Abstract
Background
Cytochrome P450 enzymes catalyze a wide range of reactions in plant metabolism. Besides their physiological functions on primary and secondary metabolites, P450s are also involved in herbicide detoxification via hydroxylation or dealkylation. Ginseng as a perennial plant offers more sustainable solutions to herbicide resistance.
Methods
Tissue-specific gene expression and differentially modulated transcripts were monitored by quantitative real-time polymerase chain reaction. As a tool to evaluate the function of PgCYP736A12, the 35S promoter was used to overexpress the gene in Arabidopsis. Protein localization was visualized using confocal microscopy by tagging the fluorescent protein. Tolerance to herbicides was analyzed by growing seeds and seedlings on Murashige and Skoog medium containing chlorotoluron.
Results
The expression of PgCYP736A12 was three-fold more in leaves compared with other tissues from two-year-old ginseng plants. Transcript levels were similarly upregulated by treatment with abscisic acid, hydrogen peroxide, and NaCl, the highest being with salicylic acid. Jasmonic acid treatment did not alter the mRNA levels of PgCYP736A12. Transgenic lines displayed slightly reduced plant height and were able to tolerate the herbicide chlorotoluron. Reduced stem elongation might be correlated with increased expression of genes involved in bioconversion of gibberellin to inactive forms. PgCYP736A12 protein localized to the cytoplasm and nucleus.
Conclusion
PgCYP736A12 does not respond to the well-known secondary metabolite elicitor jasmonic acid, which suggests that it may not function in ginsenoside biosynthesis. Heterologous overexpression of PgCYP736A12 reveals that this gene is actually involved in herbicide metabolism.
Keywords: Cytochrome P450, Gibberellin, Herbicide, Panax ginseng, Phenylurea
1. Introduction
Cytochrome P450 monooxygenases (P450s) are a large group of heme-thiolate proteins expressed ubiquitously in microorganisms, plants, and animals [1]. The number of P450 genes is estimated to be up to 1% of total genes annotated in each plant species [2]. P450s catalyze a wide variety of monooxygenation/hydroxylation reactions. As monooxygenases, P450s insert one atom of oxygen into inert hydrophobic molecules and make them more reactive and hydrosoluble [1], [3]. Beside their physiological roles in hormone biosynthesis, lipids, and secondary metabolites, P450 enzymes function to tolerate harmful exogenous chemicals such as herbicides [3], [4].
Herbicides are one class of xenobiotics that are commonly used to control the growth and reproduction of unwanted vegetation [4]. Herbicide resistance mechanisms are divided into three phases: phase (I), structural changes of the herbicide's target site; phase (II), metabolic detoxification (conjugation) and alternation of uptake; and phase (III), compartmentalization of herbicides [4]. P450s play a primary role in the phase I metabolism of herbicides. The phenylurea inhibitor of photosystem II, the herbicide chlorotoluron, has been used for selective weed control in wheat and can be detoxified via ring-methyl hydroxylation and N-demethylation [3], [5].
After the first report of Frear et al[6], who demonstrated metabolism of the herbicide monuron by a mixed-function oxidase from microsomal fractions of cotton seedlings, an extensive number of reports support the involvement of P450s in the detoxification of herbicides [1]. Modern herbicides contribute to global food production by easily removing weeds [7]. The enzymatic activity of P450s is believed to represent one mechanism that enables certain crop plants to be more herbicide tolerant than other crops or weed species. Up to now, there have been several reports that P450s are involved in herbicide detoxification. A trans-cinnamic acid hydroxylase (CYP73A1) isolated from wound-induced Jerusalem artichoke (Helianthus tuberosus) tuber and expressed in yeast catalyzed the ring-methyl hydroxylation of chlorotoluron [8]. CYP71A10 isolated from soybean leaves conferred enhanced tolerance to linuron and chlorotoluron when overexpressed in tobacco [1]. CYP76B1 isolated from Jerusalem artichoke actively metabolized the phenylurea herbicides, chlorotoluron and isoproturon [9], [10]. CYP81B1, which was also isolated from Jerusalem artichoke, exhibited relatively low levels of chlorotoluron ring-methyl hydroxylase activity [11]. CYP76C1 derived from Arabidopsis also conferred tolerance to chlorotoluron and isoproturon, when overexpressed in native Arabidopsis plants [12].
To shed more light on our understanding of the molecular mechanisms by which plants use P450s to metabolize phenylurea herbicides, expressed sequence tag (EST) clones obtained from methyl jasmonate (MeJA)–treated adventitious roots from ginseng [13] were used to retrieve full protein sequences from the Panax ginseng genome database (http://ginsengdb.snu.ac.kr/blast/blast.php). Overexpression of a ginseng-derived P450 gene designated PgCYP736A12 in Arabidopsis plants showed herbicide-metabolizing activity. In this study, we describe the molecular cloning and expression of PgCYP736A12 that is able to metabolize the phenylurea herbicide chlorotoluron. We also analyzed abiotic stress responsiveness of PgCYP736A12 and effects of its overexpression on growth and development.
2. Materials and methods
2.1. Plant materials and growth conditions
The National Institute of Horticultural and Herbal Science of Rural Development Administration in Eumsung, Korea, provided seeds and two-year-old plants of Korean ginseng (P. ginseng Meyer) ‘Chun-Poong’. Ginseng tissues (stems, leaves, roots, petioles, and rhizomes) were collected from the two-year-old plants. The Columbia ecotype (Col-0) of Arabidopsis thaliana was used as a heterologous model system. Mature Arabidopsis seeds were surface sterilized for 2 min with 70% (v/v) ethanol and rinsed three times with sterile water. Sterilized seeds were sown on 1/2 Murashige and Skoog (MS) medium (Duchefa Biochemie, Haarlem, Netherlands) containing 1% sucrose, 0.5 g/L 2-[N-morpholino] ethanesulfonic acid, and 0.8% phytoagar, of pH 5.7, adjusted with 1 M KOH. Seeds that were cold-treated for two days were germinated under long-day conditions of 16 h light/8 h dark at 23°C in a growth chamber.
2.2. Sequence analysis and construction of a phylogenetic tree
The full cDNA sequences derived from EST clones [13] were confirmed by basic local alignment search tool (BLAST) analysis against the ginseng genome database (http://ginsengdb.snu.ac.kr/blast/blast.php) and by homology-based polymerase chain reaction (PCR) using previously reported CYP gene sequences [14]. The amino acid sequence was analyzed using online programs: (https://blast.ncbi.nlm.nih.gov/Blast.cgi) for gene search, (https://www.ebi.ac.uk/Tools/psa/emboss_needle/) for pairwise sequence alignment, and the ProtParam tool (https://web.expasy.org/protparam/). Multiple sequence alignment was performed using the BioEdit program (version 7.1.9, Copyright ©1997-2017 Tom Hall). A phylogenetic tree was generated by the neighbor-joining method using the MEGA6 (version 6.06) program.
2.3. Abiotic stresses and phytohormone treatment in ginseng
Three-week-old ginseng plantlets (‘Chun-Poong’) were treated with several abiotic stresses and two defense-modulating plant hormones, as reported previously [15], [16]. The ginseng plantlets were placed on a petri dish by dipping the roots in a 60-mL solution containing the following concentrations of chemicals: 100 μM of abscisic acid (ABA), 10 mM of hydrogen peroxide (H2O2), 0.2 mM of jasmonic acid (JA), 5 mM of salicylic acid (SA), and 100 mM of NaCl. The following were used as solvents: methanol for JA and distilled water for SA, ABA, NaCl, and H2O2. Chilling stress (4°C) was induced by soaking the roots in tap water. All the treated plantlets with the corresponding control sample at 0 h treatment were collected at intervals of 1 h, 4 h, 12 h, 24 h, and 48 h after treatments. The samples were immediately frozen in liquid nitrogen and stored at −70°C.
2.4. RNA isolation and quantitative real-time polymerase chain reaction
Total plant RNA was isolated using the RNeasy plant mini kit (Qiagen, Valencia, CA, USA) as per the manufacturer's instructions with modifications. DNase I (Takara, Japan) treatment was included before the washing step in a total reaction volume of 100 μL for 1 h, to remove contaminated DNA. The RNA concentration and quantity were determined using a Nano-MD UV-Vis spectrophotometer (Scinco, Seoul, Korea). First-strand cDNA was synthesized using 4 μg of total RNA and RevertAid Reverse transcriptase (Thermo, USA) following the manufacturer's instructions. Quantitative real-time polymerase chain reaction (q-PCR) was carried out using the Thermal Cycle Dice real-time PCR system (Takara, Shiga, Japan) where 2 μL of cDNA was used in a total reaction volume of 20 μL, as per the manufacturer's instructions. Thermal cycler conditions were as follows: initial denaturation at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 50°C for 30 s, and additional 95°C for 15 s, 60°C for 30 s, and 95°C for 15 s for dissociation. A dissociation curve was produced to evaluate the generation of by-products at the end of the q-PCR. To determine the absolute fold difference in expression for each sample, the threshold cycle (Ct) value of each sample was normalized to that of β-actin (DC03005B05) and calculated relative to a calibrator, using the 2–△△Ct method. Experiments were conducted at least three independent times. The gene-specific primers for PgCYP736A12 were 5ʹ-TCTTAG TGCAAAGGTGGCGA-3′ (forward) and 5′-TTGTCAAGTCTCTTGCCGGT-3′ (reverse). Control primers for ginseng β-actin (DC03005B05) were 5′-AGAGATTCCGCTGTCCAGAA-3′ (forward) and 5′-ATCAGCGATACCAGGGAACA-3′ (reverse). Other primer sequences that were used are listed in supplementary Table 1.
2.5. Vector construction and in planta transformation
Full-length cDNA sequence of PgCYP736A12 was overexpressed under the control of a 35S promoter using modified pCAMBIA1390 where enhanced cyan fluorescence protein (ECFP) was integrated [17]. Full-length PgCYP736A12 cDNA was amplified using primers with Sal I (for forward) and EcoRI (for reverse) restriction sites that are underlined: 5′-GC GTC GAC ATG TTC CCC TTA GCA TAT-3′ and 5′-CG GAA TTC AAG GCG ATA TTT AGG CAC-3′. The purified PCR product of PgCYP736A12 was cloned into the pCAMBIA1390 vector using Sal I and Eco RI restriction enzyme sites. The sequences of all transgene constructs were confirmed by nucleotide sequencing, before being used to transform Arabidopsis. These constructs were transformed into Arabidopsis Col-0 by dipping floral buds, using Agrobacterium tumefaciens C58C1 (pMP90) [18]. Transformants were selected on hygromycin-containing plates (50 μg/mL), and more than 20 independent T1 lines were obtained. Homozygous transgenic lines carrying one copy of the insertion and following a Mendelian segregation ratio were further characterized.
2.6. Confocal microscopy analysis
For subcellular localization of PgCYP736A12-ECFP, the fluorescence emitted from reporter proteins was observed by confocal laser scanning microscopy (TCS SP5 AOBS/Tandem; Leica Germany). ECFP was detected using 436/20 and 480/40 nm excitation/emission filter sets. Images were acquired at the Korea Basic Science Institute, Gwangju.
2.7. Phenylurea herbicide resistance test
Surface-sterilized seeds were sown on 1/2 MS media containing two phenylurea herbicides, chlorotoluron (chemical abstracts service (CAS) Registry No. 15545-48-9; Cayman, USA) and isoproturon (CAS Registry No. 34123-59-6; Tokyo Chemical Industry, Japan). After two days of cold treatment at 4°C, seedlings were further grown at 23°C under long-day photoperiod conditions of 16 h light/8 h dark. Chlorotoluron and isoproturon were added to the medium at different concentrations for preliminary tests, and a physiological concentration was chosen to show a clear-cut difference in tolerance.
3. Results and discussion
3.1. Isolation of PgCYP736A12 from P. ginseng‘Chun-Poong’
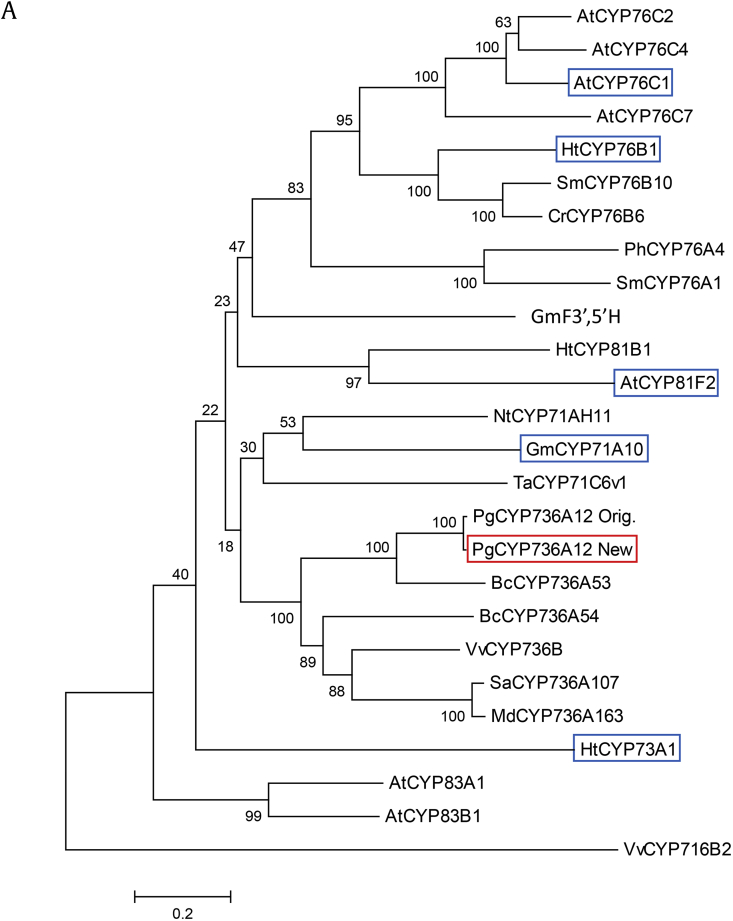
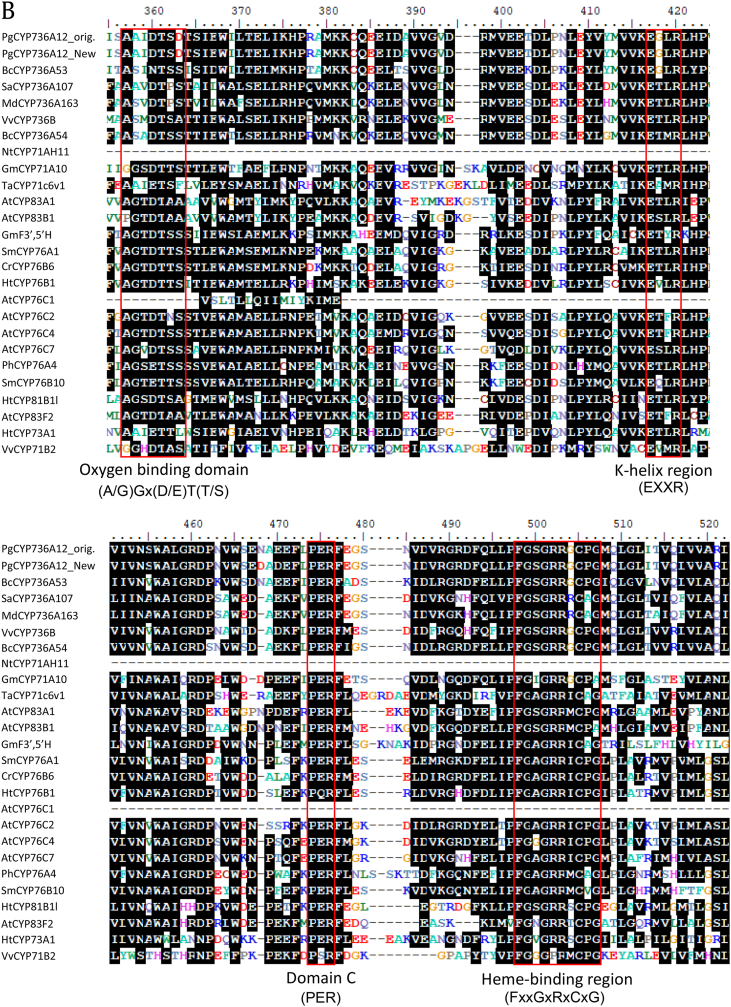
Previously, gene sequences of nine cytochromes P450 (CYP) from P. ginseng Meyer were reported to identify CYP genes involved in ginsenoside biosynthesis [14]. Among nine of them, PgCYP736A12 was not responsive to a major ginsenoside elicitor, MeJA; therefore, it was not selected for further characterization. Because xenobiotics metabolism such as herbicide detoxification is one of the major targets of the P450 family, a full-length cDNA was obtained by homology-based PCR amplification, using ginseng Chun-Poong cultivar as a template. The amino acid sequence of newly amplified PgCYP736A12 (PgCYP736A12 New, hereafter PgCYP736A12) shows 98.8% identity and 99.4% similarity to the previously reported sequence (PgCYP736A12 Orig.) [14], where three amino acids are different (Fig. 1). Full-length cDNA of PgCYP736A12 is 1,500 bp in length, encoding 500 amino acids. The PgCYP736A12 enzyme is predicted to have a molecular weight of 56 kDa and a pI of 7.23, using the ProtParam program [19]. The closest enzyme from the NCBI database was BcCYP736A53, which displays 81% similarity and 65% identity to PgCYP736A12 (Fig. 1A), but the function of the enzyme was not known. Among closely clustered enzymes with PgCYP736A12, functionally characterized CYPs are CYP73A1 and CYP81B1 from Jerusalem artichoke [8], [11] and CYP71A10 from soybean [1]. These three CYP enzymes display 32%, 31%, and 35% amino acid identity, respectively, with PgCYP736A12, and are reported to be involved in the metabolism of herbicides such as chlorotoluron and isoproturon. Typically, CYP enzymes share low sequence identity in their amino acid sequences except several domains [20]. The most well-conserved motifs are the domain C and heme-binding region (Fig. 1B). The heme-binding domain (FxxGxRxCxG), which is also known as the CxG motif where the C (cysteine) binds to the heme group, is well conserved. Two motifs such as the ExxR (K-helix region) and domain C (PER) form the E-R-R triad and play important roles in locking the structure of the heme pocket. The last conserved motif, (A/G)Gx(D/E)T(T/S), also known as the oxygen-binding domain, is less conserved in PgCYP736A12 by changing to (A/G)Ax(D/E)T(T/S) (Fig. 1B). Several closely related CYPs also had a G to A change. However, the biochemical significance of these motifs needs to be clarified.
Fig. 1.
Ginseng-derived cytochrome P450 PgCYP736A12 is closely related to other CYP family proteins. (A) Phylogenetic tree of PgCYP736A12 protein with other closely related cytochrome P450 proteins in plants. The phylogenetic tree was constructed by the neighbor-joining method using the ClustalX program. At, Arabidopsis thaliana; Ht, Helianthus tuberosus; Sm, Swertia mussotii; Cr, Catharanthus roseus; Ph, Petunia hybrid; Sm, Solanum melongena; Gm, Glycine max; Nt, Nicotiana tabacum; Ta, Triticum aestivum; Pg, Panax ginseng; Vv, Vitis vinifera; Bc, Bupleurum chinense; Sa, Sorbus aucuparia, Md, Malus domestica. The gene accession numbers are as follows: AtCYP76C2 (AT2G45570), AtCYP76C4 (AT2G45550), AtCYP76C1 (AT2G45560), AtCYP76C7 (AT3G61040), HtCYP76B1 (Y10098), SmCYP76B10 (GU168041), CrCYP76B6 (AJ251269), PhCYP76A4 (AB016061), SmCYP76A1 (X71658), GmF3′,5′H (GLYMA_13G072100), HtCYP81B1 (AJ000478), AtCYP81F2 (NM_125104), NtCYP71AH11 (GU590868), GmCYP71A10 (GLYMA_06G176100), TaCYP71C6v1 (BAF97103), BcCYP736A53 (JF803814), BcCYP736A54 (AFK79031), VvCYP736B (ACM89789), SaCYP736A107 (AHN09742), MdCYP736A163 (AKB94045), HtCYP73A1 (Z17369), AtCYP83A1 (NM_117451), AtCYP83B1 (NM_119299), VvCYP716B2 (VIT_00035577001). The bar represents 0.1 substitution per amino acid position. PgCYP736A12 New are marked with red square box. Blue square–boxed proteins are reported to be involved in herbicide tolerance. (B) Sequence alignment of PgCYP736A12 with other close homologs. Black boxes indicate identical residues, and similar residues are shaded in gray. The four red-box motifs represent the oxygen-binding domain (A/G)GX(D/E)T(T/S), K-helix region (EXXR), domain C (PER), and heme-binding region (FXXGXRXCXG). Gaps were inserted to maximize homology.
3.2. Organ-specific expression patterns of PgCYP736A12
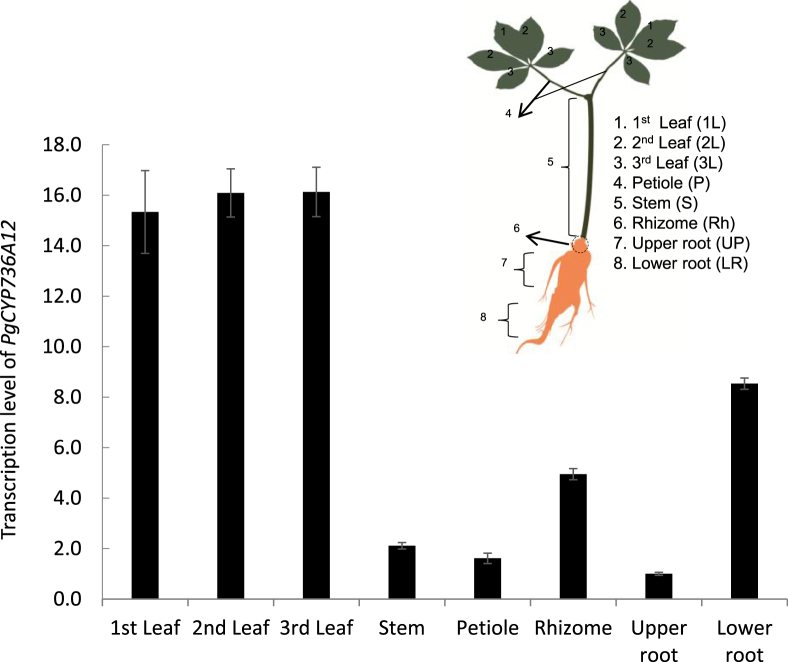
Examination of the fractions of the genome expressed in each tissue type revealed that the percentage of expressed genes varied from tissue to tissue. From two-year-old plantlets, almost all ginseng organs can be obtained [15] except floral inflorescence. To understand the mRNA expression patterns of PgCYP736A12, q-PCR was performed (Fig. 2). On average, three-fold more transcripts were observed from leaves, compared with the rest of the organs. In roots, PgCYP736A12 was expressed nine times higher in the lower part of roots than in the upper part (Fig. 2). The lower part of roots contains meristematic and elongation tissues, which suggests its function in actively dividing parts of tissues.
Fig. 2.
Tissue-specific expression patterns of PgCYP736A12 in fully grown two-year-old ginseng plants. Different levels of transcripts were quantified by q-PCR using cDNA from leaves, roots, rhizomes, and petioles. The data are presented as mean ± SD of three independent replicates. SD, standard deviation; q-PCR, quantitative real-time polymerase chain reaction.
3.3. Abiotic stress–induced differential mRNA levels of PgCYP736A12
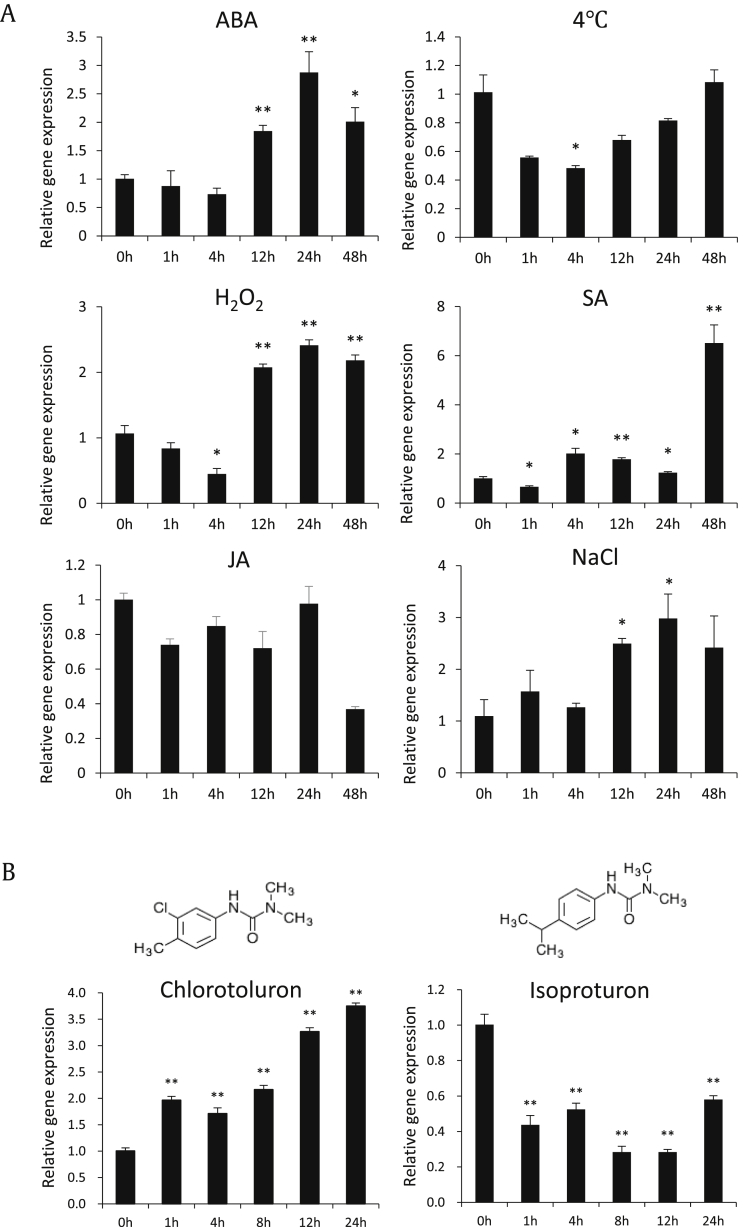
JA and/or MeJA are well known elicitors that can effectively increase the amount of secondary metabolites [21], [22]. PgCYP736A12 was not responsive to MeJA treatment; thus, it was not further selected in deciphering ginsenoside-biosynthesis–related CYP genes [14]. To understand the function of PgCYP736A12, several abiotic stresses and stress-responsive phytohormones such as ABA, SA, and JA were applied to track the responsiveness of PgCYP736A12. In our study, PgCYP736A12 also did not respond to the treatment of JA, which supports previous observations [14]. The transcripts of PgCYP736A12 started to increase at 12 h after treatment with ABA, H2O2, and NaCl, to almost similar levels (Fig. 3). Chilling stress decreased its expression only at 4 h after treatment and was not significant at the rest of the time points tested. However, by treatment with SA, the mRNA level of PgCYP736A12 initially decreased by 40% but increased by an average of 1.7-fold more at 4, 12, and 24 h after treatment (Fig. 3A). At 48 h after SA treatment, transcripts of PgCYP736A12 reached the highest expression levels, 6.5-fold more than those of the control. Taken together, these gene expression studies against various stresses indicate that PgCYP736A12 may be involved in SA-related signaling pathways.
Fig. 3.
Modulation of transcript levels of PgCYP736A12 in response to abiotic stresses and phenylurea herbicides. (A) Transcript levels of PgCYP736A12 for the treatment of abiotic stresses. Three-week-old ginseng plantlets were exposed to abscisic acid (ABA, 100 μM), H2O2 (10 mM), jasmonic acid (JA, 0.2 mM), salicylic acid (SA, 5 mM), NaCl (100 mM), and chilling conditions (4°C), for the time intervals indicated. (B) Transcript levels of PgCYP736A12 against herbicides. Four-week cultivated ginseng adventitious roots were treated with 10 μM of each chlorotoluron and isoproturon for the time intervals indicated and used for the quantification of gene expression. The data are presented as mean ± standard error (SE) of three independent replicates at P < 0.05 (*) and P < 0.01 (**) by Student's t-test, respectively.
3.4. Transcript levels of PgCYP736A12 are differentially regulated by phenylurea herbicides
Based on the previous reports that P450s are involved in herbicide detoxification, transcript levels of PgCYP736A12 were evaluated by treatment of two known phenylurea herbicides, chlorotoluron and isoproturon. These are two substituted phenylureas, chlorinated and unchlorinated, respectively (Fig. 3B, inset). The herbicidal action of these chemicals is based on their ability to inhibit photosynthesis. The mRNA levels of PgCYP736A12 were all upregulated by chlorotoluron during 1 to 24 h of treatment (Fig. 3B), whereas its expression was 58% downregulated in average by isoproturon (Fig. 3B). It indicates that PgCYP736A12 is differentially regulated by two different phenylurea herbicides and may confer herbicide resistance.
3.5. Overexpression of PgCYP736A12 reduces plant height and leads to protein localization in the cytoplasm and nucleus
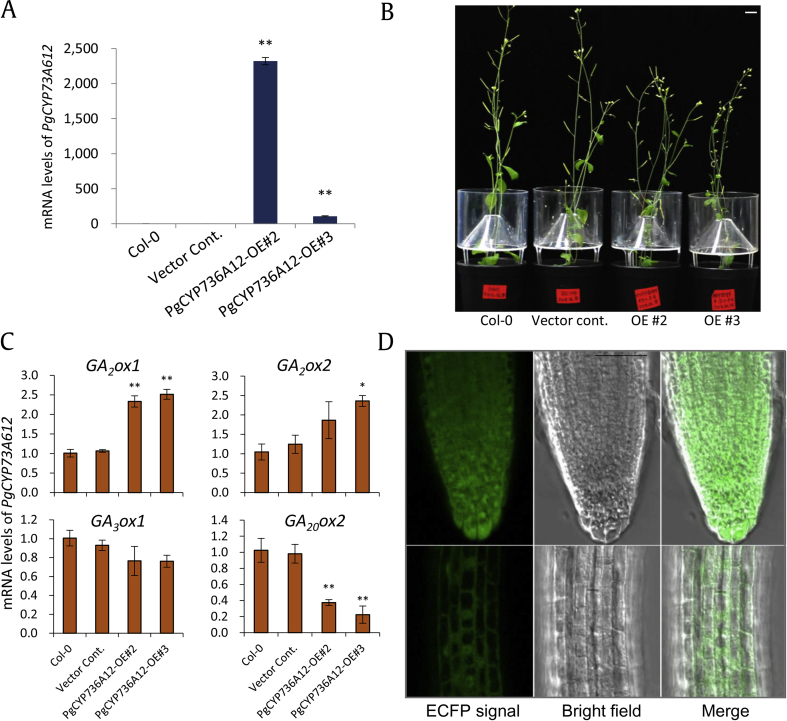
Ginseng transformation has previously been performed using adventitious roots when the final purpose is focused on secondary metabolite accumulation [22], [23]. Here, we used an alternative approach to study the function of ginseng PgCYP736A12. We used the well-known model plant, Arabidopsis. The full-length cDNA sequence obtained from Chun-Poong cultivar was overexpressed in Arabidopsis using a 35S CaMV promoter. Among several transgenic plants selected, two lines that followed Mendelian inheritance were chosen for further analysis. Two independent lines displayed differential heterologous overexpression, compared with the zero levels of two controls (Col-0 and empty vector control) (Fig. 4A). Although the threshold level of transcripts was different (Fig. 4A), both independent transgenic lines displayed similarly reduced plant height (Fig. 4B). However, root elongation was not altered in four-day-old and seven-day-old seedlings (Supplementary Fig. 1A and B). Hypocotyl length in the 4-day-old seedling stage also displayed 11% reduced height compared with that of controls (Supplementary Fig. 1C), which might be resulted in shorter plant height (Fig. 4B). Gibberellins (GAs) are one of the well-studied plant hormones focused on plant stem elongation [24]. Furthermore, cytochrome P450 genes that encode one of key enzymes catalyze steps of the GA biosynthesis pathway from ent-kaurene to GA12 and GA53[25]. Thus, we could not ignore the possible function of PgCYP736A12 in GA biosynthesis pathway based on the stem elongation phenotype. GAs can be stored in plant cells as bioactive and inactive forms. To unravel the possible link with gibberellin biosynthesis, four representative GA monooxygenase genes were analyzed. The expression of two genes GA2ox1 and GA2ox2 that play a role in the conversion of bioactive gibberellins into an inactive form was increased (Fig. 4C). However, GA20ox1 and GA3ox1 genes involved in catalyzing bioactive GA biosynthesis were significantly downregulated (Fig. 4C). These data indicate that the PgCYP736A12-mediated reduction in plant height might be via the reduction of more bioactive GAs. Of course, the possible function of PgCYP736A12 whether its encoding enzyme use biosynthetic intermediates or products of GA catabolism remains to be confirmed by biochemical analysis.
Fig. 4.
Heterologous overexpression of PgCYP736A12-ECFP in Arabidopsis. (A) Transcript levels of PgCYP736A12 overexpressing lines compared with that of Col-0 and vector control. Leaf samples were used for RNA extraction. Data represent the mean ± SE of three independent replicates at P < 0.05 (*) and P < 0.01 (**) by Student's t-test, respectively. (B) Fully grown individual plants representing two transgenic lines are shorter than controls. Bar = 1 cm. (C) Relative gene expression patterns of four gibberellin oxidases (GA2ox1, GA2ox2, GA3ox1, and GA20ox1) in 14-day-old seedlings. Data represent the mean ± standard error (SE) of three independent replicates at P < 0.05(*) and P < 0.01(**) by Student's t-test. (D) Subcellular localization of PgCYP736A12-ECFP in the cytoplasm and nucleus. Confocal microscopy images of enhanced cyan fluorescence protein (ECFP) are in the left panel, bright-field images of the same cells in the middle panel, and the merged images in the right panel. Bar = 500 μm.
The subcellular localization of proteins is very likely indicative of the sites of their catalytic functions. Thus, C-terminal fluorescence protein (ECFP) tagging of PgCYP736A12 protein was analyzed. PgCYP736A12 localized mostly in the cytoplasm and nucleus in the meristematic and elongation zones of Arabidopsis root cells (Fig. 4D). The nuclear localization was more apparent in the elongation zone of Arabidopsis roots (Fig. 4B).
3.6. Overexpression of CYP736A12 confers herbicide tolerance
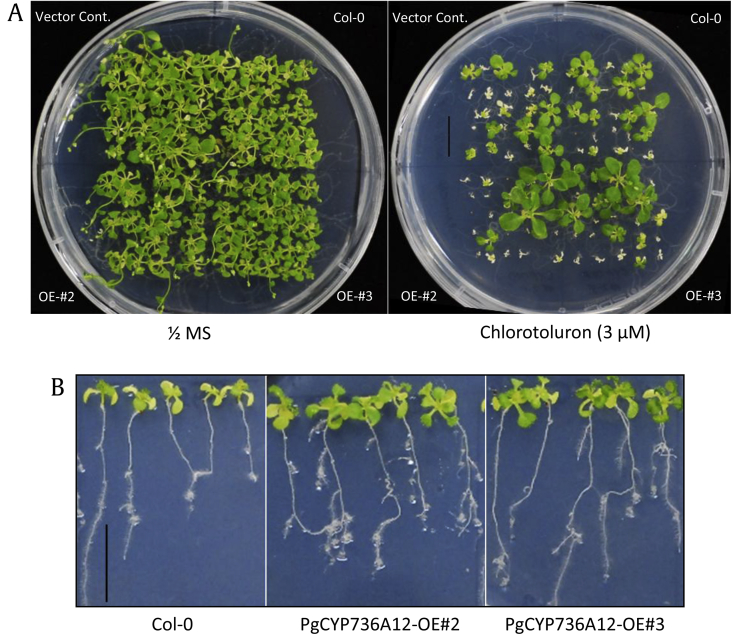
The P450 monooxygenase superfamily is responsible for the metabolism of herbicides representing several classes of organic compounds [4]. Because CYP736A12 might not be involved in ginsenoside biosynthesis [14], possible links with several abiotic stresses was not that apparent, and its mRNA levels were modulated by two substituted phenylureas (Fig. 3B), we tested its function in herbicide tolerance. The normal herbicide detoxification mechanism follows several steps: (1) oxidation by P450 and hydrolysis by carboxylesterases, (2) bonding with reduced glutathione or glucose, catalyzed by glutathione transferases, (3) conjugating transportation into the vacuole, and (4) conjugating degradation in the vacuole or secondary conjugation and incorporation into cell wall components [4], [26]. Two overexpressing lines displayed apparently more tolerance to the phenylurea herbicide, chlorotoluron (Fig. 5). The tolerance was observed from germinated seedlings, which were grown on 1/2 MS media for 25 days, containing 3 μM of chlorotoluron (Fig. 5A). When five-day-old seedlings were transferred onto 1/2 MS media containing 4 μM chlorotoluron, they also displayed tolerance (Fig. 5B), whereas we did not observe any significant phenotypic differences by the treatment of isoproturon. These results explain that the upregulation of PgCYP736A12 transcripts positively involved in chlorinated chlorotoluron tolerance mechanism, but no significant function on unchlorinated phenylurea, isoproturon. Taken together, it is plausible that PgCYP736A12 might have dual function on GA metabolism and herbicide chlorotoluron detoxification. Weeding can improve the health of planting by allowing for better air circulation, reducing competition for resources and reducing cover for pests. Ginseng is a slow-growing perennial oriental herb that grows for more than six years in humus-enriched soils. Thus, special care is required so weeding does not damage the roots of ginseng plants, especially at the seedling stages. In East Asia, ginseng has been cultivated covered with rice straw not only to protect the seedling and small roots during the cold winter but also to minimize weed growth. Because there are no selective herbicides known for use on ginseng, engineering cytochrome P450 can be one of the key determinants of herbicide tolerance and selectivity [3]. Therefore, increases in its expression levels can be responsible for the appearance of herbicide resistance in ginseng, which ultimately led to skip the laborious step in ginseng cultivation by covering rice straw all over the field. Engineering ginseng-derived CYP736A12 in other useful crops also can be utilized for enhanced herbicide tolerance traits.
Fig. 5.
Overexpression of PgCYP736A12 shows herbicide tolerance. (A) Arabidopsis Col-0 and transgenic overexpressing lines of PgCYP736A12 were germinated on 1/2 MS media containing 3 μM of chlorotoluron for 25 days. (B) Col-0 and OE lines were first grown for five days on 1/2 MS media and transferred onto 1/2 MS media in the presence or absence of 4 μM of chlorotoluron. Transferred plants were grown for a further 10 days. Scale bars = 1 cm. MS, Murashige and Skoog.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This study was supported by grants from Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT, and Future Planning, South Korea (grant number: 2016R1A2B4012016 and 2019R1A2C1004140) and from Chonnam National University (grant number: 2018-3380), South Korea.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jgr.2019.04.005.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Siminszky B., Corbin F.T., Ward E.R., Fleischmann T.J., Dewey R.E. Expression of a soybean cytochrome P450 monooxygenase cDNA in yeast and tobacco enhances the metabolism of phenylurea herbicides. Proc Natl Acad Sci. 1999;96:1750–1755. doi: 10.1073/pnas.96.4.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizutani M., Ohra D. Diversification of P450 genes during land plant evolution. Annu Rev Plant Biol. 2010;61:291–315. doi: 10.1146/annurev-arplant-042809-112305. [DOI] [PubMed] [Google Scholar]
- 3.Werck-Reichhart D., Hehn A., Didierjean L. Cytochrome P450 for engineering herbicide tolerance. Trends Plant Sci. 2000;5:116–123. doi: 10.1016/s1360-1385(00)01567-3. [DOI] [PubMed] [Google Scholar]
- 4.Siminszky B. Plant cytochrome P450-mediated herbicide metabolism. Phytochem Rev. 2006;5(2–3):445–458. [Google Scholar]
- 5.Ohkawa H., Tsujii H., Ohkawa Y. The use of cytochrome P450 genes to introduce herbicide tolerance in crops: a review. Pestic Sci. 1999;55(9):867–874. [Google Scholar]
- 6.Frear D.S., Swanson H.R., Tanaka F.S. N-Demethylation of substituted 3-(phenyl)-1-methylureas – isolation and characterization of a microsomal mixed function oxidase from cotton. Phytochem. 1969;8:2157–2169. [Google Scholar]
- 7.Powles S.B., Yu Q. Evolution in action: plant resistant to herbicides. Annu Rev Plant Biol. 2010;61:317–347. doi: 10.1146/annurev-arplant-042809-112119. [DOI] [PubMed] [Google Scholar]
- 8.Pierrel M.A., Batard Y., Kazmaier M., Mignottevieux C., Durst F., Werck-Reichhart D. Catalytic properties of the plant cytochrome-P450 CYP73 expressed in yeast-substrate-specificity of a cinnamate hydroxylase. Eur J Biochem. 1994;224:835–844. doi: 10.1111/j.1432-1033.1994.00835.x. [DOI] [PubMed] [Google Scholar]
- 9.Robinau T., Batard Y., Nedelkina S., Cabello-Hurtado F., LeRet M., Sorokine O., Didierjean L., Werck-Reichhart D. The chemically inducible plant cytochrome P450 CYP76B1 actively metabolizes phenylureas and other xenobiotics. Plant Physiol. 1998;118(3):1049–1056. doi: 10.1104/pp.118.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Didierjean L., Gondet L., Perkins R., Lau S.M., Schaller H., O'Keefe D.P., Werck-Reichhart D. Engineering herbicide metabolism in tobacco and Arabidopsis with CYP76B1, a cytochrome P450 enzyme from Jerusalem artichoke. Plant Physiol. 2002;130:179–189. doi: 10.1104/pp.005801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cabello-Hurtado F., Batard Y., Salaun J.P., Durst F., Pinot F., Werck-Reichhart D. Cloning, expression in yeast and functional characterization of CYP81B1, a plant P450 which catalyzes in-chain hydroxylation of fatty acids. J Biol Chem. 1998;273:7260–7267. doi: 10.1074/jbc.273.13.7260. [DOI] [PubMed] [Google Scholar]
- 12.Höfer R., Boachon B., Renault H., Gavira C., Miesch L., Iglesias J., Ginglinger J.F., Allouche L., Miesch M., Grec S. Dual function of the CYP76 family from Arabidopsis thaliana in the metabolism of monoterpenols and phenylurea herbicides. Plant Physiol. 2014;166:1149–1161. doi: 10.1104/pp.114.244814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim M.K., Lee B.S., In J.G., Sun H., Yoon J.H., Yang D.C. Comparative analysis of expressed sequence tags (ESTs) of ginseng leaf. Plant Cell Rep. 2006;25:599–606. doi: 10.1007/s00299-005-0095-0. [DOI] [PubMed] [Google Scholar]
- 14.Han J.Y., Kim H.J., Kwon Y.S., Choi Y.E. The Cyt P450 enzyme CYP716A47 catalyzes the formation of protopanaxadiol from dammarenediol-II during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 2011;52:2062–2073. doi: 10.1093/pcp/pcr150. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen N.Q., Lee O.R. Overexpression of ginseng UGT72AL1 causes organ fusion in the axillary leaf branch of Arabidopsis. J Ginseng Res. 2017;41:419–427. doi: 10.1016/j.jgr.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen N.Q., Lee S.C., Yang T.J., Lee O.R. cis-Prenyltransferase interacts with a Nogo-B receptor homolog for dolichol biosynthesis in Panax ginseng Meyer. J Ginseng Res. 2017;41:403–410. doi: 10.1016/j.jgr.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee O.R., Kim S.J., Kim H.J., Hong J.K., Ryu S.B., Lee S.H., Cho H.T. Phospholipase A2 is required for PIN-FORMED protein trafficking to the plasma membrane in the Arabidopsis root. Plant Cell. 2010;22:1812–1825. doi: 10.1105/tpc.110.074211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bechtold N., Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- 19.Wilkins M.R., Gasteiger E., Bairoch A., Sanchez J.C., Williams K.L., Appel R.D., Hochstrasser D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;122:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 20.Chen W., Lee M.K., Jefcoate C., Kim S.C., Chen F., Yu J.H. Fungal cytochrome p450 monooxygenases: their distribution, structure, functions, family expansion, and evolutionary origin. Genome Biol Evol. 2014;6:1620–1634. doi: 10.1093/gbe/evu132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizukami H., Tabira Y., Ellis B.E. Methyl jasmonate-induced rosmarinic acid biosynthesis in Lithospermum erythrorhizon cell suspension cultures. Plant Cell Rep. 1993;12(12):706–709. doi: 10.1007/BF00233424. [DOI] [PubMed] [Google Scholar]
- 22.Kim Y.J., Lee O.R., Oh J., Jang M.G., Yang D.C. Functional analysis of 3-hydroxy-3-methylglutaryl coenzyme A reductase encoding genes in triterpene saponin-producing ginseng. Plant Physiol. 2014;165:373–387. doi: 10.1104/pp.113.222596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee O.R., Han J.H., Kim Y. Agrobacterium-mediated transformation of mature ginseng embryos. Bio-protocol. 2014;4:e1362. [Google Scholar]
- 24.Lester D.R., Ross J.J., Davies P.J., Reid J.B. Mendel's stem length gene (Le) encodes a gibberellin 3 β-hydroxylase. Plant Cell. 1997;9:1435–1443. doi: 10.1105/tpc.9.8.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davidson S.E., Reid J.B., Helliwell C.A. Cytochromes P450 in gibberellin biosynthesis. Phytochem Rev. 2006;5:405–419. [Google Scholar]
- 26.Riechers D.E., Kreuz K., Zhang Q. Detoxification without intoxication: herbicide safeners activate plant defense gene expression. Plant Physiol. 2010;153(1):3–13. doi: 10.1104/pp.110.153601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.