
Cognitive and clinical associations
Cognitive and clinical associations were tested in subsets based on data availability and were performed in clinical groups only (excluding controls) as described in the main text. Using linear models accounting for age, age2, sex, scanning site and Euler number we associated brain age gaps with scores of the Global Assessment of Functioning scale49 (GAF), the Positive and Negative Syndrome Scale50 (PANSS), the Expanded Disability Status Scale51 (EDSS) and Mini Mental State Examination scores52 (MMSE). The t-statistics of the linear models were transformed to r, thus the correlation coefficients depicted in Fig 2d essentially reflect a partial correlation between full brain / regional brain age gaps and clinical/cognitive scores, controlling for confounding effects of age, sex, site and image quality.
Supplementary Material
Acknowledgements
The author list between Ingrid Agartz and Mathias Zink is in alphabetic order. The authors were funded by the Research Council of Norway (276082 LifespanHealth (T.K.), 213837 (O.A.A), 223273 NORMENT (O.A.A.), 204966 (L.T.W.), 229129 (O.A.A.), 249795 (L.T.W.), 273345 (L.T.W.), 283798 SYNSCHIZ (O.A.A.)), the South-Eastern Norway Regional Health Authority (2013-123 (O.A.A.), 2014-097 (L.T.W.), 2015-073 (L.T.W.), 2016083 (L.T.W.)), Stiftelsen Kristian Gerhard Jebsen, the European Research Council (ERC StG 802998 BRAINMINT (L.T.W.)), NVIDIA Corporation GPU Grant (T.K.), and the European Commission 7th Framework Programme (602450, IMAGEMEND (A.M.-L.)). The data used in this study were gathered from various sources. A detailed overview of the included cohorts and acknowledgement of their respective funding sources and cohort-specific details is provided in Suppl. Table 1. Data used in preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu), from the AddNeuroMed consortium, and from the Pediatric Imaging, Neurocognition and Genetics Study (PING) database (www.chd.ucsd.edu/research/ping-study.html, now shared through the NIMH Data Archive (NDA)). The investigators within the ADNI and PING contributed to the design and implementation of ADNI/PING and/or provided data but did not participate in analysis or writing of this report. This publication is solely the responsibility of the authors and does not necessarily represent the views of the National Institutes of Health or PING investigators. Complete listings of participating sites and study investigators can be found at http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf and https://ping-dataportal.ucsd.edu/sharing/Authors10222012.pdf. The AddNeuroMed consortium was led by Simon Lovestone, Bruno Vellas, Patrizia Mecocci, Magda Tsolaki, Iwona Kłoszewska, Hilkka Soininen.
Footnotes
Code availability.
Code needed to run brain age prediction models is available at github.com/tobias-kaufmann (see Data availability). Additional R statistics53 code is available from the authors upon request.
Data availability
The raw data incorporated in this work were gathered from various resources. Material requests will need to be placed with individual PIs. A detailed overview of the included cohorts is provided in Suppl. Table 1. GWAS summary statistics for the brain age gaps as well as the models needed to predict brain age in independent cohorts are available at github.com/tobias-kaufmann.
Author contributions
T.K. and L.T.W. conceived the study; T.K., N.T.D. and L.T.W. pre-processed all data in Freesurfer; N.T.D., M.J.L., C.L.B, L.B.N., L.T.W. and T.K. performed quality control of the data; T.K. performed the analysis with contributions from L.T.W. and D.v.d.M.; T.K., L.T.W., N.T.D., D.v.d.M. and O.A.A. contributed to interpretation of the results. All remaining authors were involved in data collection at various sites as well as cohort-specific tasks. T.K. and L.T.W. wrote the first draft of the paper and all authors contributed to and approved the final manuscript.
Competing financial interests
Some authors received educational speaker’s honorarium from Lundbeck (O.A. Andreassen, A. Bertolino, T. Elvsåshagen, M. Zink, N. I. Landrø), Sunovion (O.A. Andreassen), Shire (B. Franke), Medice (B. Franke), Otsuka (A. Bertolino, M. Zink) and Jannsen (A. Bertolino), Roche (M. Zink), Ferrer (M. Zink), Trommsdorff (M. Zink), Servier (M. Zink), all of these unrelated to this work. A. Bertolino is a stockholder of Hoffmann-La Roche Ltd and has received consultant fees from Biogen Idec. E. G. Celius and H. F. Harbo have received travel support, honoraria for advice and lecturing from Almirall (Celius), Biogen Idec (both), Genzyme (both), Merck (both), Novartis(both), Roche (both), Sanofi-Aventis (both) and Teva (both). They have received unrestricted research grants from Novartis (Celius, Harbo), Biogen Idec (Celius) and Genzyme (Celius). G. Pergola has been the academic supervisor of a Roche collaboration grant (years 2015-16) that funds his salary. None of the mentioned external parties had any role in the analysis, writing or decision to publish this work. Other authors declare no competing financial interests.
Members of the Karolinska Schizophrenia Project (KaSP)
Lars Farde5, Lena Flyckt5, Göran Engberg71, Sophie Erhardt71, Helena Fatouros-Bergman5, Simon Cervenka5, Lilly Schwieler71, Fredrik Piehl72, Ingrid Agartz1,4,5, Karin Collste5, Pauliina Victorsson5, Anna Malmqvist71, Mikael Hedberg71, Funda Orhan71
71 Department of Physiology and Pharmacology, Karolinska Institutet, Stockholm, Sweden.
72 Neuroimmunology Unit, Department of Clinical Neuroscience, Karolinska Institutet, Stockholm, Sweden
References
- 1.WHO. World Health Statistics 2016. 2016. [Google Scholar]
- 2.Insel TR, Cuthbert BN. Brain disorders? Precisely. Science. 2015;348:499–500. doi: 10.1126/science.aab2358. [DOI] [PubMed] [Google Scholar]
- 3.Prince M, et al. No health without mental health. Lancet. 2007;370:859–877. doi: 10.1016/S0140-6736(07)61238-0. [DOI] [PubMed] [Google Scholar]
- 4.Parikshak NN, Gandal MJ, Geschwind DH. Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat Rev Genet. 2015;16:441–458. doi: 10.1038/nrg3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marin O. Developmental timing and critical windows for the treatment of psychiatric disorders. Nat Med. 2016;22:1229–1238. doi: 10.1038/nm.4225. [DOI] [PubMed] [Google Scholar]
- 6.Insel TR. Rethinking schizophrenia. Nature. 2010;468:187–193. doi: 10.1038/Nature09552. [DOI] [PubMed] [Google Scholar]
- 7.Aubert-Broche B, et al. Onset of multiple sclerosis before adulthood leads to failure of age-expected brain growth. Neurology. 2014;83:2140–2146. doi: 10.1212/WNL.0000000000001045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masters CL, et al. Alzheimer's disease. Nat Rev Dis Primers. 2015;1 doi: 10.1038/nrdp.2015.56. 15056. [DOI] [PubMed] [Google Scholar]
- 9.Dosenbach NU, et al. Prediction of individual brain maturity using fMRI. Science. 2010;329:1358–1361. doi: 10.1126/science.1194144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franke K, Ziegler G, Kloppel S, Gaser C. Alzheimer's Disease Neuroimaging, I. Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: exploring the influence of various parameters. Neuroimage. 2010;50:883–892. doi: 10.1016/j.neuroimage.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Cole JH, Franke K. Predicting Age Using Neuroimaging: Innovative Brain Ageing Biomarkers. Trends Neurosci. 2017;40:681–690. doi: 10.1016/j.tins.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Ritchie SJ, et al. Sex Differences in the Adult Human Brain: Evidence from 5216 UK Biobank Participants. Cereb Cortex. 2018;28:2959–2975. doi: 10.1093/cercor/bhy109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole JH, et al. Predicting brain age with deep learning from raw imaging data results in a reliable and heritable biomarker. Neuroimage. 2017;163:115–124. doi: 10.1016/j.neuroimage.2017.07.059. [DOI] [PubMed] [Google Scholar]
- 14.Bansal V, et al. Genome-wide association study results for educational attainment aid in identifying genetic heterogeneity of schizophrenia. Nature Communications. 2018;9:3078. doi: 10.1038/s41467-018-05510-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellison-Wright I, Bullmore E. Anatomy of bipolar disorder and schizophrenia: a meta-analysis. Schizophrenia research. 2010;117:1–12. doi: 10.1016/j.schres.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 16.Jernigan TL, Salmon DP, Butters N, Hesselink JR. Cerebral structure on MRI, Part II: Specific changes in Alzheimer's and Huntington's diseases. Biological psychiatry. 1991;29:68–81. doi: 10.1016/0006-3223(91)90211-4. [DOI] [PubMed] [Google Scholar]
- 17.Wolfers T, et al. Mapping the Heterogeneous Phenotype of Schizophrenia and Bipolar Disorder Using Normative Models. Jama Psychiat. 2018;75:1146–1155. doi: 10.1001/jamapsychiatry.2018.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ecker C, Bookheimer SY, Murphy DG. Neuroimaging in autism spectrum disorder: brain structure and function across the lifespan. Lancet Neurol. 2015;14:1121–1134. doi: 10.1016/S1474-4422(15)00050-2. [DOI] [PubMed] [Google Scholar]
- 19.Faraone SV, et al. Attention-deficit/hyperactivity disorder. Nature Reviews Disease Primers. 2015;1 doi: 10.1038/nrdp.2015.20. 15020. [DOI] [PubMed] [Google Scholar]
- 20.Andreassen OA, et al. Genetic pleiotropy between multiple sclerosis and schizophrenia but not bipolar disorder: differential involvement of immune-related gene loci. Molecular psychiatry. 2015;20:207. doi: 10.1038/mp.2013.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischl B, et al. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/S0896-6273(02)00569-X. [DOI] [PubMed] [Google Scholar]
- 22.Rosen AFG, et al. Quantitative assessment of structural image quality. Neuroimage. 2018;169:407–418. doi: 10.1016/j.neuroimage.2017.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith SM, Nichols TE. Statistical Challenges in "Big Data" Human Neuroimaging. Neuron. 2018;97:263–268. doi: 10.1016/j.neuron.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 24.Glasser MF, et al. A multi-modal parcellation of human cerebral cortex. Nature. 2016;536:171–178. doi: 10.1038/nature18933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho D, Imai K, King G, Stuart E. A. MatchIt: Nonparametric Preprocessing for Parametric Causal Inference. Journal of Statistical Software. 2011;42:1–28. [Google Scholar]
- 26.Chen T, Guestrin C. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; San Francisco, California, USA: ACM; 2016. pp. 785–794. [Google Scholar]
- 27.Tianqi C, Tong H, Benesty M, Khotilovich V, Tang Y. R package v0.4-2. 2015. Xgboost: extreme gradient boosting. [Google Scholar]
- 28.Le TT, et al. A Nonlinear Simulation Framework Supports Adjusting for Age When Analyzing BrainAGE. Front Aging Neurosci. 2018;10:317. doi: 10.3389/fnagi.2018.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuber V, Strimmer K. Care. R package v 1.1.10. 2017. Care. R package v 1.1.10.
- 30.Cole JH, et al. Brain age predicts mortality. Molecular psychiatry. 2018;23:1385–1392. doi: 10.1038/mp.2017.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakagawa S, Cuthill IC. Effect size, confidence interval and statistical significance: a practical guide for biologists. Biol Rev Camb Philos Soc. 2007;82:591–605. doi: 10.1111/j.1469-185X.2007.00027.x. [DOI] [PubMed] [Google Scholar]
- 32.Viechtbauer W. Conducting meta-analysis in R with the metafor package. Journal of Statistical Software. 2010;36:1–48. [Google Scholar]
- 33.Warnes GR, et al. R Package gplots: Various R Programming Tools for Plotting Data. 2016. [Google Scholar]
- 34.Gu Z. R Package circlize: Circular Visualization. 2017. [Google Scholar]
- 35.Bycroft C, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nature genetics. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nature genetics. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nichols T, Brett M, Andersson J, Wager T, Poline J-B. Valid conjunction inference with the minimum statistic. Neuroimage. 2005;25:653–660. doi: 10.1016/j.neuroimage.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 40.Andreassen OA, et al. Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. The American Journal of Human Genetics. 2013;92:197–209. doi: 10.1016/j.ajhg.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grove J, et al. Identification of common genetic risk variants for autism spectrum disorder. Nature genetics. 2019;51:431–444. doi: 10.1038/s41588-019-0344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Demontis D, et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nature genetics. 2018 doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schizophrenia Working Group of the PGC et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stahl EA, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nature genetics. 2019;51:793–803. doi: 10.1038/s41588-019-0397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patsopoulos N, et al. The Multiple Sclerosis Genomic Map: Role of peripheral immune cells and resident microglia in susceptibility. bioRxiv. 2017 doi: 10.1101/143933. 143933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wray NR, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nature genetics. 2018;50:668–681. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lambert J-C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nature genetics. 2013;45:1452. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McLaren W, et al. The ensembl variant effect predictor. Genome biology. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pedersen G, Karterud S. The symptom and function dimensions of the Global Assessment of Functioning (GAF) scale. Comprehensive Psychiatry. 2012;53:292–298. doi: 10.1016/j.comppsych.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 50.Kay SR, Fiszbein A, Opfer LA. The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophrenia Bull. 1987;13:261. doi: 10.1093/schbul/13.2.261. [DOI] [PubMed] [Google Scholar]
- 51.Kurtzke JF. Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS) Neurology. 1983;33:1444–1444. doi: 10.1212/wnl.33.11.1444. [DOI] [PubMed] [Google Scholar]
- 52.Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”: a practical method for grading the cognitive state of patients for the clinician. Journal of psychiatric research. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 53.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.