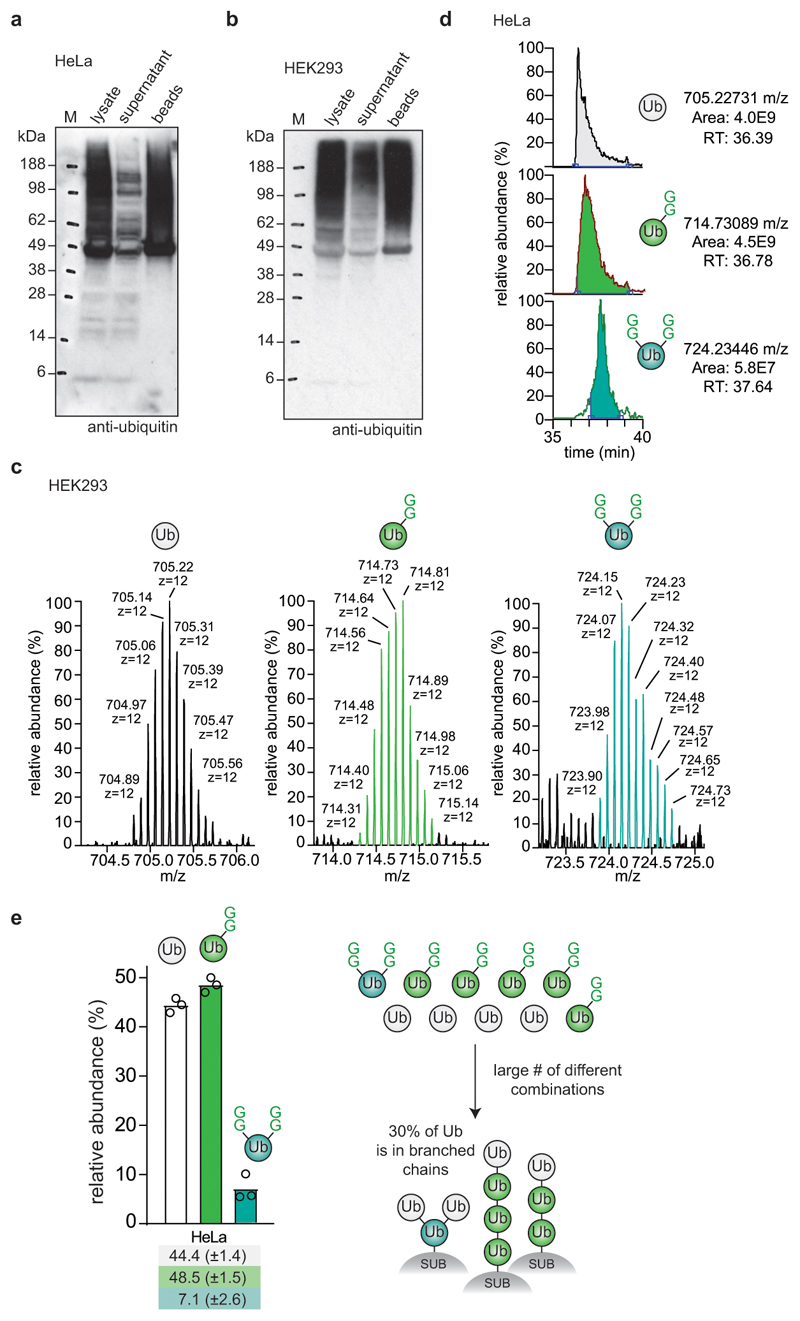
Extended Data Figure 8. Characterisation of TUBE pull-downs from cells.
a, b Efficiency of polyubiquitin depletion by TUBE pull-down from (a) HeLa and (b) HEK293 cells. Prior to the addition of the glutathione affinity resin, an input sample was taken (lysate). Samples of the supernatant after incubation with the affinity resin and of the beads after washing with PBS were analysed by ubiquitin Western blotting. Experiments in a and b were performed in triplicate. c, Intact MS analysis of TUBE-purified ubiquitin species. The isotopic distribution of the charge state z = +12 is shown. Distinct GlyGly-modified samples are easily distinguishable by mass. d, Peak integration of each ubiquitin species’ nominal mass (unmodified, 1xGG, 2xGG) from LC-MS analysis. Distinct GlyGly-modified samples are also distinguishable by chromatographic behaviour. Experiments in c and d were performed in triplicate. Results are consistent with previous data using limited trypsinolysis18; we consistently identified ~2-3% more branched ubiquitin, possibly due to differences in ubiquitin enrichment strategies or owing to partial digestion of ubiquitin by trypsin. e, Data from HeLa cells (Fig. 3f), when applied to a finite pool of 10 ubiquitin moieties, correspond roughly to a collection of 4 x unmodified ubiquitin (white), 5 x GlyGly modified ubiquitin (green) and 1 x double-GlyGly modified, branch-point ubiquitin. With these ratios multiple distinct species can be assembled, some of which are depicted. It is clear that a single branch-point ubiquitin in this case requires 3 of the 10 ubiquitin molecules (i.e. 30%) to be in a branched chain. This number can be higher if the branched chain also incorporates mono-GlyGly-modified species. For expanded systems (with e.g. double the number of ubiquitins, and 2 x double-GlyGly-modified ubiquitin) this number can also be lower when e.g. multiple branched species exist in the same polymer (in which case a chain can be built from 5 out of 20 ubiquitin molecules, i.e. 25% of ubiquitin is in branched architectures). More definitive assessments of individual architectures would be possible once individual lengths of chains, e.g. tetra-ubiquitin, could be analysed with this method. Centred values correspond to the mean of independent experiments performed in triplicate. Error values represent s.d. from the mean.