Abstract
Metabolic profiling, metabonomics and metabolomics are terms coined in the late 1990s as they emerged as the newest ‘omics’ technology at the time. This line of research enquiry uses spectroscopic analytical platforms, which are mainly nuclear magnetic resonance spectroscopy and mass spectrometry (MS), to acquire a snapshot of metabolites, the end products of a complex biological system. Metabolic profiling enables the detection, quantification and characterisation of metabolites in biofluids, cells and tissues. The source of these compounds can be of endogenous, microbial or exogenous origin, such as dietary or xenobiotic. This results in generating extensive, multivariate spectroscopic data that require specific statistical manipulation, typically performed using chemometric and pattern recognition techniques to reduce its dimensions, facilitate its biological interpretation and allow sample classification and biomarker discovery. Consequently, it is possible to study the dynamic metabolic changes in response to disease, intervention or environmental conditions. In this review, we describe the fundamentals of MS so that clinicians can be literate in the field and are able to interrogate the right scientific questions.
Keywords: mass spectroscopy, metabolomics, metabolic profiling, mass-charge ratio, targeted profiling
Abbreviations: CID, collision-induced dissociation; DC, direct current; ESI, electrospray ionisation; FC, fold change; GC, gas chromatography; HILIC, hydrophilic interaction liquid chromatography; LC, liquid chromatography; MS, mass spectrometry; MWA, metabolome-wide association; NMR, nuclear magnetic resonance; OPLS-DA, orthogonal partial least squared-discriminant analysis; PC, principal component; PCA, principal components analysis; Q-TOF, quadrupole coupled with time-of-flight; RF, radio frequency; RP, reversed-phase; UPLC, ultra-performance liquid chromatography; VIP, variable importance of projection
Metabonomic studies are applied generally at an individual or an epidemiological level. Metabolome-wide association studies are based on large-scale metabolic profiling of specific populations and aim to discover novel markers associated with health-related conditions. Population-based studies have successfully demonstrated an association between candidate metabolites and risk of disease, such as cancer, diabetes and hypertension.1, 2 Metabonomics in medical science has diverse applications; it can be applied in vivo (using imaging or live cells) or in vitro (using extracts or biofluids), and the technology is capable of analysing a wide range of bioproducts, including solids (tissues), liquids (blood, urine, fecal water, bile and cell extracts) and gases (breath).3
Comprehensive metabolome coverage requires a broad array of instrumentation as each spectroscopic platform provides coverage of different classes of organic compounds. The most popular techniques in metabonomics are nuclear magnetic resonance spectroscopy and mass spectrometry (MS) (coupled with liquid chromatography [LC] or gas chromatography [GC]); each of these technologies has their own strengths and weaknesses. Numerous studies have shown the particular use of each technology, and various studies reported standardised protocols to ensure high reproducibility.4, 5
Global and targeted profiling
Metabolic profiling studies are generally divided into targeted and untargeted analytical designs. The global or untargeted approach attempts to detect and identify as many molecular compounds as possible and is primarily hypothesis generating. It aims to capture the overall molecular signal patterns and disrupted pathways.6 Targeted analysis focuses on known, small groups of metabolites of interest, such as lipids, carnitines and bile acids or a mixed panel of distinct compounds. The approach tends to be driven by a specific biochemical question or hypothesis that directs further perusal of a particular biological pathway. The method is highly robust and offers absolute quantification of molecules. However, the initial method development requires extensive work to cater to the specific requirements of the targeted compounds.6
Global and targeted metabonomics are complementary and can be used simultaneously to discover and validate disease biomarkers. Ideally, comprehensive profiling of specimens of interest is generated using both global and targeted approaches, as well as multiple spectroscopic platforms.5
Metabolic profiling in biomarker discovery
Interest in metabolites as biomarkers of disease progression has gained wide popularity in the last decade. The discovery of biomarker candidates for cancer screening and diagnosis in particular is no exception; a search in the PubMed database for articles including the keywords ‘metabolomics’, ‘cancer’, and ‘biomarker’ found 1 article in 2003, 58 articles in 2010, 154 articles in 2015 and a total of 787 articles at the time of writing. The basic concept of the approach is that cancer has several years of asymptomatic latency before it is clinically diagnosed. During this phase, there is deregulation of cellular energetics and a profound state of metabolic dysregulation in various pathways. The pathogenesis of cancer can lead to alterations in metabolite levels in the body, which in turn, influence the metabolite levels in biological fluids. Hence, metabolite levels can reflect the pathophysiological condition of the host.7
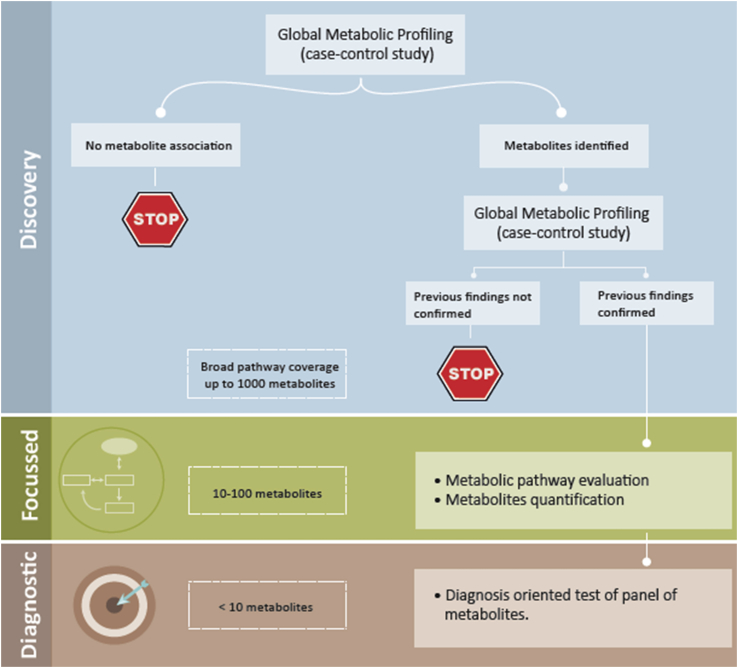
The pipeline of metabonomics in biomarker discovery is illustrated in Figure 1. Initially, the discovery phase aims to establish the disease metabolic signature and trace key metabolic pathways altered in response to the disease. This can be followed by an in-depth quantitative analysis on validated compounds, and ultimately, a targeted, diagnostic assay that is focused on a subset of biomarkers can be designed.
Figure 1.
Metabolic profiling pipeline.
Mass spectrometry
MS is a powerful analytical technique that is widely used in areas such as food safety, forensic sciences and environmental and biomedical science. Its origin dates back to the early 1900s, when it was mainly in the domain of physicists and physical chemists. The technology underwent progressive improvements and revolutionary changes that earned several Nobel Prizes.
Mass spectrometers can give qualitative information (structure) and quantitative information (mass-to-charge ratio, m/z, and relative abundance) on the elemental, isotopic and molecular composition of organic and inorganic samples.8 The principle is simple. The sample is first ionised, and magnetic and/or electric fields are used to separate ions by virtue of their different trajectories in vacuum. Unfortunately, the details are complicated. The sample may be in the solid, liquid or gaseous phase. Different ionisation methods are required for each phase; ions may be positively or negatively charged, and ionisation may occur at different pressures and may be continuous or pulsed. Fortunately, some simplification has arisen. In biomedical applications, where samples are predominantly liquids containing large molecules, continuous soft ionisation methods that avoid fragmentation such as electrospray ionisation (ESI) are preferred. However, these operate at atmospheric pressure, and a mechanism is then needed to transfer the ions into vacuum.
Analysis may be based on predominantly magnetic or electric fields or a combination of the two; the fields may be static or time varying, and operation may again be continuous or cyclic. The main variants of mass analysers (the magnetic sector, Fourier-transform ion cyclotron resonance, quadrupole, ion trap and time-of-flight mass spectrometer) and their broad characteristics are detailed in Table 1. Again, some simplification has occurred: electric fields are preferred because they avoid the need for a large, heavy magnet. Consequently, the quadrupole, ion trap and time-of-flight mass spectrometer are the most common. All offer high performance and advantages that mainly depend on the application. The most important performance parameters are sensitivity, mass resolution and mass range. The ion flux and space charge effects set the limit to sensitivity, at low and high fluxes, respectively. The thermal spread in ion velocity and the precision of the applied fields limit mass resolution, whereas the magnitude of the field limits mass range.
Table 1.
Comparison of Mass Spectrometer Types.
| Analysis method | Magnet | Operation | Resolution | Mass range |
|---|---|---|---|---|
| Magnetic sector | Y | Continuous | High | Medium |
| Fourier-transform ion cyclotron resonance | Y | Cyclic | High | Medium |
| Quadrupole mass spectrometer | N | Continuous | Low | Low |
| Ion trap | N | Cyclic | Medium | Medium |
| Time-of-flight | N | Cyclic | Medium | High |
Separation
Mass spectrometers are best suited for the identification of single molecules. However, biomedical samples are almost universally present as mixtures. The problem is addressed by first passing the sample through a stationary phase, using variations in mobility to separate the different components in time. In the late 1960s, the commercialisation of the interfacing of GC with MS in GC–MS enabled the separation of complex gas mixtures in a packed column so that individual molecules could be admitted sequentially into the mass spectrometer.8 GC–MS was followed in the 1980s by LC (LC–MS), allowing separation of liquid analytes before analysis, and increases in pressure led to a high- and ultrahigh-performance variants, high-performance liquid chromatography (HPLC)–MS and ultra-performance LC (UPLC)–MS, respectively. Other so-called ‘hyphenated systems’ including ion-mobility spectrometry–MS and capillary electrophoresis–MS combining different forms of separation with MS then followed; however, UPLC–MS is now the de facto standard for biomedical analysis.
Tandem MS
Even well-separated samples may still present identification problems because a variety of different large molecules may have a similar m/z ratio. The solution is provided by tandem MS (MS–MS or MS2), which was also invented in the late 1970s and provides greater certainty in the identification of heavy ions by analysing their fragmentation. Once again, a large number of different systems may perform a similar task, depending on the type of the analyser used to perform the initial ion selection, the methods of fragmentation and subsequent fragment analysis. The first article using MS in a metabonomic context was by Robinson and Pauling in 1971, entitled ‘Quantitative Analysis of Urine Vapour and Breath by Gas-Liquid Partition Chromatography’.9
Example system
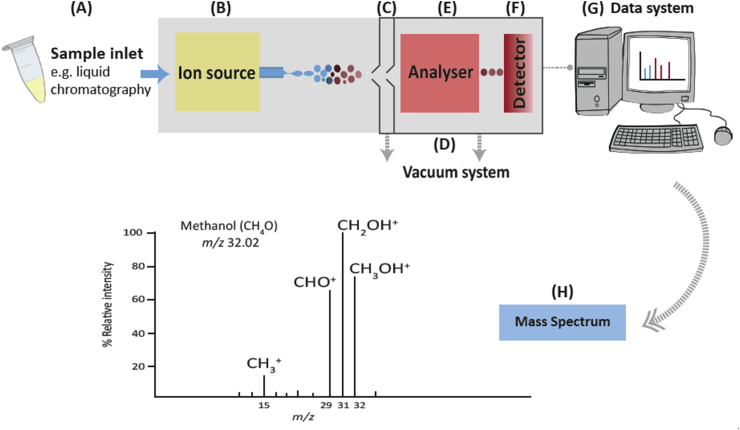
The most common approaches to biomedical analysis will be illustrated in terms of an example system, and detailed alternatives will be discussed later. A typical hyphenated mass spectrometer (Figure 2) consists of a number of major components: an initial separation system (such as LC or GC), ); an ion source; , a differentially pumped interface between atmospheric pressure and vacuum, a high vacuum chamber, one or more mass analysers; , a detector, and an instrument controller and data handling system, which allow the instrument to perform the following processes:
-
1.
Molecules are first introduced into the liquid handling system, and separated as appropriate (Figure 2A).
-
2.
Ions are created from the sample at atmospheric pressure in the ionisation source (Figure 2B).
-
3.
Ions pass through a specialised vacuum interface (Figure 2C) into the vacuum system (Figure 2D).
-
4.
Ions enter a first mass analyser (Figure 2E) where they are separated according to their mass-to-charge ratio.
-
5.
A collision cell fragments the selected ions, and the fragments are analysed in a second analyser (if present).
-
6.
Ions are detected as electrical signals in proportion to their abundance and charge as they emerge from the mass analyser (Figure 2F).
-
7.
The signals are passed to a data system (Figure 2G), and a mass spectrum of the molecule is produced (Figure 2H).
Figure 2.
Basic sequence of events in a mass spectrometer. m/z, mass-to-charge ratio.
Sample introduction/LC
LC is a popular MS separation technique used in metabolic profiling studies; biological samples are aqueous, composed of involatile, polar compounds and are often of high molecular mass, preventing their analysis by GC–MS. In addition, LC–MS requires a small sample volume and minimal sample preparation, and unlike GC–MS, it does not require sample derivatisation.10
The basic function of the sample introduction system is to inject a sample from the autosampler into a moving stream of the mobile phase (solvent mixture) and transfer it to the chromatographic column or the stationary phase. The separation is generally based on an analyte's relative affinity for the mobile phase versus the stationary phase. LC separation uses different types of chromatographic columns.
Normal-phase chromatography
In normal-phase columns, the mobile phase is a nonpolar solvent, and the stationary phase is polar. Molecules elute according to their polarity; polar analytes are retained by the stationary phase, whereas hydrophobic analytes move faster with the nonpolar mobile phase.
Reversed-phase chromatography
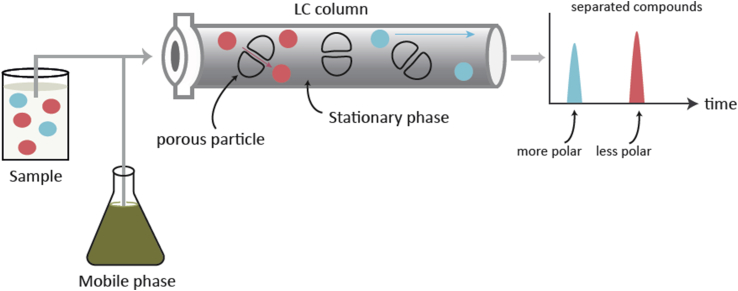
Reversed-phase (RP) chromatography is the opposite of normal-phase chromatography. A polar mobile phase and a nonpolar stationary phase are used (Figure 3). RP is more popular than normal-phase because it is efficient and stable and has retention power for polar analytes, which are typically present in biological fluids. A C18-bonded silica column is commonly used for low-molecular-weight analytes (<5 kDa molecules).
Figure 3.
Separation in reversed-phase liquid chromatography. LC, liquid chromatography.
Such a column is tightly packed with coating material. A high pressure pump (high or ultrahigh pressure) is used to sustain constant liquid flow at high pressure through the small porous particle bed coating the column, hence the name UPLC, which produces higher resolution chromatograms, compared with the traditional LC system (operating at a pressure of 15,000 psi vs. 5800 psi, respectively).11 The size of particles is another important factor that can maintain the column at high pressure.
Gradient elution is used to deliver solvents at varying proportions to elute molecules at different times. The eluotropic strength of the solvent is progressed linearly as the analysis progresses, starting at a high aqueous rate (99–100% water) and increasing gradually to high organic content, typically over 5–30 min. The separated mixture is then carried into the ionisation source.10
Hydrophilic interaction LC
Highly polar metabolites are poorly retained in RP chromatography; they elute very early with the polar solvent and are separated ineffectively from each other. In hydrophilic interaction LC (HILIC), similar to normal-phase chromatography, the stationary phase is polar. However, the mobile phase comprises a relatively hydrophilic solvent. Thus, it enhances the retention and separation of extremely polar analytes. Together, RP and HILIC provide complementary metabolite information and better metabolome coverage.
The ion source
Chromatographic separation ensures that a constant stream of molecules enters the ion source, rather than a large bundle at the same time. In the ion sources, the molecules are ionised before entering the mass spectrometer because they cannot be manipulated by electrical or magnetic fields in their neutral state. There are various forms of ionisation methods compatible with the LC–MS system. The selection is mainly based on the physicochemical characteristics of the analyte of interest. Some ionisation types are highly energetic and cause extensive fragmentation of the chemical bonds (hard ionisation). Other methods do not have strong means of separation; the energy used is only sufficient to produce adduct ions and not cause fragmentation.8 ESI is a soft ionisation technique. ESI transfers the ions gently from the solution (sample plus solvent mixture) to the gas phase via the following process: (i) a high voltage is applied between a capillary tip and a counter electrode, creating a high electric field, (ii) the sample is dispersed from a fine jet extending from the Taylor cone created by the field into an aerosol of highly charged droplets, (iii) a stream of drying gas evaporates the solvent from the charged droplets, (iv) the high density of the charged droplet and the repulsive forces between the ions cause the droplet to disintegrate into smaller droplets and (v) eventually, vapour phase analyte ions are released from the droplet and left free in an atmosphere of drying gas.8
The vacuum interface
Efficient transfer of ions created at atmospheric pressure into a high vacuum system for analysis is a difficult problem, which was solved in 1980 by John Fenn of Yale University, based on his knowledge of jet engines. Fenn was awarded the Nobel Prize in chemistry in 2002 for this work, which has transformed MS. The most obvious interface, an extremely small hole in the chamber wall, cannot be used because this leads to an effusive source that overwhelms the high vacuum pump while coupling poorly into the mass analyser. Instead, the ions and entraining gas are passed through a larger sampling orifice into an intermediate chamber through a free jet expansion, an isentropic process that trades thermal energy for kinetic energy, simultaneously cooling and collimating the ions. The collimated beam is then passed through a further orifice—a small hole at the tip of a conical ‘skimmer’—into the high vacuum system. With careful design, the loads on the intermediate and high vacuum pumps can be balanced, and shocks can be avoided. Additional components such as RF ion guides can be used to increase the axial ion flux density, the key to high sensitivity.
Mass analyser
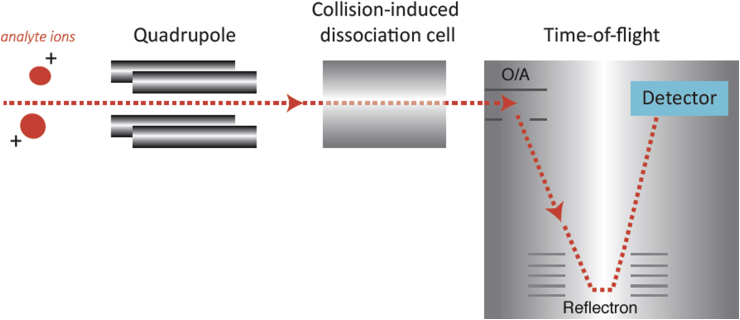
The basic function of the mass analyser is to separate the ions according to their mass-to-charge ratio (m/z) and transfer these ions into a detector. Different mass analysers separate the ions by a variety of methodologies, the choice depending on the type of data required (structural or quantitative), resolution and mass accuracy. A quadrupole coupled with a time-of-flight (Q-TOF) mass analyser (Figure 4) is regarded commercially as the most successful of hybrid analysers.12 A hybrid instrument combines different types of mass analysers in a single instrument.
Figure 4.
Principle of the quadrupole time-of-flight instrument. O/A, orthogonal extraction.
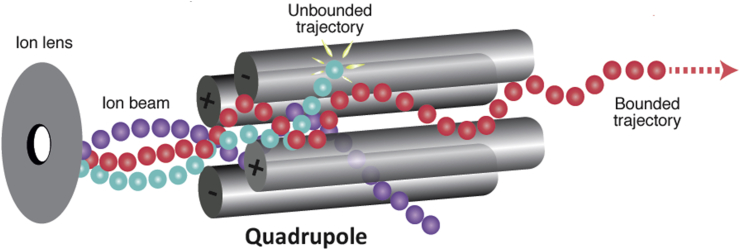
The quadrupole (which was invented by Wolfgang Pauli, who won the Nobel Prize in physics in 1989) consists of four cylindrical-shaped metal rods set parallel to each other at a well-defined spacing (Figure 5). It separates ions based on the stability of their trajectories when a combination of direct current (DC) and radio frequency (RF) voltages is applied to the rods, creating a time-varying quadrupolar field. With a particular ratio of DC to RF voltage, ions near only one given value of m/z will have bounded trajectories and hence pass through the length of the quadrupole without discharging on the rods.
Figure 5.
Quadrupole mass analyser.
The quadrupole is a workhorse of mass analysis, but its mass selectivity is limited by its length and constructional precision and by the frequency of the RF voltage, and its mass range is limited by the amplitude of the RF voltage. Consequently, its main function is often to select ions for a more accurate analyser. In the RF-only mode, when filled with an inert gas at moderate pressure, a quadrupole may also act as a so-called ‘collision cell’ in which selected analyte ions are fragmented.
Improved mass accuracy and range are provided by the TOF analyser, which can in principle identify a m/z value simply based on the time taken by a pulse of ions to traverse a known distance before striking a detector. If the pulse is short, the axial velocity is low, and the distance is large; the mass range can in principle be very high. In practice, however, uncertainty in the initial position and velocity of the ions caused by thermal energy requires complicated additional correction. The most common techniques are orthogonal extraction (O/A), used to shorten a pulse of ions selected from a quadrupole, and reflection by a stacked electrode system known as a ‘reflectron’, used to correct for thermal spread.
The quadrupole can be used in narrow or wide band pass mode to determine which ions are passed into the collision region and then into the TOF analyser. In full-scan mode, the quadrupole is set in wide band pass mode so that all ions are transferred into the TOF analyser, and there is no gas in the collision cell. The narrow pass mode is operated when fragmentation of a selected ion with a known m/z value is required. The quadrupole will act as a filter, allowing the passage of ions with a particular m/z value.
The fragmentation of an ion of a known m/z value in tandem MS allows ions of the same mass to be differentiated on the basis of their fragmentation pattern. A collision-induced dissociation cell, usually placed after the quadrupole, is filled with an inert gas such as argon. Ions are selectively passed into the cell, whereupon a voltage is applied, which results in fragmenting the ion into its constituents. Most ions will produce a signature fragmentation pattern, which may be identifiable using databases or chemical standards. The Q-TOF system features high mass accuracy together with the possibility of tandem MS, which is ideal for nontargeted profiling applications.
Ion detector and vacuum system
After emerging from the mass filter, ions are converted into electrons and thence into a usable electrical signal in an ion detector. Detection and amplification are generally carried out in a single component, known as a ‘channeltron’ detector and based on a tube that has been internally coated with a layer of high secondary electron emissivity. The detector is heavily biased to attract ions, which strike with high velocity. The kinetic energy thus liberated creates a shower of secondary electrons, which emit further electrons in a cascade along the tube. The tube is normally curved to prevent a direct line of sight path between the entrance and final collection electrode.
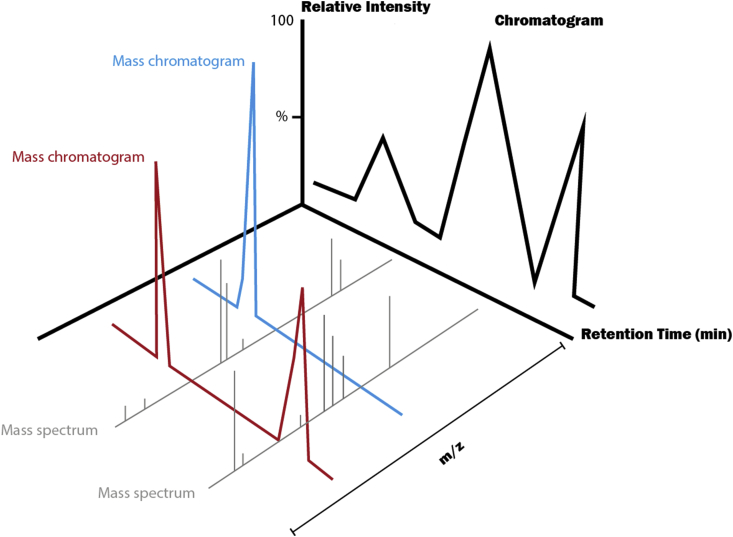
The chamber housing the aforementioned components is normally maintained at high vacuum, using a low capacity, high vacuum pump such as a turbopump backed by a high capacity, low vacuum pump such as a rotary pump. The low pressure is required to maintain a sufficiently large mean free path, allowing ions to pass through analyser components without collision. Because ultimate sensitivity is determined by ion flux and hence by gas flow into the system, pumps are typically large and floor mounted. However, recent developments have seen the emergence of ‘desktop’ ESI–MS systems, based on miniaturised components fabricated by machining of silicon and with the entire pump set contained in an instrument of the size of a personal computer (Figure 6). Performance is moderate but steadily improving.
Figure 6.
Three-dimensional visualisation of the electrospray ionisation chromatogram, mass chromatogram and mass spectrum. m/z, mass-to-charge ratio.
Preprocessing of raw spectral data
Preprocessing of spectral data produced by the LC–MS system is a necessary step before any statistical analysis. The process aims to transform the raw data in the analogue format to a tabular matrix, to correct for instrumental variation, to filter the data set (reduce noise) and to extract the molecular features from each sample. XCMS software (XCMS Online, La Jolla, California, USA) provides a relatively automated processing pipeline tailored for untargeted LC–MS metabolic profiling data. The software is a robust and a very popular platform (+1500 citations in the literature).
The outcome of the MS analysis is the generation of a spectrum for each sample. The software initially identifies peaks detected in each sample and then extracts and reports their m/z values. Peak integrals are then created by grouping and binning the peaks. The corresponding peaks from run to run are then matched and grouped. The drift in analyte retention time from run to run is calculated and corrected by ‘peak alignment’. The peak alignment algorithm corrects for retention time drifts by calculating a median retention time from ‘well-behaved’ peaks. Some groups may be missing peaks from some of the samples because an analyte is not present or was not detected in the previous peak detection step (because of its low concentration, the bad peak shape or a mistake in the identification algorithm). This is corrected for by recovering the missing signals from raw data via a ‘peak filling’ method.
Finally, the data are normalised to refine confounding variation arising from experimental sources, while preserving relevant biological variation. The normalisation process scales the data so that different samples in a study can be compared with each other. This is a key step for the urinary matrix; urine volume changes result in significant variation in the dilution of analyte concentrations across samples.13 After preprocessing the spectral data, XCMS generates a peak intensity table, which is then imported to statistical analysis software.
Multivariate analysis
Similar to the other ‘omics’ disciplines, metabolic profiling generates large and complex data sets that require sophisticated statistical approaches to reduce its dimension and account for intrasample and intersample variance. Typically, hundreds or even thousands of multicollinear metabolic features are produced by LC–MS for each sample. Each metabolite detected can generate several isotopes, adducts and fragments. Moreover, multiple correlations are present among the metabolites that are biochemically interrelated.14
The data set requires reduction of variables, with a selection and visualisation statistical approach. The statistical method used needs to account for correlation as the data are collinear. Dimension reduction methods summarise and transform thousands of features into a few key components that capture the variance or discriminatory covariance in the data. Usually, the data are initially explored with principal components analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA) for feature selection.
PCA is a statistical technique for finding patterns in high-dimensional data. The method is unsupervised; it does not require any input related to class information and is used to project the data to highlight the maximum sources of variance using a minimal number of principal components (PCs). This approach has the advantage of reducing the number of dimensions, without much loss of the biological information by removing the noise and redundancy in the data. The new PCs correspond to an uncorrelated, linear combination of the original variables. The model uses the variability and correlation of each variable (metabolites) to create a simplified set of new variables (PCs). PCA reduces the number of variables in a data set. However, it still accounts for as much of the total variation in the original data as possible.15
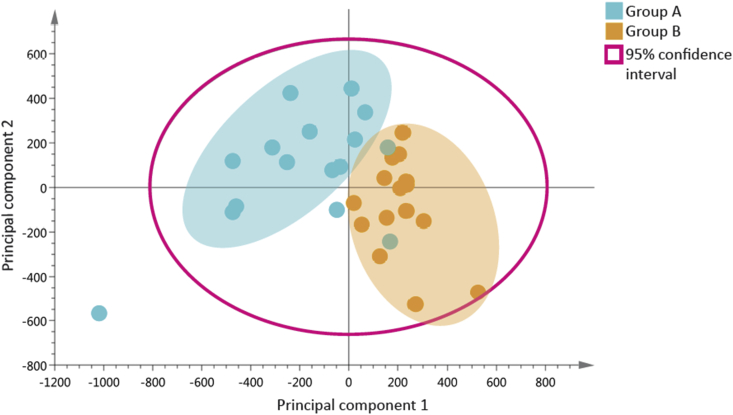
PC analysis
The data are visualised as score and loading plots. In the score plot (Figure 7), which maps the distribution of samples and allows inherent similarities between samples to be detected, the horizontal axis represents the first PC, which explains the largest variation. The vertical axis represents the second-most variance or PC2, and it is orthogonal to the PC1 axis. If there are x variables in a data set, it is possible to add up to x PCs. Usually, the most interesting observation can be seen in the first few components; the remaining PCs are either uninteresting or noise.15
Figure 7.
Principal components analysis score plot.
The PCA is plotted with colour codes assigned to each group to elucidate relationship between observations. Close clustering trends between observations within the same group indicate similar underlying metabotypes. Observations outside the 95% confidence interval (the magenta circle in Figure 7) are identified as potential outliers and require further examination of their original spectra.
Orthogonal projections to latent structures discriminant analysis
Supervised modelling using OPLS-DA is mainly used to maximise separation between predefined sample classes to view discriminatory metabolites. The inbuilt orthogonal signal correction filter removes confounding variations not related to class separation (within group variations, age, gender, diet and so on), which may be observed in PCA models.
Variation that is unrelated to the class response is described in the orthogonal component(s).16 Similar to PCA, OPLS-DA models take into account the joint effect of all metabolites and use their correlation structure with the additional aim of maximising the variance related to the biological class (or y variable). A corresponding ‘loading plot’ can explain which variables (or loadings) are different between samples, thus causing separation between groups (i.e., cancer vs. control).
Model evaluation and cross-validation is critical to avoid ‘overfitting’ the OPLS-DA model and prevent false positive results. Overfitting the model can show excellent results, but the outcome might not be reproducible. Internal and external validation is required to overcome this issue. The goodness of fit and prediction can both be evaluated by assessing R2 and Q2 model statistics, respectively. In addition, a permutation test, which is a leave-one-out cross validation procedure can assess the model validity. The permutation test compares the goodness of fit of the original model with the goodness of fit of several randomly permuted models (resampling Y observations, while the X matrix is intact). Analysis of variance testing of cross-validated predictive residuals (CV-ANOVA; P-value<0.05) can also be performed to determine significant differences between groups in the OPLS-DA models.17 Feature selection only proceeds if the models are robust to identify relevant and putative biomarkers. An OPLS-DA with Q2Y less than 20% and CV-ANOVA greater than 0.05 is considered a nondiscriminatory model.
Variable importance of projection (VIP) scores are used to select the most differential metabolites from the supervised OPLS-DA model. VIP scores are used to estimate the importance of each variable in the projection used in the OPLS model, which allow the X variables to be classified according to their explanatory power of Y (class information). A variable with a VIP score close to or greater than 1 can be considered important in a given model. Univariate significance tests and fold change are then performed on the selected features to estimate the magnitude of change between the groups.
Metabolite annotations
The data obtained by LC–MS are merely a series of peaks without any chemical identity. Therefore, following the selection of important, discriminant features, their interpretation in a biological context is limited only by the extent to which their precise chemical identity is known. A single metabolite can produce multiple features; its identification is not always straightforward because the feature is not always observed as a protonated or deprotonated ion, but rather as adducts or fragments. Initial examination of the metabolite chromatographic data is key to determining the true monoisotopic parent in a cluster of masses detected at the same retention time.
The identification of metabolites is labour-intensive, time-consuming and a major bottleneck in the interpretation of MS results. The gold standard for assignment of metabolites is to use internal isotope-labelled standards, or ‘spiked-in’ nonlabelled authentic standards. However, for untargeted metabolic profiling, the comparison of each detected metabolite with the compound standard is not feasible because of the cost and commercial availability of standards.
Currently, the best approach for metabolite annotations is to use the molecular feature information (m/z, retention time and MS/MS) to determine its chemical identity against a number of freely available online databases, such as the human metabolome database (HMDB),18 METLIN,19 Lipid Maps20 and MS data available from earlier publications.
The potential assignments returned from the database search of a mass are then carefully examined for molecular formula and structure match, as well as the biological relevance of the molecule to the human metabolome. Nevertheless, many of the peaks cannot be assigned to a metabolite. They are labelled as ‘unknown’ and are not characterised further.
Conclusions
Although seemingly complex, knowledge of the principles of mass spectroscopy allows the right research questions to be asked for translational research and the correct methodologies to be selected for maximum discrimination of the desired metabolites under scrutiny. Such an approach allows the clinician to be mindful of what is possible and what is not and to tailor investigation appropriately.
Conflicts of interest
The authors have none to declare.
Acknowledgements
The authors have been funded by grants from the Wellcome Trust ISSF Fund at Imperial College London and AMMF—the Cholangiocarcinoma Charity (Stansted, Essex, UK). M.A. was funded by the StratiGrad PhD programme at Imperial College London. All authors acknowledge the support of the United Kingdom National Institute for Health Research Biomedical Research Centre at Imperial College London for infrastructure support.
References
- 1.Bictash M., Ebbels T., Chan Q. ‘Opening up the” black box”: metabolic phenotyping and metabolome-wide association studies in epidemiology.’. J Clin Epidemiol. 2010;63(9):970–979. doi: 10.1016/j.jclinepi.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holmes E., Loo R.L., Stamler J. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453(7193):396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wishart D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat Rev Drug Discov. 2016;15(7):473–484. doi: 10.1038/nrd.2016.32. [DOI] [PubMed] [Google Scholar]
- 4.Beckonert O., Keun H.C., Ebbels T.M. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc. 2007;2(11):2692–2703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- 5.Lenz E.M., Wilson I.D. Analytical strategies in metabonomics. J Proteome Res. 2007;6(2):443–458. doi: 10.1021/pr0605217. [DOI] [PubMed] [Google Scholar]
- 6.Patti G.J., Yanes O., Siuzdak G. Innovation: metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13(4):263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vermeersch K., Styczynski M. Applications of metabolomics in cancer research. J Carcinog. 2013;12(1):9. doi: 10.4103/1477-3163.113622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Hoffmann E., Stroobant V. John Wiley and Sons Ltd; Chichester, West Sussex, England: 2007. Mass Spectrometry Principles and Applications. [Google Scholar]
- 9.Pauling L., Robinson A.B., Teranishi R., Cary P. Quantitative analysis of urine vapor and breath by gas-liquid partition chromatography. Proc Natl Acad Sci Unit States Am. 1971;68(10):2374–2376. doi: 10.1073/pnas.68.10.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Want E.J., Wilson I.D., Gika H. Global metabolic profiling procedures for urine using UPLC-MS. Nat Protoc. 2010;5(6):1005–1018. doi: 10.1038/nprot.2010.50. [DOI] [PubMed] [Google Scholar]
- 11.Fountain K.J. UPLC versus UHPLC: comparison of loading and peak capacity for small molecule drugs. Waters Appl Notes. 2011 720003869EN IH-PDF. [Google Scholar]
- 12.Gross J. Springer; Heidelberg, Germany: 2004. Mass Spectrometry, A Textbook. [Google Scholar]
- 13.Veselkov K.A., Vingara L.K., Masson P. Optimized preprocessing of ultra-performance liquid chromatography/mass spectrometry urinary metabolic profiles for improved information recovery. Anal Chem. 2011;83(15):5864–5872. doi: 10.1021/ac201065j. [DOI] [PubMed] [Google Scholar]
- 14.Xi B., Gu H., Baniasadi H., Raftery D. Springer New York; New York, NY: 2014. Statistical Analysis and Modeling of Mass Spectrometry-Based Metabolomics Data; pp. 333–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartel J., Krumsiek J., Theis F.J. Statistical methods for the analysis of high-throughput metabolomics data. Comput Struct Biotechnol J. 2013;4(5):1–9. doi: 10.5936/csbj.201301009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trygg J., Holmes E., Lundstedt T. Chemometrics in metabonomics. J Proteome Res. 2007;6(2):469–479. doi: 10.1021/pr060594q. [DOI] [PubMed] [Google Scholar]
- 17.Eriksson L., Trygg J., Wold S. Cv-ANOVA for significance testing of PLS and OPLSR models. J Chemom. 2008;22(11–12):594–600. [Google Scholar]
- 18.Wishart D.S., Jewison T., Guo A.C. Nucleic Acids Research; 2012. ‘HMDB 3.0—the Human Metabolome Database in 2013’; p. gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith C.A., Want E.J., Qin C. METLIN: a metabolite mass spectral database. Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 20.Sud M., Fahy E., Cotter D. LMSD: lipid maps structure database. Nucleic Acids Res. 2007;35:D527–D532. doi: 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]