Abstract
Complex immune dysregulation is a hallmark of sepsis. The occurring phases of immunosuppression and hyperinflammation require rapid detection and close monitoring. Reliable tools to monitor patient’s immune status are yet missing. Currently, microRNAs are being discussed as promising new biomarkers in sepsis. However, no suitable internal control for normalization of miRNA expression by qPCR has been validated so far, thus hampering their potential benefit. We here present the first evaluation of endogenous controls for miRNA analysis in human sepsis. Novel candidate reference miRNAs were identified via miRNA microArray. TaqMan qPCR assays were performed to evaluate these microRNAs in T-cells and whole blood cells of sepsis patients and healthy controls in two independent cohorts. In T-cells, U48 and miR-320 proved suitable as endogenous controls, while in whole blood cells, U44 and miR-942 provided best stability values for normalization of miRNA quantification. Commonly used snRNA U6 exhibited worst stability in all sample groups. The identified internal controls have been prospectively validated in independent cohorts. The critical importance of housekeeping gene selection is emphasized by exemplary quantification of imuno-miR-150 in sepsis patients. Use of appropriate internal controls could facilitate research on miRNA-based biomarker-use and might even improve treatment strategies in the future.
Subject terms: Translational research, Molecular medicine, miRNAs, Experimental models of disease
Introduction
Sepsis is a dysregulated immune response to host infection causing severe organ dysfunction1. To date, it remains a life-threatening condition with high mortality rates2,3. Although originally considered to present with a biphasic immune response, it is nowadays accepted that sepsis is characterized by a complex immune dysregulation of both innate and adaptive immunity, with transitory immunosuppression and hyperinflammation emerging alternately or simultaneously4,5. Although various biomarkers have been proposed6, effective tools for monitoring patients’ immune status are still missing, thus hampering targeted therapy of sepsis. In this situation, microRNAs have been suggested as biomarkers in sepsis7.
MicroRNAs are a class of small non-coding RNA molecules that exert pivotal biological functions through post-transcriptional regulation of cellular gene expression8. By base-pairing to specific recognition sites within the 3′ untranslated region (UTR) of their respective target mRNAs they either repress translation or enable mRNA degradation9,10. MicroRNAs are remarkably stable and easily measurable7,11. They are tissue- and cell-type-dependently expressed and specific changes in miRNA expression due to cellular damage create disease-specific miRNA signatures4,12,13. These features render microRNAs ideal biomarkers14.
Innate and adaptive immune cells are regulated by specific miRNAs -so called immuno-miRs- that create a versatile balance of pro- and anti-inflammatory signalling circuitries15. Consequently, immuno-miRs have gained attraction in sepsis research and numerous studies have been conducted to identify immuno-miRs that could serve as biomarkers to determine the inflammatory state of the individual sepsis patient7,16. Results, however, were heterogeneous thus currently impeding the use of miRNA as biomarkers. This lack of consensus might -at least partially- be due to the fact that valid reference genes for quantification of miRNAs in sepsis have not been established, yet17–23. Moreover, in many cases either plasma or serum samples were used, which bears the risk of detecting contaminating miRNAs induced or released by co-morbidities or environmental influences.
We here present the first validation of internal controls for qPCR-based miRNA quantification in immune cells in sepsis. Importantly, to address the divergent miRNAomes of immune cell types, we have investigated reference miRNAs for two relevant set-ups: T-cells and whole blood cells. We analyzed native and stimulated T-cells from healthy volunteers and T cells isolated from septic patients and newly identified two reliable internal reference miRNAs. To expand these findings to PAXgene bed-side application, whole blood cells from a second, independent cohort of sepsis patients and healthy controls were isolated, and two valid reference miRNAs were identified also for this scenario.
Results
Selection of candidate miRNA reference genes
An optimal housekeeping gene should fulfil several essential demands: (1) it is highly expressed in the target cell, (2) it shows no differential regulation thus exhibiting high expression stability over time, (3) it is unaffected by any experimental condition and (4) it is easily detectable by use of available assays24,25. Taking into account the highly cell- and tissue-specific expression of miRNAs, it is intuitively clear that universal reference genes do not exist. Instead, optimal internal controls according to these requirements have to be determined for each utilized type of cell or tissue and every experimental approach20,26,27.
To identify potential miRNAs/snRNAs/snoRNAs that could serve as endogenous controls for quantification of miRNA expression in T-cells and whole blood cells in sepsis, we first evaluated data from a miRNA microArray performed with RNA from T-cells of septic patients and unstimulated/stimulated T-cells of healthy controls. Expression levels of candidate miRNAs are shown in Fig. S1. Requirements for suitable miRNAs were: (i) Expression detectable in at least 19/21 samples, (ii) not differentially regulated in sepsis (p-value > 0.05) and (iii) TaqMan miRNA assay commercially available28. With respect to these criteria, three microRNAs fulfilled all preconditions and thus were included into subsequent analyses: miR-30c-1*, miR-320a and miR-942. In addition, the commonly used albeit not properly evaluated normalizers U6, U44, U47 and U48 were analysed (Table 1). These seven candidate reference genes where then further assessed in two independent cohorts: (1) T-cells of septic patients and age-adjusted healthy controls (native and activated via anti-CD3/CD28 microBeads) and (2) whole blood cells of septic patients and age-adjusted healthy controls. Patient characteristics are depicted in Supplementary Tables 2–5.
Table 1.
TaqMan miRNA assays.
Evaluation of candidate miRNA reference genes in T-cells
Recently, paralysis of adaptive immunity has evolved as the major cause of death in sepsis29,30. On the search for effective biomarkers for early detection and consecutive monitoring of immunosuppression, miRNAs have entered the field16. However, valid internal normalizers to enable reliable miRNA quantification have not been determined so far.
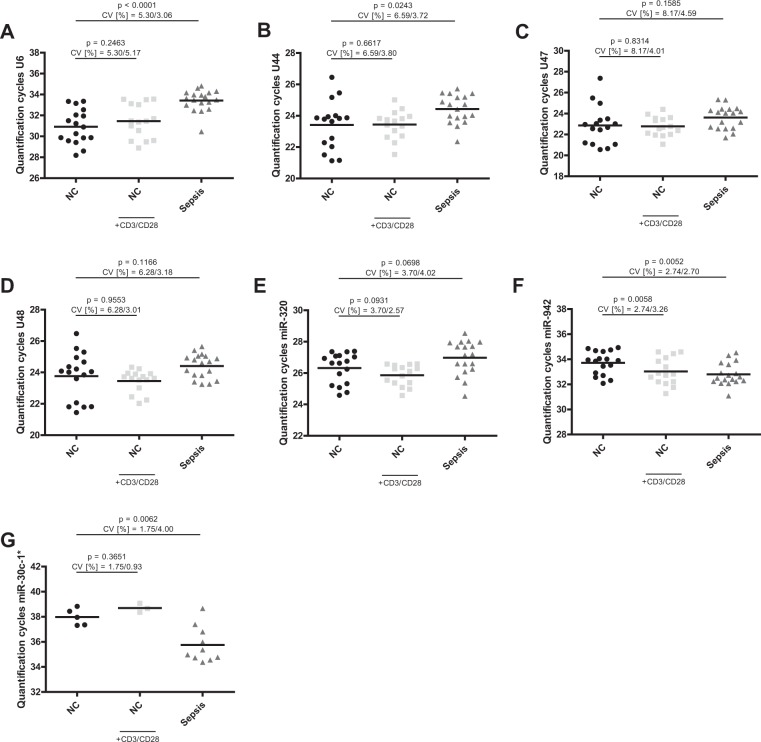
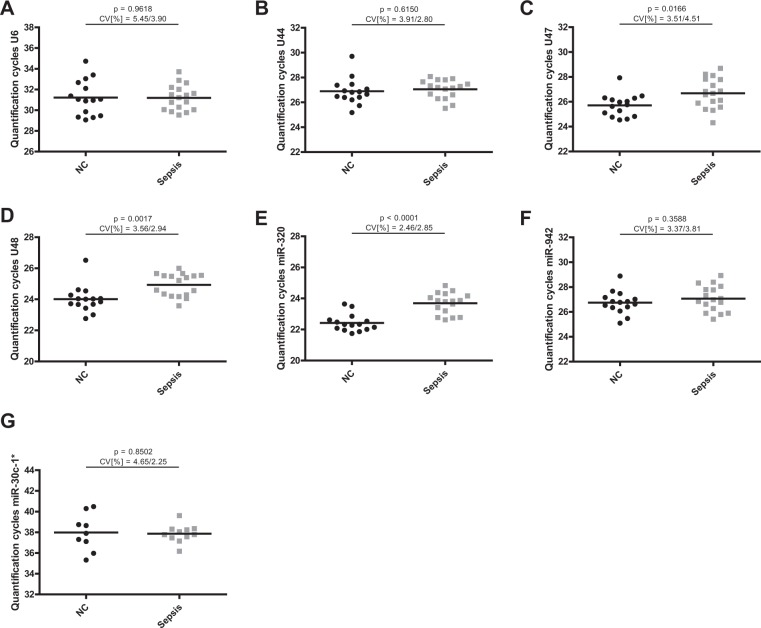
To address this important issue, we evaluated expression levels of the seven previously defined candidate miRNAs in T-cells from septic patients (n = 18) and in both native and stimulated T-cells from healthy controls (n = 17) (Fig. 1). Raw quantification cycles (Cq) ranged from 23 to 34 (Table 2 shows raw Cq values and standard deviation (SD) of each subset, amplification efficiencies are depicted in Supplementary Table 1). Only MicroRNA-30c-1* exhibited higher quantification cycles (in the range of 36–38), indicating only unspecific amplification, and thus was excluded from further analyses.
Figure 1.
Quantification cycles of candidate internal normalizers in T-cells. Quantification cycles of (A) U6, (B) U44, (C) U47, (D) U48, (E) miR-320a, (F) miR-942 and (G) miR-30c-1* were assessed in T-cells of sepsis patients and native and CD3/CD28-activated T-cells from age-adjusted healthy controls. Data are shown as mean with range. n = 18/17 (Sepsis/NC), miR-30c-1* n = 10/5 (Sepsis/NC).
Table 2.
Mean raw Cq and SD for candidate miRNA internal normalizers in T-cells.
| miRNA | Healthy controls | Healthy controls anti-CD3/CD28 |
Sepsis patients | |||
|---|---|---|---|---|---|---|
| Mean Cq | SD | Mean Cq | SD | Mean Cq | SD | |
| U6 | 30.92 | 1.639 | 31.45 | 1.626 | 33.42 | 1.023 |
| U44 | 23.42 | 1.543 | 23.44 | 0.8913 | 24.43 | 0.9093 |
| U47 | 22.87 | 1.867 | 22.78 | 0.9135 | 23.61 | 1.083 |
| U48 | 23.77 | 1.493 | 23.45 | 0.7055 | 24.41 | 0.7760 |
| miR-30c-1* | 37.98 | 0.6651 | 38.70 | 0.3607 | 35.76 | 1.429 |
| miR-320a | 26.32 | 0.9726 | 25.86 | 0.6649 | 26.97 | 1.083 |
| miR-942 | 33.71 | 0.9237 | 33.03 | 1.076 | 32.80 | 0.8868 |
Mean Cq = mean quantification cycle; SD = standard deviation of Cq values.
Reference genes for miRNA quantification in native vs. stimulated primary human T-cells
As ex vivo stimulation of primary T-cells is a commonly used and generally accepted method to mimic global activation of T-cells in response to infection, we first investigated expression stability of the remaining six candidate miRNAs in unstimulated and in anti-CD3/CD28 stimulated primary T-cells of healthy donors. To this end, we used the open-source algorithms NormFinder, BestKeeper and geNorm. Standard deviations and gene stability values of these analyses are shown in Table 3. Fold change of miRNA expression is depicted in Fig. S4 and Supplementary Table 6. Rankings of all applied algorithms are depicted in Table 4.
Table 3.
Results of BestKeeper, NormFinder and geNorm analysis for candidate microRNA internal controls in naive and stimulated primary T-cells.
| miRNA | U6 | U44 | U47 | U48 | miR-320 | miR-942 |
|---|---|---|---|---|---|---|
| geo Mean [Cq]* | 31.19 | 23.40 | 22.78 | 23.50 | 26.07 | 33.33 |
| ar Mean [Cq]* | 31.23 | 23.43 | 22.82 | 23.53 | 26.08 | 33.35 |
| min Cq [Cq]* | 28.18 | 21.13 | 20.55 | 21.45 | 24.57 | 31.27 |
| max Cq [Cq]* | 33.57 | 26.45 | 27.37 | 25.53 | 27.39 | 34.93 |
| SD [±Cq]* | 1.36 | 0.96 | 1.06 | 0.85 | 0.73 | 0.89 |
| CV [% Cq]* | 4.34 | 4.12 | 4.65 | 3.59 | 2.82 | 2.66 |
| stability value# | 0.343 | 0.125 | 0.153 | 0.095 | 0.125 | 0.190 |
| best combination# | U44 and U48 | combined stability value: 0.084 | |||||
| M stability value | 1.910 | 0.951 | 1.119 | 0.957 | 0.949 | 1.039 |
geo Mean = geometric mean of Cq values; ar Mean = arithmetic mean of Cq values; min Cq = minimal Cq; max Cq = maximal Cq; SD = standard deviation of Cq values; CV = coefficient variation. *BestKeeper results. #NormFinder results. M stability value = geNorm results.
Table 4.
Stability ranking of candidate reference genes in naive and stimulated primary T-cells.
| Rank | NormFinder | BestKeeper | geNorm |
|---|---|---|---|
| 1 | U48 | miR-320 | miR-320 |
| 2 |
miR-320/ U44 |
U48 | U44 |
| 3 | miR-942 | U48 | |
| 4 | U47 | U44 | miR-942 |
| 5 | miR-942 | U47 | U47 |
| 6 | U6 | U6 | U6 |
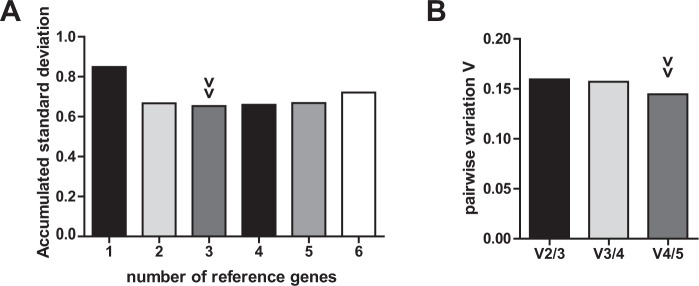
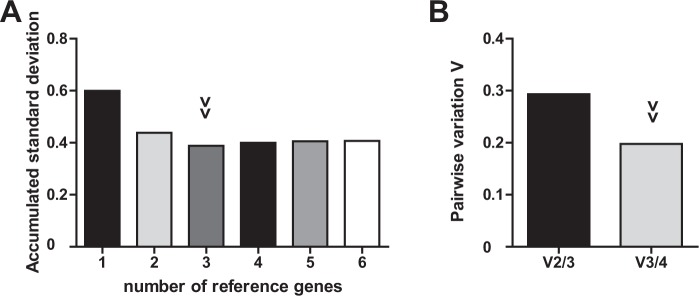
MicroRNA-320 exhibited best performance according to BestKeeper and geNorm analysis. NormFinder identified U48 as most stable gene and U48/U44 as best combination of genes. However, both miR-320 and U48 proved as reliable with only marginal differences and high performance throughout all analyses. Notably, commonly used U6 was ranked worst by all applied algorithms. The combined use of three or even two reference genes resulted in only marginally higher yet similar Vn/Vn+1 ratios. In accordance to these findings, accumulated standard deviations are comparably low for two to four reference genes (Fig. 2). Taken together, miR-320 or the combination of miR-320 and U48 are suitable as internal controls of miRNA expression analysis in naive and stimulated primary human T-cells.
Figure 2.
Optimal number of reference genes for normalization of stimulated primary T-cells. (A) Accumulated standard deviation and (B) pairwise variation calculations (Vn/Vn+1) of native and CD3/CD28-activated primary T-cells of healthy donors according to geNorm and GenEx software applications. Arrowheads indicate optimal number of reference genes.
Reference genes for miRNA quantification in septic T-cells vs. healthy controls
In clinical settings, evaluation of T-cell miRNAs in sepsis is based on comparison of sepsis samples to healthy controls. We thus determined Cq values of all candidate miRNA internal normalizers in T-cells of sepsis patients and in healthy controls and analysed their expression stability. Results are shown in Table 5, respective rankings are listed in Table 6. While U48 exhibited best stability values proposed by NormFinder and geNorm, miR-942 showed superior performance according to BestKeeper analysis. NormFinder combination analysis revealed miR-320 and U48 as best performing genes. Overall, values of U48 and miR-320 were in close proximity throughout all calculations. Again, U6 was consistently ranked worst. The combined use of two internal controls exhibited the best stability values (Fig. 3). In summary, applying U48 or the combination of miR-320 and U48 provides best internal normalization for miRNA quantification when comparing primary T-cells of sepsis patients to those of healthy controls.
Table 5.
Results of BestKeeper, NormFinder and geNorm analysis for candidate microRNA internal controls in healthy and septic primary T-cells.
| miRNA | U6 | U44 | U47 | U48 | miR-320 | miR-942 |
|---|---|---|---|---|---|---|
| geo Mean [Cq]* | 32.24 | 26.63 | 23.21 | 24.00 | 26.63 | 33.18 |
| ar Mean [Cq]* | 32.39 | 23.95 | 23.26 | 24.03 | 26.65 | 33.19 |
| min Cq [Cq]* | 28.18 | 21.13 | 20.55 | 21.45 | 24.53 | 31.08 |
| max Cq [Cq]* | 34.81 | 26.45 | 27.37 | 25.65 | 28.54 | 34.93 |
| SD [±Cq]* | 1.50 | 1.00 | 1.20 | 0.88 | 0.88 | 0.86 |
| CV [% Cq]* | 4.65 | 4.16 | 5.15 | 3.68 | 3.28 | 2.59 |
| stability value# | 0.743 | 0.176 | 0.226 | 0.079 | 0.099 | 0.697 |
| best combination# | miR-320 and U48|combined stability value: 0.076 | |||||
| M stability value | 1.814 | 0.994 | 1.178 | 0.973 | 1.016 | 1.525 |
geo Mean = geometric mean of Cq values; ar Mean = arithmetic mean of Cq values; min Cq = minimal Cq; max Cq = maximal Cq; SD = standard deviation of Cq values; CV = coefficient variation. *BestKeeper results. #NormFinder results. M stability value = geNorm results.
Table 6.
Stability ranking of candidate reference genes in healthy and septic primary T-cells.
| Rank | NormFinder | BestKeeper | geNorm |
|---|---|---|---|
| 1 | U48 | miR-942 | U48 |
| 2 | miR-320 | miR-320 | U44 |
| 3 | U44 | U48 | miR-320 |
| 4 | U47 | U44 | U47 |
| 5 | miR-942 | U47 | miR-942 |
| 6 | U6 | U6 | U6 |
Figure 3.
Optimal number of reference genes for normalization of septic T-cells. (A) Accumulated standard deviation and (B) pairwise variation calculations (Vn/Vn+1) of septic T-cells and age-adjusted healthy controls according to geNorm and GenEx software applications. Arrowheads indicate optimal number of reference genes.
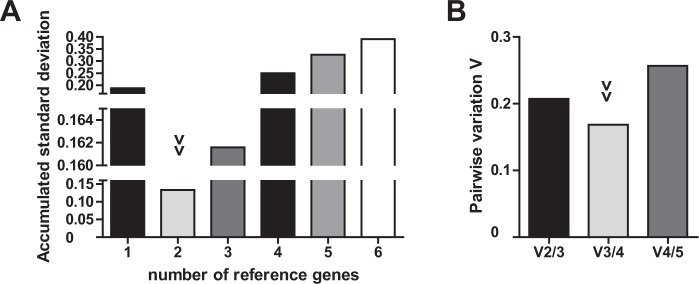
Since both analyses identified miR-320 and U48 as best performing internal normalizers, we next validated if they are indeed suitable “universal” reference genes for primary human T cells in the context of sepsis and thus usable in experimental as well as in clinical settings. Standard deviations and gene stability values of the respective analyses are given in Table 7, integrated rankings of all applied algorithms in Table 8. Both NormFinder and BestKeeper showed miR-320 and U48 as top performing candidate genes exhibiting comparable results. GeNorm analysis found best stability values for U44, U48 and miR-320, all exhibiting similar performance. A combination of two reference genes depicted lowest accumulated standard deviation and sufficient pairwise variation ratio (Fig. 4).
Table 7.
Results of BestKeeper, NormFinder and geNorm analysis for candidate microRNA internal controls in primary human T-cells.
| miRNA | U6 | U44 | U47 | U48 | miR-320 | miR-942 |
|---|---|---|---|---|---|---|
| geo Mean [Cq]* | 31.93 | 23.85 | 23.21 | 23.88 | 26.40 | 33.16 |
| ar Mean [Cq]* | 31.98 | 23.89 | 23.27 | 23.90 | 26.42 | 33.18 |
| min Cq [Cq]* | 28.18 | 21.13 | 20.55 | 21.45 | 24.53 | 31.08 |
| max Cq [Cq]* | 34.81 | 28.34 | 30.94 | 26.47 | 28.54 | 34.93 |
| SD [±Cq]* | 1.53 | 0.92 | 1.07 | 0.81 | 0.84 | 0.88 |
| CV [% Cq]* | 4.78 | 3.85 | 4.64 | 3.38 | 3.17 | 2.65 |
| stability value# | 0.603 | 0.147 | 0.184 | 0.092 | 0.130 | 0.545 |
| best combination# | U44/miR-320 | combined stability value: 0.095 | |||||
| M stability value | 1.791 | 0.939 | 1.076 | 0.940 | 0.961 | 1.404 |
geo Mean = geometric mean of Cq values; ar Mean = arithmetic mean of Cq values; min Cq = minimal Cq; max Cq = maximal Cq; SD = standard deviation of Cq values; CV = coefficient variation. *BestKeeper results. #NormFinder results. M stability value = geNorm results. Comprehensive gene stability = RefFinder results.
Table 8.
Integrated stability ranking of candidate reference genes in primary human T-cells.
| Rank | NormFinder | BestKeeper | geNorm |
|---|---|---|---|
| 1 | U48 | U48 | U44 |
| 2 | miR-320 | miR-320 | U48 |
| 3 | U44 | miR-942 | miR-320 |
| 4 | U47 | U44 | miR-942 |
| 5 | miR-942 | U47 | U47 |
| 6 | U6 | U6 | U6 |
Figure 4.
Optimal number of reference genes for normalization of T-cells. (A) Accumulated standard deviation and (B) pairwise variation calculations (Vn/Vn+1) of septic T-cells and native/CD3CD28-activated age-adjusted healthy controls according to geNorm and GenEx software applications. Arrowheads indicate optimal number of reference genes.
Collectively, miR-320 and U48 proved as suitable internal controls for miRNA expression analysis in primary human T-cells in the context of sepsis.
Evaluation of candidate miRNA internal normalizers in whole blood samples
In a clinical setting, analysis of whole blood cells is a straightforward approach as the purification of single immune cell populations usually is not feasible. However, reliable references for miRNA quantification in this context are also lacking. We therefore examined expression levels of the above-mentioned candidate endogenous controls in total RNA purified from whole blood cells in an independent cohort of septic patients (n = 17) and healthy controls (n = 15, Fig. 5). Quantification cycles ranged from 23 to 32 (Table 9 shows raw Cq values and standard deviation (SD) of each subset). Similar to T-cells, miR-30c-1* was excluded from further analyses because of unspecific amplification (Cq values in the range of 37–38).
Figure 5.
Quantification cycles of candidate internal normalizers in whole blood cells. Quantification cycles of (A) U6, (B) U44, (C) U47, (D) U48, (E) miR-320a, (F) miR-942 and (G) miR-30c-1* were assessed in whole blood cell RNA of sepsis patients and age-adjusted healthy controls. Data are shown as mean with range. n = 17/15 (Sepsis/NC), miR-30c-1* n = 11/9 (Sepsis/NC).
Table 9.
Mean raw Cq and SD for candidate miRNA internal normalizers in whole blood cells.
| miRNA | Healthy controls | Sepsis patients | ||
|---|---|---|---|---|
| Mean Cq | SD | Mean Cq | SD | |
| U6 | 32.22 | 1.701 | 31.20 | 1.218 |
| U44 | 26.89 | 1.050 | 27.05 | 0.7577 |
| U47 | 25.71 | 0.9022 | 26.68 | 1.203 |
| U48 | 24.01 | 0.8550 | 24.93 | 0.7320 |
| miR-30c-1* | 37.98 | 1.766 | 37.87 | 0.8536 |
| miR-320a | 22.42 | 0.5513 | 23.69 | 0.6741 |
| miR-942 | 26.75 | 0.9012 | 27.07 | 1.032 |
Mean Cq = mean quantification cycle; SD = standard deviation of Cq values.
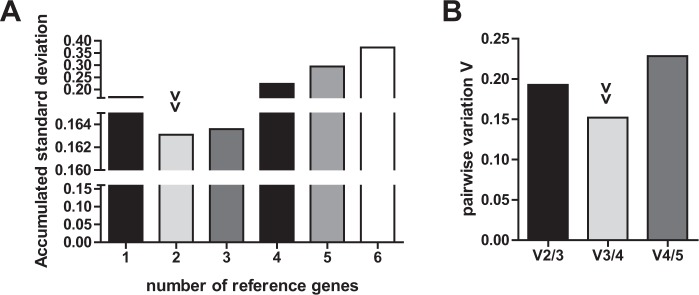
Results of expression stability displayed high consensus between all software applications: NormFinder proposes miR-942 as best internal control, whereas U44 is most suitable according to BestKeeper and geNorm. Performance of both candidate genes U44 and miR-942 is highly comparable throughout all algorithms, geNorm results are almost identical (Tables 10 and 11). Similar to T-cell analysis, U6 exhibited worst gene stability in nearly all applied software algorithms. Considering calculation of accumulated standard deviations, at least two internal controls are required (Fig. 6).
Table 10.
Results of BestKeeper, NormFinder and geNorm analysis in whole blood cells.
| U6 | U44 | U47 | U48 | miR-320 | miR-942 | |
|---|---|---|---|---|---|---|
| geo Mean [Cq]* | 31.18 | 26.96 | 26.20 | 25.47 | 23.08 | 26.90 |
| ar Mean [Cq]* | 31.21 | 26.98 | 26.22 | 25.51 | 23.10 | 26.92 |
| min Cq [Cq]* | 29.07 | 25.18 | 24.31 | 22.77 | 21.74 | 25.10 |
| max Cq [Cq]* | 34.74 | 29.70 | 28.69 | 28.89 | 24.83 | 28.93 |
| SD [±Cq]* | 1.15 | 0.68 | 0.92 | 1.20 | 0.78 | 0.75 |
| CV [% Cq]* | 3.70 | 2.52 | 3.51 | 4.70 | 3.38 | 2.79 |
| stability value# | 0.338 | 0.232 | 0.292 | 0.261 | 0.321 | 0.188 |
| best combination# | miR-942 and U47 | combined stability value: 0.136 | |||||
| M stability value | 1.620 | 1.189 | 1.377 | 1.545 | 1.257 | 1.190 |
geo Mean = geometric mean of Cq values; ar Mean = arithmetic mean of Cq values; min Cq = minimal Cq; max Cq = maximal Cq; SD = standard deviation of Cq values; CV = coefficient variation. *BestKeeper results. #NormFinder results. M stability value = geNorm results. Comprehensive gene stability = RefFinder results.
Table 11.
Integrated stability ranking of candidate reference genes in whole blood cells.
| Rank | NormFinder | BestKeeper | geNorm |
|---|---|---|---|
| 1 | miR-942 | U44 | U44 |
| 2 | U44 | miR-942 | miR-942 |
| 3 | U48 | miR-320 | miR-320 |
| 4 | U47 | U47 | U47 |
| 5 | miR-320 | U6 | U48 |
| 6 | U6 | U48 | U6 |
Figure 6.
Optimal number of reference genes for normalization of whole blood cells. (A) Accumulated standard deviation and (B) pairwise variation calculations (Vn/Vn+1) of whole blood cells of septic patients and age-adjusted healthy controls according to geNorm and GenEx software applications. Arrowheads indicate optimal number of reference genes.
Taken together, our evaluation revealed miR-942 and U44 as best suitable internal controls for miRNA quantification of whole blood cell RNA in the context of sepsis.
Validation of the identified miRNA endogenous controls
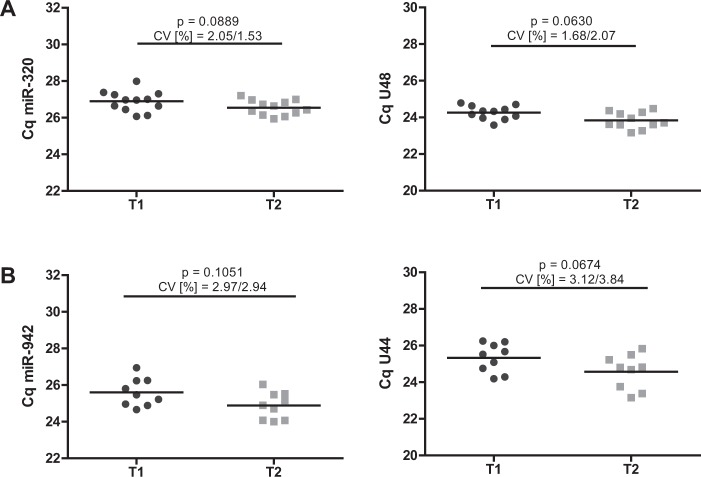
For further validation of the identified reference genes, we prospectively analysed two additional independent cohorts of patients undergoing elective cardiopulmonary bypass (CPB) surgery (patient characteristics are shown in Supplementary Tables 8 and 9). A pronounced immune dysfunction affecting both innate and adaptive immunity has been demonstrated for these patients immediately after surgical intervention31–35. As we have recently published, both septic patients and patients after CPB display comparable T-cell phenotypes with upregulation of immunosuppressive markers, rendering these cohorts ideal for validation of the identified reference genes16,31. As depicted in Fig. 7, expression of miR-320/U48 in T-cells and miR-942/U44 in whole blood cells remained stable before (T1) and after (T2) CPB, thus confirming their suitability to serve as endogenous controls.
Figure 7.
Validation of identified miRNA endogenous controls. Quantification cycles of (A) miR-320/U48 in T-cells and (B) miR-942/U44 in whole blood cells of patients before (T1) and after (T2) elective cardiopulmonary bypass surgery. Data are shown as mean with range. n = 11/9 (T-cells/whole blood cells).
Impact of the choice of reference genes on miRNA quantification
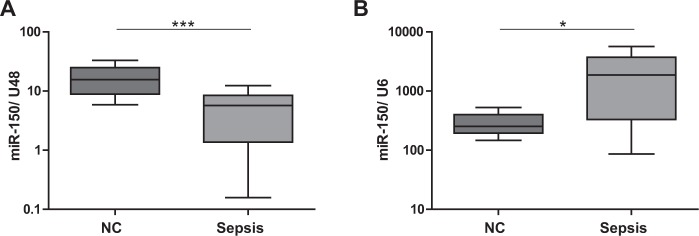
To evaluate the effects of different reference genes on miRNA expression analysis, we quantified immuno-miR-150 in sepsis using best-performing U48 and worst ranked U6 as exemplary housekeeping genes. Consistent with recently published data16, miR-150 was significantly downregulated in septic patients as compared to healthy controls in case of using U48 for normalization (Fig. 8A). Remarkably, using snRNA U6 resulted in the opposite: a significant upregulation of miR-150 expression in sepsis was found (Fig. 8B). These divergent results highlight the crucial importance of using validated internal controls for qPCR-based miRNA expression analysis.
Figure 8.
Effects of internal controls on microRNA quantification in sepsis. MiRNA-150 expression in primary human T-cells of septic patients as compared to healthy controls. Expression level of miR-150 T-cells of septic patients and healthy controls was measured by TaqMan miRNA assays relative to (A) U48 and (B) U6. Data are shown as median, 25th and 75th percentile and outliers, n = 5/16 (NC/Sepsis), performed in duplicates. *p < 0.05, ***p < 0.001. Quantification cycle (Cq) values were in the range of 21 (NC) and 23 (Sepsis) for miR-150, 30 (NC) and 33 (Sepsis) for U6 and 25 (NC/Sepsis) for U48, respectively.
Discussion
Sepsis is a clinical syndrome characterized by complex immune dysfunction with states of hyperinflammation, largely driven by innate immune cells, and immunosuppression, mainly affecting adaptive immunity36. Recent studies have pointed out that T-cell driven immunoparalysis is the leading cause of death in sepsis29. While the commonly used biomarkers, e.g. C-reactive Protein, Interleukin-6, and Procalcitonin, are able to detect hyperinflammation, effective tools for early detection and consecutive monitoring of immunosuppression are lacking37. At this point, miRNAs are being discussed to fill the gap16. MiRNAs have emerged as essential regulators of human immune responses. Moreover, they exhibit essential features -like cell-type and disease-specific expression and high stability- to render them ideal biomarkers12,15. Even though miRNAs have evolved as promising diagnostic tools in sepsis in recent years, their implementation into clinical routine is still pending7. This is -at least partially- due to the lack of valid reference genes, which impedes reliable miRNA quantification in sepsis.
To date, qPCR is the method of choice for quantification of miRNA expression, enabling a fast, accurate and reliable detection25,38. Suitable normalization strategies, however, are a pivotal prerequisite to generate valid results. To adjust for even slight sampling differences in RNA quantity and quality or reverse transcription efficiency, normalization to stably expressed reference genes is indispensable to actually identify variations in gene expression due to experimental conditions24,39–42. For miRNA quantification, small RNAs such as small nuclear RNA (<350 nt, e.g. U6) and small nucleolar RNA (~60–200 nt, e.g. U44/SNORD44, U47/SNORD47, U48/SNORD48), or other miRNAs are mainly used as endogenous controls17–23,43–50. We here validated endogenous controls for miRNA quantification in sepsis to provide reliable internal controls for current and future sepsis studies, which -in the majority- use healthy subjects as controls (Supplementary References 3–20). To this end, four widely used and commonly “accepted” yet not validated reference genes (U6, U44, U47, U48) as well as three potentially “new” miRNAs (miR-30c-1*, miR-320a, miR-942) were validated in T-cell and in whole blood cell samples both of healthy subjects and of septic patients. MiRNAs were identified via array analysis and have not been assigned to any regulatory role in immune cells, thus implying stable expression.
For T-cell miRNA quantification, miR-320 and U48 revealed as the two best reference genes, performing equally well. The most suited endogenous controls for whole blood cell miRNA quantification were miR-942 and U44, displaying stability values in close proximity. For these candidate genes, high consensus was obtained in all applied algorithms. The next placed candidate genes showed markedly inferior results. Importantly, the frequently used reference gene U6 was identified as the least stable expressed candidate gene in all cells and conditions tested, clearly showing that its use for normalization in sepsis is inadequate20,51,52. In order to further clarify this crucial point, we quantified immuno-miR-150 and could display that completely misleading results were obtained when U6 was applied for normalization.
Notably, our study could not find “universal” endogenous controls suitable for both T-cells and whole blood cells in sepsis. This is not surprising as whole blood samples are composed of a variety of different cell types exhibiting substantial differences in their transcriptome and thus miRNAome profiles. Our findings emphasize the necessity to use distinct miRNA internal reference genes for miRNA quantification in T-cell and/or whole blood samples in sepsis.
Several algorithms for analysis of qPCR results and identification of suitable endogenous controls are currently available, based on various principles and using distinct equations25. As a consequence, results may differ considerably depending on the respective algorithm53. Additionally, BestKeeper software is not specifically designed to construct a hierarchy of genes, although SD values and coefficient of variation are commonly used to rank reference candidates. Therefore, the use of multiple validation principles should be attempted42,54. Strikingly, we obtained similar results in all software algorithms used, which additionally supports our findings. Remarkably, the two best internal controls for T-cells (miR-320/U48) and whole blood cells (miR-942/U44) exhibit nearly 100% consensus amongst all normalization programs (i.e. being ranked first or second). Further prospective validation of the here-identified reference genes in independent cohorts of patients undergoing CPB surgery confirmed expression stability of miR-320/U48 and miR-942/U44 in T-cells and whole blood cells, respectively, in a highly controlled setting of immune dysfunction.
Regarding the optimal number of reference genes, pairwise variation calculations and accumulated standard deviations significantly improved in both sample groups when a second internal control was applied. Use of more than two reference genes had - if any - only minor benefits. The use of only one endogenous control may be acceptable if its use is restricted to the here-analysed experimental environment. Two reference genes may significantly reduce errors and should generally be recommended25,27. For individual studies it may be necessary to reasonably compromise on adequate normalization, sample availability, economical issues and technical execution.
Several studies have been performed to identify suitable normalizers for quantification of -mostly circulating- miRNA in different diseases. Using multiple algorithms and recommending at least two housekeepers is a common ground for most of these investigations50,52,55–57. To date, only one study has evaluated reference genes in plasma in a very small number of patients with septic shock58. However, both the small sample size and a potential distortion of results by miRNAs induced or released by co-morbidities may hamper the validity of this approach.
Thus, to the best of our knowledge, we here provide for the first time reliable identification of suitable controls for miRNA quantification in immune cells in sepsis. All patient samples have been acquired at the onset of sepsis. As the validated miRNAs remain stable even in the initial activation of immune cells -which profoundly alters T-cell transcriptional activity- it can be assumed that these internal controls also remain stable in the course of sepsis4.
Taken together, we recommend the use of miR-320 and U48 in T-cells, and miR-942 and U44 in whole blood cells as suitable endogenous controls for miRNA quantification in sepsis. These combinations feature important advantages: (i) they displayed outstanding stability values, (ii) they both combine different classes of RNAs - miRNA and snoRNA - thus ameliorating potential biases due to biochemical differences, and (iii) they do not comprise of miRNAs of the same miRNA family or miRNA cluster, avoiding similar regulation mechanisms in the context of experimental conditions.
Consensus of applied housekeepers for normalization might enable reliable research on miRNAs in sepsis, especially with respect to their use as biomarkers. In this regard, miRNAs might even prove suitable for point-of-care applications and thus could improve current treatment strategies in the future.
Methods
Blood sampling
Blood samples from septic patients were withdrawn immediately after diagnosis of sepsis or septic shock (according to the Sepsis-2 criteria59) and admission to the intensive care unit, before antibiotic or steroid treatment was initiated. Retrospective analysis also confirmed that all patients were meeting the Sepsis-3 definitions1,60. Patient analysis has been performed between 2011 and 2017, healthy age-adjusted controls (similar mean age of the compared groups) were sampled in 2011 and 2018. Two independent patient and age-adjusted control groups have been evaluated for primary T-cell and whole blood cell analysis, respectively. Previous antibiotic or steroid treatment, age younger than 18 years, history of malignant diseases, pre-existing immunodeficiency or autoimmune diseases have been defined as patient exclusion criteria. Research has been performed in accordance to the Declaration of Helsinki (ethical principles for medical research involving human subjects). Informed consent was obtained from all patients and healthy volunteers. The protocol for this prospective study was approved by the Institutional Ethics Committees of the Ludwig-Maximilian-University Munich, Germany (No. 107-11 and No. 287-13; approved in 2006 and in 2013, respectively), of the University Hospital of Jena (No. 2007-004333-42, local amendment for Munich University Hospital 2242-03/08), and by the Ethics Committee of ATTIKON University Hospital (approval 5/2008). The study protocol for the independent validation cohorts was also approved by the Institutional Ethics Committees of the Ludwig-Maximilians-University Munich (No. 17–241).
T-cell isolation, culture and stimulation
Peripheral blood mononuclear cells (PBMCs) were obtained by density centrifugation (Histopaque 1077, Sigma-Aldrich, St. Louis, MO, USA). T-cell isolation has been performed by untouched negative magnetic cell separation (Pan T Cell Isolation Kit, Cat. # 130-096-535, Miltenyi Biotec, Bergisch Gladbach, Germany), using an AutoMACS Pro Separator (Cat. # 130-092-545, Miltenyi Biotec, Bergisch-Gladbach, Germany) according to the manufacturer’s instructions. A ViCell analyzer (Beckman Coulter, Fullerton, CA, USA) was utilized to assess cell number and viability. Only samples exceeding a cell viability of 90% were included into the analysis. T-cells from healthy donors were cultured in RPMI-1640 (Sigma-Aldrich, St-Louis, MO, USA) supplemented with 10% heat-inactivated fetal calf serum (Biochrom, Berlin, Germany), 1% HEPES (Sigma-Aldrich, St. Louis, MO) and 1% L-glutamine (Life Technologies, Carlsbad, CA, USA) for 24 hours. For T-cell stimulation, cells have been incubated using anti-CD3/CD28 Dynabeads (Thermo Fisher Scientific, Waltham, MA, USA) at a bead-to-cell ratio of 1:1 for 24 hours. Figure S2 displays efficiency of T-cell stimulation and provides data to exclude non-specific activation of naive T-cells.
Whole blood analysis
For whole blood analysis, blood samples were collected using the PAXgene Blood RNA Tube (PreAnalytiX, Hombrechtikon, Switzerland) as to the manufacturer’s instructions.
RNA-Isolation
Total RNA was isolated from primary T-cells using the mirVana miRNA Isolation Kit (Thermo Fisher Scientific, Waltham, MA, USA) with subsequent DNase treatment (Turbo DNase, Thermo Fisher Scientific, Waltham, MA, USA). Total RNA from whole blood samples was purified using the PAXgene Blood RNA Kit (PreAnalytiX, Hombrechtikon, Switzerland). All isolation procedures have been performed according to the respective manufacturer’s instructions. RNA quantity and purity were measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Only samples exhibiting A260/A280 ratios between 1.70 and 2.10 have been further analysed. RNA storage has been performed at −80 °C. No difference in RNA quality of older samples has been detected over time. For additional validation of sample integrity, Fig. S3 depicts U47 quantification cycles of samples independently analysed in 2011, 2013, 2017 and 2018.
miRNA Microarray
Total (T-cell derived) RNA from seven patients and from seven controls was used for miRNA Microarray analysis (miRCURY LNA™ microRNA Array, Exiqon A/S, Vedbaek, Denmark), as to the manufacturer’s recommendations.
miRNA quantification
Four commonly applied miRNA housekeeping genes and three newly identified miRNAs were selected as candidate reference genes (Table 1). Expression of miRNAs was quantified using a LightCycler 480 instrument (Roche Diagnostics, Mannheim, Germany). In all reactions, identical amounts of extracted total RNA (6 ng) were reversely transcribed using TaqMan miRNA assays (Thermo Fisher Scientific, Waltham, MA, USA). Negative controls using only purified water were included to prevent any contamination. To account for inter-run and intra-run variations, all experiments have been performed in technical duplicates and additional calibration has been applied, respectively. The cycling conditions comprised initial denaturation a 95 °C for 10 min followed by 50 amplification cycles at 95 °C for 15 s, 60 °C for 60 s and 40 °C for 30 sec. Determination of raw quantification cycles (Cq) has been performed by the LightCycler software using the second derivative maximum method. A quantification cycle (Cq) cut-off has been defined (Cq 40) and all Cq values beyond this cut-off have been considered unspecific. For all TaqMan miRNA assays, amplification efficiencies have been determined by calculating calibration curves from 10-fold dilution series using the equation E = −1 + 10(−1/slope) (See Table S1 for efficiency values and amplification factors for each TaqMan miRNA assay).
Statistical analysis
Student’s t-test or Mann-Whitney U tests, as appropriate, served for comparisons. Normal distribution was tested using the Kolmogorov-Smirnov test. The quantified array signals were background corrected (Normexp with offset value 10 - Convolution model described by Ritchie et al.61) and normalized using the global Lowess (LOcally WEighted Scatterplot Smoothing) regression algorithm. The obtained values were further analyzed using two-sided Student’s t-test. Analysis of expression stability of potential reference genes was evaluated by Excel-based software tools BestKeeper41, NormFinder40 and geNorm24. NormFinder add-in calculates intergroup and intragroup variations by applying a model-based approach. Both variations are integrated into a gene stability value. BestKeeper algorithm displays expression stability based on Cq variations and pair-wise correlation analyses. Ranking of genes was performed based on SD values. In case of equal SD, coefficient variation served as additional ranking value. GeNorm gene expression stability is indicated by the average pairwise variation of each reference gene with all other candidate genes and additional ranking of genes by repeated stepwise exclusion of the worst performing gene and recalculation of stability values. For NormFinder and geNorm analysis, raw Cq data has been transformed into relative quantities by the comparative ∆CT method62. To calculate accumulated standard deviations of each candidate gene GenEx 6.1 (MultiD Analyses AB, Gothenburg, Sweden) was used according to the developer’s instructions. Statistical analysis of qPCR results was performed using Microsoft Excel 365 (Microsoft Corporation, Redmond, WA, USA) and GraphPad Prism 5.01 (GraphPad Software, Inc., USA). Vector artwork has been designed using Adobe Illustrator CS5.1 (Adobe Systems Inc., San Jose, CA, USA). If not stated otherwise, no missing values are reported.
Supplementary information
Acknowledgements
We kindly thank Roland Tomasi for collecting the samples of both validation cohorts.
Author contributions
S.H. and S.K. designed the study, analysed the data and wrote the manuscript. S.H. performed the experiments. M.H., D.E., G.S., M.B., S.W. and E.G. collected samples and participated in interpretation of all experiments.
Data availability
The complete data generated and analysed for this study is implemented into this publication and the Supplementary Files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-51782-w.
References
- 1.Singer M, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zimmerman JE, Kramer AA, Knaus WA. Changes in hospital mortality for United States intensive care unit admissions from 1988 to 2012. Crit. Care. 2013;17:R81. doi: 10.1186/cc12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayr FB, Yende S, Angus DC. Epidemiology of severe sepsis. Virulence. 2014;5:4–11. doi: 10.4161/viru.27372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao W, et al. A genomic storm in critically injured humans. J. Exp. Med. 2011;208:2581–2590. doi: 10.1084/jem.20111354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hotchkiss RS, Monneret G, Payen D. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat. Rev. Immunol. 2013;13:862–874. doi: 10.1038/nri3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pierrakos C, Vincent J-L. Sepsis biomarkers: a review. Critical Care. 2010;14:R15. doi: 10.1186/cc8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kreth S, Hübner M, Hinske LC. MicroRNAs as Clinical Biomarkers and Therapeutic Tools in Perioperative Medicine. Anesth. Analg. 2018;126:670–681. doi: 10.1213/ANE.0000000000002444. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilczynska A, Bushell M. The complexity of miRNA-mediated repression. Cell Death Differ. 2015;22:22–33. doi: 10.1038/cdd.2014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benz, F., Roy, S., Trautwein, C., Roderburg, C. & Luedde, T. Circulating MicroRNAs as Biomarkers for Sepsis. Int. J. Mol. Sci. 17 (2016). [DOI] [PMC free article] [PubMed]
- 12.Weiland M, Gao X-H, Zhou L, Mi Q-S. Small RNAs have a large impact: circulating microRNAs as biomarkers for human diseases. RNA Biol. 2012;9:850–859. doi: 10.4161/rna.20378. [DOI] [PubMed] [Google Scholar]
- 13.Leidinger P, Backes C, Meder B, Meese E, Keller A. The human miRNA repertoire of different blood compounds. BMC Genomics. 2014;15:474. doi: 10.1186/1471-2164-15-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol. Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Hirschberger S, Hinske LC, Kreth S. MiRNAs: dynamic regulators of immune cell functions in inflammation and cancer. Cancer Lett. 2018;431:11–21. doi: 10.1016/j.canlet.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 16.Möhnle P, et al. MicroRNAs 143 and 150 in whole blood enable detection of T-cell immunoparalysis in sepsis. Mol. Med. 2018;24:54. doi: 10.1186/s10020-018-0056-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodwin AJ, et al. Plasma levels of microRNA are altered with the development of shock in human sepsis: an observational study. Crit. Care. 2015;19:440. doi: 10.1186/s13054-015-1162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roderburg C, et al. Circulating microRNA-150 serum levels predict survival in patients with critical illness and sepsis. PLoS One. 2013;8:e54612. doi: 10.1371/journal.pone.0054612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Müller V, et al. Changes in serum levels of miR-21, miR-210, and miR-373 in HER2-positive breast cancer patients undergoing neoadjuvant therapy: a translational research project within the Geparquinto trial. Breast Cancer Research and Treatment. 2014;147:61–68. doi: 10.1007/s10549-014-3079-3. [DOI] [PubMed] [Google Scholar]
- 20.Schwarzenbach H, da Silva AM, Calin G, Pantel K. Data Normalization Strategies for MicroRNA Quantification. Clin. Chem. 2015;61:1333–1342. doi: 10.1373/clinchem.2015.239459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Möhnle P, et al. MicroRNA-146a controls Th1-cell differentiation of human CD4+ T lymphocytes by targeting PRKCε. Eur. J. Immunol. 2015;45:260–272. doi: 10.1002/eji.201444667. [DOI] [PubMed] [Google Scholar]
- 22.Heide Vvander, et al. Down-regulation of MicroRNA-31 in CD4 T Cells Contributes to Immunosuppression in Human Sepsis by Promoting TH2 Skewing. Anesthesiology. 2016;124:908–922. doi: 10.1097/ALN.0000000000001031. [DOI] [PubMed] [Google Scholar]
- 23.Zhou J, et al. Dysregulation in microRNA expression in peripheral blood mononuclear cells of sepsis patients is associated with immunopathology. Cytokine. 2015;71:89–100. doi: 10.1016/j.cyto.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozera B, Rapacz M. Reference genes in real-time PCR. Journal of Applied Genetics. 2013;54:391–406. doi: 10.1007/s13353-013-0173-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005;6:279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- 27.Thellin O, et al. Housekeeping genes as internal standards: use and limits. J. Biotechnol. 1999;75:291–295. doi: 10.1016/S0168-1656(99)00163-7. [DOI] [PubMed] [Google Scholar]
- 28.Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14:844–852. doi: 10.1261/rna.939908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boomer JS, et al. Immunosuppression in patients who die of sepsis and multiple organ failure. JAMA. 2011;306:2594–2605. doi: 10.1001/jama.2011.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jensen IJ, Sjaastad FV, Griffith TS, Badovinac VP, Sepsis-Induced T. Cell Immunoparalysis: The Ins and Outs of Impaired T Cell Immunity. J. Immunol. 2018;200:1543–1553. doi: 10.4049/jimmunol.1701618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hübner M, et al. Myeloid-Derived Suppressor Cells Mediate Immunosuppression After Cardiopulmonary Bypass. Crit. Care Med. 2019;47:e700–e709. doi: 10.1097/CCM.0000000000003820. [DOI] [PubMed] [Google Scholar]
- 32.Taylor KM. SIRS—The Systemic Inflammatory Response Syndrome after cardiac operations. The Annals of Thoracic Surgery. 1996;61:1607–1608. doi: 10.1016/0003-4975(96)00225-1. [DOI] [PubMed] [Google Scholar]
- 33.Paparella D, Yau TM, Young E. Cardiopulmonary bypass induced inflammation: pathophysiology and treatment. An update. Eur. J. Cardiothorac. Surg. 2002;21:232–244. doi: 10.1016/S1010-7940(01)01099-5. [DOI] [PubMed] [Google Scholar]
- 34.Stoppelkamp S, et al. Identification of Predictive Early Biomarkers for Sterile-SIRS after Cardiovascular Surgery. PLoS One. 2015;10:e0135527. doi: 10.1371/journal.pone.0135527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hadley JS, et al. Alterations in inflammatory capacity and TLR expression on monocytes and neutrophils after cardiopulmonary bypass. Shock. 2007;27:466–473. doi: 10.1097/01.shk.0000245033.69977.c5. [DOI] [PubMed] [Google Scholar]
- 36.Boomer JS, Green JM, Hotchkiss RS. The changing immune system in sepsis: is individualized immuno-modulatory therapy the answer? Virulence. 2014;5:45–56. doi: 10.4161/viru.26516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samraj RS, Zingarelli B, Wong HR. Role of biomarkers in sepsis care. Shock. 2013;40:358–365. doi: 10.1097/SHK.0b013e3182a66bd6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gachon C, Mingam A, Charrier B. Real-time PCR: what relevance to plant studies? J. Exp. Bot. 2004;55:1445–1454. doi: 10.1093/jxb/erh181. [DOI] [PubMed] [Google Scholar]
- 39.Bustin SA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 40.Andersen CL, Jensen JL, Ørntoft TF. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 41.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004;26:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 42.Ledderose C, Heyn J, Limbeck E, Kreth S. Selection of reliable reference genes for quantitative real-time PCR in human T cells and neutrophils. BMC Res. Notes. 2011;4:427. doi: 10.1186/1756-0500-4-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Serafin A, et al. Identification of a set of endogenous reference genes for miRNA expression studies in Parkinson’s disease blood samples. BMC Res. Notes. 2014;7:715. doi: 10.1186/1756-0500-7-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scott MS, Ono M. From snoRNA to miRNA: Dual function regulatory non-coding RNAs. Biochimie. 2011;93:1987–1992. doi: 10.1016/j.biochi.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delpu, Y. et al. Noncoding RNAs. Drug Discovery in Cancer Epigenetics 305–326, 10.1016/b978-0-12-802208-5.00012-6 (2016).
- 46.MacFarlane L-A, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Current Genomics. 2010;11:537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 48.Mahdipour M, van Tol HTA, Stout TAE, Roelen BAJ. Validating reference microRNAs for normalizing qRT-PCR data in bovine oocytes and preimplantation embryos. BMC Dev. Biol. 2015;15:25. doi: 10.1186/s12861-015-0075-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roth C, et al. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010;12:R90. doi: 10.1186/bcr2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song J, et al. Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Dig. Dis. Sci. 2012;57:897–904. doi: 10.1007/s10620-011-1981-7. [DOI] [PubMed] [Google Scholar]
- 51.Benz F, et al. U6 is unsuitable for normalization of serum miRNA levels in patients with sepsis or liver fibrosis. Exp. Mol. Med. 2013;45:e42. doi: 10.1038/emm.2013.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiang M, et al. U6 is not a suitable endogenous control for the quantification of circulating microRNAs. Biochemical and Biophysical Research Communications. 2014;454:210–214. doi: 10.1016/j.bbrc.2014.10.064. [DOI] [PubMed] [Google Scholar]
- 53.Lee JM, Roche JR, Donaghy DJ, Thrush A, Sathish P. Validation of reference genes for quantitative RT-PCR studies of gene expression in perennial ryegrass (Lolium perenne L.) BMC Mol. Biol. 2010;11:8. doi: 10.1186/1471-2199-11-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakayama T, et al. Assessment of suitable reference genes for RT-qPCR studies in chronic rhinosinusitis. Sci. Rep. 2018;8:1568. doi: 10.1038/s41598-018-19834-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lou G, et al. Differential distribution of U6 (RNU6-1) expression in human carcinoma tissues demonstrates the requirement for caution in the internal control gene selection for microRNA quantification. Int. J. Mol. Med. 2015;36:1400–1408. doi: 10.3892/ijmm.2015.2338. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y, Tang W, Peng L, Tang J, Yuan Z. Identification and validation of microRNAs as endogenous controls for quantitative polymerase chain reaction in plasma for stable coronary artery disease. Cardiol. J. 2016;23:694–703. doi: 10.5603/CJ.2016.0109. [DOI] [PubMed] [Google Scholar]
- 57.Popov A, Szabo A, Mandys V. Small nucleolar RNA U91 is a new internal control for accurate microRNAs quantification in pancreatic cancer. BMC Cancer. 2015;15:774. doi: 10.1186/s12885-015-1785-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schlosser K, McIntyre LA, White RJ, Stewart DJ. Customized Internal Reference Controls for Improved Assessment of Circulating MicroRNAs in Disease. PLoS One. 2015;10:e0127443. doi: 10.1371/journal.pone.0127443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dellinger RP, et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2012. Crit. Care Med. 2013;41:580–637. doi: 10.1097/CCM.0b013e31827e83af. [DOI] [PubMed] [Google Scholar]
- 60.Giamarellos-Bourboulis EJ, et al. Validation of the new Sepsis-3 definitions: proposal for improvement in early risk identification. Clin. Microbiol. Infect. 2017;23:104–109. doi: 10.1016/j.cmi.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 61.Ritchie ME, et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23:2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 62.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nature Protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete data generated and analysed for this study is implemented into this publication and the Supplementary Files.