Abstract
Purpose
To identify genomic imbalances and candidate loci in idiopathic male infertility.
Methods
Affymetrix CytoScan 750K Array was used to analyze genomic imbalances and candidate loci in 34 idiopathic infertile cases of different phenotypes (hypo-spermatogenesis, n = 8; maturation arrest, n = 7; and Sertoli cell-only syndrome, n = 13, severe oligozoospermia, n = 6, and 10 normozoospermic fertile men). Ten ethnically matched controls were screened for comparison.
Results
The cytogenetic array analysis detected a genomic gain at the 19p13.3 region in 9 (26.47%) cases, with the highest frequency in patients with Sertoli cell-only syndrome (SCOS) (38%). Its complete absence in the control group suggests its likely pathogenic nature. In addition to Y-classical, micro, and partial deletions, the duplication in 19p13.3 could serve as a unique biomarker for evaluation of infertility risk. The common region across the individuals harboring the duplication identified STK11, ATP5D, MIDN, CIRBP, and EFNA2 genes which make them strong candidates for further investigations. The largest duplicated region identified in this study displayed a major network of 7 genes, viz., CIRBP, FSTL3, GPX4, GAMT, KISS1R, STK11, and PCSK4, associated with reproductive system development and function. The role of chance was ruled out by screening of ethnically matched controls.
Conclusion
The result clearly indicates the significance of 19p13.3 duplication in infertile men with severe testicular phenotypes. The present study underlines the utility and significance of whole genomic analysis in the cases of male infertility which goes undiagnosed due to limitations in the conventional cytogenetic techniques and for identifying genes that are essential for spermatogenesis.
Electronic supplementary material
The online version of this article (10.1007/s10815-019-01547-1) contains supplementary material, which is available to authorized users.
Keywords: Copy number variations, Genomic imbalances, Cytogenetic microarray, Infertility, Spermatogenesis
Introduction
Infertility refers to the inability of a couple to attain pregnancy after 1 year of regular intercourse without any contraception (WHO) [1]. It affects around 10–15% of all couples and approximately 50% of all infertility cases are attributed to “male factor.” Human male infertility is a complex multifactorial disorder, commonly identified by disorders of sperm production and function. The etiology of human male infertility includes a diverse range of genetic factors such as cytogenetic abnormalities and Y chromosome deletions [2]; however, in most of the cases, the underlying cause remains unidentified or idiopathic [3]. It is largely recognized that, like many other complex traits, human male infertility is often genetic in origin [4, 5]. Evidences suggest that idiopathic male infertility, clinically described as azoospermia (nil sperm count) and severe oligozoospermia (sperm count < 5 million/mL), is the most frequent phenotype among infertile men [6]. Despite its high prevalence, the genetic origin of such phenotype remains largely unknown in humans.
The emergence of copy number variations (CNVs) has been recognized as a major source of genomic variation and genetic diversity [7, 8]. CNVs are the sub-microscopic chromosomal deletions or duplications that involve > 1-kb stretch of DNA that can considerably vary in size and prevalence [3]. The presence of CNVs has been shown to widely correlate with various complex diseases such as autism, schizophrenia, mental retardation, and cancer [9–12]. Recently, CNVs have been documented as essential determinants of human male infertility [13–16]. Identification of the DNA-based markers for infertility can have a significant impact on early detection of infertility risk and hence timely management. Y chromosome deletions have already been subjected to clinical testing for inclusion in the diagnosis of male infertility. Apart from SNPs, CNVs can be easily genotyped for routine diagnosis of male infertility, which can provide additional markers for screening.
Cytogenetic array is a powerful genetic tool that allows the genome-wide detection of submicroscopic duplications/deletions across the genome by screening thousands of loci at the same time [16]. It is instigated as a first-tier test for diagnosis of genomic imbalances [17, 18]. Additionally, the cytogenetic array allows the determination of copy-neutral loss of heterozygosity (cnLOH) [19, 20]. Chromosome 19 has been recognized to have the highest gene density of all chromosomes with sets of genes of evolutionary significance [21]. The disease-associated deletions and duplications within the terminal band 19p13.3 are sparsely described in the medical literature and those too exhibit substantial phenotypic variability. 19p duplications have been reported in correlation with developmental delay, macrocephaly, and hypotonia [22–24]. However, since most of these disorders come into picture in childhood, there is no report of infertility in cases with 19p duplication. In the present study, we used cytogenetic microarray to identify genomic imbalances in the form of CNVs that are associated with “idiopathic” human male infertility. Normozoospermic fertile controls were analyzed for comparison. We found a common genomic gain (duplication) at 19p13.3 locus in 9 (26.47%) cases, which was completely absent in controls. The present study is the first report of a duplication in chromosome 19 in association with male infertility. We also identified genomic duplications at Yp11 locus in 16 (47%) cases and 4 (40%) controls and cnLOH at the 3p21.31 region in 6 (17.6%) cases.
Materials and methods
The present study was approved by the Institutional Human Ethics Committee of the Institute of Science, Banaras Hindu University, Varanasi (Approval letter No. Dean/2012-13/546). All the subjects were informed about the purpose of sample collection and written consent was obtained. Study subjects were recruited from the Department of Urology, Sir Sunderlal Hospital, Institute of Medical Sciences, Banaras Hindu University, Varanasi. Before sample collection, all subjects underwent meticulous medical and physical examinations. Patients with endocrine abnormalities (hypogonadism) and those with gross dysmorphic abnormalities, history of pelvic/spinal injuries, acquired and congenital structural defects of the urogenital system (cystic fibrosis, Young’s syndrome, etc.), karyotype abnormalities, and AZF microdeletions were excluded. Patients with the history of genital tract obstruction or/and dysfunction (varicocele, obstructive azoospermia) were also excluded. Infertile individuals used to extreme alcohol consumption, smoking, and drug abuse (marijuana and recreational substances), and those having been exposed to radiation, were also excluded. The possibility of involvement of female factors was excluded after exhaustive clinical examinations undertaken on the female partner of each patient.
All the patients recruited in this study had infertility persisting for longer than 1 year. Collection of semen samples was performed as per the WHO criteria for semen collection and analysis [25]. Semen analyses were carried out thrice, after 3 to 4 days of sexual abstinence to ascertain infertility status of the patients. Five milliliters of peripheral blood and testicular biopsy samples was obtained from each patient. Semen was collected in a clean, well-labeled, wide-mouthed, and non-toxic semen collection container followed by incubation at 37 °C until liquefaction. The control group consisted of healthy fertile normozoospermic males who had fathered at least one child and had no history of chronic illness. The controls belonged to the same age group (between 20 and 45 years) and had the same ethnicity as the patients. Hormonal profiling using quantitative analysis of LH, FSH, and testosterone was carried out for all the patients. All the patients indicated hormone values within the normal range. In total, 44 infertile patients with different histologic phenotypes were recruited (normozoospermic controls (NS), n = 10; hypo-spermatogenesis (HS), n = 8; maturation arrest (MA), n = 7; Sertoli cell-only syndrome (SCOS), n = 13; and severe oligozoospermia (SO), n = 6).
Plus/minus-polymerase chain reaction for Y chromosome deletion mapping
Genomic DNA was extracted from peripheral blood using DNA/RNA extraction kits (Illumina, San Diego, CA, USA) per the manufacturer’s protocol. The quality and quantity of DNA were estimated using a NanoDrop 2000 instrument (Thermo Fisher Scientific, Waltham, MA, USA). DNA was subjected to AZF-polymerase chain reaction (PCR) simplex using 11 sets of sequence-tagged site (STS) markers (STSs: sY83, sY84, sY69, sY117, sY152, sY255, sY254, sY157, sY158, sY159, and sY160) on both sides of the euchromatic region of the Y chromosome from the centromere to interval 7. A DNA sample from a normal fertile male was used as a fertile control; and two negative controls, (1) female DNA sample and (2) water sample (without template), were used in every PCR assay. The PCR assay was performed thrice for confirmation.
Histologic analysis
Histology analysis was performed using the standard method. Testicular biopsies obtained from the patients were fixed with Bouin’s fixative for 24 h at room temperature. After dehydration in ascending alcohol concentrations, the specimens were embedded in paraffin, and testicular blocks were prepared. Five-micrometer sections were mounted onto poly-l-lysine (Sigma) pre-coated slides. The sections were deparaffined, rehydrated, and further stained with hematoxylin (HiMedia) and eosin (Merck) for light microscopic examination and phenotype determination.
Chromosome microarray analysis
To identify the genomic imbalances in infertile patients (n = 34) vs fertile controls (n = 10), we utilized the CytoScan™ 750K Array (Affymetrix, USA) for genomic hybridization as per the manufacturer’s instruction. The array is characterized by more than 750,000 CNV markers and 200,000 genotype-able SNP probes, which provides high-resolution copy number, accurate breakpoint estimation, and loss of heterozygosity (LOH) detection. The data were visualized and analyzed using the Chromosome Analysis Suite (ChAS) software package (Affymetrix, USA).
Further, the software for the 750K Array was set to a cutoff of ≥ 5 Mb for displaying loss of heterozygosity. Quality control (QC) thresholds for all the data included a SNP quality control (SNPQC) of ≥ 15, median of the absolute values of all pairwise differences (MAPD) of ≤ 0.25, and waviness standard deviation (SD) of ≤ 0.12.
Bioinformatics analysis
To analyze and characterize the canonical pathways, biological functions, and potential molecular interactions between the candidate genes identified by chromosome microarray analysis (CMA), pathway analysis was carried out using the Ingenuity Pathway Analysis (IPA) software package (http://www.ingenuity.com). Ingenuity Pathway Knowledge Base (IPKB) (Krämer et al. 2014) presently serves as the world’s largest database of knowledge on biological networks. The genes identified from the CNV region were uploaded into Qiagen’s Ingenuity Pathway Analysis (IPA) suite to classify networks of genes that are altered in infertile samples. The analysis was carried out to ascertain the biological interactions of these genes. The network described that the functional relationship between gene products is based on information from the Ingenuity Pathways Knowledge Base or known interactions in the literature.
Results
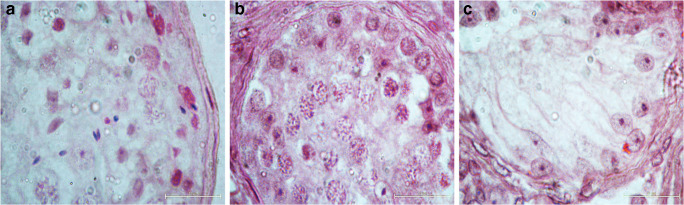
Semen analysis revealed that 28 patients were azoospermic and 6 were severe oligozoospermic. Histology analysis (Fig. 1) of testicular biopsies from azoospermia patients revealed different phenotypes (hypo-spermatogenesis (HS), n = 8; maturation arrest (MA), n = 7; and Sertoli cell-only syndrome (SCOS), n = 13). Clinical features of all the patients are described in Table 1. None of the patients displayed any Y chromosome microdeletions.
Fig. 1.
Histologic patterns of the seminiferous tubules observed in patients with a hypo-spermatogenesis (HS), b maturation arrest (MA), and c Sertoli cell-only syndrome (SCOS)
Table 1.
Clinical characteristic of the study group
| Characteristic | HS (n = 8) | MA (n = 7) | SCOS (n = 13) | SO (n = 6) | P value (analysis of variance) |
|---|---|---|---|---|---|
| Age (years) | 29.12 ± 1.7 | 27.28 ± 1.75 | 27.92 ± 2.09 | 27.83 ± 2.26 | 0.39 |
| Infertility (years) | 2.5 ± 0.75 | 2.57 ± 0.77 | 2.5 ± 0.83 | 2.75 ± 0.75 | 0.93 |
| LH (mlU/mL) | 4.27 ± 0.63 | 4.51 ± 1.68 | 4.66 ± 1.46 | 4.80 ± 1.01 | 0.89 |
| FSH (mlU/mL) | 5.60 ± 0.60 | 5.0 ± 1.63 | 4.49 ± 2.25 | 5.48 ± 2.8 | 0.64 |
| T (ng/dL) | 512 ± 87.75 | 494.7 ± 98.41 | 511.3 ± 92.51 | 500.2 ± 55.72 | 0.97 |
| Testicular volume (mL) | 34.25 ± 8.23 | 37.71 ± 7.74 | 36.0 ± 8.33 | 37.33 ± 8.29 | 0.86 |
Values are presented as mean ± SEM. NS, normal spermatogenesis; HS, hypo-spermatogenesis; MA, maturation arrest; SCOS, Sertoli cell-only syndrome; SO, severe oligozoospermia
The cytogenetic array analysis detected a genomic gain at the 19p13.3 region in 9 (26.47%) cases (Supplementary figure 1). Though the locus of duplication was the same in all nine cases, the genomic coordinates were variable, which ranged from Chr19: 633,754—1,709,958 (genome assembly hg19, NCBI build 37) (Supplementary data 1). These 9 cases belonged to different testicular phenotypes with 5 cases (38%) having Sertoli cell-only syndrome, 2 cases having hypo-spermatogenesis (25%), and 2 having severe oligozoospermia (33%). We did not find any association between the extent of duplication and the severity of the testicular phenotype. We also found a genomic duplication at Yp11 loci in 16 (47%) cases and 4 (40%) controls (Supplementary data 2). Out of these 16 cases, 8 cases had duplication of both the 19p13.3 and Yp11 regions. Additionally, a genomic loss was observed at the 14q11.2 region in 4 cases, but the region was devoid of any gene. Interestingly, all of these cases had a duplication in the 19p13.3 region.
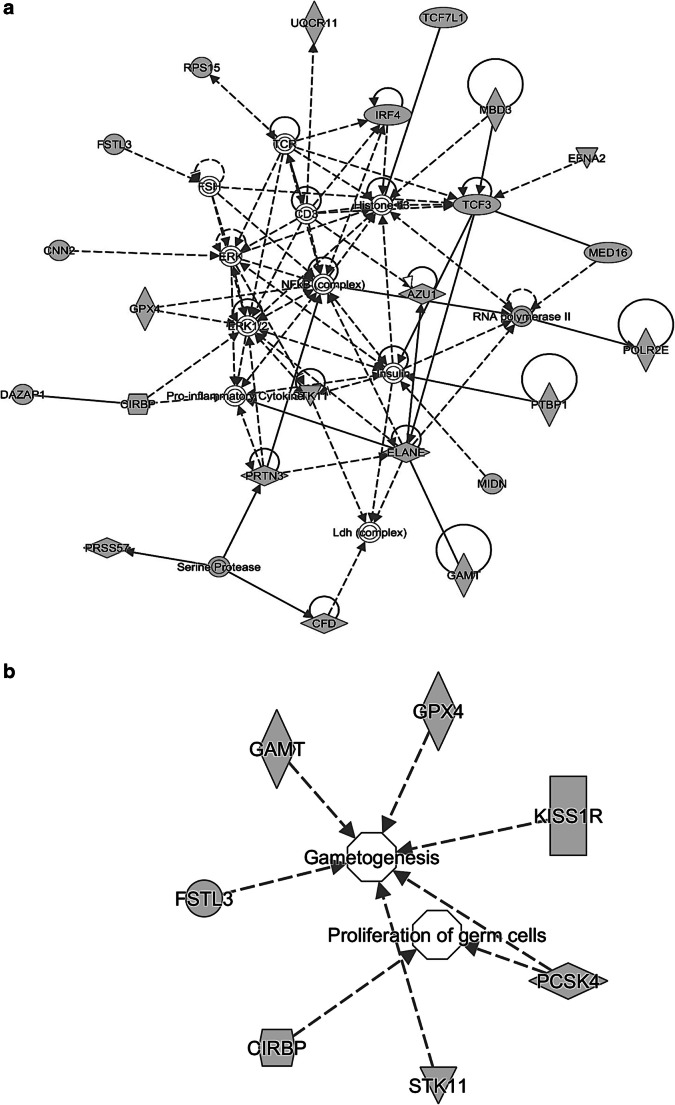
Using the IPA software, network analysis was carried out for 51 genes from the largest duplicated 19p13.3 region (633,754–1,709,95). The list of genes is described in Supplementary data 3. This revealed a network of 23 genes that were functionally correlated (Fig. 2a). Out of 23 genes, we identified a major network of 7 genes, viz., CIRBP (cold-inducible RNA-binding protein, OMIM 602649), FSTL3 (follistatin like 3, OMIM 605343), GPX4 (glutathione peroxidase 4, OMIM 138322), GAMT (guanidinoacetate N-methyltransferase, OMIM 601240), KISS1R (KISS1 receptor, OMIM 604161), STK11 (serine/threonine kinase 11, OMIM 602216), and PCSK4 (proprotein convertase subtilisin/kexin type 4, OMIM 600487) associated with reproductive system development and function (Fig. 2b). The functional annotation of these genes along with their corresponding P values was calculated using the right-tailed Fisher exact test (Supplementary data 4). P value less than 0.05 was considered to be statistically significant.
Fig. 2.
a Molecular network generated by Ingenuity Pathway Analysis (IPA) of 51 genes from the largest duplicated 19p13.3 region, which identified a network of 23 genes that were functionally correlated. b Out of 23 genes, IPA analysis highlighted a major network of 7 genes, viz., CIRBP, FSTL3, GPX4, GAMT, KISS1R, STK11, and PCSK4, associated with reproductive system development and function. Gene products are represented as nodes and biological relationships between two nodes as a line. Continuous lines show direct interactions, while dashed lines indicate indirect connections
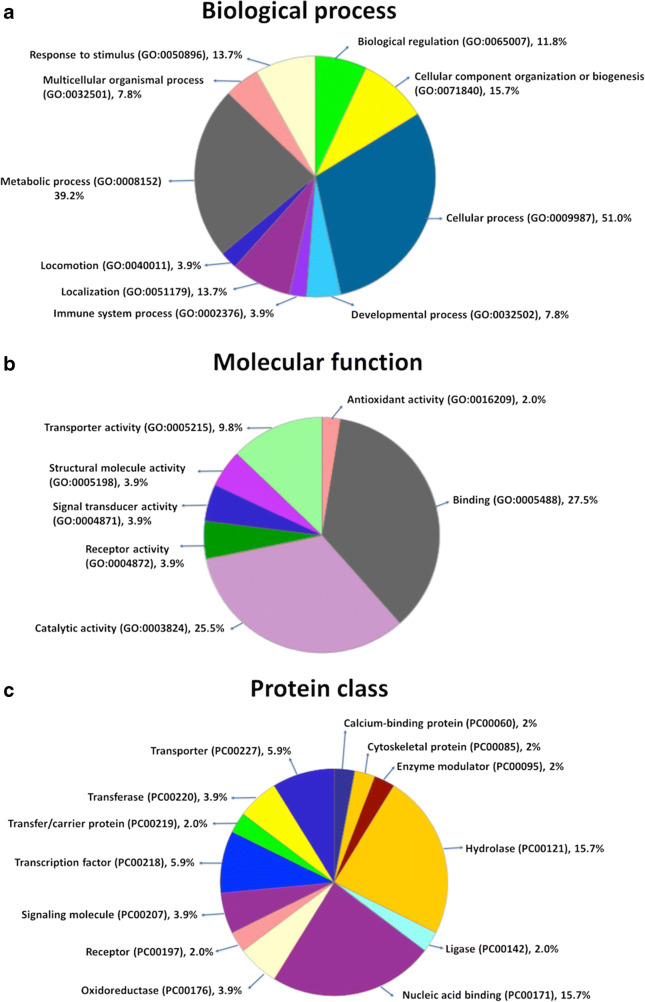
Gene ontology analysis
Those 51 target genes from the duplicated 19p13.3 region were classified for the three categories: biological process, molecular function, and signaling pathways by the PANTHER classification system (PANTHER9.0) (http://www.pantherdb.org/). Eighty-six biological processes, 39 molecular functions, and 34 protein classes were hit. The duplicated genes were essentially enriched in the biological process terms such as cellular component organization or biogenesis (GO:0071840, 8, 15.7%), cellular process (GO:0009987, 26, 51.0%), localization (GO:0051179, 7, 13.7%), biological regulation (GO:0065007, 6, 11.8%), response to stimulus (GO:0050896, 7, 13.7%), developmental process (GO:0032502, 4, 7.8%), multicellular organismal process (GO:0032501, 4, 7.8%, locomotion), (GO:0040011, 2, 3.9%), metabolic process (GO:0008152, 20, 39.2%), and immune system process (GO:0002376, 2, 23.9%) (Fig. 3a). Among the genes enriched in molecular function, binding (GO:0005488, 14, 27.5%), receptor activity (GO:0004872, 2, 3.9%), structural molecule activity (GO:0005198, 2, 3.9%), signal transducer activity (GO:0004871, 2, 3.9%), catalytic activity (GO:0003824, 13, 25.5%), antioxidant activity (GO:0016209, 1, 2.0%), and transporter activity (GO:0005215), 5, 9.8%) covered a high number of hits (Fig. 3b). In classification of the panther protein class, transporter (PC00227, 3, 5.9%), hydrolase (PC00121, 8, 15.7%), oxidoreductase (PC00176, 2, 3.9%), enzyme modulator (PC00095, 1, 2.0%), transfer/carrier protein (PC00219, 1, 2.0%), transferase (PC00220, 2, 3.9%), transcription factor (PC00218, 3, 5.9%), ligase (PC00142, 1, 2.0%), nucleic acid binding (PC00171, 8, 15.7%), receptor (PC00197, 1, 2.0%), calcium-binding protein (PC00060, 1, 2.0%), cytoskeletal protein (PC00085,1, 2.0%), and signaling molecule (PC00207, 2, 3.9%) displayed high numbers (Fig. 3c).
Fig. 3.
Gene ontology analysis of 19p13.3 harboring genes according to a molecular function, b chemical components, and c biological processes
Loss of heterozygosity
In addition to the identification of CNVs, the SNP-based arrays also provide a possibility to detect regions with copy-neutral loss of heterozygosity (cnLOH). In the present study, we observed a cnLOH at the 3p21.31 region in 6 (17.6%) of the cases (Supplementary Fig. 2). The details of cases with LOH are provided in Supplementary data 5.
Discussion
The genetic analysis of infertility risk has identified that Y chromosome deletions are the foremost cause of infertility. In a number of studies, autosomal candidate genes have been studied for correlation with infertility risk, with some of them showing promising correlation with male infertility [26, 27]. Exhaustive analysis of the human genome for infertility markers utilized copy number variation (CNV) analysis, which has identified promising markers [13]. In the present study, we utilized the cytogenetic array to identify candidate CNV regions that correlate with infertility. We identified a number of duplication and deletion events across the whole human genome, with a duplication at 19p13.3 appearing to be the most interesting. This duplication was observed in 26.47% of the individuals with the highest frequency in Sertoli cell-only syndrome cases. Its complete absence in the control group suggests its likely pathogenic nature. Chromosome 19 has the highest gene density of all chromosomes [21]. Reports of the disease-associated genomic imbalances within the terminal band, 19p13.3, are rare and are described with substantial phenotypic variability. Most of these records belong to the patients with features such as developmental delay, macrocephaly, and hypotonia [22–24]. One of the studies reported a 7.2-Mb terminal duplication of 19p13.3 (90,897–7,300,043) in a 7-year-old male, born prematurely with intrauterine growth retardation and clinical features such as neurological impairment, facial abnormalities, and urogenital malformations [28]. As most of these disorders come into picture in childhood, therefore, infertility has not been reported in association with this duplication. It is not known if such individuals would be infertile in adulthood. This is the first report of duplication in chromosome 19 in association with male infertility. In addition to Y-classical, micro, and partial deletions, the duplication at 19p13.3 could serve as a unique biomarker for the evaluation of infertility risk.
It remains to be investigated if a duplication in the 19p13.3 region is sufficient to cause infertility. Additional cases must be investigated to understand the mechanisms of duplication and if genomic architecture in this region predisposes to genomic rearrangements. Nevertheless, its correlation with infertility appears to be very interesting. Among various possibilities into the pathogenicity of this duplication, the disruption of the genes due to breakpoint remains a low plausibility due to the variation in the location of breakpoint across the samples. Another possibility into the pathogenic nature could be the high dosage of the genes encompassed in the duplicated region. For this, we identified the smallest common region across the individuals harboring the duplication, which identified STK11 (OMIM: 602216), ATP5D (OMIM: 603150), MIDN (OMIM: 606700), CIRBP (OMIM: 602649), and EFNA2 (OMIM: 602756) genes. It would be very interesting to investigate if the duplication in one candidate gene is sufficient to cause infertility or if they exert quantitative effects. Further analysis of the smallest duplication region has the potential of identifying the underlying gene(s) for infertility. The largest duplicated region identified in this study was also investigated for gene content, which identified a major network of 7 genes, viz., CIRBP (cold-inducible RNA-binding protein, OMIM: 602649), FSTL3 (follistatin like 3, OMIM: 605343), GPX4 (glutathione peroxidase 4, OMIM: 138322), GAMT (guanidinoacetate N-methyltransferase, OMIM: 601240), KISS1R (KISS1 receptor, OMIM: 604161), STK11 (serine/threonine kinase 11, OMIM: 602216), and PCSK4 (proprotein convertase subtilisin/kexin type 4, OMIM: 600487) associated with reproductive system development and function (Fig. 2b). The functional significance of these genes in reproduction has been discussed in detail in the following paragraphs.
Investigations based on RNA-binding protein immunoprecipitation-microarray (Chip) profiling (RIP-Chip) suggest that CIRBP-binding mRNAs in the testis are mostly associated with biological processes regulating male fertility [29]. Most of the CIRBP-binding mRNAs in the testis belong to genes from the azoospermia factor (AZF) region, such as RBMY1, HSFY2, EIF1AY, and DDX3Y. FSTL3 is known to specifically inhibit testicular activin [30]. FSTL3 knockout mice display increased testicular size and Sertoli cell numbers, allowing for increased spermatogenesis but otherwise showing normal testicular function and prolonged testicular life [31]. The expression of GPx4 is particularly enriched during spermatogenesis in both humans and mice and its depletion results in reduced sperm count and infertility in mice [32].
GAMT-deficient male mice display impaired spermatogenesis and absence of spermatozoa in the lumen of seminiferous tubules [33]. The kisspeptin receptor (KISS1R), also known as GPR54, plays an essential role in initiating and maintaining fertility in both humans and other animals [34]. Loss of function mutations in the GPR54 gene result in hypogonadotropic hypogonadism in humans and mice [34]. Mutations in the STK11 gene are associated with pathogenesis of Peutz–Jeghers syndrome, a condition which displays increased Sertoli cell tumors and defective spermatogenesis [35]. Furthermore, conditional knockout of both the isoforms of LKB1 (Lkb1S and Lkb1L) in germ cells results in progressive germ cell loss and sterility in male mice [35]. PCSK4 encodes for a calcium-dependent serine endoproteinase, which is primarily localized to the testicular germ cells and sperm [36]. Consistent with its distribution in reproductive tissues, inactivation of this gene in mice causes infertility in males and subfertility in females [37].
Y chromosome genes have already been established as functionally relevant for sex determination and various testicular functions. Large attention has been bestowed upon the understanding of the association of Y chromosome microdeletions with infertility. However, interestingly, upcoming evidences suggest that not just deletions but gene duplications on the Y chromosome are of interest, as they can potentially alter the gene dosage and their biological functions [38, 39]. Various upcoming evidences have suggested that Y chromosome variants can associate with impaired spermatogenesis and male infertility. In the present study, we have identified a genomic duplication at Yp11 locus in 16 cases and 4 controls. The frequency of duplications at this loci was comparatively higher in infertile men (47%) compared with that in fertile controls (40%), underlining its importance in human spermatogenesis and fertility.
Furthermore, we also identified a unique cnLOH at the 3p21.31 region in 6 (17.6%) of the cases. A region was considered a cnLOH event if loss of heterozygosity was observed without any copy number change. IPA analysis of genes from this highlighted important candidate genes, such as CSPG5, PRKDC, CDC25A, LTF, PTPN23, and BAP1, which are involved in embryonic development and function. However, the significance of cnLOH in the 3p21.31 region remains unknown. This highlights the complexity of genetic aberrations in severe infertile cases and the significance of array technology, used in combination with conventional cytogenetic methods, for a better understanding of the pathogenesis. Further studies are needed to test the utility and limits of this technology in clinical management of infertile patients.
In view of the fact that genomic imbalances in many genes are associated with male infertility, the present high-resolution genomic analysis provided an understanding of the pathophysiology of infertility in severe infertile cases. The genomic gain at the 19p13.3 region in 26.47% of cases with the highest frequency in SCOS cases highlights its significance in human spermatogenesis and fertility. Further, the study summarizes the frequency of Yp11 duplications in infertile and fertile men. In addition, identification of a copy-neutral loss of heterozygosity (cnLOH) at the 3p21.31 region opens a new horizon for further investigation of this in association with human reproduction and fertility.
To the best of our knowledge, this is the first report that highlights the significance of 19p13.3 duplication in infertile men with severe testicular phenotypes. The present study underlines the utility and significance of whole genomic analysis in the cases of male infertility which goes undiagnosed due to limitations in the conventional cytogenetic techniques and for identifying genes that are involved or essential for spermatogenesis. It is however imperative to analyze these CNVs in a larger cohort of patients along with functional analyses to derive any definitive conclusion. Together, our data along with other studies on animal models and infertile men raise the fact that these imbalances may contribute to impaired spermatogenesis and human male infertility.
Electronic supplementary material
a Microarray results for 19p13.3 duplication using CytoScan™ 750K Array (Affymetrix, USA). Analysis using the ChAS software (Affymetrix, USA) showed a common genomic gain at the 19p13.3 region in 9 (26.47%) cases (P1 to P9). The locus and extent of duplication in all the nine cases are represented by bold blue lines. For comparison, one control (CONTROL2) is used which shows an absence of duplication. b Representative microarray profile of patient with the largest 19p13.3 duplication in comparison with that of the control. The dots indicate individual markers in that region. Genomic gain is detected as an increase in the weighted log2 ratio and copy number state. The lower lines show the copy number in both cases (PNG 16.1 mb)
Graphical representation of cnLOH at the 3p21.31 region observed in 6 (17.6%) of the cases by ChAS software (Affymetrix, USA). Further, the software was set to a cutoff of ≥ 5 Mb for displaying loss of heterozygosity. The extent of cnLOH is represented using bold purple lines. For comparison, one control is represented showing absence of cnLOH (PNG 2.50 mb)
(PDF 569 kb)
(PDF 227 kb)
(XLSX 26 kb)
(XLSX 9 kb)
(PDF 286 kb)
Acknowledgments
The authors thank the patients for providing blood samples and their consent for genetic analysis. We would like to acknowledge the Interdisciplinary School of Life Sciences (ISLS), Banaras Hindu University for Affymetrix Microarray Facility. The first author thanks CSIR for the Senior Research Fellowship.
Funding
The study was funded by the Board of Research in Nuclear Sciences (BRNS), Govt. of India, with sanction number 2013/37B/27/BRNS.
Compliance with ethical standards
This study was approved by the Institutional Human Ethics Committee of the Institute of Science, Banaras Hindu University, Varanasi, approved this study (Approval letter No. Dean/2011-12/119).
Informed consent
Informed consent was obtained from all individual participants included in the study.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee.
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Holden CA, McLachlan RI, Pitts M, Cumming R, Wittert G, Agius PA, Handelsman DJ, de Kretser DM. Men in Australia Telephone Survey (MATeS): a national survey of the reproductive health and concerns of middle-aged and older Australian men. Lancet. 2005;366:218–224. doi: 10.1016/S0140-6736(05)66911-5. [DOI] [PubMed] [Google Scholar]
- 2.Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med. 2008;14:1197–1213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker G, Barak S (2012) Clinical management of male infertility. wwwENDOTEXTorg Chapter 7: MDTEXT. COM. Inc, South Dartmouth, MA, USA.
- 4.Pastuszak AW, Lamb DJ. The genetics of male fertility—from basic science to clinical evaluation. J Androl. 2012;33:1075–1084. doi: 10.2164/jandrol.112.017103. [DOI] [PubMed] [Google Scholar]
- 5.McLachlan RI, O’bryan MK. State of the art for genetic testing of infertile men. J Clin Endocrinol Metab. 2010;95:1013–1024. doi: 10.1210/jc.2009-1925. [DOI] [PubMed] [Google Scholar]
- 6.Diemer T, Desjardins C. Developmental and genetic disorders in spermatogenesis. Hum Reprod Update. 1999;5:120–140. doi: 10.1093/humupd/5.2.120. [DOI] [PubMed] [Google Scholar]
- 7.Yang L, Xu L, Zhou Y, Liu M, Wang L, Kijas JW, Zhang H, Li L, Liu GE. Diversity of copy number variation in a worldwide population of sheep. Genomics. 2018;110:143–148. doi: 10.1016/j.ygeno.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 8.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 9.Jaillard S, Drunat S, Bendavid C, Aboura A, Etcheverry A, Journel H, Delahaye A, Pasquier L, Bonneau D, Toutain A. Identification of gene copy number variations in patients with mental retardation using array-CGH: novel syndromes in a large French series. Eur J Med Genet. 2010;53:66–75. doi: 10.1016/j.ejmg.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Kumaran M, Cass CE, Graham K, Mackey JR, Hubaux R, Lam W, Yasui Y, Damaraju S. Germline copy number variations are associated with breast cancer risk and prognosis. Sci Rep. 2017;7:14621. doi: 10.1038/s41598-017-14799-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, Antaki D, Shetty A, Holmans PA, Pinto D. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet. 2017;49:27–35. doi: 10.1038/ng.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie Y, Yuan H, Wang M, Zhong L, Zhou J, Song B, Yin Q, Sun X. Copy number variations independently induce autism spectrum disorder. Biosci Rep 2017;37:BSR20160570. [DOI] [PMC free article] [PubMed]
- 13.Eggers S, DeBoer KD, van den Bergen J, Gordon L, White SJ, Jamsai D, McLachlan RI, Sinclair AH, O’Bryan MK. Copy number variation associated with meiotic arrest in idiopathic male infertility. Fertil Steril. 2015;103:214–219. doi: 10.1016/j.fertnstert.2014.09.030. [DOI] [PubMed] [Google Scholar]
- 14.Stouffs K, Vandermaelen D, Massart A, Menten B, Vergult S, Tournaye H, Lissens W. Array comparative genomic hybridization in male infertility. Hum Reprod. 2012;27:921–929. doi: 10.1093/humrep/der440. [DOI] [PubMed] [Google Scholar]
- 15.White S, Ohnesorg T, Notini A, Roeszler K, Hewitt J, Daggag H, Smith C, Turbitt E, Gustin S, van den Bergen J. Copy number variation in patients with disorders of sex development due to 46, XY gonadal dysgenesis. PLoS One. 2011;6:e17793. doi: 10.1371/journal.pone.0017793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaiswal D, Singh V, Dwivedi US, Trivedi S, Singh K. Chromosome microarray analysis: a case report of infertile brothers with CATSPER gene deletion. Gene. 2014;542:263–265. doi: 10.1016/j.gene.2014.03.055. [DOI] [PubMed] [Google Scholar]
- 17.Vermeesch JR, Brady PD, Sanlaville D, Kok K, Hastings RJ. Genome-wide arrays: quality criteria and platforms to be used in routine diagnostics. Hum Mutat. 2012;33:906–915. doi: 10.1002/humu.22076. [DOI] [PubMed] [Google Scholar]
- 18.Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, Church DM, Crolla JA, Eichler EE, Epstein CJ. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saeki H, Kitao H, Yoshinaga K, Nakanoko T, Kubo N, Kakeji Y, Morita M, Maehara Y. Copy-neutral loss of heterozygosity at the p53 locus in carcinogenesis of esophageal squamous cell carcinomas associated with p53 mutations. Clin Cancer Res. 2011;17:1731–1740. doi: 10.1158/1078-0432.CCR-10-1996. [DOI] [PubMed] [Google Scholar]
- 20.Saare M, Soritsa D, Vaidla K, Palta P, Remm M, Laan M, Karro H, Soritsa A, Salumets A, D’Hooghe T. No evidence of somatic DNA copy number alterations in eutopic and ectopic endometrial tissue in endometriosis. Hum Reprod. 2012;27:1857–1864. doi: 10.1093/humrep/des125. [DOI] [PubMed] [Google Scholar]
- 21.Grimwood J, Gordon LA, Olsen A, Terry A, Schmutz J, Lamerdin J, Hellsten U, Goodstein D, Couronne O, Tran-Gyamfi M. The DNA sequence and biology of human chromosome 19. Nature. 2004;428:529–535. doi: 10.1038/nature02399. [DOI] [PubMed] [Google Scholar]
- 22.Ishikawa A, Enomoto K, Tominaga M, Saito T, Nagai J, Furuya N, Ueno K, Ueda H, Masuno M, Kurosawa K. Pure duplication of 19p13. 3. Am J Med Genet A. 2013;161:2300–2304. doi: 10.1002/ajmg.a.36041. [DOI] [PubMed] [Google Scholar]
- 23.Nevado J, Rosenfeld JA, Mena R, Palomares-Bralo M, Vallespín E, Mori MÁ, Tenorio JA, Gripp KW, Denenberg E, Del Campo M. PIAS4 is associated with macro/microcephaly in the novel interstitial 19p13. 3 microdeletion/microduplication syndrome. Eur J Hum Genet. 2015;23:1615. doi: 10.1038/ejhg.2015.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Archer HL, Gupta S, Enoch S, Thompson P, Rowbottom A, Chua I, Warren S, Johnson D, Ledbetter DH, Lese-Martin C. Distinct phenotype associated with a cryptic subtelomeric deletion of 19p13. 3-pter. Am J Med Genet A. 2005;136:38–44. doi: 10.1002/ajmg.a.30774. [DOI] [PubMed] [Google Scholar]
- 25.Cooper TG, Noonan E, von Eckardstein S, Auger J, Baker HWG, Behre HM, Haugen TB, Kruger T, Wang C, Mbizvo MT, Vogelsong KM. World Health Organization reference values for human semen characteristics. Hum Reprod Update. 2010;16:231–245. doi: 10.1093/humupd/dmp048. [DOI] [PubMed] [Google Scholar]
- 26.Singh V, Bansal SK, Singh R, Singh K. Autosomal genes in male infertility. In: Male infertility: understanding, causes treat. Springer; 2017. pp 231–252.
- 27.Jedidi I, Ouchari M, Yin Q. Autosomal single-gene disorders involved in human infertility. Saudi J Biol Sci. 2018;25:881–887. doi: 10.1016/j.sjbs.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seidel MG, Duerr C, Woutsas S, Schwerin-Nagel A, Sadeghi K, Neesen J, Uhrig S, Santos-Valente E, Pickl WF, Schwinger W. A novel immunodeficiency syndrome associated with partial trisomy 19p13. J Med Genet. 2014;51:254–263. doi: 10.1136/jmedgenet-2013-102122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia Z, Zheng X, Zheng H, Liu X, Yang Z, Wang X. Cold-inducible RNA-binding protein (CIRP) regulates target mRNA stabilization in the mouse testis. FEBS Lett. 2012;586:3299–3308. doi: 10.1016/j.febslet.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 30.Xia Y, Sidis Y, Schneyer A. Overexpression of follistatin-like 3 in gonads causes defects in gonadal development and function in transgenic mice. Mol Endocrinol. 2004;18:979–994. doi: 10.1210/me.2003-0364. [DOI] [PubMed] [Google Scholar]
- 31.Oldknow KJ, Seebacher J, Goswami T, Villen J, Pitsillides AA, O’shaughnessy PJ, Gygi SP, Schneyer AL, Mukherjee A. Follistatin-like 3 (FSTL3) mediated silencing of transforming growth factor β (TGFβ) signaling is essential for testicular aging and regulating testis size. Endocrinology. 2013;154:1310–1320. doi: 10.1210/en.2012-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider M, Förster H, Boersma A, Seiler A, Wehnes H, Sinowatz F, Neumüller C, Deutsch MJ, Walch A, Hrabé de Angelis M. Mitochondrial glutathione peroxidase 4 disruption causes male infertility. FASEB J. 2009;23:3233–3242. doi: 10.1096/fj.09-132795. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt A, Marescau B, Boehm EA, Renema WKJ, Peco R, Das A, Steinfeld R, Chan S, Wallis J, Davidoff M. Severely altered guanidino compound levels, disturbed body weight homeostasis and impaired fertility in a mouse model of guanidinoacetate N-methyltransferase (GAMT) deficiency. Hum Mol Genet. 2004;13:905–921. doi: 10.1093/hmg/ddh112. [DOI] [PubMed] [Google Scholar]
- 34.Clarke H, Dhillo WS, Jayasena CN. Comprehensive review on kisspeptin and its role in reproductive disorders. Endocrinol Metab. 2015;30:124–141. doi: 10.3803/EnM.2015.30.2.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong F, Wang M, Huang X, Yue Q, Wei X, Dou X, Peng X, Jia Y, Zheng K, Wu T. Differential regulation of spermatogenic process by Lkb1 isoforms in mouse testis. Cell Death Dis. 2017;8:e3121. doi: 10.1038/cddis.2017.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gyamera-Acheampong C, Tantibhedhyangkul J, Weerachatyanukul W, Tadros H, Xu H, van de Loo J-W, Pelletier R-M, Tanphaichitr N, Mbikay M. Sperm from mice genetically deficient for the PCSK4 proteinase exhibit accelerated capacitation, precocious acrosome reaction, reduced binding to egg zona pellucida, and impaired fertilizing ability. Biol Reprod. 2006;74:666–673. doi: 10.1095/biolreprod.105.046821. [DOI] [PubMed] [Google Scholar]
- 37.Mbikay M, Tadros H, Ishida N, Lerner CP, De Lamirande E, Chen A, El-Alfy M, Clermont Y, Seidah NG, Chrétien M. Impaired fertility in mice deficient for the testicular germ-cell protease PC4. Proc Natl Acad Sci. 1997;94:6842–6846. doi: 10.1073/pnas.94.13.6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johansson MM, Van Geystelen A, Larmuseau MHD, Djurovic S, Andreassen OA, Agartz I, Jazin E. Microarray analysis of copy number variants on the human Y chromosome reveals novel and frequent duplications overrepresented in specific haplogroups. PLoS One. 2015;10:e0137223. doi: 10.1371/journal.pone.0137223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Connallon T, Clark AG. Gene duplication, gene conversion, and the evolution of the Y chromosome. Genetics. 2010;186:277–286. doi: 10.1534/genetics.110.116756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
a Microarray results for 19p13.3 duplication using CytoScan™ 750K Array (Affymetrix, USA). Analysis using the ChAS software (Affymetrix, USA) showed a common genomic gain at the 19p13.3 region in 9 (26.47%) cases (P1 to P9). The locus and extent of duplication in all the nine cases are represented by bold blue lines. For comparison, one control (CONTROL2) is used which shows an absence of duplication. b Representative microarray profile of patient with the largest 19p13.3 duplication in comparison with that of the control. The dots indicate individual markers in that region. Genomic gain is detected as an increase in the weighted log2 ratio and copy number state. The lower lines show the copy number in both cases (PNG 16.1 mb)
Graphical representation of cnLOH at the 3p21.31 region observed in 6 (17.6%) of the cases by ChAS software (Affymetrix, USA). Further, the software was set to a cutoff of ≥ 5 Mb for displaying loss of heterozygosity. The extent of cnLOH is represented using bold purple lines. For comparison, one control is represented showing absence of cnLOH (PNG 2.50 mb)
(PDF 569 kb)
(PDF 227 kb)
(XLSX 26 kb)
(XLSX 9 kb)
(PDF 286 kb)