Abstract
Background
20(S)-Protopanaxadiol (PPD), the aglycone part of 20(S)-protopanaxadiol ginsenosides, possesses antidepressant activity among many other pharmacological activities. It is currently undergoing clinical trial in China as an antidepressant.
Methods
In this study, an ultra-performance liquid chromatography coupled with triple quadrupole time-of-flight mass tandem mass spectrometry method was established to identify the metabolites of PPD in human plasma and urine following oral administration in phase IIa clinical trial.
Results
A total of 40 metabolites in human plasma and urine were identified using this method. Four metabolites identified were isolated from rat feces, and two of them were analyzed by NMR to elucidate the exact structures. The structures of isolated compounds were confirmed as (20S,24S)-epoxydammarane-12,23,25-triol-3-one and (20S,24S)-epoxydammarane-3,12,23,25-tetrol. Both compounds were found as metabolites in human for the first time. Upon comparing our findings with the findings of the in vitro study of PPD metabolism in human liver microsomes and human hepatocytes, metabolites with m/z 475.3783 and phase II metabolites were not found in our study whereas metabolites with m/z 505.3530, 523.3641, and 525.3788 were exclusively detected in our experiments.
Conclusion
The metabolites identified using ultra-performance liquid chromatography coupled with triple quadrupole time-of-flight mass spectrometry in our study were mostly hydroxylated metabolites. This indicated that PPD was metabolized in human body mainly through phase I hepatic metabolism. The main metabolites are in 20,24-oxide form with multiple hydroxylation sites. Finally, the metabolic pathways of PPD in vivo (human) were proposed based on structural analysis.
Keywords: Human plasma, Human urine, Metabolite, 20(S)-Protopanaxadiol, Ultra-performance liquid chromatography coupled with triple quadrupole time-of-flight
1. Introduction
Ginseng is one of the most frequently used herbs in Traditional Chinese Medicine. It has been widely used in China for thousands of years, both as medicine and as an ingredient in several kinds of functional food. Besides, it is also one of the most common ingredients in various categories of natural products worldwide. Ginsenosides are a major group of pharmacologically active components in ginseng [1]. Ginsenosides can be subdivided into four classes based on the aglycone part: 20(S)-protopanaxadiol, 20(S)-protopanaxatriol, oleanic acid, and ocotillol ginsenosides.
20(S)-protopanoxadiol ginsenosides, such as ginsenoside Rb1, Rb2, Rb3, Rc, Rd, Rg3, and Rh2, contain 20(S)-protopanaxadiol (PPD) as the aglycone part. This class of ginsenosides can be deglycosylated by gastric acid and intestinal bacteria in vivo, yielding the active metabolite, PPD. Because the PPD has no glucosidic bond, it cannot be hydrolyzed in stomach acidic condition, leading to absorption in original chemical form after oral administration. PPD has a wide range of pharmacologic activities, including antiestrogenic, antiinflammatory, antitumor, vasodilating, cardioprotecting, and antidepressive effects [2], [3], [4]. In a previous study, PPD was found to significantly increase the levels of norepinephrine and 5-hydroxytryptamine in the brain of rats with symptoms of depression [3]. Recently, PPD capsule formulation, Yuxintine, was approved by the Chinese State Food and Drug Administration to undergo clinical trials as an antidepressant in China (Clinical trial license number: 2009L01193).
Metabolite identification is an important part of drug discovery and development. Understanding the metabolism of the drug candidate can help to optimize its pharmacokinetics and to avoid toxicity [5]. Furthermore, the discovery of active metabolites may lead to a new or improved drug candidate. Therefore, it is necessary to determine the metabolic profile of PPD in human before marketing it as a new drug. To date, the information about the metabolism of PPD is based on studies in rats and in vitro [human liver microsomes (HLMs) and human hepatocytes]. The study of PPD metabolism in rats indicated that PPD underwent monooxygenation, deoxygenation, dehydrogenation, cysteine conjugation, and glucuronidation [6]. Small amount of PPD was found in rat plasma, whereas monooxygenated and deoxygenated derivatives represented major circulating metabolites [7]. However, because of the species differences, the metabolic profile of most drugs in humans and rodent animals are not comparable.
In vitro study of PPD metabolism in HLMs and human hepatocytes by Li et al has served as a guide throughout our human in vivo study of PPD metabolism [8]. The in vitro study suggested that PPD was mainly metabolized by oxidation of the 24,25-double bond to yield 24,25-epoxides, followed by hydrolysis and rearrangement to form the corresponding 24,25-vicinal diol derivatives and the 20,24-oxide form. Our study found that phase I hepatic metabolism was the main metabolic process of PPD in human and the metabolites detected in human plasma and urine were very similar to those detected in vitro with a few exceptions. This finding showed that even though the study in HLMs and human hepatocytes could largely reflect the metabolic pathways of PPD in human, it could not demonstrate the continuous metabolic process of PPD in human body. Our research is the first human in vivo study of PPD metabolism.
Our previous studies found that human plasma concentration of PPD after oral administration was extremely low (6.25 mg/d, 0.37 ± 0.91 ng/mL; 100 mg/d, 4.56 ± 3.74 ng/mL) because of the drug's low bioavailability. However, in consideration of clinical efficacy and safety, the oral dosage could not be increased. Therefore, the method to detect and identify the metabolites of PPD was required to have very high sensitivity. An ultra-performance liquid chromatography coupled with triple quadrupole time-of-flight mass tandem mass spectrometry (UPLC-Q-TOF-MS/MS) system with high speed and high detection sensitivity has exhibited an excellent performance for metabolite detection [9]. Full-scan MS spectra and product ion spectral data sets for each compound provided by UPLC-Q-TOF-MS/MS system enabled the collection of MS spectra for the identification of both target and nontarget metabolites [10]. Q-TOF-MS/MS not only provided accurate masses of ions but also gave valuable structural information from the MS/MS spectra. By analyzing the MS/MS data described before, the MS/MS fragmentation behaviors of the metabolites of PPD were demonstrated, allowing the determination of the basic structure of unknown metabolites [11]. Moreover, MetabolitePolet™ software from AB Sciex Company provided multiple data processing technologies including mass defect filtering, extracted ion chromatogram (XIC), product ion filtering, and neutral loss filtering which were successfully applied in our study for the tentative identification of PPD metabolites in human [12], [13]. Although UPLC-Q-TOF-MS/MS analysis allowed rapid identification and partial structural characterization of the metabolites, the elucidation of metabolite structures still depended on the collision-induced dissociation of the protonated molecular ions; therefore, the structures could only be elucidated tentatively. To confirm the elucidated structures of the metabolites, 500 MHz NMR with CryoProbes from Bruker Company was used to provide more detailed structural information that helped verify the structures.
In the present study, a total of 40 metabolites of PPD in human plasma and urine were identified based on the accurate mass measurement and the fragmentation patterns. Four metabolites identified in humans were extracted and isolated from rat feces, and two of them were structurally confirmed with 500 MHz CryoProbes NMR. The metabolic pathways of PPD in human and the fragmentation patterns of PPD metabolites were also proposed.
2. Materials and methods
2.1. Chemicals
PPD was kindly provided by the Shanghai Innovative Research Center of Traditional Chinese Medicine (SIRC-TCM, Shanghai, China). HPLC-grade methanol, HPLC-grade acetonitrile, and formic acid used for UPLC analysis were purchased from Merck Company (Darmstadt, Germany). Deionized water was freshly processed through a Milli-Q water purification system (Millipore, USA). Ethyl acetate, petroleum ether, dichloromethane, and diethyl ether used for extraction and column chromatography were of analytical grade (Sinopharm Chemical Reagent Co.,Ltd, Shanghai, China). Silica gel (200–300 mesh; Sinopharm Chemical Reagent Co.,Ltd, Shanghai, China) was used for column chromatography, and precoated silica gel GF254 plates (Qingdao Merine Chemical Plant, Qingdao, China) were used for thin-layer chromatography.
2.2. Clinical study design and sample collection
This phase IIa clinical trial was conducted by SIRC-TCM (Shanghai, China) following the good clinical practice guidelines, the guiding principles of the Declaration of Helsinki. The protocol was approved by local ethics committees. One hundred twenty patients diagnosed with depression by Diagnostic and Statistical Manual of Mental Disorders-IV-TR diagnostic criteria were randomly recruited from six psychiatric units in China. Each patient or the nearest relative of the patient signed an informed consent. Eligible subjects were randomly assigned to five dosage groups of PPD (6.25, 12.5 25, 50, and 100 mg) or a placebo group at 0 week. After 6 weeks of treatment, the plasma and urine samples were obtained from each patient. The plasma and urine samples from patients in 100 mg group were analyzed by the established method to identify metabolites of PPD.
2.3. Sample preparation
The plasma samples were prepared as follows: plasma samples were thawed at room temperature before analysis. Four hundred microliters of acetonitrile was added to 200 μL of plasma sample and vortex-mixed for 3 minutes allowing protein precipitation. Then, the samples were centrifuged at 10,000 g for 10 minutes. After that, the supernatant was transferred to another tube and dried at 30°C under a gentle stream of nitrogen. The dried residue was reconstituted with 50 μL of methanol followed by vortex-mixing and centrifugation at 10,000 g for 5 minutes and filtered through a 0.22 μm membrane. An aliquot of 5 μL of supernatant was used for UPLC-Q-TOF-MS/MS analysis.
For the preparation of urine, the samples were mixed with methanol and centrifuged at 10,000 g for 15 minutes before filtering through a 0.22 μm membrane for further UPLC-Q-TOF-MS/MS analysis.
2.4. UPLC-Q-TOF-MS/MS condition
Metabolites of PPD in biological samples were identified by UPLC-Q-TOF-MS/MS analysis which was conducted on Agilent 1290 UPLC system coupled to AB Triple-TOF 5600 + system (a hybrid quadrupole time-of-flight tandem mass spectrometer equipped with Turbo V sources and Turbo ion spray interface).
Chromatographic analysis was performed with an Eclipse plus C18 reversed phase LC column (2.1 × 50 mm i.d.; 1.8 μm, Agilent, USA) at a column temperature of 30°C. The mobile phase consisted of 0.5% formic acid aqueous solution (v/v, solvent A) and acetonitrile (solvent B) with a flow rate of 0.3 mL/min. The gradient elution program was optimized for improved separation. The conditions were as follows: 0–1.0 min (5–5% B), 1.0–1.5 min (5–25% B), 1.5–6.0 min (25–75% B), 6.0–12.0 min (75–100% B), 12.0–13.0 min (100-100% B), and 13.0–15.0 min (100-5% B). The sample injection volume was 5 μL.
Mass spectrometer was equipped with an electrospray ion source operating in positive ion mode. The conditions of the MS/MS detector were as follows: source temperature, 121°C; ion spray voltage, 5.5 kV; declustering potential, 80 V; and collision energy, 10 eV. Nitrogen, as the nebulizer gas (Gas 1), the heater gas (Gas 2), and the curtain gas were set at 50, 50, and 30 psi, respectively. The mass range of m/z 100 to 1000 amu with 0.25 sec accumulation time was set in positive full-scan mode. Following the information dependant acquistion (IDA) criteria, six most intense fragment ions ranged from 50 to 1000 amu of each analyte that exceeded 50 cps counts were selected for further production scan with a 0.1 sec accumulation time. The raw data were acquired and processed using the Analyst® TF 1.6 software (AB Sciex).
2.5. Isolation of the metabolites
Ten rats were raised in the metabolic cage and were given 50 mg/kg of PPD every day by gavage administration. Rat feces collected every day for 40 days (2.0 kg) were smashed and then extracted with six liters of methanol at room temperature for 2 days. Then, the solvent was evaporated. The residue was dissolved in ethyl acetate, and liquid–liquid extraction was performed with water. The ethyl acetate–soluble portion (31.0 g) was subjected to column chromatography on silica gel using 0% to 100% ethyl acetate in petroleum ether for gradient elution, producing 12 fractions (fraction 1-12). The metabolites in each extract fraction were preliminarily identified using mass spectrometer. The extract fractions with high concentration of the target metabolites were concentrated by vacuum-rotary evaporation. Fraction 8 (830 mg) was proved to contain four valuable metabolites using UPLC-Q-TOF-MS/MS and was chromatographed on preparative LC-MS with methanol–water gradient elution. With MS-guiding, metabolites that had protonated molecular weight of 475.3783 (1, 1.3 mg), 477.3938 (2, 1.6 mg), 491.3731 (3, 1.7 mg), and 493.3887(4, 3.9 mg) were finally obtained.
2.6. Data analysis
Postacquisition analyses were performed using the MasterView® program (v1.1, AB Sciex), which compared the retention time and mass spectra of a series of XICs of experimental samples with those of control samples to identify potential metabolites. The parameters of confidence settings were set as follows: mass error < 8.0 ppm, retention time error < 5.2%, and signal-to-noise ratio > 10. The collision-induced dissociation (CID) was performed on metabolites to obtain fragmentation patterns. The XIC peak intensity threshold was set at 1500 cps with minimum MS and MS/MS peak intensities of 400 and 100 cps, respectively. The structures of potential PPD metabolites were determined based on mass shift, fragmentation patterns, and retention time. The proposed chemical structure of each metabolite was investigated using assign function of MetabolitePilot™ software where a score for evaluation of the matching correctness was given. The results with score greater than 250 were considered as a credible metabolite structure matching results. The peaks in MS/MS spectra were automatically marked by MetabolitePilot software to match the assigned metabolite structure.
2.7. NMR spectroscopy
NMR spectra including 1H, 13C, heteronuclear singular quantum correlation, heteronuclear multiple-bond correlation (HMBC), 1H-1H correlation spectroscopy (COSY), and nuclear overhauser enhancement spectroscopy (NOESY) were measured using a Bruker AVANCE III spectrometer (500 MHz proton frequency) at 301.8 K equipped with a 5 mm 13C Bruker CPDCH. 1H and 13C shifts were referenced internally to the solvent signals at 2.5 ppm and 5.4 ppm, respectively. Each metabolite sample was dissolved in deuterium chloroform (CDCl3) before being transferred into a 1.7 mm NMR tube.
3. Results and discussion
PPD capsule is currently undergoing phase IIb clinical trial as an antidepressant. As a result, the information about its metabolic pathways in human becomes very crucial. Here, 40 metabolites of PPD in human were identified using UPLC-Q-TOF, and the structures of two isolated metabolites were confirmed by NMR analysis for the first time. Finally, the metabolic pathways in human were proposed.
3.1. Structural characterization of PPD and tentative identification of its metabolites
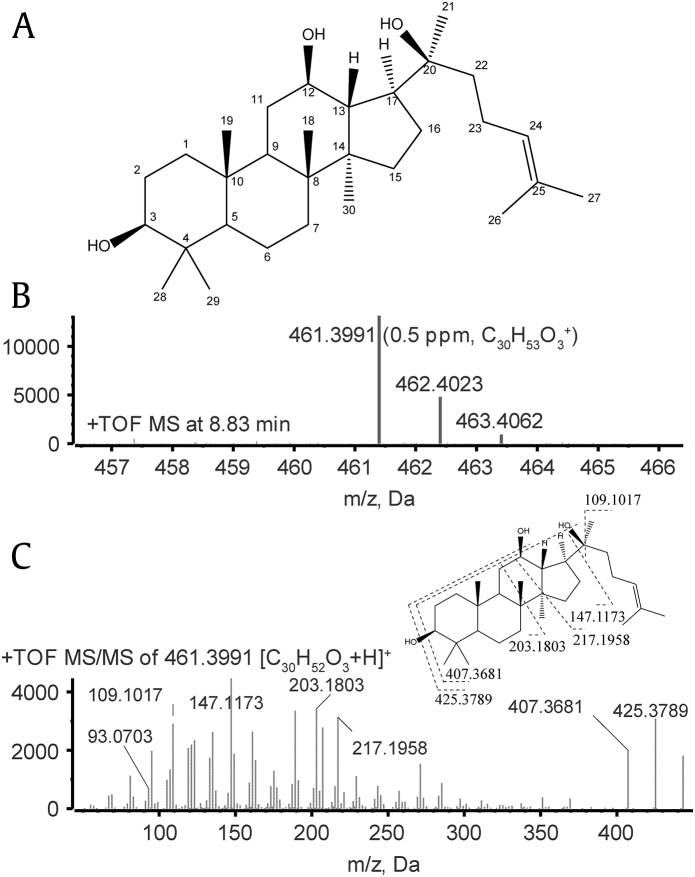
At source temperature of 121°C, soft ionization was observed. PPD had the protonated molecular ion at m/z 461.3989, and the retention time was 8.860 minutes. It is helpful to understand the fragmentation behavior of the parent compound for the sake of metabolite identification. The CID product ion spectrum of PPD (Fig. 1) showed prominent fragment ions at m/z 443 (−18), 425 (−36) and 407 (−54), which were products of the loss of one to three molecules of H2O (18 Da) from the three hydroxy groups.
Fig. 1.
(A) Chemical structure of 20(S)-protopanaxadiol (PPD). (B) Full-scan mass spectrum of PPD. (C) The TOF MS/MS spectrum of PPD at m/z 461.3991. TOF, time of flight.
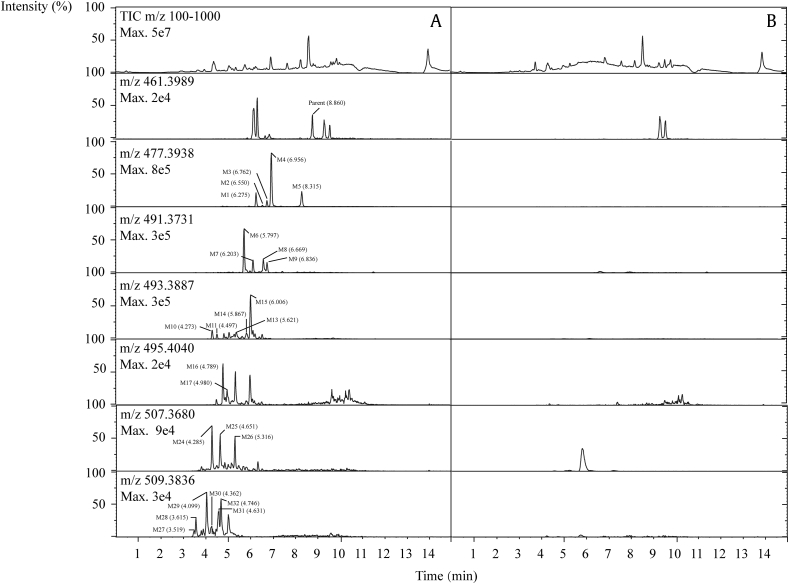
Fig. 2 shows the chromatograms of total ion and extracted ion of PPD and its metabolites in human plasma after oral administration. Twenty-five metabolites were observed by comparing the samples with blanks. These metabolites were classified into six groups and were numbered consecutively according to their molecular weights and chromatographic retention time. The information about elemental composition and accuracy error of PPD and its metabolites has been listed in Table S1. The structures of metabolites were assigned by analyzing their mass spectral fragments and proposed metabolic process. All MS/MS spectra and assigned structures are shown in Fig. 3.
Fig. 2.
The extracted ion chromatograms [M+H]+ of PPD and its metabolites in human plasma. (A) After oral administration of PPD. (B) Before oral administration of PPD. Twenty five metabolites and the parent drug were detected in human plasma after oral administration of PPD by comparing (A) with (B) and labeled as M1 (m/z 477.3938), M2 (m/z 491.3731), M3(m/z 493.3887), M4 (m/z 495.4040), M6 (m/z 507.3680), M7 (m/z 509.3836) and Parent, respectively. PPD, 20(S)-protopanaxadiol; TIC, total ion chromatogram.
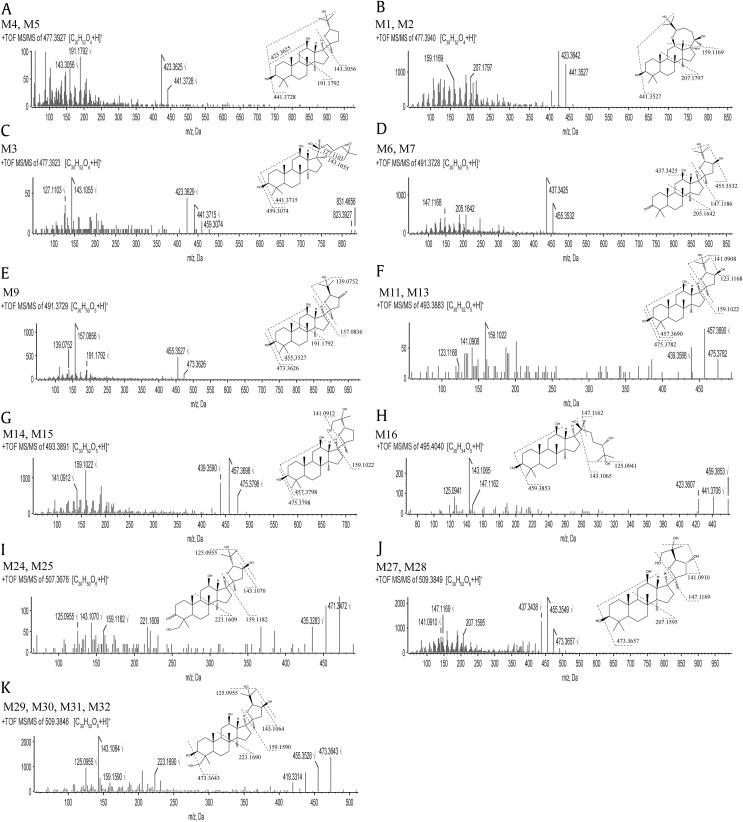
Fig. 3.
The TOF MS/MS spectra of PPD metabolites in plasma were obtained from the respective [M+H]+ ions as the precursors for collision-induced dissociation. The structures were elucidated by analyzing the fragmentation patterns and confirmed by structure matching software MetabolitePolet™. PPD, 20(S)-protopanaxadiol; TOF, time-of-flight.
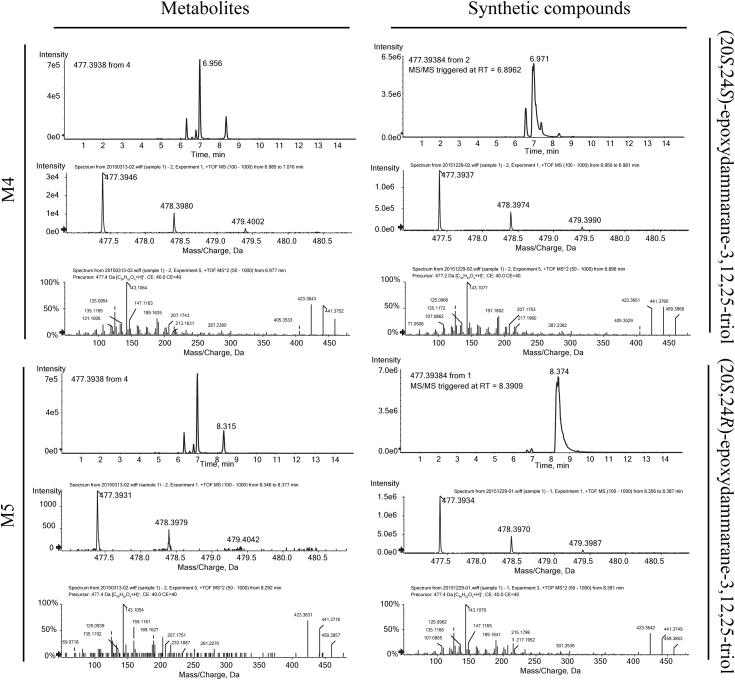
2Metabolite Group 1 (metabolites M1, M2, M3, M4, and M5): addition of one oxygen atom (+16 from the parent compound). The accurate mass of the protonated molecular ion of the metabolites mentioned above was 477.3938, which indicated the addition of one oxygen atom, with molecular formula C30H52O4. The metabolites M1, M2, M3, M4, and M5 were eluted at 6.275, 6.550, 6.762, 6.956, and 8.315 minutes, respectively. The MS/MS spectra (Fig. 3A) of M3 and M4 showed prominent ions at m/z 441 (−36), 423 (−54), 207, 191, and 143, which were the same as those of metabolites at m/z 477 (M2 and M3) in Li et al's research work [8]. In addition, these two compounds mentioned above, (20S,24S)-epoxydammarane-3,12,25-triol and (20S,24R)-epoxydammarane-3,12,25-triol, were synthesized for metabolite identification. By comparing retention time and MS/MS spectra under the same analytical condition, M3 and M4 were identified as (20S,24S)-epoxydammarane-3,12,25-triol and (20S,24R)-epoxydammarane-3,12,25-triol, respectively (Fig. S1). Li et al also found that most of the PPD metabolites were in 20,24-oxide form and possessed a hydroxy group attached to the C-25 positon of the side chain. Fig. 3B shows that the CID product ion spectra of M1 and M2 displayed identical fragment ions at m/z 441 (−36), 423 (−54), 207, and 159. According to the fragmentation behavior of PPD, the C-3, C-12, and C-20 hydroxy groups normally undergo loss of H2O one by one producing fragment ions at m/z 443 (−18), 425 (−36), and 407 (−54). The fact that fragment ion of m/z 459 (−18) was absent in the spectra of M1 and M2 indicated that one of the three hydroxy groups of PPD was modified, most possibly by ring formation. The fragment ion of m/z 143 was attributed to the five-membered ether ring side chain, containing 25-hydroxy group (Fig. 3A). Therefore, the fragment ion of m/z 159 (143 + 16) was supposed to be the side chain with one more oxygen atom. However, no more exogenous oxygen atom could be added, suggesting that the bond at C-24 position on the side chain reacted with C-12 hydroxyl to form seven-membered heterocyclic ring. As a result, M1 and M2 were tentatively assigned as 12,24-oxide metabolites of PPD. The MS/MS spectra (Fig. 3C) of M5 showed prominent ions at m/z 459 (−18), 441 (−36), 423 (−54), 143, and 127. The presence of fragment ion m/z of 459 (−18), 441 (−36), and 423 (−54) indicated that all three hydroxy groups of PPD were present. The fragment ion of m/z of 143 was attributed the oxidation of PPD's side chain. Therefore, M5 was tentatively assigned as 24,25-epoxy metabolite.
Metabolite Group 2 (metabolites M6, M7, M8, and M9): addition of two oxygen atoms with dehydrogenation (+30 from the parent compound). The accurate mass of the protonated molecular ion of the metabolites mentioned above was 491.3731, which indicated the addition of two oxygen atoms and dehydrogenation, with molecular formula C30H50O5. The retention time of M6, M7, M8, and M9 were 5.797, 6.203, 6.669, and 6.836 minutes, respectively. Fig. 3D shows that the CID product ion spectra of M6 and M7 displayed identical fragment ions at m/z 455 (−36), 437 (−54), 205, and 147. The fragment ion of m/z 147 (C7H15O3) was attributed to the dihydroxylated five-membered ether ring side chain, while fragment ion m/z 205 (C14H23O) was attributed to dehydrogenated AB ring fragment. This suggested that the dehydrogenation could most possibly occur at 3-hydroxy group, forming 3-keto metabolites. The chromatographic retention time and mass spectral fragmentation patterns of M7 were identical to those of the isolated metabolite (20S,24S)-epoxydammarane-12,23,25-triol-3-one. Thus, M7 was confirmed as (20S,24S)-epoxydammarane-12,23,25-triol-3-one. The MS/MS spectra (Fig. 3E) of M9 showed prominent ions at m/z 473 (−18), 455 (−36), 191, 157, and 139. The fragment ion of m/z 191 (C14H23) was attributed to AB ring fragment, while the fragment ion of m/z 157 was attributed to the dehydrogenation of dihydroxylated five-membered ether ring side chain, suggesting that the dehydrogenation could most possibly occur at 23-hydroxy group.
Metabolite Group 3 (metabolites M10, M11, M12, M13, M14, and M15): addition of two oxygen atoms (+32 from the parent compound). These metabolites were detected with a protonated molecular weight of 493.3887, which indicated the addition of two oxygen atoms. The retention time of M10, M11, M13, M14, and M15 were 4.273, 4.497, 5.621, 5.867, and 6.006 minutes, respectively. The CID product ion spectra of M11 and M13 (Fig. 3F) showed fragment ions at m/z 475 (−18), 457 (−36), 439 (−54), 159, 141, and 123. The fragment ion of m/z 159 was attributed to the dihydroxylated five-membered ether ring side chain, m/z 141 was attributed to single dehydroxylation of the dihydroxylated five-membered ether ring side chain, and m/z 123 was attributed to dual dehydroxylation of the dihydroxylated five-membered ether ring side chain. This suggested the hydroxylation occurred on the side chain. The chromatographic retention time and mass spectral fragmentation patterns of M13 were identical to those of the isolated metabolite (20S,24S)-epoxydammarane-3,12,23,25-tetrol. Thus, M13 was confirmed as (20S,24S)-epoxydammarane-3,12,23,25-tetrol. The MS/MS spectra (Fig. 3G) of M14 and M15 showed prominent ions at m/z 475 (−18), 457 (−36), 439 (−54), 159, and 141. The absence of fragment ion at m/z 123 indicated that the two hydroxy groups on the side chain were in adjacent positions. Therefore, the other hydroxy group was supposed to bind at C-26 or C-27 position, instead of C-23 position.
Metabolite Group 4 (metabolites M16 and M17): addition of two oxygen atoms with hydrogenation (+34 from the parent compound). In the plasma, M16 and M17 were detected at 4.789 and 4.980 minutes, respectively. The accurate mass of the protonated molecular ion of M4 was 495.4040, which indicated the addition of two oxygen atoms and hydrogenation, with molecular formula C30H54O5. The MS/MS spectra (Fig. 3H) of M16 showed prominent ions at m/z 459 (−36), 441 (−54), 423 (−72), 405 (−90), 147,143, and 125. The fragment ion of m/z 459 (−36), 441 (−54), 423 (−72), and 405 (−90) suggested that M16 contained five hydroxy groups. The presence of fragment m/z 495.4040 suggested that the side chain did not undergo ring closure reactions to form five-membered ether ring. In consideration of metabolic pathways, M16 and M17 were tentatively assigned as 24,25-dihydroxy product of PPD, formed by the addition of two hydroxy groups to the parent drug's double bond. The structure was consistent with the fragment ions by MetabolitePilot software.
Metabolite Group 6 (metabolites M22, M23, M24, M25, and M26): addition of three oxygen atoms with dehydrogenation (+46 from the parent compound). These metabolites had a protonated molecular ion at m/z 507.3680, 16 Da more than that of metabolites in Group 6, suggesting that it is an oxidation product of metabolites in Group 6. The M24, M25, and M26 were observed at 4.285, 4.651, and 5.316 minutes, respectively. The CID product ion spectra of M24 and M25 (Fig. 3I) showed prominent ions at m/z 471 (−36), 453 (−54), 435 (−72), 221, 159, 143, and 125. The fragment ion of m/z 471 (−36), 453 (−54), and 435 (−72) suggested that M24 and M25 both contained four hydroxy groups. The fragment ion of m/z 159 was attributed to the dihydroxylated five-membered ether ring side chain, while the fragment ion of m/z 221 was attributed to the dehydrogenation of hydroxylated AB ring. The dehydrogenation most possibly occurred at 3-hydroxy group, and in consideration of steric hindrance, the additional hydroxylation to M2 could most possibly occurred at C-28 or C-29 position.
Metabolite Group 7 (metabolites M27, M28, M29, M30, M31, and M32): addition of three oxygen atoms (+48 from the parent compound). These metabolites were detected with a protonated molecular weight of 509.3836, which was 16 Da higher than that of metabolites in Group 3, indicating the introduction of one more oxygen atom to metabolites in Group 3. Six chromatographic peaks with selected mass range at m/z 509.3836 were observed in the plasma samples. The retention time of M27 and M28 were 3.519 and 3.615 minutes, respectively. The MS/MS spectra (Fig. 3J) of M27 and M28 showed prominent ions at m/z 473 (−36), 455 (−54), 437 (−72), 207, 147, and 141. According to the molecular formula, M27 and M28 should contain five hydroxy groups. However, the fragment ions at 473 (−36), 455 (−54), and 437 (−72) showed that M27 and M28 could only lose four molecules of H2O. This indicated that two of the hydroxy groups were adjacent to each other. The fragment ion m/z 207 was attributed to the fragment of AB ring, suggesting that the additional hydroxy groups were on the side chain. Based on MS/MS data and metabolic pathways, the structure of M27 and M28 were supposed to be as shown in Fig. 3. M29, M30, M31, and M32 were detected at 4.099, 4.362, 4.631, and 4.746 minutes, respectively. The CID product ion spectra of M29, M30, M31, and M32 (Fig. 3K) showed prominent ions at m/z 473 (−36), 455 (−54), 437 (−72), 419 (−90), 223, 159, 143, and 125. The fragment ions of m/z 473 (−36), 455 (−54), 437 (−72), and 419 (−90) indicated the presence of five hydroxy groups. The fragment ion of m/z 223 suggested that the hydroxylated AB ring contained two hydroxy groups. The fragment ions of m/z 159, 143, and 125 suggested that the side chain underwent ring closure reactions and contained two hydroxy groups. Therefore, in consideration of steric hindrance, the additional hydroxylation could have most possibly occurred at C-28 or C-29 position (Fig. 3).
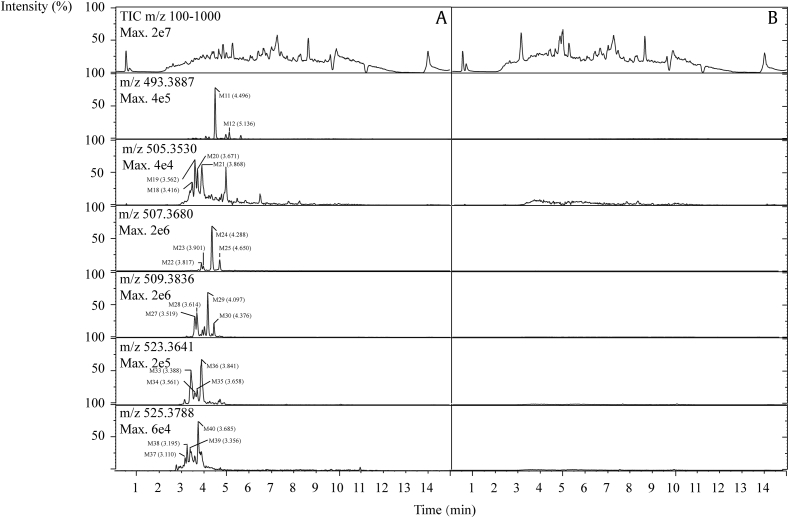
XICs for each possible biotransformation in human urine are displayed in Fig. 4. Following oral administration of PPD, the parent drug was not detected in human urine. Meanwhile, 22 metabolites were observed by comparing the urine samples with blanks. These metabolites were classified into six groups and were numbered consecutively according to their molecular weights and chromatographic retention time, respectively. The structures of metabolites were assigned by analyzing their mass spectral fragments and proposed metabolic process. All MS/MS spectra and assigned structures are shown in Fig. 5.
Fig. 4.
The extracted ion chromatograms [M+H]+ of PPD metabolites in human urine. (A) After oral administration of PPD. (B) Before oral administration of PPD. Twenty-two metabolites were detected in human urine after oral administration of PPD by comparing (A) with (B) and labeled as M3 (m/z 493.3887), M5 (m/z 505.3530), M6 (m/z 507.3680), M7 (m/z 509.3836), M8 (m/z 523.3641), and M9 (m/z 525.3788), respectively. PPD, 20(S)-protopanaxadiol; TIC, total ion chromatogram.
Fig. 5.
The TOF MS/MS spectra of PPD metabolites in urine were obtained from the respective [M+H]+ ions as the precursors for collision-induced dissociation. The structures were elucidated by analyzing the fragmentation patterns and confirmed by structure matching software MetabolitePolet™. PPD, 20(S)-protopanaxadiol; TOF, time-of-flight.
Metabolite Group 3 (Metabolite M10, M11, M12, M13, M14, and M15): addition of two oxygen atoms (+32 from the parent compound). M11 and M12 were detected in human urine with the protonated molecular weight of 493.3887, which was 32 Da higher than that of PPD, suggesting the introduction of two oxygen atoms. The retention time of M11 and M12 were 4.496 and 5.136 minutes, respectively. The CID product ion spectrum of M11 showed fragment ions at m/z 475 (−18), 457 (−36), 439 (−54), 159, 141, and 123. The structure elucidation is similar to that of M11 in human plasma.
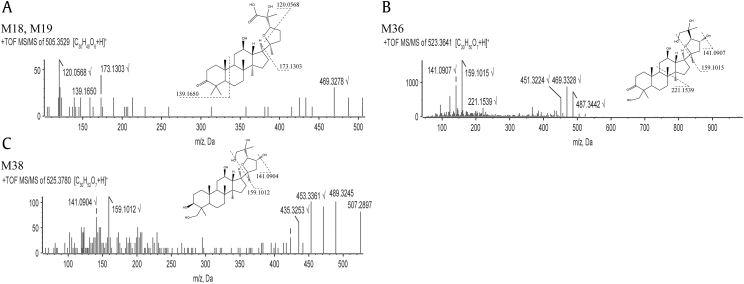
Metabolite Group 5 (metabolites M18, M19, M20, and M21): addition of three oxygen atoms and dual dehydrogenation (+44 from the parent compound). The accurate mass of the protonated molecular ion of M5 was 505.3530, which indicated the addition of three oxygen atoms with dual dehydrogenation, with molecular formula C30H48O6. The retention time of M18, M19, M20, and M21 were 3.416, 3.562, 3.671, and 3.868 minutes, respectively. The CID product ion spectra of M18 and M19 (Fig. 5A) showed fragment ions at m/z 487 (−18), 469 (−36), 173, 139, and 120. According to the molecular formula, M18 and M19 should contain three hydroxy groups. However, fragment ion of m/z 469 (−36) suggested that M18 and M19 could only lose two molecules of H2O. This result indicated that the other hydroxy group was adjacent to a quaternary carbon, most possibly a carbonyl carbon. The fragment ion at m/z 173 was attributed to the dehydogenated and hydroxylated five-membered ether ring side chain, while m/z 120 was attributed to a fragment of the side chain. This suggested that the five-membered ether ring side chain contained one hydroxy group and one carboxyl group (Fig. 5A). The fragment ion of m/z 139 was attributed to the dehydrogenation of A ring, suggesting that the other dehydrogenation site could most possibly be at 3-hydroxy group. The MS/MS spectra of M20 and M21 were not obtained. Based on molecular weight, retention time, and metabolic pathways, the structures of M20 and M21 were supposed to be as shown in Fig. 6.
Fig. 6.
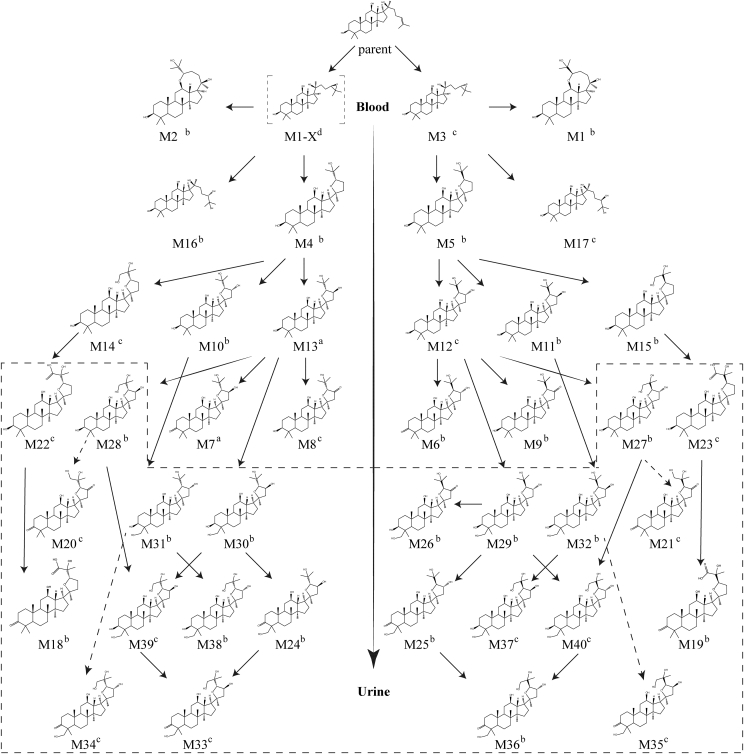
Proposed metabolic pathways of PPD in human body (in vivo). a The metabolites were detected in extracted ion chromatograms. The structures were elucidated by analyzing the fragmentation patterns and confirmed by NMR analysis. b The metabolites were detected in extracted ion chromatograms. The structures were elucidated by analyzing the fragmentation patterns and confirmed by structure matching software MetabolitePolet™. c The metabolites were detected in extracted ion chromatograms. No MS/MS spectra obtained. d No chromatographic or MS/MS data obtained. The structures were elucidated based on the proposed metabolic pathways. PPD, 20(S)-protopanaxadiol.
Metabolite Group 6 (metabolites M22, M23, M24, and M25): addition of three oxygen atoms and dehydrogenation (+46 from the parent compound). Metabolites in Group 6 had a protonated molecular ion at m/z 507.3680, 16 Da more than metabolites in Group 2, indicating the addition of one more oxygen atom to metabolites in Group 2. The M22, M23, M24, and M25 were observed at 3.817, 3.901, 4.288, and 4.650 minutes, respectively. The CID product ion spectra of M24 and M25 showed prominent ions at m/z 471 (−36), 453 (−54), 435 (−72), 221, 159, 143, and 125. The structure elucidation is similar to that of M24 and M25 in human plasma.
Metabolite Group 7 (metabolites M27, M28, M29, M30, M31, and M32): addition of three oxygen atoms (+48 from the parent compound). The metabolites in Group 7 were detected with a protonated molecular weight of 509.3836, which was 16 Da higher than metabolites in Group 3, indicating the introduction of one more oxygen atom to the metabolites in Group 3. Four chromatographic peaks with selected mass range at m/z 509.3836 were observed in the urine samples. The retention time of M27 and M28 were 3.519 and 3.614 minutes, respectively. M29 and M30 were detected at 4.097 and 4.376 minutes, respectively. The structure elucidation is similar to that of the metabolites in Group 7 in human plasma.
Metabolite Group 8 (metabolites M33, M34, M35, and M36): addition of four oxygen atoms and dehydrogenation (+62 from the parent compound). The accurate mass of the protonated molecular ion of M8 was 523.3641, which indicated the addition of four oxygen atoms and dehydrogenation, with molecular formula C30H50O7. The retention time of M33, M34, M35, and M36 were 3.388, 3.561, 3.658, and 3.841 minutes, respectively. The CID product ion spectra of M36 (Fig. 5B) showed fragment ions at m/z 487 (−36), 469 (−54), 451 (−72), 221, 159, and 141. According to the molecular formula, M36 should contain five hydroxy groups. However, the presence of fragment ions at m/z 487 (−36), 469 (−54), and 451 (−72) indicated that M36 could only lose four molecules of H2O. This suggested that two of the hydroxy groups were adjacent to each other. The fragment ions of m/z 159 and 141 were the side chain fragments. Therefore, the hydroxylation was supposed to occur at C-23, C-25, and C-26/27 position on the side chain. The fragment ion of m/z 221 was attributed to the dehydrogenation of hydroxylated AB ring, suggesting that the dehydrogenation occurred on the parent structure, most possibly at 3-hydroxy group. In consideration of steric hindrance, the other hydroxylation site was supposed to be C-28/29 position. The MS/MS spectra of M33, M34, and M35 were not obtained. Based on molecular weight, retention time, and metabolic pathways, the structures of M33, M34, and M35 were supposed to be as shown in Fig. 6.
Metabolite Group 9 (metabolites M37, M38, M39, and M40): addition of four oxygen atoms (+64 from the parent compound). The accurate mass of the protonated molecular ion of the metabolites in Group 9 was 525.3788, which indicated the addition of four oxygen atoms, with molecular formula C30H52O7. The retention time of M37, M38, M39, and M40 were 3.110, 3.195, 3.356, and 3.685 minutes, respectively. The CID product ion spectrum of M38 (Fig. 5C) showed fragment ions at m/z 507 (−18), 489 (−36), 471 (−54), 453 (−72), 435 (−90), 159, and 141. According to the molecular formula, M38 should contain six hydroxy groups. However, the presence of fragment ions at m/z 507 (−18), 489 (−36), 471 (−54), 453 (−72), and 435 (−90) indicated that M38 could only lose five molecules of H2O. This suggested that two of the hydroxy groups were in adjacent positions. The fragment ions of m/z 159 and 141 were the side chain fragments. Therefore, the hydroxylation was supposed to occur at C-23, C-25 and C-26/27 position on the side chain. In consideration of steric hindrance, the other hydroxylation was supposed to occur at C-28/29 position. The MS/MS spectra of M37, M39, and M40 were not obtained. Based on molecular mass, retention time, and metabolic pathways, the structures of M37, M39, and M40 were supposed to be as shown in Fig. 6.
3.2. Structure elucidation of two isolated metabolites
In fact, four metabolites of PPD were isolated from rat feces. However, two of the metabolites with protonated molecular weight of 475.3783 (1) and 477.3938 (2) had been reported before in the in vitro study, thus, were not the subject of our analysis. Because of the low content of the other two metabolites with protonated molecular weight of 491.3731 (3) and 493.3887 (4), 500 MHz NMR with Bruker's CryoProbes was selected to perform a highly sensitive and extensive NMR spectroscopic analysis. The NMR data are shown in Table 1. After NMR spectroscopic analysis, UPLC-Q-TOF-MS/MS was also performed. By comparing the retention time and MS/MS data, two novel metabolites with protonated molecular weight of 491.3731 (3) and 493.3887 (4) were confirmed to be M7 and M13 respectively. Finally, the structures of both compounds were elucidated.
Table 1.
NMR data of metabolites M7 (3) and M13 (4) in CDCl3
| No. |
3 |
4 |
||
|---|---|---|---|---|
| δH(J in Hz) | δC | δH(J in Hz) | δC | |
| 1 | 1.95,m1) | 39.71 | 1.71,m1) | 38.93 |
| 1.50,m1) | 1.00,m1) | |||
| 2 | 2.51,m | 33.91 | 1.64,m1) | 28.73 |
| 2.44,m | 1.58,m1) | |||
| 3 | 217.91 | 3.20,dd(11.6,4.9) | 78.90 | |
| 4 | 47.34 | 39.70 | ||
| 5 | 1.40,m1) | 55.26 | 0.74,dd(11.2,2.0) | 55.94 |
| 6 | 1.54,2H,m1) | 19.68 | 1.56,m1) | 18.29 |
| 1.46,m1) | ||||
| 7 | 1.52,m1) | 34.31 | 1.45,m1) | 34.71 |
| 1.34,m1) | 1.31,m1) | |||
| 8 | 39.61 | 39.73 | ||
| 9 | 1.58,m1) | 49.60 | 1.451) | 50.28 |
| 10 | 36.86 | 37.17 | ||
| 11 | 1.92,m1) | 31.86 | 1.93,m1) | 32.41 |
| 1.16,m1) | 1.70,m1) | |||
| 12 | 3.57,ddd(10.3,10.3,4.7) | 70.47 | 3.55,ddd(10.9,10.9,5.0) | 70.66 |
| 13 | 1.58,m1) | 49.10 | 1.60,m1) | 48.95 |
| 14 | 52.23 | 52.25 | ||
| 15 | 1.53,m1) | 32.38 | 1.50,m1) | 31.48 |
| 1.10,m1) | 1.10,m1) | |||
| 16 | 1.99,m1) | 28.70 | 1.86,m1) | 28.10 |
| 1.26,m1) | 1.31,m1) | |||
| 17 | 2.25,ddd(11.3,11.3,5.3) | 49.95 | 2.23,m | 49.96 |
| 18 | 1.03,s | 15.12 | 1.00,s | 18.00 |
| 19 | 0.98,s | 16.12 | 0.88,s | 15.30 |
| 20 | 88.01 | 88.06 | ||
| 21 | 1.55,s | 30.06 | 1.54,s | 30.04 |
| 22 | 2.03,2H1) | 40.98 | 2.07,m1) | 40.97 |
| 1.94,m1) | ||||
| 23 | 4.57,ddd(3.0,3.0,3.0) | 73.54 | 4.55,ddd(2.90,2.90,2.90) | 73.55 |
| 24 | 3.77,d(3.7) | 87.18 | 3.76,d(3.60) | 87.15 |
| 25 | 72.65 | 72.66 | ||
| 26 | 1.39,s | 24.22 | 1.39,s | 24.20 |
| 27 | 1.31,s | 29.06 | 1.31,s | 29.07 |
| 28 | 1.09,s | 26.76 | 0.98,s | 28.00 |
| 29 | 1.05,s | 20.92 | 0.79,s | 15.45 |
| 30 | 0.93,s | 17.89 | 0.92,s | 16.32 |
)Overlapping signals.
Compound 3 (M7). Compound 3 (M7) was canary yellow amorphous powder. The accurate mass of the protonated molecular ion of Compound 3 (M7) was 491.3731. Comparing the 13C NMR spectral data of Compound 3 (M7) with that of PPD, the olefinic carbon signals of PPD at δ124.8 and δ132.0 were not seen in that of Compound 3 (M7), suggesting that the double bond at Δ(24,25) on the side chain was absent. Because the degree of unsaturation was calculated to be 5, it was concluded that the side chain underwent ring formation. In addition, a carbonyl group bearing secondary carbon signal at δ217.9 was observed whereas the proton signal of Compound 3 (M7) at δ3.20 (H-3) was absent, indicating that the dehydrogenation reaction occurred at C-3 position. 1H-1H COSY data showed H-24 (δH 3.75) adjacent to H-23 (δH 4.55), suggesting the introduction of hydroxy group occurred at C-23 position. The assigned structure for Compound 3 (M7) was confirmed by 2D NMR spectra (Fig. 7).
Fig. 7.
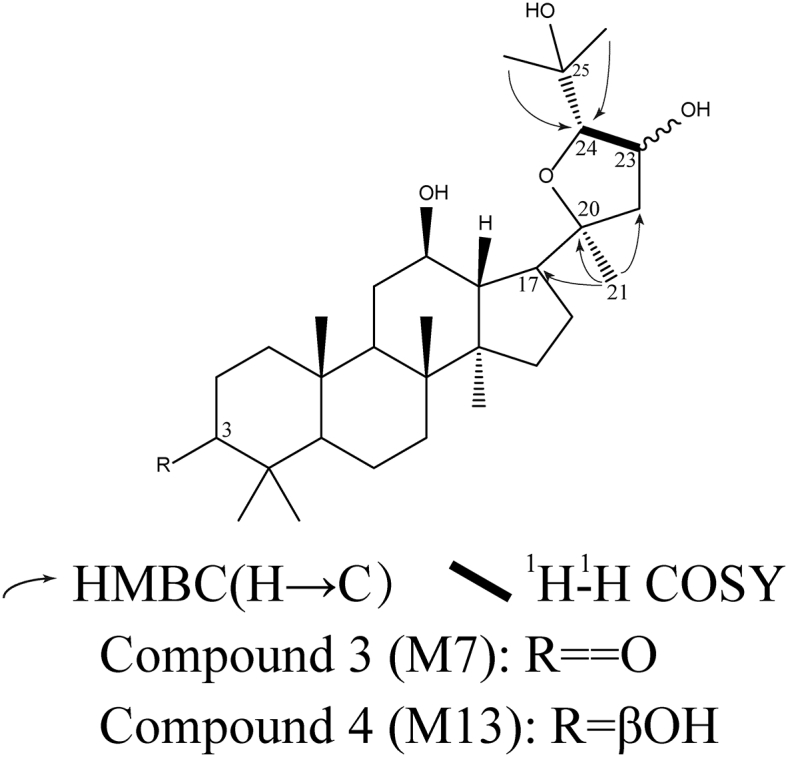
The verified chemical structures of Compound 3 and Compound 4. The C-23 hydroxy group and the substituted tetrahydrofuran side chain were elucidated by selected HMBC and 1H-1H COSY correlations. COSY, correlation spectroscopy; HMBC, heteronuclear multiple-bond correlation.
Compound 4 (M13). Compound 4 (M13) was white amorphous powder. The accurate mass of the protonated molecular ion of Compound 4 (M13) was 493.3887. According to 1H and 13C NMR spectral data, H-3 (δH 3.20), H-12 (δH 3.54), H-23 (δH 4.55), H-24 (δH 3.75), C-3 (δC 78.9), C-12 (δC 70.6), C-20 (δC 88.1), C-23 (δC 73.6), C-24 (δC 87.2), and C-25 (δC 72.7) of Compound 4 (M13) were observed, indicating that the side chain underwent ring formation with 23-hydroxylation. The assigned structure of Compound 4 (M13) was confirmed by 1H-1H COSY, NOESY and HMBC experiments (Fig. 7).
Accurate and reliable NMR data played an important role in confirming chemical structures of metabolic products. In the present study, thorough NMR analyses, including 1H-1H COSY, heteronuclear singular quantum correlation, HMBC, and NOESY, were performed on the two isolated metabolites, Compound 3 and Compound 4. The structures of the two compounds were completely elucidated and confirmed by 2D NMR. Compound 3 was confirmed as (20S,24S)-epoxydammarane-12,23,25-triol-3-one or neoalsogenin B, and Compound 4 was confirmed to be (20S,24S)-epoxydammarane-3,12,23,25-tetrol. In 1995, Fujita et al isolated neoalsogenin B from the aerial parts of Neoalsomitra integrifoliola [14], [15]. In 2004, Liu et al isolated Gynoside D, a glycoside containing Compound 4 as the aglycone part, from the aerial parts of Gynostemma pentaphyllum [16]. However, this was the first time that these two compounds were found as metabolites in human. The 1H and 13C NMR data of neoalsogenin B and Gynoside D were reported in the corresponding studies, but the 2D NMR data of the two compounds had never been reported before.
3.3. Metabolic pathways
Thanks to structure matching software MetabolitePolet™, the structures of 40 metabolites of PPD were deduced by analysis of their mass spectral fragmentation patterns obtained from UPLC-Q-TOF-MS/MS. In the light of the metabolic pathways reported in Li et al's article [8] and the structures previously elucidated, the proposed metabolic pathways of PPD in human are shown in Fig. 7. In human, carbon–carbon double bond epoxidation, giving 24,25-epoxide metabolites (M5 and its isomer), followed by hydrolysis and rearrangement to form the 20,24-oxide derivatives (M3 and M4) was the critical metabolic process, similar to that in HLMs and human hepatocytes. Apart from the main 20,24-oxide metabolites, the conversion of 24,25-epoxide intermediates to the 12,24-oxide forms (M1 and M2) also occurred in small amount, presumably by the intramolecular nucleophilic attack of the oxygen atom of C-12 hydroxy group at C-24 position. After ring formation of the side chain with 25-hydroxylation, further hydroxylation at C-23, C-26/27, and C-28/29 position and/or dehydrogenation at 3-hydroxyl or 23-hydroxy group were the main metabolic reactions.
In addition, assigned structures of compounds detected in urine samples are shown in the dashed line (Fig. 6). It was observed that only metabolites with the number of hydroxy groups and/or carbonyl groups equal to or greater than five were excreted from human body via urine, presumably because of their relatively high polarity. Thus, it could be assumed that the parent drug, PPD, was not detected in urine because of its low polarity.
3.4. Metabolic differences between in vivo and in vitro
In vitro study on the metabolism of PPD had been performed in HLMs and human hepatocytes. In the in vitro study, 24 metabolites were found in HLMs and human hepatocytes. Carbon–carbon double bond epoxidation, which gave 24,25-epoxide metabolites, was the primary metabolic reaction observed, followed by hydrolysis and rearrangement to form the corresponding 24,25-vincinal diol derivatives and the main 20,24-oxide metabolites [8]. However, the metabolism of PPD in human appeared to be different from that in rats and in vitro.
In our research, 25 metabolites in plasma and 22 metabolites in urine were identified. By analyzing and comparing the metabolites present in human plasma and urine samples (shown in Fig. 8), our study revealed that metabolites in groups 1, 2, and 4 were only detected in plasma; metabolites in groups 3, 6, and 7 were detected both in plasma and urine; metabolites in groups 5, 8, and 9 were only detected in urine. These findings suggested that PPD was first metabolized by hepatic enzymes into metabolites in groups 1, 2, and 4. Then, these metabolites were further metabolized into metabolites in groups 3, 6, and 7 and eliminated through urine. Moreover, phase II metabolites of PPD (Group 10) were not detected in any of the sample in our study. This result was in contrast with the result of the previous study in human hepatocytes, in which two glucuronide conjugates of PPD were detected. However, it is noteworthy that even though PPD concentration used for incubation in the in vitro study (6 × 108) was much higher than the plasma concentration of PPD following oral administration, only tiny amount of phase II metabolites were detected (1 × 106). This indicated that, in human, PPD was mainly metabolized by hydroxylation under the effect of cytochrome P450 enzymes and eliminated through urine but might be metabolized through phase II metabolism if phase I metabolism was saturated and the drug started to accumulate in the body. Moreover, metabolites in Group 0 with m/z 475.3783, 3-keto metabolites of PPD, discovered during in vitro study were also absent in our study, indicating that metabolites in Group 1 with m/z 477.3938 was further metabolized by hydroxylation to form metabolites in Group 2 with m/z 493.3887. This emphasized the proposition that PPD was mainly metabolized by sequential hydroxylation in human body. Furthermore, in the in vitro study, three metabolites in Group 1 were separated by chromatography whereas five metabolites were detected in our study. This might be due to decreased activity of hepatic enzymes in vitro relative to in vivo. The fact that metabolites in groups 5, 8, and 9 were detected only in urine while absent in both plasma, and in vitro study indicated that they were the products of renal metabolism. Although these metabolites in groups 5, 8, and 9 were detected at low concentration, they provided a very important piece of information concerning metabolism of PPD in human. The finding suggested that PPD, apart from hepatic metabolism, also underwent renal metabolism. These three metabolite groups might be the products of renal metabolism of metabolite groups 3, 6, and 7, once the renal elimination of these metabolites was saturated. This might indicate that at the oral dosage of 100 mg, renal elimination process of PPD began to saturate. If the dosage was increased, phase II metabolism might be required to compensate the saturated metabolism as mentioned above. This might also lead to toxicity of the drug. In addition, the study of metabolism of ginsenoside Rh2 reported that ginsenoside Rh2 was metabolized in gastrointestinal tract by intestinal flora [17]. Given the fact that PPD is the active metabolite of ginsenoside Rh2, it is highly possible that oral PPD is also metabolized in the same way. However, to confirm this hypothesis, human bile and feces samples must be obtained and analyzed.
Fig. 8.
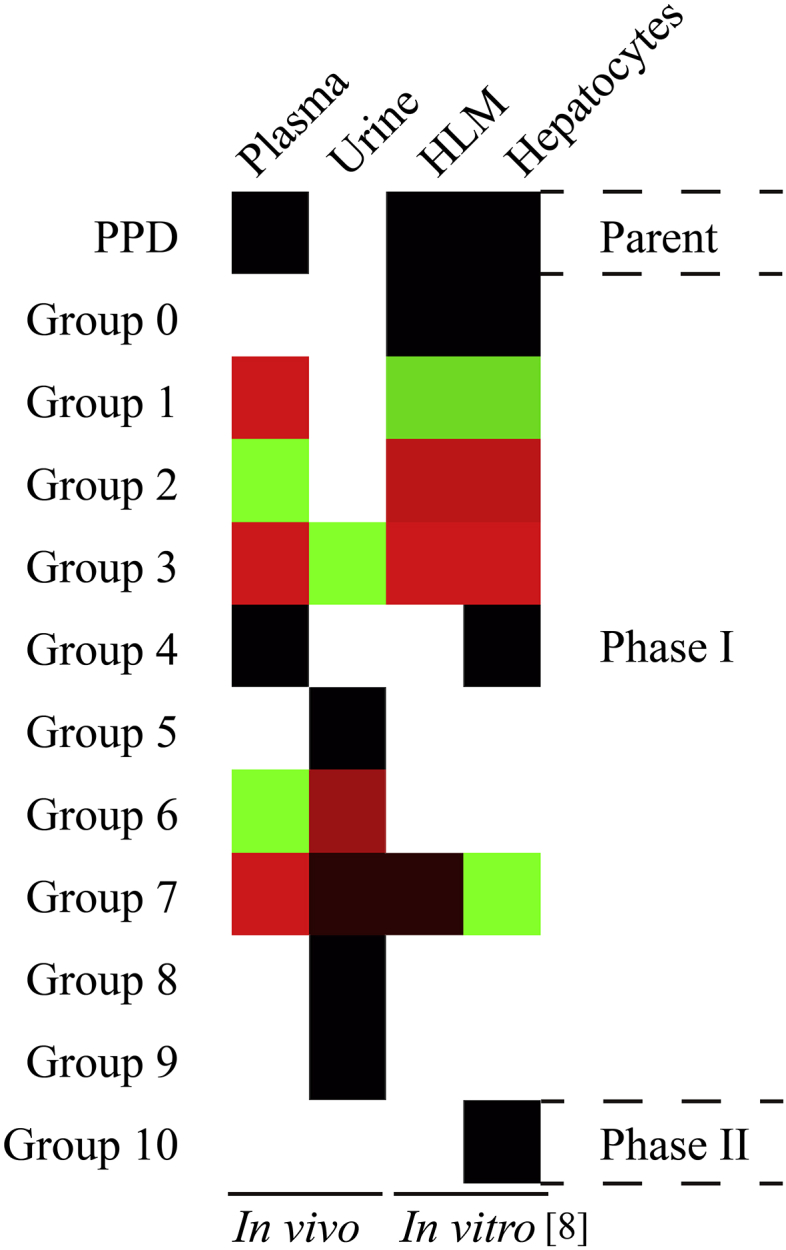
Colored heat map showing the presence of PPD and its metabolites in human plasma, human urine, human liver microsomes, and human hepatocytes. The number of isomers of each metabolite was categorized using the unweighted pair group method with an arithmetic mean in Hierarchical Clustering Explorer 3.0. Metabolites with the number of isomers equal to the arithmetic mean are shown in black; metabolites with the number of isomers higher than the arithmetic mean are shown in red; metabolites with the number of isomers lower than the arithmetic mean are shown in green. HLM, human liver microsome; PPD, 20(S)-protopanaxadiol.
As mentioned above, the results of in vitro study could only reflect the effects of hepatic enzymes on PPD under certain conditions but could not demonstrate the continuous process of its metabolism and elimination in human body. Although most drugs are metabolized in human body mainly by hepatic metabolism, other paths of metabolism such as renal metabolism and gastrointestinal (GI) metabolism still cannot be overlooked.
Conflicts of interest
The authors have no conflicts of interest to declare.
Acknowledgments
This study was supported by National Science and Technology Major Projects for “New Drugs Innovation and Development” (No. 2013ZX09101109) and National Natural Science Foundation (No. 81473175).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.jgr.2018.03.005.
Contributor Information
Yingjia Yu, Email: yuyj@fudan.edu.cn.
Changjiang Xu, Email: cjxush@163.com.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Fig. S1.
The extracted ion chromatograms [M+H]+, TOF MS spectra, and TOF MS/MS spectra of M3, M4, (20S,24S)-epoxydammarane-3,12,25-triol and (20S,24R)-epoxydammarane-3,12,25-triol, respectively.
References
- 1.Lee P.S., Han J.Y., Song T.W., Sung J.H., Kwon O.S., Song S., Chung Y.B. Physicochemical characteristics and bioavailability of a novel intestinal metabolite of ginseng saponin (IH901) complexed with beta-cyclodextrin. Int J Pharm. 2006;316:29–36. doi: 10.1016/j.ijpharm.2006.02.035. [DOI] [PubMed] [Google Scholar]
- 2.Yu Y., Zhou Q., Hang Y., Bu X., Jia W. Antiestrogenic effect of 20S-protopanaxadiol and its synergy with tamoxifen on breast cancer cells. Cancer. 2007;109:2374–2382. doi: 10.1002/cncr.22659. [DOI] [PubMed] [Google Scholar]
- 3.Xu C., Teng J., Chen W., Ge Q., Yang Z., Yu C., Yang Z., Jia W. 20(S)-protopanaxadiol, an active ginseng metabolite, exhibits strong antidepressant-like effects in animal tests. Prog Neuropsychopharmacol Biol Psychiatry. 2010;34:1402–1411. doi: 10.1016/j.pnpbp.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 4.Liu J.H., Wang X., Liu P., Deng R.X., Lei M., Chen W.T., Hu L. 20(S)-Protopanaxadiol (PPD) analogues chemosensitize multidrug-resistant cancer cells to clinical anticancer drugs. Bioorgan Med Chem. 2013;21:4279–4287. doi: 10.1016/j.bmc.2013.04.067. [DOI] [PubMed] [Google Scholar]
- 5.Xie C., Zhou J., Guo Z., Diao X., Gao Z., Zhong D., Jiang H., Zhang L., Chen X. Metabolism and bioactivation of famitinib, a novel inhibitor of receptor tyrosine kinase, in cancer patients. Br J Pharmacol. 2013;168:1687–1706. doi: 10.1111/bph.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin X., Li S.L., Zhang Z.H., Zhu F.X., Sun E., Wei Y.J., Jia X.B. Characterization of metabolites of 20(S)-protopanaxadiol in rats using ultra-performance liquid chromatography/quadrupole-time-of-flight mass spectrometry. J Chromatogr B. 2013;933:59–66. doi: 10.1016/j.jchromb.2013.03.028. [DOI] [PubMed] [Google Scholar]
- 7.He C., Li J., Wang R., Li Z., Bligh S.W., Yang L., Wang Z. Metabolic profiles of 20(S)-protopanaxadiol in rats after oral administration using ultra-performance liquid chromatography/quadrupole time-of-flight tandem mass spectrometry. Rapid Commun Mass Spectrom. 2014;28:595–604. doi: 10.1002/rcm.6813. [DOI] [PubMed] [Google Scholar]
- 8.Li L., Chen X., Li D., Zhong D. Identification of 20(S)-protopanaxadiol metabolites in human liver microsomes and human hepatocytes. Drug Metab Dispos. 2011;39:472–483. doi: 10.1124/dmd.110.036723. [DOI] [PubMed] [Google Scholar]
- 9.Deng R., Xu Y., Feng F., Liu W. Identification of poliumoside metabolites in rat feces by high performance liquid chromatography coupled with quadrupole time-of-flight tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2014;969:285–296. doi: 10.1016/j.jchromb.2014.08.032. [DOI] [PubMed] [Google Scholar]
- 10.Zhou X., Li L., Deng P., Chen X., Zhong D. Characterization of metabolites of GLS4 in humans using ultrahigh-performance liquid chromatography/quadrupole time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2013;27:2483–2492. doi: 10.1002/rcm.6710. [DOI] [PubMed] [Google Scholar]
- 11.Uutela P., Monto M., Iso-Mustajarvi I., Madetoja M., Yliperttula M., Ketola R.A. Identification of metabolites of fosinopril produced by human and rat liver microsomes with liquid chromatography-mass spectrometry. Eur J Pharm Sci. 2014;53:86–94. doi: 10.1016/j.ejps.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 12.Liu M., Zhao S., Wang Z., Wang Y., Liu T., Li S., Wang C., Wang H., Tu P. Identification of metabolites of deoxyschizandrin in rats by UPLC-Q-TOF-MS/MS based on multiple mass defect filter data acquisition and multiple data processing techniques. J Chromatogr B Analyt Technol Biomed Life Sci. 2014;949–950:115–126. doi: 10.1016/j.jchromb.2013.12.022. [DOI] [PubMed] [Google Scholar]
- 13.Tang Y.N., Pang Y.X., He X.C., Zhang Y.Z., Zhang J.Y., Zhao Z.Z., Yi T., Chen H.B. UPLC-QTOF-MS identification of metabolites in rat biosamples after oral administration of Dioscorea saponins: a comparative study. J Ethnopharmacol. 2015;165:127–140. doi: 10.1016/j.jep.2015.02.017. [DOI] [PubMed] [Google Scholar]
- 14.Fujita S., Kasai R., Ohtani K., Yamasaki K., Chiu M.H., Nie R.L., Tanaka O. Dammarane glycosides from aerial parts of neoalsomitra-integrifoliola. Phytochemistry. 1995;38:465–472. doi: 10.1016/0031-9422(95)00020-8. [DOI] [PubMed] [Google Scholar]
- 15.Minghua C., Ruilin N., Nagasawa H., Isogai A., Jun Z., Suzuki A. A dammarane saponin from Neoalsomitra integrifoliola. Phytochemistry. 1992;31:2451–2453. doi: 10.1016/0031-9422(92)83297-c. [DOI] [PubMed] [Google Scholar]
- 16.Liu X., Ye W.C., Mo Z.Y., Yu B., Zhao S.X., Wu H.M., Jiang R., Mak T.C., Hsiao W.L. Five new ocotillone-type saponins from Gynostemma pentaphyllum. J Nat Prod. 2004;67:1147–1151. doi: 10.1021/np034018+. [DOI] [PubMed] [Google Scholar]
- 17.Li L., Chen X., Zhou J., Zhong D. In vitro studies on the oxidative metabolism of 20(s)-ginsenoside Rh2 in human, monkey, dog, rat, and mouse liver microsomes, and human liver s9. Drug Metab Dispos. 2012;40:2041–2053. doi: 10.1124/dmd.112.046995. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.