Abstract
Evidence in support of a gut-muscle axis has been reported in rodents, but studies in older adult humans are limited. Accordingly, the primary goals of the present study were to compare gut microbiome composition in older adults that differed in terms of the percentage of whole body lean mass and physical functioning (high-functioning, HF, n=18; low-functioning, LF, n=11), and to evaluate the causative role of the gut microbiome on these variables by transferring fecal samples from older adults into germ-free mice. Family-level Prevotellaceae, genus-level Prevotella and Barnesiella, and the bacterial species Barnesiella intestinihominis were higher in HF older adults at the initial study visit, at a 1-month follow-up visit, in HF human fecal donors, and in HF-colonized mice, when compared with their LF counterparts. Grip strength was significantly increased by 6.4% in HF-, when compared with LF-colonized mice. In contrast, despite significant differences for the percentage of whole body lean mass and physical functioning when comparing the human fecal donors, the percentage of whole body lean mass and treadmill endurance capacity were not different when comparing human microbiome-containing mice. In sum, these data suggest a role for gut bacteria on the maintenance of muscle strength, but argue against a role for gut bacteria on the maintenance of the percentage of whole body lean mass or endurance capacity, findings that collectively add to elucidation of the gut-muscle axis in older adults.
Keywords: microbiome, body composition, sarcopenia, physical function, muscle strength, aging
Introduction
Emerging evidence supports a role for the gut microbiome on the maintenance of lean body mass and physical functioning (Grosicki et al., 2018). In support of this, lean mass is decreased when germ-free mice are colonized with cecal microbiota from conventional mice (Backhed et al., 2004). Similarly, lean mass is increased following antibiotic treatment in mice (Nobel et al., 2015). Although these studies suggest that factors within the microbiome may limit lean body mass, conversely, selectively increasing gut bacterial levels with pre- or pro-biotics increases muscle mass. For example, lean and skeletal muscle muscle mass were increased in obese mice that were fed the prebiotic, oligofructose (Cani et al., 2009; Everard et al., 2011), in association with alterations in gut microbiome composition, including increases in family-level Prevotellaceae, and genus-level Prevotella and Barnesiella (Everard et al., 2011). In addition, Barnesiella are decreased in association with the age-related reduction in the percentage of whole body lean mass in rats (Siddharth et al., 2017). Investigating further, probiotic supplementation with Lactobacillus reuteri and Lactobacillus gasseri decreased muscle atrophy gene expression (i.e. Atrogin-1, MuRF1, LC3, Cathepsin L) and increased muscle mass in a mouse model of leukemia (Bindels et al., 2012), and probiotic supplementation with Lactobacillus plantarum increased lean mass, grip strength, and swimming endurance in young mice (Chen et al., 2016).
A limited number of studies have investigated associations between gut bacteria with lean mass or physical function in humans. In young adults, elevated gut bacterial diversity was found in lean, when compared with their corresponding obese co-twin (Turnbaugh et al., 2009a) and in athletes, when compared with sedentary controls (Clarke et al., 2014). Calf circumference (as an indirect index of muscle mass) was reduced in frail, when compared with less frail older adults in association with elevated abundances of family-level Enterobacteriaceae and Erysipelotrichaceae, whereas genus-level Barnesiella were decreased (Claesson et al., 2012). Similarly, Enterobacteriaceae were increased, whereas Bacteroides/Prevotella and the bacterial species Faecalibacterium prausnitzii were decreased in frail, when compared with less frail older adults (van Tongeren et al., 2005; Jackson et al., 2016). Although the frailty indices in these studies included questions about physical functioning, studies aimed at identification of associations between the gut microbiome with direct measures of lean mass or physical function have yet to be reported in older adults. Accordingly, the first goal of the present study was to identify bacterial taxa and functions that differentiate older adults that had higher values for the percentage of whole body lean mass and for measures of physical function (defined as high-functioning, HF), when compared with older adults that had lower levels of these variables (defined as low-functioning, LF). Furthermore, the reproducibility of these associations was evaluated by comparing gut microbiome composition and functions in HF and LF older adults one-month after the initial study visit.
Although identification of bacterial taxa and functions that are different when comparing older adults are important observations, transplanting intact, uncultured human fecal samples into recipient germ-free mice permits the donors’ microbiota to be replicated and the causative role of their microbiota on outcome measures to be discerned. With use of this approach, a causative role for the gut microbiome on immunosenescence (Fransen et al., 2017), adiposity (Turnbaugh et al., 2009b), obesity (Ridaura et al., 2013), and muscle composition (Yan et al., 2016) has been reported. Accordingly, the second goal of the present study was to examine the causative role of the gut microbiome on the percentage of whole body lean mass and physical function by transferring fecal samples from older adult humans into germ-free mice.
Materials and Methods
Study design
Sedentary older adults (70–85y), defined as the absence of structured exercise during the previous 6 months, were recruited by the Nutrition, Exercise Physiology, and Sarcopenia (NEPS) Laboratory at the Jean Mayer USDA Human Nutrition Research Center on Aging (HNRCA) at Tufts University. Participants were recruited from a database of older adults (65y+) that were screened by the NEPS laboratory for the VIVE2 clinical trial (Kirn et al., 2015), and via advertisements posted on HNRCA bulletin boards and online media. Interested individuals were prescreened over the telephone based on a BMI ≤ 35 kg/m2. Qualified individuals were invited to an onsite screening visit where they signed an IRB-approved informed consent form, had their height, weight, and vital signs measured, completed a medical history questionnaire and the Mini-Mental State Examination (MMSE), and had their short physical performance battery (SPPB) score measured. The SPPB score includes standing balance, a 4-m walk, and a chair stand test (Guralnik et al., 1994). Subjects qualified for the study as low (LF)- or high (HF)-functioning if their BMI was ≤ 35 kg/m2 and if their SPPB score was ≤ 8 or ≥ 11, respectively. Exclusion criteria included a SPPB score of 9 or 10, a BMI ≥ 35 kg/m2, acute or terminal illness, myocardial infarction in the past 6 months, unstable cardiovascular disease, MMSE score < 23, blood pressure > 150/90 mmHg, neuromuscular disease or drugs affecting neuromuscular function, androgen therapy in males, estrogen therapy in females, any significant immune disorder, kidney failure, pancreatic disease, or diabetes. Moreover, subjects that had gastrointestinal or malabsorption disease, a history of cholecystectomy, or that used of probiotics, prebiotics or antibiotics in the 3 months prior to the screening visit were excluded. Eligible subjects were provided with a stool collection kit and were instructed to produce a stool sample within the 18-hour period that preceded their study visits. The Tufts University Health Sciences Campus Institutional Review Board approved this study.
Stool sample collection and processing
Stool samples were collected in specimen containers, placed into a plastic bag, surrounded by frozen gel packs, and delivered to the Tufts HNRCA in coolers. Subjects that were not able to bring their stool sample to the HNRCA provided an in-person stool sample at their respective baseline or 1-month study visits.
Dietary assessment
Older adults recorded their 3-day food intake prior to production of a stool sample at the baseline visit in concert with the Dietary Assessment Unit (DAU) at the HNRCA. Dietary intake data, including energy intake, macro- and micro-nutrients was analyzed by the DAU with use of Nutrition Data System for Research (Schakel, 2001).
Animals, fecal transfer into germ-free mice
Germ-free male and female C57BL/6J mice (GFM) were obtained from the Center for Metagenomics Gnotobiotic Core Facility at Brigham and Women’s Hospital (BWH) for use in the fecal transplantation study. Mice were housed in the Massachusetts Host-Microbiome Center on a 12 h-light/dark cycle in a 22°C temperature-controlled room. Stool samples from 6 LF and 6 HF human donors (3 males, 3 females in each respective group) were obtained from their baseline visit and gavaged into sex-matched GFM based on the methods reported by (Ussar et al., 2015). One fecal sample from each human donor was used to colonize 5 GFM, which were then housed within the same cage. One month following gavage, three of the five mice were used for measurements of body composition and physical function, and stool was collected for gut microbiome analysis. The remaining two colonized mice were used to collect blood and to harvest skeletal muscle. Mice were fed ad libitum with an autoclaved LabDiet 5021 (LabDiet). Animal care and study protocols were in accordance with National Institutes of Health guidelines, and were approved by the Institutional Animal Care and Use Committee at BWH.
Measurement of body composition
Whole body lean, fat, and bone mineral content was measured in older adults at the baseline and 1-month visits with dual-energy X-ray absorptiometry (DXA; Hologic Inc.). DXA scan acquisition and analysis was performed according to manufacturer guidelines, with three passes over the subject to acquire the full DXA image. Scans were analyzed using Hologic QDR software version 12.3 in array mode.
Quantitative magnetic resonance imaging (qMRI) was used to determine the amount of whole body lean and fat mass in mice colonized with human fecal samples. Live mice were placed into a thin-walled plastic cylinder, which was then placed into the qMRI machine (EchoMRI, Echo Medical Systems) for measurements of lean and fat mass. Percentages of lean and fat mass were determined by dividing lean or fat mass by body weight, followed by multiplication by 100%.
Physical function measures
Measures of lower extremity function (SPPB), mobility, and strength were evaluated in older adult humans at the baseline and 1-month study visits. Mobility was assessed with the time needed to complete a 400-meter walk. Muscle strength was determined with use of a bilateral leg press (K400, Keiser Sports Health Equipment Inc) one-repetition maximum (1 RM) test. The 1 RM is defined as the maximum load that can be moved only one time throughout the full range of motion (ROM) while maintaining proper form. To attain the 1 RM value, the examiner progressively increased the resistance for each repetition until the subject could no longer move the lever arm one time through the full ROM.
Measures of physical function in mice included grip strength and treadmill endurance capacity, and were performed in a blinded manner. Forelimb gripping force by mice that were colonized with human fecal samples was measured with a computerized grip-strength meter (Columbus Instruments) based on the method of (Guo et al., 2016). Ten measurements were taken from each animal, and the average grip strength value was used for statistical analysis. To determine the endurance capacity of mice that were colonized with human fecal samples, mice were run on a treadmill (Exer-6, Columbus Instruments) on a 15% incline, as previously reported (Lustgarten et al., 2009). Sequentially, mice were run at 7 m/min for 5 min, 10 m/min for 15 minutes, 12 m/min for 10 minutes, 14 m/min for 10 minutes, 16 m/min for 10 minutes, 18 m/min for 15 minutes, 20 m/min for 10 minutes, 21 m/min for 10 minutes, 22 m/min for 15 minutes, and 24 m/min for any mouse that reached more than 100 minutes. Exhaustion was determined by a failure to engage the treadmill in the presence of a mild shock and by physical prodding.
Stool DNA extraction and high-throughput sequencing
DNA extraction and high-throughput sequencing was performed on stool samples obtained from humans and mice based on the method of (Thorpe et al., 2018). DNA was extracted from stool with the QIAquick FAST DNA Stool kit (QIAGEN). 250 mg of frozen stool was suspended in 1.4 mL of Inhibitex buffer (QIAGEN) supplemented with 50 μg of lysozyme (Sigma) and 13.5U of lysostaphin (Sigma), followed by 5 minutes of bead-beating with 500 mg of 0.1mm silica/zirconium beads (BioSpec Corp) on a Vortex Genie with a MoBio adapter. The supernatant was incubated at 70°C for 10 minutes with 100 μg Proteinase K (QIAGEN) and 400μg RNase A (Sigma), microfuged for 1 minute, and transferred to a clean tube, after which the manufacturer’s protocol was resumed. DNA was eluted in 100 μL of buffer. If the measured DNA concentration on a Nanodrop 2000 was <10 μg/mL, the extraction was repeated. A 50 μL portion of the extracted DNA was removed for 16S rDNA amplicon generation.
Amplicons of the V4 region of bacterial 16S rDNA were generated from extracted DNA using primers as described by (Caporaso et al., 2012). Triplicate PCR amplifications on 50 ng of DNA were performed using a 5’ HotStarTaq Master Mix kit (Thermo Fisher) with the following conditions: 94°C for 3 minutes, 35 cycles at 94°C for 45 seconds, 50°C for 60 seconds, 72°C for 90 seconds, and a final extension at 72°C for 10 min. Amplicon DNA concentrations were determined by Quant-iT assay (Invitrogen, Carlsbad, CA), pooled in equimolar concentrations, purified (Qiaquick PCR Purification Kit, QIAGEN), and eluted from Agencourt Ampure XP beads (Beckman-Coulter). Amplicon pools were sequenced on an Illumina MiSeq at the Tufts University Core Facility with a standard 250bp paired-end Illumina protocol. Sequencing results for the 16S amplicon libraries were processed using QIIME Version 1.8.0. After demultiplexing and joining of the reads, operational taxonomic unit (OTU) tables were created using a closed-reference OTU assignment approach. OTUs were defined by 99% identity; taxonomic assignment was performed by closed reference with GreenGenes 13_8_99.
Statistical Analysis
OTU tables were uploaded to the web application MicrobiomeAnalyst.ca (Dhariwal et al., 2017), which was used to determine between-group differences for α- and β-diversity, taxonomic abundance, and KEGG IDs (functions). A total of 5,970,814 read counts were obtained via the analysis of 58 human (n=29 each at the baseline and 1-month study visits) and 36 mouse samples, with an average of 63,519 counts per sample (range: 21,235 to 95,594). These sequences were assigned to 5,484 OTUs containing ≥ 2 counts. To evaluate between-group differences in α-diversity and binary β-diversity measures (Jaccard Index, Unweighted Unifrac), filtering based on prevalence or variance, and normalization based on rarefying, scaling, or transformation was not performed. Abundance-coverage estimator (ACE) and Chaol were used to evaluate between-group α-diversity differences in richness, whereas Simpson and Shannon indices were used as measures of evenness. Between-group differences in α-diversity were compared based on ANOVA/t-test (p < .05). To evaluate β-diversity, binary and abundance-based (Bray-Curtis, Weighted Unifrac) measures were used in conjunction with principal coordinates analysis. Between-group differences for β-diversity measures were compared based on permutational MANOVA (p < .05).
To evaluate between-group differences for abundance-based measures of β-diversity, taxonomic relative abundance, and KEGG IDs, the minimum count for the low count filter contained within MicrobiomeAnalyst.ca was set at 3, the mean abundance value was set at 20% of the samples, and the low variance filter was set at 30%, with filtering based on the coefficient of variation (CV). Based on these filtering steps, 4372, 4405, 4416, 4897 low abundance features were removed based on mean and 334, 324, 321, 177 low variance features were removed based on CV, with 778, 755, 747, and 410 features remaining for the 29 human subjects at baseline, for the 29 human subjects at the 1-month visit, for the 12 human fecal donors, and for the 36 human microbiome-containing mice. The remaining features were not rarefied or scaled. Data were transformed with use of relative log expression. Differential abundance analysis was performed with use of DESeq2. DESeq2 provides increased sensitivity on small datasets (n < 20 samples per group)(Weiss et al., 2017).
The gut bacterial functional potential in older adult humans and in their recipient mice was derived from their respective OTU tables with use of PICRUSt (Langille et al., 2013), as contained within Microbiomeanalyst.ca. Functional data were generated in the absence of filtering. Between-group differences for KEGG IDs were determined with use of DESeq2 with the same filtering steps that were used on the taxonomic data. Based on filtering, 1564, 1498, 1005, 378 low abundance KEGG IDs were removed based on mean and 888, 889, 943, 831 low variance KEGG IDs were removed based on CV, with 2071, 2074, 2197, and 1936 KEGG IDs remaining for the 29 human subjects at baseline, for the 29 human subjects at the 1-month visit, for the 12 human fecal donors, and for the 36 human microbiome-containing mice. Percent relative abundance for KEGG IDs was determined by dividing the abundance value for each KEGG ID by the sum of all of the KEGG IDs for a given subject, followed by multiplication by 100%. KEGG IDs were mapped to functional pathways with use of KEGG Mapper (https://www.genome.jp/kegg/tool/map_pathway2.html). KEGG IDs that were not mapped to functional pathways with use of KEGG Mapper were manually added to pathways after investigation of their individual KEGG website pages.
Statistical significance for between-group differences in taxonomy or functions was set at a p-value ≤ 0.05 and the false discovery rate at (FDR) ≤ 0.30. A FDR of 0.30 indicates that the result is likely to be valid 7 out of 10 times, which we suggest is reasonable in the setting of exploratory discovery (Lustgarten and Fielding, 2017).
Results
Subject characteristics
Subject characteristics at the baseline visit are shown in Table 1. 29 subjects were enrolled into the study, including 18 HF and 11 LF older adults. The two groups were not significantly different in terms of gender (% female), age, height, or BMI. HF were prescribed fewer medications, when compared with LF. HF had a a more favorable body composition, including a higher percentage of lean mass and a lower percentage of fat mass, and better physical functioning, including higher values for the SPPB, 400-meter (m) gait speed, and the leg press 1 RM, when compared with LF. Similar findings were identified when comparing HF with LF older adults at the 1-month follow up visit (Supplementary Table 1). Dietary energy intake and macronutrient composition were not different when comparing HF with LF at the baseline visit (Supplementary Table 2).
Table 1.
Subject characteristics at baseline.
| HF | LF | p-value | |
|---|---|---|---|
| n | 18 | 11 | |
| % Female | 44.4 | 45.5 | 0.48 |
| Age | 75.9 ± 3.42 | 77.0 ± 4.43 | 0.24 |
| Height | 1.64 ± 0.11 | 1.69 ± 0.08 | 0.09 |
| Weight | 73.8 ± 12.4 | 82.1 ± 10.4 | 0.04* |
| BMI | 27.6 ± 3.71 | 28.8 ± 3.40 | 0.18 |
| Number of prescribed medications | 1.94 ± 1.83 | 4.09 ± 2.87 | 0.01** |
| Body Composition | |||
| % Lean | 67.3 ± 6.39 | 62.2 ± 6.43 | 0.02* |
| % Fat | 29.7 ± 6.66 | 35.1 ± 6.60 | 0.02* |
| % BMC | 3.03 ± 0.45 | 2.97 ± 0.34 | 0.35 |
| Physical Function | |||
| SPPB | 11.5 ± 0.74 | 7.64 ± 1.29 | 3E-11*** |
| Balance | 3.69 ± 0.52 | 2.09 ± 1.04 | 4E-06*** |
| 4-m Gait speed | 4.00 ± 0.00 | 3.55 ± 0.52 | 4E-04*** |
| Chair stand | 3.83 ± 0.51 | 2.00 ± 1.34 | 8E-06*** |
| 400-m gait speed (m/s) | 1.13 ± 0.16 | 0.96 ± 0.16 | 0.01** |
| Leg press 1 RM (N) | 1877.8 ± 667.4 | 1406.8 ± 535.3 | 0.03* |
Values shown represent means ± SD. Statistical significance was determined with use of the student’s t-test.
= p ≤ 0.05,
= p ≤ 0.01,
= p ≤ 0.001.
Diversity, taxonomy, and functions when comparing HF with LF older adults
Measures of α- and β-diversity were not significantly different when comparing HF with LF older adults at the baseline or 1-month study visits (Supplementary Figures 1, 2). It is important to note that although β-diversity is a global measure that can detect relatively large differences in microbiota structure, in contrast, it does not provide information about between-group differences for bacterial relative abundance. Relative abundance at each taxonomic level was then compared between HF and LF older adults.
28 and 35 OTUs were significantly different when comparing HF with LF older adults at baseline and at the 1-month visits, respectively (data not shown). Bacterial percent relative abundance at the phyla, class, and order taxonomic levels were not significantly different when comparing HF with LF at baseline or at the 1-month visits (Supplementary Tables 3, 4). In contrast, significant differences at the family, genus, and species-levels were identified when comparing HF with LF at both study visits (family, Figure 1; Table 2).
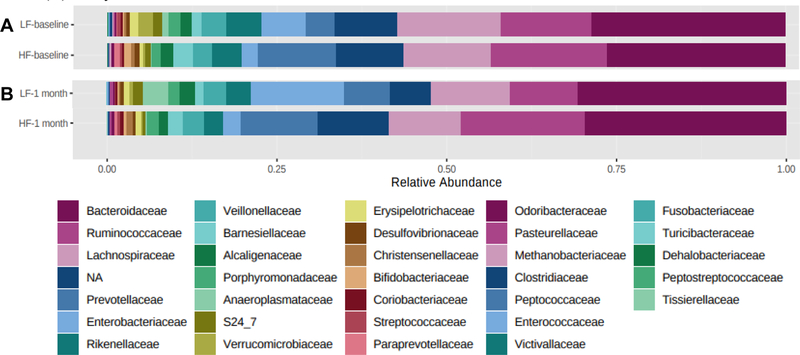
Figure 1.
Average family-level relative abundance in HF and LF older adults at baseline (A) and at the 1-month (B) study visits.
Table 2.
Percent relative abundance for family, genus, and species-level bacteria that are significantly different when comparing with HF with LF older adults at baseline and at the 1-month study visits.
| HF (% ± SD) | LF (% ± SD) | Log2FC (± SE) | p-value | FDR | |
|---|---|---|---|---|---|
| Baseline visit: Family | |||||
| S24_7 | 0.72 ± 1.81 | 1.01 ± 3.34 | −24.7 ± 3.04 | 4.7E-16 | 1.7E-14 |
| Prevotellaceae | 7.60 ± 19.8 | 3.76 ± 12.5 | −8.72 ± 1.51 | 7.5E-09 | 1.3E-07 |
| Rikenellaceae | 0.53 ± 1.03 | 2.18 ± 4.26 | 2.81 ± 1.00 | 0.005 | 0.05 |
| Enterobacteriaceae | 1.89 ± 5.61 | 5.82 ± 12.7 | 3.55 ± 1.28 | 0.006 | 0.05 |
| Peptostreptococcaceae | 0.09 ± 0.13 | 0.10 ± 0.25 | −2.25 ± 0.92 | 0.01 | 0.11 |
| Genus | |||||
| Barnesiella | 1.85 ± 4.67 | 0.00 ± 0.00 | −25.3 ± 2.74 | 2.4E-20 | 1.3E-18 |
| Paraprevotella | 0.62 ± 1.64 | 0.00 ± 0.00 | −23.6 ± 3.04 | 7.7E-15 | 2.1E-13 |
| Prevotella | 7.60 ± 19.8 | 3.76 ± 12.5 | −8.53 ± 1.51 | 1.4E-08 | 2.7E-07 |
| Clostridium | 0.05 ± 0.05 | 0.41 ± 1.03 | 2.36 ± 0.74 | 0.001 | 0.02 |
| Enterobacter | 0.00 ± 0.00 | 0.02 ± 0.06 | 4.80 ± 2.17 | 0.03 | 0.25 |
| Alistipes | 0.34 ± 0.87 | 0.04 ± 0.05 | −2.64 ± 1.22 | 0.03 | 0.25 |
| Adlercreutzia | 0.04 ± 0.09 | 0.08 ± 0.19 | 1.60 ± 0.75 | 0.03 | 0.25 |
| Species | |||||
| Barnesiella intestinihominis | 1.85 ± 4.67 | 0.00 ± 0.00 | −25.7 ± 3.04 | 2.7E-17 | 9.9E-16 |
| 1-month visit: Family | |||||
| Paraprevotellaceae | 0.37 ± 1.05 | 0.00 ± 0.00 | −23.7 ± 3.04 | 6.6E-15 | 2.2E-13 |
| Prevotellaceae | 6.93 ± 21.1 | 5.79 ± 19.2 | −8.32 ± 1.44 | 6.8E-09 | 1.1E-07 |
| S24_7 | 0.38 ± 1.03 | 1.11 ± 3.69 | −7.18 ± 2.56 | 0.005 | 0.06 |
| Genus | |||||
| Prevotella | 6.93 ± 21.1 | 5.79 ± 19.2 | −9.00 ± 1.43 | 3.2E-10 | 1.8E-08 |
| Barnesiella | 1.03 ± 2.47 | 0.00 ± 0.00 | −9.02 ± 3.04 | 0.003 | 0.08 |
| Phascolarctobacterium | 0.37 ± 0.91 | 0.04 ± 0.14 | −8.64 ± 3.04 | 0.004 | 0.08 |
| Species | |||||
| Eubacterium biforme | 0.05 ± 0.14 | 0.21 ± 0.47 | 22.3 ± 3.01 | 1.4E-13 | 5.2E-12 |
| Desulfovibrio D168 | 0.05 ± 0.19 | 0.22 ± 0.48 | 21.1 ± 3.02 | 2.6E-12 | 4.7E-11 |
| Faecalibacterium prausnitzii | 1.00 ± 1.04 | 0.24 ± 0.29 | −2.29 ± 0.76 | 0.003 | 0.02 |
| Barnesiella intestinihominis | 1.03 ± 2.47 | 0.00 ± 0.00 | −8.79 ± 2.93 | 0.003 | 0.02 |
| Bacteroides caccae | 0.01 ± 0.02 | 0.00 ± 0.01 | −3.12 ± 1.13 | 0.006 | 0.04 |
| Escherichia coli | 0.48 ± 1.03 | 4.32 ± 9.23 | 2.32 ± 1.20 | 0.05 | 0.29 |
| Clostridium citroniae | 0.03 ± 0.05 | 0.02 ± 0.03 | −1.45 ± 0.76 | 0.05 | 0.29 |
Percent relative abundance (± SD) for family, genus, and bacterial species that are significantly different when comparing HF with LF older adults at baseline and at the 1-month study visits are shown in order of significance with log 2-fold change (Log2FC) values ± standard error (SE).
At baseline, family-level Prevotellaceae, genus-level Barnesiella, Paraprevotella, Prevotella, and Alistipes, and species-level Barnesiella intestinihominis were higher, whereas family-level S24_7, Rikenellaceae, Enterobacteriaceae, and Peptostreptococcaceae, and genus-level Clostridium, Enterobacter, and Adlercreutzia were lower in HF, when compared with LF. At the 1-month visit, family-level Prevotellaceae and Paraprevotellaceae, genus-level Prevotella, Barnsiella, and Phascolarctobacterium, and species-level Faecalibacterium prausnitzii, Barnesiella intestinihominis, Bacteroides caccae, and Clostridium citroniae were higher, whereas family-level S24_7, and species-level Eubacterium biforme, Desulfovibrio D168, and Escherichia coli were lower in HF, when compared with LF.
Bacteria that were significantly different when comparing HF with LF older adults at both study visits included family-level S24_7 and Prevotellaceae, genus-level Prevotella and Barnesiella, and species-level Barnesiella intestinihominis. Non-significant associations when comparing HF with LF older adults at the family, genus, and species-levels at both study visits are shown in Supplementary Tables 5–10.
41 and 256 KEGG IDs were significantly different when comparing HF with LF older adults at the baseline and 1-month study visits, respectively (data not shown). 26 functions were significantly different when comparing HF with LF older adults at both visits (Supplementary Table 11). Expression of glutathione peroxidase (K00432; GPx) was higher, whereas the remaining 25 functions were lower in HF, when compared with LF. Moreover, GPx was the most highly expressed function (2 to 20-fold increased) when compared with all other significant KEGG IDs.
Functional pathways that are represented by the 26 significant, overlapping KEGG IDs are shown in Supplementary Table 12. 17 functions mapped to enzymes (oxidoreductases, transferases, lyases, hydrolases), 8 mapped to metabolism (carbohydrate, amino acid, fatty acid, lipopolysaccharide biosynthesis), 7 mapped to energy metabolism (nitrogen, methane, sulfur, carbon fixation in prokaryotes), 5 mapped to genetic information processing (transcription factors, DNA repair and recombination proteins, ribosome biogenesis, transfer RNA biogenesis), 4 mapped to environmental information processing (signal transduction with two-component systems), 4 functions were poorly characterized, 3 were classified as structural proteins, and 2 mapped to cellular processes (biofilm formation).
Fecal transfer from older adult humans into germ-free mice
To evaluate a causative role for the gut microbiome on the percentage of whole body lean mass or physical functioning, fecal samples from 6 HF and 6 LF (3 males, 3 females in each group) older adult humans were obtained from the baseline visit and transplanted into young (8–9 weeks), sex-matched C57/BL6J germ-free mice. Four weeks after fecal transfer, measurements of body composition, physical function, and gut microbiome composition were performed. Subject characteristics for the human fecal donors are shown in Supplementary Table 13. HF and LF human fecal donors were not different in terms of age or height. Although differences for BMI were close to statistical significance (p = 0.07), it is important to note that body composition for the human fecal donors was similar to the larger human group, including a higher percent lean mass and a decreased percent fat mass in HF fecal donors, when compared with their LF counterparts. In addition, HF fecal donors had better physical function (higher values for the SPPB, 400-m gait speed, and the leg press 1 RM) when compared with LF. Dietary energy intake and macronutrient composition were not different when comparing HF with LF fecal donors (Supplementary Table 14).
In terms of the reproducibility of bacterial transplantation from older adult human fecal donors into germ-free mice, 99.1 and 99.9% of phyla, 98.8 and 98.3% of classes, 98.8 and 97.9% of orders, 83.8 and 90.1% of (assigned) families, and 53.4 and 60.7% of (assigned) genera that were present in HF and LF human fecal donors, respectively, were represented in the microbiota of their recipient mice (Supplementary Table 15). From the phyla to the family taxonomic levels, these data are similar to the transfer reproducibility reported in the fecal transfer studies of (Turnbaugh et al., 2009b; Ridaura et al., 2013).
Diversity, taxonomy, and functions when comparing HF with LF older adult human fecal donors and their recipient mice
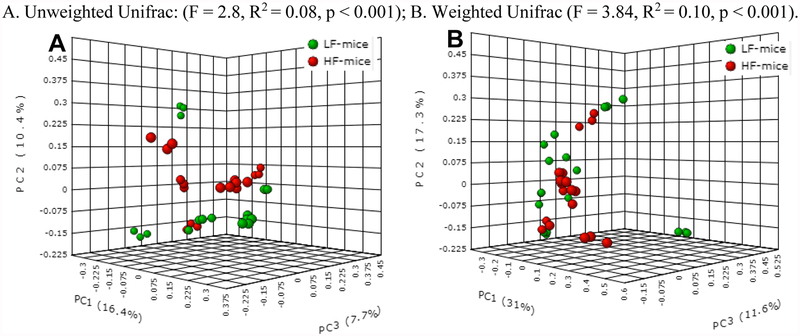
α-diversity was not significantly different when comparing HF with LF human fecal donors, and HF- with LF-colonized mice (Supplementary Figure 3). β-diversity was not significantly different when comparing the 12 human fecal donors (Supplementary Figure 4). In contrast, each of the 4 measures of β-diversity were significantly different when comparing HF- with LF-colonized mice (unweighted, weighted Unifrac, Figure 2; Jaccard, Bray-Curtis, Supplementary Figure 5). As a possible explanation for the absence of differences in β-diversity for the human fecal donors but significant differences in β-diversity for their respectively colonized mice, it is possible that the sample size used for the human fecal donors (n=12) was underpowered to detect a significant effect, when compared with the larger sample size used for human microbiome-containing mice (n=36).
Figure 2.
β-diversity measures when comparing HF- with LF-colonized mice.
A. Unweighted Unifrac: (F = 2.8, R2 = 0.08, p < 0.001); B. Weighted Unifrac (F = 3.84, R2 = 0.10, p < 0.001).
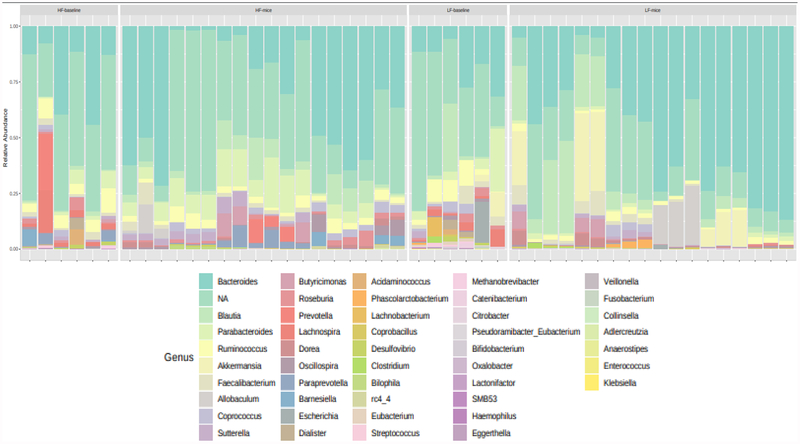
200 and 123 OTUs were significantly different when comparing the human fecal donors and HF- with LF-colonized mice, respectively (data not shown). Bacterial relative abundance for the human fecal donors and their recipient mice are visually represented at the genus-level in Figure 3.
Figure 3.
Genus-level relative abundance in HF and LF human fecal donors and in their respectively colonized mice.
Investigating further, bacteria were identified that were significantly different at each taxonomic level in both the human fecal donors and in their respectively colonized mice (Table 3). Of note, family-level Prevotellaceae, genus-level Prevotella and Barnesiella, and species-level Barnesiella intestinihominis were higher in HF human fecal donors and in HF-colonized mice, and were also higher in the larger HF group at baseline and at the 1-month study visits, when compared with their LF counterparts.
Table 3.
Percent relative abundance for bacteria that are significantly different when comparing HF with LF human fecal donors, and HF- with LF-colonized mice.
| Human fecal donors | Human-microbiome containing mice | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HF (% ± SD) | LF (% ± SD) | Log2FC (± SE) | p-value | FDR | HF-mice (% ± SD) | LF-mice (% ± SD) | Log2FC (± SE) | p-value | FDR | |
| Phyla | ||||||||||
| Verrucomicrobia | 0.06 ± 0.04 | 0.83 ± 3.27 | 6.47 ± 1.64 | 8.0E-05 | 0.0008 | 0.01 ± 0.03 | 6.12 ± 10.1 | 10.3 ± 1.83 | 1.9E-0.08 | 1.1E-07 |
| Cyanobacteria | 0.37 ± 0.55 | 0.00 ± 0.00 | −8.68 ± 2.94 | 0.003 | 0.002 | 2.22 ± 4.04 | 0.00 ± 0.00 | −9.10 ± 1.72 | 1.3E-07 | 4.0E-07 |
| Class | ||||||||||
| 4C0d_2 | 0.37 ± 0.55 | 0.00 ± 0.00 | −9.67 ± 2.53 | 0.0001 | 0.001 | 2.22 ± 4.04 | 0.00 ± 0.00 | −9.84 ± 1.28 | 1.9E-14 | 2.1E-13 |
| Gammaproteobacteria | 0.13 ± 0.16 | 9.54 ± 14.1 | 5.46 ± 1.44 | 0.0002 | 0.001 | 0.02 ± 0.03 | 0.19 ± 0.47 | 5.10 ± 1.46 | 0.0005 | 0.002 |
| Verrucomicrobiae | 0.06 ± 0.04 | 1.83 ± 3.27 | 5.70 ± 1.65 | 0.0005 | 0.003 | 0.01 ± 0.03 | 6.12 ± 0.00 | 10.4 ± 1.6 | 2.7E-10 | 1.5E-09 |
| Order | ||||||||||
| YS2 | 0.37 ± 0.55 | 0.00 ± 0.00 | −8.77 ± 2.55 | 0.0006 | 0.004 | 2.22 ± 4.04 | 0.00 ± 0.00 | −9.84 ± 1.29 | 2.1E-14 | 2.6E-13 |
| Verrucomicrobiales | 0.06 ± 0.04 | 1.83 ± 3.27 | 6.45 ± 1.75 | 0.002 | 0.002 | 0.01 ± 0.03 | 6.12 ± 10.1 | 10.7 ± 1.84 | 5.6E-9 | 3.3E-08 |
| Enterobacteriales | 0.06 ± 0.05 | 9.33 ± 14.0 | 6.53 ± 1.67 | 9.2E-05 | 0.002 | 0.02 ± 0.03 | 0.19 ± 0.47 | 5.09 ± 1.46 | 0.0005 | 0.002 |
| Family | ||||||||||
| Verrucomicrobiaceae | 0.06 ± 0.04 | 1.83 ± 3.27 | 7.63 ± 1.83 | 2.9E-05 | 0.0002 | 0.01 ± 0.03 | 6.12 ± 10.1 | 9.48 ± 1.93 | 8.9E-07 | 6.8E-14 |
| Prevotellaceae | 4.72 ± 11.6 | 0.00 ± 0.00 | −28.3 ± 3.00 | 4.6E-21 | 8.1E-20 | 1.11 ± 2.75 | 0.00 ± 0.00 | −5.58 ± 1.94 | 0.004 | 0.02 |
| Enterobacteriaceae | 0.06 ± 0.05 | 9.33 ± 14.0 | 7.18 ± 1.72 | 3.0E-05 | 0.0002 | 0.02 ± 0.03 | 0.19 ± 0.47 | 3.77 ± 1.53 | 0.01 | 0.04 |
| S24_7 | 2.05 ± 0.00 | 0.00 ± 0.00 | 27.1 ± 2.85 | 1.9E-21 | 6.6E-20 | 11.4 ± 19.3 | 1.23 ± 2.93 | −4.27 ± 1.88 | 0.02 | 0.06 |
| Erysipelotrichaceae | 0.15 ± 0.26 | 1.50 ± 2.04 | 2.83 ± 1.30 | 0.03 | 0.15 | 1.25 ± 2.86 | 3.69 ± 7.58 | 1.49 ± 0.68 | 0.03 | 0.06 |
| Genus | ||||||||||
| Paraprevotella | 1.78 ± 2.76 | 0.00 ± 0.00 | −27.6 ± 3.00 | 3.9E-20 | 1.1E-18 | 1.40 ± 2.70 | 0.00 ± 0.00 | −27.3 ± 2.34 | 2.1E-31 | 6.9E-30 |
| Akkermansia | 0.06 ± 0.04 | 1.83 ± 3.27 | 5.67 ± 1.80 | 0.002 | 0.01 | 0.01 ± 0.03 | 6.12 ± 10.1 | 9.91 ± 1.82 | 5.1E-08 | 5.6E-07 |
| Barnesiella | 1.62 ± 3.46 | 0.00 ± 0.00 | −26.3 ± 2.95 | 4.9E-19 | 8.9E-18 | 0.79 ± 1.63 | 0.00 ± 0.00 | −8.37 ± 1.65 | 3.6E-07 | 3.0E-06 |
| Eubacterium | 0.00 ± 0.00 | 0.72 ± 0.84 | 6.98 ± 2.01 | 0.0005 | 0.004 | 0.01 ± 0.02 | 0.07 ± 0.07 | 4.44 ± 1.08 | 4.1E-05 | 0.0002 |
| Prevotella | 4.72 ± 11.6 | 0.00 ± 0.00 | −28.7 ± 3.00 | 1.3E-21 | 7.0E-20 | 1.11 ± 2.75 | 0.00 ± 0.00 | −6.58 ± 2.05 | 0.001 | 0.005 |
| Coprobacillus | 1.18 ± 1.26 | 2.16 ± 2.16 | 5.90 ± 3.01 | 0.05 | 0.15 | 0.12 ± 0.12 | 0.23 ± 0.22 | 1.46 ± 0.58 | 0.01 | 0.04 |
| Species | ||||||||||
| Barnesiella intestinihominis | 1.62 ± 3.46 | 0.00 ± 0.00 | −28.7 ± 2.90 | 3.8E-23 | 1.2E-21 | 0.79 ± 1.63 | 0.00 ± 0.00 | −25.6 ± 1.71 | 1.9E-50 | 4.1E-49 |
| Akkermansia muciniphila | 0.06 ± 0.04 | 1.83 ± 3.27 | 5.02 ± 1.87 | 0.007 | 0.03 | 0.01 ± 0.03 | 6.12 ± 10.1 | 8.40 ± 1.88 | 7.8E-06 | 4.3E-05 |
Percent relative abundance (± SD) for bacteria that are significantly different when comparing both the human fecal donors and their respectively colonized mice are shown with log 2-fold change (Log2FC) values ± standard error (SE).
Bacteria that were significantly different when comparing the human fecal donors but that were not different when comparing their respectively colonized mice are shown in Supplementary Table 16. Conversely, bacteria that were significantly different when comparing HF- with LF-colonized mice but that were not different when comparing HF with LF human fecal donors are shown in Supplementary Table 17. Non-significant associations at each taxonomic level for the human fecal donors or their recipient mice are shown in Supplementary Tables 18–23.
158 and 820 KEGG IDs were significantly different when comparing HF with LF human fecal donors, and HF- with LF-colonized mice, respectively (data not shown). 16 KEGG IDs were significantly different when comparing both the human fecal donors and and their recipient mice (Supplementary Table 24). Eight KEGG IDs were higher, whereas seven KEGG IDs were lower in HF human donors and in their recipient mice, when compared with LF. One KEGG ID (K01133) was higher in HF donors but was lower in its recipient mice, and was removed from further analysis based on the hypothesis that functions in the human fecal donors and in their recipient mice should be found in the same direction. K03315, a gene that encodes a Na+:H+ antiporter, was the most highly expressed function (1.3 to 41-fold higher in LF) when compared with all other significant KEGG IDs.
Functional pathways that are represented by the 15 significant, overlapping KEGG IDs are shown in Supplementary Table 25. Of the functions that mapped to more than 1 pathway and that were higher in HF humans and in their recipient mice, 5 functions mapped to enzymes (hydrolases, isomerases, transferases, ligases), 3 mapped to metabolism (glycosaminoglycan degradation, phosphonate and phosphinate metabolism, fatty acid degradation/glycerophospholipid metabolism), and 2 mapped to protein families: genetic information processing (DNA repair and recombination proteins, chromosome partitioning). Of the functional pathways that were lower in HF humans and in their recipient mice, 4 mapped to protein families: signaling and cellular processes (drug transporters, electrochemical potential-driven transporters, unclassified transporter), 3 functions mapped to enzymes (oxidoreductases, hydrolases), and 2 mapped to metabolism (fatty acid degradation/glycerophospholipid metabolism).
Effect of fecal transfer from older adult humans on body composition and physical function in recipient mice:
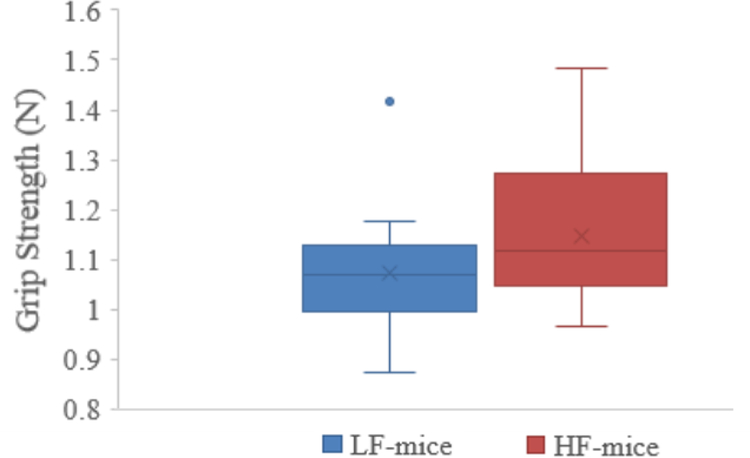
Grip strength was significantly increased by 6.4% in mice colonized with fecal samples from HF, when compared with LF (HF vs LF mice, β ± SD: 1.15 ± 0.14, 1.07 ± 0.12 N, p = 0.05; Figure 4).
Figure 4.
Grip strength when comparing HF- with LF-colonized mice.
Body weight, percent lean or fat mass, and treadmill endurance capacity were not significantly different when comparing HF- with LF-colonized mice (Supplementary Table 26).
Discussion
The goals of the present study were to compare gut microbiome composition and functions in older adults that were different in terms of the percentage of whole body lean mass and physical functioning, and to examine a causative role for the gut microbiome on these variables by transferring fecal samples from older adults into germ-free mice. Primary findings include higher amounts of family-level Prevotellaceae, genus-level Prevotella and Barnesiella, and species-level Barnesiella intestinihominis in HF older adults at the baseline and 1-month study visits, in HF human fecal donors, and in HF-colonized mice, when compared with their LF counterparts. Moreover, grip strength was increased in HF-colonized mice, a finding that suggests a causative role for these bacterial taxa on the maintenance of muscle strength in older adults.
Prevotella (and their corresponding family-level taxa, Prevotellaceae) are elevated in young professional athletes (Clarke et al., 2014), and Prevotella and Barnesiella are higher in less frail, when compared with more frail older adults (van Tongeren et al., 2005; Claesson et al., 2012; Verdi et al., 2018), evidence that supports the hypothesis that these bacteria may be involved in mechanisms related to the maintenance of physical function. Although Prevotella have been reported as a marker of a diet that is rich in polysaccharides and fiber (Gorvitovskaia et al., 2016), in contrast, levels of the polysaccharides starch and pectin (data not shown), and dietary fiber were not significantly different when comparing HF with LF older adults, evidence that argues against the role of diet on Prevotella in the present study. In terms of how these bacteria may impact physical function, Barnesiella and Prevotellaceae contain genes that produce the short-chain fatty acids (SCFA) acetate, propionate, and butyrate (Morotomi et al., 2008; Chen et al., 2017; Esquivel-Elizondo et al., 2017; Louis and Flint, 2017), a potentially important finding because feeding a mixture of these SCFAs to germ-free mice was recently shown to improve muscle strength (Lahiri et al., 2019).
Previously published literature and our findings for S24_7, Akkermansia, and Erysipelotrichaceae would be expected to suggest a role for these bacteria on the maintenance of muscle strength, body composition, or endurance capacity in older adults. First, S24_7 were higher in HF human fecal donors and in HF-colonized mice, when compared with LF, a finding that suggests a positive role for S24_7 on the maintenance of muscle strength. In support of this, S24_7 are elevated in athletes (Clarke et al., 2014). However, S24_7 were lower in HF older adults in the larger human study (n=29) at both study visits, evidence that argues against the role of S24_7 on the maintenance of muscle strength. Second, Akkermansia are increased in young elite athletes that have a favorable body composition (higher percent lean mass, lower percent fat mass) (Clarke et al., 2014). In contrast with these findings, Akkermansia were higher in LF human fecal donors and in their recipient mice, when compared with HF, evidence that argues against the role of Akkermansia on the maintenance of a favorable body composition and physical function in older adults. Third, Erysipelotrichaceae are positively associated with cardiorespiratory fitness (Estaki et al., 2016), a finding that would suggest higher levels of Erysipelotrichaceae in HF older adults. In contrast, Erysipelotrichaceae were higher in LF human fecal donors and in LF mice, when compared with HF, in conjunction with no difference in mouse treadmill endurance capacity.
In terms of bacterial functions that were significantly different when comparing with HF with LF older adults at both study visits, glutathione peroxidase was higher in HF, and was the most highly expressed function among all significantly different KEGG IDs. Serum levels of glutathione peroxidase are increased in microbiome-containing, when compared with germ-free mice, in conjunction with improved physical function (Hsu et al., 2015). Moreover, when considering that circulating and intestinal mucosal GPx activity decreases during aging in rats and humans (Rao et al., 1990; Espinoza et al., 2008), findings from the present study suggest that the gut microbiome may be an important source for the production of glutathione peroxidase, and potentially, for the maintenance of a favorable body composition and higher physical function. In contrast, three KEGG IDs for nitrate reductase and one for lipopolysaccharide biosynthesis were higher in LF, when compared with HF, evidence that may suggest a role for intestinal permeability and microbial burden on body composition and physical functioning. Nitrate reductase is involved in the conversion of nitrate to nitrite, a precursor metabolite for the formation of ammonia. Ammonia readily combines with water to produce ammonium ions (Bourke et al., 1966), which are involved in mechanisms that increase intestinal epithelial cell permeability (Lytton et al., 2005). Moreover, muscle mass and strength are decreased in the presence of LPS (Morales et al., 2015). Although systemic markers of intestinal permeability or circulating microbial burden (i.e. LPS) were not measured in the present study, future studies aimed at quantifying these variables in HF, when compared with LF older adults, are of interest.
Contrary to expectation, overlap was not identified for bacterial functions that were significantly different in the larger human group at both study visits with human fecal donors and their recipient mice. Of the bacterial functions that were significantly different when comparing the human fecal donors and their recipient mice, the most highly expressed function was K03315, a gene that encodes a Na+:H+ antiporter. In bacteria, Na+:H+ antiporters aid in the maintenance of an intracellular acidic pH by extruding sodium ions while taking up protons from the extracellular space (Pragai et al., 2001). Whether this bacterial function is elevated enough to impact colonic acid-base balance, which is involved in the regulation of microbiome composition (Duncan et al., 2009) is unknown, but it raises the possibility that colonic alkalization may be associated with worse physical function in older adults.
Study limitations include a small sample size, an inefficiency of fecal bacterial transplantation for Enterobacteriaceae, and the absence of additional time points for the fecal transfer study. First, Enterobacteriaceae are elevated in frail, when compared with less frail older adults (van Tongeren et al., 2005; Claesson et al., 2012). In support of this finding, Enterobacteriaceae were significantly higher in LF older adults at the baseline visit, but we may have been underpowered to detect a significant difference for Enterobacteriaceae (despite being 4.9-fold higher in LF) at the 1-month visit. The means ± SD reported for Enterobacteriaceae in Supplementary Table 8 generate a Cohen’s d equal to 0.51, a value that yields a significant effect at 80% power (p < 0.05) with a total study sample size of 96 subjects. Accordingly, with a larger study sample size, we may have identified significant differences for Enterobacteriaceae at both study visits. Second, although Enterobacteriaceae were 156-fold elevated when comparing HF with LF human fecal donors (0.06% vs 9.33% relative abundance), the magnitude of this difference, albeit statistically significant was far less when comparing HF- with LF-colonized mice (0.02% vs 0.19%). Interestingly, in human microbiome-colonized mice, Enterobacteriaceae levels may have peaked earlier than at the 1-month time point, then decreased. In support of this, when young rats are fed a high fat-high sucrose diet, Enterobacteriaceae levels peak 1 week after diet onset, but return to the levels found in control-fed animals after 4 weeks on the diet (Collins et al., 2016). In further support of the incorporation of a 1-week time point, fecal sample transfer from aged conventional mice into young germ-free mice revealed that gut microbiota composition in the recipient mice was most similar to the aged donor 1-week after fecal transfer, but at later time points the gut microbiota composition adapted to the host (Fransen et al., 2017). When considering these findings, greater differences in grip strength and potentially, differences in the percentage of whole body lean mass or treadmill endurance capacity may have been identified with the inclusion of a 1-week time point.
Additional limitations include potential roles for medication use, recreational physical activity, or constipation on gut microbiome composition, and use of 3-day diet records as a proxy for long-term dietary intake. First, the number of prescribed medications was higher in LF, when compared with HF older adults. Statins were the most frequently prescribed medication, and a higher percentage of LF subjects (6/11) were prescribed statins, when compared with HF (6/18). Statins have been reported to negatively affect physical function in older adults (Scott et al., 2009), and are associated with alterations in gut microbiome composition (Zhernakova et al., 2016; Bedarf et al., 2017). However, statin usage has not been shown to affect levels of bacteria that were significantly different when comparing HF with LF older adults or HF- with LF-colonized mice, including Prevotellaceae, Prevotella, Barnesiella, or Barnesiella intestinihominis (Zhernakova et al., 2016; Bedarf et al., 2017). Second, although all subjects were required to be sedentary as a study inclusion criterion, it is possible that recreational physical activity was increased in HF older adults, thereby leading to higher levels of Prevotella. In support of this, Prevotella are associated with weekly exercise duration in cyclists (Petersen et al., 2017). In addition, Prevotella levels are reduced in constipated older adults (Huang et al., 2018). However, as study limitations, levels of recreational physical activity and constipation status were not quantified. Third, alterations in gut bacterial composition occur within 24-hours in response to a change in diet (Wu et al., 2011). Although a dietary change was not implemented in the present study, the 3-day diet records that were collected were expected to represent subjects’ habitual diet. However, whether 3-day diet records are indicative of subjects’ long-term diet is unknown, an important finding because long-term dietary habits impact the longitudinal stability of the gut microbiome (Wu et al., 2011).
In summary, findings from the present study suggest that family-level Prevotellaceae, genus-level Barnesiella and Prevotella, and species-level Barnesiella intestinihominis may be involved in mechanisms related to the maintenance of muscle strength in older adults. Furthermore, despite differences in the percentage of whole body lean mass and physical function in the human fecal donors, the percentage of whole body lean mass and treadmill endurance capacity were not different when comparing HF with LF-colonized mice, evidence that argues against the role of the gut microbiome on these variables in older adults. However, when considering that gut microbiome composition in mice that are colonized with fecal samples from aged mice has been reported to be most similar to the donor 1-week after transplantation, studies aimed at reproducing our findings with earlier time points are of interest.
Supplementary Material
Funding
This work was supported by National Institute on Aging (K01AG050700) and Boston Claude D. Pepper Older Americans Independence Center (5P30AG031679) grants to MSL, and a USDA grant (58-1950-4-003) to RAF.
Footnotes
Conflict of Interest
None declared.
References
- Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al. (2004). The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A 101(44), 15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedarf JR, Hildebrand F, Coelho LP, Sunagawa S, Bahram M, Goeser F, et al. (2017). Functional implications of microbial and viral gut metagenome changes in early stage L-DOPA-naive Parkinson’s disease patients. Genome Med 9(1), 39. doi: 10.1186/s13073-017-0428-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindels LB, Beck R, Schakman O, Martin JC, De Backer F, Sohet FM, et al. (2012). Restoring specific lactobacilli levels decreases inflammation and muscle atrophy markers in an acute leukemia mouse model. PLoS One 7(6), e37971. doi: 10.1371/journal.pone.0037971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourke E, Milne MD, and Stokes GS (1966). Caecal pH and ammonia in experimental uraemia. Gut 7(5), 558–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani PD, Possemiers S, Van de Wiele T, Guiot Y, Everard A, Rottier O, et al. (2009). Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut 58(8), 1091–1103. doi: 10.1136/gut.2008.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6(8), 1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Long W, Zhang C, Liu S, Zhao L, and Hamaker BR (2017). Fiber-utilizing capacity varies in Prevotella-versus Bacteroides-dominated gut microbiota. Sci Rep 7(1), 2594. doi: 10.1038/s41598-017-02995-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YM, Wei L, Chiu YS, Hsu YJ, Tsai TY, Wang MF, et al. (2016). Lactobacillus plantarum TWK10 Supplementation Improves Exercise Performance and Increases Muscle Mass in Mice. Nutrients 8(4), 205. doi: 10.3390/nu8040205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, et al. (2012). Gut microbiota composition correlates with diet and health in the elderly. Nature 488(7410), 178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- Clarke SF, Murphy EF, O’Sullivan O, Lucey AJ, Humphreys M, Hogan A, et al. (2014). Exercise and associated dietary extremes impact on gut microbial diversity. Gut 63(12), 1913–1920. doi: 10.1136/gutjnl-2013-306541. [DOI] [PubMed] [Google Scholar]
- Collins KH, Paul HA, Hart DA, Reimer RA, Smith IC, Rios JL, et al. (2016). A High-Fat High-Sucrose Diet Rapidly Alters Muscle Integrity, Inflammation and Gut Microbiota in Male Rats. Sci Rep 6, 37278. doi: 10.1038/srep37278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhariwal A, Chong J, Habib S, King IL, Agellon LB, and Xia J (2017). MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res 45(W1), W180–W188. doi: 10.1093/nar/gkx295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Louis P, Thomson JM, and Flint HJ (2009). The role of pH in determining the species composition of the human colonic microbiota. Environ Microbiol 11(8), 2112–2122. doi: 10.1111/j.1462-2920.2009.01931.x. [DOI] [PubMed] [Google Scholar]
- Espinoza SE, Guo H, Fedarko N, DeZern A, Fried LP, Xue QL, et al. (2008). Glutathione peroxidase enzyme activity in aging. J Gerontol A Biol Sci Med Sci 63(5), 505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquivel-Elizondo S, Ilhan ZE, Garcia-Pena EI, and Krajmalnik-Brown R (2017). Insights into Butyrate Production in a Controlled Fermentation System via Gene Predictions. mSystems 2(4). doi: 10.1128/mSystems.00051-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estaki M, Pither J, Baumeister P, Little JP, Gill SK, Ghosh S, et al. (2016). Cardiorespiratory fitness as a predictor of intestinal microbial diversity and distinct metagenomic functions. Microbiome 4(1), 42. doi: 10.1186/s40168-016-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Lazarevic V, Derrien M, Girard M, Muccioli GG, Neyrinck AM, et al. (2011). Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes 60(11), 2775–2786. doi: 10.2337/db11-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen F, van Beek AA, Borghuis T, Aidy SE, Hugenholtz F, van der Gaast-de Jongh C, et al. (2017). Aged Gut Microbiota Contributes to Systemical Inflammaging after Transfer to Germ-Free Mice. Front Immunol 8, 1385. doi: 10.3389/fimmu.2017.01385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorvitovskaia A, Holmes SP, and Huse SM (2016). Interpreting Prevotella and Bacteroides as biomarkers of diet and lifestyle. Microbiome 4, 15. doi: 10.1186/s40168-016-0160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosicki GJ, Fielding RA, and Lustgarten MS (2018). Gut Microbiota Contribute to Age-Related Changes in Skeletal Muscle Size, Composition, and Function: Biological Basis for a Gut-Muscle Axis. Calcif Tissue Int 102(4), 433–442. doi: 10.1007/s00223-017-0345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Miller AD, Pencina K, Wong S, Lee A, Yee M, et al. (2016). Joint dysfunction and functional decline in middle age myostatin null mice. Bone 83, 141–148. doi: 10.1016/j.bone.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guralnik JM, Simonsick EM, Ferrucci L, Glynn RJ, Berkman LF, Blazer DG, et al. (1994). A short physical performance battery assessing lower extremity function: association with self-reported disability and prediction of mortality and nursing home admission. J Gerontol 49(2), M85–94. [DOI] [PubMed] [Google Scholar]
- Hsu YJ, Chiu CC, Li YP, Huang WC, Huang YT, Huang CC, et al. (2015). Effect of intestinal microbiota on exercise performance in mice. J Strength Cond Res 29(2), 552–558. doi: 10.1519/JSC.0000000000000644. [DOI] [PubMed] [Google Scholar]
- Huang LS, Kong C, Gao RY, Yan X, Yu HJ, Wen B, et al. (2018). Analysis of fecal microbiota in patients with functional constipation undergoing treatment with synbiotics. Eur J Clin Microbiol Infect Dis 37(3), 555–563. doi: 10.1007/s10096-017-3149-7. [DOI] [PubMed] [Google Scholar]
- Jackson MA, Jeffery IB, Beaumont M, Bell JT, Clark AG, Ley RE, et al. (2016). Signatures of early frailty in the gut microbiota. Genome Med 8(1), 8. doi: 10.1186/s13073-016-0262-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirn DR, Koochek A, Reid KF, von Berens A, Travison TG, Folta S, et al. (2015). The Vitality, Independence, and Vigor in the Elderly 2 Study (VIVE2): Design and methods. Contemp Clin Trials 43, 164–171. doi: 10.1016/j.cct.2015.06.001. [DOI] [PubMed] [Google Scholar]
- Lahiri S, Kim H, Garcia-Perez I, Reza MM, Martin KA, Kundu P, et al. (2019). The gut microbiota influences skeletal muscle mass and function in mice. Sci Transl Med 11(502). doi: 10.1126/scitranslmed.aan5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31(9), 814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P, and Flint HJ (2017). Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 19(1), 29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- Lustgarten MS, and Fielding RA (2017). Metabolites related to renal function, immune activation, and carbamylation are associated with muscle composition in older adults. Exp Gerontol 100, 1–10. doi: 10.1016/j.exger.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustgarten MS, Jang YC, Liu Y, Muller FL, Qi W, Steinhelper M, et al. (2009). Conditional knockout of Mn-SOD targeted to type IIB skeletal muscle fibers increases oxidative stress and is sufficient to alter aerobic exercise capacity. Am J Physiol Cell Physiol 297(6), C1520–1532. doi: 10.1152/ajpcell.00372.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytton SD, Fischer W, Nagel W, Haas R, and Beck FX (2005). Production of ammonium by Helicobacter pylori mediates occludin processing and disruption of tight junctions in Caco-2 cells. Microbiology 151(Pt 10), 3267–3276. doi: 10.1099/mic.0.28049-0. [DOI] [PubMed] [Google Scholar]
- Morales MG, Olguin H, Di Capua G, Brandan E, Simon F, and Cabello-Verrugio C (2015). Endotoxin-induced skeletal muscle wasting is prevented by angiotensin-(1–7) through a p38 MAPK-dependent mechanism. Clin Sci (Lond) 129(6), 461–476. doi: 10.1042/CS20140840. [DOI] [PubMed] [Google Scholar]
- Morotomi M, Nagai F, Sakon H, and Tanaka R (2008). Dialister succinatiphilus sp. nov. and Barnesiella intestinihominis sp. nov., isolated from human faeces. Int J Syst Evol Microbiol 58(Pt 12), 2716–2720. doi: 10.1099/ijs.0.2008/000810-0. [DOI] [PubMed] [Google Scholar]
- Nobel YR, Cox LM, Kirigin FF, Bokulich NA, Yamanishi S, Teitler I, et al. (2015). Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nature Communications. doi: 10.1038/ncomms8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen LM, Bautista EJ, Nguyen H, Hanson BM, Chen L, Lek SH, et al. (2017). Community characteristics of the gut microbiomes of competitive cyclists. Microbiome 5(1), 98. doi: 10.1186/s40168-017-0320-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pragai Z, Eschevins C, Bron S, and Harwood CR (2001). Bacillus subtilis NhaC, an Na+/H+ antiporter, influences expression of the phoPR operon and production of alkaline phosphatases. J Bacteriol 183(8), 2505–2515. doi: 10.1128/JB.183.8.2505-2515.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao G, Xia E, and Richardson A (1990). Effect of age on the expression of antioxidant enzymes in male Fischer F344 rats. Mech Ageing Dev 53(1), 49–60. [DOI] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. (2013). Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341(6150), 1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schakel SF (2001). Maintaining a nutrient database in a changing marketplace: Keeping pace with changing food products - A research perspective. J Food Comp Anal 14, 315–322. doi: 10.1006/jfca.2001.0992. [DOI] [Google Scholar]
- Scott D, Blizzard L, Fell J, and Jones G (2009). Statin therapy, muscle function and falls risk in community-dwelling older adults. QJM 102(9), 625–633. doi: 10.1093/qjmed/hcp093. [DOI] [PubMed] [Google Scholar]
- Siddharth J, Chakrabarti A, Pannerec A, Karaz S, Morin-Rivron D, Masoodi M, et al. (2017). Aging and sarcopenia associate with specific interactions between gut microbes, serum biomarkers and host physiology in rats. Aging (Albany NY) 9(7), 1698–1720. doi: 10.18632/aging.101262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe CM, Kane AV, Chang J, Tai A, Vickers RJ, and Snydman DR (2018). Enhanced preservation of the human intestinal microbiota by ridinilazole, a novel Clostridium difficile-targeting antibacterial, compared to vancomycin. PLoS One 13(8), e0199810. doi: 10.1371/journal.pone.0199810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. (2009a). A core gut microbiome in obese and lean twins. Nature 457(7228), 480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, and Gordon JI (2009b). The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 1(6), 6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ussar S, Griffin NW, Bezy O, Fujisaka S, Vienberg S, Softic S, et al. (2015). Interactions between Gut Microbiota, Host Genetics and Diet Modulate the Predisposition to Obesity and Metabolic Syndrome. Cell Metab 22(3), 516–530. doi: 10.1016/j.cmet.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tongeren SP, Slaets JP, Harmsen HJ, and Welling GW (2005). Fecal microbiota composition and frailty. Appl Environ Microbiol 71(10), 6438–6442. doi: 10.1128/AEM.71.10.6438-6442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdi S, Jackson MA, Beaumont M, Bowyer RCE, Bell JT, Spector TD, et al. (2018). An Investigation Into Physical Frailty as a Link Between the Gut Microbiome and Cognitive Health. Front Aging Neurosci 10, 398. doi: 10.3389/fnagi.2018.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A, et al. (2017). Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 5(1), 27. doi: 10.1186/s40168-017-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science 334(6052), 105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Diao H, Xiao Y, Li W, Yu B, He J, et al. (2016). Gut microbiota can transfer fiber characteristics and lipid metabolic profiles of skeletal muscle from pigs to germ-free mice. Sci Rep 6, 31786. doi: 10.1038/srep31786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, et al. (2016). Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352(6285), 565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.