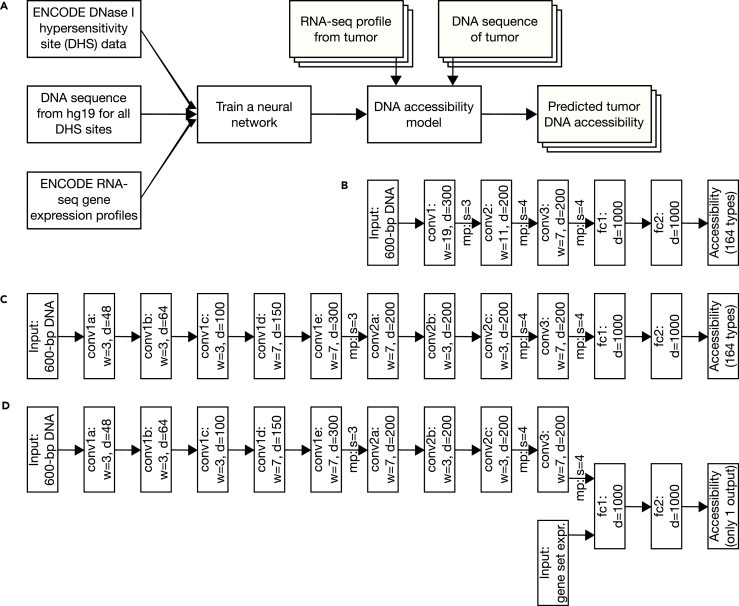
Figure 1.
Overview of Our Pipeline from Training to Application
(A) DHS, hg19 DNA, and RNA-seq information are all used to train the neural network. With tumor RNA-seq and DNA-seq data input the DNA accessibility model can then be used to predict chromatin state in tumors.
(B–D) (B) The neural network architectures for the tissue-specific baseline model, (C) the tissue-specific factorized convolutions model, and (D) the expression-informed model are shown. Depth (d) is provided for all fully connected (fc) layers. Convolution (conv) layers also list their width (w). Max pooling (mp) is indicated where present between layers and is always applied with equal size and stride (s).
See also Figures S1, S3, and S4.