Development of drug resistance is a challenge in tamoxifen-based breast cancer therapy. By assessing the differential expression of kinase proteins in drug-resistant and parental breast cancer cells, we discover that elevated expression of hexokinase 2 (HK2) and the ensuing inhibition of mTOR contribute to tamoxifen resistance, suggesting HK2 as a potential target for overcoming tamoxifen resistance.
Keywords: Kinases, breast cancer, drug resistance, SILAC, quantification, hexokinase, parallel-reaction monitoring, quantitative proteomics
Graphical Abstract
Highlights
LC-PRM-based targeted kinome analysis led to the quantification of 315 kinases in parental and tamoxifen-resistant MCF-7 breast cancer cells.
Hexokinase 2 and mTOR were up-regulated in tamoxifen-resistant MCF-7 cells, which was accompanied with elevated glycolysis rate.
Augmented expression of HK2 promotes autophagy through inhibition of the mTOR-S6K signaling pathway and results in resistance of MCF-7 cells to tamoxifen.
HK2 is a potential drug target for overcoming tamoxifen resistance.
Abstract
Tamoxifen has been clinically used in treating estrogen receptor (ER)-positive breast cancer for over 30 years. The most challenging aspect associated with tamoxifen therapy is the development of resistance in initially responsive breast tumors. We applied a parallel-reaction monitoring (PRM)-based quantitative proteomic method to examine the differential expression of kinase proteins in MCF-7 and the isogenic tamoxifen-resistant (TamR) cells. We were able to quantify the relative protein expression levels of 315 kinases, among which hexokinase 2 (HK2) and mTOR were up-regulated in TamR MCF-7 cells. We also observed that the TamR MCF-7 cells exhibited elevated rate of glycolysis than the parental MCF-7 cells. In addition, we found that phosphorylation of S6K - a target of mTOR - was much lower in TamR MCF-7 cells, and this phosphorylation level could be restored upon genetic depletion or pharmacological inhibition of HK2. Reciprocally, the level of S6K phosphorylation was diminished upon overexpression of HK2 in MCF-7 cells. Moreover, we observed that HK2 interacts with mTOR, and this interaction inhibits mTOR activity. Lower mTOR activity led to augmented autophagy, which conferred resistance of MCF-7 cells toward tamoxifen. Together, our study demonstrates that elevated expression of HK2 promotes autophagy through inhibiting the mTOR-S6K signaling pathway and results in resistance of MCF-7 breast cancer cells toward tamoxifen; thus, our results uncovered, for the first time, HK2 as a potential therapeutic target for overcoming tamoxifen resistance.
Breast cancer is the most frequently diagnosed cancer (excluding non-melanoma skin cancers) among women in 140 out of 180 countries worldwide (1). Accounting for an estimated 522,000 deaths and ∼6.4% of total cancer deaths (1), breast cancer ranks as the fifth most common cause of cancer death in women and constitutes one of the most expensive malignancies for treatment (2).
The past several decades of research showed that breast cancer is a multifaceted disease with different subtypes and stages (3). According to the receptors on the cell surface and in their cytoplasm and nucleus, breast cancer can be categorized into four main classes, namely, basal-like, luminal A, luminal B and human epidermal growth factor receptor 2 (HER2)-positive cancers (4, 5). Hormone therapy, chemotherapy, and targeted therapy have been developed for clinical treatment of breast cancer (5). Among them, tamoxifen, as a partial estrogen receptor (ER)1 antagonist (6, 7), has been clinically used for ∼30 years as a chemotherapeutic agent for patients with ER-positive breast cancer, which represents nearly 70% of all breast cancer cases (8).
Resistance to tamoxifen remains one of the major hurdles in the effective management of breast cancer (9, 10). In this vein, chemotherapeutic drug resistance may arise from different mechanisms, including aberrant metabolic activation of tamoxifen, loss of ER function/expression, as well as alterations in crosstalk between ER and growth factor/kinase-mediated signaling pathways and responses to oxidative stress (10). Kinome reprogramming, e.g. aberrant expression and/or activation of kinases, has been found to be a major mechanism through which cancer cells acquire resistance toward chemotherapy (11). However, it remains poorly understood how kinases contribute to tamoxifen resistance. We reason that a proteome-wide profiling of alterations in kinase protein expression associated with the development of drug resistance may provide mechanistic insights into therapeutic resistance and reveal new targets for endocrine therapy.
Several quantitative proteomic methods have been developed for the interrogation of the whole human kinome. For instance, affinity resin immobilized with multiple kinase inhibitors, termed kinobeads, was employed to selectively enrich protein kinases over other types of ATP-binding proteins (12, 13). In addition, ATP acyl phosphate probes have been used for the enrichment of kinase proteins or their component peptides for subsequent mass spectrometry studies (14, 15). The efficiencies of both enrichment methods are modulated by the protein expression levels of kinases and sometimes may also be affected by the alterations in activities of kinases. We recently developed a parallel-reaction monitoring (PRM)-based targeted proteomic method to assess the levels of kinase protein expression at the entire proteome scale, and we also applied successfully the method for assessing the reprogramming of the human kinome upon treatment with kinase inhibitors (16, 17). We established a Skyline kinome library for LC-PRM analysis based on shotgun proteomic data acquired from in-depth LC-MS/MS analyses of tryptic digestion mixtures of protein lysates from multiple human cell lines (16). The library encompassed 1050 tryptic peptides originated from 478 kinase proteins, including 395 protein kinases (16).
In this study, we employed the LC-PRM method to profile the differential expression of kinase proteins in parental and tamoxifen-resistant MCF-7 cells. We were able to quantify the relative expression levels of 315 unique kinases and identify HK2 as a driver for tamoxifen resistance.
EXPERIMENTAL PROCEDURES
Compounds
(Z)-4-Hydroxytamoxifen (4-OHT) and 2-deoxy-d-glucose (2-DG) were purchased from Sigma-Aldrich (St. Louis, MO). Bafilomycin A1 was obtained from Cayman Chemical (Ann Arbor, MI).
Cell Culture
Parental MCF-7 and tamoxifen-resistant MCF-7 (TamR) human breast cancer cells were kindly provided by Dr. David Eastmond at UC Riverside and Dr. Guangdi Wang at Xavier University (18), respectively. The cells were maintained in Dulbecco's Modified Eagle Medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA) and 1% penicillin/streptomycin (10,000 U/ml penicillin and 10,000 U/ml streptomycin, Thermo Fisher Scientific, Waltham, MA). 4-OHT (1 μm) was included in the culture medium for maintaining the tamoxifen-resistant subline. The cells were cultured at 37 °C in a humidified atmosphere containing 5% CO2. For SILAC labeling experiments, the cells were cultured in SILAC medium containing unlabeled lysine and arginine or [13C6, 15N2]-lysine and [13C6]-arginine for at least five cell doublings (19).
Tryptic Digestion of the Whole Cell Lysates and LC-PRM Analysis
The above-mentioned lysates from the two cell lines were incubated with 8 m urea for protein denaturation, and then treated with dithiothreitol and iodoacetamide for cysteine reduction and alkylation, respectively. The proteins were subsequently digested with modified MS-grade trypsin (Pierce, Waltham, MA) at an enzyme/substrate ratio of 1:100 in 50 mm NH4HCO3 (pH 8.5) at 37 °C overnight. Samples from four biological replicates (two forward and two reverse SILAC labeling experiments) of lysates from the MCF-7/TamR pair were prepared for LC-PRM analyses. The peptide mixtures (500 ng each) were subsequently dried in a Speed-vac, desalted with OMIX C18 pipette tips (Agilent Technologies, Santa Clara, CA), and analyzed by LC-MS/MS on a Q Exactive Plus quadruple-Orbitrap mass spectrometer (Thermo Fisher Scientific) coupled with an EASY-nLC 1200 system in the scheduled PRM mode. The samples were automatically loaded onto a 4-cm trapping column (150 μm i.d.) packed with ReproSil-Pur 120 C18-AQ resin (5 μm in particle size and 120 Å in pore size, Dr. Maisch GmbH HPLC, Ammerbuch-Entringen, Germany) at a flow rate of 3 μl/min. The trapping column was coupled to a 20-cm fused silica analytical column (PicoTip Emitter, New Objective, Woburn, MA, 75 μm i.d.) packed with ReproSil-Pur 120 C18-AQ resin (3 μm in particle size and 120 Å in pore size, Dr. Maisch GmbH HPLC). The peptides were then resolved using a 140-min linear gradient of 9–38% acetonitrile in 0.1% formic acid and at a flow rate of 300 nL/min. The spray voltage was 1.8 kV. Target ions were sequentially isolated and collisionally activated in the HCD cell at a collision energy of 29 to yield the MS/MS, which were acquired in the Orbitrap analyzer at a resolution of 17500 and with an AGC target of 1 × 105.
The linear predictor of empirical retention time (RT) from normalized retention time (iRT) for targeted kinase peptides was determined by the linear regression of RTs of BSA standard peptides obtained for the current chromatography setup. This RT-iRT linear relationship was re-established between every eight LC-PRM runs by injecting another tryptic digestion mixture of BSA (20). The targeted precursor ions for the kinase peptides were monitored in eight separate injections for each sample in scheduled PRM mode with an 8-min retention time window.
All raw files were processed using Skyline (version 19.1) (21) for the generation of extracted-ion chromatograms and peak integration. Cysteine carbamidomethylation and methionine oxidation were set as fixed and variable modifications, respectively. Six most abundant y ions found in previously published MS/MS acquired from shotgun proteomic analysis (16) were chosen for peptide identification and quantification, where a mass accuracy of 20 ppm or less was imposed for fragment ions during the identification of peptides in the Skyline platform. The targeted peptides were first manually checked to ensure the overlaid chromatographic profiles of multiple fragment ions derived from the light and heavy forms of the same peptide. The data were subsequently processed to ascertain that the distribution of the relative intensities of multiple transitions associated with the same precursor ion is correlated with the theoretical distribution in the kinome MS/MS spectral library. The sum of peak areas from all transitions of light or heavy peptides was used for quantification.
Experimental Design and Statistical Rationale
The type of our targeted quantitative proteomic experiments is a Tier 3 assay, according to previously published guidelines (22), and we employed Skyline (version 19.1) (21) for our data analysis. For each kinase protein, up to four most abundant unmodified peptides including at most one tryptic mis-cleavage site were included in the Skyline library, and all the peptides in the library were unique to specific kinase proteins (16). The LC-PRM experiments were conducted in four biological replicates, where tryptic digestion mixture of 500 ng of the whole-cell protein lysate was injected in each LC-PRM run, to determine the relative levels of expression of the kinase proteins in parental and TamR MCF-7 cells. Up to six most abundant y ions were used for peptide quantification (supplemental Table S2). The detailed quantification data for the relative levels of peptides and proteins in TamR MCF-7 over parental MCF-7 cells, including mean and standard deviation (S.D.) for peptide quantification from different replicates, and mean, S.D., and relative S.D. (R.S.D.) calculated from quantification results for different peptides derived from the same kinase proteins are listed in supplemental Table S1.
Extracellular Acidification Rate Measurement
Extracellular acidification rates were measured on a Seahorse XF96 Extracellular Flux Analyzer (Agilent Technologies). MCF-7 and TamR cells were seeded into an XF96 microplate at a final density of 20,000 cells per well. The cells were plated in growth medium and maintained overnight in a tissue culture incubator (37 °C, 5% CO2). On the day of the experiment, the cells were washed twice with freshly prepared assay medium (Seahorse XF Base Medium supplemented with 10 mm glucose, 2 mm l-glutamine and 1 mm sodium pyruvate, pH 7.4) and brought to a final volume of 175 μl per well. The XF96 plate was placed in a 37 °C incubator without CO2 for 30 min prior to loading the plate into the instrument. Oligomycin (at a final concentration of 2 μm), a mitochondrial ATP synthase inhibitor, was injected during the assay. At the conclusion of the assay, the cells were fixed with 4% paraformaldehyde, stained with Hoechst, and cell number per well was determined based on nuclei number using an Operetta High-Content Imaging System (PerkinElmer, Waltham, MA). Extracellular acidification rates were normalized to cell number per well.
shRNA and Plasmids
The sequences for shHK2 were 5′-CCG GCC AAA GAC ATC TCA GAC ATT GCT CGA GCA ATG TCT GAG ATG TCT TTG GTT TTT TG-3′ and 5′-CCG GCC AGA AGA CAT TAG AGC ATC TCT CGA GAG ATG CTC TAA TGT CTT CTG GTT TTT TG-3′. Control shRNA sequence was 5′-CCT AAG GTT AAG TCG CCC TCG CTC TAG CGA GGG CGA CTT AAC CTT AGG-3′ (Addgene, Watertown, MA) (23). The sequences for HK2 primers were 5′-AAA GCT AGC ATG ATT GCC TCG CAT CTG CTT G-3′ and 5′-AAA GAA TTC CTA TCG CTG TCC AGC CTC ACG GAT GC-3′. All oligodeoxynucleotides were purchased from Integrated DNA Technologies. All shRNAs were cloned into the Age I/EcoR I site of the pLKO.1 vector (Addgene, plasmid # 10878). HK2 plasmid was cloned into the NheI and EcoR I restriction sites of the pLJM1-EGFP vector (Addgene, plasmid # 19319). All constructs were confirmed by Sanger sequencing.
Lentivirus Production and Stable Cell Line Generation
HEK293T cells were transfected with pLKO.1/puro-shRNAs and pLJM1-EGFP-HK2 plasmids together with pLTR-G (Addgene plasmid #17532) envelope plasmid and pCMV-dR8.2 dvpr (Addgene plasmid #8455) package plasmid using PolyFect Transfection Reagent (Qiagen, Germantown, MD). Viral particles were collected at 48 h following transfection and then filtered through a 0.45 μm sterile filter. Tamoxifen-resistant MCF-7 cells were infected with a 1:1 mixture of viral particles and DMEM containing 1 μm 4-OHT for 48 h. The cells were selected with 2 μg/ml puromycin for 1 week and cultured in complete DMEM supplemented with 1 μm 4-OHT and 1 μg/ml puromycin.
Western Blotting
MCF-7 and tamoxifen-resistant MCF-7 cells were cultured in a 6-well plate and, at 40–50% confluency, were treated with 2-DG or 4-OHT for 24 h. The cells were lysed with CelLyticTM M Cell Lysis reagent (Sigma Aldrich) and the supernatant was used for Western blot analysis. Antibodies recognizing human HK2 (Santa Cruz Biotechnology, Dallas, TX, sc-374091), mTOR (Cell Signaling Technology, Danvers, MA, #2972), phospho-p70 S6 kinase (Thr389) (Cell Signaling Technology, #9209), p70 S6 kinase (Cell Signaling Technology, #2708) and MAP LC3β (Santa Cruz Biotechnology, sc-376404) were employed as primary antibodies for Western blot analysis. Goat Anti-Rabbit IgG (whole molecule)-peroxidase antibody (Sigma, #A0545) and m-IgGκ BP-HRP (Santa Cruz Biotechnology, sc-516102) were used as secondary antibodies. Membranes were also probed with anti-GAPDH antibody (Santa Cruz Biotechnology, sc-32233) to confirm equal protein loading.
Proliferation Assay
Cells were seeded in 96-well culture plates (7000 cells/well), and compounds at various concentrations were added into the wells 24 h later. Cell proliferation was determined at 72 h after compound addition. Cell viability was measured using the Cell Counting Kit-8 (Dojindo Molecular Technologies, Inc. Rockville, MD) according to the manufacturer's instructions, and luminescence was measured using a Synergy H1 Hybrid Multi-Mode Microplate Reader (BioTek, Winooski, VT). The data were normalized to control groups (DMSO or ethanol) and represented by the mean and S.D. of results from three independent measurements using Prism 7.0 (GraphPad Software).
Immunoprecipitation
The cells were washed with ice-cold PBS twice and lysed in 0.5% NP40 lysis reagent supplemented with a complete protease inhibitor mixture (Sigma-Aldrich). After lysis on ice for 30 min, the samples were centrifuged at 13000 rpm for 10 min and the supernatant was collected. HK2 and mTOR were immuno-precipitated using antibodies recognizing HK2 and mTOR, respectively. Cell lysates were pre-cleared with Protein A/G Plus-agarose beads at 4 °C for 2 h, and 150–300 μg total protein was then incubated with HK2 or mTOR antibody (4 μg) at 4 °C overnight. After addition of protein A/G agarose beads, incubation was continued for another 2 h. The resulting immune-complexes were washed with ice-cold lysis buffer for 4 times, and the beads were boiled in 2× loading buffer to elute the captured proteins, which were subjected to Western blot analysis. Band intensities were analyzed using Image J, where background was subtracted followed by normalization to the loading control obtained from the same gel (GAPDH for whole cell lysate and target molecule of immunoprecipitation for the study of HK2-mTOR interaction), and a percentage relative to the control was obtained.
Bioinformatic Analysis of Publicly Available Data Sets
The prognostic values of HK2 in breast cancer patients (n = 3951 for all patients, n = 809 for patients with tamoxifen only therapy) were analyzed by using the Km Plotter Database (http://kmplot.com/analysis/) derived from the Gene Expression Omnibus (GEO) repository. Kaplan-Meier survival analysis was conducted with patient data stratified by median HK2 mRNA expression levels and the hazard ratio (HR), 95% confidence intervals (95% CI) and log-rank p values were calculated and displayed.
RESULTS
Differential Expression of Kinase Proteins in Tamoxifen-resistant and Parental MCF-7 Cells
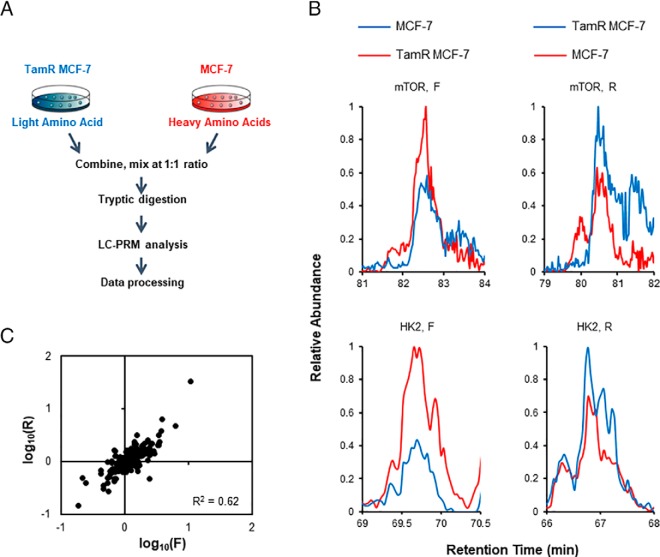
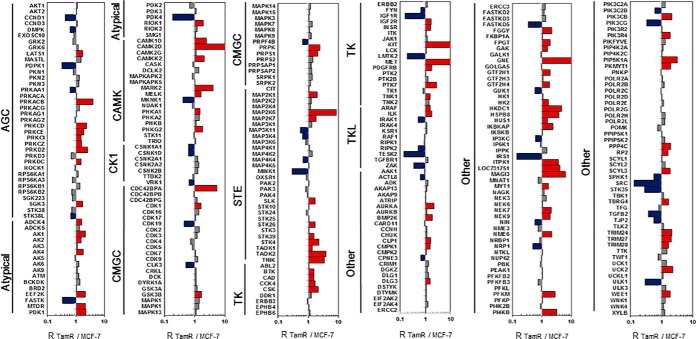
The primary objective of the present study is to assess systematically the roles of kinases in resistance toward tamoxifen in breast cancer cells. Hence, we set out to employ our recently developed LC-PRM method (16, 17), in combination with SILAC (stable isotope labeling by amino acids in cell culture), to examine the differential expression of kinase proteins in the paired tamoxifen-resistant/parental MCF-7 cells (18) (Fig. 1A). The analysis led to the quantification of 315 kinases, which cover approximately one half of the human kinome (Fig. 2). All quantified peptides for the kinases in our library exhibited an excellent linear fit between the observed retention time and iRT. Additionally, all 4–6 transitions employed for quantification of each peptide displayed the same retention time with a dot product (dotp) (24) value of > 0.7 when compared with the same fragment ions found in the full-scan MS/MS acquired from shotgun proteomic analyses (16), suggesting that our method is highly reliable for peptide identification. Furthermore, ratios of the quantified peptides obtained from forward and reverse SILAC labeling experiments exhibited an excellent linear fit and a strong correlation, indicating good reproducibility of our analytical method (Fig. 1C). Moreover, the reproducibility of the method is reflected by the observation that consistent ratios were obtained for different tryptic peptides derived from the same kinases (supplemental Table S1). In this regard, the average R.S.D. for the quantification results obtained from the different tryptic peptides of the same kinase proteins were 13.2% (supplemental Table S1).
Fig. 1.
LC-PRM for interrogating the differential expression of kinase proteins in tamoxifen-resistant and parental MCF-7 cells. A, A schematic diagram showing the targeted proteomics workflow for human kinome analysis, and depicted here is the forward SILAC labeling experiment. B, Representative PRM traces for mTOR and HK2 proteins. C, Correlation between the ratios obtained from LC-PRM analysis together with forward and reverse SILAC labeling experiments.
Fig. 2.
Differential expression of kinase proteins in tamoxifen-resistant and parental MCF-7 cells. The data represent the mean of the results obtained from two forward and two reverse SILAC labeling results (supplemental Table S1 lists the ratios obtained from individual measurements).
TamR MCF-7 Cells Exhibit Elevated Expression of HK2 Protein and Augmented Rate of Glycolysis
The quantitative proteomic data showed the differential expression of many kinases in the TamR and parental MCF-7 cells. Among these kinases, hexokinase II (HK2) has been implicated in different types of cancer (25, 26) and its expression level is correlated with prognosis of breast cancer patients undergoing tamoxifen therapy (vide infra). Therefore, we decided to focus our functional and mechanistic study on this kinase.
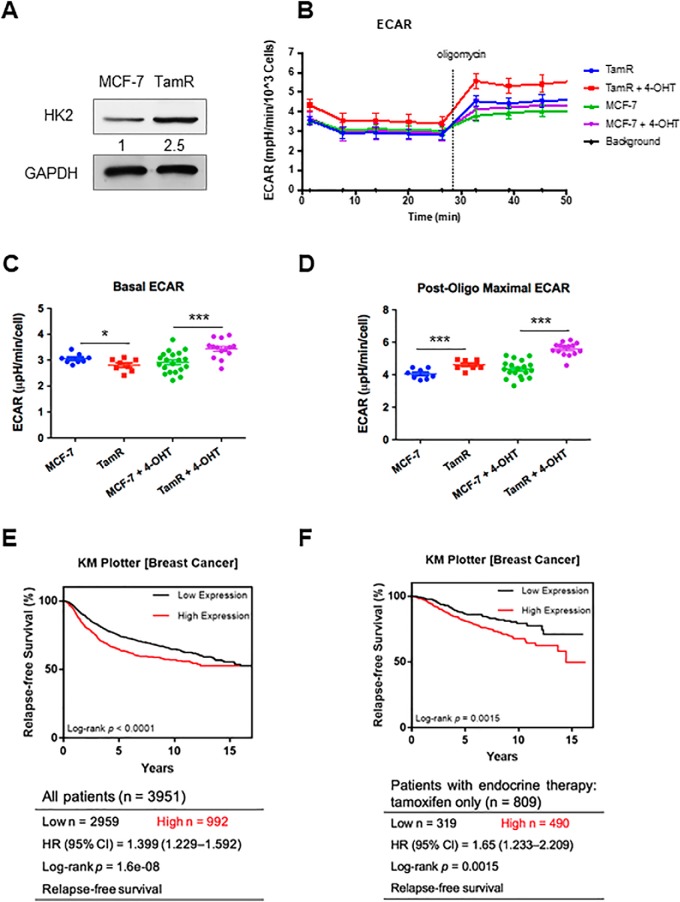
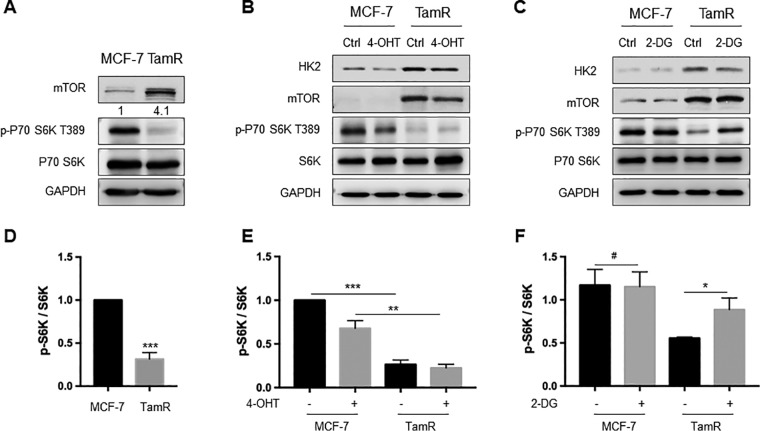
Analysis of the PRM data showed that the expression level of HK2 protein is elevated in TamR MCF-7 cells relative to the parental MCF-7 cells, which we confirmed by Western blot analysis (Fig. 1B, 3A). Because HK2 catalyzes the first and rate-limiting step of glycolysis, we asked whether higher HK2 levels contributed to elevated glycolysis in TamR MCF-7 cells. Indeed, we found that TamR MCF-7 cells exhibit much higher basal and post-oligomycin extracellular acidification rates, underscoring elevated rate of glycolysis in TamR relative to parental MCF-7 cells (Fig. 3B–3D).
Fig. 3.
TamR MCF-7 cells display elevated expression of HK2 and augmented rate of glycolysis, and elevated expression of HK2 is correlated with poorer survival of breast cancer patients. A, Western blot analysis confirmed the increased expression of HK2 protein in TamR MCF-7 cells. B, Extracellular acidification rates (ECARs) of TamR and parental MCF-7 cells before and after administration of oligomycin and with or without 4-OHT treatment. C, Basal ECAR of TamR cells and parental MCF-7 cells with or without 4-OHT treatment. D, Post-oligomycin maximal ECARs of TamR cells and parental MCF-7 cells with or without 4-OHT treatment. E, Kaplan-Meier survival analysis revealed a significant correlation between higher level of mRNA expression of HK2 gene and poorer relapse-free survival (RFS) in a large cohort of breast cancer patients (n = 3951). F, Kaplan-Meier survival analysis showed a significant correlation between higher HK2 mRNA expression and poorer RFS rate in a subgroup of breast cancer patients who received only tamoxifen treatment (n = 809). The data represent the mean ± S.D. of the quantification results (n = 3). The p values were calculated based on unpaired, two-tailed Student's t test: *, 0.01 ≤ p < 0.05; ***, p < 0.001.
Increased Expression of HK2 is Correlated with Poorer Survival of Breast Cancer Patients
We explored whether the finding made for HK2 has prognostic values for breast cancer patients, particularly those who have undergone tamoxifen therapy. Kaplan-Meier survival analysis based on the Gene Expression Omnibus (GEO) database for breast cancer patients (n = 3951) showed that higher levels of mRNA expression of HK2 gene were significantly correlated with poorer breast cancer patient survival (hazard ratio, HR = 1.399; 95% confidence interval, 95% CI = 1.299–1.592; Logrank p = 1.6e-08) (Fig. 3E). Moreover, among those breast cancer patients with only tamoxifen chemotherapy (n = 809), higher levels of HK2 mRNA expression were significantly correlated with poorer survival (HR = 1.65; 95% CI = 1.233–2.209; Logrank p = 0.0015) (Fig. 3F).
Inhibition of HK2 Diminishes 4-OHT-mediated Autophagy and Resensitizes TamR MCF-7 Cells to 4-OHT
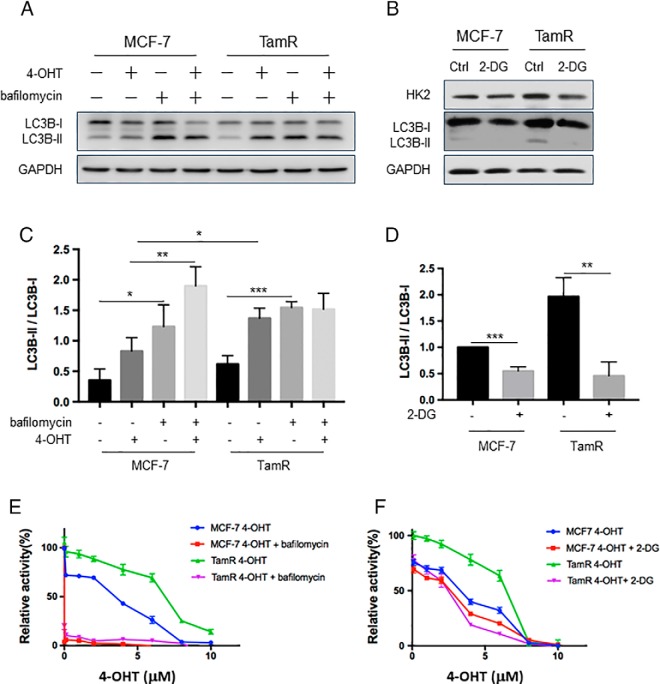
Previous studies showed that HK2 can induce autophagy by inhibiting mTOR, and treatment with tamoxifen can elicit autophagy (27, 28). Hence, we investigated if these factors function together to constitute a potential mechanism for conferring drug resistance. Our Western blotting results showed that autophagy, as reflected by the ratio of LC3B-II over LC3B-I, was higher in TamR MCF-7 than parental MCF-7 cells (Fig. 4A). Additionally, the ratio of LC3B-II/LC3B-I in MCF-7 cells was increased upon treatment with 4-OHT, which is consistent with previously reported results (29). Exposure with bafilomycin A1 led to significantly increased ratio of LC3B-II/LC3B-I (Fig. 4A, 4C) and sensitized both parental and TamR MCF-7 cells to 4-OHT (Fig. 4E), suggesting that autophagy promotes the survival of cells upon tamoxifen treatment (30).
Fig. 4.
2-DG-mediated inhibition of HK2 could reduce autophagy induction by 4-OHT and re-sensitize TamR MCF-7 cells to 4-OHT. A, Parental and TamR MCF-7 cells were treated with 2 μm 4-OHT for 24 h. At the end of the incubation, 20 nm bafilomycin A1 was added, and after an additional 1 h, autophagy flux was measured, where the expression of LC3B was monitored by Western blotting. B, MCF-7 and TamR MCF-7 cells were treated with 2 μm 2-DG for 24 h, and the lysates were used to monitor the expression level of HK2 protein and autophagy marker LC3B by Western blotting. C, D, The ratios of LC3B-II/LC3B-I were quantified from band intensity using ImageJ and the results in D are displayed relative to the level in parental MCF-7 cells without 2-DG treatment. The data were presented as the mean ± S.D. (n = 3). The p values were calculated using unpaired, two-tailed Student's t test: *, 0.01 ≤ p < 0.05; **, 0.001 ≤ p < 0.01; ***, p < 0.001. E, F, Parental and TamR MCF-7 cells were treated with serially diluted 4-OHT and 20 nm bafilomycin A1 or 1 mm 2-DG for 72 h, and cell viability was measured using the CCK8 and luminescence was monitored with a BioTek reader. Data were normalized to control groups (DMSO) and represented by the mean and S.D. of results from three independent measurements.
HK2 was previously shown to enhance cisplatin-induced autophagy, thereby conferring resistance to the anti-cancer drug (31). Therefore, we next asked whether augmented autophagy in TamR MCF-7 cells arises from elevated expression of HK2. To this end, we monitored the autophagy flux in TamR MCF-7 cells after treatment with an HK2 inhibitor, 2-DG. The results showed that 2-DG could significantly decrease autophagy in TamR MCF-7 cells (Fig. 4B, 4D). Proliferation data showed that inhibition of HK2 by 2-DG could re-sensitize TamR MCF-7 cells to 4-OHT (Fig. 4F). In contrast, autophagy was reduced to a lesser degree in MCF-7 cells upon 2-DG treatment, and 2-DG only exerted a slight effect on the sensitivity of MCF-7 cells toward 4-OHT (Fig. 4F), suggesting that higher levels of HK2 contribute to 4-OHT resistance.
HK2 Inhibition Could Restore mTOR Activity and Suppress Autophagy
A previous study showed that the mTOR-S6K signaling axis was involved in endocrine resistance in breast cancer cells (32). Thus, we assessed the expression and activity of mTOR in TamR and parental MCF-7 cells. Our results showed that mTOR was up-regulated in TamR MCF-7 cells relative to MCF-7 cells (Fig. 1B, 5A). By contrast, the phosphorylation level of S6K, a downstream kinase of mTOR, was decreased significantly in TamR MCF-7 cells (Fig. 5A, 5D). These results indicate that the magnitude of increase in mTOR protein expression cannot fully compensate for the extent of attenuation in its kinase activity. Treatment with 4-OHT resulted in marked diminution in the phosphorylation level of S6K (Fig. 5B, 5E), suggesting that exposure to 4-OHT results in attenuated mTOR activity.
Fig. 5.
HK2 inhibition could restore mTOR activity and inhibit autophagy. A, Western blot analysis confirmed the elevated expression of mTOR and decreased S6K phosphorylation in TamR MCF-7 cells. B, Parental and TamR MCF-7 cells were treated with 2 μm 4-OHT for 24 h, and the protein expression levels of HK2, mTOR and phospho-S6K were monitored by Western blotting. C, The cells were treated with 2 μm 2-DG for 24 h, and the lysates were used to monitor the levels of HK2, mTOR and phospho-S6K by Western blotting analyses. D–F, The ratio of phospho-S6K over S6K was quantified from band intensity using Image J and the values in D and E are displayed relative to levels observed in the parental MCF-7 cells. The data were presented as mean ± S.D. (n = 3). The p values were calculated based on unpaired, two-tailed Student's t test: *, 0.01 ≤ p < 0.05; **, 0.001 ≤ p < 0.01; ***, p < 0.001.
It was previously reported that, in response to glucose deprivation, HK2 binds to and inhibits mTOR to facilitate autophagy (33–36). Therefore, we next asked whether HK2 could modulate mTOR activity and confer tamoxifen resistance. It turned out that inhibition of HK2 restored S6K phosphorylation in TamR MCF-7 cells, whereas no appreciable change in the phosphorylation of S6K was observed in parental MCF-7 cells (Fig. 5C, 5F). This finding agrees with the decreased autophagy elicited by HK2 inhibition in TamR MCF-7 cells (Fig. 4B). On the grounds that mTOR protein level was elevated and its activity was diminished in TamR MCF-7 cells, our results demonstrate that HK2 inhibits mTOR activity, thereby stimulating autophagy.
Genetic Depletion of HK2 Led to Increased mTOR Activity and Attenuated Autophagy, and Resensitized Tamoxifen-resistant MCF-7 Cells to 4-OHT
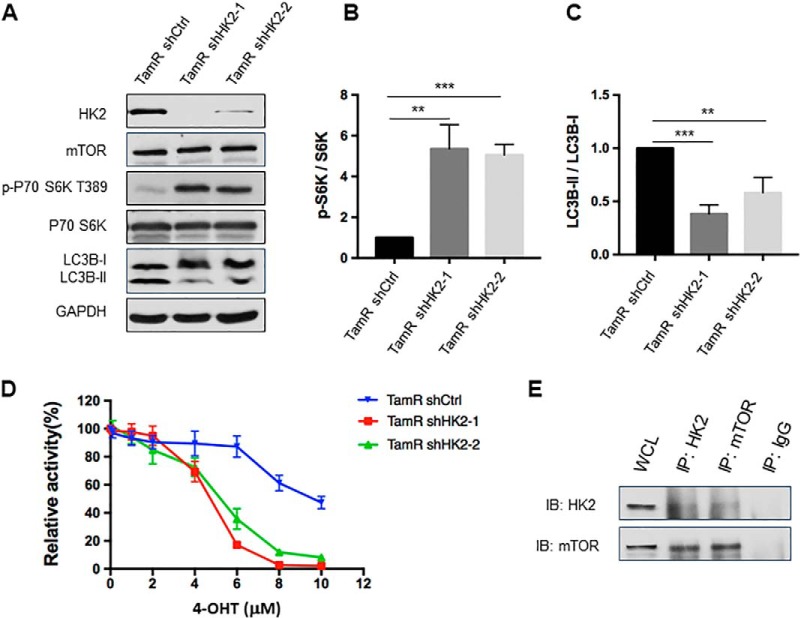
We generated TamR MCF-7 cells with stable knockdown of HK2 using two separate shRNAs, which gave rise to the depletion of HK2 protein by 97 and 80%, respectively (Fig. 6A). After stable knockdown of HK2, the phosphorylation level of S6K was significantly increased, which was accompanied with diminished autophagy compared with TamR MCF-7 cells stably expressing control shRNA (Fig. 6B–6C). Results from proliferation assay showed that HK2 knockdown re-sensitized TamR MCF-7 cells to 4-OHT (Fig. 6D).
Fig. 6.
Genetic depletion of HK2 led to increased mTOR activity and attenuated autophagy, and re-sensitized tamoxifen-resistant MCF-7 cells to 4-OHT. A, Validation of HK2 and mTOR expression levels and phospho-S6K, LC3B-II/LC3B-I changes after knocking down HK2 in TamR MCF-7 cells with two different sequences of shRNA. B, C, Quantification of changes in p-S6K and LC3B-II/LC3B-I in TamR MCF-7 cells upon shRNA-mediated knockdown of HK2. The values for p-S6K and LC3B-II were normalized against the band intensities of S6K and LC3B-I, respectively, The data were presented as the mean ± S.D. (n = 3). The p values were calculated using unpaired, two-tailed Student's t test: **, 0.001 ≤ p < 0.01; ***, p < 0.001. D, TamR MCF-7 shHK2 stable knockdown cells were treated with serially diluted 4-OHT for 72 h and cell viability was measured using CCK8. The data were normalized to control groups (ethanol) and represented by the mean and S.D. of results from three independent measurements. E, TamR MCF-7 cells were collected and cell lysates were immuno-precipitated using an antibody for either HK2 or mTOR, and the captured proteins were eluted and Western blotting was employed to monitor the interaction between HK2 and mTOR.
We next examined whether HK2 interacts with mTOR in TamR MCF-7 cells. Reciprocal immunoprecipitation followed by Western blot analysis revealed the interaction between HK2 and mTOR (Fig. 6E), suggesting that the binding of HK2 with mTOR suppresses its activity in tamoxifen-resistant cells. These results substantiated our hypothesis that HK2 stimulates autophagy through inhibiting mTOR activity, thereby leading to the 4-OHT resistance.
Overexpression of HK2 in MCF-7 Cells Led to Decreased mTOR Activity, Elevated Autophagy, and Augmented Resistance to 4-OHT
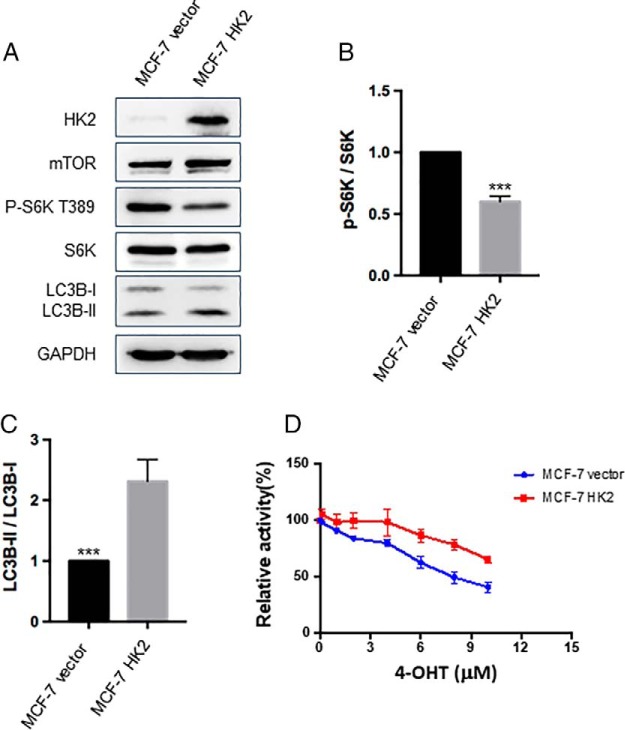
We also asked whether overexpression of HK2 in parental MCF-7 cells would mimic the tamoxifen-resistant environment. We found that stable overexpression of HK2 resulted in a significant decrease in the phosphorylation level of mTOR downstream kinase S6K and an induction of autophagy (Fig. 7). In addition, our results from proliferation assay showed that overexpression of HK2 in MCF-7 cells could confer resistance to 4-OHT (Fig. 7D). These results further strengthen the notion that HK2 inhibits mTOR activity and stimulates autophagy, thereby rendering MCF-7 cells resistant to tamoxifen.
Fig. 7.
Overexpression of HK2 in MCF-7 cells led to decreased mTOR activity, elevated autophagy, and augmented resistance to 4-OHT. A, Images from Western blotting for monitoring the changes in levels of HK2, mTOR, phospho-S6K, S6K, LC3B-II/LC3B-I in MCF-7 cells after overexpression of HK2. B–C, The quantification data obtained from the Western analyses. The data were presented as the mean ± S.D. (n = 3). The p values were calculated based on unpaired, two-tailed Student's t test: ***, p < 0.001. D, MCF-7 cells with stable overexpression of HK2 were treated with serially diluted 4-OHT for 72 h and cell viability was measured with CCK8. The data were normalized to control groups (ethanol) and represented by the mean and S.D. of results from three independent measurements.
DISCUSSION
Tamoxifen resistance has been the most challenging issue in breast cancer clinical treatment (9, 10). Hexokinases catalyze the first and rate-limiting step of glucose metabolism and these proteins are expressed at high levels in cancer cells, where their functions are required for tumor initiation and maintenance (25, 26). Our results obtained from cell-based assays and from bioinformatic analyses of publicly available data showed that HK2 expression level was up-regulated in tamoxifen-resistant MCF-7 breast cancer cells, supporting the potential role of HK2 in promoting tamoxifen resistance in ER-positive breast cancer. Along this line, it is worth noting that curcumin was shown to suppress resistance of breast cancer cells toward tamoxifen through a pathway involving HK2 (37).
Recently, dysfunction of autophagy was found to be a potential mechanism contributing to tamoxifen resistance (38–40). Silencing autophagy-related genes, such as ATG5, ATG7 or BECN1/Beclin1, re-sensitizes breast cancer cells to tamoxifen (41). Autophagy was found to be a key mechanism of cell survival in ER-positive breast cancer cells undergoing treatment with 4-OHT (40), though the molecular mechanism through which autophagy modulates tamoxifen resistance remained unclear.
We confirmed that tamoxifen-resistant MCF-7 cells exhibited elevated autophagy flux (Fig. 4A). Our results of extracellular acidification rate showed that tamoxifen-resistant MCF-7 cells exhibit higher glycolysis rate than parental MCF-7 cells (Fig. 3B–3D), providing a direct line of evidence to link elevated glycolysis rate with tamoxifen resistance. In ovarian cancer cells, HK2 confers resistance to cisplatin by enhancing drug-induced autophagy (31). Co-targeting HK2 with 2-DG and ULK1-dependent autophagy with chloroquine selectively kills cancer cells through intrinsic apoptosis, thereby leading to a nearly complete tumor suppression and pronounced extension in patient survival (39). However, it remains unknown whether HK2 functionally contributes to tamoxifen resistance in breast cancer through regulating autophagy. Here we observed decreased autophagy in TamR MCF-7 cells upon genetic depletion or pharmacological inhibition of HK2, underscoring a potent role of HK2 in modulating autophagy in drug-resistant breast cancer cells.
HK2 and mTORC1 were shown to cooperate to sense loss of homeostasis between glucose uptake and utilization, and to regulate anabolism and catabolism for the appropriate use of cellular resources (33, 35). In response to glucose deprivation, HK2 positively regulates glucose starvation-induced autophagy through mTORC1 inhibition (35). Inhibition of autophagy by mTOR was found to be a determinant of drug resistance in breast cancer (42). Investigation of mTOR-directed endocrine resistance mechanism has focused primarily on pathway cross-talk, where phosphorylation by S6K, a target of mTORC1, was shown to trigger endocrine-independent activation of ERα (32). Here we explored the relationship between HK2 and mTOR-S6K signaling axis and its implication in tamoxifen resistance. We observed that S6K phosphorylation was restored upon knock-down of HK2 or treatment with 2-DG. A previous study showed that HK2 binds with and inhibits mTOR activity in neonatal rat ventricular myocytes (35). Our reciprocal pull-down experiments showed the interaction between HK2 and mTOR in tamoxifen-resistant MCF-7 cells. Moreover, knockdown of HK2 led to elevated mTOR activity, diminished autophagy, and re-sensitized TamR MCF-7 cells to 4-OH tamoxifen (Fig. 6). On the other hand, overexpression of HK2 suppressed mTOR activity, which resulted in elevated autophagy and augmented resistance to tamoxifen in MCF-7 cells (Fig. 7).
It is worth discussing the limitation of our study. Only one pair of TamR and parental breast cancer cell lines were employed for the proteomic experiment and the subsequent functional/mechanistic investigations. Owing to differences in genetic and environmental factors, it is expected that some of the differentially expressed kinase proteins may not be generally associated with tamoxifen resistance in breast cancer patients. Nevertheless, our observation of the strong correlation between HK2 expression level and prognosis of breast cancer patients with tamoxifen therapy suggests that our observation made for HK2 should be valid at least for a significant subset of breast cancer patients.
Taken together, our study reveals the up-regulation of HK2 in tamoxifen-resistant MCF-7 cells. Additionally, elevated expression of HK2 promoted autophagy by inhibiting the mTOR-S6K signaling pathway, thereby rendering MCF-7 breast cancer cells resistant to 4-OHT. Hence, our work suggests HK2 as a novel target for developing drugs that overcome tamoxifen resistance in breast cancer therapy. Our proteomic data also revealed many other kinases exhibiting differential protein expression in the paired MCF-7 and TamR cells, which provide a wealth of information for future assessment about the roles of these kinases in resistance to tamoxifen therapy. Along this line, such studies can be leveraged by those small-molecule inhibitors whose selectivity and specificity in modulating kinase activity at the kinome-wide scale were recently examined (43).
DATA AVAILABILITY
Excel files containing the processed LC-PRM data are provided in Supplementary Materials. The raw files and the Skyline files for LC-PRM analyses of kinases for the paired breast cancer cells were deposited into PeptideAtlas with the identifier number of PASS01373 (http://www.peptideatlas.org/PASS/PASS01373). The kinome library was previously published and deposited into PeptideAtlas with the identifier number of PASS01178 (http://www.peptideatlas.org/PASS/PASS01178).
Supplementary Material
Acknowledgments
The authors would like to thank Dr. David Eastmond at UC Riverside and Dr. Guangdi Wang at Xavier University for providing the cell lines employed for the present study.
Footnotes
* This work was supported by the National Institutes of Health (R01 CA210072), and M.H. was supported by an NRSA T32 training grant (T32 ES018827).
 This article contains supplemental material. The authors declare that they have no conflict of interest.
This article contains supplemental material. The authors declare that they have no conflict of interest.
1 The abbreviations used are:
- ER
- estrogen receptor
- PRM
- parallel reaction monitoring
- RT
- retention time
- RSD
- relative standard deviation.
REFERENCES
- 1. Boyle P., and Howell A. (2010) The globalisation of breast cancer. Breast Cancer Res. 12, S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sullivan R., Peppercorn J., Sikora K., Zalcberg J., Meropol N. J., Amir E., Khayat D., Boyle P., Autier P., Tannock I. F., Fojo T., Siderov J., Williamson S., Camporesi S., McVie J. G., Purushotham A. D., Naredi P., Eggermont A., Brennan M. F., Steinberg M. L., De Ridder M., McCloskey S. A., Verellen D., Roberts T., Storme G., Hicks R. J., Ell P. J., Hirsch B. R., Carbone D. P., Schulman K. A., Catchpole P., Taylor D., Geissler J., Brinker N. G., Meltzer D., Kerr D., and Aapro M. (2011) Delivering affordable cancer care in high-income countries. Lancet Oncol. 12, 933–980 [DOI] [PubMed] [Google Scholar]
- 3. Diaby V., Tawk R., Sanogo V., Xiao H., and Montero A. J. (2015) A review of systematic reviews of the cost-effectiveness of hormone therapy, chemotherapy, and targeted therapy for breast cancer. Breast Cancer Res. Treat. 151, 27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Perou C. M., Sorlie T., Eisen M. B., van de Rijn M., Jeffrey S. S., Rees C. A., Pollack J. R., Ross D. T., Johnsen H., Akslen L. A., Fluge O., Pergamenschikov A., Williams C., Zhu S. X., Lonning P. E., Borresen-Dale A. L., Brown P. O., and Botstein D. (2000) Molecular portraits of human breast tumours. Nature. 406, 747–752 [DOI] [PubMed] [Google Scholar]
- 5. Sotiriou C., and Pusztai L. (2009) Gene-expression signatures in breast cancer. N. Engl. J. Med. 360, 790–800 [DOI] [PubMed] [Google Scholar]
- 6. Snigdha B., Neela S., Krishanu S., and Banerjee S. K. (2003) 17alpha-estradiol-induced VEGF-A expression in rat pituitary tumor cells is mediated through ER independent but PI3K-Akt dependent signaling pathway. Biochem. Biophys. Res. Commun. 300, 209–215 [DOI] [PubMed] [Google Scholar]
- 7. Chang X. Z., Li D. Q., Hou Y. F., Wu J., Lu J. S., Di G. H., Jin W., Ou Z. L., Shen Z. Z., and Shao Z. M. (2007) Identification of the functional role of peroxiredoxin 6 in the progression of breast cancer. Breast Cancer Res. 9, R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chang M. (2012) Tamoxifen resistance in breast cancer. Biomol Ther. 20, 256–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ali S., Rasool M., Chaoudhry H., P N. P., Jha P., Hafiz A., Mahfooz M., Abdus Sami G., Azhar Kamal M., Bashir S., Ali A., and Sarwar Jamal M. (2016) Molecular mechanisms and mode of tamoxifen resistance in breast cancer. Bioinformation 12, 135–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Early Breast Cancer Trialists' Collaborative, G. (2005) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet 365, 1687–1717 [DOI] [PubMed] [Google Scholar]
- 11. Duncan J. S., Whittle M. C., Nakamura K., Abell A. N., Midland A. A., Zawistowski J. S., Johnson N. L., Granger D. A., Jordan N. V., Darr D. B., Usary J., Kuan P. F., Smalley D. M., Major B., He X., Hoadley K. A., Zhou B., Sharpless N. E., Perou C. M., Kim W. Y., Gomez S. M., Chen X., Jin J., Frye S. V., Earp H. S., Graves L. M., and Johnson G. L. (2012) Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell 149, 307–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bantscheff M., Eberhard D., Abraham Y., Bastuck S., Boesche M., Hobson S., Mathieson T., Perrin J., Raida M., Rau C., Reader V., Sweetman G., Bauer A., Bouwmeester T., Hopf C., Kruse U., Neubauer G., Ramsden N., Rick J., Kuster B., and Drewes G. (2007) Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol. 25, 1035–1044 [DOI] [PubMed] [Google Scholar]
- 13. Xiao Y., and Wang Y. (2016) Global discovery of protein kinases and other nucleotide-binding proteins by mass spectrometry. Mass Spectrom. Rev. 35, 601–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barglow K. T., and Cravatt B. F. (2007) Activity-based protein profiling for the functional annotation of enzymes. Nat. Methods. 4, 822–827 [DOI] [PubMed] [Google Scholar]
- 15. Nomura D. K., Dix M. M., and Cravatt B. F. (2010) Activity-based protein profiling for biochemical pathway discovery in cancer. Nat. Rev. Cancer. 10, 630–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miao W., Guo L., and Wang Y. (2019) Imatinib-induced changes in protein expression and ATP-binding affinities of kinases in chronic myelocytic leukemia cells. Anal. Chem. 91, 3209–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miao W., and Wang Y. (2019) Quantitative interrogation of the human kinome perturbed by two BRAF inhibitors. J. Proteome Res. 18, 2624–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou C., Zhong Q., Rhodes L. V., Townley I., Bratton M. R., Zhang Q., Martin E. C., Elliott S., Collins-Burow B. M., Burow M. E., and Wang G. (2012) Proteomic analysis of acquired tamoxifen resistance in MCF-7 cells reveals expression signatures associated with enhanced migration. Breast Cancer Res. 14, R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., and Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 20. Miao W. L., Xiao Y. S., Guo L., Jiang X. G., Huang M., and Wang Y. S. (2016) A high-throughput targeted proteomic approach for comprehensive profiling of methylglyoxal-induced perturbations of the human kinome. Anal. Chem. 88, 9773–9779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., and MacCoss M. J. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carr S. A., Abbatiello S. E., Ackermann B. L., Borchers C., Domon B., Deutsch E. W., Grant R. P., Hoofnagle A. N., Huttenhain R., Koomen J. M., Liebler D. C., Liu T., MacLean B., Mani D. R., Mansfield E., Neubert H., Paulovich A. G., Reiter L., Vitek O., Aebersold R., Anderson L., Bethem R., Blonder J., Boja E., Botelho J., Boyne M., Bradshaw R. A., Burlingame A. L., Chan D., Keshishian H., Kuhn E., Kinsinger C., Lee J. S., Lee S. W., Moritz R., Oses-Prieto J., Rifai N., Ritchie J., Rodriguez H., Srinivas P. R., Townsend R. R., Van Eyk J., Whiteley G., Wiita A., and Weintraub S. (2014) Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol. Cell. Proteomics 13, 907–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sarbassov D. D., Guertin D. A., Ali S. M., and Sabatini D. M. (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 24. Sherwood C. A., Eastham A., Lee L. W., Risler J., Vitek O., and Martin D. B. (2009) Correlation between y-type ions observed in ion trap and triple quadrupole mass spectrometers. J. Proteome Res. 8, 4243–4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Patra K. C., Wang Q., Bhaskar P. T., Miller L., Wang Z. B., Wheaton W., Chandel N., Laakso M., Muller W. J., Allen E. L., Jha A. K., Smolen G. A., Clasquin M. F., Robey R. B., and Hay N. (2013) Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell 24, 399–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shinohara Y., Yamamoto K., Kogure K., Ichihara J., and Terada H. (1994) Steady state transcript levels of the type II hexokinase and type 1 glucose transporter in human tumor cell lines. Cancer Lett. 82, 27–32 [DOI] [PubMed] [Google Scholar]
- 27. Cho K. S., Yoon Y. H., Choi J. A., Lee S. J., and Koh J. Y. (2012) Induction of autophagy and cell death by tamoxifen in cultured retinal pigment epithelial and photoreceptor cells. Invest. Ophth. Vis. Sci. 53, 5344–5353 [DOI] [PubMed] [Google Scholar]
- 28. Graham C. D., Kaza N., Klocke B. J., Gillespie G. Y., Shevde L. A., Carroll S. L., and Roth K. A. (2016) Tamoxifen induces cytotoxic autophagy in glioblastoma. J. Neuropath. Exp. Neur. 75, 946–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nagelkerke A., Sieuwerts A. M., Bussink J., Sweep F. C. G. J., Look M. P., Foekens J. A., Martens J. W. M., and Span P. N. (2014) LAMP3 is involved in tamoxifen resistance in breast cancer cells through the modulation of autophagy. Endocr-Relat. Cancer 21, 101–112 [DOI] [PubMed] [Google Scholar]
- 30. Yonekawa T., and Thorburn A. (2013) Autophagy and cell death. Essays Biochem. 55, 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang X. Y., Zhang M., Cong Q., Zhang M. X., Zhang M. Y., Lu Y. Y., and Xu C. J. (2018) Hexokinase 2 confers resistance to cisplatin in ovarian cancer cells by enhancing cisplatin-induced autophagy. Int. J. Biochem. Cell B 95, 9–16 [DOI] [PubMed] [Google Scholar]
- 32. Yamnik R. L., and Holz M. K. (2010) mTOR/S6K1 and MAPK/RSK signaling pathways coordinately regulate estrogen receptor alpha serine 167 phosphorylation. FEBS Lett. 584, 124–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tan V. P., and Miyamoto S. (2015) HK2/hexokinase-II integrates glycolysis and autophagy to confer cellular protection. Autophagy 11, 963–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roberts D. J., and Miyamoto S. (2015) Hexokinase II integrates energy metabolism and cellular protection: Akting on mitochondria and TORCing to autophagy. Cell Death Differ. 22, 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Roberts D. J., Tan-Sah V. P., Ding E. Y., Smith J. M., and Miyamoto S. (2014) Hexokinase-II positively regulates glucose starvation-induced autophagy through TORC1 inhibition. Mol. Cell 53, 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kundu M. (2014) Too sweet for autophagy: hexokinase inhibition of mTORC1 activates autophagy. Mol. Cell 53, 517–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Geng C., Li J., Ding F., Wu G., Yang Q., Sun Y., Zhang Z., Dong T., and Tian X. (2016) Curcumin suppresses 4-hydroxytamoxifen resistance in breast cancer cells by targeting SLUG/Hexokinase 2 pathway. Biochem. Biophys. Res. Commun. 473, 147–153 [DOI] [PubMed] [Google Scholar]
- 38. Lee M. H., Koh D., Na H., Ka N. L., Kim S., Kim H. J., Hong S., Shin Y. K., Seong J. K., and Lee M. O. (2018) MTA1 is a novel regulator of autophagy that induces tamoxifen resistance in breast cancer cells. Autophagy 14, 812–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qadir M. A., Kwok B., Dragowska W. H., To K. H., Le D., Bally M. B., and Gorski S. M. (2008) Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res. Treat. 112, 389–403 [DOI] [PubMed] [Google Scholar]
- 40. Samaddar J. S., Gaddy V. T., Duplantier J., Thandavan S. P., Shah M., Smith M. J., Browning D., Rawson J., Smith S. B., Barrett J. T., and Schoenlein P. V. (2008) A role for macroautophagy in protection against 4-hydroxytamoxifen-induced cell death and the development of antiestrogen resistance. Mol. Cancer Ther. 7, 2977–2987 [DOI] [PubMed] [Google Scholar]
- 41. Wang L., Wang J., Xiong H., Wu F. X., Lan T., Zhang Y. J., Guo X. L., Wang H. N., Saleem M., Jiang C., Lu J. X., and Deng Y. B. (2016) Co-targeting hexokinase 2-mediated Warburg effect and ULK1-dependent autophagy suppresses tumor growth of PTEN- and TP53-deficiency-driven castration-resistant prostate cancer. Ebiomedicine 7, 50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Geter P. A., Ernlund A. W., Bakogianni S., Alard A., Arju R., Giashuddin S., Gadi A., Bromberg J., and Schneider R. J. (2017) Hyperactive mTOR and MNK1 phosphorylation of eIF4E confer tamoxifen resistance and estrogen independence through selective mRNA translation reprogramming. Gene Dev. 31, 2235–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Klaeger S., Heinzlmeir S., Wilhelm M., Polzer H., Vick B., Koenig P. A., Reinecke M., Ruprecht B., Petzoldt S., Meng C., Zecha J., Reiter K., Qiao H., Helm D., Koch H., Schoof M., Canevari G., Casale E., Depaolini S. R., Feuchtinger A., Wu Z., Schmidt T., Rueckert L., Becker W., Huenges J., Garz A. K., Gohlke B. O., Zolg D. P., Kayser G., Vooder T., Preissner R., Hahne H., Tonisson N., Kramer K., Gotze K., Bassermann F., Schlegl J., Ehrlich H. C., Aiche S., Walch A., Greif P. A., Schneider S., Felder E. R., Ruland J., Medard G., Jeremias I., Spiekermann K., and Kuster B. (2017) The target landscape of clinical kinase drugs. Science 358, eaan4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Excel files containing the processed LC-PRM data are provided in Supplementary Materials. The raw files and the Skyline files for LC-PRM analyses of kinases for the paired breast cancer cells were deposited into PeptideAtlas with the identifier number of PASS01373 (http://www.peptideatlas.org/PASS/PASS01373). The kinome library was previously published and deposited into PeptideAtlas with the identifier number of PASS01178 (http://www.peptideatlas.org/PASS/PASS01178).