Abstract
Background
Korean Red Ginseng (KRG) has been widely used as an herbal medicine to normalize and strengthen body functions. Although many researchers have focused on the biological effects of KRG, more studies on the action mechanism of red ginseng are still needed. Previously, we investigated the proteomic changes of the rat spleen while searching for molecular signatures and the action mechanism of KRG. The proteomic analysis revealed that differentially expressed proteins (DEPs) were involved in the increased immune response and phagocytosis. The aim of this study was to evaluate the biological activities of KRG, especially the immune-enhancing response of KRG.
Methods
Rats were divided into 4 groups: 0 (control group), 500, 1000, and 2000 mg/kg administration of KRG powder for 6 weeks, respectively. Isobaric tags for relative and absolute quantitation was performed with Q-Exactive LC-MS/MS to compare associated proteins between the groups. The putative DEPs were identified by a current UniProt rat protein database search and by the Gene Ontology annotations.
Results
The DEPs appear to increase the innate and acquired immunity as well as immune cell movement. These results suggest that KRG can stimulate immune responses. This analysis refined our targets of interest to include the potential functions of KRG. Furthermore, we validated the potential molecular targets of the functions, representatively LCN2, CRAMP, and HLA-DQB1, by Western blotting.
Conclusion
These results may provide molecular signature candidates to elucidate the mechanisms of the immune response by KRG. Here, we demonstrate a strategy of tissue proteomics for the discovery of the molecular function of KRG.
Keywords: Immune response, Korean Red Ginseng, LC-MS/MS, Molecular signature, Proteomics
1. Introduction
Panax ginseng Meyer (Araliaceae) is a representative indigenous plant known as Korean ginseng. It has been the most highly recognized medicinal plant throughout the long history of oriental medicine [1], [2]. Korean Red Ginseng (KRG) is a processed Korean ginseng which through a processing method enables the ginseng to be stored longer and increases its efficacy and safety [3]. Adaptogen is a unique class of healing plants that help the body to normalize in times of increased stress [4]. KRG is one of the common adaptogens considered to be relatively safe even in large amounts and over a long-term administration. KRG can improve the response to stress with its adaptogenic activities, enhancing immune function [5] and antioxidant activity [6], improving memory [7] and blood circulation [8], and fatigue recovery [9]. According to diverse pharmacological research and clinical trials, KRG received approval for 6 claims as a health functional food from the Korean Food and Drug Administration.
Recently, challenges and opportunities related to ginseng were suggested with diverse pharmacological activities. It was highlighted that a high-throughput, multidisciplinary approach should be developed to bring new insights into the molecular actions of KRG and how these multiple, distinct signaling networks are interconnected [10]. Although many studies have examined the immunomodulatory properties of ginseng, most of them are limited to the phenotypic changes at the cellular level, and only a few studies have investigated the alterations at the molecular level [11]. It was because natural products have multiple components and targets, and it probably is difficult to understand what really is happening in a biological system. Even though many KRG studies on immune responses have been performed, most of them were based on a disease model related to immune suppression. Only one study evaluated the immune-related function of KRG in a normal state [12]. Therefore, further in-depth studies on the molecular level are needed. With the advent of the development of mass spectrometry technology, proteomic studies have helped researchers to examine what is actually happening inside the cell [13]. Currently, multiomic strategy systems have been used to study natural products [14], [15], [16], [17], [18].
Previously, we investigated the proteomic changes of the rat spleen by the administration of 300 mg/kg KRG for 6 weeks. The proteomic analysis revealed that deferentially expressed proteins (DEPs) were involved in the increased immune response and phagocytosis (data not shown). The aim of this study was to evaluate the biological activities of KRG, especially the immune-enhancing response. Rats were divided into 4 groups: 0 (the control group) and 500, 1000, and 2000 mg/kg administration of KRG for 6 weeks, respectively.
In this study, we used proteomics to identify the molecular signatures and functionality of KRG during the course of understanding its underlying mechanisms. Isobaric tags for relative and absolute quantitation (iTRAQ) was performed with Q-Exactive LC-MS/MS for comparison of associated proteins between the groups. The putative DEPs were identified through the current UniProt rat protein database search and Gene Ontology annotations. Furthermore, we utilized the analysis of functional enrichments and Gene Ontology annotations to evaluate the biological effects of KRG using bioinformatic analysis.
2. Materials and methods
2.1. Preparation of the KRG water extract and general chemicals
The extraction procedure for the KRG followed the international standard production process (ISO 19610). The six-year-old P. ginseng root extract (body 75% and root 25%) was prepared by repeated stemming and drying process from the Korea Ginseng Corporation (Daejeon, Republic of Korea). The extract was freeze-dried, and finally, we obtained a dark-brown powder (KRG). Ammonium bicarbonate, dithiothreitol (DTT), formic acid (FA), trifluoroacetic acid, ammonium formate, and urea were purchased from Sigma-Aldrich (St Louis, MO, USA). The HPLC-grade acetonitrile (ACN) and water were purchased from JT Baker (Phillipsburg, NJ, USA). Lyophilized trypsin and lys-C were obtained from Promega (Madison, WI, USA) and Waco (Osaka, Japan). Antibodies against HLA-DQB1/B2, MPO, vimentin, LCN2, BANF1, and GAPDH were purchased from Abcam (Cambridge, MA, USA) as well as HLA-DQB1 from Mybiosource (San Diego CA, USA). Antibodies directed against CRAMP (CAMP), RT1-B, and actin were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
2.2. Animal model
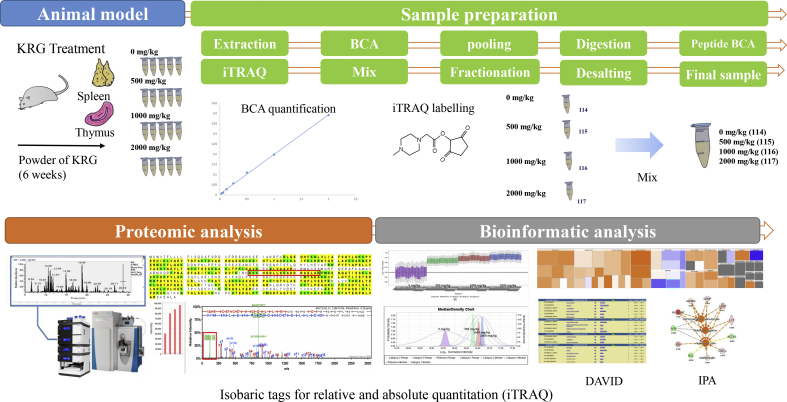
All experiments were conducted with the approval of the Institutional Animal Care and Use Committee of the Korean Ginseng Research Institute (Daejeon, Republic of Korea) in accordance with the Guide for the Care and Use of Laboratory Animals. The male Sprague–Dawley rats (7 weeks old, 200 ± 20 mg, Samtaco, Gyeonggi, Republic of Korea) were acclimatized at the Korean Ginseng Research Institute animal facility for 1 week before the experiment. They were housed at room temperature under humidity (36.2 ∼ 56.3 %) in a standard 12-h light/dark cycle. The 8-week-old male rats were randomly divided into 6 rats per groups. The rats were administered KRG (0, 500, 1000, and 2000 mg/kg) for 6 weeks. Only a vehicle was used for the negative control group (0 mg/kg) with a diet of animal chow and tap water. The body weight of each animal was measured at the start of the administration, weekly thereafter, and finally on the day of their scheduled sacrifice. After dissection, the weight of the rat spleen and thymus tissues were measured, and the tissues were stored at −70°C. An overview of the methods and procedures used is shown Fig. 1.
Fig. 1.
Schematic of the experiments: procedures and methods used for the molecular signature proteomics for the biological effects by Korean Red Ginseng. iTRAQ, isobaric tags for relative and absolute quantitation; BCA, bicinchoninic acid; KRG, Korean Red Ginseng; DAVID, Database for Annotation, Visualization and Integrated Discovery; IPA, Ingenuity Pathway Analysis.
2.3. Protein extraction
The spleen and thymus tissues each were washed with 1X phosphate buffered saline (PBS) (Welgene, Daegu, Republic of Korea) in 1X protease inhibitor (Thermo Fisher Scientific, Rockford, IL, USA) 2 times for 5 min. While on dry ice, each tissue was minced using a scalpel, and the pieces were transferred to 1 mL of 1X RIPA buffer (Thermo Fisher Scientific) containing 1X protease inhibitor (Thermo Fisher Scientific) and 1X phosphatase inhibitor (Roche, PhosSTOP Easy pack, Mannheim, Germany). Then, each sample was ultrasonicated. Next, the supernatant was obtained by centrifugation at 4°C and 8,000 g for 10 minutes. Bicinchoninic acid (BCA) quantification was performed with the Micro BCA Protein Assay Kit (Thermo Fisher Scientific).
2.4. Protein digestion and iTRAQ labeling
Pooled samples of the same amount of protein were treated as previously described [19]. Briefly, DTT (final concentration 10 mM) was added to the sample and then incubated at 70°C for 1 h. Iodoacetamide was added to the sample (final concentration 30 mM), and the mixture was incubated for 30 minutes at room temperature in the dark. After incubation, the peptide mixtures were diluted to 1:10 with 50 mM ammonium bicarbonate, and lys-C solution was added. Trypsin digestion took place at 37°C for additional overnight incubation (1:100). The desalting step was performed by activating a Macro SpinColumn (C-18; Harvard Apparatus, Holliston, MA, USA) according to the manufacturer's instructions. The sample was dried under a vacuum in the Centrivap (Labconco, Kansas City, MO, USA), and peptide quantification was performed with BCA before the iTRAQ labeling. One hundred micrograms of the calculated peptide for each group were used in the 4-plex iTRAQ (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions and a previous report [19]. Subsequently, the iTRAQ-labeled peptide sample was loaded onto a column for High pH Reversed-Phase Fractionation using a Dionex UltiMate 3000 UHPLC+ system (Thermo Scientific, Germering, Germany) following the method [20]. Briefly, XBridge C-18 column (ZORBAX, 4.6 × 250 mm, 5 μC, 300 Å; Waters, Milford, MA, USA) was equilibrated with 10 mM ammonium formate in water (pH 10). The digested peptides were separated into 12 fractions by a linear gradient fractionation (0 to 40 %) at a flow rate of 0.5 mL/min with 10 mM ammonium formate in 90% ACN (pH 10). The eluted samples were taken to dryness in the Centrivap.
2.5. Protein identification using nano-LC mass spectrometry
The peptide samples were analyzed using Q-Exactive mass spectrometry (Thermo Fisher Scientific, Bremen, Germany) equipped with a nano-UHPLC Dionex system (Thermo Scientific) using an Easy nanospray source. Peptides were resuspended in Solvent A (0.1% FA in water) and loaded onto 75 μm ID × 50 cm EASY-Spray column (Thermo Fisher Scientific, San Jose, CA, USA) equilibrated with Solvent A and separated with a linear gradient from 2 to 35% Solvent B (0.1% FA in ACN). The time program was as follows: 0-12 min, 2%; 12-14 min, 2-5%; 14-164 min, 5-35%; 164-167 min, 35-70%; 167-187 min, 70%; 187-190 min, 70-2%; 190-210 min, 2% Solvent B. Full scan of mass spectra was acquired over 350-1400 m/z at 70,000 resolution (maximum IT: 100 ms, automatic gain control (AGC) target: 3 Ⅹ 106). Data-dependent acquisition was performed with the top 10 most abundant precursor ions by the higher energy collision-induced dissociation (HCD) fragmentation mode using an isolation width of 2.0 Da, collision energy of 32, and a resolution of 17,500. First mass was fixed at 100 m/z for iTRAQ analysis with dynamic exclusion option (20 s) to minimize redundant MS/MS collection. Spray voltage was 2.0 kV.
2.6. Data search, statistical analysis, and bioinformatic analysis
Collected raw files were converted into mzXML files through the Trans Proteomic Pipeline (Seattle Proteomic Center, Seattle, WA, USA). The peptides were assigned by the SEQUEST algorithm (Thermo Fisher Scientific) against the decoy UniProt database (UniProt Release June 2016; 75052 entries; UniProt, http://www.uniprot.org/) under Sorcerer (Sage-N Research, Milpitas, CA, USA). All searches were carried out based on trypsin specificity, allowing two missed cleavages. Carbamidomethylation of cysteine was set as fixed, and oxidation of methionine, and N-terminal and lysine iTRAQ modifications were set as variable modifications. The search considered a precursor ion mass tolerance of 10 ppm, and the fragment ion mass tolerance was set to 1.0 Da. Peptide identifications were accepted if they could be established at greater than 95.0% probability by the PeptideProphet algorithm [21] with Scaffold Q+ (Proteome Software, Portland, OR, USA) delta-mass correction, (probability: protein ≧ 95∼99.9%, peptide ≧ 95%, 2 minimum peptide). Scaffold Q+ was also used to calculate the quantification of the values for each protein. The intensities of the multiplexed iTRAQ reporter ions from triplicate experiments were normalized in the program. To identify DEPs, a permutation test with the Benjamini–Hochberg correction was applied to assess whether the protein abundance is differentially expressed between the different quantitative categories. Through the use of the normalized intensities of the iTRAQ reporter ions, KRG-treated group/vehicle ratios of the peptides at 500 mg/kg (115/114), 1000 mg/kg (116/114), and 2000 mg/kg (117:114) were calculated for each replicate. At the end, we applied the Mann–Whitney test for the statistical evaluation. Based on the iTRAQ quantification values, the annotation of the protein cellular localization and the evaluation of the biological function were performed using the Ingenuity Pathway Analysis (IPA) tool (Ingenuity Systems; Redwood City, CA, USA). To identify functional annotations represented by the DEPs, we performed functional annotation analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david-d.ncifcrf.gov/).
2.7. Validation of molecular signature candidate proteins by western blot analysis
According to the animal and protein extraction section in the Materials and methods section, we recollected the same tissues from male rats. The proteins were extracted for validation of the molecular signature candidates. Thirty micrograms of the protein samples were dissolved in lithium dodecyl sulfate (LDS) sample buffer (Invitrogen, Carlsbad, CA, USA) containing 50 mM DTT and heated to 95°C for 5 min. The samples (5 samples for each group) were loaded onto a 4-12% Bis-Tris Bolt gel (Invitrogen) and electrophoresed at 100 V for 60 min, at room temperature. Proteins were transferred to a nitrocellulose membrane at room temperature (RT) using the Invitrogen iBlot Dry Blotting System blot module based on the manufacturer's recommendations (Invitrogen). The transfer efficiency was confirmed by Ponceau S (GenDEPOT, TX, USA) staining of the membrane. Membranes were blocked for one hour at RT or overnight at 4 °C using blocking buffer with 5% skim milk (Sigma-Aldrich) in tris-buffered saline containing 0.1% Tween 20 (TBST, Thermo). Membranes were incubated in the primary antibody according to the manufacturer's recommendations at 1:100 ∼ 1000 dilution in blocking buffer overnight at 4°C. The blots were washed 3 times with TBST for 5 min and then incubated in the appropriate secondary antibody conjugated to horseradish peroxidase (HRP) for one hour at room temperature. Blots were then washed with TBST again 3 times, 5 min each. Chemiluminescent detection was accomplished using the ECL kit (GenDEPOT) and a Davinch-Chemi imaging system digital imager (Davinch-K, Seoul, Republic of Korea).
3. Results
3.1. Overall characteristics of the biological changes
No mortality and notable clinical signs were observed during the 6 weeks, and there were no statistically significant changes in body weight and food consumption compared with the control group. No significant organ weight changes were observed in either the absolute or the relative organ weights (Supplemental Table 1).
3.2. Identification of proteins in rats administered KRG
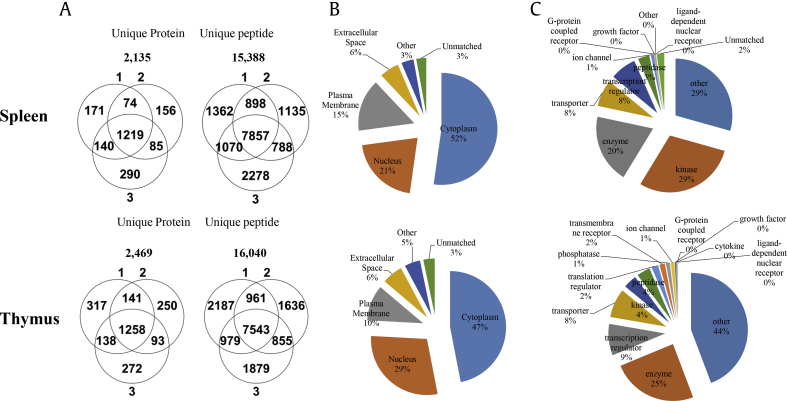
To understand the possible mechanisms and molecular signatures responsible for the effects of KRG, we performed global proteome profiling of the spleen and thymus tissues obtained from the rat model to identify differentially expressed proteins (DEPs) between 4 groups. For quantitative analysis with protein identification, we labeled the aforementioned samples using iTRAQ agents: control samples (vehicle, 114) and 500, 1000, and 2000 mg/kg after oral administration of KRG (115, 116, and 117, respectively) once every day in the morning during the 6 weeks. A total of 2,135 unique proteins were identified by the MS analysis for the spleen and 2,469 for the thymus (protein: probability>99.9%; peptide: probability>95%) with the target-decoy database (UniProt rat database) using the Sequest search engine in the Sorcerer system (Fig. 1). To understand the cellular distribution and functions of the identified proteins, we performed an analysis for both the spleen and thymus using the IPA tool. It was found that most proteins were associated with either the cytoplasm (47-52%) or nucleus (21-29%), with fewer associated with the plasma membrane (10∼15%) shown in Fig. 2B. Analysis of the molecule types revealed that they belong to kinases (29%) in the spleen and enzymes (25%) in the thymus using the IPA tool. (Fig. 2C and Supplemental Table 2-1–2-2) Next, the quantitative values of the iTRAQ were determined for the DEPs. (Supplemental Table 3-1–3-2) Among the DEP pool between the normal and KRG-treated samples, we further selected DEPs whose changes showed upregulated/downregulated trends which were greater than those of the log2 values in Table 1. (|log2x| > 0.2 in spleen and > 0.13 in the thymus).
Fig. 2.
Distribution of the identified proteins in the spleen and thymus in the rat. (A) Total intersection of unique proteins and peptides for all the groups. (B) Subcellular localization of the identified proteins and (C) molecule types of the identified proteins by the Ingenuity Pathway Analysis (IPA) tool.
Table 1.
Selected biosignature candidate proteins identified in the spleen and thymus of the rat
| Identified proteins of the spleen | Accession No. | Log2 value (mg/kg) |
Identified proteins of the thymus | Accession No. | Log2 value (mg/kg) |
||||
|---|---|---|---|---|---|---|---|---|---|
| 500 | 1000 | 2000 | 500 | 1000 | 2000 | ||||
| Ifi47 protein | Q6NYB8 | 0.24 | 0.24 | Myosin-4 | Q29RW1 | 1.05 | 0.21 | ||
| Protein Mpo | A0A0G2K1A2 | 0.25 | 0.30 | Tropomyosin beta chain | P58775 | 0.88 | 0.61 | ||
| Neutrophilic granule protein (predicted) | D3ZY96 | 0.26 | 0.27 | Myosin light chain 1/3, skeletal muscle isoform | P02600 | 0.87 | 0.44 | ||
| Cytochrome c, somatic | P62898 | 0.26 | 0.57 | Creatine kinase M-type | A0A0G2JSP8 | 0.56 | 0.38 | ||
| Hemoglobin subunit alpha-1/2 | P01946 | 0.23 | 0.28 | Parvalbumin alpha | P02625 | 0.33 | 0.53 | ||
| Neutrophil gelatinase–associated lipocalin | P30152 | 0.21 | 0.47 | 0.57 | Protein S100-A11 | Q6B345 | 0.24 | 0.57 | 0.26 |
| Isoform 2 of adenylate kinase 2, mitochondrial | P29410-2 | 0.37 | 0.24 | 0.71 | Niemann–Pick type C2 | F7FJQ3 | 0.23 | 0.21 | |
| Oxygen-dependent coproporphyrinogen-III oxidase, mitochondrial | Q3B7D0 | 0.22 | 0.21 | 0.24 | Myosin light chain 4 | M0R4E1 | 0.19 | 0.30 | |
| Rano class II histocompatibility antigen, B-1 beta chain | P29826 | 0.26 | 0.20 | ATP synthase-coupling factor 6, mitochondrial | P21571 | 0.15 | 0.39 | ||
| MHC class Ia protein | O19446 | 0.30 | 0.22 | Rano class II histocompatibility antigen, B-1 beta chain | P29826 | 0.14 | 0.21 | 0.14 | |
| Vimentin | P31000 | 0.37 | 0.24 | 0.71 | Prolargin | Q9EQP5 | 0.14 | 0.16 | 0.12 |
| Galectin-5 | P47967 | 0.45 | 0.35 | 0.55 | Protein Prkcsh | B1WC34 | 0.12 | 0.31 | 0.16 |
| Ubiquitin-40S ribomal protein S27a | P62982 | 0.20 | 0.35 | Regulator of G-protein signaling 10 | P49806 | 0.10 | 0.35 | 0.17 | |
| Histone H4 | P62804 | 0.21 | 0.30 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial | D3ZCZ9 | 0.10 | 0.20 | 0.14 | |
| Protein Wdr75 | A0A0G2K9A7 | 0.43 | 0.27 | Thymosin beta-4 | P62329 | 0.46 | 0.19 | ||
| Arachidonate 5-lipoxygenase–activating protein | P20291 | 1.88 | 0.26 | Metallothionein | D3ZHV3 | 0.45 | 0.21 | ||
| Carbonic anhydrase 1 | B0BNN3 | 0.33 | 0.26 | Myristoylated alanine-rich C-kinase substrate | P30009 | 0.37 | 0.19 | ||
| Isoform A2 of Heterogeneous nuclear ribonucleoproteins A2/B1 | A7VJC2-2 | 0.23 | 0.26 | High-mobility group nucleosome binding domain 1 | Q5U1W8 | 0.34 | 0.24 | ||
| RCG45246 | A0A0G2K654 | 0.51 | 0.25 | Protein Sh3bgrl3 | B2RZ27 | 0.26 | 0.28 | ||
| Carbonic anhydrase 2 | P27139 | 0.26 | 0.23 | Granulin, isoform CRA_c | G3V8V1 | 0.26 | 0.20 | ||
| Protein RoBo-1 | O55006 | 0.25 | 0.28 | 0.21 | MARCKS-related protein | Q9EPH2 | 0.23 | 0.18 | |
| Lysozyme f2 | F1M8E9 | 0.29 | 0.21 | Protein Rbm17 | Q6AY02 | 0.21 | 0.17 | ||
| Protein LOC684828 | M0R7B4 | 0.45 | 0.62 | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1 | Q64244 | 0.30 | 0.16 | ||
| Structural maintenance of chromosome protein 1A | Q9Z1M9 | 0.31 | 0.47 | Atrial natriuretic peptide-converting enzyme | Q80YN4 | 0.21 | 0.41 | ||
| Barrier-to-autointegration factor | Q9R1T1 | 0.34 | 0.44 | Isoform 2 of coiled-coil and C2 domain-containing protein 1A | Q66HA5-2 | 0.20 | 0.15 | ||
| Mast cell protease 9 (fragment) | P97595 | 0.22 | 0.25 | D-dopachrome decarboxylase | P80254 | 0.18 | 0.38 | ||
| CRAMP (fragment) | Q71KM5 | 0.25 | 0.22 | Tropomyosin alpha-1 chain | P04692 | 0.15 | 0.43 | ||
| Chromosome segregation 1-like (Saccharomyces cerevisiae) (predicted) | D3ZPR0 | 0.21 | 0.21 | Protein S100-G | P02634 | 0.11 | 0.49 | 0.24 | |
| Beta-arrestin-2 | P29067 | 0.20 | 0.21 | PYD and CARD domain containing | G3V8L1 | 0.10 | 0.45 | 0.21 | |
| CD44 antigen | P26051 | 0.97 | 0.30 | ||||||
| Mitochondrial import inner membrane translocase subunit Tim13 | P62076 | 0.54 | 0.17 | ||||||
| Histone H2A type 1 | P02262 | 0.19 | 0.27 | ||||||
| Lon protease homolog, mitochondrial | Q924S5 | −0.22 | −0.20 | Caveolin-1 | P41350 | −0.10 | −0.33 | −0.17 | |
| Elongation factor Tu, mitochondrial | P85834 | −0.21 | −0.21 | Lamin B receptor | O08984 | −0.12 | −0.18 | −0.17 | |
| ADP/ATP translocase 2 | Q09073 | −0.21 | −0.27 | Dihydrolipoyl dehydrogenase, mitochondrial | Q6P6R2 | −0.12 | −0.19 | −0.16 | |
| Actin, aortic smooth muscle | P62738 | −0.23 | −0.21 | −0.28 | Cytochrome b-c1 complex subunit 8 | Q7TQ16 | −0.13 | −0.38 | −0.34 |
| Isoform 2 of tropomyin beta chain | P58775-2 | −0.20 | −0.27 | −0.59 | Long-chain-fatty-acid–CoA ligase 1 | P18163 | −0.14 | −0.18 | |
| Uncharacterized protein | M0RCH6 | −0.23 | −0.28 | Fatty acid synthase | P12785 | −0.15 | −0.15 | ||
| Serum albumin | P02770 | −0.22 | −0.39 | Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic | O35077 | −0.16 | −0.20 | ||
| Protein SET | A0A0G2JSU3 | −0.50 | −0.43 | Myosin-6 | P02563 | −0.20 | −0.15 | ||
| Apolipoprotein C–I | P19939 | −0.25 | −0.46 | Succinyl-CoA ligase subunit beta (fragment) | B2RZ24 | −0.24 | −0.14 | ||
| Protein Cd209b | F1LM87 | −0.46 | −0.44 | −0.57 | Isoform 2 of Echinoderm microtubule–associated protein-like 2 | Q6P6T4-2 | −0.12 | −0.25 | −0.26 |
| Nucleolin | P13383 | −0.25 | −0.25 | Signal recognition particle 54 kDa protein | Q6AYB5 | −0.14 | −0.21 | −0.11 | |
| Histone H2B | D3ZWM5 | −0.14 | −0.26 | −0.19 | |||||
| Protein Ncapg | D3ZD72 | −0.18 | −0.24 | −0.10 | |||||
| NADH dehydrogenase (ubiquinone) Fe–S protein 7 | Q5RJN0 | −0.18 | −0.31 | −0.28 | |||||
| Protein Ptges3l1 | Q5PQL9 | −0.18 | −1.54 | −1.30 | |||||
| Period circadian protein homolog 3 | G3V8D9 | −1.02 | −0.48 | −0.99 | |||||
| Proteasome activator complex subunit 2 | Q63798 | −0.17 | −0.14 | ||||||
| Pyruvate dehydrogenase E1 component subunit beta, mitochondrial | P49432 | −0.17 | −0.17 | ||||||
| Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | P08461 | −0.18 | −0.15 | ||||||
| Succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial | P21913 | −0.20 | −0.15 | ||||||
| DNA helicase | D3ZP96 | −0.20 | −0.14 | ||||||
| Actin, aortic smooth muscle | P62738 | −0.29 | −0.26 | ||||||
| Histone H1t | P06349 | −2.32 | −2.60 | ||||||
Representatives of selected upregulated or downregulated DEPs∗ from the spleen and thymus of the KRG-treated rat model. The intensities of proteins were calculated by Scaffold Q+ within triplicate analysis
Log2 % value = Log2 fold change (FC) of the concentration among 500, 1000, and 2000 mg/kg, respectively
∗DEP (differentially expressed protein): Proteins that showed greater |log2x| values than 0.2 and 0.13 in the spleen and thymus, respectively, compared to control (0 mg/kg). These proteins exhibit the same increase or decrease tendency within at least 2 different concentrations among 500, 1000, and 2000 mg/kg (permutation test p < 0.01, Mann–Whitney test p < 0.01), except thymus proteins in gray highlighted box below (permutation test p < 0.01, Mann–Whitney test p < 0.05)
3.3. Functional analysis of the proteins showing a trend
To identify the relevant biological function with the proteins showing upregulated /downregulated trends, we performed enrichment analysis using DAVID (https://david.ncifcrf.gov/home.jsp) for functional annotation, and then, further detailed analysis was executed with IPA. Among DEPs, we selected proteins that showed upregulated or downregulated trends in the spleen and thymus in at least 2 KRG concentration groups. (Table 1) For example, the upregulated proteins in the spleen showed a function associated with gram-positive bacteria in the GOTERM_BP_DIRECT (p-value 7.5 × 10−3) based on the functional annotation clustering tool, which decreased the redundancy of similar annotations: IFI47, MPO, and NPG in the spleen. Furthermore, IPA top disease and biofunction analysis was used for both protein groups. The results identified that DEPs were involved in cellular movement, and cell death and survival in molecular and cellular functions, and hematological system development and function and immune cell trafficking in physiological system development and function. Among the top analysis, we representatively focused on the main disease and biofunctions, which predicted as upregulated/downregulated by the IPA Z-score. The leukocyte migration and immune response of cells were predicted to be increased, whereas bacterial infection function and infection of mammalia were predicted to be decreased. In the thymus, the DAVID analysis showed muscle proteins, and myosin-related proteins (MYL4, Tpm2, MYH4 and MYL1, and PVALB) were upregulated. The IPA analysis showed that the DEPs were involved in lipid metabolism and small molecule biochemistry in the molecular and cellular function analysis. Especially, lipid metabolism was associated with a decreased synthesis of fatty acids. IPA analysis also showed that immune response of cells was predicted to be increased, whereas viral infection and accumulation of lipid were predicted to be decreased. (Table 2)
Table 2.
Selected DAVID and IPA functional analysis of the identified proteins
|
A. | |||
| Spleen | Selected DAVID analysis for functional annotation | p-value | Proteins |
| Defense response | 1.79E-03 | IFI47, MPO, NGP | |
| Defense response to gram-positive bacterium | 6.41E-03 | CAMP, LYZ2, MPO | |
| Defense response to bacterium | 1.30E-02 | CAMP, LYZ2, MPO | |
| Molecular and cellular functions | p-value range | Main related function | |
| Cellular movement | 1.60*10−2 – 2.52*10−6 | Leukocyte migration, cell movement of cell, cell movement of granulocytes | |
| Cellular function and maintenance | 1.78*10−2 – 4.27*10−5 | Phagocytosis, engulfment of cells, cellular homeostasis | |
| Main disease and biofunction | Activation z-score* | Related proteins | |
| Leukocyte migration | 2.194 | LYZ, ALOX5AP, CRAMP, ARRB2, LCN2, ALB | |
| Immune response of cells | 1.274 | VIM, HLA-DQB1, Cd209b, CAMP, LCN2 | |
| Infection of mammalia | −1.246 | HLA-DQB1, Cd209b, CAMP, Ifi47, LCN2 | |
| Bacterial Infections |
−1.225 |
LYZ, ALOX5AP, Cd209b, CAMP, LCN2 |
|
|
B. | |||
| Thymus | Selected DAVID analysis for functional annotation | p-value | Genes |
| Muscle protein | 4.10E-08 | MYH4, MYL1, MYL4, PVLB, TPM2 | |
| Myosin | 6.10E-04 | MYH4, MYL1, MYL4 | |
| Actin binding | 8.10E-03 | MYH4, MARCKS, MARCKS1, TMSB4X, TPM2 | |
| Molecular and cellular functions | p-value range | Main related function | |
| Lipid metabolism | 1.25*10−2 – 1.72*10−8 | accumulation of lipid, synthesis of fatty acid | |
| Small molecule biochemistry | 1.25*10−2 – 1.72*10−8 | metabolism of ATP, synthesis of acetyl-coenzyme A | |
| Main disease and biofunction | Activation z-score | Related proteins | |
| Immune response of cells | 1.699 | PYCARD, CC2D1A, PSME2, CD44, CD38, HLA-DQB1 | |
| Immune response of leukocytes | 1.432 | PYCARD, PSME2, CD44, CD38, HLA-DQB1 | |
| Accumulation of lipid | −1.690 | SDHB,CAV1,CD44,ACSL1,NPC2 | |
| Viral Infection | −1.474 | HIST2H2AA3/HIST2H2AA4, RBM17, PVALB, CKM, NDUFS7, PYCARD, PSME2, HLA-DQB1, HIST1H2BN, PER3, ACTA2, CD44, CAV1, CD38, FGA, ACSL1 | |
Functional annotations of proteins in Table 1 were obtained from DAVID (https://david.ncifcrf.gov/home.jsp) and further detailed analysis was performed with IPA. P-value, which is a probability of associated molecules from experimental data sets, was also calculated by DAVID and IPA, respectively. Z-score is a statistical measure of the match between expected relations and observed protein expression. The Z-score indicates overall increase (+) or decrease (−) of identified proteins in pathway database of IPA
DAVID, Database for Annotation, Visualization and Integrated Discovery; IPA, Ingenuity Pathway Analysis.
3.4. A more detailed functional analysis of the proteins
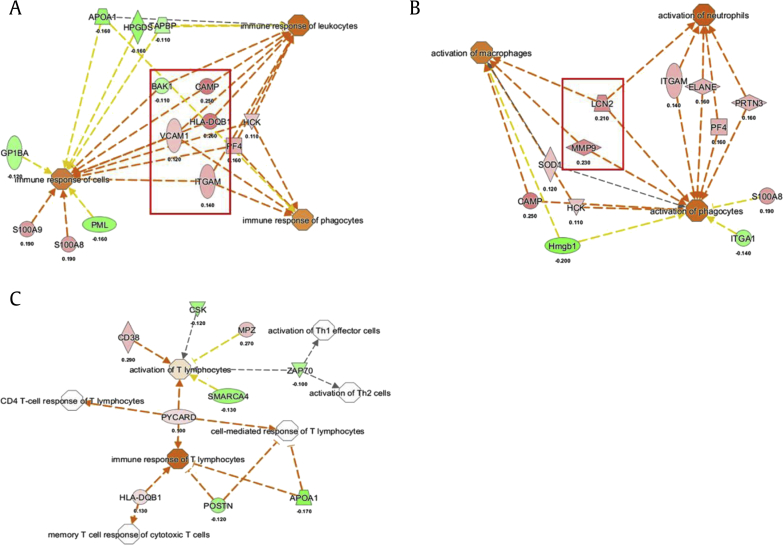
To determine the overall trend and changes of an involved disease and biofunction by KRG, we enlarged the pool of DEPs for each KRG concentration group for the spleen and thymus (permutation test p < 0.01, Mann–Whitney test p < 0.05, log2 value (|log2x| > 0.1) IPA test). In the next step, we analyzed the pools using the IPA tool in two different conditions: (a) focusing on immune function–related immune organs and cell lines because the spleen and thymus are the organs related to the immune system and (b) all organs and cell lines for the overall changes. In a previous study, KRG extract was orally administered to male Sprague Dawley (SD) rats at a dose of 500 mg/kg/day [22] corresponding to the human usual daily dose of KRG [23]. Therefore, we focused on the results of 500 mg/kg/day for the functional analysis of KRG. As a result, the overall obtained immune functions were related to cell movement, recruitment of cells, immune responses, and phagocytosis in the spleen. Especially, VACAM1, ITGAM, HLA-DQB1, HCK, and PF4 were commonly upregulated according to the function of the immune response in the spleen from the 500 mg/kg group (Fig. 3A). CARMP is commonly related to the immune response of leukocytes and phagocytes. In addition, CARMP and HLA-DQB1 showed an increased trend. In addition, MMP9 and LCN2 were commonly identified as activation of immune cells, and CRAMP, and PRTN3 with the related function of immune cells were upregulated as well (Fig. 3B). In the thymus, the IPA analysis showed that DEPs were related to lipid metabolism, and nucleic acid metabolism and small molecule biochemistry. In addition, HLA-DQB1 and PYCARD associated with the immune response of T lymphocytes and CD 38 for the activation of T lymphocytes were upregulated in the 500 mg/kg group. These changes are visualized by a line plot of the log2 fold changes (Fig. 3C).
Fig. 3.
Analysis of immune-related activation components. (A) Selected increased immunological functions and (B) function related to activating immune cells in the spleen and (C) in the thymus by KRG. Orange color, the biological process or disease is trending toward an increase. Blue color, the biological process or disease is trending toward a decrease. The values represent the log2 fold change. KRG, Korean Red Ginseng.
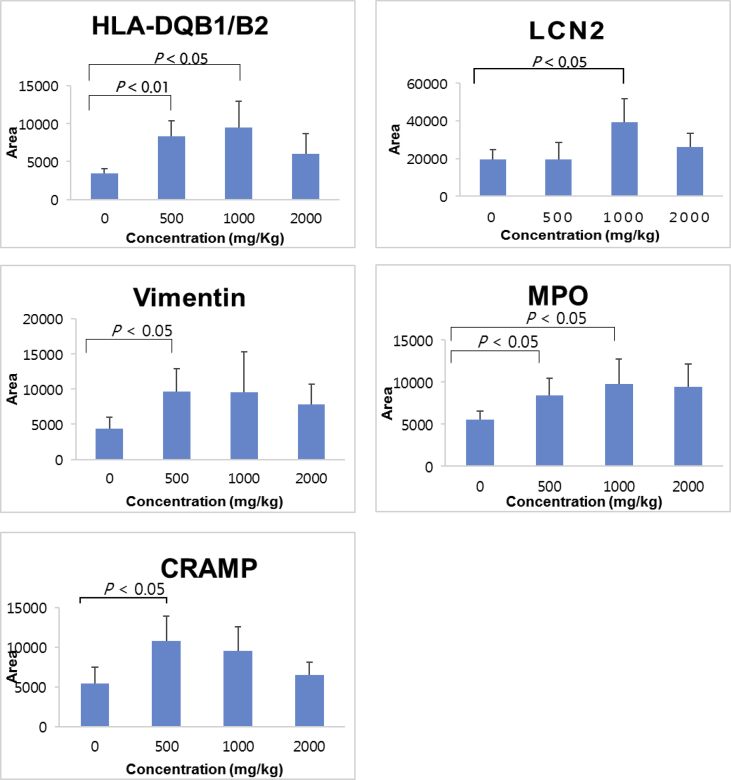
3.5. Verification of the protein changes by Western blotting
We chose as validation target proteins those that were commonly increased in immune-related function (Table 1). CARMP and HLA-DQB1 were selected for validation due to their common increase among the immune response of cells. The LCN2-associated activation of macrophages, neutrophils, and phagocytes and the MPO-associated defense response to bacterium were selected. BANF1 protein was also selected because it is related to immune function. Although vimentin was not related to the immune response directly, vimentin was selected for further validation because of its increase among all the KRG concentrations. We validated the proteomic identification of the proteomic results using antibodies against several biosignature candidates for KRG. Our particular interest was to compare the identification and quantification of the iTRAQ analysis with the results from Western blotting. Through these validation studies, we mainly identified the upregulated proteins from the spleen and the thymus tissues. (Fig. 4) The results indicated that HLA-DQB1/B2, MPO, vimentin, LCN2, and BANF1 in the spleen were increased. Vimentin and CRAMP were significantly increased in the 500 mg/kg group, and HLA-DQB1/B2 and MPO were also increased in both the 500 and 1000 mg/kg groups. LCN2 was increased in the 1000 mg/kg group. Even though all the tested proteins were not significantly increased in all the KRG concentration groups, the tendency of an increasing pattern seemed to increase in both the 500 and 1000 mg/kg groups, while the patterns seemed to be decreased a little bit in the 2000 mg/kg group.
Fig. 4.
Western blot validation of the representative biosignature candidates in the rat spleen. Western blot bands were quantified by Image J (NIH), and the protein levels of the spleen (normalized to actin) from the experiment were expressed as an arbitrary unit. (Quantitative analysis n = 5) HLA-DQB1/B2, LCN2, vimentin, MPO, and CRAMP were tested. Actin was used as a loading control (See Supplemental Fig. 1).
4. Discussion
iTRAQ analysis of the spleen and thymus tissues using LC-MS/MS resulted in the identification of unique proteins. We selected possible candidates for molecular biosignatures which were upregulated or downregulated, respectively, at least in 2 concentration groups. The DAVID analysis revealed the function of defense against gram-positive bacteria in the spleen, and muscle proteins and myosin-related proteins were upregulated. Furthermore, we enlarged the pool of DEPs from the tissues for each KRG concentration, and then, we identified the proteins that were upregulated for immune function. Overall, we identified potential biosignature candidates in the spleen and thymus. Of these, we validated LCN2, CARMP, HLA-DQB1, vimentin, BANF1, and MPO by Western blotting. Our purpose of Western blotting was to validate the identification and quantification results from proteomic profiling. The results demonstrated that proteomic analysis using iTRAQ represented a tendency of positive correlation with that from Western blotting with an increasing pattern in most cases. For example, the result of the Western blot for HLA-DQB1 in Fig. 4 was increased in both the 500 and 2000 mg/kg groups, matching the proteomic analysis of Table 1.
KRG is a well-known immune modulator, and its extracts have been used to maintain immune homeostasis and immune function. Although many studies have observed the immunomodulatory properties of KRG, only a few studies have focused on the alterations caused by KRG at the molecular level [11]. This research is the initial step to investigate the global molecular level of the alterations induced by KRG. Our results also show that KRG might stimulate both innate and acquired immune responses. First, LCN2, MPO, and CRAMP associated with innate immunity were upregulated. LCN2 is a secretory protein and known as a mediator of the innate response to bacterial infection [24]. During infection, bacteria acquire their iron from the host, but LCN2 limits bacterial growth. In addition, macrophages control iron recycling and availability to pathogens [25]. Therefore, LCN2 seems to be important in the innate immune response to bacterial infection. Cathelicidin is a protein-related antimicrobial peptide in polymorphonuclear leukocytes and in monocytes/macrophages [26], [27], [28], [29], [30]. Cathelicidin-related antimicrobial peptide (CARMP) have has a critical role in mammalian innate immune defense and maintenance against invasive bacterial infection [29], [30].
In addition, KRG can also stimulate acquired immunity to stimulate T-cell activation. The major histocompatibility complex (MHC) is essential for the acquired immune system. The main function of the MHC is to bind to antigens derive from pathogens and display them on the cell surface for recognition by the appropriate T cells [31]. The human MHC is called the human leukocyte antigen (HLA) complex and has 3 types. HLA-DQB1 (HLA, class II, DQ beta 1) is one of the proteins in the MHC class II, a cell surface receptor expressed in antigen-presenting cells. This protein is related to immune response [32], peptide antigen binding [33], and so on.
In this experiment, we identified RT1-B, the MHC in rats, which contains genes that code for two class II histocompatibility antigens. The rat RT1-B is equivalent to the HLA-DQ [34]. Even though HLA-DQB1 was not identified directly, the IPA results suggest that HLA-DQB1 instead of RT-B1 is one of the molecular signature candidates for immune response, which is specific for mammal species. The Western blot results show that HLA-DQB1 was increased in the spleen. In addition, HLA-DQB1 was also associated with the immune response in the thymus. The protein was also upregulated as well as RT-B1 in the thymus. However, RT-B1 in the spleen showed no significant changes in the level. These results are probably related to the sequence homology. The Basic Local Alignment Search Tool analysis revealed regions of local similarity between HLA-DQB1 (A0A0P6JUN1) and RT-1B which had 77.4% similarity, and the HLA-DQB1 and the human (A0A0D5XQ77) had 75% similarity. (Supplemental Fig. 2)
The thymus is a primary organ of the immune system in which T cells mature. The thymus is the most active at preadolescence, and the immunological functions decline with aging. Concomitantly, the level of thymic hormones declines in blood as we age [35]. The gradual loss of thymic functions leads to an increased susceptibility to infections, cancer, and autoimmune diseases as we age [36]. It was reported that the thymic attenuation with increasing aging is also reflected in the quantity of peripheral naïve T cells in humans [37]. Our result shows that muscle and filament proteins were increased. These results suggest that KRG might attenuate the aging process by decreasing the thymic stroma.
Meanwhile, several DEPs were downregulated in the spleen and thymus, and the proteins are listed in Table 1. Nucleoin is a phosphoprotein involved in the synthesis and maturation of ribosomes [38]. It plays a vital role in promoting replication in hepatitis delta virus and poliovirus [39]. Lon protease homolog, mitochondrial (LONP1) is a conserved serine peptidase that contributes an important role in the degradation of misfolded and damaged proteins. It was demonstrated that LON1 knockdown inhibits tumor and metastasis formation [40]. Lamin B receptor is an inner nuclear membrane protein associated with the nuclear lamina. It was reported that the lamin B receptor plays an important role in cholesterol biosynthesis, which is essential to both myeloid cell growth and functional maturation [41]. Fatty acid synthase (FAS) is an enzyme that catalyzes the synthesis of palmitate [42]. It was reported that inhibition of FAS prevents the proinflammatory response in macrophages [43]. In addition, it has been investigated as a potential oncogene because of its upregulation in cancers [44], [45], [46]. For the function of KRG, further in-depth studies need to be conducted in the near future.
Granulocyte-macrophage colony-stimulating factor (CSF2) was also predicted to be increased which is a cytokine that stimulates the growth and differentiation of hematopoietic precursor cells [47] by the IPA analysis. It was reported that gene expression of CSF2 was upregulated in the response of peripheral blood mononuclear cell to an aqueous North American ginseng extract in a microarray analysis. In addition, the IFN-γ gene, coding for the primary Th1 cytokine, was found to be the most up-regulated gene in response to the North American ginseng extract [48]. The results of this study are in good agreement with those of the studies mentioned previously. Even though many KRG studies on immune responses have been conducted, only one study has reported on the immune-related function of KRG in a normal state which suggested that KRG can induce infection-relevant immune responses and may activate CD8+ T-cell [12]. Further investigation should be performed to confirm the immune-related function of KRG.
Conflicts of interest
All authors declare no conflicts of interest.
Acknowledgments
The authors would like to thank Dr. Min Jueng Kang at Samsung Bioepis for assistance with the experiments.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jgr.2019.05.001.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Yang H., Son G.W., Park H.R., Lee S.E., Park Y.S. Effect of Korean Red Ginseng treatment on the gene expression profile of diabetic rat retina. J Ginseng Res. 2016;40:1–8. doi: 10.1016/j.jgr.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim S.J., Shin J.Y., Ko S.K. Changes in the contents of prosapogenin in Red ginseng (Panax ginseng) depending on the extracting conditions. J Ginseng Res. 2016;40:86–89. doi: 10.1016/j.jgr.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie Y.Y., Luo D., Cheng Y.J., Ma J.F., Wang Y.M., Liang Q.L. Steaming-induced chemical transformations and holistic quality assessment of red ginseng derived from Panax ginseng by means of HPLC-ESI-MS/MS(n)-based multicomponent quantification fingerprint. J Agric Food Chem. 2012;60:8213–8224. doi: 10.1021/jf301116x. [DOI] [PubMed] [Google Scholar]
- 4.Provino R. The role of adaptogens in stress management. Australian Journal of Medical Herbalism. 2010;22:41. [Google Scholar]
- 5.Suh S.O., Kim J., Cho M.Y. Prospective study for Korean red ginseng extract as an immune modulator following a curative gastric resection in patients with advanced gastric cancer. J Ginseng Res. 2004;28:104–110. [Google Scholar]
- 6.Kim J.Y., Park J.Y., Kang H.J., Kim O.Y., Lee J.H. Beneficial effects of Korean red ginseng on lymphocyte DNA damage, antioxidant enzyme activity, and LDL oxidation in healthy participants: a randomized, double-blind, placebo-controlled trial. Nutr J. 2012;11:1. doi: 10.1186/1475-2891-11-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee S.T., Chu K., Sim J.Y., Heo J.H., Kim M. Panax ginseng enhances cognitive performance in Alzheimer disease. Alzheimer Dis Assoc Disord. 2008;22:222–226. doi: 10.1097/WAD.0b013e31816c92e6. [DOI] [PubMed] [Google Scholar]
- 8.Shin K.S., Lee J.J., Kim Y.I., Yu J.Y., Park E.S., Im J.H. Effect of Korean red ginseng extract on blood circulation in healthy volunteers: a randomized, double-blind, placebo-controlled trial. J Ginseng Res. 2007;31:109–116. [Google Scholar]
- 9.Kennedy D., Reay J., Scholey A. Effects of 8 weeks administration of Korean Panax ginseng extract on the mood and cognitive performance of healthy individuals. J Ginseng Res. 2007;31:34. [Google Scholar]
- 10.Choi R., Wong A.S., Jia W., Chang I.M., Wong R.N., Fan T.P. Ginseng: a panacea linking East Asia and North America? Science. 2015;350:S54–S56. [Google Scholar]
- 11.Kang S., Min H. Ginseng, the 'immunity boost': the effects of panax ginseng on immune system. J Ginseng Res. 2012;36:354–368. doi: 10.5142/jgr.2012.36.4.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee B.J., Heo H., Oh S.C., Lew J.H. Comparison study of Korean and Chinese ginsengs on the regulation of lymphocyte proliferation and cytokine production. J Ginseng Res. 2008;32:250–256. [Google Scholar]
- 13.Hart A. iUniverse; 2005. Predictive medicine for rookies: consumer watchdogs, reviews, & genetics testing firms online. [Google Scholar]
- 14.Leung E.L., Cao Z.W., Jiang Z.H., Zhou H., Liu L. Network-based drug discovery by integrating systems biology and computational technologies. Brief Bioinform. 2013;14:491–505. doi: 10.1093/bib/bbs043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X., Xu X., Tao W., Li Y., Wang Y., Yang L. A systems biology approach to uncovering pharmacological synergy in herbal medicines with applications to cardiovascular disease. Evid Based Complement Alternat Med. 2012;2012:519031. doi: 10.1155/2012/519031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ru J., Li P., Wang J., Zhou W., Li B., Huang C. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y., Xu A., Zheng A systems biology approach to diagnosis and treatments. Science. 2014;346:S13–S15. [Google Scholar]
- 18.Leung E.L., Wong V.K., Jiang Z.H., Li T., Liu L. Integrated network-based medicine: the role of traditional Chinese medicine in developing a new generation of medicine. Science. 2014;346:S16–S18. [Google Scholar]
- 19.Cha M.Y., Kwon Y.W., Ahn H.S., Jeong H., Lee Y.Y., Moon M. Protein-induced pluripotent stem cells ameliorate cognitive dysfunction and reduce abeta deposition in a mouse model of Alzheimer's disease. Stem Cells Transl Med. 2017;6:293–305. doi: 10.5966/sctm.2016-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stein D.R., Hu X., McCorrister S.J., Westmacott G.R., Plummer F.A., Ball T.B. High pH reversed-phase chromatography as a superior fractionation scheme compared to off-gel isoelectric focusing for complex proteome analysis. Proteomics. 2013;13:2956–2966. doi: 10.1002/pmic.201300079. [DOI] [PubMed] [Google Scholar]
- 21.Keller A., Nesvizhskii A.I., Kolker E., Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 22.Kim Y.S., Woo J.Y., Han C.K., Chang I.M. Safety analysis of panax ginseng in randomized clinical trials: a systematic review. Medicines (Basel) 2015;2:106–126. doi: 10.3390/medicines2020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin Y.R., Yu J.Y., Lee J.J., You S.H., Chung J.H., Noh J.Y., Im J.H., Han X.H., Kim T.J., Shin K.S. Antithrombotic and antiplatelet activities of Korean red ginseng extract. Basic Clin Pharmacol Toxicol. 2007;100:170–175. doi: 10.1111/j.1742-7843.2006.00033.x. [DOI] [PubMed] [Google Scholar]
- 24.Sultan S., Ahmad S., Pascucci M., Ramadori G. Changes of LCN–2 gene expression in different organs in a rat model of tissue damage. Zeitschrift für Gastroenterologie. 2012;50:P2_34. [Google Scholar]
- 25.Nairz M., Ferring-Appel D., Casarrubea D., Sonnweber T., Viatte L., Schroll A. Iron regulatory proteins mediate host resistance to Salmonella infection. Cell Host Microbe. 2015;18:254–261. doi: 10.1016/j.chom.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Annane D., Bellissant E., Cavaillon J.M. Septic shock. Lancet. 2005;365:63–78. doi: 10.1016/S0140-6736(04)17667-8. [DOI] [PubMed] [Google Scholar]
- 27.Zanetti M., Gennaro R., Romeo D. Cathelicidins: a novel protein family with a common proregion and a variable C-terminal antimicrobial domain. FEBS Lett. 1995;374:1–5. doi: 10.1016/0014-5793(95)01050-o. [DOI] [PubMed] [Google Scholar]
- 28.Larrick J.W., Hirata M., Balint R.F., Lee J., Zhong J., Wright S.C. Human CAP18: a novel antimicrobial lipopolysaccharide-binding protein. Infect Immun. 1995;63:1291–1297. doi: 10.1128/iai.63.4.1291-1297.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zanetti M. Cathelicidins, multifunctional peptides of the innate immunity. J Leukoc Biol. 2004;75:39–48. doi: 10.1189/jlb.0403147. [DOI] [PubMed] [Google Scholar]
- 30.Bu H.F., Wang X., Zhu Y.Q., Williams R.Y., Hsueh W., Zheng X., Rozenfeld R.A., Zuo X.L., Tan X.D. Lysozyme-modified probiotic components protect rats against polymicrobial sepsis: role of macrophages and cathelicidin-related innate immunity. J Immunol. 2006;177:8767–8776. doi: 10.4049/jimmunol.177.12.8767. [DOI] [PubMed] [Google Scholar]
- 31.Janeway C.A., Travers P., Walport M., Shlomchik M. 5th ed. Garland Science; New York: 2001. Immunobiology: the immune system in health and disease. [Google Scholar]
- 32.Tonnelle C., DeMars R., Long E.O. DO beta: a new beta chain gene in HLA-D with a distinct regulation of expression. EMBO J. 1985;4:2839–2847. doi: 10.1002/j.1460-2075.1985.tb04012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dai S., Murphy G.A., Crawford F., Mack D.G., Falta M.T., Marrack P. Crystal structure of HLA-DP2 and implications for chronic beryllium disease. Proc Natl Acad Sci U S A. 2010;107:7425–7430. doi: 10.1073/pnas.1001772107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Easterfield A.J., Bradley J.A., Bolton E.M. Complementary DNA sequences encoding the rat MHC class II RT1-Bu and RT1-Du alpha and beta chains. Immunogenetics. 2003;55:344–350. doi: 10.1007/s00251-003-0584-7. [DOI] [PubMed] [Google Scholar]
- 35.Montecino-Rodriguez E., Berent-Maoz B., Dorshkind K. Causes, consequences, and reversal of immune system aging. J Clin Invest. 2013;123:958–965. doi: 10.1172/JCI64096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goya R.G., Console G., Herenu C., Brown O., Rimoldi O.J. Thymus and aging: potential of gene therapy for restoration of endocrine thymic function in thymus-deficient animal models. Gerontology. 2002;48:325–328. doi: 10.1159/000065258. [DOI] [PubMed] [Google Scholar]
- 37.Gruver A.L., Hudson L.L., Sempowski G.D. Immunosenescence of ageing. J Pathol. 2007;211:144–156. doi: 10.1002/path.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spurr N.K., Leppert M. Report of the committee on the genetic constitution of chromosome 2. Cytogenet Cell Genet. 1990;55:86–91. doi: 10.1159/000132998. [DOI] [PubMed] [Google Scholar]
- 39.Tajrishi M.M., Tuteja R., Tuteja N. Nucleolin: the most abundant multifunctional phosphoprotein of nucleolus. Commun Integr Biol. 2011;4:267–275. doi: 10.4161/cib.4.3.14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quiros P.M., Espanol Y., Acin-Perez R., Rodriguez F., Barcena C., Watanabe K. ATP-dependent Lon protease controls tumor bioenergetics by reprogramming mitochondrial activity. Cell Rep. 2014;8:542–556. doi: 10.1016/j.celrep.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 41.Tsai P.L., Zhao C., Turner E., Schlieker C. The Lamin B receptor is essential for cholesterol synthesis and perturbed by disease-causing mutations. Elife. 2016;5 doi: 10.7554/eLife.16011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alberts A.W., Strauss A.W., Hennessy S., Vagelos P.R. Regulation of synthesis of hepatic fatty acid synthetase: binding of fatty acid synthetase antibodies to polysomes. Proc Natl Acad Sci U S A. 1975;72:3956–3960. doi: 10.1073/pnas.72.10.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carroll R.G., Zaslona Z., Galvan-Pena S., Koppe E.L., Sevin D.C., Angiari S. An unexpected link between fatty acid synthase and cholesterol synthesis in proinflammatory macrophage activation. J Biol Chem. 2018;293:5509–5521. doi: 10.1074/jbc.RA118.001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baron A., Migita T., Tang D., Loda M. Fatty acid synthase: a metabolic oncogene in prostate cancer? J Cell Biochem. 2004;91:47–53. doi: 10.1002/jcb.10708. [DOI] [PubMed] [Google Scholar]
- 45.Hunt D.A., Lane H.M., Zygmont M.E., Dervan P.A., Hennigar R.A. MRNA stability and overexpression of fatty acid synthase in human breast cancer cell lines. Anticancer Res. 2007;27:27–34. [PubMed] [Google Scholar]
- 46.Gansler T.S., Hardman W., 3rd, Hunt D.A., Schaffel S., Hennigar R.A. Increased expression of fatty acid synthase (OA-519) in ovarian neoplasms predicts shorter survival. Hum Pathol. 1997;28:686–692. doi: 10.1016/s0046-8177(97)90177-5. [DOI] [PubMed] [Google Scholar]
- 47.Lee F., Yokota T., Otsuka T., Gemmell L., Larson N., Luh J., Arai K., Rennick D. Isolation of cDNA for a human granulocyte-macrophage colony-stimulating factor by functional expression in mammalian cells. Proc Natl Acad Sci U S A. 1985;82:4360–4364. doi: 10.1073/pnas.82.13.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lemmon H.R. Modulatory effects of North American ginseng extracts on human innate and adaptive immune responses. Electronic Thesis and Dissertation Repository. 2012;399 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.