Abstract
Introduction
Despite initial effectiveness of ALK tyrosine kinase inhibitors (TKIs) in patients with ALK+ non-small cell lung cancer (NSCLC), therapeutic resistance will ultimately develop. Serial tracking of genetic alterations detected in circulating tumor DNA (ctDNA) can be an informative strategy to identify response and resistance. This study evaluated the utility of analyzing ctDNA as a function of response to ensartinib, a potent second-generation ALK TKI.
Methods
Pre-treatment plasma was collected from 76 patients with ALK+ NSCLC who were ALK TKI naïve or had received prior ALK TKI, and analyzed for specific genetic alterations. Longitudinal plasma samples were analyzed from a subset (N=11) of patients. Analysis of pre-treatment tumor biopsies from 22 patients was compared with plasma.
Results
Disease-associated genetic alterations were detected in 74% (56/76) of patients, the most common being EML4-ALK. Concordance of ALK fusion between plasma and tissue was 91% (20/22). Twenty-four ALK kinase domain mutations were detected in 15 patients, all had previously received an ALK TKI; G1269A was the most prevalent (4/24). Patients with a detectable EML4-ALK variant 1 (V1) fusion had improved response (9/17, 53%) to ensartinib compared to patients with EML4-ALK V3 fusion (1/7, 14%). Serial changes in ALK alterations were observed during therapy.
Conclusions
Clinical utility of ctDNA was demonstrated, both at pre-treatment by identifying a potential subgroup of ALK+ NSCLC patients that may derive more benefit from ensartinib and longitudinally by tracking resistance. Prospective application of this technology may translate to improved outcomes for NSCLC patients treated with ALK TKIs.
Keywords: NSCLC, ctDNA, next-generation sequencing, ALK TKI, liquid biopsies
Introduction
Chromosomal rearrangements involving the gene that encodes for the anaplastic lymphoma kinase (ALK) are detected in 3–8% of NSCLCs1–3, and testing for ALK rearrangements is considered standard clinical practice for patients with metastatic disease. Successful clinical trials have led to FDA approval of multiple ALK tyrosine kinase inhibitors (TKIs) in both the first and later lines of therapy.4–8 Despite achieving initial responses, patients eventually progress, typically within 1–2 years. Mechanisms of acquired resistance to ALK TKIs include both ‘on-target’ genomic alterations, i.e., mutations in the ALK tyrosine kinase domain (KD) and amplification of the ALK fusion, as well as activation of bypass signaling networks.9–11 Thus, accurate identification of tumor molecular evolution during therapy could have significant impact on the selection of therapy likely to have the greatest impact on patient outcomes.
The gold standard for detecting specific tumor mutations is analyzing tumor tissue, yet the lack of sufficient tissue available from certain biopsy procedures (e.g., fine needle aspirations) and the threat of complications arising from tissue biopsies remain obstacles for obtaining genetic testing in some cases.12 Additionally, biopsy of a single lesion may not provide a true sample of the heterogeneous molecular landscape of the tumor.13 As a result, “liquid biopsies” that noninvasively quantify the molecular profile of tumors from circulating-tumor DNA (ctDNA) are an area of growing interest.14–17 The question remains, however, whether analyzing ctDNA represents an adequate surrogate to tissue biopsies and a feasible way to monitor disease response and resistance, especially within the scope of identifying ALK fusions and development of resistance mechanisms during ALK TKI therapy.
In this study, plasma was collected from ALK+ NSCLC patients enrolled on the phase I/II eXalt2 trial investigating safety and efficacy of ensartinib, a second-generation ALK inhibitor.18,19 A preliminary report from this trial demonstrated that ensartinib was generally well tolerated, with rash being the most common toxicity.19 Additionally, ensartinib had good clinical activity in patients who were ALK TKI naïve or those who had received prior crizotinib, as well as in patients with lesions in central nervous system. Results from the phase I/II study formed the basis for the ongoing eXalt3 study () comparing ensartinib to crizotinib in first line therapy. The purpose of the current study was to evaluate the feasibility of ctDNA next-generation sequencing (NGS) to identify actionable genomic alterations, as well as monitor response and development of resistance to ensartinib. Additionally, we analyzed the concordance between ctDNA and paired tumor tissue, as well as the association of total detected ctDNA with progression-free survival (PFS).
Material and Methods
Serial plasma samples were collected from 76 ALK+ NSCLC patients enrolled on the phase I/II multi-cohort eXalt2 trial ().19 Cohorts included patients who: a) were ALK TKI naïve, b) received prior crizotinib only, and c) received crizotinib and at least one second-generation ALK TKI. Archival tumor tissue was analyzed in a subset of patients (n=25), as tissue collection was not mandated for patients enrolled on the study. This study was approved by the review boards at all participating institutions, and all patients provided written informed consent. The study was conducted in accordance with good clinical practice and the Declaration of Helsinki and its amendments.
Targeted NGS analysis
Materials and methods describing the isolation of DNA from plasma and tissue are included in the Supplementary Methods. Analysis of tumor mutations using the Resolution Bioscience targeted hybrid-capture system has been described previously.20,21 Briefly, tissue and plasma samples collected at the time of enrollment (baseline) were hybridized to a panel of probes targeting single nucleotide variants (SNVs) and insertions and deletions (indels) in ALK, BRAF, EGFR, KRAS, MAP2K1, MET, NRAS, NTRK1, PIK3CA, RET, ROS1, and TP53. Specifically for ALK, the assay covers fusions in intron 19 and SNVs and indels in exons 20 through 29. This panel also targets gene fusion regions within ALK, RET, ROS1, and NTRK1, and contains additional probes for enhanced detection of copy variation in MET, as well as control probes on each autosome. After analysis of the corresponding tissue and the initial plasma time point, longitudinal plasma samples were analyzed with a reduced panel targeting ALK and TP53 only. Additional details can be found in the Online-only Supplement.
Statistical analysis
The concordance between genetic alterations detected in the plasma and tissue was calculated in the subset of patients that had matched plasma and tissue samples at baseline. Kaplan-Meier method was used to estimate PFS, and a Log-rank test was used to evaluate difference between groups. A Cox proportional hazards model was used to assess the association between PFS and ctDNA levels at baseline, which were measured as genomic equivalents (GEs) in the plasma. A Wilcoxon rank sum test was used to compare the GEs between responders and non-responders; responders included any patient that experienced at least a partial tumor response to ensartinib. Two-sided p<0.05 was considered statistically significant. As the eXalt2 trial was still ongoing at the time this manuscript was prepared, the data cutoff for all analyses was April 1, 2018.
In vitro ALK inhibitor sensitivity
Methods for the in vitro ALK sensitivity experiments are presented in the Supplementary Methods. Primers for site-directed mutagenesis and Sanger Sequencing are presented in Supplemental Tables 2 and 3, respectively.
Results
Patient characteristics
We collected and analyzed baseline plasma samples from 76 patients with ALK+ NSCLC (Table 1, Supplemental Figure 1). Of note, pre-ensartinib plasma samples analyzed on cycle 1 day 1 were either from pooled pharmacokinetic samples (Supplemental Table 3, n=60, pre-dose and two to three hours post-dose) or one biomarker sample collected pre-dose (n=16). We also collected and analyzed longitudinal samples from 11 patients, who were selected based on availability of samples, response to ensartinib, and/or the presence of pre-treatment genetic alterations of interest. The median age was 56 years and 53% of patients were female. Of the entire cohort, 22% (17/76) were ALK-TKI naïve, 49% (37/76) had received prior crizotinib, 20% (15/76) had received prior crizotinib plus one second-generation ALK TKI (ceritinib or alectinib), and 9% (7/76) had received prior crizotinib plus two second-generation ALK TKIs (ceritinib and either alectinib or brigatinib).
Table 1:
Patient baseline characteristics
| Characteristic | Patients (%) n=76 |
|---|---|
| Sex − no. (%) | |
| Male | 36 (47%) |
| Female | 40 (53%) |
| Age − no. (%) | |
| Median | 56 |
| Range | 22–82 |
| Race − no. (%) | |
| White | 58 (76%) |
| Black/African American | 2 (2%) |
| Asian | 12 (16%) |
| Other/Unknown | 4 (5%) |
| Smoking History − no. (%) | |
| Current | 3 (4%) |
| Former | 26 (34%) |
| Never | 47 (62%) |
| No. of prior treatments − no. (%) | |
| 0 | 14 (18%) |
| 1 | 17 (22%) |
| 2 | 18 (24%) |
| 3 | 10 (13%) |
| ≥ 4 | 17 (22%) |
| Prior ALK TKI Treatment − no. (%) | |
| ALK TKI Naïve | 17 (22%) |
| Prior Crizotinib only | 37 (49%) |
| Prior Crizotinib and Ceritinib | 9 (12%) |
| Prior Crizotinib and Alectinib | 6 (8%) |
| Prior Crizotinib, Ceritinib, and Alectinib | 6 (8%) |
| Prior Crizotinib, Ceritinib, and Brigatinib | 1 (1%) |
no: number; ALK: anaplastic lymphoma kinase; TKI: tyrosine kinase inhibitor
Detection of disease-associated variants in baseline plasma samples
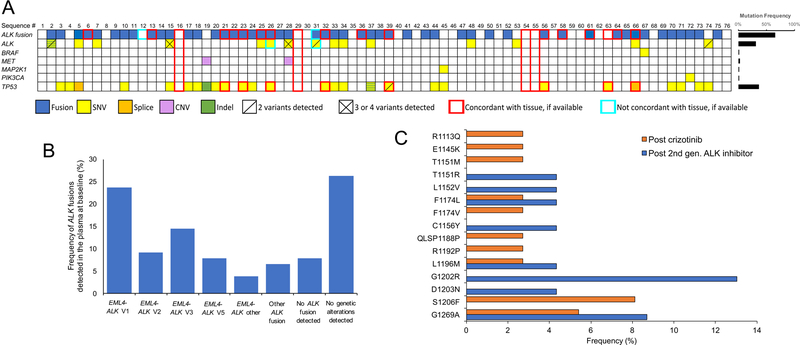
In total, we detected disease-associated genomic alterations in the ctDNA of 56 (74%) of 76 patients (Figure 1A), including SNVs, indels, and fusions, with ALK fusions being the most common alteration detected (50/76, 66%). Sufficient input cell-free DNA was obtained and sequenced but no somatic variants were detected for 20 patients. Individual patient information regarding ALK fusion (variant and breakpoints), ALK KD mutations, and details of prior ALK TKIs and response to ensartinib are listed in Supplemental Tables 4 and 5. An EML4-ALK fusion was detected in 45 patients, with a range of variants (Figure 1B). A PRKAR1A-ALK fusion was detected in one patient, which was confirmed via tissue NGS. Another patient had an AKAP8L-ALK fusion. Three other patients had a noncoding-ALK rearrangement that could not be mapped to a specific fusion partner, but each patient responded to ensartinib (Supplemental Table 4).
Figure 1. Detection of molecular alterations in plasma from patients with ALK+ NSCLC enrolled in the eXALT2 trial.
(A) Summary of all mutations identified by individual patient at the beginning of treatment with ensartinib. Alterations are color-coded per the figure legend below the image. The red or brown line around a particular mutation (or lack thereof) means that the mutation is either confirmed or not confirmed in the tissue, respectively. The mutation frequencies for each gene are graphed in the right panel, with the denominator equal to total number of patients (i.e., 76). Boxes with yellow and green alternating strips represent a gene for which both an SNV (yellow) and an indel (green) were detected. KRAS, NRAS, NTRK1, RET, and ROS1 were included in the NGS panel; however, no alterations in these genes were detected. ALK, anaplastic lymphoma kinase gene; MET, mesenchymal-epithelial transition gene; MAP2K, mitogen-activated protein kinase 2 gene; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha gene; TP53, tumor protein p53 gene. (B) Frequency of various ALK fusions detected in the plasma at the time of study entrance. “EML4-ALK Other” includes three patients with EML4-ALK fusions that had breakpoints in regions that could not be mapped to a specific variant. An ALK fusion of any kind was not detected in six patients. Note: frequencies in the bar graphs are expressed based upon the total number of patients (n=76). (C) Frequency and distribution of ALK kinase domain mutations detected across the study population. Orange bars represent samples from patients who had received prior crizotinib. Blue bars represent samples from patients who had received crizotinib and at least one second-generation ALK inhibitor. Frequencies are expressed based upon the total number of patients that received post-crizotinib (n=37) or received post-crizotinib and a second-generation ALK inhibitor (n=23).
Twenty-four ALK KD mutations were observed in 15 patients, with the most prevalent being G1269A (Figure 1C). Overall mutation frequency in patients who received prior crizotinib as their only prior ALK TKI was 24% (9/37). In the patients that received crizotinib and at least one second-generation ALK TKI, ALK KD mutations were detected in 27% (6/22). ALK fusions were not identified in ctDNA of two patients with a secondary ALK mutation detected in the plasma, despite tumor samples from these patients being positive for an ALK rearrangement by FISH; one patient was previously treated with crizotinib and the other received crizotinib followed by alectinib. We also detected alterations in five other genes (TP53, BRAF, MET, MAP2K1, and PIK3CA) within our patient population (Supplemental Table 6). Most notably, TP53 mutations were detected in plasma from 27/76 patients (35%). Additionally, a copy number variant in MET was detected in two patients. KRAS, NRAS, NTRK1, RET, and ROS1 were included in the NGS panel; however, no alterations in these genes were detected.
Concordance between tissue and plasma genotypes
NGS was also performed on pre-ensartinib tumor tissue in 25 patients; however, NGS analysis of tumor tissue from three patients failed due to insufficient tissue. Tumor and blood NGS testing results for detection of an ALK fusion matched in 20/22 (91%); 15/20 being fusion positive and 5/20 being fusion negative. The patient with the PRKAR1A-ALK fusion in ctDNA was confirmed in the tissue; this patient also had a TP53 SNV that was confirmed in tissue. One of the five fusion negative patients had one non-ALK mutation (#63, TP53) detected in tissue and plasma; notably, this patient was ALK TKI naive and subsequently progressed on ensartinib at the first imaging time point (i.e., 8 weeks), suggesting a false positive ALK FISH result. The other four fusion negative (in both ctDNA and tissue NGS) patients did not have any disease-associated genetic alterations detected in the plasma (#16, #29, #54, #55); all four patients, however, experienced a partial response (PR) to ensartinib. Two of these four patients were ALK TKI naïve (#16, #55), while the other two (#29, #54) had previously received crizotinib as their only ALK TKI. All eight patients with TP53 mutations detected in pre-treatment tissue had the same mutation detected in ctDNA.
Of the 22 patients with tissue NGS, four patients had ALK KD mutations detected in plasma that were not detected in the tissue. One patient (#25, post crizotinib) had a G1269A mutation (allelic frequency [AF] 0.1%) detected in the plasma, while a L1196M mutation (AF 0.04%) mutation was detected in tissue (Supplemental Table 4); this patient demonstrated a PR to ensartinib that lasted for six months. Another patient (#66, post crizotinib and ceritinib) who experienced a PR to ensartinib lasting four months had a G1202R mutation detected only in ctDNA (Supplemental Table 4). One patient who received prior crizotinib and ceritinib had D1203N (AF 0.81%) and C1156Y (AF 0.47%) mutations detected in plasma but not in tumor; this patient progressed on ensartinib at the first imaging time point (patient #31, Supplemental Table 4). S1206F (AF 0.28%) was detected in the plasma of the fourth patient following crizotinib; however, aTP53 and mutation detected in ctDNA were detected in both plasma and tissue (patient #26, Supplemental Table 6). Collectively, these data suggest the notion that single-site biopsies may not fully capture the spatial heterogeneity of mutations that might exist in sub-clones of tumors.
Correlation between baseline ctDNA findings and response to ensartinib
As previous retrospective studies have demonstrated differences in sensitivity to ALK TKIs according to EML4-ALK fusion variant (as defined by different genomic breakpoints within EML4),22–24 we also examined the relationship between the detected variant in ctDNA and clinical response to ensartinib. Within our cohort, 39 patients had an EML4-ALK fusion detected in plasma and were efficacy-evaluable, which, similar to the parent clinical study19, is defined as patients who received an effective dose of ensartinib (doses ≥200mg) and had at least one post-baseline imaging assessment. EML4-ALK variant 1 (V1) was detected in 17/39 (44%) patients, EML4-ALK variant 3 (V3) was detected in 7/39 (18%), and other EML4-ALK variants (i.e., non-V1, non-V3) were detected in 12/39 (31%) of patients (Figure 1B, Supplemental Tables 4–5). The response rate (RR) in the efficacy-evaluable subcohort with an EML4-ALK V1 fusion was 53% (9/17). However, the RR was only 14% (1/7) in patients with a V3 fusion, despite one V3 fusion patient being on study >12 months with stable disease (SD). In patients that had received crizotinib as their only prior ALK TKI, RR was 78% (7/9) and 25% (1/4) for V1 and V3 fusion cohorts, respectively. Pooled ORR for patients with other EML4-ALK variants was 58% (7/12); RR for those patients that had received crizotinib as their only prior ALK TKI was 67% (2/3). Excluding those ALK TKI naïve patients, the RRs for the V1 and V2/V5 fusion cohorts were 53% (8/15) and 38% (3/8), respectively. In this study, the EML4-ALK V3 fusion was not detected in any of the ALK TKI naïve patients.
PFS curves for patients with EML4-ALK fusion variants detected in the plasma at baseline are presented in Supplemental Figure 2. Within the efficacy-evaluable cohort, median PFS of patients exhibiting V1 and V3 fusion variants was 8.2 months (95% confidence interval [CI]: 2.1–11.7) and 1.9 months (95% CI: 1.8-not estimable), respectively. Pooled median PFS for patients with V2 or V5 fusion variants was 5.5 months (95% CI: 1.7–23.8). In patients that had received crizotinib as their only prior ALK TKI, the median PFS for the V1 and V3 cohorts was 10.9 months (95% CI: 5.4–16.4) and 2.2 months (95% CI: 1.8-not estimable), respectively. Excluding ALK TKI naïve patients, the median PFS for the V1 and V2/V5 cohorts was 7.4 months (95% CI: 1.8–11.6) and 4.4 months (95% CI: 1.5-not estimable), respectively.
Next, we evaluated whether clinical outcomes stratified with amount of ctDNA detected in baseline plasma samples. A Cox regression analysis showed that for every 1000 unit increase in GEs detected at baseline, the risk of disease progression significantly increased by 11% (hazard ratio = 1.11, 95% CI: 1.03–1.19). Additionally, a Wilcoxon rank sum test showed that the total plasma GEs in the patients that responded to ensartinib was significantly lower at baseline (p=0.02, Supplemental Figure 3).
Lastly, we questioned whether baseline mutations in genes other than ALK portended response to ensartinib (Supplemental Table 6). Patients with a TP53 mutation at the start of the trial experienced significantly shorter median PFS compared with patients without TP53 mutation (3.7 months vs. 11.6 months, respectively, p=0.006). MET amplifications were observed in plasma from two patients prior to ensartinib therapy (patient #19 and patient #28, Figure 1A). MET amplification has previously been demonstrated as a mechanism of resistance to second generation ALK TKIs, such as alectinib.25 One of these patients was unevaluable for response and the other had progressive disease as best response to ensartinib, suggestive of primary resistance to ensartinib therapy. Of note, plasma from this patient failed to reveal the presence of any ALK alteration.
Evolution of response and resistance during molecular surveillance
We sought to understand the activity of ensartinib in patients with baseline ALK KD mutations. Fifteen patients had at least one baseline ALK mutation (Supplemental Table 4); 9/12 efficacy-evaluable patients had SD or PR to ensartinib. Activity was seen in patients whose plasma harbored T1151M, L1152V, F1174V, L1196M, and G1269A; variable activity was observed between two patients that harbored a G1202R mutation (Supplemental Table 4). The difference in sensitivity to ensartinib between these two patients is thought to be influenced by the different AFs detected; the patient with a PR exhibited a G1202 mutation at an AF of 0.7% whereas the mutation was detected at an AF of 2.1% for the patient with a PD. Furthermore, as stated above, the G1202R mutation was only detected in the plasma of the patient with the PR and not the tumor tissue, suggesting that this subclone represented a small portion of the tumor.
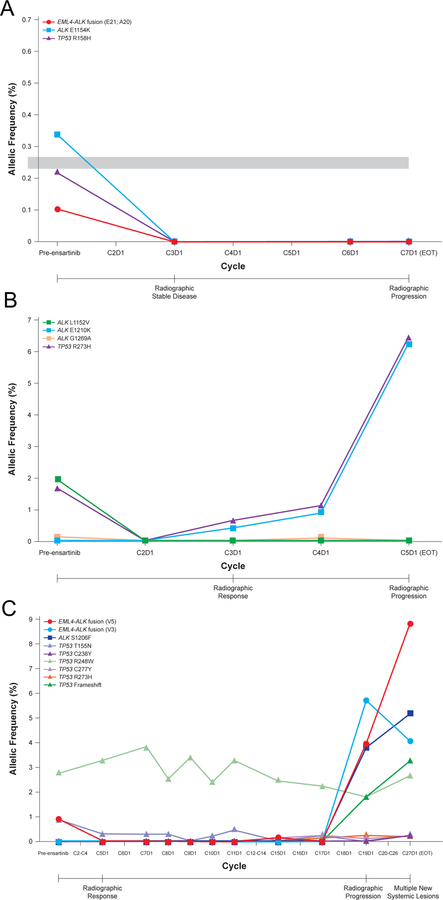
From our entire cohort, longitudinal plasma samples were analyzed from 11 patients (Supplemental Figures 4–11). These cases were selected based on sample availability, the patient’s response to ensartinib, and/or the presence of interesting ALK fusions or mutations detected at baseline. For example, an ALK E1154K mutation was detected in the plasma from patient #34, who had received prior crizotinib (Figure 2A). To the best of our knowledge, this mutation has not previously been reported in clinical samples. We verified in vitro that ALK E1154K is sensitive to ensartinib (Supplemental Figure 12). In agreement with these findings, the E1154K mutation was undetectable after two cycles of ensartinib and did not reappear upon ensartinib progression.
Figure 2. Longitudinal assessment of molecular alterations detected in plasma during ensartinib treatment.
Graphs illustrate the change in allelic fraction of ALK and TP53 alterations for patient #34 (A), patient #74 (B), and patient #39 (C) during treatment with ensartinib. The time of radiographic response and progression are annotated below the x-axis. An x-axis point with a missing marker designates that plasma was not analyzed at that time point. The duration of each treatment cycle is 28 days.
Next, we sought to understand the mutations that emerge upon ensartinib progression. Patient #74 received prior treatment with crizotinib and ceritinib before enrolling in the ensartinib trial. At baseline (prior to the start of ensartinib), ALK L1152V (AF: 1.9%) and G1269A (AF: 0.13%) mutations were present, both of which disappeared with ensartinib treatment. The patient experienced a PR on ensartinib. However, a new ALK mutation, E1210K, was present at the time of radiographic progression (Figure 2B). Glutamic acid (E) 1210 maps to the ribose-binding pocket of the ALK tyrosine kinase domain26 and has been reported in patients with crizotinib and brigatinib resistance.9 In vitro data (Figure 3) corroborates E1210K as a potential ensartinib vulnerability. Interestingly, this mutation appeared in plasma prior to radiographic progression, suggesting the potential utility of ctDNA analysis in monitoring the emergence of putative resistance mechanisms.
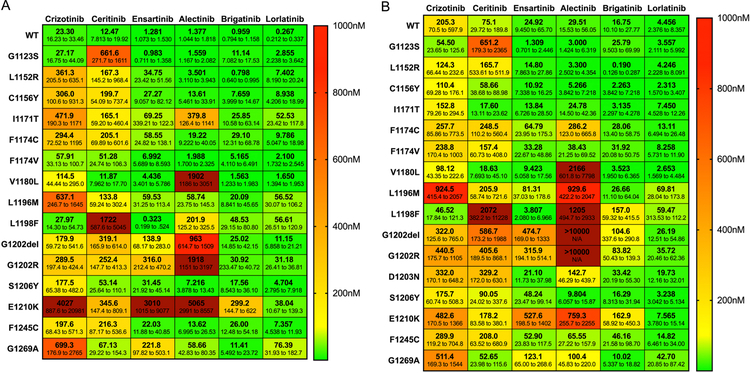
Figure 3. Efficacy of ensartinib against ALK kinase domain mutations.
BA/F3 cells expressing the indicated ALK kinase domain mutations within the context of EML4-ALK variant 1 (A) or EML4-ALK variant 3 (B) were treated with increasing concentrations of crizotinib, ceritinib, ensartinib, alectinib, brigatinib, and lorlatinib (0–10 µmol/L) for 72 hours. Cell titer blue assays were performed to assess cell viability. Experiments were performed with six replicates per drug concentration and repeated three times. IC50 values with 95% confidence intervals were generated with the data from the dose response curves using GraphPad Prism 7. All variants were tested three independent times, except for V1 E1210K, V3 D1203N, and V3 E1210K, which were tested two independent times.
As an additional example, an EML4-ALK V5 fusion was detectable in both the plasma and tissue of the ALK TKI naïve patient #39 (Figure 2C). As expected, no ALK KD mutations were detected at the start of the trial, as the patient had not previously been exposed to an ALK TKI. This patient experienced a PR lasting 11 cycles, coinciding with a decrease in the EML4-ALK V5 AF. At the time of disease progression, the original EML4-ALK V5 fusion re-emerged in the plasma, and interestingly, there was also emergence of an EML4-ALK V3 fusion and an ALK S1206F mutation at AF 5.2%. Ensartinib is active against S1206F mutation in vitro (IC50<50nM for V1 and V3, Figure 3). It is unclear from this case whether S1206F mutation was truly driving ensartinib resistance, or if there were other genetic alterations present that were not evaluated on our panel. Interestingly, a new TP53 frameshift mutation was also detectable at progression, with six distinct TP53 variants found in the plasma from this patient. Overall, these results suggest underlying tumor heterogeneity within this individual patient.
Comparison of patient ctDNA with in vitro ALK inhibitor sensitivity analysis
We examined the activity of crizotinib, ensartinib, and other second- and third-generation ALK TKIs (ceritinib, alectinib, brigatinib, and lorlatinib) against “wild-type” EML4-ALK V1 (E12;A20) and V3 (E6;A20), as well as both EML4-ALK fusions harboring various ALK mutations (Figures 3A,B and Supplemental Figure 13). Compared to the other ALK TKIs, ensartinib was the most potent inhibitor against G1123S and L1198F mutations; G1123S has previously been reported in a patient who exhibited resistance to ceritinib.27 Our in vitro data would suggest that samples exhibiting either of these mutations would not be sensitive to ceritinib, but could potentially be sensitive to ensartinib (IC50s<1nM). Conversely, our in vitro data suggest that ensartinib would be less active against G1202R (IC50=316nM) and G1269A (IC50=222nM) mutations. Indeed, one patient (patient #7) with a G1202R mutation at baseline did not respond to ensartinib. G1269A was present at baseline in another patient (Supplemental Figure 6, patient #25), fluctuated in the plasma throughout ensartinib, and then was again present at the time of radiographic progression. Additionally, one patient (Supplemental Figure 10, patient #66) had a G1202R mutation at baseline that was not detected after starting ensartinib but was again present along with a G1269A mutation at progression. It should be noted, however, that in vitro studies do not always predict results that will be observed clinically, which may explain why a few patients with G1202R or G1269A mutations responded to ensartinib. Although these patients responded, the response was not durable and these patients eventually developed disease progression on ensartinib.
Discussion
Personalized medicine offers the ability to tailor a patient’s cancer treatment based on a number of factors, including molecular status. This is especially true for patients with ALK+ NSCLC, where clinical outcome is significantly improved after treatment with an ALK TKI compared to traditional chemotherapy.4,28 Moreover, the responses of ALK+ NSCLC patients to specific ALK TKIs vary even though they share a molecular driver.6,8,19 Thus, tracking the evolution of resistance is imperative in this patient population. Serial tissue sampling remains the gold-standard for quantifying the genomic landscape of tumors, however it often fails to capture the dynamic and heterogeneous nature of acquired resistance. Here, we present, as proof of concept, findings from the eXalt2 study that highlight the potential clinical utility of hybrid-capture NGS of ctDNA at predicting response and acquired resistance to the ALK TKI ensartinib. Of note, this is the first study to highlight treatment response and resistance mechanisms to ensartinib, both in vivo and in vitro.
In our cohort of 76 patients, genetic alterations were detected in 74% (56/76) of plasma samples, which is similar to prior reports investigating the clinical utility of ctDNA.17,29,30 We observed plasma ALK mutations in 24% of patients who had prior crizotinib, and in 27% of patients receiving prior crizotinib and a second-generation ALK TKI (i.e., ceritinib or alectinib). These data are in accord with previous studies of crizotinib resistance, however, we detected a lower frequency of ALK KD mutations post second-generation ALK TKIs (ceritinib, alectinib) than previously reported in tissue biopsies by Gainor et al.9 or in ctDNA by McCoach et al31. This may be a sampling issue, as only 22 of 76 patients in this study had received a prior second-generation ALK TKI, or could be an artifact of the original study design that did not include standard collection and processing protocols for all plasma samples. However, of the 13 patients that did not have an ALK mutation detected in the plasma, 12 had an ALK fusion detected and the other patient had non-ALK mutations (TP53 and MAP2K1) detected, suggesting that these plasma samples contained sufficient ctDNA for analysis.
Overall, there was a high degree (91%) of fusion concordance between plasma and tissue NGS analyses, which is similar to the previous study by Dagogo-Jack et al. that reported a fusion concordance of 86% between plasma and tissue.30 Of note, tissue sequencing failed to detect ALK genetic alterations detected in the plasma of four patients, thereby providing supporting evidence that tissue sampling may not always fully capture the genetic heterogeneity of tumors.
In our cohort, 35% (27/76) of patients had TP53 mutations detected in the plasma at baseline. Similar percentage (36%) was observed in patients that had received at least one prior ALK TKI. Specifically in the cohort of patients that had prior crizotinib and at least one 2nd generation ALK TKI, 50% (11/22) of patients had TP53 mutations detected in the plasma at baseline. Our results were slightly higher than two previous studies by Gainor et al. and McCoach et al. that reported 36% (9/25, post ceritinib or alectinib) and 41% (36/88, prior ALK status could not be confirmed) of patients exhibited a TP53 mutation in their tumor or plasma, respectively.9,31
ORR and PFS were improved, albeit not significantly, in patients with an EML4-ALK V1 fusion compared to those with a V3 fusion. Even the pooled clinical outcome data with patients exhibiting either a V2 or V5 fusion variant was better than the V3 patients; however, these results were also not statistically different. The lack of statistical significance could be due to the small numbers of patients in each of these categories. Nonetheless, these results collectively suggest that ensartinib is more active against the EML4-ALK V1 variant, and the differential improvement in anti-tumor activity does not appear to depend on prior treatment with ALK TKIs. These results are especially important given the emerging studies that have reported differential sensitivities to ALK TKIs according to ALK variant.22–24 Prospective use of ctDNA analysis, specifically quantifying the ALK fusion variant, would be useful, for example, when defining eligibility criteria for future clinical trials evaluating ensartinib, as well as other ALK TKIs.
In addition to providing potential prognostic information at baseline, serial analysis of ctDNA could provide information regarding specific genetic determinants of acquired resistance. Although this study was limited by the small number of patients with longitudinal ctDNA analysis, we show that serial changes in ALK mutation AFs were detected while on treatment with ensartinib, demonstrating that analyzing ctDNA has the potential to track tumor response and the evolution of acquired resistance. We are not the first group to serially quantify ctDNA as a function of ALK TKI therapy. Dagogo-Jack et al. analyzed plasma ALK mutation kinetics during treatment and observed decreases in AFs of the ALK fusion and KD mutation with response to the ALK TKI, and then resurgence of the same genetic alteration at the time of disease progression.30 Wang et al. analyzed longitudinal blood samples from seven patients undergoing crizotinib and observed similar trends in the mutant AF of ALK and ctDNA concentration.17 Unique to our study, however, is the analysis of plasma samples collected during ensartinib where the resurgence or development of new genetic alterations that are driving tumor resistance could be detected prior to radiographic progression. Collectively, results from these studies suggest that serial monitoring of genetic biomarkers that are associated with acquired resistance is feasible. Application of this technology would allow for a physician to change therapy before progression, thereby reducing the time a patient receives an ineffective therapy and potentially improving clinical outcome by switching to another, more effective treatment. However, validation of such an approach would require evaluation in a prospective clinical trial. Additionally, the targeted NGS analysis performed on the longitudinal samples in this study only included two genes, thus limiting the number of potential mechanisms of resistance that could be detected; future studies will include a more comprehensive targeted NGS panel.
In conclusion, clinical utility of ctDNA was demonstrated in this study, both at pre-treatment by demonstrating a potential subgroup of ALK+ NSCLC patients who may derive more clinical benefit from ensartinib, and serially tracking genetic determinants of resistance. Prospective application of this technology could translate to improved outcomes for NSCLC patients treated with ALK TKIs.
Supplementary Material
Acknowledgements
The authors would like to thank the patients and their families for participating in this study. We also thank Yingjun Yan for her technical assistance, and Merrida Childress, PhD for thoughtful discussion regarding the interpretation of results.
Funding: This work was supported by Xcovery Holdings and Resolution Biosciences. Work in the CML laboratory is supported through the National Institutes of Health (NIH) and National Cancer Institute (NCI) R01CA121210 and P01CA129243, the Damon Runyon Foundation, the LUNGevity Foundation, the V Foundation, the Lung Cancer Foundation of America, and the International Association for the Study of Lung Cancer. CML was also supported through P30CA6848.
LH is a consultant/advisory board member for AbbVie, AstraZeneca, Bristol-Myers Squibb, Lilly, Incyte, EMD Serono, Merck, Roche-Genentech, Tessaro, and Xcovery, and reports receiving commercial research support from Boehringer Ingelheim. HAW is a consultant/advisory board member for AstraZeneca, Genentech/Roche (uncompensated), Merck (uncompensated), Novartis (uncompensated), Ariad (uncompensated) and has received commercial research grants to her institution for conduct of clinical trial work from Genentech/Roche, Novartis, Pfizer, Lilly, Celgene, Astrazeneca/Medimmune, Exelixis, Clovis Oncology, BMS, Gilead, Pharmacyclics, ACEA biosciences, Merck and Xcovery. KLR is a consultant/advisory board member for ARIAD Takeda, Exelixis, Guardant, Loxo, Genentech, and reports receiving commercial research grants from Xcovery, AbbVie, Acea, Adaptimmune, Boehringer Ingelheim, Bristol Myers Squibb, Genentech, Guardant, Janssen, Loxo Oncology, Seattle Genetics, Takeda, Zeno. TAL is a consultant/advisory board member for Roche-Genentech, Ariad, Takeda, AstraZeneca, Novartis, AbbVie, Bristol-Myers Squibb. GRB has served as a consultant for Abbvie, Adicet, Amgen, ARIAD, Bayer, Bristol-Myers Squibb, Celgene, Clovis, Merck, Novartis & Xcovery and has received research funding from Adaptimmune, Bayer, Bristol-Myers Squibb, Celgene, Exelixis, Genetech, GlaxoSmithKline, Hoffman-La Roche, Immatics, Incyte, Macrogenetics, MedImmune, Merck, Novartis, Torque & Xcovery. SPP is a scientific advisory board member for AstraZeneca, BMS, Illumina, Tempus, and Novartis, and receives research funding from BMS, Eli Lilly, Fate, Incyte, AstraZeneca/MedImmune, Merck, Pfizer, Roche/Genentech, Xcovery, Fate Therapeutics, Genocea, and Iovance. Research funds were distributed through University of California San Diego. GRO is a consultant for Takeda, AstraZeneca, Loxo, Inivata, and GRAIL. RES is a consultant/advisory board member for ARIAD and Takeda. JH, TS, KG, LPL, and ML are employees and shareholders of Resolution Biosciences. AH is an employee of Xcovery Holdings, Inc. KH and CL are employees of and have ownership interests (including patents) in Xcovery Holdings, Inc. CML has served as a consultant for Pfizer, Novartis, Astra Zeneca, Genoptix, Sequenom, Ariad, Takeda, Foundation Medicine, Blueprints Medicine, and Cepheid and has received research funding from Novartis, Astra Zeneca, and Xcovery (the funds were distributed through Vanderbilt University, not given to Dr. Lovly directly).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interests: All other authors do not have any conflicts to disclose.
References:
- 1.Perner S, Wagner PL, Demichelis F, Mehra R, Lafargue CJ, Moss BJ, Arbogast S, Soltermann A, Weder W, Giordano TJ, Beer DG, Rickman DS, Chinnaiyan AM, Moch H, Rubin MA. EML4-ALK fusion lung cancer: a rare acquired event. Neoplasia 2008;10:298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boland JM, Erdogan S, Vasmatzis G, Yang P, Tillmans LS, Johnson MR, Wang X, Peterson LM, Halling KC, Oliveira AM, Aubry MC, Yi ES. Anaplastic lymphoma kinase immunoreactivity correlates with ALK gene rearrangement and transcriptional up-regulation in non-small cell lung carcinomas. Hum Pathol 2009;40:1152–8. [DOI] [PubMed] [Google Scholar]
- 3.Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba II, Varella-Garcia M, Franklin WA, Aronson SL, Su PF, Shyr Y, Camidge DR, Sequist LV, Glisson BS, Khuri FR, Garon EB, Pao W, Rudin C, Schiller J, Haura EB, Socinski M, Shirai K, Chen H, Giaccone G, Ladanyi M, Kugler K, Minna JD, Bunn PA. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014;311:1998–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw AT, Kim DW, Nakagawa K, Seto T, Crino L, Ahn MJ, De Pas T, Besse B, Solomon BJ, Blackhall F, Wu YL, Thomas M, O’Byrne KJ, Moro-Sibilot D, Camidge DR, Mok T, Hirsh V, Riely GJ, Iyer S, Tassell V, Polli A, Wilner KD, Janne PA. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385–2394. [DOI] [PubMed] [Google Scholar]
- 5.Soria JC, Tan DS, Chiari R, Wu YL, Paz-Ares L, Wolf J, Geater SL, Orlov S, Cortinovis D, Yu CJ, Hochmair M, Cortot AB, Tsai CM, Moro-Sibilot D, Campelo RG, McCulloch T, Sen P, Dugan M, Pantano S, Branle F, Massacesi C, de Castro G Jr. First-line ceritinib versus platinum-based chemotherapy in advanced ALK-rearranged non-small-cell lung cancer (ASCEND-4): a randomised, open-label, phase 3 study. Lancet 2017;389:917–929. [DOI] [PubMed] [Google Scholar]
- 6.Peters S, Camidge DR, Shaw AT, Gadgeel S, Ahn JS, Kim DW, Ou SI, Perol M, Dziadziuszko R, Rosell R, Zeaiter A, Mitry E, Golding S, Balas B, Noe J, Morcos PN, Mok T, Investigators AT. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2017;377:829–838. [DOI] [PubMed] [Google Scholar]
- 7.Shaw AT, Felip E, Bauer TM, Besse B, Navarro A, Postel-Vinay S, Gainor JF, Johnson M, Dietrich J, James LP, Clancy JS, Chen J, Martini JF, Abbattista A, Solomon BJ. Lorlatinib in non-small-cell lung cancer with ALK or ROS1 rearrangement: an international, multicentre, open-label, single-arm first-in-man phase 1 trial. Lancet Oncol 2017;18:1590–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim DW, Tiseo M, Ahn MJ, Reckamp KL, Hansen KH, Kim SW, Huber RM, West HL, Groen HJM, Hochmair MJ, Leighl NB, Gettinger SN, Langer CJ, Paz-Ares Rodriguez LG, Smit EF, Kim ES, Reichmann W, Haluska FG, Kerstein D, Camidge DR. Brigatinib in Patients With Crizotinib-Refractory Anaplastic Lymphoma Kinase-Positive Non-Small-Cell Lung Cancer: A Randomized, Multicenter Phase II Trial. J Clin Oncol 2017;35:2490–2498. [DOI] [PubMed] [Google Scholar]
- 9.Gainor JF, Dardaei L, Yoda S, Friboulet L, Leshchiner I, Katayama R, Dagogo-Jack I, Gadgeel S, Schultz K, Singh M, Chin E, Parks M, Lee D, DiCecca RH, Lockerman E, Huynh T, Logan J, Ritterhouse LL, Le LP, Muniappan A, Digumarthy S, Channick C, Keyes C, Getz G, Dias-Santagata D, Heist RS, Lennerz J, Sequist LV, Benes CH, Iafrate AJ, Mino-Kenudson M, Engelman JA, Shaw AT. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov 2016;6:1118–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiao H, Lovly CM. Cracking the Code of Resistance across Multiple Lines of ALK Inhibitor Therapy in Lung Cancer. Cancer Discov 2016;6:1084–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCoach CE, Le AT, Gowan K, Jones K, Schubert L, Doak A, Estrada-Bernal A, Davies KD, Merrick DT, Bunn PA Jr., Purcell WT, Dziadziuszko R, Varella-Garcia M, Aisner DL, Camidge DR, Doebele RC. Resistance Mechanisms to Targeted Therapies in ROS1(+) and ALK(+) Non-small Cell Lung Cancer. Clin Cancer Res 2018;24:3334–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sholl LM, Aisner DL, Varella-Garcia M, Berry LD, Dias-Santagata D, Wistuba II, Chen H, Fujimoto J, Kugler K, Franklin WA, Iafrate AJ, Ladanyi M, Kris MG, Johnson BE, Bunn PA, Minna JD, Kwiatkowski DJ, Investigators L. Multi-institutional Oncogenic Driver Mutation Analysis in Lung Adenocarcinoma: The Lung Cancer Mutation Consortium Experience. J Thorac Oncol 2015;10:768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Bruin EC, McGranahan N, Swanton C. Analysis of intratumor heterogeneity unravels lung cancer evolution. Mol Cell Oncol 2015;2:e985549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson JC, Yee SS, Troxel AB, Savitch SL, Fan R, Balli D, Lieberman DB, Morrissette JD, Evans TL, Bauml J, Aggarwal C, Kosteva JA, Alley E, Ciunci C, Cohen RB, Bagley S, Stonehouse-Lee S, Sherry VE, Gilbert E, Langer C, Vachani A, Carpenter EL. Detection of Therapeutically Targetable Driver and Resistance Mutations in Lung Cancer Patients by Next-Generation Sequencing of Cell-Free Circulating Tumor DNA. Clin Cancer Res 2016;22:5772–5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwaederle M, Patel SP, Husain H, Ikeda M, Lanman R, Banks KC, Talasaz A, Bazhenova L, Kurzrock R. Utility of genomic assessment of blood-derived circulating tumor DNA (ctDNA) in patients with advanced lung adenocarcinoma. Clin Cancer Res 2017;23:5101–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yanagita M, Redig AJ, Paweletz CP, Dahlberg SE, O’Connell A, Feeney N, Taibi M, Boucher D, Oxnard GR, Johnson BE, Costa DB, Jackman DM, Janne PA. A Prospective Evaluation of Circulating Tumor Cells and Cell-Free DNA in EGFR-Mutant Non-Small Cell Lung Cancer Patients Treated with Erlotinib on a Phase II Trial. Clin Cancer Res 2016;22:6010–6020. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Tian PW, Wang WY, Wang K, Zhang Z, Chen BJ, He YQ, Li L, Liu H, Chuai S, Li WM. Noninvasive genotyping and monitoring of anaplastic lymphoma kinase (ALK) rearranged non-small cell lung cancer by capture-based next-generation sequencing. Oncotarget 2016;7:65208–65217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lovly CM, Heuckmann JM, de Stanchina E, Chen H, Thomas RK, Liang C, Pao W. Insights into ALK-driven cancers revealed through development of novel ALK tyrosine kinase inhibitors. Cancer Res 2011;71:4920–4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horn L, Infante JR, Reckamp KL, Blumenschein GR, Leal TA, Waqar SN, Gitlitz BJ, Sanborn RE, Whisenant JG, Du L, Neal JW, Gockerman JP, Dukart G, Harrow K, Liang C, Gibbons JJ, Holzhausen A, Lovly CM, Wakelee HA. Ensartinib (X-396) in ALK-Positive Non-Small Cell Lung Cancer: Results from a First-in-Human Phase I/II, Multicenter Study. Clin Cancer Res 2018;24:2771–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paweletz CP, Sacher AG, Raymond CK, Alden RS, O’Connell A, Mach SL, Kuang Y, Gandhi L, Kirschmeier P, English JM, Lim LP, Janne PA, Oxnard GR. Bias-Corrected Targeted Next-Generation Sequencing for Rapid, Multiplexed Detection of Actionable Alterations in Cell-Free DNA from Advanced Lung Cancer Patients. Clin Cancer Res 2016;22:915–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raymond CK, Hernandez J, Karr R, Hill K, Li M. Collection of cell-free DNA for genomic analysis of solid tumors in a clinical laboratory setting. PLoS One 2017;12:e0176241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida T, Oya Y, Tanaka K, Shimizu J, Horio Y, Kuroda H, Sakao Y, Hida T, Yatabe Y. Differential Crizotinib Response Duration Among ALK Fusion Variants in ALK-Positive Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3383–3389. [DOI] [PubMed] [Google Scholar]
- 23.Zheng D, Wang R, Zhang Y, Pan Y, Cheng X, Cheng C, Zheng S, Li H, Gong R, Li Y, Shen X, Sun Y, Chen H. Prevalence and clinicopathological characteristics of ALK fusion subtypes in lung adenocarcinomas from Chinese populations. J Cancer Res Clin Oncol 2016;142:833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin JJ, Zhu VW, Yoda S, Yeap BY, Schrock AB, Dagogo-Jack I, Jessop NA, Jiang GY, Le LP, Gowen K, Stephens PJ, Ross JS, Ali SM, Miller VA, Johnson ML, Lovly CM, Hata AN, Gainor JF, Iafrate AJ, Shaw AT, Ou SI. Impact of EML4-ALK Variant on Resistance Mechanisms and Clinical Outcomes in ALK-Positive Lung Cancer. J Clin Oncol 2018;36:1199–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gouji T, Takashi S, Mitsuhiro T, Yukito I. Crizotinib can overcome acquired resistance to CH5424802: is amplification of the MET gene a key factor? J Thorac Oncol 2014;9:e27–8. [DOI] [PubMed] [Google Scholar]
- 26.Zhang S, Wang F, Keats J, Zhu X, Ning Y, Wardwell SD, Moran L, Mohemmad QK, Anjum R, Wang Y, Narasimhan NI, Dalgarno D, Shakespeare WC, Miret JJ, Clackson T, Rivera VM. Crizotinib-resistant mutants of EML4-ALK identified through an accelerated mutagenesis screen. Chem Biol Drug Des 2011;78:999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toyokawa G, Inamasu E, Shimamatsu S, Yoshida T, Nosaki K, Hirai F, Yamaguchi M, Seto T, Takenoyama M, Ichinose Y. Identification of a Novel ALK G1123S Mutation in a Patient with ALK-rearranged Non-small-cell Lung Cancer Exhibiting Resistance to Ceritinib. J Thorac Oncol 2015;10:e55–7. [DOI] [PubMed] [Google Scholar]
- 28.Kiura K, Imamura F, Kagamu H, Matsumoto S, Hida T, Nakagawa K, Satouchi M, Okamoto I, Takenoyama M, Fujisaka Y, Kurata T, Ito M, Tokushige K, Hatano B, Nishio M. Phase 3 study of ceritinib vs chemotherapy in ALK-rearranged NSCLC patients previously treated with chemotherapy and crizotinib (ASCEND-5): Japanese subset. Jpn J Clin Oncol 2018;48:367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Almodovar K, Iams WT, Meador CB, Zhao Z, York S, Horn L, Yan Y, Hernandez J, Chen H, Shyr Y, Lim LP, Raymond CK, Lovly CM. Longitudinal Cell-Free DNA Analysis in Patients with Small Cell Lung Cancer Reveals Dynamic Insights into Treatment Efficacy and Disease Relapse. J Thorac Oncol 2018;13:112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dagogo-Jack I, Brannon AR, Ferris LA, Campbell CD, Lin JJ, Schultz KR, Ackil J, Stevens S, Dardaei L, Yoda S, Hubbeling H, Digumarthy SR, Riester M, Hata AN, Sequist LV, Lennes IT, Iafrate AJ, Heist RS, Azzoli CG, Farago AF, Engelman JA, Lennerz JK, Benes CH, Leary RJ, Shaw AT, Gainor JF. Tracking the Evolution of Resistance to ALK Tyrosine Kinase Inhibitors through Longitudinal Analysis of Circulating Tumor DNA. JCO Precis Oncol 2018;2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCoach CE, Blakely CM, Banks KC, Levy B, Chue BM, Raymond VM, Le AT, Lee CE, Diaz J, Waqar SN, Purcell WT, Aisner DL, Davies KD, Lanman RB, Shaw AT, Doebele RC. Clinical Utility of Cell-Free DNA for the Detection of ALK Fusions and Genomic Mechanisms of ALK Inhibitor Resistance in Non-Small Cell Lung Cancer. Clin Cancer Res 2018;24:2758–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.