Abstract
The synapse is a complex cellular module crucial to the functioning of neurons. It evolved largely through the exaptation of pre-existing smaller submodules, each of which are comprised of ancient sets of proteins that are conserved in modern animals and other eukaryotes. Although these ancient submodules themselves have non-neural roles, it has been hypothesized that they may mediate environmental sensing behaviors in aneural animals, such as sponges. Here we identify orthologues in the sponge Amphimedon queenslandica of genes encoding synaptic submodules in neural animals, and analyse their cell-type specific and developmental expression to determine their potential to be co-regulated. We find that genes comprising certain synaptic submodules, including those involved in vesicle trafficking, calcium-regulation and scaffolding of postsynaptic receptor clusters, are co-expressed in adult choanocytes and during metamorphosis. Although these submodules may contribute to sensory roles in this cell type and this life cycle stage, total synaptic gene co-expression profiles do not support the existence of a functional synapse in A. queenslandica. The lack of evidence for the co-regulation of genes necessary for pre- and post-synaptic functioning in A. queenslandica suggests that sponges, and perhaps the last common ancestor of sponges and other extant animals, had the ability to promulgate sensory inputs without complete synapse-like functionalities. The differential co-expression of multiple synaptic submodule genes in sponge choanocytes, which have sensory and feeding roles, however, is consistent with the metazoan ancestor minimally being able to undergo exo- and endocytosis in a controlled and localized manner.
Subject terms: Evolutionary developmental biology, Gene expression
Introduction
Understanding the origin and the evolution of the nervous system and the neuron has remained an unresolved challenge despite research and debates spanning over a century1–9. Over the last decade, genomic and transcriptomic data, particularly from non-bilaterian metazoans (sponges, placozoans, cnidarians and ctenophores)10–14 and closely related unicellular holozoans (choanoflagellates, filastereans and ichthyosporeans)15–17 have shed light on the evolution of regulatory and structural gene families involved in neuron formation and function. However, the evolutionary gain and loss of neural features in early-branching metazoan phyla has been difficult to reconstruct8,18–30.
As with nested hierarchies in other biological systems31–33, one approach to reconstruct the origin of the neuron is to examine operational modules that contribute to its functionality, such as the synapse and its constitutive submodules34,35, in aneural (sponges and placozoans) and neural (ctenophores, cnidarians) non-bilaterian animals. Modules and their constituent submodules are composed of an assembly of biomolecules collectively performing a particular function. These collective performances are supported by the co-expression of gene products that participate in these common functions (e.g. signaling pathways and subcellular structures). As these gene products are often under the control of a shared transcriptional regulatory regime32,36,37, analysis and comparison of the expression of the genes comprising such modules provides a potential way to reconstruct the evolution of the neuron.
The origin of the synapse as an operational module of the neuron is critical to understanding the evolution of the nervous system28,38,39. Essential for building neural networks, the functional synapse is defined by a well-characterised set of co-regulated genes that can be assigned to specific synaptic submodules, including the post-synaptic density, synaptic vesicle and vacuolar-ATPase36. As is often the case, these and other synaptic submodules served other, often more ancient, biological functions prior to be being co-opted into the regulatory network underlying the functioning of the synapse31,33,36,40. These preexisting modules are able to retain their ancient functions as evolutionary selective pressure occurs primarily on the interactions between modules; internal connections within more ancient modules are typically more constrained and less evolvable31,32,41.
Most synaptic genes are present in non-bilaterian aneural animals and closely related unicellular holozoans15,42–44 and have been collectively termed the “protosynapse”28,44–46. However, there is a limited understanding of how these genes are expressed in these taxa36,46,47. Thus it has been difficult to gain insight into the regulatory relationship of synaptic submodules in aneural animals and how the synapse may have evolved. Here, we use a reassembled genome of the demosponge Amphimedon queenslandica, extensive developmental and cell type transcriptomes, and knowledge of cell type sensory functionality in larvae, juveniles and adults to infer the presence of synaptic submodules based on gene co-expression. Specifically, we target specific cell types and developmental stages with putative sensory functioning, including the adult choanocytes and pinacocytes that interface with the external environment, and the larval stage expressing neural genes46,48,49 and displaying phototactic behaviour50 and metamorphic cue detection51. This approach can allow insights into evolutionary and regulatory settings that may have shaped the evolution of the synapse and the neuron. In this process, we also compiled an updated list of orthologues of synaptic genes in A. queenslandica46.
Results
Synaptic genes in Amphimedonqueenslandica
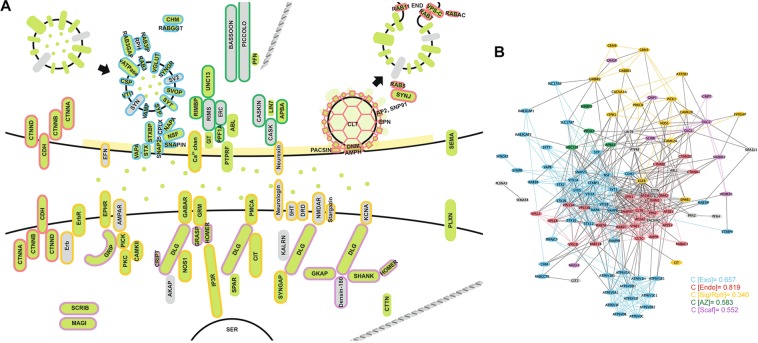
Based on sequence similarity, phylogenetics, domain architecture and the presence of conserved motifs, we determined that Amphimedon queenslandica has 125 genes that are orthologues of the canonical human synaptome (Supplementary Fig. 1). These A. queenslandica genes provide a near-complete coverage of a functional synapse and largely can be categorised into one of the five functional synaptic groups: exocytosis; endocytosis; signaling/receptor system; active zone; and post-synaptic scaffolding (Fig. 1A). Synaptic gene orthologues apparently absent from the A. queenslandica genome include genes encoding trans-synaptic adhesion molecules neurexin and neuroligin, NMDA and AMPA ionotropic glutamate receptors, presynaptic adaptor CASK, vesicle surface protein synapsin and vesicle priming protein RIMS. Genes encoding serotonin and dopamine receptors, as well as voltage-gated potassium channels, are also absent from the A. queenslandica genome. The final merged synaptome generated from protein interaction databases BioGrid, STRING and APID consists of a total of 108 gene products (15 of the 125 Amphimedon synaptic genes are paralogues and two have no documented non-self interactions) connected by 691 interactions (Fig. 1B).
Figure 1.
The synapse and its core submodules. (A) A diagram of the bilaterian synapse, signal transmitting and receiving cells, top and bottom respectively. Synaptic genes present in Amphimedon queenslandica are shaded green; genes not present are grey. Gene products are clustered into synaptic functions (submodules) and outlined by colour: exocytosis, blue; endocytosis, red; cell surface signals and receptors, yellow; active zone, dark green; and post-synaptic scaffolding, purple; gene products not outlined do not comprise these five submodules. (B) Evidence-based interactome for the human synaptome based on a non-redundant merging of BioGrid, STRING and APID databases. Genes (nodes) and associated interactions (edges/connecting lines) falling under the five core synaptic submodules are coded with the same color scheme as in (A). Clustering coefficients (C) for these submodules are shown.
Cell type and developmental co-expression of synaptic genes
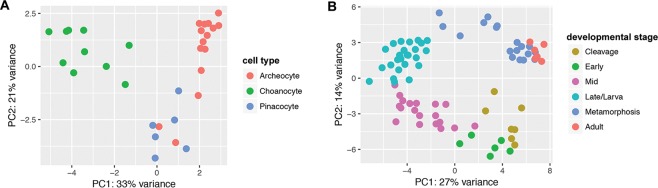
Using CEL-Seq2, we assessed the expression of Amphimedon synaptic genes between stages of the sponge life cycle – embryonic, larval, postlarval, juvenile and adult52 and three types of manually isolated adult cells53. Principle component analysis (PCA) shows that the expression profiles of these synaptic genes cluster according to both developmental stage and cell type (Fig. 2).
Figure 2.
PCA of synaptic gene expression in Amphimedon queenslandica. Transcript counts cluster by (A) adult cell types and by (B) developmental stage; see Table 1 for descriptions of these.
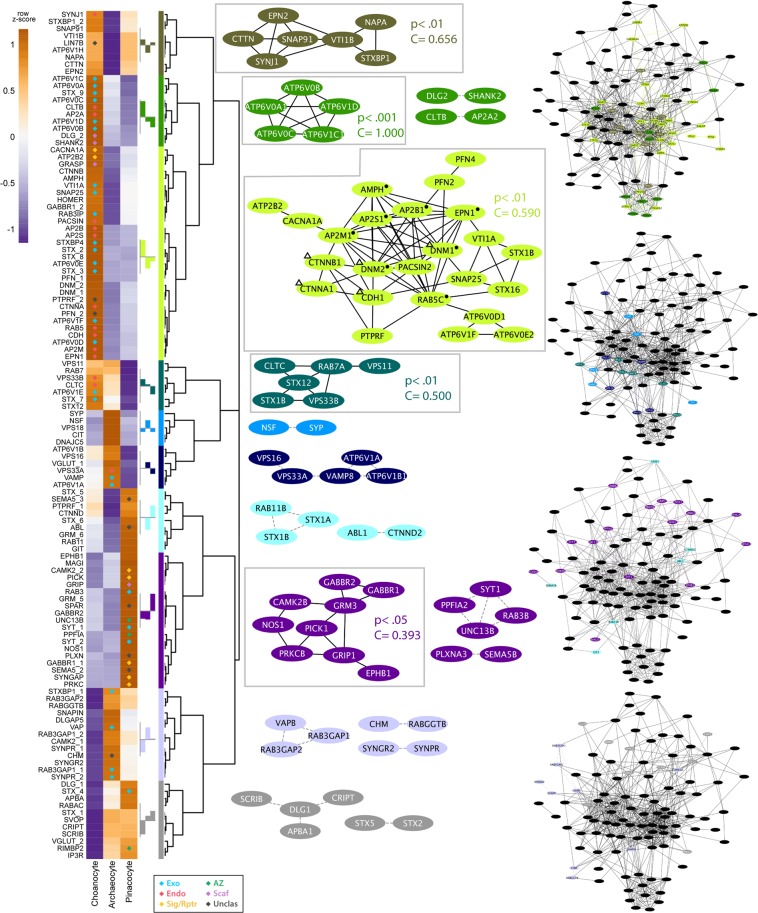
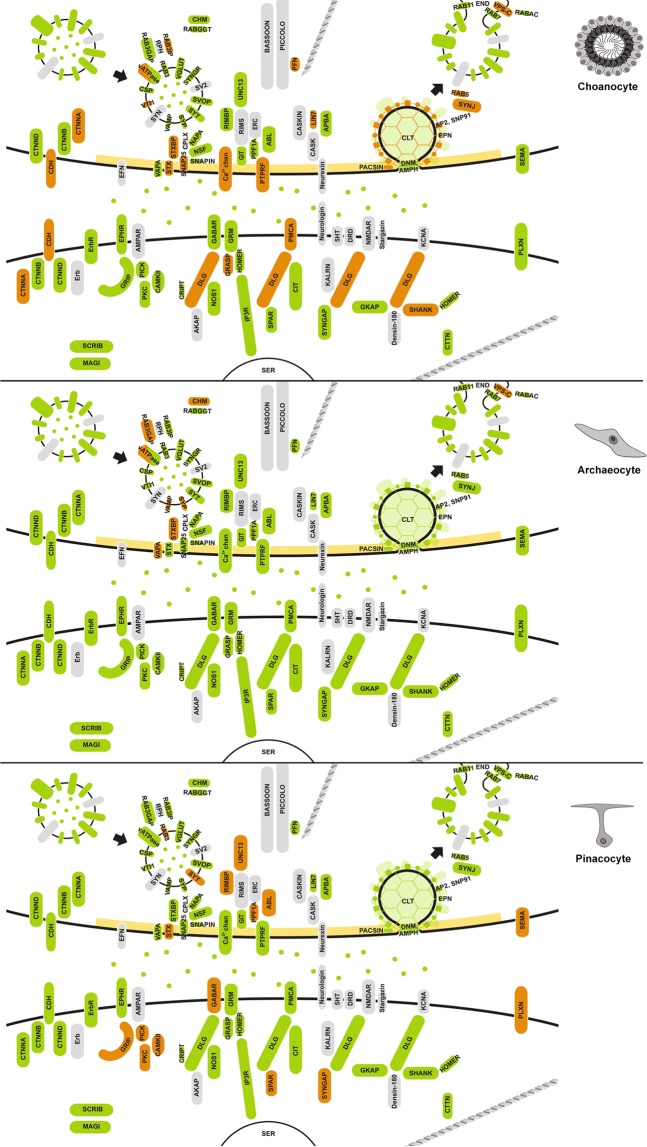
The majority of Amphimedon synaptic genes are upregulated during metamorphosis and in the adult, with the most substantial increase in expression of synaptic genes (37.6%) being when metamorphosis commences. Given this, we first focused on the expression of synaptic genes in adult cell types. We targeted three cell types that are essential for the sponge body plan: (i) choanocytes, internal epithelial feeding cells that form chambers that pump water through the sponge and capture exogenous microbes54,55; (ii) pinacocytes, epithelial cells that line internal canals and the outside of the sponge; and (iii) archeocytes, pluripotent stem cells that inhabit the middle collagenous mesohyl layer56,57. Both choanocytes and pinacocytes first appear during metamorphosis and directly interact with the external environment; archeocytes are present from embryogenesis onwards. The number of synaptic genes that are significantly upregulated in choanocytes (38; 30.4%) is more than double that of either pinacocytes (18; 14.4%) or archeocytes (8; 6.4%) (Fig. 3).
Figure 3.
Cell type expression of synaptic genes in Amphimedon queenslandica. Heatmap to the left shows synaptic gene expression profiles across three adult cell types, with diamonds indicating statistically significant (p < 0.05) gene upregulation in corresponding cell type; colour-coding of the diamonds is in relation to synaptic function as per Fig. 1 – blue, exocytosis; red, endocytosis; yellow, cell surface signals and receptors; dark green, active zone; purple, post-synaptic scaffolding. Z-scores reflect expression levels (variance stablising transformed (vst) counts), scaled by rows. Genes are divided into ten clades (colour-codes) based on expression profile similarities. The consensus expression profile for each clade is shown to the left of the colour bar. For each clade, all inferred interactions are shown based on the human synaptome in Fig. 1B. Non-interacting nodes are not shown. Significant co-regulating modules supported by Monte Carlo analysis have black edges and are are shown with corresponding p-values and clustering coefficients (C); edges are otherwise grey dashed. To the right are the complete human synaptic interactome decorated with the genes from the four major clades (i.e. genes with co-localised expressions) grouped in the same synaptome. Symbols in the largest clade (lime) indicate genes mapped to the enriched pathways of Endocytosis (●) and Bacterial invasion of epithelial cells (Δ). See Supplementary Table 1 for a complete list of mapped pathways.
Based on cell type specific co-expression profiles, Amphimedon synaptic genes can be divided into ten clades, from which interactive network modules were generated (Fig. 3). Of these, five co-expression network modules were deemed significant. Four of these correspond to networks of genes that are highly expressed in choanocytes, most of which are significantly upregulated (olive, green, lime and teal modules, Fig. 3); the other significant co-expression network (purple) corresponds to genes upregulated in pinacocytes. In both cell types, the genes comprising the significant networks encode proteins that are part of multiple synaptic submodules (Fig. 3).
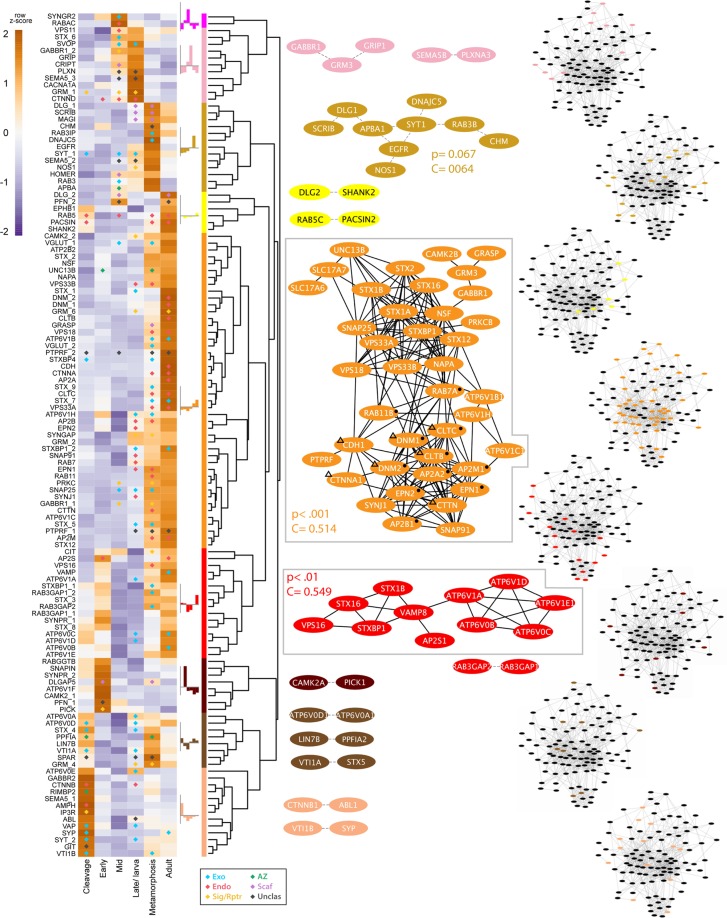
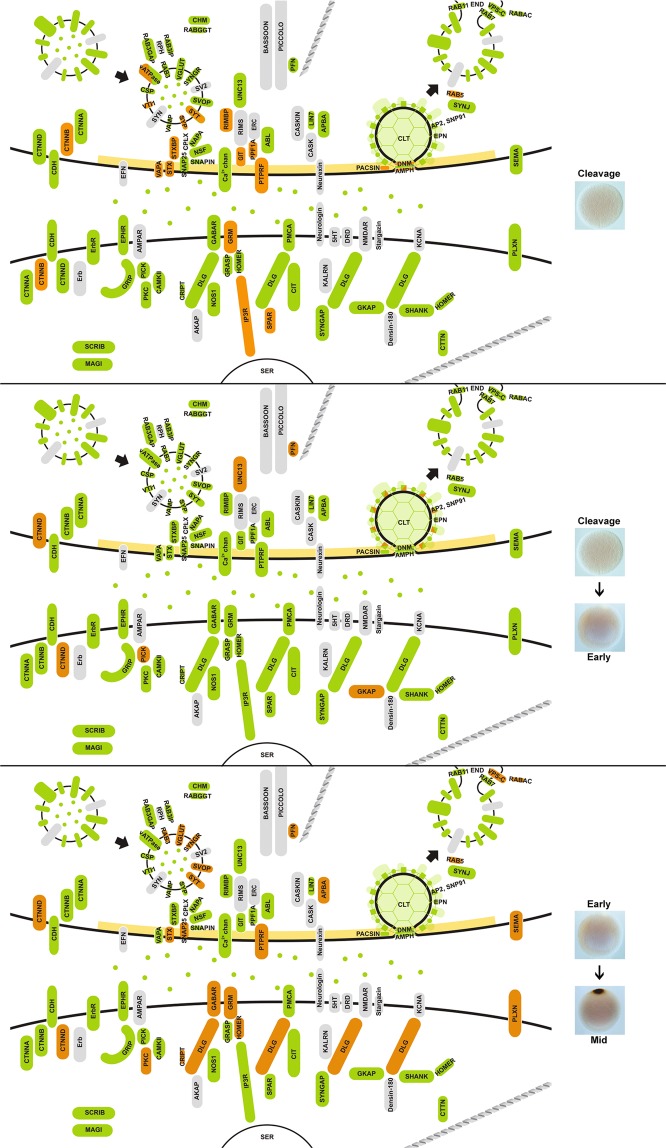
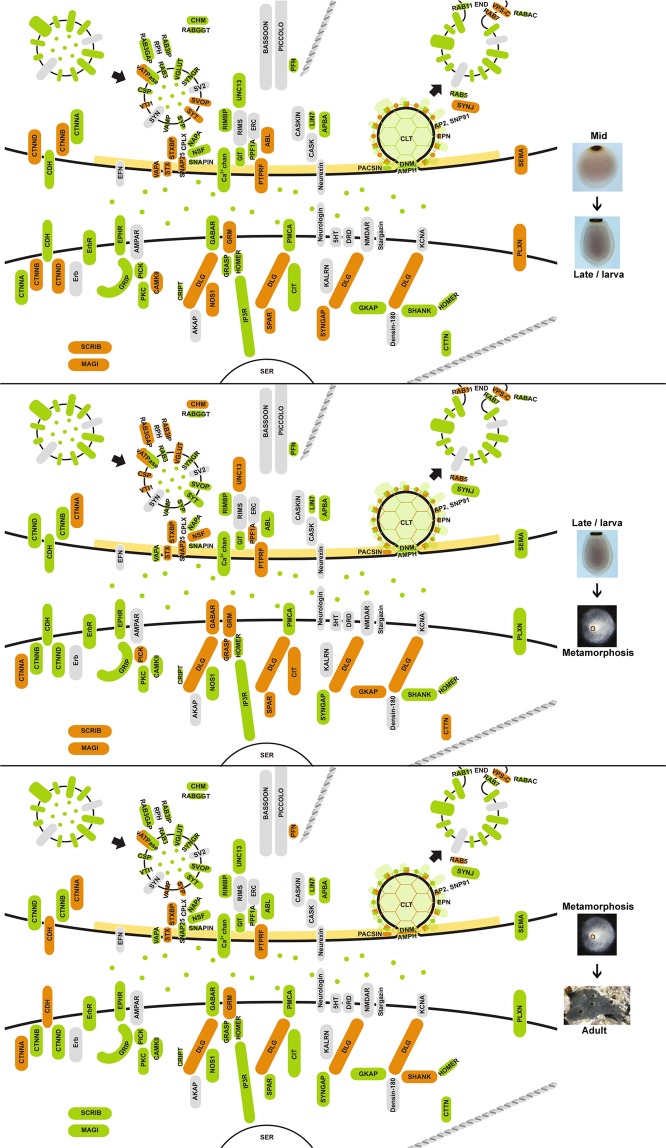
More synaptic genes are significantly upregulated in late developmental stages (late embryogenesis/larva, 46; metamorphosis, 47; adult, 32) than in early developmental stages (cleavage, 25; early embryogenesis, 9; mid-embryogenesis, 25) (Fig. 4). Based on developmental co-expression profile, genes were divided into eight clades (Fig. 4). Interactive network modules were generated for all but one clade (magenta), which consists of non-interacting genes. Two co-expression networks were significant and consisted of genes that are co-expressed at high levels during metamorphosis and in adults (orange and red modules, Fig. 4). These co-expressed genes comprise primarily exocytosis and endocytosis modules, and are highly expressed in choanocytes or pinacocytes (Fig. 3).
Figure 4.
Developmental expression of synaptic genes in Amphimedon queenslandica. Heatmap shows synaptic gene expression profiles over six developmental stages, with diamonds indicating statistically significant (p < 0.05) gene upregulation compared to the previous stage (except for cleavage stage, where upregulation is with respect to the early embryogenesis stage); colour-coding is in relation to synaptic function as per Fig. 1 - blue, exocytosis; red, endocytosis; yellow, cell surface signals and receptors; dark green, active zone; purple, post-synaptic scaffolding. The generation of dendrogram, colour modules, mapped pathways and clustering coefficients are as described in Fig. 3. Symbols in the largest clade (orange) indicate genes mapped to the enriched pathways of Endocytosis (●) and Bacterial invasion of epithelial cells (Δ). See Supplementary Table 1 for a complete list of mapped pathways.
All cell type and developmental co-expression clades mapped to multiple biological pathways (Supplementary Table 1).
Co-expression of synaptic submodule genes
Endocytosis
The majority of proteins involved in the endocytosis pathway in humans58, including genes not associated with the synapse, are present in the Amphimedon genome (Supplementary Fig. 2). A subset of these genes are tightly co-expressed at high levels in choanocytes (AMPH, AP2B1, AP2M1, AP2S1, DNM1, DNM2, EPN1, RAB5C; lime module, Figs 3 and 5) and metamorphosing and adult life cycle stages (AP2A, AP2B, AP2M, CLTB, CLTC, DNM1, DNM2, EPN1, EPN2, RAB11B, RAB7A; orange module, Figs 4, 6, 7). Monte-Carlo (MC) samplings show that these gene sets significantly co-express both locally (Chi-square H(2) = 41672, p < 0.001) and developmentally (H(2) = 43616, p < 0.001), with degrees of connectivity (C = 0.590 and 0.514 respectively) comparable to those of synaptic submodules (0.340–0.819; Fig. 1B). Other endocytic genes (AP2A2, AP2B1, AP2M1, CLTB, CTTN, DNM1, DNM2, EPN1, EPN2, PACSIN, RAB5, SNAP91) are also co-expressed and upregulated in choanocytes (olive and green modules, Fig. 3 [H(2) = 36732, p < 0.001]; Fig. 5) and in late development (yellow and red modules, Fig. 4 [H(2) = 38511, p < 0.001]; Figs 6, 7). In total, most Amphimedon endocytic genes are developmentally co-expressed with the 11 synaptic genes mapped to the endocytic pathway (Supplementary Fig. 2), suggesting that co-expression of synaptic endocytic genes in Amphimedon are part of the endocytosis pathway conserved between sponge and bilaterians.
Figure 5.
Upregulated synaptic genes in Amphimedon queenslandica cell types. Significantly upregulated synaptic genes (p-adj < 0.05) in each cell type, based on pairwise comparisons, are in orange. Green, orthologues of synaptic genes in Amphimedon; grey, genes not present in Amphimedon; yellow, active zone. Genes coding the endocytosis pathway (e.g. CLT, EPN, PACSIN), scaffolding and adhesion (e.g. DLG, GRASP, CDH) are enriched in choanocytes, while signaling/receptor genes (e.g. GABAR, CAMKII, SYNGAP) are upregulated in pinacocytes.
Figure 6.
Upregulated synaptic genes during Amphimedon queenslandica embryogenesis. Significantly upregulated genes were identified by pairwise comparisons between life stages (p-adj < 0.05) and are coloured orange. Green, orthologues of synaptic genes in Amphimedon; grey, genes not present in Amphimedon; yellow, active zone.
Figure 7.
Upregulated synaptic genes during Amphimedon queenslandica larval development, metamorphosis and in the adult. Significantly upregulated genes were identified by pairwise comparisons between life stages (p-adj < 0.05) and are coloured orange. Green, orthologues of synaptic genes in Amphimedon; grey, genes not present in Amphimedon; yellow, active zone.
Exocytosis
Genes involved in vesicle exocytosis (SLC17A6, SLC17A7, STX1a, STX1b, STX2, STXBP, SNAP25, NSF, NAPA, UNC13, ATP6V0/V1) are also co-regulated during metamorphosis and in adults (orange module, Fig. 4 [H(2) = 45455, p < 0.001]; Figs 6, 7); these genes are co-expressed with some of the endocytosis genes (orange module, Fig. 4). However, these genes are expressed differentially across cell types, and are not enriched in a single adult cell type in this sponge (Figs 3, 5). Genes encoding the SNARE complex (STX1, STXBP/UNC18, SNAP25), which facilitates the fusion of presynaptic vesicles to the plasma membrane during synaptic transmission59,60, are co-expressed and upregulated at metamorphosis (orange module, Fig. 4 [H(2) = 42275, p < 0.001]; Fig. 7) and in choanocytes together with calcium channel subunit and cellular calcium flux regulator (CACNA1, PMCA) (lime module, Fig. 3 [H(2) = 41075, p < 0.001]; Fig. 5). Although the SNARE complex contributes to a viable plasma membrane vesicle “dock”61, the gene encoding the vesicle-tethering protein VAMP, which is essential for proper SNARE-mediated vesicle fusion, is not co-expressed or significantly upregulated in choanocytes (Figs 3, 5).
Zipper mechanism (phagocytosis)
Another pathway that is enriched in metamorphosis is the zipper mechanism of bacterial invasion of epithelial cells, where invasive bacteria interact with receptors on non-phagocytic host cells to activate signaling cascades leading to cytoskeletal rearrangement and bacteria engulfment (orange module, Figs 4, 6, 7; Supplementary Fig. 2)62. Although Amphimedon has homologues for most of the curated pathway, co-expressed genes (CDH, CLTB, CLTC, CTNNA, CTTN, DNM1, DNM2 [H(2) = 41596, p < 0.001]) primarily contribute to upstream components of the pathway63. Genes of this pathway are expressed across all three cell types (Figs 3, 5; Supplementary Fig. 2).
Receptors and the post-synaptic density
Scaffolding and signaling molecules in the post-synaptic density (PSD), a key feature of the synapse, play a major role in clustering receptors and are known to have deep unicellular eukaryote origins42,64–66. Genes encoding components of the pathways that mediate neurotransmitter receptor functions, stability and trafficking are co-expressed at significantly high levels in the pinacocyte (CAMK2B, GABBR1, GABBR2, GRIP1, PICK1, PRKCB; purple module, Fig. 3 [H(2) = 39293, p < 0.001]; Fig. 5) and co-regulated in late development (AP2A2, AP2B1, AP2M1, CAMK2B, GABBR1, NSF, PRKCB; orange module, Fig. 4 [H(2) = 39482, p < 0.001]; Figs 6, 7). Genes encoding the core scaffolding proteins in PSD (DLG, SHANK, GRASP) are significantly upregulated in choanocytes (green and lime modules, Figs 3, 5).
Cell type- and stage-specific co-expression of genes with other synaptic functions
In addition to co-expressed synaptic genes corresponding to functional modules or pathways, we find evidence for the upregulation of small sets of synaptic genes in specific cell types and developmental stages. For instance, in addition to genes encoding endocytosis, exocytosis and scaffolding/adhesion proteins being upregulated in choanocytes, the plasma membrane calcium ATPase (PMCA) and the pore-forming α subunit of calcium channel (CACNA1A) are also upregulated in this cell type (Figs 3, 5), suggesting potential electrochemically mediated activities in these cells. A number of signaling genes (CAMK, GABAR, PICK, PRKC, SYNGAP) are upregulated in pinacocytes. In addition, the trans-synaptic pair semaphorin (SEMA) and its receptor plexin (PLXN), known to regulate cell communication and epithelial morphogenesis beyond axon-specific roles67, are also upregulated in pinacocytes and during late embryogenesis (Figs 3–7). Several other signaling genes (EGFR, CAMK, GRM, NOS, SYNGAP) are also upregulated in late embryogenesis (Figs 4, 6, 7), just prior to the swimming larval stage that has a capacity to detect light and other environmental signals68, including exogenous cues associated with inductive benthic substrata69.
Discussion
A functional synapse evolved through the exaptation of ancient genes with pre-exisiting non-neuronal functions, and the evolution and diversification of new gene families36,42,44,46,48,70,71. Together, ancient and more recently evolved synaptic genes were co-opted into a neuronal gene regulatory network that directed the co-expression of all the components necessary for the transmission and reception of synaptic signals; a single cell could have both these functionalities. In this study, we used developmental and cellular gene expression profiles from the sponge Amphimedon queenslandica to determine if there is evidence for the co-regulation of genes comprising the modern animal synapse; co-expression is used as a proxy for co-regulation.
To undertake this analysis, we first reassessed the annotation of synaptic genes in the Amphimedon genome by extending previous BLAST-based predictions10,72 to incorporate other lines of structural and phylogenetic evidence. Although most of our updated gene annotations support previous ones, we do not find sufficient support for the presence of the serotonin and dopamine receptors reported by Srivastava et al.10. These GPCRs do not clade with eumetazoan representatives73 and are therefore regarded as sponge-specific innovations. We also do not find strong support for voltage-gated potassium channels, kalirin and complexin, which were previously reported as present in Amphimedon10,36,42,65. In contrast, we confirm the absence of ionotrophic glutmate receptors (iGluR)10; iGluRs are present in other sponge species74,75, suggesting secondary loss of this receptor family in A. queenslandica.
Using 82 transcriptomes from individual embryos, larvae, metamorphosing postlarvae and adults condensed into six developmental stages, and 31 manually isolated pools of adult choanocytes, archeocytes and pinacocytes53,54, our analyses markedly expand on previous studies that focused on few life cycle stages and on expression of individual genes in specific cell types46,48,72,76, and allow for the identification of genes that are differentially co-expressed in specific cell types or at particular developmental stages. We find that co-expressed synaptic genes in Amphimedon largely comprise ancient cellular pathways that predate the divergence of metazoans from unicellular holozoans, including exocytotic, endocytotic, induced-phagocytotic and signaling pathways.
In contrast to the global co-expression of synaptic genes in neural animals36, we find that these submodules are often separated in Amphimedon, displaying cell type- and developmental stage-specific expression profiles. However, amongst the three cell types analysed for this study, the choanocyte co-expresses the most genes. These are associated with three synaptic submodules that predate the divergence of metazoans from choanoflagellates – endocytosis, vesicle docking, and PSD anchoring and scaffolding proteins – along with genes involved with calcium signaling. These functionalities are consistent with the primary role of choanocytes in phagocytic feeding, nutrient sorting and trafficking, and waste elimination, but also may be related to intercellular communication. The ability of PSD proteins to form a complex scaffold predates neural evolution46, hence the upregulation of transcripts encoding both PSD proteins and calcium regulators in choanocytes supports the view that the ancestral PSD served to link calcium signaling to cytoskeletal regulation42, although this role in choanocytes remains largely unexplored.
The co-upregulation of these multiple synaptic submodules in choanocytes may suggest that this sponge cell type evolved from an ancestral cell that also gave rise to neurons in other animal lineages, but may also be consistent with the choanocyte being a remnant neuron and thus support the proposal that poriferans have secondarily lost a nervous system21,25,77–79. Although these data cannot unequivocally support one scenario over the other, it is worth noting that choanocytes share some features with neurons, including an apical microvillar collar and cilium, basal cytoplasmic projections and a raft of dynamic phagosomes and vesicles, and are known to respond rapidly to external stimuli56,57. In contrast, nearly all the synaptic components expressed in choanocytes predate metazoans, thus lending support to the “protosynapse” theory28, with vesicle-trafficking modules being an aneural neurosecretory apparatus that has been co-opted early in neural evolution80–82.
Other synaptic components are co-upregulated in epithelial pinacocytes, namely genes related to the transduction of external signals. The co-expression of receptor-supporting active zone scaffold and receptor-interacting proteins (GRIP, PICK) further supports this epithelial-like cell type being able to sense and respond to exogenous signals. The co-upregulation of trans-synaptic pair SEMA and PLXN in this epithelial cell type suggests that semaphorin–plexin signaling is involved in non-neural cellular processes such as cell movement, migration and proliferation67.
Although most synaptic genes are upregulated during metamorphosis and highly expressed in adults, there are smaller subsets that are upregulated during embryogenesis and in larvae. Analysis of synaptic genes in larvae does not support a synapse-like function in known photo- and chemosensory systems underlying larval swimming behavior and responses to exogenous settlement cues50,51,69,83. Larval cell types that may contribute to these sensory systems are known to express individual synaptic genes, including globular cells which expressed PSD genes DLG, GKAP, GRIP, HOMER and CRIPT, proneural transcription factor related to atonal/neurogenin-bHLH gene families, and Notch-Delta signaling, and larval pigment ring cells which express a number of neurogenic transcription factors and signaling ligands46,48,76,84. Despite the A. queenslandica larva having a number of sensory capabilities, there is little evidence for substantial co-regulation of synaptic genes.
The synaptic submodules that were found to be co-expressed in A. queenslandica are comprised of genes present also in choanoflagellates65,82. This raises the possibility that cell-level sensory behaviours in sponges are akin, and perhaps homologous, to those observed in unicellular eukaryotes85–87, although sponges also exhibit tissue- and organismal-level responses to external stimuli50,83,88. The lack of strong support for the integration of synaptic submodules under a common regulatory framework in A. queenslandica is consistent with sponges not having an integrated synapse or synapse-like function. Analysis of another aneural lineage of animals, the placozoans, provides a means to compare co-expression within and between genes of synaptic submodules to address the origin – and the potential subsequent loss – of neurons at the base of the animal kingdom. Interestingly, however, recent single cell RNA-Seq analysis did not detect strong support for neural gene co-expression in the ctenophore Mnemiopsis leidyi89, suggesting that module analysis may not adequately resolve the earliest metazoan cladogenic events.
Conclusions
Analysis of developmental and cell type-specific expression of orthologues of genes encoding human synaptic proteins in Amphimedon queenslandica does not find evidence for a near-complete synapse in this sponge. Thus sensory systems and intercellular signaling in this sponge appear to function without synapse-like capabilities. Ancient submodules that comprise the modern synaptome are expressed in specific cell types and life cycle stages, which is consistent with sponges using these submodules as in other eukaryotes. However, the enrichment of multiple submodules and other synaptic genes in choanocytes (i.e. vesicle trafficking, scaffolding, and calcium signaling) suggests the common ancestor of sponges and bilaterians may have possessed a protosynapse involved in localized intercellular communication using exo- and endocytosis.
Materials and Methods
Identification of orthologues of synaptic genes in Amphimedon queenslandica
A list of synaptic genes was compiled from the canonical human synapse35,90–95. These genes partake in the following functions: (i) vesicle exocytosis (including synaptic vesicle surface proteins and vesicle docking machinery); (ii) vesicle recycling via clathrin-mediated endocytosis; (iii) signal transduction (including membrane receptors and some adhesion proteins); (iv) active zone scaffolding; and (v) post-synaptic scaffolding (Fig. 1).
Orthologues of these synaptic genes were identified from the latest version of the Amphimedon queenslandica genome, Aqu2.110,96 by: (i) using the human peptide sequences (downloaded from UniProtKB) in a local BLAST at a cut-off value of 1e−06; (ii) undertaking a reciprocal-BLAST of potential A. queenslandica sequences back to NCBI97, with a criterion that at least three of the top five hits must be the relevant synaptic protein; and (iii) examining domain arrangement of candidates with Pfam98 and HMMER99, with identified domains retained at a cut-off value of 1e−03. When putative orthologues had uncertain hits and domain variations between invertebrates and vertebrates, hidden Markov models (HMM) were re-built in-house using only invertebrate sequences, and the putative orthologues reassessed. For some families — dopamine/serotonin receptors, metabotropic glumate receptors (mGluR), γ-aminobutyric acid receptors (GABAR), membrane-associated guanylate kinases (MAGUK), Ras GTPases, cadherins and ion channels — additional information was taken into consideration, including conserved motifs, and structural, functional and phylogenetic analyses73,78,93,100–120. The domain arrangements of all Amphimedon synaptic were collated and presented using DoMosaics v1.0121.
Amphimedon synaptic genes identified as above were entered into the interactome databases BioGrid122, STRING123 and APID124 to retrieve evidence-based interactions documented for corresponding orthologues in Homo sapiens. All interactions were visualised in Cytoscape v.3.4.0125, with duplicate edges, directionality and self-interactions removed. Interactomes from the three databases were non-redundantly merged to produce a “synaptome”, representing all known protein-protein interactions within a functional bilaterian synapse (Fig. 1). The clustering coefficient C, where larger values are indicative of modularity in real-world networks126–128, was generated from Cytoscape’s in-built network analysis for each module.
CEL-Seq RNA datasets
Transcriptomes of Amphimedon were previously sequenced and processed using CEL-Seq2129 for choanocyte, archeocyte and pinacocyte cell types isolated (under the microscope with a micromanipulator) from adults53,130, and CEL-Seq131 for 82 developmental samples (whole animals) including embryonic, larval, postlarval, juvenile and adult stages52,132 (Table 1). For developmental transcriptomes, the “basic linear index determination of transcriptomes” (BLIND) method was performed52 on the 82 developmental samples, which allowed classification into six stages with strong within-group correlation: cleavage; early embryogenesis; mid-embryogenesis; late embryogenesis/larval development; metamorphosis; and adult. These stages were used to compare developmental gene expression profiles (Table 1). Genes with overall CEL-Seq read counts of less than 50 across the three cell types, or 100 across the 82 developmental stages, were discarded. Counts were variance stabilizing transformed (vst) using the Bioconductor package DESeq2133 and subjected to principal component analysis (PCA) in R to visualise differences in transcriptome profiles across sample types.
Table 1.
Developmental and cell-type sampling of Amphimedon queenslandica for transcriptome sequencing, by life stages52 and cell types53.
| Life stage/cell type | No. of samples | Sampling notes | |
|---|---|---|---|
| Life stage | Cleavage | 7 | Individual embryos |
| Early embryogenesis | 6 | Individual embryos | |
| Mid embryogenesis | 19 | Individual embryos | |
| Late embryogenesis/Larva | 26 | Individual embryos & larvae | |
| Metamorphosis | 18 | Individual postlarvae | |
| Adult | 6 | Individual sponge biopsies | |
| Cell type | Choanocyte | 15 (from 3 animals) | Chambers, 40–60 cells |
| Archaeocyte | 15 (from 3 animals) | Pools of 5–6 cells | |
| Pinacocyte | 9 (from 3 animals) | Pools of 5–6 cells |
Analysis of functional modules via pathway mapping
Cell type and developmental expression heatmaps were generated with the R packages pheatmap134 and RColorBrewer135, using the complete linkage method to cluster expression profiles. Genes were classified into 10 (approximate and arbitrary) clades based on expression profile similarities; each profile group was assigned a unique colour. For each group (colour module), all within-group gene interactions were determined using the human-based synaptome built in Cytoscape. Non-interacting genes were removed from the module network. Each module with more than three interacting genes (nodes) was mapped to curated human pathways using the Cytoscape plugin ReactomeFIViz136, incorporating data from Reactome137, KEGG58 and Panther138. Filtering was set at a false discovery rate (FDR) of <0.05 and a p-value of < 0.05. Schematic diagrams of selected pathways of interest were downloaded directly from corresponding databases.
Co-expression validation via Monte Carlo sampling
Co-expressing genes were statistically validated by Monte Carlo (MC) sampling139 over 10,000 dendrograms generated by genes of interest (GOIs) and randomly selected Aqu2.1 protein coding genes, with total number of selected genes being equal to that in the original dendrogram generated for synaptic genes. The number of clades GOIs appear in were contrasted against that of five sets of randomly selected control genes (GOCs) sampled in the same manner over 10,000 runs, using the Kruskal-Wallis test and visualising in boxplots (Supplementary Fig. 3). Dunn’s test is used as post-hoc comparison between selected pairs of gene sets to establish signifcant difference. Confidence cut-off is set at p = 0.05.
For selected clusters of co-expressing genes, clustering coefficient C was generated (as described above for the bilaterian synapse). The significance of node connectivities in this cluster is then tested by MC sampling with 10,000 sub-networks induced from an equal number of randomly selected nodes of the synaptome. A cluster is confirmed as a functional submodule of the synapse if the number of interacting edges is within top 5 percentile of the distribution of edge numbers in the 10,000 randomly induced subgraphs (p < 0.05).
All analyses are performed in R. Scripts for analyses are deposited on Github (https://github.com/AquSensory/SciRep2019).
Differential gene expression analyses
Differential gene expression analyses were performed using the Bioconductor package DESeq2133 in R. Differentially expressed genes (DEGs) were extracted by conducting pairwise comparisons between each cell type and between each pair of consecutive developmental stages (p-adj < 0.05). DEGs for each cell type and stage comparison were manually mapped to a custom-made synapse figure to help visualise the synaptic usage of upregulated genes.
Supplementary information
Acknowledgements
We thank W. Hatleberg and J. Rykr for their advice on bioinformatics analyses and illustrations, S.R. Williams for discussions, and two anonymous reviewers whose suggestions have greatly improved this manuscript.
Author contributions
E.W., S.M.D. and B.M.D. designed the study; E.W. compiled data; E.W. and J.M. analysed data with critical input from S.M.D., V.A. and B.M.D.; E.W. wrote the manuscript. All authors reviewed and edited the manuscript.
Data availability
The datasets generated during the current study are available in the Github repository, https://github.com/AquSensory/SciRep2019.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-51282-x.
References
- 1.Parker GH. The origin and significance of the primitive nervous system. Proc. Am. Philos. Soc. 1911;50:217–225. [Google Scholar]
- 2.Pantin CFA. The origin of the nervous system. Pubbl. della Stn. Zool. di Napoli. 1956;28:171–181. [Google Scholar]
- 3.Passano LM. Primitive nervous systems. Proc. Natl. Acad. Sci. USA. 1963;50:306–313. doi: 10.1073/pnas.50.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mackie GO. Neuroid conduction and the evolution of conducting tissues. Q. Rev. Biol. 1970;45:319–332. doi: 10.1086/406645. [DOI] [PubMed] [Google Scholar]
- 5.Hensen VDV. Zur Entwickehng des Nervensystems. Arch. pathol. anat. physiol. klin. med. 1864;30:176–186. doi: 10.1007/BF02280894. [DOI] [Google Scholar]
- 6.Hertwig, O. & Hertwig, R. Das Nervensystem und die Sinnesorgane der Medusen. (Vogel, 1878).
- 7.Keijzer F. Moving and sensing without input and output: early nervous systems and the origins of the animal sensorimotor organization. Biol. Philos. 2015;30:311–331. doi: 10.1007/s10539-015-9483-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lichtneckert, R. & Reichert, H. Origin and evolution of the first nervous systems. in Evolution of Nervous Systems (ed. Kaas, J. H.) 289–315 (Academic Press, 2007).
- 9.Arendt D, Tosches MA, Marlow H. From nerve net to nerve ring, nerve cord and brain — evolution of the nervous system. Nat. Rev. Neurosci. 2016;17:61–72. doi: 10.1038/nrn.2015.15. [DOI] [PubMed] [Google Scholar]
- 10.Srivastava M, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srivastava M, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- 12.Putnam NH, et al. Sea anemone genome reveals the gene repertoire and genomic organization of the eumetazoan ancestor. Science (80-.). 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 13.Moroz LL, et al. The ctenophore genome and the evolutionary origins of neural systems. Nature. 2014;510:109–14. doi: 10.1038/nature13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryan JF, et al. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science (80-.). 2013;342:1242592. doi: 10.1126/science.1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fairclough SR, et al. Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta. Genome Biol. 2013;14:1–15. doi: 10.1186/gb-2013-14-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suga, H. et al. The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat. Commun. 4 (2013). [DOI] [PMC free article] [PubMed]
- 17.Grau-Bové X, et al. Dynamics of genomic innovation in the unicellular ancestry of animals. Elife. 2017;6:1–35. doi: 10.7554/eLife.26036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burkhardt P, Sprecher SG. Evolutionary origin of synapses and neurons – Bridging the gap. BioEssays. 2017;39:1–10. doi: 10.1002/bies.201700024. [DOI] [PubMed] [Google Scholar]
- 19.Marlow H, Arendt D. Evolution: ctenophore genomes and the origin of neurons. Curr. Biol. 2014;24:R757–R761. doi: 10.1016/j.cub.2014.06.057. [DOI] [PubMed] [Google Scholar]
- 20.Whelan NV, Kocot KM, Moroz LL, Halanych KM. Error, signal, and the placement of Ctenophora sister to all other animals. Proc. Natl. Acad. Sci. USA. 2015;112:5773–8. doi: 10.1073/pnas.1503453112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moroz LL. Convergent evolution of neural systems in ctenophores. J. Exp. Biol. 2015;218:598–611. doi: 10.1242/jeb.110692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moroz LL, Kohn AB. Unbiased view of synaptic and neuronal gene complement in ctenophores: Are there pan-neuronal and pan-synaptic genes across Metazoa? Integr. Comp. Biol. 2015;55:1028–1049. doi: 10.1093/icb/icv104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moroz LL, Kohn AB. Independent origins of neurons and synapses: insights from ctenophores. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2016;371:20150041. doi: 10.1098/rstb.2015.0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halanych KM, Whelan NV, Kocot KM, Kohn AB, Moroz LL. Miscues misplace sponges. Proc. Natl. Acad. Sci. 2016;113:E946–E947. doi: 10.1073/pnas.1525332113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryan JF, Chiodin M. Where is my mind? How sponges and placozoans may have lost neural cell types. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2015;370:20150059–20150059. doi: 10.1098/rstb.2015.0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jékely, G., Paps, J. & Nielsen, C. The phylogenetic position of ctenophores and the origin(s) of nervous systems. Evodevo6 (2015). [DOI] [PMC free article] [PubMed]
- 27.Kosik KS. Exploring the early origins of the synapse by comparative genomics. Biol. Lett. 2009;5:108–111. doi: 10.1098/rsbl.2008.0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryan TJ, Grant SGN. The origin and evolution of synapses. Nat. Rev. Neurosci. 2009;10:701–712. doi: 10.1038/nrn2717. [DOI] [PubMed] [Google Scholar]
- 29.Watanabe H, Fujisawa T, Holstein TW. Cnidarians and the evolutionary origin of the nervous system. Dev. Growth Differ. 2009;51:167–183. doi: 10.1111/j.1440-169X.2009.01103.x. [DOI] [PubMed] [Google Scholar]
- 30.Simion P, et al. A large and consistent phylogenomic dataset supports sponges as the sister group to all other animals. Curr. Biol. 2017;27:958–967. doi: 10.1016/j.cub.2017.02.031. [DOI] [PubMed] [Google Scholar]
- 31.Wagner GP. Homologues, natural kinds and the evolution of modularity. Am. Zool. 1996;36:36–43. doi: 10.1093/icb/36.1.36. [DOI] [Google Scholar]
- 32.Arendt D, et al. The origin and evolution of cell types. Nat. Publ. Gr. 2016;17:744–757. doi: 10.1038/nrg.2016.127. [DOI] [PubMed] [Google Scholar]
- 33.Riedl, R. Die Ordnung des Lebendigen. Systembedingungen der Evolution. (Verlag Paul Parey, Hamburg und Berlin, 1975).
- 34.He Z, Yu Q. Identification and characterization of functional modules reflecting transcriptome transition during human neuron maturation. BMC Genomics. 2018;19:1–11. doi: 10.1186/s12864-017-4368-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng W, Zhang M. Organization and dynamics of PDZ-domain-related supramodules in the postsynaptic density. Nat. Rev. Neurosci. 2009;10:87–99. doi: 10.1038/nrn2540. [DOI] [PubMed] [Google Scholar]
- 36.Conaco C, et al. Functionalization of a protosynaptic gene expression network. Proc. Natl. Acad. Sci. 2012;109:10612–10618. doi: 10.1073/pnas.1201890109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stefanakis N, Carrera I, Hobert O. Regulatory logic of pan-neuronal gene expression in C. elegans. Neuron. 2015;87:733–750. doi: 10.1016/j.neuron.2015.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parker, G. H. The elementary nervous system, 10.1017/CBO9781107415324.004 (1919).
- 39.Nickel M. Evolutionary emergence of synaptic nervous systems: What can we learn from the non-synaptic, nerveless Porifera? Invertebr. Biol. 2010;129:1–16. doi: 10.1111/j.1744-7410.2010.00193.x. [DOI] [Google Scholar]
- 40.Achim K, Arendt D. Structural evolution of cell types by step-wise assembly of cellular modules. Curr. Opin. Genet. Dev. 2014;27:102–108. doi: 10.1016/j.gde.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 41.Espinosa-Soto, C. & Wagner, A. Specialization can drive the evolution of modularity. PLoS Comput. Biol. 6 (2010). [DOI] [PMC free article] [PubMed]
- 42.Alié A, Manuel M. The backbone of the post-synaptic density originated in a unicellular ancestor of choanoflagellates and metazoans. BMC Evol. Biol. 2010;10:34–43. doi: 10.1186/1471-2148-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burkhardt P. The origin and evolution of synaptic proteins — choanoflagellates lead the way. J. Exp. Biol. 2015;218:506–514. doi: 10.1242/jeb.110247. [DOI] [PubMed] [Google Scholar]
- 44.Emes RD, et al. Evolutionary expansion and anatomical specialization of synapse proteome complexity. Nat. Neurosci. 2008;11:799–806. doi: 10.1038/nn.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emes RD, Grant SGN. Evolution of synapse complexity and diversity. Annu. Rev. Neurosci. 2012;35:111–131. doi: 10.1146/annurev-neuro-062111-150433. [DOI] [PubMed] [Google Scholar]
- 46.Sakarya, O. et al. A post-synaptic scaffold at the origin of the animal kingdom. PLoS One2 (2007). [DOI] [PMC free article] [PubMed]
- 47.Varoqueaux, F. et al. High cell diversity and complex peptidergic signalling underlie placozoan behaviour. Curr. Biol. (2018). [DOI] [PubMed]
- 48.Richards GS, et al. Sponge genes provide new insight into the evolutionary origin of the neurogenic circuit. Curr. Biol. 2008;18:1156–1161. doi: 10.1016/j.cub.2008.06.074. [DOI] [PubMed] [Google Scholar]
- 49.Fortunato S, et al. Genome-wide analysis of the sox family in the calcareous sponge Sycon ciliatum: multiple genes with unique expression patterns. Evodevo. 2012;3:14. doi: 10.1186/2041-9139-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leys SP, Degnan BM. The cytological basis of photoresponsive behavior in a sponge larva. Biol. Bull. 2001;201:323–338. doi: 10.2307/1543611. [DOI] [PubMed] [Google Scholar]
- 51.Nakanishi N, Stoupin D, Degnan SM, Degnan BM. Sensory flask cells in sponge larvae regulate metamorphosis via calcium signaling. Integr. Comp. Biol. 2015;55:1018–1027. doi: 10.1093/icb/icv014. [DOI] [PubMed] [Google Scholar]
- 52.Anavy L, et al. BLIND ordering of large-scale transcriptomic developmental timecourses. Development. 2014;141:1161–1166. doi: 10.1242/dev.105288. [DOI] [PubMed] [Google Scholar]
- 53.Sogabe S, et al. Pluripotency and the origin of animal multicellularity. Nature. 2019;570:519–522. doi: 10.1038/s41586-019-1290-4. [DOI] [PubMed] [Google Scholar]
- 54.Taylor MW, Radax R, Steger D, Wagner M. Sponge-associated microorganisms: Evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 2007;71:295–347. doi: 10.1128/MMBR.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reiswig HM. Particle feeding in natural populations of three marine demosponges. Biol. Bull. 1971;141:568–591. doi: 10.2307/1540270. [DOI] [Google Scholar]
- 56.Funayama N. The stem cell system in demosponges: Suggested involvement of two types of cells: Archeocytes (active stem cells) and choanocytes (food-entrapping flagellated cells) Dev. Genes Evol. 2013;223:23–38. doi: 10.1007/s00427-012-0417-5. [DOI] [PubMed] [Google Scholar]
- 57.Simpson, T. L. The Cell Biology of Sponges. (Springer-Verlag, 1984).
- 58.Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47:D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sudhof TC. The synaptic vesicle cycle. Annu. Rev. Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 60.Han, J., Pluhackova, K. & Böckmann, R. A. The multifaceted role of SNARE proteins in membrane fusion. Front. Physiol. 8 (2017). [DOI] [PMC free article] [PubMed]
- 61.Verhage M, Sørensen JB. Vesicle docking in regulated exocytosis. Traffic. 2008;9:1414–1424. doi: 10.1111/j.1600-0854.2008.00759.x. [DOI] [PubMed] [Google Scholar]
- 62.Cossart, P. & Helenius, A. Endocytosis of viruses and bacteria. Cold Spring Harb. Perspect. Biol. 6 (2014). [DOI] [PMC free article] [PubMed]
- 63.Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y. & Morishima, K. KEGG Pathway: Bacterial invasion of epithelial cells - Homo sapiens (human). Available at: https://www.genome.jp/dbget-bin/www_bget?hsa05100. (Accessed: 19th November 2018) (2018).
- 64.de Mendoza A, Suga H, Ruiz-trillo I. Evolution of the MAGUK protein gene family in premetazoan lineages. BMC Evol. Biol. 2010;10:1–10. doi: 10.1186/1471-2148-10-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burkhardt P, et al. Evolutionary insights into premetazoan functions of the neuronal protein Homer. Mol. Biol. Evol. 2014;31:2342–2355. doi: 10.1093/molbev/msu178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sakarya O, et al. Evolutionary expansion and specialization of the PDZ domains. Mol. Biol. Evol. 2010;27:1058–1069. doi: 10.1093/molbev/msp311. [DOI] [PubMed] [Google Scholar]
- 67.Xia J, et al. Semaphorin-plexin signaling controls mitotic spindle orientation during epithelial morphogenesis and repair. Dev. Cell. 2015;33:299–313. doi: 10.1016/j.devcel.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 68.Wanninger Andreas., editor. Evolutionary Developmental Biology of Invertebrates 1. Vienna: Springer Vienna; 2015. [Google Scholar]
- 69.Say, T. E. & Degnan, S. M. Interdependent photo- and chemosensory systems regulate larval settlement in a marine sponge. BioRxiv, 10.1101/519512 (2019). [DOI] [PubMed]
- 70.Grant SGN. A general basis for cognition in the evolution of synapse signaling complexes. Cold Spring Harb. Symp. Quant. Biol. 2009;74:249–257. doi: 10.1101/sqb.2009.74.033. [DOI] [PubMed] [Google Scholar]
- 71.Noda AO, Ikeo K, Gojobori T. Comparative genome analyses of nervous system-specific genes. Gene. 2006;365:130–136. doi: 10.1016/j.gene.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 72.Conaco C, et al. Transcriptome profiling of the demosponge Amphimedon queenslandica reveals genome-wide events that accompany major life cycle transitions. BMC Genomics. 2012;13:209. doi: 10.1186/1471-2164-13-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Krishnan A, et al. The GPCR repertoire in the demosponge Amphimedon queenslandica: insights into the GPCR system at the early divergence of animals. BMC Evol. Biol. 2014;14:270–283. doi: 10.1186/s12862-014-0270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Riesgo A, Farrar N, Windsor PJ, Giribet G, Leys SP. The analysis of eight transcriptomes from all poriferan classes reveals surprising genetic complexity in sponges. Mol. Biol. Evol. 2014;31:1102–1120. doi: 10.1093/molbev/msu057. [DOI] [PubMed] [Google Scholar]
- 75.Francis, W. R. et al. The genome of the contractile demosponge Tethya wilhelma and the evolution of metazoan neural signalling pathways. BioRxiv, 10.1101/120998 (2017).
- 76.Adamska, M. et al. Wnt and TGF-β expression in the sponge Amphimedon queenslandica and the origin of metazoan embryonic patterning. PLoS One2 (2007). [DOI] [PMC free article] [PubMed]
- 77.Moroz LL. The genealogy of genealogy of neurons. Commun. Integr. Biol. 2014;7:e993269. doi: 10.4161/19420889.2014.993269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moran Y, Barzilai MG, Liebeskind BJ, Zakon HH. Evolution of voltage-gated ion channels at the emergence of Metazoa. J. Exp. Biol. 2015;218:515–525. doi: 10.1242/jeb.110270. [DOI] [PubMed] [Google Scholar]
- 79.Ryan JF. Did the ctenophore nervous system evolve independently? Zoology. 2014;117:225–226. doi: 10.1016/j.zool.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 80.Senatore A, Reese TS, Smith CL. Neuropeptidergic integration of behavior in Trichoplax adhaerens, an animal without synapses. J. Exp. Biol. 2017;220:3381–3390. doi: 10.1242/jeb.162396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jorgensen EM. Animal evolution: Looking for the first nervous system. Curr. Biol. 2014;24:R655–R658. doi: 10.1016/j.cub.2014.06.036. [DOI] [PubMed] [Google Scholar]
- 82.Burkhardt P, et al. Primordial neurosecretory apparatus identified in the choanoflagellate Monosiga brevicollis. Proc. Natl. Acad. Sci. 2011;108:15264–15269. doi: 10.1073/pnas.1106189108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ueda N, et al. An ancient role for nitric oxide in regulating the animal pelagobenthic life cycle: Evidence from a marine sponge. Sci. Rep. 2016;6:1–14. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Richards GS, Degnan BM. The expression of Delta ligands in the sponge Amphimedon queenslandica suggests an ancient role for Notch signaling in metazoan development. Evodevo. 2012;3:1–15. doi: 10.1186/2041-9139-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Greenspan, R. J. An Introduction to Nervous Systems. (Cold Spring Harbor Laboratory Press, 2007).
- 86.Shigeno Shuichi, Murakami Yasunori, Nomura Tadashi., editors. Brain Evolution by Design. Tokyo: Springer Japan; 2017. [Google Scholar]
- 87.Van Houten JLV, Cote BL, Zhang J, Baez J, Gagnon ML. Studies of the cyclic adenosine monophosphate chemoreceptor of Paramecium. J. Membr. Biol. 1991;119:15–24. doi: 10.1007/BF01868536. [DOI] [PubMed] [Google Scholar]
- 88.Leys SP, Meech RW. Physiology of coordination in sponges. Can. J. Zool. 2006;84:288–306. doi: 10.1139/z05-171. [DOI] [Google Scholar]
- 89.Sebé-Pedrós A, et al. Early metazoan cell type diversity and the evolution of multicellular gene regulation. Nat. Ecol. Evol. 2018;2:1176–1188. doi: 10.1038/s41559-018-0575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ziv NE, Garner CC. Cellular and molecular mechanisms of presynaptic assembly. Nat. Rev. Neurosci. 2004;5:385–399. doi: 10.1038/nrn1370. [DOI] [PubMed] [Google Scholar]
- 91.Kim E, Sheng M. PDZ domain proteins of synapses. Nat. Rev. Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- 92.Dalva MB, Mcclelland AC, Kayser MS. Cell adhesion molecules: signalling functions at the synapse. Nat. Rev. Neurosci. 2007;8:206–220. doi: 10.1038/nrn2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Collins MO, et al. Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J. Neurochem. 2006;97:16–23. doi: 10.1111/j.1471-4159.2005.03507.x. [DOI] [PubMed] [Google Scholar]
- 94.Zhang W, et al. SynDB: A synapse protein database based on synapse ontology. Nucleic Acids Res. 2007;35:737–741. doi: 10.1093/nar/gkl876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Croning MDR, Marshall MC, McLaren P, Armstrong JD, Grant SGN. G2Cdb: The genes to cognition database. Nucleic Acids Res. 2009;37:846–851. doi: 10.1093/nar/gkn700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fernandez-Valverde SL, Calcino AD, Degnan BM. Deep developmental transcriptome sequencing uncovers numerous new genes and enhances gene annotation in the sponge Amphimedon queenslandica. BMC Genomics. 2015;16:1–11. doi: 10.1186/s12864-015-1588-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wheeler DL, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Finn RD, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 100.Senatore, A., Raiss, H. & Le, P. Physiology and evolution of voltage-gated calcium channels in early diverging animal phyla: Cnidaria, Placozoa, Porifera and Ctenophora. Front. Physiol. 7 (2016). [DOI] [PMC free article] [PubMed]
- 101.Catterall WA. Voltage-gated calcium channels. Cold Spring Harb. Perspect. Biol. 2011;3:a003947. doi: 10.1101/cshperspect.a003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ferraguti F, Shigemoto R. Metabotropic glutamate receptors. Cell Tissue Res. 2006;326:483–504. doi: 10.1007/s00441-006-0266-5. [DOI] [PubMed] [Google Scholar]
- 103.Nollet F, Kools P, van Roy F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J. Mol. Biol. 2000;299:551–572. doi: 10.1006/jmbi.2000.3777. [DOI] [PubMed] [Google Scholar]
- 104.Oliva C, Escobedo P, Astorga C, Molina C, Sierralta J. Role of the MAGUK protein family in synapse formation and function. Dev. Neurobiol. 2012;72:57–72. doi: 10.1002/dneu.20949. [DOI] [PubMed] [Google Scholar]
- 105.Tompkins-MacDonald GJ, et al. Expression of a poriferan potassium channel: insights into the evolution of ion channels in metazoans. J. Exp. Biol. 2009;212:761–767. doi: 10.1242/jeb.026971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Proft J, Weiss N. G protein regulation of neuronal calcium channels: back to the future. Mol. Pharmacol. 2015;87:890–906. doi: 10.1124/mol.114.096008. [DOI] [PubMed] [Google Scholar]
- 107.Bettler B, et al. Molecular structure and physiological functions of GABAB receptors. Physiol. Rev. 2004;84:835–867. doi: 10.1152/physrev.00036.2003. [DOI] [PubMed] [Google Scholar]
- 108.Bennett MK, et al. The syntaxin family of vesicular transport receptors. Cell. 1993;74:863–873. doi: 10.1016/0092-8674(93)90466-4. [DOI] [PubMed] [Google Scholar]
- 109.Abedin, M. Cadherin evolution and the origin of animals. (UC Berkeley, 2010).
- 110.Murray PS, Zaidel-bar R. Pre-metazoan origins and evolution of the cadherin adhesome. Biol. Open. 2014;3:1183–1195. doi: 10.1242/bio.20149761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hulpiau P, van Roy F. Molecular evolution of the cadherin superfamily. Int. J. Biochem. Cell Biol. 2009;41:349–369. doi: 10.1016/j.biocel.2008.09.027. [DOI] [PubMed] [Google Scholar]
- 112.Nichols SA, Roberts BW, Richter DJ, Fairclough SR, King N. Origin of metazoan cadherin diversity and the antiquity of the classical cadherin/β-catenin complex. Proc. Natl. Acad. Sci. 2012;109:13046–13051. doi: 10.1073/pnas.1120685109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Colicelli J. Human RAS superfamily proteins and related GTPases. Sci STKE. 2004;2004:1–53. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J. Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 115.Te Velthuis AJW, Admiraal JF, Bagowski CP. Molecular evolution of the MAGUK family in metazoan genomes. BMC Evol. Biol. 2007;7:1–10. doi: 10.1186/1471-2148-7-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhu J, Shang Y, Chen J, Zhang M. Structure and function of the guanylate kinase-like domain of the MAGUK family scaffold proteins. Front. Biol. (Beijing). 2012;7:379–396. doi: 10.1007/s11515-012-1244-9. [DOI] [Google Scholar]
- 117.Tyson JR, Snutch TP. Molecular nature of voltage-gated calcium channels: Structure and species comparison. Wiley Interdiscip. Rev. Membr. Transp. Signal. 2013;2:181–206. doi: 10.1002/wmts.91. [DOI] [Google Scholar]
- 118.Liebeskind BJ. Evolution of sodium channels and the new view of early nervous system evolution. Commun. Integr. Biol. 2011;4:679–683. doi: 10.4161/cib.17069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jeziorski MC, Greenberg RM, Anderson PAV. The molecular biology of invertebrate voltage-gated Ca2+ channels. J. Exp. Biol. 2000;203:841–856. doi: 10.1242/jeb.203.5.841. [DOI] [PubMed] [Google Scholar]
- 120.Moran Y, Zakon HH. The evolution of the four subunits of voltage-gated calcium channels: ancient roots, increasing complexity, and multiple losses. Genome Biol. Evol. 2014;6:2210–2217. doi: 10.1093/gbe/evu177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Moore AD, Heldy A, Terrapon N, Weiner J, Bornberg-Bauer E. DoMosaics: software for domain arrangement visualization and domain-centric analysis of proteins. Bioinformatics. 2014;30:282–283. doi: 10.1093/bioinformatics/btt640. [DOI] [PubMed] [Google Scholar]
- 122.Chatr-Aryamontri A, et al. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017;45:D369–D379. doi: 10.1093/nar/gkw1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Szklarczyk D, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Alonso-López D, et al. APID interactomes: providing proteome-based interactomes with controlled quality for multiple species and derived networks. Nucleic Acids Res. 2016;44:W529–W535. doi: 10.1093/nar/gkw363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi A-L. Hierarchical organization of modularity in metabolic networks. Science (80-.). 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- 127.Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 128.Newman MEJ. The structure and function of complex networks. SIAM Rev. 2003;45:167–256. doi: 10.1137/S003614450342480. [DOI] [Google Scholar]
- 129.Hashimshony T, et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 2016;17:1–7. doi: 10.1186/s13059-016-0938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sogabe, S. et al. Amphimedon queenslandica cell-type transcriptomes. NCBI BioProject PRJNA412708 Available at: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA412708 (2019).
- 131.Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 132.Anavy, L. et al. A high-resolution Amphimedon queenslandica transriptomic timecourse. NCBI Gene Expression Omnibus GSE54364 Available at: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE54364 (2014).
- 133.Love M, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kolde, R. Pheatmap: pretty heatmaps (R package). (2015).
- 135.Neuwirth, E. RColorBrewer (R package). (2014).
- 136.Wu G, Dawson E, Duong A, Haw R, Stein L. ReactomeFIViz: the Reactome FI Cytoscape app for pathway and network-based data analysis. F1000 Res. 2014;3:146. doi: 10.12688/f1000research.4431.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fabregat A, et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2016;44:D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mi H, et al. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ulam S, von Neumann J. On Combination of Stochastic and Deterministic Processes. Bull. Am. Math. Soc. 1947;53:1120. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available in the Github repository, https://github.com/AquSensory/SciRep2019.