Abstract
Canine distemper virus (CDV) is the cause of a multisystem disease in domestic dogs and wild animals, infecting more than 20 carnivore and non-carnivore families and even infecting human cell lines in in vitro conditions. Phylogenetic classification based on the hemagglutinin gene shows 17 lineages with a phylogeographic distribution pattern. In Medellín (Colombia), the lineage South America-3 is considered endemic. Phylogenetic studies conducted in Ecuador using fragment coding for the fusion protein signal peptide (Fsp) characterized a new strain belonging to a different lineage. For understanding the distribution of the South America-3 lineage in the north of the South American continent, we characterized CDV from three Colombian cities (Medellín, Bucaramanga, and Bogotá). Using phylogenetic analysis of the hemagglutinin gene and the Fsp region, we confirmed the circulation of CDV South America-3 in different areas of Colombia. We also described, for the first time to our knowledge, the circulation of a new lineage in Medellín that presents a group monophyletic with strains previously characterized in dogs in Ecuador and in wildlife and domestic dogs in the United States, for which we propose the name “South America/North America-4” due its intercontinental distribution. In conclusion, our results indicated that there are at least four different CDV lineages circulating in domestic dogs in South America: the Europe/South America-1 lineage circulating in Brazil, Uruguay, and Argentina; the South America-2 lineage restricted to Argentina; the South America-3 lineage, which has only been reported in Colombia; and lastly an intercontinental lineage present in Colombia, Ecuador, and the United States, referred to here as the “South America/North America-4” lineage.
Subject terms: Viral epidemiology, Viral evolution
Introduction
Canine distemper virus (CDV) belongs to the Paramyxoviridae family, genus Morbillivirus, which includes viruses with epidemiological relevance to human and animal populations1,2. The etiological agent of a highly prevalent viral infectious disease of domestic and wild carnivores, CDV poses a conservation threat to endangered species worldwide3,4.
Clinical symptoms in all affected species are influenced by strain virulence, environmental conditions, host age, and immune status. The gastrointestinal tract and the respiratory and nervous systems are the most affected in all species5,6. Clinical signs of CDV in dogs include the onset of a cutaneous rash, serous nasal and ocular discharge, conjunctivitis, and anorexia, followed by gastrointestinal and respiratory signs, which are often complicated by secondary bacterial infections and neurological disorders7.
Of the six encoded viral proteins, the hemagglutinin (H) and fusion (F) glycoproteins are responsible for virus attachment to and fusion with the host cells8. The H protein is of principal importance as it determines viral tropism in vivo and in vitro and thus determines host-specific immunity9.
Due to external pressures on the host immune system, the H gene displays higher genetic variability when compared with other CDV genes9, which makes it suitable for lineage identification and phylogenetic analysis. Moreover, researchers have established that a short region of the F gene, which encodes the signal peptide of the F protein (Fsp), is extremely variable, suggesting that this region could also be a useful marker for evolutionary studies as it allows for the straightforward identification of CDV lineages10–14.
Phylogenetic studies based on the complete sequence of the H gene or the Fsp-coding region of several CDV viruses worldwide have revealed a geographical pattern of genetic diversity. According to this pattern, there are multiple distinct lineages/genotypes, most of which follow a geographical pattern of distribution. These lineages are known as America-1 (which includes the commercially available vaccines), America-2-5, Arctic-like, Asia-1-4, Africa-1 and -2, European Wildlife, Europe/South America-1, and South America-2 and -39,15–24.
Although immunization with attenuated vaccines has been widely used for preventing CDV, researchers have hypothesized that the strong genetic diversity and wide variability of the H gene could mean that the antigenic profile of these new genetic variants is altered relative to the vaccine strain if specific sites on the H protein associated with immune neutralization are affected9,25,26. In fact, recent analysis has suggested the need for developing an updated CDV vaccine due to differences in cross-neutralization assays revealing wide antigenic differences among wild-type CDV isolates and the vaccine strains currently used in the U.S.27.
Different publications have shown a worldwide increase in the incidence of the disease, even in vaccinated dog populations12,17,28,29 and as most CDV commercial vaccines are formulated with strains belonging to the America-1 lineage21, antigenic differences could possibly explain the worldwide increase in the incidence of the disease even in vaccinated dogs.
Researchers have suggested that the South American continent has one of the highest CDV antigenic variabilities in the world13. Phylogenetic analyses based on the CDV H gene from South America have been performed for establishing the evolutionary patterns of the virus in the region and have revealed multiple circulating lineages of CDV, each differing in prevalence. In Brazil, Uruguay, and Argentina, the most prevalent lineage is Europe/South America-120,28; the second-most prevalent, known as South America-2, is restricted to Argentinian canine populations and appears to be associated with strains isolated from wild carnivore species in Europe30.
In the northern part of South America, the circulation of different lineages has been reported; in Colombia, a third lineage (South America-3) that causes disease even in vaccinated dogs was identified on the basis of complete H gene sequencing17. In the same year, a possible different CDV lineage was described based on analysis of the Fsp-coding region of Ecuadorian strains11. Owing to differences in methodologies, complete H gene sequences are not comparable with Fsp-coding region sequences available from this region of the continent. It is possible that a genetic relationship exists between CDV strains from Colombia and Ecuador because they have similar geographic distributions in the northern region of South America.
For determining the genetic diversity of CDV from Colombia in relation to CDV from other regions of South America, we analyzed the genetic diversity within the Fsp-coding region and H gene of CDV strains currently circulating in different regions of Colombia and compared it with that of wild-type CDV circulating throughout America and worldwide in addition to vaccine strains.
Materials and Methods
Ethical considerations
This study was approved by the Ethics Committee for Animal Experimentation of the Universidad Cooperativa de Colombia in Bucaramanga. All experiments were performed in accordance with relevant guidelines and regulations. Dog owners signed informed consent forms approved by the ethics committee. Viral samples are subject of contract for access to genetic resources and derived products No. 132 of 2016 RGE0177 signed with the Ministry of Environment and Sustainable Development of the Republic of Colombia. In addition, the authors declare that the implementation of this work followed all scientific, technical, and administrative rules for animal research.
Clinical specimens and vaccine strains
A total of 86 clinical samples from dogs exhibiting signs suggestive of CDV were obtained from three main Colombian provinces (Supplemental Material Fig. S1). Of these, 48 clinical specimens were taken from Bogotá D.C. between January 2014 and June 2015, 11 were taken from Bucaramanga between June 2016 and November 2016, and 27 were taken from Medellín between May and September 2017. Samples were taken in different veterinary hospitals and basic data were gathered at the time of sampling, including the dogs’ age, sex, breed, vaccination status, municipality and/or neighborhood of origin, and clinical signs. Clinical specimens included serum and ocular discharge.
RNA extraction
Total RNA was extracted from 140 µl of the supernatant of ocular discharge, serum, and four commercial CDV vaccines using QIAamp Viral RNA Mini Kit (QIAGEN®, Hilden, Germany) in accordance with the manufacturer’s instructions. The quality and quantity of the RNA was determined using spectrophotometric analysis with a NanoDrop™ One UV-Vis Spectrophotometer (Thermo Scientific, Wilmington, Delaware, USA), and RNA aliquots were stored at −80 °C until use.
Complementary cDNA synthesis
Complementary DNA (cDNA) was synthesized using RevertAid™ Premium First Strand cDNA Synthesis Kit (Thermo Scientific®, Glen Burnie, MD) in accordance with the manufacturer´s instructions. A denaturation mix consisting of 1 µl (100 pmol/µl) random hexamers, 1 µl dNTP Mix (10 mM) and 13 µl (0.02–4.6 µg) total RNA was initially denatured at 65 °C for 5 min and immediately incubated on ice. The RT mix solution consisted of 4 µl 5X Reverse Transcriptase Buffer and 1 µl RevertAid™ Premium Enzyme Mix. The RT mix was added to the denaturation mix and reverse transcription was performed in a total volume of 20 µl in a ProFlex™ PCR Thermal Cycler (Applied Biosystems®, Foster city, California, USA) for 10 min at 25 °C followed by 30 min at 50 °C; the reaction was terminated by heating to 85 °C for 5 min. The reaction product was stored at −80 °C until use. Commercially available vaccines were used as positive controls for RT-PCR reactions.
PCR and sequencing
Next, cDNAs from clinical specimens were screened by PCR of the phosphoprotein (P) gene using the Maxima Hot Start PCR Master Mix (2X) (Thermo Scientific®) reagent kit in accordance with the manufacturer’s instructions. Viral cDNA was detected using morbillivirus universal primers31 for amplifying a 429 bp fragment of the phosphoprotein gene. For PCR, 4 µl cDNA was added to a reaction mix, which consisted of 25 µl Maxima Hot Start PCR Master Mix (2X), 15 µl nuclease-free water, and 3 µl (10 µM) of each of the forward and reverse primers. PCR was performed on a ProFlex™ PCR Thermal Cycler (Applied Biosystems®) under the following conditions: initial denaturation at 95 °C for 4 min followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 50.8 °C for 30 s, extension at 72 °C for 1 min, and a final extension at 72 °C for 5 min. Ultrapure water was used as a negative control and cDNA from one of the vaccines as positive control.
In all samples that tested positive for the P gene, the full-length H gene and the Fsp-coding region were amplified using Maxima Hot Start PCR Master Mix Kit in accordance with the manufacturer’s instructions. The H gene was detected using the primers CDVff1 and HS232 for amplifying a 2099 bp fragment of the CDV genome that includes the H gene and flanking regions at both ends. The Fsp-coding region was amplified using the primers CDV-F4854 and CDV-R553510 or F5/R521 flanking the Fsp-coding region. In all cases, 4 µl cDNA was added to a PCR reaction mix, which consisted of 25 µl Maxima Hot Start PCR Master Mix (2X), 15 µl nuclease-free water and 3 µl (10 µM) of each of the primers (Table 1). PCR was performed on ProFlex™ PCR Thermal Cycler (Applied Biosystems®) under the following conditions: initial denaturation at 95 °C for 4 min followed by 35 cycles of denaturation at 95 °C for 30 s, annealing for 30 s, extension at 72 °C for 2 min, and a final extension at 72 °C for 10 min. The annealing temperature for H gen was 48.2 °C, and for the Fsp-coding region the temperature was 50.8 °C for the F5/R5 primers and 58 °C for the CDV-F4854/R5535 primers.
Table 1.
Oligonucleotides used for CDV P gene detection and for full length H gene and Fsp-coding region amplification and sequencing.
| Oligonucleotide label | Oligonucleotide sequence | Genomic position* | Reference |
|---|---|---|---|
| Amplification of P gene | |||
| CDV Universal (forward) | ATGTTTATGATCACAGCGGT | 2132–2151 | Daly et al., 2006 |
| CDV Universal (reverse) | ATTGGGTTGCACCACTTGTC | 2541–2560 | Daly et al., 2006 |
| Amplification and sequencing of H gene | |||
| CDVff1 (forward) | TCGAAATCCTATGTGAGATCACT | 6897–6919 | Lan et al.32 |
| CDVHS2 (reverse) | ATGCTGGAGATGGTTTAATTCAATCG | 8994–8969 | Lan et al.32 |
| CDVHS1 (forward) | AACTTAGGGCTCAGGTAGTCC | 7054–7074 | Lan et al.32 |
| CDVHforD (forward) | GACACTGGCTTCCTTGTGTGTAG | 7948–7970 | Lan et al.32 |
| CDVHr2 (reverse) | GTTCTTCTTGTTTCTCAGAGG | 8198–8178 | Lan et al.32 |
| CDVP2F (forward) | ACTTCCGCGATCTCCACT | 7372–7389 | Pardo et al.33 |
| CDVP3R (reverse) | ACACTCCGTCTGAGATAGC | 7760–7742 | Pardo et al.33 |
| CDVP5R (reverse) | GTGAACTGGTCTCCTCTA | 8395–8378 | Pardo et al.33 |
| Amplification and sequencing of Fsp-coding region | |||
| F5 (forward) | TGTTACCCGCTCATGGAGAT | 4272–4292 | Riley and Wilkes, 2015 |
| R5 (reverse) | CCAAGTACTGGTGACTGGGTCT | 5411–5433 | Riley and Wilkes, 2015 |
| CDV-F4854 (Forward) | TCCAGGACATAGCAAGCCAACA | 4854–4875 | Sarute et al., 2013 |
| CDV-R5535 (Reverse) | GGTTGATTGGTTCGAGGACTGAA | 5513–5535 | Sarute et al., 2013 |
*Reference genome AF164967 (A75/17).
Following PCR, 5 µl amplicons were analyzed by gel electrophoresis on a 1.5% agarose gel (AGAROSE I™, Amresco, Solon, OH, USA) at 110 V for 60 min. The gels were stained using EZ-VISION™ dye (Amresco1 Solon, OH, USA) and viewed by transillumination with UV light using the Molecular Imager® GelDoc TM XR + System with the image acquisition software ImageLab™ (Bio-Rad, Hercules, CA, USA). Amplification product sizes were estimated using a 100–3000 bp molecular weight ladder (GeneRuler™ 100 bp Plus DNA Ladder, Thermo Scientific®).
PCR amplicons of the H gene and Fsp-coding region were submitted to Macrogen Inc. (Seoul, Korea) for purification and sequencing. An additional set of eight primers, published elsewhere, for the H gene were used for sequencing32,33 (Table 1) using ABI3711™ automatic sequencer (Macrogen™).
Phylogenetic analysis
Sequence data were assembled and edited using SeqMan program (DNAStar Lasergene™ V15.0 software package, Madison, Wisconsin, USA). Nucleotide BLAST (Basic Local Alignment Search Tool) was used for exploring similarity between Colombian CDV strain sequences and all CDV sequences available in the NCBI nucleotide databases. For the H gene, a total length of 1824 nucleotides and corresponding deduced amino acid sequences were obtained only from dogs from Medellín and Bucaramanga, and for the Fsp-coding region (405 nucleotides and the corresponding deduced amino acid sequence), samples were obtained from all three studied cities (Bogotá DC, Medellín, and Bucaramanga). Phylogenetic analyses were carried out with at least two sequences for each reported lineage and vaccine strains from different geographical regions using MEGA™ 734 and the MUSCLE algorithm, and nucleotide and amino acid differences were assessed as uncorrected (p) distances.
Phylogenetic relationships based on the nucleotide alignment of complete H gene sequences were inferred using distance-based (neighbor-joining) and character-based (maximum likelihood, Bayesian) approaches implemented in MEGA™ 7 and MrBayes 3.2.635 software, respectively. The best-fit model for nucleotide substitution was identified by MEGA™ 7 as Tamura 3-parameter with gamma-distributed rate heterogeneity (T92 + G) according to the Bayesian information criterion for the H gene and Hasegawa-Kishino-Yano with gamma distribution (G) (HKY + G) for the Fsp-coding region. Maximum likelihood analysis was performed using the latter model; however, Bayesian inference analysis was performed with a general time-reversible model plus gamma-distributed rate heterogeneity (GTR + G) because the T92 + G model cannot be implemented in MrBayes 3.2.6. For this method, two parallel analyses were run for 1,000,000 generations with a 25% burn-in period. The convergence of the Markov chain Monte Carlo (MCMC) chains was assessed by the standard deviation of split frequencies, which fall below 0.01. The America-1 lineage was used as an outgroup to root the phylogenetic trees. The consensus trees were edited in FigTree software version 1.436.
Amino acid analysis of the H protein and the Fsp peptide
The deduced amino acid sequences of the H protein (607 aa) and the Fsp peptide (135 aa) of the Colombian wild-type CDV strains were aligned with multiple CDV protein sequences from different geographical regions using MEGA™ 7 for exploring their amino acid profiles and potential differences between vaccine and wild-type strains of known CDV lineages. Potential N-linked glycosylation sites were predicted using NetNGlyc 1.037.
Sites under positive selection
To identify amino acid sites under positive selection in the CDV H protein and Fsp peptide, the ratio of non-synonymous (dN) to synonymous (dS) substitutions was calculated by ML phylogenetic reconstruction using the general reversible nucleotide substitution model available through the Datamonkey web server. To detect non-neutral selection, Fast Unconstrained Bayesian AppRoximation (FUBAR) within the HyPhy software package was implemented in Datamonkey38. The range of significance for the posterior probability was 0–1. Generally, posterior probabilities > 0.9 are strongly suggestive of positive selection. Finally, a Bayes factor = 50 was used for estimating the rates of dN and dS within each codon. Values of dN/dS > 1, dN/dS = 1, and dN/dS < 1 were used for defining positive selection (adaptive molecular evolution), neutral mutations, and negative selection (purifying selection), respectively.
H gene and Fsp phylogeography
The mean substitution rate (substitutions per site per year), the time to the most recent common ancestor (TMRCA), the geographic origin, and the overall spatial dynamics of the major CDV clades were inferred using the Bayesian approach of the MCMC implemented in the BEAUti/BEAST v1.8.4 package39. The analysis was implemented using a strict molecular clock with a constant population size, and 3E07 generations were run in order to ensure an effective population size greater than 200 for the evaluated parameters using the Tracer v1.7 program39. The initial 10% of the MCMC, which corresponds with low probability states at the beginning of the chain, was eliminated. The tree of maximum credibility of the MCC clades was built with TreeAnnotator and visualized with FigTree v1.4.340.
Results
Detection of P gene and clinical features
A fragment of 429 bp of the phosphoprotein gene was detected in 68 (79.1%) clinical specimens from the 86 dogs sampled. In total, 44.1% of the CDV positive animals were male and 55.9% were female. Young dogs from one to six months old were the most affected (46.9%), although the disease also presented in dogs older than 12 months (21.9%). Concerning clinical manifestations in affected dogs, nervous and respiratory symptoms accounted for 25% of the cases, closely followed by the presentation of respiratory signs alone (21.4%). An equal proportion (17.9%) of clinically ill dogs presented with tegumentary/respiratory/nervous signs or respiratory/digestive symptoms alone.
Sequence analysis of the H gene and the Fsp-coding region
We only were able to amplify and sequence a fragment of 2099 bp of the H gene of six clinical specimens. Next, 405 bp of the Fsp-coding region was assessed, and we were able to obtain positive amplifications and sequence 23 clinical samples out of the 68 P gene positives Samples. Information regarding the age, gender, breed, vaccination status, and clinical signs of the dogs, as well as outcome and accession numbers from H gene–positive samples, is summarized in Table 2.
Table 2.
Clinical features of Colombian dogs infected with canine distemper virus (CDV).
| Samplea | Sexb | Agec | Clinical signsd | Vaccination status | Outcome | Gene | Region | Code GenBank | |
|---|---|---|---|---|---|---|---|---|---|
| MDE 2a/CO/2017 | NS | M | 2Y | R, GI | Unknown | Euthanized | H,F | MEDELLIN | MK617353/MK599333 |
| MDE 2aP/CO/2017 | CS | F | 6M | R,N | Unknown | Euthanized | H,F | MEDELLIN | MK617350/MK599334 |
| MDE 1aM/CO/2017 | CS | M | 8Y | R | Incomplete vacination | Euthanized | H,F | MEDELLIN | MK617348/MK599332 |
| MDE 13b/CO/2017 | CS | M | 7Y | R,N | Not vaccinated | Euthanized | H,F | MEDELLIN | MK617351/MK599328 |
| MDE 16a/CO/2017 | CS | F | 9M | N | Not vaccinated | Euthanized | H,F | MEDELLIN | MK617352/MK599329 |
| MDE 18 a/CO/2017 | CS | M | 7M | R,N | Incomplete vacination | Euthanized | H | MEDELLIN | MK617349 |
| BUCA 12/CO/2016 | CS | F | 3M | R,N | Incomplete | Died | F | BUCARAMANGA | MK599327 |
| MDE 19a/CO/2017 | CS | F | 5M | R | Incomplete | Unknown | F | MEDELLIN | MK599331 |
| MDE 9a/CO/2017 | CS | F | 7Y | R,N | Not vaccinated | Euthanized | F | MEDELLIN | MK599337 |
| MDE 18/CO/2012 | CS | M | Unknown | R,N | Unknown | Unknown | F | MEDELLIN | MK599330 |
| MDE 42e/CO/2012 | CS | M | 3M | GI,N | Not vaccinated | Unknown | F | MEDELLIN | MK599335 |
| MDE 44/CO/2012 | CS | M | 5M | R,N | Unknown | Unknown | F | MEDELLIN | MK599336 |
| CM-14–027 | BLOOD | Unknown | 1M | R,GI,N | Unknown | Unknown | F | BOGOTÁ | MK617339 |
| CM-14-160 | URINE | Unknown | 2M | R,GI,N | F | BOGOTÁ | MK617343 | ||
| CM-15-001 | URINE | Unknown | Unknown | R | Unknown | Unknown | F | BOGOTÁ | MK617344 |
| CM-15-061 | URINE | Unknown | 8M | R | Unknown | Unknown | F* | BOGOTÁ | — |
| CM-15-089 | BLOOD | Unknown | 4M | NS | Unknown | Unknown | F | BOGOTÁ | MK617340 |
| CM-15-079 | BLOOD | Unknown | 4M | NS | Unknown | Unknown | F | BOGOTÁ | MK617341 |
| CM-15-018 | URINE | Unknown | 3M | Unknown | Unknown | Unknown | F | BOGOTÁ | MK617345 |
| CM-15-052 | URINE | Unknown | 14M | Unknown | Unknown | Unknown | F* | BOGOTÁ | — |
| CM-15-061 | URINE | Unknown | 8M | R | Unknown | Unknown | F* | BOGOTÁ | — |
| CM-15-066 | URINE | Unknown | 6M | R,GI,N | Unknown | Unknown | F | BOGOTÁ | MK617346 |
| CM-15-069 | BLOOD | Unknown | 5M | NS | Unknown | Unknown | F | BOGOTÁ | MK617347 |
| CM-15-078 | BLOOD | Unknown | 4M | NS | Unknown | Unknown | F* | BOGOTÁ | — |
| CM-15-135 | NS | Unknown | 4Y | R | Unknown | Unknown | F* | BOGOTÁ | — |
| CM-15-171 | BLOOD | Unknown | 3M | GI,T | Unknown | Unknown | F | BOGOTÁ | MK617342 |
a S: Serum; CS: Conjunctival swab – b F: female; M: male – c M: months; Y: Years – d R: Respiratory; GI: Gastrointestinal; O: Ocular; N: Neurological; T: Tegumentary; NS: Without Signs * Sequences don´t include in the phylogenetic analysis.
The Fsp-coding region sequences of the strains from Medellín, D/Mde_19a/CO/2017, D/Mde_2aP/CO/2017, D/Mde_13b/CO/2017, and D/Mde_16a/CO/2017, were 100% identical. Likewise, the sequences from Bogotá, D/Bog-4/CO/2015, D/Bog-5/CO/2015, and D/Bog-6/CO/2015, were 100% identical to each other. Consequently, only one of those sequences (D/Bog-4/CO/2015) were included with the rest of the Colombian strains in the phylogenetic analysis. Vaccines used as positive controls in the PCR reactions were also sequenced.
As we previously reported17, BLAST analysis of H sequence data from commercial Colombian vaccines used as positive controls in the present study revealed that vaccines had 99–100% identity with the vaccine strains from the North America-1 lineage, and one vaccine showed 99% identity with a Rockborn vaccine strain.
Colombian H sequences subjected to analysis displayed high identity with each other (93.5–99.9% nt; 93–99.9% aa) with an overall mean distance of 0.039. Alignment of the H gene of Colombian CDV strains and the Onderstepoort vaccine strain (AF378705) showed an identity that varied between 89.6% and 91.1% at the aa level and between 90.8% and 91.8% at the nt level. As expected, higher variability was found in the Fsp-coding region (81.39–99.01% nt, 63.91–97.7% aa) with an overall mean distance of 0.1014. Moreover, the Colombian Fsp-coding region sequences displayed very low identity with those of the Onderstepoort vaccine strain (80.9–83.6% nt, 57.6–67.4% aa).
Phylogenetic relationships based on the nucleotide alignment of complete H gene sequences inferred by distance (neighbor-joining) and character approaches (maximum likelihood and Bayesian inference) resulted in trees with similar topology. The phylogenetic tree of the H gene showed 16 lineages with a defined geographical distribution pattern (the Asia-3 lineage was grouped with strains of the America-1 lineage), while the Fsp tree only showed 15 lineages, primarily because there are no available Fsp sequences for the European Wildlife lineage.
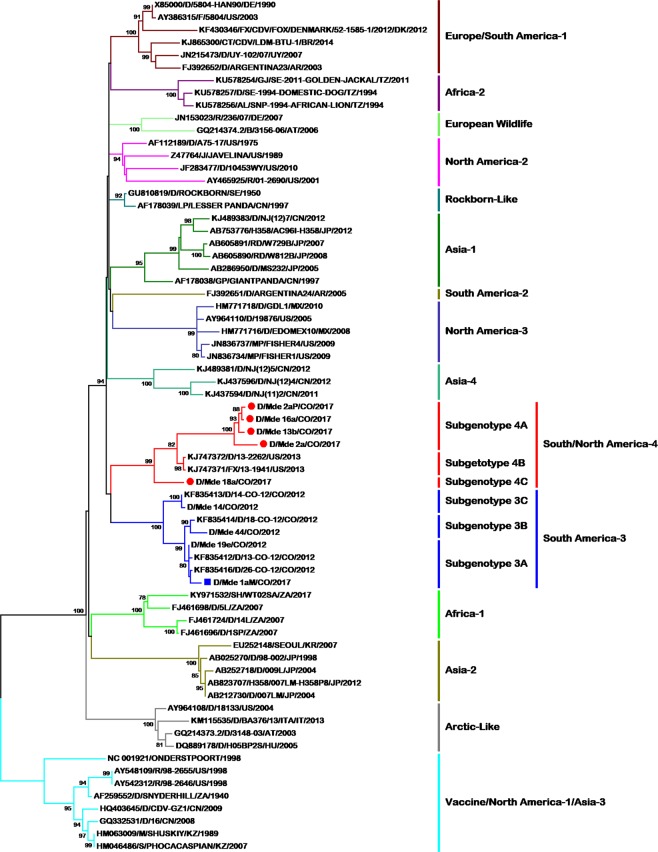
Interestingly, we showed that Colombian CDV sequences cluster in two different branches in both the Fsp and H gene trees (Figs 1 and 2); one group of Colombian Fsp-coding sequences cluster in the same clade as Ecuadorian strains (Fig. 1), and also, interestingly, with two recently reported North America-4 lineage sequences21 (97.1% identity). Other Colombian CDV sequences cluster with the South America-3 lineage previously reported in Colombia (Fig. 2). Unfortunately, no Ecuadorian H sequences have been reported to date.
Figure 1.
Phylogenetic relationships between 64 CDV strains based on Fsp fragment gene sequences. The phylogenetic tree was inferred by the maximum likelihood method using 1000 replicates. GenBank accession numbers, the species from which each isolate was obtained, name of the strain, country of origin, and year of isolation are indicated in the tip labels if available. Numbers at the nodes are bootstrap values for the clade. Abbreviations for animal species: AL: African lion (Panthera leo), AWD: African wild dog (Lycaon pictus), B: badger (Meles meles), BS: Baikal seal (Pusa sibirica), D: dog (Canis lupus familiaris), F: ferret (Mustela putorius furo), FX: fox (Vulpes urocyon), H358: human lung cells, M: mink (Neovison vison), R: raccoon (Procyon lotor), RD: raccoon dog (Nyctereutes procyonoides), S: seal (Phoca vitulina), SH: spotted hyena (Crocuta crocuta). Abbreviations for countries: AR: Argentina, BR: Brazil, CN: China, CO: Colombia, DE: Germany, EC: Ecuador, GA: Gabon, IT: Italy, JP: Japan, KZ: Kazakhstan, RU: Russia, TZ: Tanzania, US: United States, UY: Uruguay, ZA: South Africa.
Figure 2.
Phylogenetic relationships between 68 CDV strains based on H gene sequences. The phylogenetic tree was inferred by the maximum likelihood method using 1000 replicates. GenBank accession numbers, the species from which each isolate was obtained, name of the strain, country of origin, and year of isolation are indicated in the tip labels if available. Numbers at the nodes are bootstrap values for the clade. Abbreviations for animal species: AL: African lion (Panthera leo), B: badger (Meles meles), CT: Cerdocyon thous, D: dog (Canis lupus familiaris), F: ferret (Mustela putorius furo), FX: fox (Vulpes urocyon), GJ: golden jackal (Canis aureus), GP: giant panda (Ailuropoda melanoleuca), J: javelina (Tayassu pecari), LP: lesser panda (Ailurus fulgens), M: mink (Neovison vison), MP: Martes pennanti, R: raccoon (Procyon lotor), RD: raccoon dog (Nyctereutes procyonoides), S: seal (Phoca vitulina), SH: spotted hyena (Crocuta crocuta). H358: human lung cells. Abbreviations for countries: AR: Argentina, AT: Austria, BR: Brazil, CN: China, CO: Colombia, DE: Germany, DK: Denmark, HU: Hungary, IT: Italy, JP: Japan, KR: South Korea, KZ: Kazakhstan, MX: Mexico, SE: Sweden, TZ: Tanzania, US: United States, UY: Uruguay, ZA: South Africa.
Keeping in mind that Colombian viruses showed high variability at both the nucleotide and amino acid levels, we analyzed the identity of Colombian CDV H sequences and the previously reported CDV lineages (Table 3). By analysis of uncorrected (p) distances, we found that the amino acid sequences of Colombian and Ecuadorian CDV strains of the South America-4 lineage differed by less than 4% to those of strains of the previously reported North America-4 lineage. According to this, we propose that this lineage should be termed “South America/North America-4” due its intercontinental distribution. Remarkably, we observed high variation (approximately 10%) between Colombian CDV lineages and the North America-1 lineage, which includes most of the commercial vaccine strains (Table 3).
Table 3.
Uncorrected distances (p) observed in pairs of amino acid sequences of the Fsp and H genes between CDV lineages.
| NA-4 | SA-4 | SA-3 | VAC | SA-2 | NA-1 | NA-2 | NA-3 | EU/SA-1 | ARC-L | EU-WL | AFR-1 | AFR-2 | ASIA-1 | ASIA-2 | ASIA-3 | RCK-L | ASIA-4 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NA-4 | 0,024 | 0,035 | 0,039 | 0,036 | 0,038 | 0,037 | 0,036 | 0,032 | 0,38 | NA | 0,038 | 0,037 | 0,035 | 0,038 | 0,041 | 0,033 | NA | |
| 0,01 | 0,018 | 0,022 | 0,017 | 0,021 | 0,013 | 0,016 | 0,015 | 0,02 | 0,017 | 0,017 | 0,017 | 0,016 | 0,02 | 0,022 | 0,013 | 0,017 | ||
| SA-4 | 0,124 | 0,034 | 0,038 | 0,034 | 0,037 | 0,035 | 0,036 | 0,032 | 0,036 | NA | 0,036 | 0,036 | 0,034 | 0,036 | 0,039 | 0,031 | NA | |
| 0,028 | 0,018 | 0,022 | 0,017 | 0,021 | 0,014 | 0,016 | 0,016 | 0,02 | 0,018 | 0,017 | 0,017 | 0,016 | 0,02 | 0,022 | 0,014 | 0,017 | ||
| SA-3 | 0,292 | 0,314 | 0,036 | 0,033 | 0,033 | 0,03 | 0,033 | 0,028 | 0,035 | NA | 0,035 | 0,033 | 0,031 | 0,034 | 0,038 | 0,028 | NA | |
| 0,064 | 0,078 | 0,024 | 0,019 | 0,023 | 0,016 | 0,016 | 0,016 | 0,021 | 0,019 | 0,019 | 0,019 | 0,017 | 0,021 | 0,024 | 0,016 | 0,018 | ||
| VAC | 0,371 | 0,405 | 0,367 | 0,037 | 0,033 | 0,037 | 0,038 | 0,035 | 0,037 | NA | 0,036 | 0,038 | 0,037 | 0,038 | 0,017 | 0,035 | NA | |
| 0,111 | 0,12 | 0,132 | 0,02 | 0,01 | 0,02 | 0,020 | 0,019 | 0,022 | 0,022 | 0,019 | 0,021 | 0,021 | 0,022 | 0,012 | 0,02 | 0,022 | ||
| SA-2 | 0,257 | 0,262 | 0,249 | 0,332 | 0,035 | 0,033 | 0,035 | 0,029 | 0,036 | NA | 0,035 | 0,035 | 0,3 | 0,036 | 0,038 | 0,028 | NA | |
| 0,054 | 0,06 | 0,076 | 0,099 | 0,019 | 0,014 | 0,016 | 0,013 | 0,019 | 0,018 | 0,017 | 0,018 | 0,016 | 0,019 | 0,021 | 0,014 | 0,016 | ||
| NA-1 | 0,284 | 0,324 | 0,255 | 0,228 | 0,239 | 0,026 | 0,036 | 0,032 | 0,035 | NA | 0,034 | 0,035 | 0,034 | 0,037 | 0,038 | 0,03 | NA | |
| 0,105 | 0,112 | 0,127 | 0,04 | 0,09 | 0,019 | 0,020 | 0,018 | 0,021 | 0,021 | 0,019 | 0,021 | 0,021 | 0,022 | 0,009 | 0,019 | 0,022 | ||
| NA-2 | 0,246 | 0,268 | 0,206 | 0,321 | 0,194 | 0,104 | 0,033 | 0,028 | 0,035 | NA | 0,034 | 0,032 | 0,03 | 0,035 | 0,04 | 0,025 | NA | |
| 0,045 | 0,058 | 0,069 | 0,108 | 0,051 | 0,01 | 0,012 | 0,01 | 0,017 | 0,013 | 0,014 | 0,014 | 0,012 | 0,017 | 0,02 | 0,008 | 0,013 | ||
| NA-3 | 0,261 | 0,285 | 0,251 | 0,313 | 0,228 | 0,231 | 0,201 | 0,03 | 0,037 | NA | 0,035 | 0,035 | 0,034 | 0,039 | 0,041 | 0,03 | NA | |
| 0,043 | 0,059 | 0,056 | 0,1 | 0,049 | 0,094 | 0,044 | 0,01 | 0,018 | 0,016 | 0,016 | 0,016 | 0,015 | 0,019 | 0,021 | 0,012 | 0,016 | ||
| EU/SA-1 | 0,242 | 0,263 | 0,235 | 0,337 | 0,205 | 0,242 | 0,176 | 0,216 | 0,034 | NA | 0,032 | 0,03 | 0,028 | 0,031 | 0,036 | 0,022 | NA | |
| 0,048 | 0,063 | 0,059 | 0,093 | 0,042 | 0,088 | 0,046 | 0,033 | 0,017 | 0,015 | 0,014 | 0,015 | 0,014 | 0,017 | 0,019 | 0,01 | 0,013 | ||
| ARC-L | 0,355 | 0,344 | 0,318 | 0,323 | 0,296 | 0,261 | 0,259 | 0,294 | 0,287 | NA | 0,034 | 0,035 | 0,036 | 0,033 | 0,038 | 0,032 | NA | |
| 0,085 | 0,099 | 0,106 | 0,121 | 0,081 | 0,114 | 0,083 | 0,078 | 0,078 | 0,02 | 0,018 | 0,021 | 0,018 | 0,017 | 0,022 | 0,018 | 0,019 | ||
| EU-WL | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
| 0,063 | 0,076 | 0,085 | 0,118 | 0,066 | 0,111 | 0,058 | 0,06 | 0,062 | 0,094 | 0,017 | 0,017 | 0,016 | 0,02 | 0,022 | 0,014 | 0,017 | ||
| AFR-1 | 0,343 | 0,351 | 0,3 | 0,325 | 0,267 | 0,25 | 0,254 | 0,291 | 0,273 | 0,288 | NA | 0,036 | 0,032 | 0,035 | 0,038 | 0,031 | NA | |
| 0,064 | 0,08 | 0,084 | 0,099 | 0,07 | 0,093 | 0,062 | 0,062 | 0,06 | 0,074 | 0,071 | 0,018 | 0,016 | 0,018 | 0,02 | 0,014 | 0,017 | ||
| AFR-2 | 0,275 | 0,291 | 0,237 | 0,312 | 0,252 | 0,241 | 0,189 | 0,241 | 0,202 | 0,252 | NA | 0,316 | 0,033 | 0,037 | 0,04 | 0,029 | NA | |
| 0,064 | 0,079 | 0,083 | 0,109 | 0,056 | 0,107 | 0,06 | 0,061 | 0,064 | 0,1 | 0,073 | 0,078 | 0,017 | 0,021 | 0,022 | 0,014 | 0,018 | ||
| ASIA-1 | 0,274 | 0,302 | 0,245 | 0,343 | 0,215 | 0,256 | 0,204 | 0,254 | 0,223 | 0,322 | NA | 0,305 | 0,242 | 0,034 | 0,039 | 0,027 | NA | |
| 0,05 | 0,063 | 0,068 | 0,109 | 0,079 | 0,106 | 0,048 | 0,048 | 0,049 | 0,085 | 0,064 | 0,064 | 0,066 | 0,016 | 0,022 | 0,012 | 0,014 | ||
| ASIA-2 | 0,308 | 0,307 | 0,27 | 0,317 | 0,248 | 0,226 | 0,203 | 0,271 | 0,221 | 0,251 | NA | 0,27 | 0,258 | 0,273 | 0,04 | 0,031 | NA | |
| 0,079 | 0,093 | 0,099 | 0,112 | 0,09 | 0,112 | 0,078 | 0,077 | 0,073 | 0,069 | 0,088 | 0,069 | 0,089 | 0,056 | 0,023 | 0,018 | 0,019 | ||
| ASIA-3 | 0,384 | 0,408 | 0,388 | 0,071 | 0,328 | 0,254 | 0,343 | 0,321 | 0,336 | 0,352 | NA | 0,328 | 0,323 | 0,353 | 0,321 | 0,037 | NA | |
| 0,102 | 0,113 | 0,124 | 0,045 | 0,054 | 0,025 | 0,099 | 0,091 | 0,084 | 0,112 | 0,108 | 0,09 | 0,106 | 0,103 | 0,109 | 0,02 | 0,022 | ||
| RCK-L | 0,194 | 0,205 | 0,175 | 0,246 | 0,138 | 0,134 | 0,104 | 0,149 | 0,116 | 0,223 | NA | 0,205 | 0,152 | 0,164 | 0,159 | 0,246 | NA | |
| 0,03 | 0,046 | 0,052 | 0,093 | 0,036 | 0,087 | 0,028 | 0,028 | 0,03 | 0,067 | 0,045 | 0,046 | 0,046 | 0,032 | 0,061 | 0,084 | 0,013 | ||
| ASIA-4 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
| 0,056 | 0,067 | 0,069 | 0,108 | 0,054 | 0,106 | 0,054 | 0,05 | 0,046 | 0,093 | 0,071 | 0,072 | 0,072 | 0,048 | 0,081 | 0,102 | 0,131 |
NA-4: North America-4; SA-4: South America-4; SA-3: South America-3;VAC: Vaccine; SA-2: South America-2; NA-1:North America-1, NA-2:North America-2;NA-3: North America-3; EU/SA-1:Europe/South America-1; ARC-L:Arctic-Like; EU/WL: European Wildlife; AFR-1: Africa-1; AFR-2: Africa-2;RCK-L: Rockborn-Like. Fsp values are bold. Standard error estimates are shown (in italics) above the diagonal and were obtained by a bootstrap procedure with 1000 replicates. NA: values not computed due lack of Fsp sequences.
CDV subgenotype analysis
In CDV subgenotype analysis, based on criteria for measles Paramyxovirus (H amino acid identity of 98% and bootstrap values >70%), we identified at least three subgenotypes in the South America-3 lineage. Subgenotype A included strains 13-CO-12, 19-CO-2012, 26-CO-12, and 1aM-CO-2017 (aa variation 0.2–0.06%); subgenotype B included strains 18-CO-2012, 40-CO-2012, and 44-CO-2012 (aa variation 0.8–1%); and subgenotype C included strains 14-CO-2012 and 39-CO-2012 (aa variation 2.2%) (subgenotypes 3A to 3C in Fig. 2).
Also, we identified three subgenotypes within the “South America/North America-4” lineage. Subgenotype A included strains 16a-CO-2017, 2aP/CO-2017, 13b-CO-2017, and 2a-CO-2017 (aa variation 0.2–1.4%); subgenotype B included strains 13-1941-US-2013 and 13-2262-US-2013 (aa variation 2.6%) (American strains); and subgenotype C included strain 18a-CO-2017 (aa variation 4.3%) (subgenotypes 4A to 4C in Fig. 2).
For evaluating this subclassification for the Fsp-coding region, we arbitrarily extrapolated the classification and found five subgenotypes in the South America-3 lineage (3A to 3E in Fig. 1). Subgenotype A included strains 42-CO-2012, 1aM-CO-2017, and BUC-12-CO-2016 (aa variation 1.2–2.5%) and subgenotype B included strains 18-CO-2012 and 44-CO-12 (aa variation 2–3.5%); both of these subgenotypes were also found with the H gene. Two other subgenotypes were not characterized with the H gene: subgenotype D, including eight Bogotá 2015 strains (aa variation 4.7–5.7%) and subgenotype E, including strains BOG2-CO-2014 and BOG11-CO-2015 (aa variation 5.5–5.7%).
Likewise, in the Fsp analysis of the “South America/North America-4” lineage, we observed four well-defined subgenotypes (4A to 4D in Fig. 1). Subgenotype A included strains 16-CO-2017, 2aP/CO-2017, 13b-CO-2017, 9-CO-2017, and 2-CO-2017 (aa variation 0–1.5%); subgenotype B included strains 13-1941-US-2013, 13-2262-US-2013, and 19-CO-2017 (aa variation 5.5–11.2%); and subgenotype D included Ecuadorian strains not characterized with the H gene (aa variation 1.5–2%).
Amino acid analysis of the H protein
Analysis of the deduced amino acid sequences of the full-length H protein (607 amino acids) of Colombian CDV viruses showed the presence of exclusive substitutions also found in the South America-3 (N261S, G488R, T544S) and the “South America/North America-4” lineages (E333V, T348K). Remarkably, we found a set of substitutions that are common to both Colombian lineages (T193I, V198I, E333V, S343L, T348K, and A365T); however, the substitutions in the South America-3 lineage appear in the new sequences and not in the previously reported 2012 sequences.
Colombian strains showed the same residue, isoleucine, at position 506 as vaccine strains belonging to the America-1 lineage. Several substitutions in Colombian strains were also present in other wild-type strains reported in different lineages: S22R (America-2), V41I (European Wildlife), N128S (Africa-1), K281R (America-2), G314S (Asia-2 and America-2), and I315V (European Wildlife). All Colombian South America-3 lineage sequences carried asparagine at position 530, a highly variable residue linked to interspecies transmission of the virus (McCarthy et al. 2007). The South America-4 sequences included a serine at position 530, while North America-4 displayed an aspartic acid in this position. The H sequences of the 2017 “South America/North America-4” lineage displayed the following unique substitutions: Q5R, L38S, T193I, V198I, V235I, T291M, E333V, H339D, S341L, T348K, and F353I.
In the present study, three sequences from three dogs that had not been completely vaccinated were grouped in the “South America/North America-4” lineage; one strain had a very extensive branch (Fig. 1; sequence Mde-19a-CO-2017), while another was shown to be related to CDV strains circulating in North America (Fig. 2; sequence Mde-18a-CO-2017), and the final strain, 1aM-CO-2017, was shown to belong to the South America-3 lineage (Fig. 2). A comparison of the linear hemagglutinin noose epitope (HNE) between the vaccine strains and these Colombian strains showed the presence of multiple substitutions (Supplemental material Fig. S2). Sequence 18a-CO-2017 presented the following substitutions: A367V, E372D, G376N, and T386S, while strain 1aM-CO-2017 of the South America-3 lineage presented A367V, G376N, and T386S.
Amino acid analysis of the Fsp peptide
The Fsp peptide possesses 95 variable amino acids (of 135 total). We found 16 substitutions in its sequence that are exclusive to the South America-3 lineage: S9P, T32I, A35S, T40P, D54N, R55K, S58N, Y59H, M61T, R67F H80C, H83R, I102S, Q115H, C116F, and L129F. In the Fsp peptide of the “South America/North America-4” lineage, 20 exclusive substitutions were found: D28S, E29G, A35T, N62D, S71G, H80R, V94I, N108T, S112P, S114P, and K134E. Furthermore, a different set of amino acid substitutions with a specific geographic pattern were found only in South America-4: T13V, T40K, S45F, S58Q, V79I, S95P, R105W, and G113C. In the most identical strains “South America/North America-4” we only found the substitutions T13M and N76S, indicating autapomorphic characteristics.
CDV H and Fsp glycosylation analysis
Potential glycosylation sites for the CDV H protein from the South America-3 lineage have previously been reported17. No new potential glycosylation sites were found in the 2017 samples belonging to this lineage in new sampling areas in Colombia. For the “South America/North America-4” lineage, we identified the presence of eight potential glycosylation sites (NXS/T) at positions 19–21, 149–151, 309–311, 391–393, 422–424, 456–458, 587–589, and 603–605, which are common to other lineages. The previously reported Asia-1 potential glycosylation sites at positions 584–58641 were not present in the sequences of the “South America/North America-4” lineage.
Regarding the potential glycosylation analysis of the Fsp peptide of the South America-3 lineage, we found the presence of two potential glycosylation sites (NXS/T) at positions 62–64 and 108–110 that are common to the other lineages. However, no potential glycosylation sites were found in the any of the “South America/North America-4” sequences.
CDV H and Fsp sites under positive selection
We evaluated non-neutral selection using the FUBAR method. For the H gene, we found that sequences harbored three sites under positive selection: 522, 549, and 582, with a posterior probability of 0.9 and a Bayes factor of 28.3, 220.9, and 44.7 respectively. We also found 247 sites under negative selection with a posterior probability of 0.9 and Bayes factor < 1.
In the Fsp-coding region, we found nine sites under positive selection using the FUBAR method: 21, 39, 46, 51, 79, 98, 99, 101, and 102, with a posterior probability of 0.9 and a Bayes factor of 67, 306, and 88 in sites 21, 76, and 98, respectively. Conversely, we found five sites under negative selection: 45, 83, 89, 121, and 133, with a posterior probability of 0.9 and a Bayes factor < 1.
H gene and Fsp phylogeography
For this analysis, the vaccine sequences were eliminated since there were no exact dates of isolation for these strains, which would have produced biases in the substitution rates. Table 4 shows the evolutionary model for each gene. Also, Table 5 shows the estimated TMRCA for the H gene. The South America-3 and “South America/North America-4” lineages have a TMRCA corresponding to 1964 and 1925, respectively (see Table 5). The phylogeography of the Fsp and H gene is shown in Figs 3 and 4, respectively.
Table 4.
Evolutionary Parameters in Gene H and Fsp coding region.
| Evolution model | Evolution rate | Rate dN/dS | tMRCA | HDP 95% Interval | ||
|---|---|---|---|---|---|---|
| Mean | HDP 95% Interval | |||||
| H | T92 + G | 4,87 × 10−4 | 3,78 × 10−4–5,94 × 10−4 | 0,024 | 1900 | 1873–1925 |
| Fsp | HKY + G | 1,642 × 10−3 | 1,12 × 10−3–2,12 × 10−3 | 0,4 | 1942 | 1914–1964 |
Table 5.
H gene molecular clocks in different CDV lineages.
| Lineage | tMRCA | N° Sequences | |
|---|---|---|---|
| Mean | HDP 95% Interval | ||
| North America-1 | 1998 | 1996–1997 | 2 |
| North America-2 | 1957 | 1947–1965 | 3 |
| North America-3 | 2005 | n/a | 1 |
| Asia-1 | 1971 | 1961–1980 | 6 |
| Asia-2 | 1991 | 1986–1996 | 5 |
| Asia-3 | 1999 | 1994–2003 | 2 |
| Asia-4 | 1979 | 1969–1988 | 3 |
| Africa-1 | 1982 | 1973–1991 | 4 |
| Africa-2 | 1985 | 1979–1990 | 3 |
| Arctic -Like | 1992 | 1986–1997 | 4 |
| European wildlife | 1975 | 1965–1985 | 2 |
| Rockborn-Like | 1944 | 1932–1956 | 2 |
| Europe/South America-1 | 1975 | 1966–1982 | 6 |
| South America-2 | 2005 | n/a | 1 |
| South America-3 | 1964 | 1943–1983 | 8 |
| South/North America-4 | 1925 | 1891–1955 | 7 |
Figure 3.
Time-measured Bayesian MCC tree for CDV Fsp fragment. Branches are colored according to the country color code in the upper left. Colombian sequences are depicted in red.
Figure 4.
Time-measured Bayesian MCC tree for CDV H gene. Branches are colored according to the country color code in the upper left. Colombian sequences are depicted in red.
Discussion
Phylogenetic characterization of CDV is performed on the basis of the H gene sequence because this gene shows high nucleotide variability in CDV between field strains and vaccine strains in comparison with other paramyxoviruses42,43. Using this method, it is accepted that two strains belong to the same lineage when their amino acid diversity is less than 4%42. Presently, there are 17 known lineages worldwide with a geographical distribution pattern. However, a fragment that codes for the Fsp of CDV has been suggested as an alternative for the classification of CDV strains since it is highly divergent and has given similar classification results to the H gene. Using this approach, two strains are considered to belong to the same CDV lineage if their amino acid divergence is less than 19%10.
In 2012, based on samples only from Medellin city17, it was characterized the South America-3 lineage by using the full H gene sequencing approach. However, it was not possible to compare the circulation of this CDV lineage with viruses from other regions of Colombia or in neighboring countries. In the present study, the Fsp-coding region was sequenced from samples obtained in Medellín between 2012 and 2017 and from other Colombian cities such as Bucaramanga (2016) and Bogotá (2015), establishing the wide circulation of the CDV South America-3 lineage in these regions (Fig. 1). In agreement with Sarute et al.10, the amino acid divergence among the strains (1.5–6.7%) showed that they belonged to the same lineage (data not shown).
Through the phylogenetic analysis of the Fsp-coding region of the present study, we observed that the sequences of the Ecuadorian strains and the former called “America-4” lineage formed a monophyletic group, evidencing a 12% amino acid divergence, which was supported by the table of distances (Table 3). This led us to the conclusion that those viral sequences belong to the same lineage. However, for confirming this finding, we try to amplify the H gene of the same samples. In the resulting phylogenetic tree, we observed a monophyletic group of the strains of the lineage America-4 And also the Ecuadorian strain, which harbored an amino acid divergence in the H gene of 2.8%, showing that these strains belong to the same CDV lineage. For this reason, we suggest calling this lineage “South America/North America-4.”
For comparing the topologies of the phylogenetic trees, we used the North America-1/Vaccine lineage as an outgroup. We observed that although the lineages characterized in this work are sister groups, they present different ancestry. In the H gene tree, the oldest clades are the Africa-1, Arctic-like, and Asia-2 lineages. From this node, the Asia-4, South America-3, and “South America/North America-4” lineages appear as sister groups. A polytomy emerges, from which lineages emerge as sister groups: North America-3 and South America-2; North America-2, European Wildlife, Asia-1, and Rockborn-like; and Africa-2 and Europe/South America-1.
In the Fsp-coding region tree, the ancestral clades of the characterized lineages arise from a polytomy from which sister groups originate: Arctic-like and Asia-2; Africa-1; North America-1 and North America-2; and Rockborn-like (Fig. 1). From this node, another polytomy emerges, giving rise to more sister groups as follows: the Europe/South America-1, South America-2, and Africa-2; and Asia-1, North America-3, South America-3, and “South America/North America-4.” In their 2013 study, Sarute et al. described topological differences between the Fsp and H gene trees10. Although the topological structures they identified did not entirely correspond with those found in the present study (Figs 1 and 2), it should be taken into account that nowadays, there are a higher number of Fsp coding sequences available, representing most of the lineages characterized to date; besides, Fsp-coding region sequences could now be obtained from Genbank full CDV genomes. However standard phylogenetic studies still been done with the H gene, thus having a greater number of sequences available in the Genbank.
On the basis of the classification system for measles, a subgenotype consists of H gene sequences that have an amino acid identity of 98% and a high bootstrap value (>70%)28. On the basis of these criteria, both the South America-3 and “South America/North America-4” lineages present three subgenotypes. When we arbitrarily extrapolated this classification to the Fsp region, a different set of subgenotypes was found (subgenotypes A–E for South America-3 and A–C for “South America/North America-4”), although fewer than for Europe/South America-1, which reportedly contains at least eight subgenotypes (A–H), including CDV H gene sequences from at least seven different countries28.
In measles, different subgenotypes are not geographically restricted, although some appear to be mainly endemic in different areas of the world44. In the present study, it was not possible to determine the geographic pattern of CDV subgenotypes on the basis of the H gene as has been previously reported for the Europe/South America-1 and South African subgenotypes6,45 and for measles46. However, with the Fsp fragment, distribution patterns can be observed between regions (Fig. 1); subgenotype 3A circulates only in Medellín, while subgenotype 3D circulates only in Bogotá. Also, in the “South America/North America-4” lineage, subgenotype 4A was only reported in Colombian strains and subgenotype 4D in Ecuadorian strains. A higher number of CDV sequences collected from different areas within those countries would be necessary for better understanding of the circulation history of CDV subgenotypes in the Americas.
A temporary pattern of distribution has been reported for some of the Europe/South America-1 and South African subgenotypes as well as for measles virus genotypes and subgenotypes28,44,46. Our results showed a similar temporary pattern of distribution in most of the subgenotypes in both the South America-3 and the “South America/North America-4” lineages (Figs 1 and 2). These results must be carefully evaluated, as although they may show a temporary pattern of CDV distribution and a possible strain displacement pattern, sampling bias could be another possible explanation. Routine international determination of CDV lineages and subgenotypes plus molecular surveillance could be useful for gaining a more accurate epidemiological understanding of temporary CDV distribution.
The uncontrolled commercialization of puppies from South America in the USA could be the route of transmission of the “South America/North America-4” lineage in these two regions of the continent. It is important to highlight that this is the second lineage that is reportedly actively circulating in two different continental regions, the first being Europe/South America-120. It is imperative that wider phylogeographic studies of the “South America/North America-4” lineage are conducted to establish its origin and geographical spread throughout the American continent; it may have originated in Ecuador and spread through Colombia to the USA, or vice versa. Since CDV is a re-emerging infection in the USA, with at least five different lineages in circulation15, deeper phylogenetic analysis could help in gaining an understanding of the epidemiology of CDV on this continent.
In the present study, an amino acid divergence close to 11% in the H protein was observed between vaccine strains and the lineages South America-3 and “South America/North America-4” (Table 3). CDV is presently recognized as a single serotype47 as there is little evidence of antigenic divergence as a result of genetic divergence. Recently, significant differences were reported in the evaluation of neutralizing titers between “South America/North America-4” lineage strains and an America-1 type vaccine strain27. Given those results and the fact that multiple recognized CDV cases have been recorded in vaccinated animals17,21,28,48, it is necessary to perform wider, updated antigenic analyses of CDV for understanding the antigenic differences between the multiple worldwide circulating lineages and, potentially, to produce a vaccine update that includes most prevalent antigenic types.
In the positive selection analysis, we observed that the “South America/North America-4” lineage possesses a unique substitution (V79I) in the Fsp fragment at a site that is under positive selection; the South America-3 lineage also possesses a unique substitution (I102S) at a site under positive selection. This was determined using the FUBAR method, which assumes that the selection pressure for each site is constant throughout the phylogeny38. In this way, it was also determined that sites 98, 99, 101, and 102 of Fsp are under positive selection. These changes must be studied to understand the role of such substitutions in vaccine failures and interspecies host changes.
Likewise, we found in gene H sites 522, 549, and 582 under positive selection coinciding with site 549, which has been proposed as a key in the species barrier jump4, however sites 522 and 582 have not been previously reported related to pathogenicity, vaccine failure or species barrier jump, which should be deeply studied to understand the role of this sites in pathogenesis and inter-species transmission.
The linear HNE located at amino acids 364–392 of the CDV H protein is conserved among the morbilliviruses49. This is the region of the H protein that is recognized by antibodies50,51. Recently, it has been suggested that substitutions in this epitope may interfere with the ability of the vaccine to provide adequate protection against infection with wild-type viruses15. As reported recently in the previously called “North America-4” strains, we observed the presence of multiple substitutions in the HNE of CDV viruses in vaccinated animals (Supplemental Material Fig. S2). However, from the bioinformatic approach used in this study, we can only suggest that these substitutions could be interfering with the capacity of the vaccine to neutralize wild strains. For this reason, it is necessary to perform neutralization studies of vaccines with wild strains. Currently, structural biology studies of Colombian CDV viruses are underway in an effort to understand the role of structural substitutions in the HNE epitope of Colombian CDV viruses and their role in viral neutralization. On this way, it is important to consider thate glycosylation sites found in the Fsp region of the South America-3 lineage (62–64 and 108–110) could be involved in the evasion of the response or could be the result of epistatic interactions in the H gene52.
CDV has one of the highest-reported substitution rates in the Paramyxoviridae family (10.53–11.65 × 10−4 substitutions/base/year)43. Our results show that CDV circulating in Colombia exhibits high variability and includes two lineages and multiple subgenotypes (Figs 1 and 2 and Table 3). The temporary and geographical scope of our sampling was not sufficient to accurately explain the variability of CDV in the region. However, considering that the H gene has undergone genetic drift in different geographical regions26, we hypothesize that selective and nonselective processes may play important roles in the co-circulation of multiple lineages in an area, as has been reported previously52.
Phylogeographic analysis of the Fsp region and the H gene shows an evolutionary rate for the H gene similar to that reported by Fischer et al.53. However, the resulting two trees differ in their topology in such a way that the ancestry of the lineages of interest is very different (Figs 3 and 4). These differences in topology in comparison to other trees54 may be due to the fact that in the present analysis we excluded reported vaccine strains because the vaccine strains has been adapted to cell culture and have different evolutionary rates in comparison with CDV wild strains52; Also added three newly reported lineages, including “South America/North America-4.” By comparing both trees, we observed apparently, that the “South America/North America-4”lineage circulated first in Colombia and Ecuador, then in the United States, and again in Colombia (Figs 3 and 4). It is unclear if the variation in spatiotemporal sampling of the “South America/North America-4” lineage is more likely due to bias, as has been reported in other viral models55. In addition, since we suspected that most of the ancestral Colombian sequences of both trees are immune escape mutants, deeper analyses must be performed to avoid misleading results regarding the dynamics of the “South America/North America-4” CDV lineage.
Outbreaks of CDV occur in endemic and acute epidemic cycles, leading to transmission among susceptible host populations54. In the presence of full or partial vaccination, lifelong immunity could lead to the survival of the remaining coexisting lineages driven by nonselective epidemiological processes44,56. Our results, based on unvaccinated and/or incompletely vaccinated populations, support this hypothesis (Table 2).
In contrast with measles, the only natural host of which is humans, broad ranges of host species are susceptible to CDV infection, which results in complications in terms of selection pressure for this virus. It is noteworthy that the “South America/North America-4” lineage characterized in the USA was isolated from domestic dogs and foxes, indicating that this lineage has the ability to jump the species barrier3,21. In Colombia, there have been reports of CDV infection in wildlife57; however, no phylogenetic analysis has been performed in viruses from those infected animals.
Reported substitutions in circulating CDV protein H in wildlife include E276V, Q392R, R519I, I542F, and Y549H, i.e., sites that show the same substitutions in the South America-3 and South America/NorthAmerica-4 lineages, which indicates the potential of these viruses to jump the species barrier. However, there is no statistical association that demonstrates these hypotheses58.
In conclusion, we report the co-circulation of two CDV lineages in Colombia: the South America-3 lineage circulating in Medellín, Bucaramanga, and Bogotá, and the concurrent circulation of a new lineage not previously described in the country that mainly infects dogs in Medellín. The latter lineage is evolutionarily related to strains reported in domestic dogs in Ecuador and in domestic dogs and wildlife in the USA. Given the intercontinental circulation of this lineage, we propose to name it “South America/North America-4.”
Supplementary information
Acknowledgements
To the Clinics and veterinary hospitals involved in the sampling. This work was funded by the Departamento Administrativo de Ciencia, Tecnología e Investigación — COLCIENCIAS [Administrative Department of Science, Technology and Research]. Project No. 123171249669. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
J.R.S. and P.B. conceived the study. J.D.V., N.F.R.M., F.J.D., E.M., P.B. and J.R.S. were involved in all other aspects of the study, including data collection, data analysis, drafting, and editing the paper. All authors have read and approved the final manuscript.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: July Duque-Valencia and Norma R. Forero-Muñoz.
Supplementary information
is available for this paper at 10.1038/s41598-019-52345-9.
References
- 1.de Vries RD, Duprex WP, de Swart RL. Morbillivirus infections: an introduction. Viruses. 2015;7:699–706. doi: 10.3390/v7020699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beineke A, Baumgartner W, Wohlsein P. Cross-species transmission of canine distemper virus-an update. One Health. 2015;1:49–59. doi: 10.1016/j.onehlt.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez-Gutierrez M, Ruiz-Saenz J. Diversity of susceptible hosts in canine distemper virus infection: a systematic review and data synthesis. BMC Vet Res. 2016;12:78. doi: 10.1186/s12917-016-0702-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCarthy AJ, Shaw MA, Goodman SJ. Pathogen evolution and disease emergence in carnivores. Proc Biol Sci. 2007;274:3165–3174. doi: 10.1098/rspb.2007.0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.da Fontoura Budaszewski Renata, von Messling Veronika. Morbillivirus Experimental Animal Models: Measles Virus Pathogenesis Insights from Canine Distemper Virus. Viruses. 2016;8(10):274. doi: 10.3390/v8100274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loots AK, Mitchell E, Dalton DL, Kotze A, Venter EH. Advances in canine distemper virus pathogenesis research: a wildlife perspective. J Gen Virol. 2017;98:311–321. doi: 10.1099/jgv.0.000666. [DOI] [PubMed] [Google Scholar]
- 7.Deem SL, Spelman LH, Yates RA, Montali RJ. Canine distemper in terrestrial carnivores: a review. J Zoo Wildl Med. 2000;31:441–451. doi: 10.1638/1042-7260(2000)031[0441:CDITCA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 8.Sawatsky B, von Messling V. Canine distemper viruses expressing a hemagglutinin without N-glycans lose virulence but retain immunosuppression. J Virol. 2010;84:2753–2761. doi: 10.1128/JVI.01813-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwatsuki K, et al. Antigenic differences in the H proteins of canine distemper viruses. Vet Microbiol. 2000;71:281–286. doi: 10.1016/S0378-1135(99)00172-8. [DOI] [PubMed] [Google Scholar]
- 10.Sarute N, et al. The fusion protein signal-peptide-coding region of canine distemper virus: a useful tool for phylogenetic reconstruction and lineage identification. PLoS One. 2013;8:e63595. doi: 10.1371/journal.pone.0063595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarute N, et al. Molecular typing of canine distemper virus strains reveals the presence of a new genetic variant in South America. Virus Genes. 2014;48:474–478. doi: 10.1007/s11262-014-1054-z. [DOI] [PubMed] [Google Scholar]
- 12.Romanutti C, Gallo Calderon M, Keller L, Mattion N, La Torre J. RT-PCR and sequence analysis of the full-length fusion protein of Canine Distemper Virus from domestic dogs. J Virol Methods. 2016;228:79–83. doi: 10.1016/j.jviromet.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 13.Panzera Y, Sarute N, Carrau L, Aldaz J, Perez R. Genetic diversity of canine distemper virus in South America. Br J Virol. 2014;1:48. [Google Scholar]
- 14.Freitas LA, Leme RA, Saporiti V, Alfieri AA, Alfieri AF. Molecular analysis of the full-length F gene of Brazilian strains of canine distemper virus shows lineage co-circulation and variability between field and vaccine strains. Virus Res. 2019;264:8–15. doi: 10.1016/j.virusres.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Anis E, Newell TK, Dyer N, Wilkes RP. Phylogenetic analysis of the wild-type strains of canine distemper virus circulating in the United States. Virol J. 2018;15:118. doi: 10.1186/s12985-018-1027-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blixenkrone-Møller M, et al. Antigenic relationships between field isolates of morbilliviruses from different carnivores. Arch Virol. 1992;123:279–294. doi: 10.1007/BF01317264. [DOI] [PubMed] [Google Scholar]
- 17.Espinal MA, Diaz FJ, Ruiz-Saenz J. Phylogenetic evidence of a new canine distemper virus lineage among domestic dogs in Colombia, South America. Vet Microbiol. 2014;172:168–176. doi: 10.1016/j.vetmic.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 18.Haas L, et al. Analysis of the haemagglutinin gene of current wild-type canine distemper virus isolates from Germany. Virus Res. 1997;48:165–171. doi: 10.1016/S0168-1702(97)01449-4. [DOI] [PubMed] [Google Scholar]
- 19.Harder TC, et al. Canine distemper virus from diseased large felids: biological properties and phylogenetic relationships. J Gen Virol. 1996;77(Pt 3):397–405. doi: 10.1099/0022-1317-77-3-397. [DOI] [PubMed] [Google Scholar]
- 20.Panzera Y, et al. Evidence of two co-circulating genetic lineages of canine distemper virus in South America. Virus Res. 2012;163:401–404. doi: 10.1016/j.virusres.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 21.Riley MC, Wilkes RP. Sequencing of emerging canine distemper virus strain reveals new distinct genetic lineage in the United States associated with disease in wildlife and domestic canine populations. Virol J. 2015;12:219. doi: 10.1186/s12985-015-0445-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikolin VM, et al. Canine distemper virus in the Serengeti ecosystem: molecular adaptation to different carnivore species. Mol Ecol. 2017;26:2111–2130. doi: 10.1111/mec.13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao JJ, et al. Phylogenetic analysis of the haemagglutinin gene of canine distemper virus strains detected from breeding foxes, raccoon dogs and minks in China. Vet Microbiol. 2010;140:34–42. doi: 10.1016/j.vetmic.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 24.Radtanakatikanon A, et al. Genotypic lineages and restriction fragment length polymorphism of canine distemper virus isolates in Thailand. Vet Microbiol. 2013;166:76–83. doi: 10.1016/j.vetmic.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 25.Blixenkrone-Moller M, et al. Studies on manifestations of canine distemper virus infection in an urban dog population. Vet Microbiol. 1993;37:163–173. doi: 10.1016/0378-1135(93)90190-I. [DOI] [PubMed] [Google Scholar]
- 26.Martella V, et al. Heterogeneity within the hemagglutinin genes of canine distemper virus (CDV) strains detected in Italy. Vet Microbiol. 2006;116:301–309. doi: 10.1016/j.vetmic.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 27.Anis E, Holford AL, Galyon GD, Wilkes RP. Antigenic analysis of genetic variants of Canine distemper virus. Vet Microbiol. 2018;219:154–160. doi: 10.1016/j.vetmic.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 28.Budaszewski Rda F, et al. Genotyping of canine distemper virus strains circulating in Brazil from 2008 to 2012. Virus Res. 2014;180:76–83. doi: 10.1016/j.virusres.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 29.Decaro N, et al. Canine distemper and related diseases: report of a severe outbreak in a kennel. New Microbiol. 2004;27:177–181. [PubMed] [Google Scholar]
- 30.Calderon MG, et al. Detection by RT-PCR and genetic characterization of canine distemper virus from vaccinated and non-vaccinated dogs in Argentina. Vet Microbiol. 2007;125:341–349. doi: 10.1016/j.vetmic.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 31.Barrett T, et al. Dolphin and porpoise morbilliviruses are genetically distinct from phocine distemper virus. Virology. 1993;193:1010–1012. doi: 10.1006/viro.1993.1217. [DOI] [PubMed] [Google Scholar]
- 32.Lan NT, et al. Comparative analyses of canine distemper viral isolates from clinical cases of canine distemper in vaccinated dogs. Vet Microbiol. 2006;115:32–42. doi: 10.1016/j.vetmic.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 33.Pardo ID, Johnson GC, Kleiboeker SB. Phylogenetic characterization of canine distemper viruses detected in naturally infected dogs in North America. J Clin Microbiol. 2005;43:5009–5017. doi: 10.1128/JCM.43.10.5009-5017.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ronquist F, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rambaut, A., Drummond, A. J., Xie, D., Baele, G. & Suchard, M. A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol10 (2018). [DOI] [PMC free article] [PubMed]
- 37.Gupta, R., Jung, E. & Brunak, S. NetNGlyc 1.0 Server. Center for biological sequence analysis, technical university of Denmark available from, http://www.cbs.dtudk/services/NetNGlyc (2004).
- 38.Murrell B, et al. FUBAR: a fast, unconstrained bayesian approximation for inferring selection. Mol Biol Evol. 2013;30:1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rambaut, A. (2012).
- 41.Mochizuki M, Hashimoto M, Hagiwara S, Yoshida Y, Ishiguro S. Genotypes of canine distemper virus determined by analysis of the hemagglutinin genes of recent isolates from dogs in Japan. J Clin Microbiol. 1999;37:2936–2942. doi: 10.1128/jcm.37.9.2936-2942.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolt G, et al. Genetic diversity of the attachment (H) protein gene of current field isolates of canine distemper virus. J Gen Virol. 1997;78(Pt 2):367–372. doi: 10.1099/0022-1317-78-2-367. [DOI] [PubMed] [Google Scholar]
- 43.Pomeroy LW, Bjornstad ON, Holmes EC. The evolutionary and epidemiological dynamics of the paramyxoviridae. J Mol Evol. 2008;66:98–106. doi: 10.1007/s00239-007-9040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rima BK, et al. Temporal and geographical distribution of measles virus genotypes. J Gen Virol. 1995;76(Pt 5):1173–1180. doi: 10.1099/0022-1317-76-5-1173. [DOI] [PubMed] [Google Scholar]
- 45.Oliferenko S, et al. Analysis of CD44-containing lipid rafts: Recruitment of annexin II and stabilization by the actin cytoskeleton. J Cell Biol. 1999;146:843–854. doi: 10.1083/jcb.146.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riddell MA, Rota JS, Rota PA. Review of the temporal and geographical distribution of measles virus genotypes in the prevaccine and postvaccine eras. Virol J. 2005;2:87. doi: 10.1186/1743-422X-2-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harder TC, Osterhaus AD. Canine distemper virus–a morbillivirus in search of new hosts? Trends Microbiol. 1997;5:120–124. doi: 10.1016/S0966-842X(97)01010-X. [DOI] [PubMed] [Google Scholar]
- 48.Kapil S, et al. Canine distemper virus strains circulating among North American dogs. Clinical and Vaccine Immunology. 2008;15:707–712. doi: 10.1128/CVI.00005-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santibanez S, et al. Probing neutralizing-antibody responses against emerging measles viruses (MVs): immune selection of MV by H protein-specific antibodies? J Gen Virol. 2005;86:365–374. doi: 10.1099/vir.0.80467-0. [DOI] [PubMed] [Google Scholar]
- 50.Sugiyama M, Ito N, Minamoto N, Tanaka S. Identification of immunodominant neutralizing epitopes on the hemagglutinin protein of rinderpest virus. J Virol. 2002;76:1691–1696. doi: 10.1128/JVI.76.4.1691-1696.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tahara M, et al. Functional and structural characterization of neutralizing epitopes of measles virus hemagglutinin protein. J Virol. 2013;87:666–675. doi: 10.1128/JVI.02033-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ke GM, et al. Phylodynamic analysis of the canine distemper virus hemagglutinin gene. BMC Vet Res. 2015;11:164. doi: 10.1186/s12917-015-0491-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Panzera Y, Sarute N, Iraola G, Hernandez M, Perez R. Molecular phylogeography of canine distemper virus: Geographic origin and global spreading. Mol Phylogenet Evol. 2015;92:147–154. doi: 10.1016/j.ympev.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 54.Fischer CDB, et al. Phylogenetic analysis of canine distemper virus in South America clade 1 reveals unique molecular signatures of the local epidemic. Infect Genet Evol. 2016;41:135–141. doi: 10.1016/j.meegid.2016.03.029. [DOI] [PubMed] [Google Scholar]
- 55.Magee D, Taylor JE, Scotch M. The Effects of Sampling Location and Predictor Point Estimate Certainty on Posterior Support in Bayesian Phylogeographic Generalized Linear Models. Sci Rep. 2018;8:5905. doi: 10.1038/s41598-018-24264-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rendon-Marin S, da Fontoura Budaszewski R, Canal CW, Ruiz-Saenz J. Tropism and molecular pathogenesis of canine distemper virus. Virol J. 2019;16:30. doi: 10.1186/s12985-019-1136-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Varela-Arias, N., Toro-Mesa, D., Caicedo-Martínez, J. A. & Ospina-Chiriví, J. In Memorias de la Conferencia Interna en Medicina y Aprovechamiento de Fauna Silvestre, Exótica y no Convencional. 145–159.
- 58.Nikolin VM, Wibbelt G, Michler FU, Wolf P, East ML. Susceptibility of carnivore hosts to strains of canine distemper virus from distinct genetic lineages. Vet Microbiol. 2012;156:45–53. doi: 10.1016/j.vetmic.2011.10.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author upon request.