Summary
Environmental pH can be an important cue for symbiotic bacteria as they colonize their eukaryotic hosts. Using the model mutualism between the marine bacterium Vibrio fischeri and the Hawaiian bobtail squid, we characterized the bacterial transcriptional response to acidic pH experienced during the shift from planktonic to host-associated lifestyles. We found several genes involved in outer membrane structure were differentially expressed based on pH, indicating alterations in membrane physiology as V. fischeri initiates its symbiotic program. Exposure to host-like pH increased the resistance of V. fischeri to the cationic antimicrobial peptide polymixin B, which resembles antibacterial molecules that are produced by the squid to select V. fischeri from the ocean microbiota. Using a forward genetic screen, we identified a homolog of eptA, a predicted phosphoethanolamine transferase, as critical for antimicrobial defense. We used MALDI-MS to verify eptA as an ethanolamine transferase for the lipid-A portion of V. fischeri lipopolysaccharide. We then used a DNA pulldown approach to discover that eptA transcription is activated by the global regulator H-NS. Finally, we revealed that eptA promotes successful squid colonization by V. fischeri, supporting its potential role in initiation of this highly specific symbiosis.
Keywords: symbiosis, bacterial physiology, Aliivibrio fischeri, antimicrobial cationic peptides
Graphical Abstract
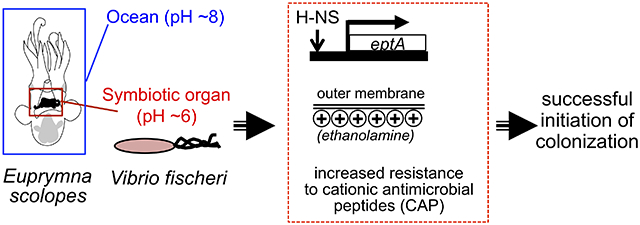
We characterize acidic pH as a cue that stimulates an anticipatory defensive response during initiation of symbiosis. This cue induces transcriptional changes that prepare Vibrio fischeri to resist host-derived antimicrobial peptides and increase colonization efficiency of its specific host, the Hawaiian bobtail squid. Using genetics and mass spectrometry, we demonstrate that the global regulator H-NS mediates this acid-cued response, and that antimicrobial resistance arises from a modification of lipopolysaccharide by the symbiont’s ethanolamine transferase.
Introduction
The host environment provides a landscape of informative chemical cues for symbiotic microbes (Wang and Ruby, 2011; Li et al., 2016). These cues often induce protective responses against host innate defenses, increasing symbiont fitness. While sub-lethal exposure to a toxic compound can cue a protective response (Merrell and Camilli, 2002), an unrelated environmental cue from the host (Palmer et al., 2007), other microbes (Ramsey and Whiteley, 2009), or the symbiont population itself (Goo et al., 2012) can also induce a protective response. Such anticipatory switching can evolve when the factors that microbes experience are predictably correlated (Mitchell et al., 2009; Brunke and Hube, 2014; Schwartzman and Ruby, 2016a; Cao and Goodrich-Blair, 2017), and predictive of each other, or where the cue predictably precedes the stress. In both examples, the ability of microbes to predict their future environments requires a period of entrainment and adaptive change. Foreseeable aspects of host biology, such as circadian regulation of inflammatory responses (Tognini et al., 2017), and conserved environmental features at sites of host-microbe interaction provide such a predictable landscape. In this way, the anticipatory regulation of protective responses by symbiotic microbes can be used to prepare for the characteristic chemistry of the host-tissue environment.
In metazoans, the mucus matrix is an ecological filter that ‘winnows’ symbionts from the surrounding environment (Fraune & Bosch, 2007; Nyholm & McFall-Ngai, 2004). A particularly well-studied example of host-microbe interactions at the mucus interface is that of the bobtail squid Euprymna scolopes and its bioluminescent symbiont Vibrio (Aliivibrio) fischeri. The first contact between host and microbe occurs in mucus that covers the symbiotic organ in this species-specific association (Schwartzman and Ruby, 2016b). The mildly acidic pH of the squid mucus is one of several initial cues presented to symbiotic cells during their transition from the planktonic state to host-association (Wang and Ruby, 2011; Kremer et al., 2013; Tischler et al., 2019). Because V. fischeri cells aggregate in this mucus immediately before encountering a multitude of chemical assaults from the squid, we hypothesized that acidic pH serves as an important cue to induce defensive responses from V. fischeri as it establishes its exclusive relationship with the host.
In this work, we describe an asymmetrical anticipatory switch in V. fischeri related to protection against cationic antimicrobial peptides like polymyxin B (PMB). Specifically, we show that in response to mild acidity, such as is encountered in the surface mucus of the host, V. fischeri alters the structure of its lipid A by attaching positively charged ethanolamine residues. We find that the modification of lipid A by V. fischeri is catalyzed by a homolog of the ethanolamine transferase, eptA, and is sufficient to confer acid-induced PMB resistance. ethanolamine transferase-catalyzed lipid A modification is a conserved mechanism for resisting host innate-immunity derived cationic antimicrobial peptides (CAPs) among diverse pathogenic and mutualistic microbes, including Vibrio spp. (Beceiro et al., 2011; Chen and Groisman, 2013; Cullen et al., 2015; Herrera et al., 2017). Moreover, we show that ethanolamine transferase activity increases the infectivity of V. fischeri.
Results
pH drives differential expression of several V. fischeri genes
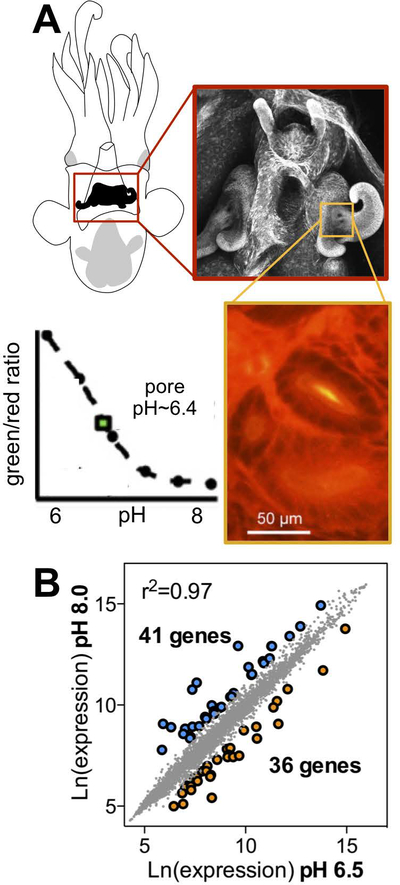
To examine the role of acidic pH in V. fischeri gene regulation during light-organ colonization of the newly hatched E. scolopes juvenile (Fig. 1A), we measured the global transcriptional profile of bacteria exposed to planktonic (seawater) or host-associated pHs (i.e., pH 8.0 and 6.5, respectively) (Fig. 1B, Table S1). We measured the pH that V. fischeri encounters in the mucus around the light organ pores using the pH sensitive fluorescent dye SNARF and found that this pH is approximately 6.4, consistent with previous findings (Kremer et al., 2013). To better mimic conditions experienced by the bacteria as they first contact the acidic regions of the squid’s light organ surface, we included soluble mucins in both treatments, simulating the acidified mucus produced by the ciliated fields of the squid light organ. Under these growth conditions, we identified a total of 77 genes that were differentially expressed based on pH; 36 of these were upregulated at a host-like acidic pH (Fig. 1B, Table S1). Acid-induced genes included those that encode several predicted outer membrane porins, including the major outer membrane proteins, OmpU and OmpC, as well as the homolog of a phosphoethanolamine transferase, EptA (also known as PmrC, or polymyxin resistance protein C). EptA homologs have previously been shown to modify the structure of the lipid A portion of lipopolysaccharide (LPS) in Gram-negative bacteria like Salmonella typhimurium and certain strains of Vibrio cholerae (Lee et al., 2004; Herrera et al., 2017). We verified the differential transcription of eptA and ompC (VF_1795) at different growth pHs using qRT-PCR (Fig. S1).
Figure 1.
V. fischeri differentially regulates many genes after exposure to marine or host-like pH. (A) Drawing (upper left) and confocal micrograph (red box) of the E. scolopes light organ. Orange box displays pH measurement of the region surrounding the light organ pore using the pH-sensitive dual-emission dye SNARF. Comparison of the emission ratio at green and red wavelengths gives a ratiometric measurement of tissue pH, relative to a calibration curve (inset graph). Plotting the tissue signal on the graph shows that the pore is a more acidic pH (yellow in the magnified image) than surrounding tissues. (B) Correlation between expression levels of V. fischeri transcripts after exposing cells to pH 8.0 or pH 6.5. Sets of genes were differentially expressed at high (blue) or low (orange) pH values.
Acid exposure increases V. fischeri’s resistance to CAPs
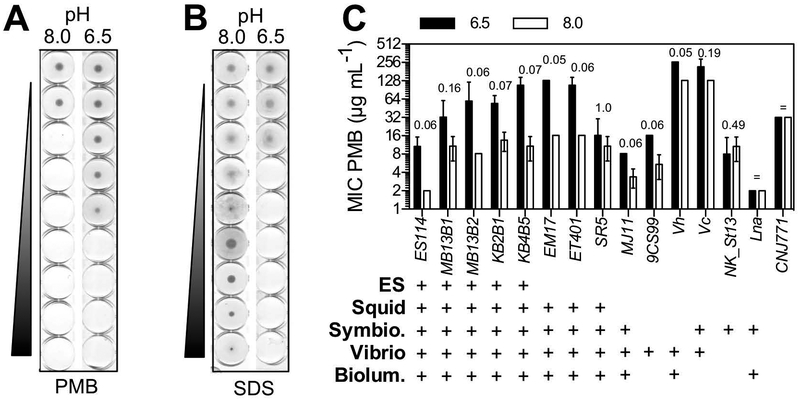
Because pH had a strong effect on the expression of eptA and other genes predicted to be involved in membrane function, we predicted that acidic conditions would lead to a shift in the cell’s sensitivity to certain membrane-targeting antimicrobials. Many eukaryotes, including the squid, produce CAPs that prevent growth of susceptible microbial partners (Chen et al., 2017; Krasity et al., 2011), and we reasoned that such a response might serve as an acidity-cued, symbiont-selection mechanism. To test this hypothesis, we exposed V. fischeri cells grown in either acidic or alkaline growth conditions to serial dilutions of PMB, a collection of CAPs that associates with negatively charged regions of the lipid A portion of LPS. This association eventually creates pores in the outer membrane, killing the cells (Orwa et al., 2001). We found that upon exposure to acidic conditions, the symbiotic V. fischeri strain ES114 became 6-fold more resistant to PMB killing (Fig. 2A). In contrast, sensitivity to sodium dodecyl sulfate (SDS) treatment increased under these conditions (Fig. 2B), unlike with previously described mutations that altered PMB sensitivity (DeLoney et al., 2002), indicating that the response does not represent a general enhancement of membrane integrity.
Figure 2.
V. fischeri isolates become more resistant to cationic antimicrobial peptides after exposure to low pH. (A) Representative PMB MIC assay of V. fischeri ES114 after exposure to seawater (pH 8.0) or host-associated (pH 6.5) conditions. (B) SDS MIC assay of strain ES114, grown under the same conditions as (A). (C) pH-mediated PMB MIC values of other marine bacteria. ‘ES’, symbiont isolated from Euprymna scolopes light organ; ‘Squid’, light-organ isolate from Euprymna or Sepiola squid species; ‘Symbio.’, host-associated strains; ‘Vibrio’, a species of the genus Vibrio; ‘Biolum.’, isolate producing bioluminescence. See Table 1 for strain descriptions. p values are reported according to a Mann-Whitney test of 3 replicates. The ‘=’ symbol indicates that all measured MIC values are equal in both conditions and, therefore, the p value was not calculated.
Acid exposure increases resistance to CAPs across different species of the Vibrionaceae
To determine how common this pH-altered sensitivity to PMB was, we grew several V. fischeri isolates, as well as other Vibrio and non-Vibrio species of marine bacteria, under the same set of acidic and alkaline conditions, before testing their resistance to PMB killing. The V. fischeri isolates were light-organ symbionts from several squid and fish hosts, as well as a single environmental seawater isolate (Table 1). We found that the increased resistance to PMB in cells grown under acidic conditions was shared across the V. fischeri clade (Fig. 2C), even in presumably non-symbiotic strains like 9CS99. This effect was more variable elsewhere in Vibrionaceae lineage: the tested strains of V. harveyi, and V. cholerae showed high basal resistance to PMB in the absence of the acidic pH cue, while Vibrio azureus and Photobacterium leiognathi showed intermediate levels of resistance in both pH environments (Fig. 2C). Similarly, growth under acidic conditions did not induce PMB resistance in the Gram-positive marine bacteria Exiguobacterium aestuarii (Fig. 2C).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Description/relevant genotype | Reference |
|---|---|---|
| V. fischeri | ||
| ES114 | Wild-type Euprymna scolopes light-organ isolate | (Boettcher and Ruby, 1990) |
| MJ11 | Wild-type isolate from Monocentris japonica light organ | (Ruby and Nealson, 1976) |
| MB13B1 | Wild-type E. scolopes light-organ isolate | (Wollenberg and Ruby, 2012) |
| MB13B2 | Wild-type E. scolopes light-organ isolate | (Wollenberg and Ruby, 2012) |
| KB2B1 | Wild-type E. scolopes light-organ isolate | ((Wollenberg and Ruby, 2012) |
| KB4B5 | Wild-type E. scolopes light-organ isolate | ((Wollenberg and Ruby, 2012) |
| SR5 | Wild-type Sepiola robusta light-organ isolate | (Wollenberg and Ruby, 2012) |
| EM17 | Wild-type E. morsei light-organ isolate | (Wollenberg and Ruby, 2012) |
| ET401 | Wild-type E. tasmanica light-organ isolate | (Wollenberg and Ruby, 2012) |
| 9CS99 | Wild-type planktonic isolate from North Atlantic Ocean | (Wollenberg et al., 2012) |
| JAS340 | ΔeptA (VF_A0210) deletion in ES114 | This work |
| MB06256 | VF_A0176 ::Tnerm in ES114 | This work |
| MB13215 | VF_2503::Tnerm in ES114 | This work |
| MB13278 | VF_2503::Tnerm in ES114 | This work |
| MB13555 | VF_2503::Tnerm in ES114 | This work |
| SLV15 | hns::Tnerm (VF_1631) in ES114 | (Lyell et al., 2010) |
| MB06859 | waaL::Tnerm (VF_0151) in ES114 | (Post et al., 2012) |
| AMJ2 | ΔarcA (VF_2120) deletion in ES114 | (Bose et al., 2007) |
| Seawater isolates | ||
| NK_St13 | Vibrio azureus-like | (Kremer et al., 2014) |
| Lna | Photobacterium leiognathi | (Dunlap, 1985) |
| CNJ-771 | Exiguobacterium aestuartii-like | (Gontang et al., 2007) |
| 0395-N1 | Vibrio cholerae isolate (ctxAB mutant) | (Mekalanos et al., 1983) |
| B392 | Vibrio harveyi isolate | (Reichelt and Baumann, 1973) |
| E. coli | ||
| DH5α-λpir | deoR hsdR17 endA1 gyrA96 pir | (Hanahan, 1983) |
| β−3914 | RP4–2 gyrA462 zei298::Tn10 ΔdapA::(erm-pir) | (Le Roux et al., 2007) |
| WM3064 | RP4–1360 rpsL hsdS ΔdapA1341::(erm-pir) | (Saltikov and Newman, 2003) |
| Plasmids | ||
| pVSV105 | Cmr, lacZα-(SphI, SalI/HincII, XbaI, SmaI/XmaI, KpnI, SacI) | (Dunn et al., 2006) |
| pVSV104 | Knr, lacZα-(SphI, SalI/HincII, XbaI, SmaI/XmaI, KpnI, SacI) | (Dunn et al., 2006) |
| pEVS104 | R6Kori RP4 oriT trb tra Knr | (Stabb and Ruby, 2002) |
| pKV363 | Mobilizable suicide vector; Cmr; pSW7848 + MCS | (Shibata et al., 2012) |
| pTM267 | Two-color fluorescent reporter, tetA’::mCherry, GFP downstream of a multiple cloning site, Cmr | (Miyashiro et al., 2010) |
| pJAS340 | ΔeptA::pKV363 | This work |
| pJAS341 | eptA::pVSV105 | This work |
| pJAS342 | eptA’::GFP in pTM267 | This work |
| pJBL301 | tetA’::eptA in pVSV104H | This work |
eptA increases the PMB resistance of V. fischeri
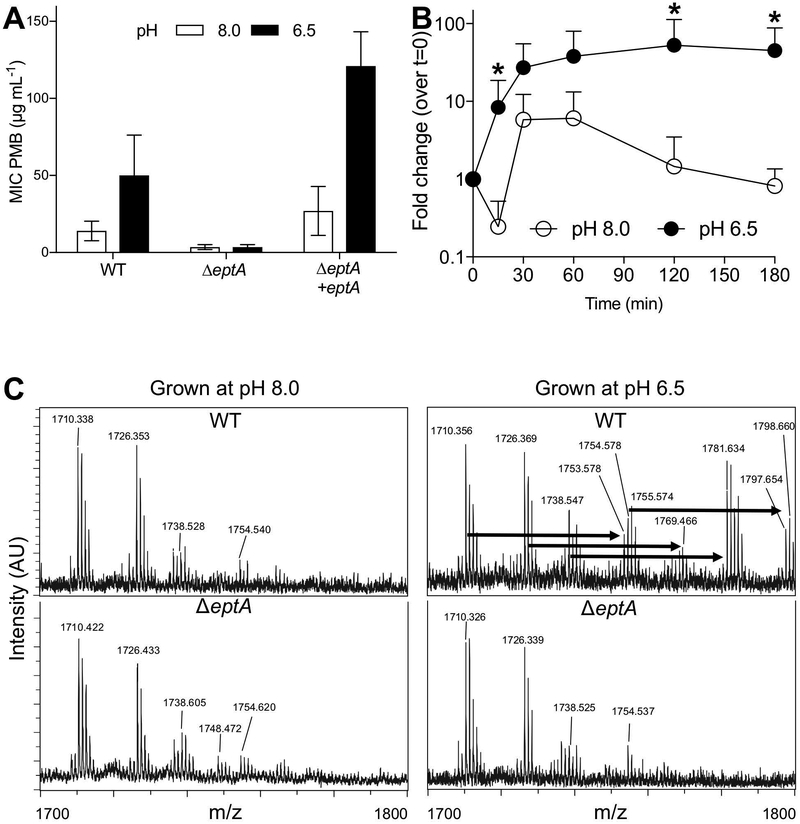
We conducted a forward genetic screen to identify genes that might contribute to PMB resistance in V. fischeri. Using a previously generated transposon mutant library (Brennan, 2013), we screened >13,000 insertions for increased pH-dependent susceptibility to PMB. We also assayed the response of these mutants to SDS exposure so that we could exclude those isolates with an increased sensitivity to both membrane-perturbing agents as mutants with broadly compromised membranes. As a result, we identified 12 mutants with diminished resistance to PMB, but not SDS, relative to wild type V. fischeri ES114 (Table S2). Of these, only one – eptA – was also differentially expressed based on pH. These results led us to predict that this gene was responsible for pH-mediated antimicrobial resistance. We deleted the entire eptA open-reading frame, and confirmed that the gene product was necessary to confer acid-induced PMB resistance (Fig. 3A). An in trans complementation of eptA, including its native promoter region, restored the pH-induced activity, further supporting this finding (Fig. 3A). Using qRT-PCR to measure transcript levels, we found that low pH activated eptA expression (Fig. 3B). This transcriptional increase occurred within 15 minutes of low pH exposure, and plateaued after approximately 30 minutes (Fig. 3B). The outer membrane protein OmpU has been shown to confer CAP resistance in V. cholerae (Mathur and Waldor, 2004; Mathur et al., 2007), but when we tested a V. fischeri ΔompU mutant for PMB sensitivity, we observed no difference between this strain and its isogenic parent (Fig. S2). In contrast, a V. fischeri waaL mutant had 2- to 4-fold increase in sensitivity to PMB that was pH-independent (Fig. S2). WaaL homologs ligate the O-antigen to LPS, and mutation of this gene in V. fischeri also conferred sensitivity to SDS (Table S2). Thus, we concluded that, while there are several activities that promote cellular resistance to PMB, eptA is the main gene responsible for pH-induced PMB resistance in V. fischeri.
Figure 3.
The eptA homolog of V. fischeri (VF_A0210) is differentially expressed under acidic conditions and acts as a lipid A ethanolamine transferase. (A) pH-dependency of PMB MIC assay performed on wild-type V. fischeri ES114 (WT) and its derivatives, with or without the eptA gene. Bars represent mean with 95% confidence intervals (CI). (B) Transcription of eptA over time after exposure to acidic or basic pH. Points represent mean + SD. Data represent at least three biological replicates, *: p≤0.1 according to Mann-Whitney test. (C) Representative mass spectrometry spectra of lipid A extracts isolated from either wild-type V. fischeri or its ΔeptA derivative, grown at pH 6.5 or 8.0. Arrows indicate 43 m/z shifts, consistent with a dehydration addition of an ethanolamine residue.
eptA is essential for the addition of a ethanolamine residue to V. fischeri lipid A
The eptA homologs in Salmonella typhimurium and Vibrio cholerae El Tor encode a protein that acts as a phosphoethanolamine transferase, altering the charge on the bacterial outer membrane and increasing CAP resistance (Lee et al., 2004; Needham and Trent, 2013; Herrera et al., 2017). Heterologous expression experiments have also demonstrated that the EptA homolog in V. fischeri is functional (Herrera et al., 2017). To confirm the biochemical role of EptA in V. fischeri, we performed a mass spectroscopy analysis of purified lipid A from wild-type and ΔeptA V. fischeri, grown at different pHs (Fig. 3C). At pH 8.0, signals with m/z corresponding to previously identified lipid A species were observed in both wild-type and mutant extracts (Phillips et al., 2011). When grown at pH 6.5, but not 8.0, we identified mass shifts of ~43 Daltons in wild-type lipid A that corresponded to the dehydration attachment of ethanolamine (Fig. 3C, top). These shifts were missing in the ΔeptA mutant (Fig. 3C, bottom) at both pHs, consistent with an acidic pH-dependent and EptA-catalyzed addition of this moiety to lipid A. Together, these data support the conclusion that EptA acts as an acid-induced ethanolamine transferase in V. fischeri.
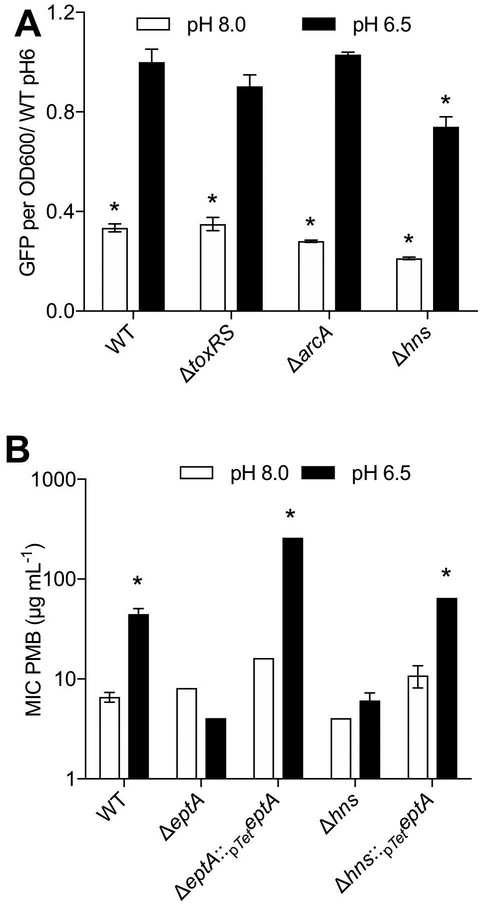
eptA expression is promoted by the dual regulator H-NS
We next sought out regulators of eptA expression that might connect pH variation to CAP resistance by V. fischeri. In S. typhimurium, eptA is transcriptionally regulated by PhoP in a magnesium- and PmrD-dependent manner (Chen and Groisman, 2013). In addition, our transposon screen for PMB-sensitivity identified several predicted transcriptional regulators that were potentially involved in V. fischeri eptA expression (Table S2). To clarify the roles of these candidates, we determined the relative levels of eptA transcription in mutants lacking two phoP homologs (VF_0526 and VF_1396) (Hussa et al., 2007) and two of our predicted regulators of eptA (VF_2503 and VF_A0176) in V. fischeri. We found no effect of the absence of any of these candidates on the activity of the eptA promoter (data not shown). We reasoned that regulators of eptA may have been missed in a loss-of-function screen either if they were required for in vitro growth of V. fischeri, or if pH-dependent transcription was mediated by a repressor. To address these issues, we performed a DNA-pulldown with the presumed promoter of eptA as bait to discover potential regulatory proteins of eptA transcription (Table 2, Table S3; see Methods). We identified eight non-ribosomal target genes, and tested strains with mutations in a subset of these genes for defects in eptA promoter activity at acidic and alkaline pHs. Mutation of three of the eight candidates (nagC, arcA, and VF_1195) showed no significant alteration in eptA expression (data not shown). We also found that ToxR, a master regulator of V. cholerae host-associated physiology, did not alter eptA expression at either pH (Fig. 4A). However, a mutant of hns (Lyell et al., 2010), displayed significantly decreased eptA expression at both low and high pH (Fig. 4A). We tested this hns mutant for pH-dependent PMB sensitivity, and found that it showed significantly diminished resistance to PMB (Fig. 4B). This strain was equally sensitive to PMB at low and high pH, similar to the ΔeptA mutant, indicating that it had lost its pH-mediated resistance. We were able to complement the PMB sensitivity defect by driving eptA expression off of the tetA promoter, showing that the increased sensitivity of the hns mutation could be overridden in trans (Fig. 4B). Interestingly, although we have previously found the TetA promoter to be insensitive to our experimental conditions (data not shown), this complemented strain still showed a pH-influenced effect, indicating that additional post-transcription regulation (e.g., acting either on mRNA or protein expression, or directly on EptA activity) may also be pH-dependent.
Table 2.
Most abundant potential regulators of eptA, identified through promoter pulldown†
| Locus Tag | Gene name | Predicted function |
|---|---|---|
| VF_2423 | tufA | Elongation factor Tu |
| VF_0806 | nagC | DNA-binding transcriptional dual regulator, repressor of N-acetylglucosamine |
| VF_0815 | seqA | Negative modulator of initiation of replication |
| VF_0252 | rplF | 50S ribosomal protein L6 |
| VF_0658 | rimK | Probable alpha-L-glutamate ligase |
| VF_1951 | fabZ | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase |
| VF_1962 | rpsB | 30S ribosomal protein S2 |
| VF_2566 | atpA | ATP synthase subunit alpha |
| VF_A0001 | Uncharacterized protein | |
| VF_1631 | hns | Global DNA-binding transcriptional dual regulator |
Ten most abundant proteins listed. For complete list of proteins pulled down with the eptA promoter region, see Table S1. Genes listed in descending order of #PSM in pulldown analysis.
Figure 4.
The V. fischeri eptA is regulated by H-NS. (A) eptA reporter expression in mutants strains of V. fischeri after exposure to alkaline or acidic pH. (B) PMB MIC assay of eptA and hns mutants. The eptA mutation is complemented by a plasmid borne copy of eptA under the control of the tetA promoter. In (A), * indicates significant (p<0.05) difference from WT at pH 6 by Dunnett’s test; in (B) * indicates significant p<0.05 difference from WT at pH 8.
eptA is important for colonization initiation
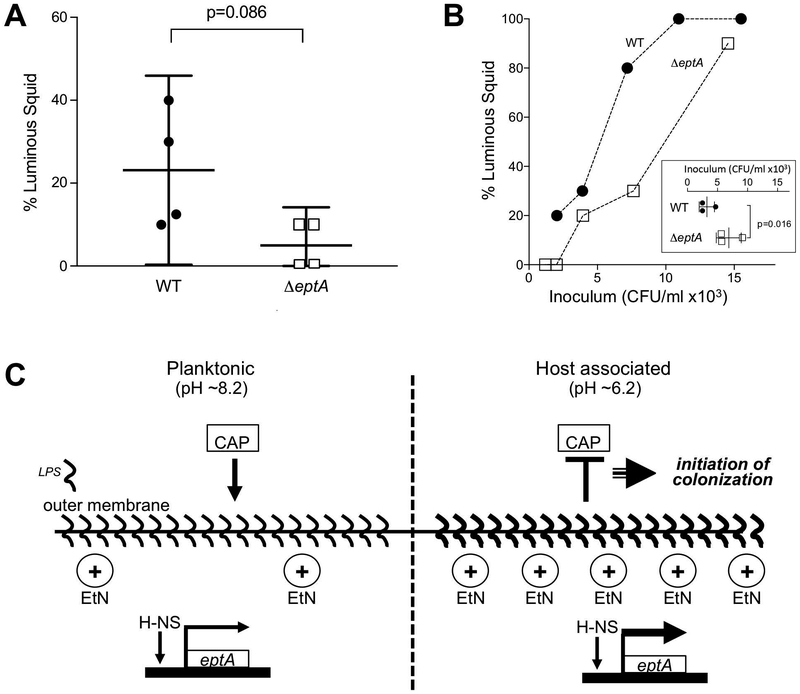
The tissues of E. scolopes present several chemical and physical challenges to bacteria transitioning from the ambient seawater to the surface of the symbiotic organ, where the cell must then traverse these assaults after exposure to the squid’s acidic mucus. We hypothesized that eptA is important for surviving these challenges and efficiently initiating colonization. To test this idea, hatchling squid were exposed for 3 h to a relatively low inoculum of either wild-type or ΔeptA V. fischeri cells, and the fraction of squid successfully colonized was determined. Although the data are spread, the ΔeptA mutant was markedly less likely to colonize squid under these conditions (Fig. 5A), consistent with our prediction. We then tested whether the colonization defect of the ΔeptA mutant could be overcome by increasing the concentration of V. fischeri that the squid were exposed to. In fact, the ΔeptA mutant could successfully colonize the squid with an efficiency similar to wild type, but it required a higher inoculum to do so; mutating eptA resulted in a >2-fold increase in ID50, from 2,500 CFU mL−1 to 5,700 CFU mL−1 (Fig. 5B). Significantly, animals successfully colonized under any of these conditions had similar bacterial loads in the light organ after 24 h (data not shown), and there was no difference in the competitive index of ΔeptA when co-colonized with wild-type V. fischeri at an initial cell density greater than the ID50. Together, these results indicate that the effect of the ΔeptA mutation was restricted to the early initiation events of colonization, and that the carriage of eptA enhances the colonization efficiency of V. fischeri.
Figure 5.
eptA is required for efficient squid colonization by V. fischeri. (A) Percentage of luminous squid after a 3 h exposure to 800 CFU mL−1 of V. fischeri. The means, with 95% CI, are indicated. (B) Fraction of colonized squid as a function of V. fischeri inoculum size. Inset: calculated ID50 for WT and the eptA mutant V. fischeri. (C) Schematic model of eptA regulation and its role in cationic antimicrobial peptide (CAP) resistance in V. fischeri by LPS modification with ethanolamine (EtN).
Discussion
In this study, we show that an acid-cued switch in V. fischeri physiology is an anticipatory modification that allows V. fischeri to effectively colonize its squid host. Our molecular characterization of acid-induced PMB resistance (Figs. 3 & 4), together with the requirement of this protective response for efficient initiation of symbiosis (Fig. 5B), contributes to an understanding of the mechanisms by which the host’s environmental filter promotes colonization of its tissues. Specifically, an H-NS-dependent transcriptional activation of eptA governs the decoration of V. fischeri LPS with ethanolamine (Fig. 5C). Interestingly, this addition differs from the predicted function of eptA homologs, in that we described lipid A mass shifts consistent with an addition of ethanolamine, not phosphoethanolamine as has been previously described (Herrera et al., 2017), potentially owing to the unique lipid A structure of V. fischeri (Phillips et al, 2011). This remodeling of the outer membrane increases the resistance of V. fischeri to host antimicrobial peptides, some of which are thought to be enhanced by charge-charge interactions, similar to PMB (Heath-Heckman et al., 2014; Chen et al., 2017). Additionally, the E. scolopes genome (Belcaid et al., 2019) has revealed a large set of potential homologs for known antimicrobials that may rely on similar interactions to either control V. fischeri populations or selectively allow V. fischeri to prosper in the light organ. By inducing eptA expression before the symbionts enter the antimicrobial environment of the light organ ducts (Nyholm and Graf, 2012; McFall-Ngai, 2014), the acidic pH encountered by V. fischeri cells in the >2 hours they spend aggregating outside the light organ (Kremer et al., 2013) provides a cue and enough time for an anticipatory switch to a symbiosis-competent physiological state (Fig. 3B, Fig. 5C). As a result, the timely expression of eptA potentiates colonization of squid by V. fischeri, even at the low cell concentrations characteristic of its natural environment (Lee and Ruby, 1994).
Despite the fact that acidic pH and antimicrobial peptides are common motifs of metazoan epithelial tissue (Wesley et al., 1985), pathogenic and mutualistic bacteria are likely to experience the host-tissue environment differently. A comparison between the PMB resistance mechanisms and acid-induced transcriptional responses described in V. fischeri and in the related gastrointestinal pathogen V. cholerae hints at these differences. Unlike V. fischeri, V. cholerae encodes an additional LPS-modifying system that confers PMB resistance: the aminoacyl lipid modifying enzymes AlmEFG. In contrast to the acid-induced modification described here, transcription of almEFG is repressed by exposure to acid via the LeuO-ToxRS-CarRS/VprAB cascade of transcription factors (Herrera et al., 2014; Ante et al., 2015; Bilecen et al., 2015; Bina et al., 2016). The V. cholerae PMB resistance phenotype is also regulated by the envelope stress response through the alternative sigma factor RpoE and the outer membrane protein OmpU (Mathur et al., 2007; Shaw, 2012). Several lines of evidence suggest that this architecture of the envelope-stress response pathway is not conserved between the V. cholerae and V. fischeri: (i) our transposon screen for PMB-sensitive mutants identified neither ompU nor rpoE (Table S2), (ii) there was no evidence that any of the four V. fischeri RpoE homologs bound upstream of eptA (Table S1) (Mandel et al., 2008), and (iii) deletion of ompU did not increase sensitivity to PMB (Fig. S1). Like V. fischeri, the El Tor biotype of V. cholerae expresses EptA in response to an acid cue (Herrera et al., 2017); however, El Tor encodes the acid-responsive regulator AphB, which lacks a homolog in V. fischeri. The AphB regulated genes pepA, tcpPH, or the cadABC, all of which are components of the acid-inducible response in V. cholerae (Merrell and Camilli, 2000; Behari et al., 2001), but are not induced by V. fischeri under our experimental conditions (Table S1). Taken together, the additional AlmEFG-dependent mechanism of acid-independent PMB resistance, the integration of PMB resistance into the envelope stress response, and the existence of a dedicated acid-stress response all suggest that V. cholerae El Tor encounters conserved, but differently presented, chemical cues than V. fischeri in their respective host tissue environments. Future comparative studies of the regulation of eptA in diverse symbiotic microbes will provide further insight into the range of adaptations by which pathogenic and mutualistic microbes sense and respond to a host environment.
Our finding that H-NS plays an activating role in eptA expression through interaction with the promoter region was surprising given H-NS’s typical role as a repressor; however, it is consistent with previous findings where H-NS promotes expression by outcompeting other repressors or organizing the genome to coordinate positive regulation (Ayala, 2017). In our case, it appears as though H-NS is involved in activity of the putative promoter region upstream of eptA, but that additional pH-sensitive regulation of the eptA-driven membrane modification occurs (Fig. 4B). H-NS activity has been found to be pH-sensitive (Liu, 2010), but as we found the hns mutant to decrease peptA activity at both tested pHs, we believe additional factors are involved in pH-mediated V. fischeri membrane structure. Specifically, H-NS may drive overall expression of eptA, while something else facilitates pH-mediated expression. Future work may explore additional interactions between H-NS and other regulators at the interface of the eptA promoter region, as well as discovering other actors that guide V. fischeri’s physiologic shifts at acidic pH.
Bacteria are highly effective at sensing changes in their environment, and rapidly adjusting their physiology and biochemistry to heighten the cell’s fitness under the new conditions. Typically, the connection between the environmental cue and the response is direct; for example, when planktonic V. fischeri cells first encounter the nascent light organ, they respond to the presence of host-derived NO by inducing the detoxifying enzyme Hmp (Wang et al., 2010), and to the host’s release of chitobiose increasing their ability to chemotax toward that sugar (Kremer et al., 2013). In both of these examples, the response is initially primed as the bacteria pause in an aggregate just outside the organ (McFall-Ngai, 2014), and failure to adapt results in a colonization defect. Here we have described a different strategy, called anticipatory adaptation, by which V. fischeri ‘bets’ that coming across an acidity cue means they have contacted host tissue, and that inducing an LPS-modifying enzyme in response will help protect them from an inevitable encounter with host-derived cationic antimicrobial peptides. This example is likely to be only the first evidence of adaptive switching in the squid-vibrio symbiosis: the NO-protective enzyme Hcr (VF_A0863) (Wang et al., 2016) also appears to be induced by an acidity cue (Table 1).
While the work presented here establishes that EptA is a significant colonization factor for symbiotic V. fischeri, it remains possible that there are context-specific costs to eptA expression that select for the ability to differentially, rather than constitutively, express this gene. Regardless, EptA restructures V. fischeri LPS in a fashion that is similar to that of several other animal-associated microorganisms, yet is regulated in a manner that has not been previously described. This difference may be related to lineage and lifestyle related factors, and highlights the benefit of comparing co-association strategies across diverse animal-microbe symbioses.
Experimental Procedures
Bacterial strains and growth conditions
Bacterial strains and plasmids used in this work are listed in Table 1. Escherichia coli was cultured at 37 °C with shaking at 250 rpm in Luria-Bertani medium (LB) (Sambrook J. et al., 1989), with (per mL) 150 μg erythromycin (Erm), 25 μg chloramphenicol (Cam) or 50 μg kanamycin (Kan), as needed. Unless otherwise noted, V. fischeri was grown in Luria-Bertani salt medium (LBS) shaking at 225 rpm at 28 °C, with (per mL) 5 μg Erm, 5 μg Cam or 100 μg Kan where indicated. All Vibrio fischeri mutant strains were derived from the light-organ isolate ES114 (Boettcher and Ruby, 1990).
Sample collection, RNA extraction and transcriptional analysis
To assess the transcription of V. fischeri in an environment approximating the squid’s mucus, strain ES114 was cultured in an artificial seawater medium (ASW: 300 mM NaCl, 50mM MgSO4, 10 mM CaCl2, 10mM KCl) containing 50 mM PIPES buffer and 0.1% hog gastric mucin (HGM, w/v; 1% bound sialic acid, Sigma). The medium was buffered at pH 6.5 or pH 8.0 to characterize transcription in response to acidic pH. Triplicate overnight cultures of ES114 in LBS were diluted 1:100 into 25 mL high osmolarity seawater tryptone medium (SWTO, (Stabb et al., 2004)) in 125 mL flasks, and grown with shaking and 28 °C. When cultures reached 0.60 +/− 0.05 optical density at 600 nm (OD600, 1 cm path length), they were centrifuged for 8 min at 6000 × RCF and 4 °C to pellet the cells. The supernatants were aspirated, and pellets were resuspended in 25 mL of 50 mM PIPES (pH 8.0) ASW in a 125 mL flask. Cultures were incubated for 1 h and were centrifuged again, and the pellet was resuspended in 1 mL of PIPES ASW. One-half milliliter of this resuspension was used to inoculate 15 mL of PIPES ASW/HGM (0.1%), buffered to either pH 6.5 or at pH 8.0. Cultures were incubated for 2 h, then centrifuged, and the pellet was resuspended in 4 mL RNAlater (Life Technologies). This final suspension contained 2.5 × 108 CFU mL−1 (+/− 5%).
The cells suspended in RNAlater were pelleted by centrifugation at 5000 × RCF and 4 °C for 5 min. An RNeasy mini kit (Qiagen) was used to extract RNA, according to the manufacturer’s instructions. Samples were eluted in RNase-free water, and treated with RQ-DNase (Promega), according to the manufacturer’s instructions. An on-column sample cleanup was performed following the protocol outlined in the kit. Samples were at least 1 μg RNA μL−1, with absorbance ratios at 260 nm:280 nm and at 260 nm:230 nm of >2.1. Samples were submitted to the University of Wisconsin-Madison Gene Expression Center for further quality assessment using an Agilent RNA NanoChip. Ten micrograms of total RNA was used for cDNA synthesis following protocols defined by NimbleGen array analysis (Roche NimbleGen). cDNA was submitted to NimbleGen for prokaryotic gene-expression microarray analysis. The expression values were processed to yield the fold change in expression between experimental conditions using ArrayStar software (DNAStar). Differential regulation was defined as >2-fold difference in gene expression between the two conditions with at least 95% confidence, which allowed us to detect potentially subtle physiological responses to each condition. The linear correlation in signal intensities between the two conditions was 0.97. BLAST2GO software (BioBam Bioinformatics Solutions) was used to perform functional enrichment analysis of the dataset based on gene ontology classification.
Quantitative RT-PCR procedures were conducted in accordance with the MIQE protocol guidelines (Bustin et al., 2009). Primers were designed by OligoAnalyzer software (Integrated DNA Technologies) with a 60 °C annealing temperature. Primer pair efficiencies were between 95 and 105%. Candidate gene expression was normalized using the geometric mean of polA, a previously defined endogenous control gene (Wang et al., 2010), whose expression was confirmed to be invariant between the conditions compared in this experiment. Mann-Whitney test, were used to compare measurements across conditions.
Minimum inhibitory concentration (MIC) assay
MIC assays were performed with V. fischeri based on previously described protocols (DeLoney et al., 2002; Kremer et al., 2014). To assay for pH-dependent resistance to various stressors, MIC assays were set up in pH 6.5 or pH 8.0 PIPES MIC buffer containing 10 mM PIPES (pH 6.5 or 8.0) in ASW. Two-fold serial dilutions of stressors of interest (starting concentrations: 0.26 mg mL−1 PMB or 1% SDS) were set up in 96-well plates containing 90 μL total volume. Bacterial cultures were started in SWT medium (Boettcher and Ruby, 1990) at 28 °C, and grown to an OD600 of 0.10 +/− 0.02. Cultures were diluted 1:100 into 3 mL PIPES (pH 8.0) in ASW, and incubated for 3 h at 28 °C with shaking. Another 1:100 dilution into SWT was performed upon completion of the 3-h incubation, and 10 μL of the bacterial culture was added to 90 μL of a MIC buffer (pH 6.5 or 8.0) plus stressor. Plates were incubated under static conditions at 28 °C and checked at 24 h for pellet formation at the bottom of the plate as well as any turbidity in the wells. Inoculation levels that fell between 10 and 100 CFUs were found to produce consistent MIC values.
Molecular techniques
In-frame deletions of V. fischeri genes were constructed by CcdB-toxin mediated double homologous recombination, using a V. fischeri-specific suicide vector pKV363 as previously described (Stabb and Ruby, 2002; Le Roux et al., 2007; Shibata et al., 2012). Briefly, segments upstream and downstream of the coding region were amplified, and ligated together using a unique restriction enzyme site. The resulting chimeric DNA was ligated directionally into the pKV363 backbone at the multiple cloning site (MCS). Conjugative transfer was used to introduce the plasmid into V. fischeri ES114, and the double crossover of the construct was selected by arabinose induction of the CcdB toxin encoded on the pKV363 plasmid backbone. Deletions were confirmed by sequencing around the target locus.
To complement eptA under control of its native promoter, the intact locus was amplified from wild-type V. fischeri, and cloned into the plasmid pVSV105: a shuttle vector carrying an origin of replication derived from a V. fischeri-specific plasmid pES213 (Dunn et al., 2006),) Cloning was carried out in the E. coli strain DH5α-λ pir. The resultant vector was introduced into V. fischeri by pEVS104-assisted conjugation (Stabb and Ruby, 2002). For complementation of eptA off a non-native, constitutive promoter, the tetA promoter from plasmid pTM267 was fused to the upstream region of eptA, and the resulting construct cloned into the MCS of pVSV104 by Goldengate cloning. This plasmid construct was transformed into E. coli WM3064, and conjugated into V. fischeri.
Construction of two-color fluorescent reporters: The regions 400 bp upstream and 200 bp downstream of the predicted translational start sites of the eptA gene were amplified, and cloned into XmaI and XbaI sites of the of plasmid pTM267. This plasmid encodes a constitutively activated copy of the red fluorescent protein mCherry, as well as the gene (gfp) encoding a green-fluorescent protein downstream of a MCS (Miyashiro et al., 2010). The insertion of the putative promoter region into the MCS of pTM267 resulted in the promoter-dependent transcription of gfp. The reporter plasmid is maintained at ~10 copies per cell.
Screening of an arrayed transposon mutant library for genetic components of PMB resistance
We screened 13,686 mutants of an existing 23,904 strain library (Brennan et al., 2013), for a loss of resistance to PMB. Briefly, a pin replicator was used to inoculate 96-well plates containing 100 μL of LBS-Erm from frozen glycerol stocks of the first-generation plates created in the original screen. After overnight growth, the pin replicator was used to inoculate fresh 96-well plates containing 100 μL pH 6.5 SWTO with the LBS-Erm overnight cultures. The plates were placed on a shaker for 3 h at 28 °C prior to being used to inoculate 100 μL MIC buffer (pH 6.5) containing 10 % SWT medium, with and without 8 μg PMB mL−1, in a round-bottomed 96-well plate. Plates were incubated under static conditions overnight at 28 °C, prior to assessment of growth in the presence and absence of PMB. Five microliters of culture material from strains of interest were combined with 95 μL of elution buffer (Qiagen), and frozen at −20 °C to create template mixes for arbitrarily primed PCR. Arbitrarily primed PCR was performed as described previously (Brennan et al., 2013), with the addition of ExoSAP-IT treatments (Affymetrix), following the manufacturer’s instructions between successive rounds of PCR to remove excess single-strand oligonucleotide primers. Amplified reactions were sent for Sanger sequencing at the University of Wisconsin-Madison Gene Expression Center.
Lipid A analysis by MALDI-TOF mass spectrometry
V. fischeri lipid A was purified as previously described (Phillips et al., 2011). Overnight cultures of wild-type and ΔeptA V. fischeri grown in LBS were diluted 1:100 into 600 mL SWTO in 2 L flasks, then grown with shaking at 28 °C. When cultures reached OD600 0.60 +/− 0.05, they were centrifuged for 20 min at 6000 × RCF and 4 °C to pellet the cells. Supernatants were aspirated, and pellets were resuspended in 600 mL of 50 mM PIPES (pH 8.0) ASW in a 2 L flask. Cultures were incubated for 1 h and were centrifuged again, and the pellets were resuspended in 10 mL of PIPES ASW. For each strain, 5 mL of this resuspension was used to inoculate 250 mL of PIPES ASW/HGM (0.1%), buffered to either pH 6.5 or pH 8.0. Cultures were incubated for 2 h, then centrifuged, and the pellets were stored at −20 °C for subsequent lipid A purification. Each lipid A pellet was then extracted twice with CHCl3/MeOH/H2O (10:5:6, v/v/v), and the pooled extract was concentrated under a gentle stream of N2. Mass spectral analysis was performed in negative ion mode using a Bruker UltraFlex III MALDI-TOF/TOF mass spectrometer equipped with a 337 nm wavelength, 50 Hz N2 laser (pulse energy: 150 μJ; 3 ns pulse width). Mass spectra were acquired and analyzed with FlexControl 3.3 and FlexAnalysis 3.3 software. Samples were dissolved in CHCl3/ CH3OH (3:1, v/v) at approximately 1–1.3 μg μL−1. A 0.5 μL aliquot of the sample was mixed with an equal volume of 20 mg mL−1 matrix (5-chloro-2-mercaptobenzothiazole (Sigma-Aldrich) in CHCl3/ MeOH/ H2O (4:4:1, v/v/v) with 50 mM ammonium citrate on a stainless-steel target, and allowed to dry at room temperature. Approximately 1500 laser shots were recorded for each sample. Calibration was performed with a mixture of angiotensin II, renin substrate tetradecapeptide, and insulin chain B (Sigma-Aldrich).
DNA pulldown and proteomic analysis
DNA pulldowns using the presumptive eptA promoter region were performed as previously described (Lynch, in prep). Briefly, 250 bp upstream of the eptA coding region were PCR amplified using biotinylated primers (Integrated DNA Technologies). This probe was then added in excess to V. fischeri cell lysates and incubated at room temperature. Probe-protein complexes were precipitated with streptavidin-coupled DynaBeads (Thermo-Fisher), and proteins were eluted with increasing NaCl concentrations (100–700 mM). Proteins were processed and identified proteomically using LC-MS/MS at the University of Vermont VGN Proteomics Facility, and aligned against the predicted V. fischeri ES114 proteome (UniProt, ID: UP000000537).
Effects of regulators on eptA activity
To monitor promoter activity in response to pH and other environmental cues, strains carrying reporter plasmids were grown to an OD600 of 0.60 +/− 0.05 in LBS with appropriate antibiotic selection. One milliliter of this culture was centrifuged for 1 min at 10,000 × RCF to pellet the cells, and the cell pellet was resuspended in 0.5 mL of PIPES MIC buffer, modified by adjusting pH or adding stressors, as indicated in the text. Duplicate 200 μL aliquots of the cell suspensions were added to a 96-well plate, and the plate was incubated with shaking for 3 h at 28 °C. A Tecan Genios Pro plate reader was used to monitor the accumulation of GFP (485 and 535 nm excitation/emission filter sets) mCherry (535 and 612 nm filter sets), and OD600 in the pTM267-based transcriptional reporter constructs. Alternatively, cells were grown to equivalent OD600 values in the defined media (50 mM PIPES, 60 mM glucose, 2% casamino acids, 300 mM NaCl, 100 mM MgSO4, 10 mM CaCl2, 10 mM KCl, 287 μM Na2PO4, 33 μM K2HPO4, 25 μM NaH2PO4 and 20 μM FeSO4) at the noted pH, and GFP levels were normalized to OD600. Non-fluorescent ES114 strain was used as a control for autofluorescence, and emission values obtained from this sample were subtracted from experimental measurements.
Squid colonization
Adult Euprymna scolopes were collected in near-shore Oahu, Hawaii, USA. Juveniles were hatched in captivity in either filter-sterilized ocean water or artificial seawater, and exposed to the indicated inoculum of wildtype or ΔeptA V. fischeri ES114 for three hours, rinsed, and maintained on a normal 12 h:12 h light: dark schedule. After 24 h, squid were individually checked for luminescence >10 AU in a TD 20/20 luminometer (Turner Designs).
Supplementary Material
Figure S2. Some outer membrane modifications alter pH-dependent PMB sensitivity in V. fischeri but, unlike in V. cholerae, deletion of OmpU does not. PMB MIC data for strains of V. fischeri with mutations in noted genes grown at the listed pH.
Figure S1. RT-qPCR verification of differential transcription in response to pH. Relative transcript levels of VF_1795 and eptA, which were identified as upregulated in low pH by transcriptomic analysis, were determined after growth at basic (pH 8.0) and acidic (pH 6.5) conditions. Data presented represent mean +/− SD of six biological replicates. Values were normalized to transcript levels of polA.
Acknowledgements
We thank the members of the Ruby and McFall-Ngai labs for helpful discussions. We thank Drs. Qing Li and Margaret Butler for providing access to the MALDI-TOF MS instrument at the UH Proteomics Core Facility. The authors were funded by an NSF Graduate Research Fellowship, Chemical Biology Training Program (UW-Madison, NIH-NIGMS T32 GM008505), and the Microbes in Health and Disease Training Program (UW-Madison NIH-NIAID T32 AI055397) to JAS, NIH-NIGMS F32 GM119238 to JBL, OD11024 and GM135254 to EGR and M McFall-Ngai, and R37 AI50661 to M McFall-Ngai and EGR.
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Aeckersberg F, Lupp C, Feliciano B, and Ruby EG (2001) Vibrio fischeri outer membrane protein OmpU plays a role in normal symbiotic colonization. J Bacteriol 183: 6590–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ante VM, Bina XR, Howard MF, Sayeed S, Taylor DL, and Bina JE (2015) Vibrio cholerae leuO transcription is positively regulated by ToxR and contributes to bile resistance. J Bacteriol 197: 3499–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala JC, Silva AJ, and Benitez JA (2017) H-NS: an overarching regulator of the Vibrio cholerae life cycle. Res Microbiol 168:16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beceiro A, Llobet E, Aranda J, Bengoechea JA, Doumith M, Hornsey M, et al. (2011) Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the PmrAB two-component regulatory system. Antimicrob Agents Chemother 55: 3370–3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behari J, Stagon L, and Calderwood SB (2001) pepA, A gene mediating pH regulation of virulence genes in Vibrio cholerae. J Bacteriol 183:178–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcaid M, Casaburi G, McAnulty SJ, Schmidbaur H, Suria AM, Moriano-Gutierrez SM, et al. (2019) Symbiotic organs shaped by distinct modes of genome evolution in cephalopods. PNAS 116: 3030–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilecen K, Fong JCN, Cheng A, Jones CJ, Zamorano-Sánchez D, and Yildiz FH (2015) Polymyxin B resistance and biofilm formation in Vibrio cholerae are controlled by the response regulator CarR. Infect Immun 83: 1199–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina XR, Howard MF, Ante VM, and Bina JE (2016) Vibrio cholerae LeuO links the ToxR regulon to expression of lipid A remodeling genes. Infect Immun 84: 3161–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettcher KJ, and Ruby EG (1990) Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol 172:3701–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose JL, Kim U, Bartkowski W, Gunsalus RP, Overley AM, Lyell NL, et al. (2007) Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Mol Microbiol 65: 538–553 [DOI] [PubMed] [Google Scholar]
- Brunke S, and Hube B (2014) Adaptive prediction as a strategy in microbial infections. PLoS Pathog 10: e1004356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55: 611–622 [DOI] [PubMed] [Google Scholar]
- Cao M, and Goodrich-Blair H (2017) Ready or not: microbial adaptive responses in dynamic symbiosis environments. J Bacteriol 199: e00883–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Krasity BC, Peyer SM, Koehler S, Ruby EG, Zhang X, and McFall-Ngai MJ (2017) Bactericidal permeability-increasing proteins shape host-microbe interactions. MBio 8: e00040–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HD, and Groisman EA (2013) The biology of the PmrA/PmrB two-component system: the major regulator of lipopolysaccharide modifications. Annu Rev Microbiol 67: 83–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen TW, Schofield WB, Barry NA, Putnam EE, Rundell EA, Trent MS, et al. (2015) Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science 347: 170–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLoney CR, Bartley TM, and Visick KL (2002) Role for phosphoglucomutase in Vibrio fischeri-Euprymna scolopes symbiosis. J Bacteriol 184: 5121–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap PV (1985) Osmotic control of luminescence and growth in Photobacterium leiognathi from ponyfish light organs. Arch Microbiol 141: 44–50 [DOI] [PubMed] [Google Scholar]
- Dunn AK, Millikan DS, Adin DM, Bose JL, and Stabb EV (2006) New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl Environ Microbiol 72: 802–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraune S, and Bosch TCG (2007) Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc Natl Acad Sci 104: 13146–13151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontang EA, Fenical W, and Jensen PR (2007) Phylogenetic diversity of gram-positive bacteria cultured from marine sediments. Appl Environ Microbiol 73: 3272–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goo E, Majerczyk CD, An JH, Chandler JR, Seo Y-S, Ham H, et al. (2012) Bacterial quorum sensing, cooperativity, and anticipation of stationary-phase stress. Proc Natl Acad Sci 109: 19775–19780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166: 557–580 [DOI] [PubMed] [Google Scholar]
- Heath-Heckman EAC, Gillette AA, Augustin R, Gillette MX, Goldman WE, and McFall-Ngai MJ (2014) Shaping the microenvironment: evidence for the influence of a host galaxin on symbiont acquisition and maintenance in the squid-vibrio symbiosis. Environ Microbiol 16: 3669–3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera CM, Crofts AA, Henderson JC, Pingali SC, Davies BW, and Trent MS (2014) The Vibrio cholerae VprA-VprB two-component system controls virulence through endotoxin modification. MBio 5: e02283–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera CM, Henderson JC, Crofts AA, and Trent MS (2017) Novel coordination of lipopolysaccharide modifications in Vibrio cholerae promotes CAMP resistance. Mol Microbiol 106: 582–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussa EA, O’Shea TM, Darnell CL, Ruby EG, and Visick KL (2007) Two-component response regulators of Vibrio fischeri: Identification, mutagenesis, and characterization. J Bacteriol 189: 5825–5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasity BC, Troll JV, Weiss JP, and McFall-Ngai MJ (2011) LBP/BPI proteins and their relatives: conservation over evolution and roles in mutualism. Biochem Soc Trans: 1039–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer N, Philipp EER, Carpentier M-C, Brennan CA, Kraemer L, Altura MA, et al. (2013) Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. Cell Host Microbe 142: 183–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer N, Schwartzman J, Augustin R, Zhou L, Ruby EG, Hourdez S, and McFall-Ngai MJ (2014) The dual nature of haemocyanin in the establishment and persistence of the squid-vibrio symbiosis. Proc R Soc B Biol Sci 281: 20140504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Hsu FF, Turk J, and Groisman EA (2004) The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J Bacteriol 186: 4124–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, and Ruby EG (1994) Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl Environ Microbiol 60: 1565–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Rivera-Cancel G, Kinch LN, Salomon D, Tomchick DR, Grishin NV, and Orth K (2016) Bile salt receptor complex activates a pathogenic type III secretion system. eLife 5: e15718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chen H, Kenney JC, and Yan J (2010) A divalent switch drives H-NS/DNA-binding conformations between stiffening and bridging modes. Genes & Dev 24: 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyell NL, Dunn AK, Bose JL, and Stabb EV (2010) Bright mutants of Vibrio fischeri ES114 reveal conditions and regulators that control bioluminescence and expression of the lux operon. J Bacteriol 19219: 5103–5114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MJ, Stabb EV, and Ruby EG (2008) Comparative genomics-based investigation of resequencing targets in Vibrio fischeri: Focus on point miscalls and artefactual expansions. BMC Genomics 91:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Davis BM, and Waldor MK (2007) Antimicrobial peptides activate the Vibrio cholerae σE regulon through an OmpU-dependent signalling pathway. Mol Microbiol 63: 848–858. [DOI] [PubMed] [Google Scholar]
- Mathur J, and Waldor MK (2004) The Vibrio cholerae ToxR-regulated porin OmpU confers resistance to antimicrobial peptides. Infect Immun 72: 3577–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai MJ (2014) The importance of microbes in animal development: lessons from the squid-vibrio symbiosis. Annu Rev Microbiol 68: 177–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekalanos JJ, Swartz DJ, Pearson GDN, Harford N, Groyne F, and Wilde M. De (1983) Cholera toxin genes: nucleotide sequence, deletion analysis and vaccine development. Nature 306: 551. [DOI] [PubMed] [Google Scholar]
- Merrell DS, and Camilli A (2000) Regulation of Vibrio cholerae genes required for acid tolerance by a member of the “ToxR-like” family of transcriptional regulators. J Bacteriol 182: 5342–5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrell DS, and Camilli A (2002) Acid tolerance of gastrointestinal pathogens. Curr Opin Microbiol 5: 51–55 [DOI] [PubMed] [Google Scholar]
- Mitchell A, Romano GH, Groisman B, Yona A, Dekel E, Kupiec M, et al. (2009) Adaptive prediction of environmental changes by microorganisms. Nature 460: 220–224 [DOI] [PubMed] [Google Scholar]
- Miyashiro T, Wollenberg MS, Cao X, Oehlert D, and Ruby EG (2010) A single qrr gene is necessary and sufficient for LuxO-mediated regulation in Vibrio fischeri. Mol Microbiol 77: 1556–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham BD, and Trent MS (2013) Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat Rev Microbiol 11: 467–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, and Graf J (2012) Knowing your friends: invertebrate innate immunity fosters beneficial bacterial symbioses. Nat Rev Microbiol 10: 815–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, and McFall-Ngai M (2004) The winnowing: establishing the squid–vibrio symbiosis. Nat Rev Microbiol 2: 632–642 [DOI] [PubMed] [Google Scholar]
- Nyholm SV, Stabb EV, Ruby EG, and McFall-Ngai MJ (2002) Establishment of an animal-bacterial association: Recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci 97: 10231–10235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, Stewart JJ, Ruby EG, and McFall-Ngai MJ (2009) Recognition between symbiotic Vibrio fischeri and the haemocytes of Euprymna scolopes. Environ Microbiol 11: 483–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orwa JA, Govaerts C, Busson R, Roets E, Schepdael A. Van, and Hoogmartens J (2001) Isolation and structural characterization of polymyxin B components. J Chromatogr A 912: 369–373 [DOI] [PubMed] [Google Scholar]
- Palmer KL, Aye LM, and Whiteley M (2007) Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol 189: 8079–8087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips NJ, Adin DM, Stabb EV, McFall-Ngai MJ, Apicella MA, and Gibson BW (2011) The lipid A from Vibrio fischeri lipopolysaccharide. J Biol Chem 286: 21203–21219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post DMB, Yu L, Krasity BC, Choudhury B, Mandel MJ, Brennan CA, et al. (2012) O-antigen and core carbohydrate of Vibrio fischeri lipopolysaccharide: Composition and analysis of their role in Euprymna scolopes light organ colonization. J Biol Chem 287: 8515–8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey MM, and Whiteley M (2009) Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proc Natl Acad Sci 106: 1578–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelt JL, and Baumann P (1973) Taxonomy of the marine, luminous bacteria. Arch Mikrobiol 94: 283–330 [Google Scholar]
- Roux F. Le, Binesse J, Saulnier D, and Mazel D (2007) Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl Environ Microbiol 73: 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CA, Mandel MJ, Gyllborg MC, Thomasgard KA, and Ruby EG. (2013) Genetic determinants of swimming motility in the squid light-organ symbiont Vibrio fischeri Microbiologyopen 2: 576–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG, and Nealson KH (1976) Symbiotic association of Photobacterium fischeri with the marine luminous fish Monocentris japonica; a model of symbiosis based on bacterial studies. Biol Bull 151: 574–586 [DOI] [PubMed] [Google Scholar]
- Saltikov CW, and Newman DK (2003) Genetic identification of a respiratory arsenate reductase. Proc Natl Acad Sci 100: 10983–10988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, E.F., F., and Maniatis T (1989) Molecular Cloning: A Laboratory Manual, 2nd ed., Vols. 1, 2 and 3. Cold spring harbor laboratory press. [Google Scholar]
- Schwartzman JA, and Ruby EG (2016a) Stress as a normal cue in the symbiotic environment. Trends Microbiol 24: 414–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzman JA, and Ruby EG (2016b) A conserved chemical dialog of mutualism: Lessons from squid and vibrio. Microbes Infect 18: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw LN (2012) Stress response in microbiology Edited by Requena José M.. ChemBioChem 13: 2455–2456 [Google Scholar]
- Shibata S, Yip ES, Quirke KP, Ondrey JM, and Visick KL (2012) Roles of the structural symbiosis polysaccharide (syp) genes in host colonization, biofilm formation, and polysaccharide biosynthesis in Vibrio fischeri. J Bacteriol 19424: 6736–6747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabb EV, Butler MS, and Adin DM (2004) Correlation between osmolarity and luminescence of symbiotic Vibrio fischeri Strain ES114. J Bacteriol 186: 2906–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabb EV, and Ruby EG (2002) RP4-based plasmids for conjugation between Escherichia coli and members of the vibrionaceae Methods Enzymol Methods in enzymology. Vol. 358 Academic Press, 2002 413–426. [DOI] [PubMed] [Google Scholar]
- Tischler AH, Hodge‐Hanson KM, and Visick KL (2019) Vibrio fischeri –squid symbiosis. eLS. 1–9 [Google Scholar]
- Tognini P, Thaiss CA, Elinav E, and Sassone-Corsi P (2017) Circadian coordination of antimicrobial responses. Cell Host Microbe 22: 185–192 [DOI] [PubMed] [Google Scholar]
- Wang J, Vine CE, Balasiny BK, Rizk J, Bradley CL, Tinajero-Trejo M, et al. (2016) The roles of the hybrid cluster protein, Hcp and its reductase, Hcr, in high affinity nitric oxide reduction that protects anaerobic cultures of Escherichia coli against nitrosative stress. Mol Microbiol 100: 877–892 [DOI] [PubMed] [Google Scholar]
- Wang Y, Dunn AK, Wilneff J, McFall-Ngai MJ, Spiro S, and Ruby EG (2010) Vibrio fischeri flavohaemoglobin protects against nitric oxide during initiation of the squid-vibrio symbiosis. Mol Microbiol 78:903–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, and Ruby EG (2011) The roles of NO in microbial symbioses. Cell Microbiol 13: 518–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley A, Mantle M, Man D, Qureshi R, Forstner G, and Forstner J (1985) Neutral and acidic species of human intestinal mucin. Evidence for different core peptides. J Biol Chem 260: 7955–7959 [PubMed] [Google Scholar]
- Wollenberg MS, Preheim SP, Polz MF, and Ruby EG (2012) Polyphyly of non-bioluminescent Vibrio fischeri sharing a lux-locus deletion. Environ Microbiol 14: 655–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenberg MS, and Ruby EG (2012) Phylogeny and fitness of Vibrio fischeri from the light organs of Euprymna scolopes in two Oahu, Hawaii populations. ISME J 6: 352–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S2. Some outer membrane modifications alter pH-dependent PMB sensitivity in V. fischeri but, unlike in V. cholerae, deletion of OmpU does not. PMB MIC data for strains of V. fischeri with mutations in noted genes grown at the listed pH.
Figure S1. RT-qPCR verification of differential transcription in response to pH. Relative transcript levels of VF_1795 and eptA, which were identified as upregulated in low pH by transcriptomic analysis, were determined after growth at basic (pH 8.0) and acidic (pH 6.5) conditions. Data presented represent mean +/− SD of six biological replicates. Values were normalized to transcript levels of polA.