Abstract
Purpose of Review
The purpose of this review is to provide an update on the recent advances in the research and clinical care of patients with the major phenotypes of inherited cardiomyopathies - hypertrophic, dilated, and arrhythmogenic. Developments in genetics, risk stratification, therapies, and disease modeling will be discussed.
Recent Findings
Diagnostic, prognostic, and therapeutic tools which incorporate genetic and genomic data are being steadily incorporated into the routine clinical care of patients with genetic cardiomyopathies. Human pluripotent stem cells are a breakthrough model system for the study of genetic variation associated with inherited cardiovascular disease.
Summary
Next generation sequencing technology and molecular-based diagnostics and therapeutics have emerged as valuable tools to improve the recognition and care of patients with hypertrophic, dilated, and arrhythmogenic cardiomyopathies. Improved adjudication of variant pathogenicity and management of genotype-positive/phenotype-negative individuals are imminent challenges in this realm of precision medicine.
Keywords: hypertrophic cardiomyopathy, dilated cardiomyopathy, arrhythmogenic right ventricular dysplasia, pluripotent stem cells, genetic testing, genomics
INTRODUCTION
In 1961, John F. Goodwin designed a classification system for cardiomyopathy based on his personal observations of cardiac structural and functional changes in 66 patients. His tripartite taxonomy identified (1) cardiac dilatation, (2) constriction, and (3) inflow or outflow obstruction. This early insight was validated through modern imaging techniques and corresponds with three phenotypes we now recognize as dilated cardiomyopathy (DCM), restrictive cardiomyopathy (RCM), and hypertrophic cardiomyopathy (HCM), respectively [1]. A fourth form of cardiomyopathy, arrhythmogenic right ventricular cardiomyopathy/dysplasia (ARVC/D), was identified later [2, 3]. Recognizing the familial segregation of these disorders facilitated our understanding of these cardiomyopathies as largely monogenic. Almost 60 years after Goodwin’s initial observations about structure and function, we can now also classify these cardiomyopathies based on their genetic and molecular alterations [4, 5].
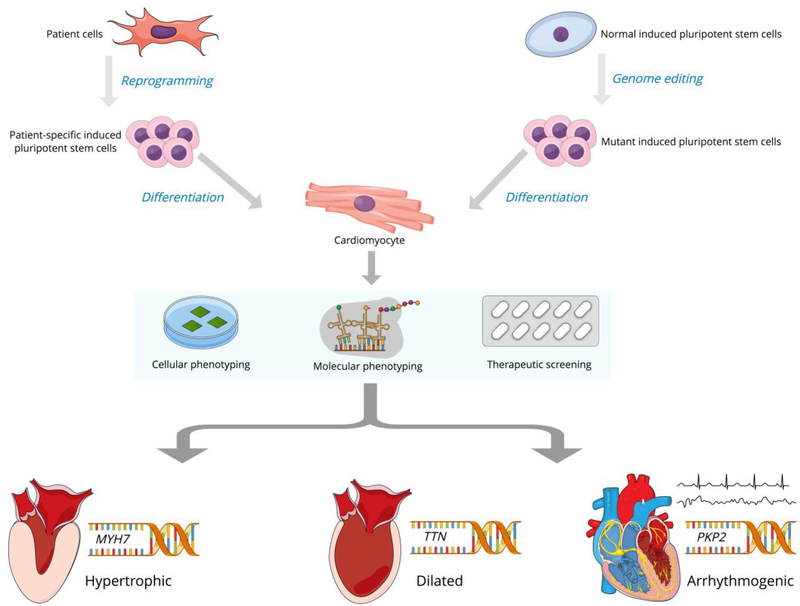
Human pluripotent stem cells (hPSCs) provide a unique model system for the study of genetic alterations associated with inherited cardiovascular disease (Figure 1). Among their advantages, they can be differentiated into any somatic cell type and can generate extremely large numbers of cell progeny. Induced pluripotent stem cells (iPSCs) are now the most widely used type of hPSCs and are produced by reprogramming human somatic cells with the heterologous expression of certain transcription factors [6]. For monogenic disorders, iPSCs are an exemplary disease modeling system because they are genetically matched to the person from whom they were derived without many of the epigenetic influences that might contribute to disease phenotype [7]. Studies performed with iPSCs that have been differentiated into cardiomyocytes (iPSC-CMs) have already proven successful in helping us understand the cellular consequences of mutations that lead to genetic cardiomyopathies [8].
Figure 1.
Human pluripotent stem cells for modeling of genetic cardiomyopathies
Recent rapid growth in genetic and genomic technologies has also transformed the clinical care of patients with inherited cardiomyopathies. Within the last 15 years, available strategies for genetic testing have advanced from targeted multigene cardiomyopathy panels to more agnostic platforms, including whole exome and whole genome sequencing [9, 10]. Defining the genetic cause of cardiomyopathy through testing provides opportunities for disease screening and risk stratification for affected individuals and their family members [11]. As our techniques to identify individuals with genetic cardiomyopathy improve, our knowledge regarding the genetic architecture of these disorders must keep pace. Here we will review the latest bench-to-bedside developments in three major phenotypes of inherited cardiomyopathies with a focus on genetics, risk stratification, therapy, and disease modeling.
Hypertrophic Cardiomyopathy
Hypertrophic cardiomyopathy (HCM) is characterized by increased left ventricular wall thickness unexplained by cardiac loading conditions and a nondilated LV with preserved or increased ejection fraction. A “phenotype-positive” individual is recognized by a maximal left ventricular wall thickness ≥ 15 millimeters with ≥ 13 millimeters considered borderline [12, 13]. Older echocardiography-based studies in diverse cohorts established the general population prevalence of HCM at 0.16–0.23% (~1 in 500) [14–19]. Enhanced phenotype detection with advanced diagnostic imaging, widely available commercial genetic testing, protocolized clinical screening of family members, and larger genetic population studies offer evidence for a revised prevalence estimate of 1 in 200 [20].
Hypertrophic cardiomyopathy is largely considered to be a monogenic disorder with autosomal dominant inheritance caused by variants in genes that encode sarcomere proteins. Variants in two genes, beta-myosin heavy chain (MYH7) and myosin-binding protein 3 (MYBPC3), are responsible for disease in about 50% of individuals with familial HCM [21–24]. Less than 10% of cases can be attributed to seven other genes that encode sarcomere proteins: cardiac troponin T (TNNT2), cardiac troponin I (TNNI3), alpha-tropomyosin (TPM1), cardiac alpha-actin (ACTC1), regulatory myosin light chain (MYL2), essential myosin light chain (MYL3), and cysteine and glycine-rich protein 3 (CSRP3) [25–31]. In addition to variants in the genes above, other genetic and environmental factors with a range of effect sizes are believed to contribute to the variable penetrance and expression of HCM. At least 15 additional sarcomere and non-sarcomere genes have been implicated in HCM, however variants in these and other “missing causal genes” occur less frequently and in smaller families resulting in weaker evidence for causality [32, 33]. In ~40% of HCM cases, the causal genes remain unknown [34].
Individuals with HCM become symptomatic due to diastolic dysfunction, left ventricular outflow tract (LVOT) or intracavitary obstruction, myocardial oxygen supply-demand mismatch, and atrial and ventricular arrhythmias. Beta-adrenergic receptor blockers remain the mainstay of pharmacologic treatment and are used to decrease obstruction, increase diastolic duration, and reduce myocardial ischemia [35]. Disopyramide, in combination with beta-blockers, is used to alleviate obstructive symptoms and to the reduce LVOT gradient [36]. In patients who do not respond to or cannot tolerate beta-blockers, symptomatic benefit can be achieved with nondihydropyridine calcium channel blockers such as diltiazem and verapamil [37]. Septal reduction strategies (surgical septal myectomy and alcohol septal ablation) are reserved for patients with severe symptomatic LVOT obstruction despite maximally tolerated or optimal pharmacologic therapy. At experienced centers, surgical myectomy now has a <1% 30-day operative mortality with a majority of patients experiencing symptom relief [38–40]. Left ventricular assist device implantation has emerged as a mechanical support strategy for patients with end-stage HCM. In the largest published series of continuous flow left ventricular assist device therapy in HCM and restrictive cardiomyopathy (RCM), overall survival of HCM and RCM patients was similar to that of traditional dilated cardiomyopathy (DCM) patients. Survival was worse for those with a preimplant left ventricular end diastolic dimension of < 5.0 centimeters [41]. HCM patients comprise ~1% of heart-only transplant recipients in the United States, and their post-transplant survival is comparable to those with non-HCM diagnoses [42, 43].
Ventricular arrhythmias and sudden cardiac death (SCD) are the most feared complications of HCM. Accordingly, risk stratification algorithms aim to identify individuals at increased risk for SCD who would benefit from prophylactic implantable cardioverter defibrillator (ICD) implantation. The 2011 American College of Cardiology Foundation/American Heart Association HCM guidelines highlighted five conventional risk factors, drawing primarily from observational studies, for the estimation of SCD risk: (1) a family history of SCD; (2) maximal left ventricular wall thickness ≥ 30 millimeters; (3) unexplained syncope; (4) nonsustained ventricular tachycardia; and (5) abnormal blood pressure response to exercise [12]. In a departure from the U.S. framework, the 2014 European Society of Cardiology guidelines added a Class I recommendation for the use of a new SCD risk prediction model – HCM Risk-SCD [13]. HCM Risk-SCD was derived from a retrospective, multi-center longitudinal cohort study of 3,675 consecutive patients with the goal to provide a 5-year individualized risk estimates of SCD. Variables in the model include age, severity of left ventricular hypertrophy, left atrium size, LVOT gradient, family history of SCD, nonsustained ventricular tachycardia, and unexplained syncope. HCM Risk-SCD was internally validated and improved risk prediction (c-statistic from 0.54 to 0.7) compared to a more traditional model using four major risk factors [44]. A few smaller external validation efforts have suggested that the HCM Risk-SCD model is superior to previous models, however the most recently published validation study found that it performed well at lower and higher levels of risk but less well at intermediate risk levels [45–49]. Late gadolinium enhancement on cardiac magnetic resonance imaging (CMR), another potential risk marker, has been shown to be associated with SCD but has not yet been incorporated into formal prediction models [50, 51]. The highly anticipated Hypertrophic Cardiomyopathy Registry, planned to conclude in 2022, aims to improve SCD prognostication with international prospective analyses of clinical, imaging, genetic, and biomarker data [52].
In addition to clinical and imaging data, incorporating genetics into SCD risk prediction has shown some correlation with clinical outcomes. In a cohort that spanned nearly 30 years, HCM phenotype-positive carriers of likely pathogenic or pathogenic sarcomeric and non-sarcomeric variants had increased risks of all-cause death, cardiovascular death, heart failure-related death, and SCD/aborted SCD [53]. Despite pathogenic sarcomere variants being associated with an increase in heart failure events, there was no difference in events between MYH7 and MYBPC3 carriers in another study [54]. Overall, genotype status has been correlated with long-term outcomes but it alone cannot predict patient-specific outcomes given the contribution of modifying genetic, epigenetic, and environmental factors.
Although it may seem logical to obtain as much genetic data as possible for incorporation into risk prediction, the method of acquisition is important. Expanded gene panel testing did not significantly increase the sensitivity of pathogenic variant detection over a smaller panel in a broad referral population [55]. Newer agnostic platforms have shown more promise. In a comparison of whole genome sequencing (WGS) to multipanel gene testing, WGS identified 19 of 20 variants called as pathogenic, likely pathogenic, or uncertain significance and provided one new diagnostic finding. However, WGS also identified more variants of uncertain significance and secondary genetic findings, emphasizing the importance of expertise in clinical genetics and genomics when translating WGS to clinical care [9]. In an Australian HCM cohort in which targeted panel testing had not previously identified causal variants, WGS found a pathogenic or likely pathogenic variant in 20% of families and identified plausible disease-causing intronic and mitochondrial variants [10]. These technologies may serve to expand the population of “genotype-positive/phenotype-negative” individuals.
To date, pharmacologic and interventional therapies for HCM have not targeted the underlying genetic defect or affected intermediary pathways. However, experimental disease-modifying and molecular therapies are under development. The VANISH (Valsartan for Attenuating Disease Evolution in Early Sarcomeric HCM) trial is a multicenter, double-blind, placebo-controlled, phase II, randomized clinical trial to assess the safety and efficacy of valsartan in attenuating HCM disease progression in unaffected or mildly affected sarcomeric variant carriers with New York Heart Association Class I-II symptoms [56]. Mavacamten is an oral small molecule that regulates cardiac myosin ATPase and was shown to prevent hypertrophy and reduce myocyte disarray and interstitial fibrosis in murine models [57]. The safety and efficacy of mavacamten in symptomatic obstructive HCM is being tested in PIONEER-HCM, a phase 2 open-label trial. Preliminary data show that mavacamten reduces post-exercise peak LVOT gradient, resting LVOT gradient, and subjective dyspnea scores while increasing peak exercise oxygen consumption [58]. Modeling with iPSC-CMs also demonstrate the potential to link sarcomere variant status with targeted pharmacologic therapy. Mutant iPSC-CMs generated from 10 affected and unaffected family members with an MYH7 missense variant exhibited contractile arrhythmia and cellular enlargement in the setting of abnormal calcium handling. A similar phenotype was displayed by iPSC-CMs with a different MYH7 missense variant. Both sets of these phenotypes could be normalized with verapamil treatment [59, 60]. Despite the preliminary nature of these results, it is evident that the genetic era of HCM is rapidly shifting focus to targeted therapeutics.
Dilated Cardiomyopathy
Dilated cardiomyopathy (DCM) is characterized by ventricular enlargement with depressed myocardial contractility. This ventricular dysfunction often progresses to overt heart failure with reduced ejection fraction, making it the most common indication for adult heart transplantation worldwide [61]. Heart failure symptoms are the most frequent clinical manifestation of DCM, however it can also present with arrhythmia, SCD, and thromboembolic events. DCM is typically diagnosed by the detection of enlarged left ventricular dimension by echocardiography or cardiac magnetic resonance imaging. In first-degree relatives of individuals with newly diagnosed idiopathic DCM, left ventricular echocardiographic deformation parameters (strain, strain rate, fractional shortening) were significantly impaired compared to age- and sex-matched controls suggesting that familial DCM may be detectable prior to the development of left ventricular cavity enlargement [62].
Clinical trials have conventionally distinguished DCM patients on the basis of ischemic versus nonischemic idiopathic etiologies, with the latter comprising 30% to 40% of participants [63]. Recent meta-analyses suggest a 23% prevalence estimate of familial DCM, indicating an important genetic contribution to these nonischemic idiopathic DCM cases [64, 65]. Familial DCM has been defined by the presence of (1) > 2 affected relatives with DCM or (2) a relative of a DCM patient with unexplained sudden death before the age of 35 years, however these definitions have not been uniformly applied across studies [66, 67]. Cases of nonfamilial or sporadic DCM have also been shown to have genetic bases, although the frequency of this finding is unknown [68]. In addition, the phenotypes of DCM attributed to nongenetic causes, such as hypertension, valvular disease, and toxin exposure, may be influenced by genetic and epigenetic factors. Overall, the true prevalence of genetically-mediated DCM remains undetermined due in part to this heterogeneity in classification.
A genetic cause of cardiomyopathy can be identified in 30% to 40% of patients with familial DCM [69]. The majority of these causes are inherited in an autosomal dominant fashion with variable penetrance and expressivity. The most commonly mutated gene in familial DCM is titin (TTN), followed by lamin-A/C (LMNA), myosin-7 and −6 (MYH7 and MYH6), sodium channel protein type 5 subunit alpha (SCN5A), MYBPC3, and TNNT2 [70]. Autosomal recessive, X-linked recessive, and mitochondrial inheritance patterns have also been described [71]. Variants in over 50 genes that regulate a broad diversity of cellular functions have been associated with familial DCM. These genes encode proteins required for myocardial force generation, force transmission, sarcomere integrity, cytoskeletal and nuclear architecture, electrolyte homeostasis, mitochondrial function, and transcription. Efforts to capture this locus and allelic heterogeneity has led to the expansion of targeted DCM testing panels offered by clinical diagnostic laboratories. The ensuing improvements in diagnostic sensitivity have been countered by a higher number of inconclusive results at a greater cost [72]. Genome sequencing is being investigated as an alternative to multigene panel sequencing and has shown high accuracy for variant detection along with the added capacity to interrogate noncoding regions of the genome [73, 74].
Despite these advances in genetic testing, there are only a few genotype-phenotype correlations that can be made in familial DCM. Truncating variants (nonsense, frameshift, splice site) in TTN, a massive sarcomeric protein, are believed to cause 20% to 25% of familial DCM [75, 76]. In an integrated analysis of TTN sequence, protein, transcriptional, and phenotypic data of more than 5,200 individuals, individuals with DCM associated with TTN truncating variants experienced worse left ventricular function, more sustained ventricular tachycardia, and poorer heart failure outcomes compared to individuals with DCM without TTN truncating variants [76]. However, TTN truncating variants have also been identified in control and general population reference datasets, although the prevalence is lower [75, 77]. There is also a high prevalence of TTN missense variants in individuals without DCM - 23 variants per individual on average in the Exome Sequencing Project [78]. The clinical significance of these variants remains unclear, but iPSC modeling has provided some mechanistic insight. iPSC-CMs generated from DCM patients with either TTN truncating or missense variants displayed deficits in contractile function and limited compensatory reserve mechanisms in response to mechanical and β-adrenergic stress [79]. These phenotypes were similarly reproduced in genome-edited wild-type iPSC-CMs into which TTN truncating variants had been introduced [79].
Pathogenic variants in LMNA are the second most common cause of inherited DCM, occurring in 5% to 8% [80, 81]. LMNA encodes 2 proteins, lamins A and C, which are involved in many cellular processes including nuclear to cytoplasmic transport, mechanosignaling, and gene expression regulation. iPSC-CMs with either a LMNA nonsense or missense mutation exhibited increased nuclear bleb formation, micronucleation, and apoptosis upon electrical stimulation [82]. Pathogenic LMNA variants are inherited in an autosomal dominant pattern and are predictive of poor arrhythmic and heart failure-related outcomes [83, 84]. The clinical signatures of LMNA-associated DCM include dysrhythmias (sinus and atrioventricular nodal dysfunction, atrial fibrillation, ventricular tachycardia, ventricular fibrillation, SCD) and progressive left ventricular systolic dysfunction, often necessitating advanced therapies. Multiple studies have described the high rate of appropriate ICD therapies for ventricular arrhythmia in DCM patients with disease-causing LMNA variants who had borderline or normal left ventricular systolic function and did not meet otherwise traditional criteria for ICD implantation [85–87]. The role of prophylactic ICD implantation in mitigating SCD risk has been addressed in both European and U.S. consensus documents, and ICD implantation should be addressed especially when individuals undergo pacemaker implantation for LMNA-associated conduction disease [88, 89].
In addition to DCM, other multisystem diseases associated with LMNA mutations, also called laminopathies, include limb-girdle muscular dystrophies, Charcot-Marie-Tooth neuropathy, autosomal Emery-Dreifuss muscular dystrophy, and lipodystrophy syndromes (e.g. Hutchinson- Gilford progeria syndrome).
While mutations in other genes such as SCN5A, filamin C (FLNC), and phospholamban (PLN) have been associated with high-risk features in DCM, the wide genetic heterogeneity and variable penetrance and expressivity has limited further translation to clinical management [90–92]. A number of studies have more specifically characterized the cellular and molecular consequences of mutations in DCM-associated genes with iPSC modeling (Table 1). The most intensively studied familial DCM iPSC lines to date were derived from a family whose affected members harbor a missense R173W variant in TNNT2. Compared to control line iPSCs generated from unaffected family members, the mutant iPSC-CMs exhibited abnormal calcium handling, reduced contractility, and myofibrillar disarray, which were exacerbated with β-adrenergic stimulation [93]. Other studies have demonstrated the potential of genome editing for phenotype correction; corrected iPSC-CMs showed reversal of calcium handling abnormalities caused by an in-frame deletion variant in PLN [94].
Table 1.
Cardiomyopathies modeled with human pluripotent stem cell cells
| Disease modeled | Mutated gene | Type of variant | Reference |
|---|---|---|---|
| Dilated cardiomyopathy | TTN | Truncating, Missense | [79] |
| TNNT2 | Missense | [93] | |
| LMNA | |||
| PLN | In-frame deletion | [94] | |
| DES | Missense | [117] | |
| Hypertrophic cardiomyopathy | MYH7 | Missense | [59, 60] |
| Arrhythmogenic right ventricular cardiomyopathy/dysplasia | PKP2 | Splicing defect, Frameshift, Missense | [114–116] |
| Duchenne muscular dystrophy | DMD | Frameshift, Nonsense | [118, 119] |
| Barth syndrome | TAZ | Frameshift, Missense | [120] |
| Left ventricular noncompaction | TBX20 | Nonsense | [121] |
| GATA4 | Missense | [122] | |
| Restrictive cardiomyopathy | FLNC | Missense | [123] |
TTN, titin; TNNT2, cardiac troponin T; LMNA, lamin A/C; PLN, phospholamban; DES, desmin; MYH7, beta-myosin heavy chain; PKP2, plakophilin 2; DMD, dystrophin; TAZ, tafazzin; TBX20, T-box 20; GATA4, GATA binding protein 4; FLNC, filamin C
Arrhythmogenic Right Ventricular Cardiomyopathy/Dysplasia
Histopathologically, ARVC/D is characterized by the replacement of right ventricular myocardium by fibrous and fatty tissue. This fibrofatty infiltration predominantly involves the right ventricular (RV) free wall leading to thinning and aneurysmal enlargement [3, 95]. Inflammatory lymphocytic and histiocytic infiltrates, focal necrosis, and apoptosis have been observed in biopsy specimens, prompting investigation into the nebulous relationship between myocarditis and ARVC/D [96, 97]. The classical clinical phenotypes of RV precordial T-wave inversions, recurrent ventricular arrhythmia, and RV enlargement and failure first described in 1982 still comprise the basis for the updated international task force criteria used to diagnose ARVC/D [3, 98]. Common but alarming clinical presentations include exercise-induced syncope and SCD. Available therapies aim to reduce SCD risk and alleviate heart failure and arrhythmic symptoms. Left ventricular involvement, manifested by inferior and/or lateral T-wave inversions, ventricular arrhythmias with right bundle branch block morphology, and pump dysfunction, has been described as a distinct pattern of disease expression, leading some to suggest a nomenclature revision to “arrhythmogenic cardiomyopathy” [99]. Similar to other genetic cardiomyopathies, pathologic, electrocardiographic, echocardiographic, and MRI phenotypes are used to make the diagnosis. However, ARVC/D is the only cardiomyopathy in which the presence of a known pathogenic variant is currently incorporated into the diagnostic framework [98].
Most often, causal genes in ARVC/D cases encode the desmosomal proteins plakophilin-2 (PKP2), desmoplakin (DSP), desmoglein-2 (DSG2), desmocollin-2 (DSC2), and junction plakoglobin (JUP) which are critical to intercellular adhesion, signal transduction, and maintenance of tissue integrity [100–104]. Variants in these genes are typically inherited in an autosomal dominant pattern with incomplete penetrance and variable expression and have been identified in 33% to 63% of probands [105]. The estimated overall rate of successful genetic screening in individuals who meet international task force diagnostic criteria is 50% [106]. In multiple cohorts, only 30% to 40% of at-risk relatives carrying identified desmosomal variants fulfill diagnostic task force criteria [107, 108]. Sex-related hypotheses for this variable penetrance are based on observations of lower disease expressivity in women carrying desmosomal gene mutations and more malignant outcomes, including SCD, in men [109, 110]. Sex differences in reproductive hormones and in rates of participation in endurance athletics, a risk factor for early manifestation and progression of disease, have been proposed as reasons for this discrepancy [111]. Digenic inheritance and compound heterozygosity are frequent and can manifest with more severe phenotypes, further complicating the narrative of simple monogenic inheritance [112]. These issues with genetic diagnosis along with the need for advanced imaging and electrophysiologic evaluation (i.e. CMR, signal-averaged electrocardiogram, electroanatomic mapping) required to diagnose ARVC/D likely contribute to underestimation of familial disease.
Non-desmosomal genes including CTNNA3 (alpha T-catenin), CDH2 (N-cadherin), TMEM43 (transmembrane protein 43), LMNA, TTN, PLN, RYR2 (ryanodine-receptor type 2), and SCN5A have been associated with ARVC/D, although with different electrical phenotypes [105]. Similar to HCM and DCM, the expansion of WES and WGS into genetic evaluation for ARVC/D has raised issues regarding the interpretation of rare desmosomal and non-desmosomal variants and of variants of unknown significance. Classification of desmosomal missense variants has been particularly problematic with some PKP2 variants being reclassified after initially being thought to be pathogenic [105]. Segregation studies to inform pathogenicity are limited in ARVC/D by small family sizes and incomplete penetrance. Five genes implicated in ARVC/D pathogenesis (PKP2, DSP, DSG2, DSC2, TMEM43) are included in the American College of Medical Genetics and Genomics list of 59 medically actionable genes recommended for return of results in clinical genomic sequencing [113]. While this genome-first approach could provide an early opportunity for disease prevention, the lack of evidence regarding the appropriate diagnostic evaluation, risk stratification, lifestyle modification, and follow-up for these presumed genotype- positive/phenotype-negative individuals should be addressed.
iPSC modeling for ARVC/D has mirrored the complexities of eliciting disease phenotypes in humans. The most intensively studied ARVC iPSC lines to date were derived from two unrelated individuals, one homozygous for a PKP2 variant that causes a splicing defect and the other heterozygous for a PKP2 frameshift variant [114]. The iPSC-CMs from these individuals manifested ARVC-related phenotypes only when they were treated with five adipogenic factors which led to increased lipogenesis and apoptosis and abnormal calcium handling. In another study, iPSC-CMs from two patients heterozygous for different PKP2 frameshift variants had increased lipid accumulation and desmosomal disruption in standard differentiation conditions, and these phenotypes became exaggerated with treatment with adipogenic factors [115]. Similar findings were observed in another study with iPSC-CMs from an individual heterozygous for a PKP2 missense mutation [116]. While certainly provocative, extrapolation of these findings to causality for disease development in native human myocardium is still premature.
CONCLUSION
Classification of the genetic cardiomyopathies has long relied upon pattern recognition of cardiac structure and function. Rapid progress in next generation sequencing technology, bioinformatics, and functional genomics has facilitated the personalization of diagnosis and management for individuals with hypertrophic, dilated, and arrhythmogenic cardiomyopathy. These tools are becoming more widely available and less expensive and hold great potential for mechanistic insight into inherited cardiovascular disorders. Standardizing and centralizing clinically relevant genomic knowledge will be imperative for accurate variant annotation, precise risk stratification, and achievement of optimal outcomes.
Acknowledgements
Conflict of Interest
Dr. Reza is supported by the NIH National Human Genome Research Institute Ruth L. Kirschstein Institutional National Research Service T32 Award in Genomic Medicine (T32 HG009495). Dr. Owens is supported by the Winkelman Family Fund in Cardiovascular Innovation. Dr. Musunuru declares no conflict of interest.
Footnotes
Compliance with Ethics Guidelines
Human and Animal Rights: All reported studies/experiments with human or animal subjects performed by the authors have been previously published and complied with all applicable ethical standards (including the Helsinki declaration and its amendments, institutional/national research committee standards, and international/national/institutional guidelines).
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
Papers Of Particular Interest, Published Recently, Have Been Highlighted As:
• Of Importance
•• Of Major Importance
- 1.Goodwin JF, Gordon H, Hollman A, Bishop MB. Clinical aspects of cardiomyopathy. British Medical Journal 1961:1(5219):69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fontaine G, Guiraudon G, Frank R, Vedel J, Grosgogeat Y, Cabrol C et al. Stimulation studies and epicardial mapping in ventricular tachycardia: study of mechanisms and selection for surgery In: Re-entrant arrhythmias: mechanisms and treatment. Baltimore: University Park Press;1977. [Google Scholar]
- 3.Marcus FI, Fontaine GH, Guiraudon G, Frank R, Laurenceau JL, Malergue C et al. Right ventricular dysplasia: a report of 24 adult cases. Circulation 1982:65(2):384–98. [DOI] [PubMed] [Google Scholar]
- 4.Maron BJ, Towbin JA, Thiene G, Antzelevitch C, Corrado D, Arnett D et al. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation 2006:113(14):1807–16. Doi: 10.1161/circulationaha.106.174287. [DOI] [PubMed] [Google Scholar]
- 5.Elliott P, Andersson B, Arbustini E, Bilinska Z, Cecchi F, Charron P et al. Classification of the cardiomyopathies: a position statement from the European Society Of Cardiology Working Group on Myocardial and Pericardial Diseases. European Heart Journal 2008:29(2):270–6. Doi: 10.1093/eurheartj/ehm342. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006:126(4):663–76. Doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 7.Musunuru K, Sheikh F, Gupta RM, Houser SR, Maher KO, Milan DJ et al. Induced Pluripotent Stem Cells for Cardiovascular Disease Modeling and Precision Medicine: A Scientific Statement From the American Heart Association. Circulation Genomic and Precision Medicine 2018:11(1):e000043 Doi: 10.1161/hcg.0000000000000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsa E, Ahrens JH, Wu JC. Human Induced Pluripotent Stem Cells as a Platform for Personalized and Precision Cardiovascular Medicine. Physiological Reviews 2016:96(3):1093–126. Doi: 10.1152/physrev.00036.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cirino AL, Lakdawala NK, McDonough B, Conner L, Adler D, Weinfeld M et al. A Comparison of Whole Genome Sequencing to Multigene Panel Testing in Hypertrophic Cardiomyopathy Patients. Circulation Cardiovascular Genetics 2017:10(5):e001768 Doi: 10.1161/circgenetics.117.001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bagnall RD, Ingles J, Dinger ME, Cowley MJ, Ross SB, Minoche AE et al. Whole Genome Sequencing Improves Outcomes of Genetic Testing in Patients With Hypertrophic Cardiomyopathy. Journal of the American College of Cardiology 2018:72(4):419–29. Doi: 10.1016/jjacc.2018.04.078.• This study demonstrates the utility of whole genome sequencing to identify causes of HCM in cases that were not diagnosed with targeted testing.
- 11.Burke MA, Cook SA, Seidman JG, Seidman CE. Clinical and Mechanistic Insights Into the Genetics of Cardiomyopathy. Journal of the American College of Cardiology 2016:68(25):2871–86. Doi: 10.1016/jjacc.2016.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gersh BJ, Maron BJ, Bonow RO, Dearani JA, Fifer MA, Link MS et al. 2011. ACCF/AHA Guideline for the Diagnosis and Treatment of Hypertrophic Cardiomyopathy: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Developed in collaboration with the American Association for Thoracic Surgery, American Society of Echocardiography, American Society of Nuclear Cardiology, Heart Failure Society of America, Heart Rhythm Society, Society for Cardiovascular Angiography and Interventions, and Society of Thoracic Surgeons. Journal of the American College of Cardiology 2011:58(25):e212–60. Doi: 10.1016/j.jacc.201L06.01L [DOI] [PubMed] [Google Scholar]
- 13.Elliott PM, Anastasakis A, Borger MA, Borggrefe M, Cecchi F, Charron P et al. 2014. ESC Guidelines on diagnosis and management of hypertrophic cardiomyopathy: the Task Force for the Diagnosis and Management of Hypertrophic Cardiomyopathy of the European Society of Cardiology (ESC). European Heart Journal 2014:35(39):2733–79. Doi: 10.1093/eurheartj/ehu284. [DOI] [PubMed] [Google Scholar]
- 14.Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation 1995:92(4):785–9. [DOI] [PubMed] [Google Scholar]
- 15.Hada Y, Sakamoto T, Amano K, Yamaguchi T, Takenaka K, Takahashi H et al. Prevalence of hypertrophic cardiomyopathy in a population of adult japanese workers as detected by echocardiographic screening. The American Journal of Cardiology 1987:59(1): 183–4. [DOI] [PubMed] [Google Scholar]
- 16.Zou Y, Song L, Wang Z, Ma A, Liu T, Gu H et al. Prevalence of idiopathic hypertrophic cardiomyopathy in China: a population-based echocardiographic analysis of 8080 adults. The American Journal of Medicine 2004:116(1): 14–8. [DOI] [PubMed] [Google Scholar]
- 17.Maron BJ, Mathenge R, Casey SA, Poliac LC, Longe TF. Clinical profile of hypertrophic cardiomyopathy identified de novo in rural communities. Journal of the American College of Cardiology 1999:33(6): 1590–5. [DOI] [PubMed] [Google Scholar]
- 18.Maron BJ, Spirito P, Roman MJ, Paranicas M, Okin PM, Best LG et al. Prevalence of hypertrophic cardiomyopathy in a Population-Based sample of American Indians aged 51 to 77 years (the Strong Heart Study). The American Journal of Cardiology 2004:93(12): 1510–4. [DOI] [PubMed] [Google Scholar]
- 19.Maro E, Janabi M, Kaushik R. Clinical and echocardiographic study of hypertrophic cardiomyopathy in Tanzania. Tropical Doctor 2006:36(4):225–7. [DOI] [PubMed] [Google Scholar]
- 20.Semsarian C, Ingles J, Maron MS, Maron BJ. New perspectives on the prevalence of hypertrophic cardiomyopathy. Journal of the American College of Cardiology 2015:65(12): 1249–54. [DOI] [PubMed] [Google Scholar]
- 21.Millat G, Bouvagnet P, Chevalier P, Dauphin C, Jouk PS, Da Costa A et al. Prevalence and spectrum of mutations in a cohort of 192 unrelated patients with hypertrophic cardiomyopathy. European Journal of Medical Genetics 2010:53(5):261–7. [DOI] [PubMed] [Google Scholar]
- 22.Kaski JP, Syrris P, Esteban MTT, Jenkins S, Pantazis A, Deanfield JE et al. Prevalence of sarcomere protein gene mutations in preadolescent children with hypertrophic cardiomyopathy. Circulation: Genomic and Precision Medicine 2009:2(5):436–41. [DOI] [PubMed] [Google Scholar]
- 23.Erdmann J, Daehmlow S, Wischke S, Senyuva M, Werner U, Raible J et al. Mutation spectrum in a large cohort of unrelated consecutive patients with hypertrophic cardiomyopathy. Clinical Genetics 2003:64(4):339–49. [DOI] [PubMed] [Google Scholar]
- 24.Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation 2003:107(17):2227–32. [DOI] [PubMed] [Google Scholar]
- 25.Watkins H, McKenna WJ, Thierfelder L, Suk HJ, Anan R, O’donoghue A et al. Mutations in the genes for cardiac troponin T and a-tropomyosin in hypertrophic cardiomyopathy. New England Journal of Medicine 1995:332(16): 1058–65. [DOI] [PubMed] [Google Scholar]
- 26.Kimura A, Harada H, Park JE, Nishi H, Satoh M, Takahashi M et al. Mutations in the cardiac troponin I gene associated with hypertrophic cardiomyopathy. Nature Genetics 1997:16(4):379–82. [DOI] [PubMed] [Google Scholar]
- 27.Thierfelder L, Watkins H, MacRae C, Lamas R, McKenna W, Vosberg H-P et al. a-Tropomyosin and cardiac troponin T mutations cause familial hypertrophic cardiomyopathy: a disease of the sarcomere. Cell 1994:77(5):701–12. [DOI] [PubMed] [Google Scholar]
- 28.Mogensen J, Klausen IC, Pedersen AK, Egeblad H, Bross P, Kruse TA et al. alpha-cardiac actin is a novel disease gene in familial hypertrophic cardiomyopathy. J Clin Invest 1999:103(10):R39–R43. Doi: 10.1172/jci6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olson TM, Doan TP, Kishimoto NY, Whitby FG, Ackerman MJ, Fananapazir L. Inherited and de novo mutations in the cardiac actin gene cause hypertrophic cardiomyopathy. J Mol Cell Cardiol 2000:32(9): 1687–94. Doi: 10.1006/jmcc.2000.1204. [DOI] [PubMed] [Google Scholar]
- 30.Poetter K, Jiang H, Hassanzadeh S, Master SR, Chang A, Dalakas MC et al. Mutations in either the essential or regulatory light chains of myosin are associated with a rare myopathy in human heart and skeletal muscle. Nature Genetics 1996:13(1):63–9. Doi: 10.1038/ng0596-63. [DOI] [PubMed] [Google Scholar]
- 31.Geier C, Gehmlich K, Ehler E, Hassfeld S, Perrot A, Hayess K et al. Beyond the sarcomere: CSRP3 mutations cause hypertrophic cardiomyopathy. Human Molecular Genetics 2008:17(18):2753–65. [DOI] [PubMed] [Google Scholar]
- 32.Marian AJ, Braunwald E. Hypertrophic Cardiomyopathy: Genetics, Pathogenesis, Clinical Manifestations, Diagnosis, and Therapy. Circulation Research 2017:121(7):749–70. Doi: 10.1161/circresaha.117.311059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marian AJ. The case of “missing causal genes” and the practice of medicine: a Sherlock Holmes approach of deductive reasoning. Circulation Research 2016:119(1):21–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li L, Bainbridge MN, Tan Y, Willerson JT, Marian AJ. A potential oligogenic etiology of hypertrophic cardiomyopathy: a classic single-gene disorder. Circulation Research 2017:120(7):1084–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nistri S, Olivotto I, Maron MS, Ferrantini C, Coppini R, Grifoni C et al. beta Blockers for prevention of exercise-induced left ventricular outflow tract obstruction in patients with hypertrophic cardiomyopathy. Am J Cardiol 2012:110(5):715–9. doi: 10.1016/j.amjcard.2012.04.051. [DOI] [PubMed] [Google Scholar]
- 36.Sherrid MV, Barac I, McKenna WJ, Elliott PM, Dickie S, Chojnowska L et al. Multicenter study of the efficacy and safety of disopyramide in obstructive hypertrophic cardiomyopathy. Journal of the American College of Cardiology 2005:45(8):1251–8. doi: 10.1016/jjacc.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 37.Gilligan DM, Chan WL, Joshi J, Clarke P, Fletcher A, Krikler S et al. A double-blind, placebocontrolled crossover trial of nadolol and verapamil in mild and moderately symptomatic hypertrophic cardiomyopathy. Journal of the American College of Cardiology 1993:21(7): 1672–9. [DOI] [PubMed] [Google Scholar]
- 38.Maron BJ, Dearani JA, Ommen SR, Maron MS, Schaff HV, Nishimura RA et al. Low Operative Mortality Achieved With Surgical Septal Myectomy: Role of Dedicated Hypertrophic Cardiomyopathy Centers in the Management of Dynamic Subaortic Obstruction. Journal of the American College of Cardiology 2015:66(11): 1307–8. Doi: 10.1016/jjacc.2015.06.1333. [DOI] [PubMed] [Google Scholar]
- 39.Maron BJ, Nishimura RA. Surgical septal myectomy versus alcohol septal ablation: assessing the status of the controversy in 2014. Circulation 2014:130(18): 1617–24. Doi: 10.1161/circulationaha.114.011580. [DOI] [PubMed] [Google Scholar]
- 40.Kim LK, Swaminathan RV, Looser P, Minutello RM, Wong SC, Bergman G et al. Hospital Volume Outcomes After Septal Myectomy and Alcohol Septal Ablation for Treatment of Obstructive Hypertrophic Cardiomyopathy: US Nationwide Inpatient Database, 2003–2011. JAMA Cardiology 2016:1(3):324–32. Doi: 10.1001/jamacardio.2016.0252. [DOI] [PubMed] [Google Scholar]
- 41.Patel SR, Saeed O, Naftel D, Myers S, Kirklin J, Jorde UP et al. Outcomes of Restrictive and Hypertrophic Cardiomyopathies After LVAD: An INTERMACS Analysis. Journal of Cardiac Failure 2017:23(12):859–67. Doi: 10.1016/j.cardfail.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 42.Maron MS, Kalsmith BM, Udelson JE, Li W, DeNofrio D. Survival after cardiac transplantation in patients with hypertrophic cardiomyopathy. Circulation Heart Failure 2010:3(5):574–9. Doi: 10.1161/circheartfailure.109.922872. [DOI] [PubMed] [Google Scholar]
- 43.Rowin EJ, Maron BJ, Abt P, Kiernan MS, Vest A, Costantino F et al. Impact of Advanced Therapies for Improving Survival to Heart Transplant in Patients with Hypertrophic Cardiomyopathy. The American Journal of Cardiology 2018:121(8):986–96. [DOI] [PubMed] [Google Scholar]
- 44.O’Mahony C, Jichi F, Pavlou M, Monserrat L, Anastasakis A, Rapezzi C et al. A novel clinical risk prediction model for sudden cardiac death in hypertrophic cardiomyopathy (HCM Risk-SCD). European Heart Journal 2014:35(30):2010–20. Doi: 10.1093/eurheartj/eht439. [DOI] [PubMed] [Google Scholar]
- 45.Vriesendorp PA, Schinkel AF, Liebregts M, Theuns DA, van Cleemput J, ten Cate FJ et al. Validation of the 2014 European Society of Cardiology guidelines risk prediction model for the primary prevention of sudden cardiac death in hypertrophic cardiomyopathy. Circulation Arrhythmia and Electrophysiology 2015:8(4):829–35. [DOI] [PubMed] [Google Scholar]
- 46.Maron BJ, Casey SA, Chan RH, Garberich RF, Rowin EJ, Maron MS. Independent assessment of the European Society of Cardiology sudden death risk model for hypertrophic cardiomyopathy. The American Journal of Cardiology 2015:116(5):757–64. [DOI] [PubMed] [Google Scholar]
- 47.Fernández A, Quiroga A, Ochoa JP, Mysuta M, Casabé JH, Biagetti M et al. Validation of the 2014 European Society of Cardiology sudden cardiac death risk prediction model in hypertrophic cardiomyopathy in a reference center in South America. The American Journal of Cardiology 2016:118(1): 121–6. [DOI] [PubMed] [Google Scholar]
- 48.Ruiz-Salas A, García-Pinilla J, Cabrera-Bueno F, Fernández-Pastor J, Peña-Hernández J, Medina-Palomo C et al. Comparison of the new risk prediction model (HCM Risk-SCD) and classic risk factors for sudden death in patients with hypertrophic cardiomyopathy and defibrillator. Europace: European pacing, arrhythmias, and cardiac electrophysiology: journal of the working groups on cardiac pacing, arrhythmias, and cardiac cellular electrophysiology of the European Society of Cardiology 2016:18(5):773. [DOI] [PubMed] [Google Scholar]
- 49.O’Mahony C, Jichi F, Ommen SR, Christiaans I, Arbustini E, Garcia-Pavia P et al. International External Validation Study of the 2014 European Society of Cardiology Guidelines on Sudden Cardiac Death Prevention in Hypertrophic Cardiomyopathy (EVIDENCE-HCM). Circulation 2018:137(10): 101523 Doi: 10.1161/circulationaha.117.030437. [DOI] [PubMed] [Google Scholar]
- 50.Chan RH, Maron BJ, Olivotto I, Pencina MJ, Assenza GE, Haas T et al. Prognostic Value of Quantitative Contrast-Enhanced Cardiovascular Magnetic Resonance for the Evaluation of Sudden Death Risk in Patients With Hypertrophic Cardiomyopathy. Circulation 2014:130(6):484–95. [DOI] [PubMed] [Google Scholar]
- 51.Mentias A, Raeisi-Giglou P, Smedira NG, Feng K, Sato K, Wazni O et al. Late Gadolinium Enhancement in Patients With Hypertrophic Cardiomyopathy and Preserved Systolic Function. Journal of the American College of Cardiology 2018: 72(8):857–70. [DOI] [PubMed] [Google Scholar]
- 52.Kramer CM, Appelbaum E, Desai MY, Desvigne-Nickens P, DiMarco JP, Friedrich MG et al. Hypertrophic Cardiomyopathy Registry: The rationale and design of an international, observational study of hypertrophic cardiomyopathy. American Heart Journal 2015:170(2):223–30.• This article reviews the design of the highly anticipated international Hypertrophic Cardiomyopathy Registry.
- 53.van Velzen HG, Vriesendorp PA, Oldenburg RA, van Slegtenhorst MA, van der Velden J, Schinkel AF et al. Value of genetic testing for the prediction of long-term outcome in patients with hypertrophic cardiomyopathy. The American Journal of Cardiology 2016:118(6):881–7. [DOI] [PubMed] [Google Scholar]
- 54.Li Q, Gruner C, Chan RH, Care M, Siminovitch K, Williams L et al. Genotype-Positive Status in Patients With Hypertrophic Cardiomyopathy Is Associated With Higher Rates of Heart Failure Events. Circulation Cardiovascular Genetics 2014:7(4):416–22. Doi: 10.1161/circgenetics.113.000331. [DOI] [PubMed] [Google Scholar]
- 55.Alfares AA, Kelly MA, McDermott G, Funke BH, Lebo MS, Baxter SB et al. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: expanded panels offer limited additional sensitivity. Genetics In Medicine 2015:17:880 Doi: 10.1038/gim.2014.205 [DOI] [PubMed] [Google Scholar]
- 56.Ho CY, McMurray JJ, Cirino AL, Colan SD, Day SM, Desai AS et al. The design of the valsartan for attenuating disease evolution in early sarcomeric hypertrophic cardiomyopathy (VANISH) trial. American Heart Journal 2017:187:145–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Green EM, Wakimoto H, Anderson RL, Evanchik MJ, Gorham JM, Harrison BC et al. A small- molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science 2016:351(6273):617–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jacoby D, Lester S, Owens A, Wang A, Young D, Tripuraneni R et al. Reduction In Left Ventricular Outflow Tract Gradient With Mavacamten (MYK-461) In Symptomatic Obstructive Hypertrophic Patients (PIONEER-HCM). Journal of the American College of Cardiology 2018:71(11 Supplement):A644 Doi: 10.1016/s0735-1097(18)31185-9. [DOI] [Google Scholar]
- 59.Lan F, Lee AS, Liang P, Sanchez-Freire V, Nguyen PK, Wang L et al. Abnormal Calcium Handling Properties Underlie Familial Hypertrophic Cardiomyopathy Pathology in Patient-Specific Induced Pluripotent Stem Cells. Cell Stem Cell 2013:12(1): 101–13. Doi: 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han L, Li Y, Tchao J, Kaplan AD, Lin B, Li Y et al. Study familial hypertrophic cardiomyopathy using patient-specific induced pluripotent stem cells. Cardiovascular Research 2014:104(2):258–69. Doi: 10.1093/cvr/cvu205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lund LH, Khush KK, Cherikh WS, Goldfarb S, Kucheryavaya AY, Levvey BJ et al. The Registry of the International Society for Heart and Lung Transplantation: thirty-fourth adult heart transplantation report—2017; Focus theme: Allograft ischemic time. The Journal of Heart and Lung Transplantation 2017:36(10): 1037–46. [DOI] [PubMed] [Google Scholar]
- 62.Sefa MO, Tuluce K, Yakar ST, Kilic S, Soner HK, Sayin A et al. Screening first-degree relatives of patients with idiopathic dilated cardiomyopathy. Herz 2017:42(7):669–76. [DOI] [PubMed] [Google Scholar]
- 63.Bozkurt B, Colvin M, Cook J, Cooper LT, Deswal A, Fonarow GC et al. Current diagnostic and treatment strategies for specific dilated cardiomyopathies: a scientific statement from the American Heart Association. Circulation 2016:134(23):e579–e646. [DOI] [PubMed] [Google Scholar]
- 64.Yancy CW, Jessup M, Bozkurt B, Butler J, Casey DE, Drazner MH et al. 2013. ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Journal of the American College of Cardiology 2013:62(16):e147–e239. [DOI] [PubMed] [Google Scholar]
- 65.Petretta M, Pirozzi F, Sasso L, Paglia A, Bonaduce D. Review and metaanalysis of the frequency of familial dilated cardiomyopathy. The American Journal of Cardiology 2011:108(8): 1171–6. [DOI] [PubMed] [Google Scholar]
- 66.Mestroni L, Maisch B, McKenna W, Schwartz K, Charron P, Rocco C et al. Guidelines for the study of familial dilated cardiomyopathies. European Heart Journal 1999:2(20):93–102. [DOI] [PubMed] [Google Scholar]
- 67.Kinnamon DD, Morales A, Bowen DJ, Burke W, Hershberger RE. Toward Genetics-Driven Early Intervention in Dilated Cardiomyopathy: Design and Implementation of the DCM Precision Medicine Study. Circulation Cardiovascular Genetics 2017:10(6). Doi: 10.1161/circgenetics.117.001826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haas J, Frese KS, Peil B, Kloos W, Keller A, Nietsch R et al. Atlas of the clinical genetics of human dilated cardiomyopathy. European Heart Journal 2014:36(18): 1123–35.• This study is a comprehensive investigation of the genetics of DCM in a large-scale cohort.
- 69.Ganesh SK, Arnett DK, Assimes TL, Basson CT, Chakravarti A, Ellinor PT et al. Genetics and genomics for the prevention and treatment of cardiovascular disease: update: a scientific statement from the American Heart Association. Circulation 2013:128(25):2813–51. [DOI] [PubMed] [Google Scholar]
- 70.Hershberger RE, Morales A. Dilated Cardiomyopathy Overview In: GeneReviews [Internet]. University of Washington, Seattle, Seattle, Washington: 1993. https://www.ncbi.nlm.nih.gov/books/NBK1309/. 2018. [Google Scholar]
- 71.Hershberger RE, Hedges DJ, Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nature Reviews Cardiology 2013:10(9):531–47. Doi: 10.1038/nrcardio.2013.105. [DOI] [PubMed] [Google Scholar]
- 72.Pugh TJ, Kelly MA, Gowrisankar S, Hynes E, Seidman MA, Baxter SM et al. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genetics in Medicine 2014:16(8):601. [DOI] [PubMed] [Google Scholar]
- 73.Golbus JR, Puckelwartz MJ, Dellefave-Castillo L, Fahrenbach JP, Nelakuditi V, Pesce LL et al. Targeted analysis of whole genome sequence data to diagnose genetic cardiomyopathy. Circulation Genomic and Precision Medicine 2014:7(6):751–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Minoche AE, Horvat C, Johnson R, Gayevskiy V, Morton SU, Drew AP et al. Genome sequencing as a first-line genetic test in familial dilated cardiomyopathy. Genetics in Medicine 2018. doi: 10.1038/s41436-018-0084-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Herman DS, Lam L, Taylor MR, Wang L, Teekakirikul P, Christodoulou D et al. Truncations of titin causing dilated cardiomyopathy. The New England Journal of Medicine 2012:366(7):619–28. Doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roberts AM, Ware JS, Herman DS, Schafer S, Baksi J, Bick AG et al. Integrated allelic, transcriptional, and phenomic dissection of the cardiac effects of titin truncations in health and disease. Science Translational Medicine 2015:7(270):270ra6–ra6. Doi: 10.1126/scitranslmed.3010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Akinrinade O, Koskenvuo JW, Alastalo T-P. Prevalence of titin truncating variants in general population. PLoS One 2015:10(12):e0145284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Norton N, Li D, Rampersaud E, Morales A, Martin ER, Zuchner S et al. Exome sequencing and genome-wide linkage analysis in 17 families illustrate the complex contribution of TTN truncating variants to dilated cardiomyopathy. Circulation Cardiovascular Genetics 2013:6(2): 144–53. Doi: 10.1161/circgenetics.111.000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hinson JT, Chopra A, Nafissi N, Polacheck WJ, Benson CC, Swist S et al. Titin Mutations in iPS cells Define Sarcomere Insufficiency as a Cause of Dilated Cardiomyopathy. Science 2015;349(6251):982–6. Doi: 10.1126/science.aaa5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Parks SB, Kushner JD, Nauman D, Burgess D, Ludwigsen S, Peterson A et al. Lamin A/C mutation analysis in a cohort of 324 unrelated patients with idiopathic or familial dilated cardiomyopathy. American Heart Journal 2008:156(1): 161–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van Tintelen JP, Hofstra RM, Katerberg H, Rossenbacker T, Wiesfeld AC, du Marchie Sarvaas GJ et al. High yield of LMNA mutations in patients with dilated cardiomyopathy and/or conduction disease referred to cardiogenetics outpatient clinics. American Heart Journal. 2007:154(6): 1130–9. [DOI] [PubMed] [Google Scholar]
- 82.Siu CW, Lee YK, Ho JC, Lai WH, Chan YC, Ng KM et al. Modeling of lamin A/C mutation premature cardiac aging using patient-specific induced pluripotent stem cells. Aging 2012:4(11):803–22. Doi: 10.18632/aging.100503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fatkin D, Macrae C, Sasaki T, Wolff MR, Porcu M, Frenneaux M et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. New England Journal of Medicine 1999:341(23): 1715–24. [DOI] [PubMed] [Google Scholar]
- 84.van Rijsingen IAW, Arbustini E, Elliott PM, Mogensen J, Hermans-van Ast JF, van der Kooi AJ et al. Risk Factors for Malignant Ventricular Arrhythmias in Lamin A/C Mutation Carriers: A European Cohort Study. Journal of the American College of Cardiology 2012:59(5):493–500. Doi: 10.1016/jjacc.2011.08.078. [DOI] [PubMed] [Google Scholar]
- 85.Meune C, Van Berlo JH, Anselme F, Bonne G, Pinto YM, Duboc D. Primary prevention of sudden death in patients with lamin A/C gene mutations. New England Journal of Medicine 2006:354(2):209–10. [DOI] [PubMed] [Google Scholar]
- 86.Anselme F, Moubarak G, Savouré A, Godin B, Borz B, Drouin-Garraud V et al. Implantable cardioverter-defibrillators in lamin A/C mutation carriers with cardiac conduction disorders. Heart Rhythm 2013:10(10): 1492–8. Doi: 10.1016/j.hrthm.2013.06.020. [DOI] [PubMed] [Google Scholar]
- 87.Kumar S, Baldinger SH, Gandjbakhch E, Maury P, Sellal J-M, Androulakis AF et al. Long-term arrhythmic and nonarrhythmic outcomes of lamin A/C mutation carriers. Journal of the American College of Cardiology 2016:68(21):2299–307.• This article contributes important natural history and prognostic information about LMNA-related heart disease.
- 88.Kusumoto FM, Calkins H, Boehmer J, Buxton AE, Chung MK, Gold MR et al. HRS/ACC/AHA expert consensus statement on the use of implantable cardioverter-defibrillator therapy in patients who are not included or not well represented in clinical trials. Journal of the American College of Cardiology 2014:64(11): 1143–77. Doi: 10.1016/jjacc.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 89.Priori SG, Blomstrom-Lundqvist C, Mazzanti A, Blom N, Borggrefe M, Camm J et al. 2015. ESC Guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: The Task Force for the Management of Patients with Ventricular Arrhythmias and the Prevention of Sudden Cardiac Death of the European Society of Cardiology (ESC). Endorsed by: Association for European Paediatric and Congenital Cardiology (AEPC). European Heart Journal 2015:36(41):2793–867. Doi: 10.1093/eurheartj/ehv316. [DOI] [PubMed] [Google Scholar]
- 90.McNair WP, Sinagra G, Taylor MR, Di Lenarda A, Ferguson DA, Salcedo EE et al. SCN5A mutations associate with arrhythmic dilated cardiomyopathy and commonly localize to the voltage-sensing mechanism. Journal of the American College of Cardiology 2011:57(21):2160–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ortiz-Genga MF, Cuenca S, Dal Ferro M, Zorio E, Salgado-Aranda R, Climent V et al. Truncating FLNC Mutations Are Associated With High-Risk Dilated and Arrhythmogenic Cardiomyopathies. Journal of the American College of Cardiology 2016:68(22):2440–51. Doi: 10.1016/jjacc.2016.09.927. [DOI] [PubMed] [Google Scholar]
- 92.Van Der Zwaag PA, Van Rijsingen IA, Asimaki A, Jongbloed JD, Van Veldhuisen DJ, Wiesfeld AC et al. Phospholamban R14del mutation in patients diagnosed with dilated cardiomyopathy or arrhythmogenic right ventricular cardiomyopathy: evidence supporting the concept of arrhythmogenic cardiomyopathy. European Journal of Heart Failure 2012:14(11): 1199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ et al. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med 2012:4(130): 130ra47 Doi: 10.1126/scitranslmed.3003552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Karakikes I, Stillitano F, Nonnenmacher M, Tzimas C, Sanoudou D, Termglinchan V et al. Correction of human phospholamban R14del mutation associated with cardiomyopathy using targeted nucleases and combination therapy. Nature Communications 2015:6:6955 Doi: 10.1038/ncomms7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Basso C, Thiene G, Corrado D, Angelini A, Nava A, Valente M. Arrhythmogenic right ventricular cardiomyopathy: dysplasia, dystrophy, or myocarditis? Circulation 1996:94(5):983–91. [DOI] [PubMed] [Google Scholar]
- 96.Asimaki A, Saffitz JE. The role of endomyocardial biopsy in ARVC: looking beyond histology in search of new diagnostic markers. Journal of Cardiovascular Electrophysiology 2011:22(1): 111–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lopez-Ayala JM, Pastor-Quirante F, Gonzalez-Carrillo J, Lopez-Cuenca D, Sanchez-Munoz JJ, Oliva-Sandoval MJ et al. Genetics of myocarditis in arrhythmogenic right ventricular dysplasia. Heart Rhythm 2015:12(4):766–73. [DOI] [PubMed] [Google Scholar]
- 98.Marcus FI, McKenna WJ, Sherrill D, Basso C, Bauce B, Bluemke DA et al. Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: proposed modification of the Task Force Criteria. European Heart Journal 2010:31(7):806–14. Doi: 10.1093/eurheartj/ehq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sen-Chowdhry S, Syrris P, Prasad SK, Hughes SE, Merrifield R, Ward D et al. Left-dominant arrhythmogenic cardiomyopathy: an under-recognized clinical entity. Journal of the American College of Cardiology 2008:52(25):2175–87. [DOI] [PubMed] [Google Scholar]
- 100.Gerull B, Heuser A, Wichter T, Paul M, Basson CT, McDermott DA et al. Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nature Genet 2004:36(11):1162–4. Doi: 10.1038/ng1461. [DOI] [PubMed] [Google Scholar]
- 101.Rampazzo A, Nava A, Malacrida S, Beffagna G, Bauce B, Rossi V et al. Mutation in human desmoplakin domain binding to plakoglobin causes a dominant form of arrhythmogenic right ventricular cardiomyopathy. American Journal of Human Genetics 2002:71(5):1200–6. Doi: 10.1086/344208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pilichou K, Nava A, Basso C, Beffagna G, Bauce B, Lorenzon A et al. Mutations in desmoglein-2 gene are associated with arrhythmogenic right ventricular cardiomyopathy. Circulation 2006:113(9): 1171–9. Doi: 10.1161/circulationaha.105.583674. [DOI] [PubMed] [Google Scholar]
- 103.Syrris P, Ward D, Evans A, Asimaki A, Gandjbakhch E, Sen-Chowdhry S et al. Arrhythmogenic right ventricular dysplasia/cardiomyopathy associated with mutations in the desmosomal gene desmocollin-2. American Journal of Human Genetics 2006:79(5):978–84. Doi: 10.1086/509122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McKoy G, Protonotarios N, Crosby A, Tsatsopoulou A, Anastasakis A, Coonar A et al. Identification of a deletion in plakoglobin in arrhythmogenic right ventricular cardiomyopathy with palmoplantar keratoderma and woolly hair (Naxos disease). Lancet 2000:355(9221):2119–24. Doi: 10.1016/s0140-6736(00)02379-5. [DOI] [PubMed] [Google Scholar]
- 105.Gandjbakhch E, Redheuil A, Pousset, Charron P, Frank R. Clinical Diagnosis, Imaging, and Genetics of Arrhythmogenic Right Ventricular Cardiomyopathy/Dysplasia. Journal of the American College of Cardiology 2018:72(7):784–804.• This is an excellent review of ARVC/D by the group involved in its early description.
- 106.Corrado D, Link MS, Calkins H. Arrhythmogenic Right Ventricular Cardiomyopathy. The New England Journal of Medicine 2017:376(1):61–72. Doi: 10.1056/NEJMra1509267. [DOI] [PubMed] [Google Scholar]
- 107.Quarta G, Muir A, Pantazis A, Syrris P, Gehmlich K, Garcia-Pavia P et al. Familial Evaluation in Arrhythmogenic Right Ventricular Cardiomyopathy Impact of Genetics and Revised Task Force Criteria. Circulation 2011:123(23):2701–U99. Doi: 10.1161/circulationaha.110.976936. [DOI] [PubMed] [Google Scholar]
- 108.Groeneweg JA, Bhonsale A, James CA, te Riele AS, Dooijes D, Tichnell C et al. Clinical Presentation, Long-Term Follow-Up, and Outcomes of 1001 Arrhythmogenic Right Ventricular Dysplasia/Cardiomyopathy Patients and Family Members. Circulation Cardiovasc Genetics 2015:8(3):437–46. Doi: 10.1161/circgenetics.114.001003. [DOI] [PubMed] [Google Scholar]
- 109.Hodgkinson K, Connors S, Memer N, Haywood A, Young TL, McKenna W et al. The natural history of a genetic subtype of arrhythmogenic right ventricular cardiomyopathy caused by a p. S358L mutation in TMEM43. Clinical Genetics 2013:83(4):321–31. [DOI] [PubMed] [Google Scholar]
- 110.Bhonsale A, Groeneweg JA, James CA, Dooijes D, Tichnell C, Jongbloed JD et al. Impact of genotype on clinical course in arrhythmogenic right ventricular dysplasia/cardiomyopathy-associated mutation carriers. European Heart Journal 2015:36(14):847–55. [DOI] [PubMed] [Google Scholar]
- 111.James CA, Bhonsale A, Tichnell C, Murray B, Russell SD, Tandri H et al. Exercise increases age- related penetrance and arrhythmic risk in arrhythmogenic right ventricular dysplasia/cardiomyopathy-associated desmosomal mutation carriers. Journal of the American College of Cardiology 2013:62(14): 1290–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xu TH, Yang Z, Vatta M, Rampazzo A, Beffagna G, Pillichou K et al. Compound and Digenic Heterozygosity Contributes to Arrhythmogenic Right Ventricular Cardiomyopathy. Journal of the American College of Cardiology 2010:55(6):587–97. Doi: 10.1016/jjacc.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kalia SS, Adelman K, Bale SJ, Chung WK, Eng C, Evans JP et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2. 0): a policy statement of the American College of Medical Genetics and Genomics. Genetics in Medicine 2017:19(2):249.•• This policy statement provides an update regarding the clinician’s responsibility of reporting of secondary findings in clinical genetic sequencing.
- 114.Kim C, Wong J, Wen J, Wang S, Wang C, Spiering S et al. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature 2013:494(7435): 105–10. Doi: 10.1038/nature11799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Caspi O, Huber I, Gepstein A, Arbel G, Maizels L, Boulos M et al. Modeling of arrhythmogenic right ventricular cardiomyopathy with human induced pluripotent stem cells. Circulation Cardiovascular Genetics 2013:6(6):557–68. Doi: 10.1161/circgenetics.113.000188. [DOI] [PubMed] [Google Scholar]
- 116.Ma D, Wei H, Lu J, Ho S, Zhang G, Sun X et al. Generation of patient-specific induced pluripotent stem cell-derived cardiomyocytes as a cellular model of arrhythmogenic right ventricular cardiomyopathy. European Heart Journal 2013:34(15): 1122–33. Doi: 10.1093/eurheartj/ehs226. [DOI] [PubMed] [Google Scholar]
- 117.Tse HF, Ho JC, Choi SW, Lee YK, Butler AW, Ng KM et al. Patient-specific induced-pluripotent stem cells-derived cardiomyocytes recapitulate the pathogenic phenotypes of dilated cardiomyopathy due to a novel DES mutation identified by whole exome sequencing. Human Molecular Genetics 2013:22(7): 1395–403. Doi: 10.1093/hmg/dds556. [DOI] [PubMed] [Google Scholar]
- 118.Dick E, Kalra S, Anderson D, George V, Ritso M, Laval SH et al. Exon skipping and gene transfer restore dystrophin expression in human induced pluripotent stem cells-cardiomyocytes harboring DMD mutations. Stem Cells and Development 2013:22(20):2714–24. Doi: 10.1089/scd.2013.0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lin B, Li Y, Han L, Kaplan AD, Ao Y, Kalra S et al. Modeling and study of the mechanism of dilated cardiomyopathy using induced pluripotent stem cells derived from individuals with Duchenne muscular dystrophy. Disease Models & Mechanisms 2015:8(5):457–66. Doi: 10.1242/dmm.019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med 2014:20(6):616–23. Doi: 10.1038/nm.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kodo K, Ong SG, Jahanbani F, Termglinchan V, Hirono K, Inanloo RK et al. iPSC-derived cardiomyocytes reveal abnormal TGF-beta signalling in left ventricular non-compaction cardiomyopathy. Nature Cell Biology 2016:18(10): 1031–42. Doi: 10.1038/ncb3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ang YS, Rivas RN, Ribeiro AJS, Srivas R, Rivera J, Stone NR et al. Disease Model of GATA4 Mutation Reveals Transcription Factor Cooperativity in Human Cardiogenesis. Cell 2016:167(7): 1734–49.e22. Doi: 10.1016/j.cell.2016.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tucker NR, McLellan MA, Hu D, Ye J, Parsons VA, Mills RW et al. Novel Mutation in FLNC (Filamin C) Causes Familial Restrictive Cardiomyopathy. Circulation Cardiovascular Genetics 2017:10(6). Doi: 10.1161/circgenetics.117.001780. [DOI] [PMC free article] [PubMed] [Google Scholar]