The LC-MS/MS-based quantitative phosphoproteomic and metabonomic approaches were used to identify changes in phosphoproteins and metabolites in soybean roots in response to rhizobia inoculation and salt stress. The GmMYB183 protein was found to regulate the transcriptional expression of the GmCYP81E11 gene, which contributes to the accumulation of ononin, a monohydroxy B-ring flavonoid negatively regulate soybean tolerance to salinity. Rhizobia could inhibit the phosphorylation of GmMYB183 and its activity in abovementioned transcriptional regulation, to enhance soybean's tolerance to salinity.
Keywords: Label-free quantification, phosphoproteome, transcription, iTRAQ, metabolomics, flavonoid, GmMYB183, phosphorylation, salt tolerance, soybean
Graphical Abstract
Highlights
Rhizobia affects flavonoid metabolism via GmMYB183 phosphorylation.
Flavonoids rebalance the cellular ROS under salt stress.
Mechanistic insights into soybean responses to salinity revealed by proteomics.
Abstract
Soybean (Glycine max (L.) Merrill) is an important component of the human diet and animal feed, but soybean production is limited by abiotic stresses especially salinity. We recently found that rhizobia inoculation enhances soybean tolerance to salt stress, but the underlying mechanisms are unaddressed. Here, we used quantitative phosphoproteomic and metabonomic approaches to identify changes in phosphoproteins and metabolites in soybean roots treated with rhizobia inoculation and salt. Results revealed differential regulation of 800 phosphopeptides, at least 32 of these phosphoproteins or their homologous were reported be involved in flavonoid synthesis or trafficking, and 27 out of 32 are transcription factors. We surveyed the functional impacts of all these 27 transcription factors by expressing their phospho-mimetic/ablative mutants in the roots of composite soybean plants and found that phosphorylation of GmMYB183 could affect the salt tolerance of the transgenic roots. Using data mining, ChIP and EMSA, we found that GmMYB183 binds to the promoter of the soybean GmCYP81E11 gene encoding for a Cytochrome P450 monooxygenase which contributes to the accumulation of ononin, a monohydroxy B-ring flavonoid that negatively regulates soybean tolerance to salinity. Phosphorylation of GmMYB183 was inhibited by rhizobia inoculation; overexpression of GmMYB183 enhanced the expression of GmCYP81E11 and rendered salt sensitivity to the transgenic roots; plants deficient in GmMYB183 function are more tolerant to salt stress as compared with wild-type soybean plants, these results correlate with the transcriptional induction of GmCYP81E11 by GmMYB183 and the subsequent accumulation of ononin. Our findings provide molecular insights into how rhizobia enhance salt tolerance of soybean plants.
Soybean (Glycine max (L.) Merrill) is becoming an important ingredient of human diet and animal feeds. It is also used as an important industrial material because of its high content of protein and oil. Hence it is not surprising that the demand for soybean is growing at an unprecedented pace (2, 3). Salinity is a major abiotic stress that limits the growth and yields of soybean (4). The excessive Na+ ions usually stimulate the accumulation of reactive oxygen species (ROS)1 and triggers oxidative stress in plant cells (5, 6). Plants develop two main strategies to rebalance the intracellular ROS, including enzymatic degradation (7) and deoxidization using the reductive capacities of small molecules such as ascorbic acid, mannitol and flavonoid (2, 8).
Flavonoids from plants constitute a huge family with over 10,000 species reported so far (9, 10). Plants synthesize various classes of flavonoids, such as chalcones, flavanones (e.g. naringenin), isoflavone (e.g. ononin,) and flavonols (e.g. quercetin) (9). These flavonoids have been reported to play a wide range of physiological functions in plant defense responses to abiotic stresses, for example, protecting plants from harmful UV radiation (11–13) and eliminating destructive hydrogen peroxide (6, 14, 15). The hydroxyl groups of flavonoids play a pivotal role in fulfilling their bioactive functions when they are oxidized to quinones during these processes (16–19).
The flavonoids in soybean also play a critical role in plant-microbe symbiosis (20). Some species of flavonoids diffuse into rhizosphere signal the presence of hosts to the rhizobia before the initiation of the rhizobia-legume interaction. Once they are perceived by rhizobia, the bacteria will be induced to express the NodD protein which forms a complex with isoflavones. This complex further activates the transcription of the bacterial nodulation genes (20), producing Nod factor which triggers a series of biological responses in the soybean roots and finally leads to the nodule formation on the roots of the legume host (21).
Microbes have been known to play significant roles in enhancing legume tolerance to salinity. Ectomycorrhizal fungi were reported to help their host plants to tolerate salt stress by reducing the accumulation of Na+ and increasing the absorption of K+ (22–24). Some rhizosphere bacteria could also enhance the host's salt tolerance using bioactive components such as indolacetic acid, phenolic compounds, flavonoids and antioxidant enzymes in plants (25). We recently found that inoculation of rhizobia could enhance soybean's tolerance to salinity (4). However, the molecular mechanism by which rhizobia regulate soybean salt responses is still unclear.
To date, three categories of enzymes, including the chalcone synthase (CHS), chalcone isomerase (CHI) and cytochrome P450 monooxygenase (CYP), are documented to play significant roles in the complicated flavonoid metabolic processes where 4-coumaroyl-CoA and malonyl-CoA are first converted to naringenin chalcone by CHS. The chalcone is then converted to flavanone and isoflavone by CHI and CYP, respectively. Interestingly, CHS, CHI and CYP have been shown to play key roles in plant responses to salt stress (26–28). For example, the RNA interference of GmFNSII (coding for CYP93B) significantly reduces the soybean root's tolerance to salinity because of the decreased flavone level and increased accumulation of H2O2 (8). Our recent study found that salt tolerance of Arabidopsis and soybean were positively regulated by CHS but negatively regulated by CHI and CYP (3).
Genes encoding CHSs, CHIs, and CYPs that are involved in various flavonoid biosynthetic steps, are regulated by MYB transcription factors (29–33). AtMYB75, AtMYB90, MYB112, MYB113, and MYB114 have been shown to play important roles in activating genes of the anthocyanin biosynthetic pathway (32, 34, 35). In addition, the Scutellaria baicalensis MYB8 was reported to improve drought stress tolerance by regulating flavonoid biosynthesis (36). Moreover, GmMYBJ2 was found to markedly improve the seed germination rate of transgenic Arabidopsis plants undergoing water-deficit stress (37).
Omics studies monitoring the proteome and metabolome which are directly related to the development of phenotype, allow us to obtain a comprehensive insight and to correlate specific nodes of the stress-related molecular networks to the establishment of stress tolerance (38). Furthermore, the quantitative analyses of these high-throughput data at the systemic level provide us with unique opportunities to investigate the precise network structure and signaling kinetics underlying physiological processes during the establishment of plant tolerance to the causative stresses (39).
In this study, we attempted to explore whether and how rhizobia inoculation affects soybean's phosphoproteome and metabonome, and how these changes help protect soybean from salinity. We identified changes in phosphorylation status of transcription factor MYB183 in response to both salt treatment and rhizobia inoculation. In addition, we documented a mechanism regarding how GmMYB183 regulates the expression of CYP and flavonoid biosynthesis using reverse genetic and metabolomic approaches.
EXPERIMENTAL PROCEDURES
Plant Materials, Growth Conditions, Rhizobia Inoculation and NaCl Treatments
Seeds of Glycine max cultivar Union85140, kindly provided by Prof. Lijuan Qiu at the Chinese Academy of Agricultural Sciences, were surface sterilized and germinated between wet filter papers as previously described (3). Two days after germination, the seedlings were inoculated with Sinorhizobium fredii 15006 (Agricultural Culture Collection of China, http://www.accc.org.cn/show.asp) as described (4), and then transplanted into a mixture of pearlite and sphagnum peat (v : v = 1 : 3). The light intensity in the growth chamber was set at 200 μmol×m−2×s−1 with a photo period of 18 h of light per day; and the temperature was set as 25/18 °C for day/night cycle. The seedlings were watered with 1/4 strength of Fahräeus medium every 4 days and deionized water was used to irrigate every 2 days after adding Fahräeus medium. At the second trifoliate leaf stage, the seedlings were treated with 200 mM of NaCl for 24 h as described by Qu et al. (4). Next, the samples were collected and immediately stored at −80 °C until further use.
Protein Extraction and Digestion with FASP
The TCA/Acetone extraction method was used to isolate the total proteins from soybean roots as previously described (3). Five grams of root tissue for each sample was ground into fine powder in liquid nitrogen. The powder was thoroughly suspended in 45 ml of pre-cooled TCA/Acetone (v: v = 1: 9) for overnight protein extraction at −20 °C. The homogenate was then centrifuged at 7000 × g for 20 min and the pellet was washed three times with 40 ml acetone. After that, the residual acetone was removed by vacuum and 50 mg of white powder from each sample was resuspended in 1 ml of SDT lysis buffer (4% SDS, 100 mM Tris-HCl, 1 mM DTT, 1 mM PMSF, pH 7.6, including 1× PhosSTOP phosphatase inhibitor mixture from Roche, Basel, Switzerland). The solution was boiled for 15 min in a thermal block, sonicated for 100 s, centrifuged at 13,400 × g for 15 min, and the pellet was discarded. The protein concentration in the supernatant was measured using the BCA (bicinchoninic acid) method and 20 μg of extracted proteins was run in an SDS-PAGE gel for quality assurance.
The protein digestion was carried out as previously described (3). Before digestion, each sample containing 200 μg of protein was processed through filter aided sample preparation (FASP) procedure to remove the residual SDS. Next, the concentrated proteins were digested with 8 μg of trypsin at 37 °C for 16 to18 h. After digestion, the peptide solution was passed through a Microcon filtration device (MWCO 10 kd) and the OD280 was measured to estimate its concentration.
All the above procedures were carried out at 4 °C unless otherwise stated.
Eight-plex iTRAQ Labeling and Phosphopeptide Enrichment
An aliquot containing 100 μg of digested peptides from each sample was subjected to AB Sciex iTRAQ labeling. The eight-plex iTRAQ labeling was performed according to the manufacturer's instructions.
TiO2 beads were used to enrich phosphopeptides as previously described (3). The labeled peptides were acidified with 50 μl of DHB buffer (3% 2, 5-dihydroxy benzoic acid, 80% acetonitrile and 0.1% TFA), and then incubated with 25 μg of TiO2 beads (10 μm in diameter, Sangon Biotech, Shanghai, China) for 40 min at room temperature. After the incubation, the TiO2 beads were spun down and the pellets were packed into 10 μl pipette tips. The peptide-TiO2 beads were washed three times with 20 μl of wash solution I (20% acetic acid, 300 mM of octanesulfonic acid sodium salt and 20 mg/ml DHB), then three times with 20 μl of wash solution II (70% water; 30% acetonitrile). The phosphopeptides were finally eluted using freshly prepared ABC buffer (50 mM of ammonium phosphate, pH 10.5). The enriched phosphopeptide solution was lyophilized and redissolved in 20 μl 0.1% TFA solution for Nano RPLC-MS/MS Analysis.
NanoRPLC-MS/MS Analysis of Phosphorylated Peptides
For NanoRPLC-MS/MS analysis, 5 μg (≤ 10 μl) phosphopeptides solution was loaded into a two dimensional EASY-nLC1000 system coupled to a Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Scientific, Waltham, Massachusetts). In the nanoLC separation system, the mobile phase A solution contained 0.1% formic acid in water, and mobile the phase B solution contained 84% acetonitrile and 0.1% formic acid. Before peptide loading, the Thermo EASY SC200 trap column (RP-C18, 3 μm, 100 mm × 75 μm) was pre-equilibrated with 95% mobile phase A for 30 min. The phosphopeptides were first transferred to the Thermo scientific EASY column (2 cm × 100 μm 5 μm-C18) and then separated through the trap column using a gradient of 0% to 55% mobile phase B for 220 min. Then the columns were rinsed with 100% mobile phase B for 8 min and re-equilibrated to the initial conditions for 12 min. The flow rate of the above procedures was set at 0.25 μl per minute.
Phosphopeptide Identification and Quantitative Analysis
The raw data were extracted by Mascot2.2 and analyzed by Proteome Discoverer1.4 (Thermo Scientific). To identify the phosphopeptides, the mascot data was searched against 74305 entries curated in the peptide database uniprot_Glycine_74305_20140429.fasta (http://www.uniprot.org/, on April 29, 2014). To generate reliable phosphopeptides, the missed cleavages and false discovery rates (FDR) values were set at less than 2 and 0.01, respectively; the mass tolerances for precursor and fragment ions were set at less than 20 ppm and 0.1 Da, respectively. In addition, the carbamidomethyl (C), Itraq8plex (N-term) and iTRAQ8plex (K) were chosen as fixed modifications, whereas the oxidation (M), phospho (ST), and phospho (Y) were selected as variable modifications.
Metabolomic Analysis
The metabolomics analysis was conducted mostly as previously described (40) with appropriate modifications. 5 g of root tissue from each sample was homogenized in liquid nitrogen. For metabolite extraction, 50 mg homogenate was dissolved in 1 ml of pre-cooled methanol: acetonitrile: water solution (2:2:1, v/v/v). The mixtures were briefly vortexed, and then sonicated for 30 min, these vortex-sonication steps were repeated twice. After 60 min incubation at −20 °C, the mixture was centrifuged at 14,000 × g for 15 min.
Two microliters of supernatant was injected into the Agilent 1290 Infinity LC system coupled to a Triple TOF 6600 Mass Spectrometer (AB SCIEX, Framingham, Massachusetts). In the UHPLC separation system, the mobile phase A solution contained 25 mM ammonium acetate and 25 mM ammonia in water, and the mobile phase B solution contained 100% acetonitrile. The ACQUITY UPLC BEH Amide separation column (Waters, 1.7 μm, 2.1 mm× 100 mm) was pre-equilibrated with 85% mobile phase B at 4 °C before metabolite loading. The metabolites were separated using a gradient of 85–65% mobile phase B for 12 min. Then, the mobile phase B was kept at 40% for 3 minuntes followed by a 5 min rinse with 85% of mobile phase B. The flow rate of the total mobile phase was set at 300 μl per minute. Ten biological replicates were analyzed for each treatment. The QC (Quality Control) sample was used to calibrate the equipment after every 5 metabolite samples were tested.
The separated metabolites were ionized with ESI positive and negative modes. The parameters of MS analysis were listed in supplemental Table S1. The raw data was transformed into mzML format via the software ProteoWizard. The calibration of retention time, extraction of peak integration and value normalization was performed using the XCMS program. The identity of each ion was searched against the METLIN database (https://metlin.scripps.edu/index.php).
Experimental Design and Statistical Rationale for Mass Spectrometric Data Analyses
In this research, four biological replicates of each treatment were analyzed. The average value of the four replicates served as a normalizing reference (REF) for the calculation of peptide content.
The MS data ranged from 350 to 1800 m/z was acquired at the resolution of 70,000. To acquire the MS/MS spectra, the 10 most abundant ions from each MS scan were subsequently dissociated by higher energy collisional dissociation (HCD) in alternating data-dependent mode with a resolution of no less than 17500.
For the spectra of a specific sample, the median of its intensity values was used for quantification analysis. The average of 8 median values of the spectra for a specific sample was used as a reference (REF in supplemental Table S2) for data normalization in one Eight-plex iTRAQ labeled sample group. A Student's t test was performed using the standard deviation to assess the changes between two samples (each had four biological replicates). The phosphopeptides that passed both FDR (False Discovery Rate) value ≤ 0.05 and p value ≤ 0.05 were significantly changed.
Bioinformatic Analysis
Peptide motifs were extracted using the motif-X algorithm (http://motif-x.med.harvard.edu/motif-x.html) (41). The width of the resulting motifs was set as seven amino acids; and S or T was selected as the central amino acid. The kinases that might use them as targets were predicted by the PhosphoMotif Finder software (http://hprd.org/PhosphoMotif_finder) using default search parameters.
The MYB binding motifs in promoters of interested genes were predicted using the JASPAR database and its associated tools (http://jaspar.genereg.net/) as previous described (53). Briefly, all known MYB binding motifs in plants were first found using the 'Search' tool. Then, the −1kb region of the CYP81E11 genomic sequence was scanned for the existence of these plant MYB binding motifs with an 80% relative profile score threshold.
RNA Extraction and Quantitative RT-PCR
RNA extraction, mRNA reverse transcription and quantitative real-time PCR were performed as previously described (3). Briefly, 100 mg of root tissue was ground to fine powder in liquid nitrogen and then suspended in 1 ml of TRIzol reagent solution (Invitrogen, Carlsbad, CA). Total RNA was isolated according to the manufacturer's instructions, and then subjected to DNA digestion with 20 units of DNase I for 15 min at 25 °C. The isolated total RNA was run on 1.5% (w/v) agarose gels for quality assurance.
About 3 μg of the total RNA template was used to synthesize cDNA using the HiFiScript first Strand cDNA Synthesis Kit (CWBiotech, Beijing, China) following the manufacturer's instructions. UltraSYBR Mixture (CWBiotech) was used for qRT-PCR on a Bio-Rad CFX96 PCR instrument. The primers (supplemental Table S3) were selected using the Primer Blast online software (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) against the Glycine max (taxid:3847) database. Each reaction mixture consisted of 10 ng of cDNA template, 1 μl each forward and reverse primers (10 μM stocks), and 10 μl of SYBR green in a final volume of 20 μl. PCR reaction was run with an initial denaturing temperature of 95 °C for 10 min followed by 40 cycles at 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 32 s. Melting curve analysis was also performed at the end of the PCR reaction to verify that only one product was amplified. PCR efficiencies and expression levels were calculated for target and reference gene from a serial dilution of the cDNA template. Target gene transcript levels were normalized to the transcript level of GmActin11 (3).
Plant Expression Constructs
Full-length cDNA sequences encoding for soybean GmMYB183 (Glyma06G187600), and GmCYP81E11 (Glyma09g05440.1) were retrieved from the Phytozome database (http://www.phytozome.net/soybean), and synthesized using RT-PCR from soybean total RNA, and the amplified fragments were cloned in pEASY-Blunt Zero (TransGen Biotech, Beijing, China). Primer sequences are listed in supplemental Table S3 and all constructs were verified by sequencing.
For gene overexpression and ChIP analysis, the full-length CDS of target genes were sub-cloned into the pDL28-HA/Ubq10::dsRed vector, a derivate of pDL28 carrying an HA-tag and a Ubq10 driven dsRed (42), between SacI and KpnI (GmMYB183) sites or BamH I single site (GmCYP81E11), downstream to the 35S promoter. The empty pDL28-HA/Ubq10::dsRed vector was used as a negative control. All these constructs were transformed into the Union85140 via Agrobacterium rhizogenes strain K599. The composite plants with transgenic roots were treated with 200 mM of NaCl in 1/4 fold Fahräeus medium for 24 h. The transgenic roots were verified by their expression of dsRed.
For subcellular localization analysis, the full-length CDS of GmMYB183 was cloned into the pDL28 vector (42) between SacI and KpnI sites, downstream to the 2×35S promoter (defined as pDL28-gene). The pDL28, a derivative of pCambia1300, carries a 2×35S promoter and an HA-tag for target gene expression and a Ubq10 driven dsRed for convenient selection of transgenic roots. Then, the RFP-tag was inserted into the pDL28-gene construct between KpnI and SalI (defined as pDL28-gene-RFP). The pDL28-gene-RFP was transformed into the Union85140 via A. rhizogenes strain K599.
For RNAi-mediated knockdown, the Oligo Engine 2.0 software was used to select an approximate 300 bp (300∼800) length fragment from the coding region of the target gene. The fragment was amplified from a cDNA library and cloned into the pHANNIBAL vector in opposite orientations on either side of a PDK intron for constructing an invert repeat (10). The RNAi construct was sub-cloned into the pDL28-HA/Ubq10::dsRed vector between SalI and SpeI (GmMYB183 and GmCYP81E11) sites downstream to the 35S promoter. The pDL28-Gene-RNAi vector was transformed into A. rhizogenes strain K599 for transformation of soybean roots.
For site-directed mutations, the primers (supplemental Table S3) were generated by QuikChange Primer Design software (http://www.genomics.agilent.com/primerDesignProgram.jsp). Full-length cDNA of the target gene was inserted into pEASY®-Blunt Zero Cloning Vector (TransGen Biotech) and mutated by PCR. The PCR product was digested with 1 U DpnI enzyme (Thermo Scientific) at 37 °C for 1 h and then used to transform the DH5α strain of E. coli. The mutated cDNA was then sub-cloned into pDL28 derivatives between SacI and KpnI sites.
Subcellular Localization
Soybean roots were mounted between glass slides and cover slips, and then imaged using an inverted Carl Zeiss LSM 710 laser scanning microscope (43).
ChIP-qPCR Assay
The transgenic roots expressing empty vector (EV), pDL28-35S::GmMYB183-HA/Ubq10::dsRed and pDL28-GmMYB183-RNAi constructs were used for ChIP-qPCR. ChIP analysis was performed as previously described (44) with minor revisions, and 1.5 g of fresh root sample was cross-linked with 1% (v/v) of formaldehyde for 10 min before the chromatin preparation. Briefly, the cross-linked roots were ground into fine powders in liquid nitrogen and incubated in 10 ml pre-cold lysis buffer (HEPES 0.05 M, NaCl 0.14 M, EDTA 0.001 M, 10% glycerol, 0.5% NP-40 and 0.25% Triton X-100, pH7.5) for 10 min. After centrifugation (4 °C, 1000 × g, 15 min), the pellets were resuspended in 10 ml extraction buffer (NaCl 0.2 M, EDTA 0.001 M, EGTA 0.0005 M and Tris 0.01 M, pH8.0) and rocked for 10 min at room temperature. The nuclei were then pelleted by centrifuging at 1000 × g for 15 min at 4 °C. The pellets were resuspended in 3 ml of ice-cold chromatin extraction buffer (EDTA 0.001 M, EGTA 0.0005 M and Tris 0.01 M, pH8.0) for chromatin sonication. The sonication was carried out at 20% power output for 160 s using a Sonics Vibra Cell Sonicator (Sonics & materials Inc, CT). The debris was discarded after centrifuging at 2000 × g for 15 min at 4 °C and the rabbit anti-HA antibody was used to precipitate the GmMYB183-HA:DNA complexes in the supernatant. The Antibody:GmMYB183-HA:DNA complexes were isolated with protein A/G at 4 °C overnight and then centrifuged at 1000 × g for 15 min at 4 °C. The DNA was extracted from the pellet for qPCR assay. Enrichments were calculated by computation of ΔΔCT [CT(EV) -CT(anti-HA)]. Primers used are listed in supplemental Table S3.
Electrophoresis Mobility Shift Assay (EMSA)
DNA fragment encoding the N-terminal 168 aa containing the DNA-binding domain (aa 74–130) of GmMYB183 were amplified using PCR and with a primer pair listed in the supplemental Table S3. The amplified fragment was cloned in pEASY-blunt (Transgene Biotech). Site-directed mutagenesis was used to generate the S61A and S61D mutants. All the constructs in pEASY-blunt were sequenced and sub-cloned into pET32a vector between EcoRI and XhoI in the reading frame to express the 6 × His-tags at both N- and C-ends of the 168 aa GmMYB183 truncated protein or its mutants. In addition, the 184 aa Trx protein with two his-tags at its C terminus expressed from the empty pET32b vector and was used as a negative control. These expression constructs were transformed into E. coli strain BL21 (DE3) pLysS, and the WT or mutated versions of recombinant GmMYB183 protein was purified through nickel-affinity chromatography (Ni-NTA, Qiagene). Probe labeling and EMSA were carried out as previously described (42). Briefly, oligo primers contain a 30 bp tested promoter sequence were synthesized and four hot (32P-labeled) “A” were added at the end of both strands using Klenow fragment of DNA polymer I. 32P-labeled CYP81E11-P1 probe (see supplemental Table S3, 104 cpm per sample) was incubated with 2 μg of tested protein at room temperature for 30 min. The reaction mixture was separated on 8% of polyacrylamide gel in 0.5 x TBE buffer, and the dried gel was exposed to X-ray film for 48 to 96 h.
In Vitro Kinase Activity Assays
The Full length cDNA of soybean casein kinase-II (GmCK2) was generated with PCR using a pair of primers with EcoRI and XhoI added to the 5′- and 3′- ends (supplemental Table S3). The fragment was cloned in pEASY-blunt (Transgene), sequenced and sub-cloned into pET32a to express the 6 × His-tags at both N- and C-ends. The purified GmCK2α was used to test whether recombinant GmMYB183 (aa 1–168, see the previous section) could be phosphorylated by GmCK2α. The wild type and S61D mutant of GmMYB183 was tested as substrate. The substrate phosphorylation assay was performed in 20 μl of kinase buffer containing HEPES (25 mM), MgCl2 (5 mM), 50 μM ATP, 1 μCi of [γ-32P] ATP, 0.4 μg GmCK2α and 0.4 μg GmMYB183-WT or GmMYB183-S61D for 30 min at 25 °C. The reactions were stopped using SDS loading buffer and electrophoresed on a 12% SDS-PAGE. The dried gel was exposed X-ray film for 72 h.
HPLC Analysis
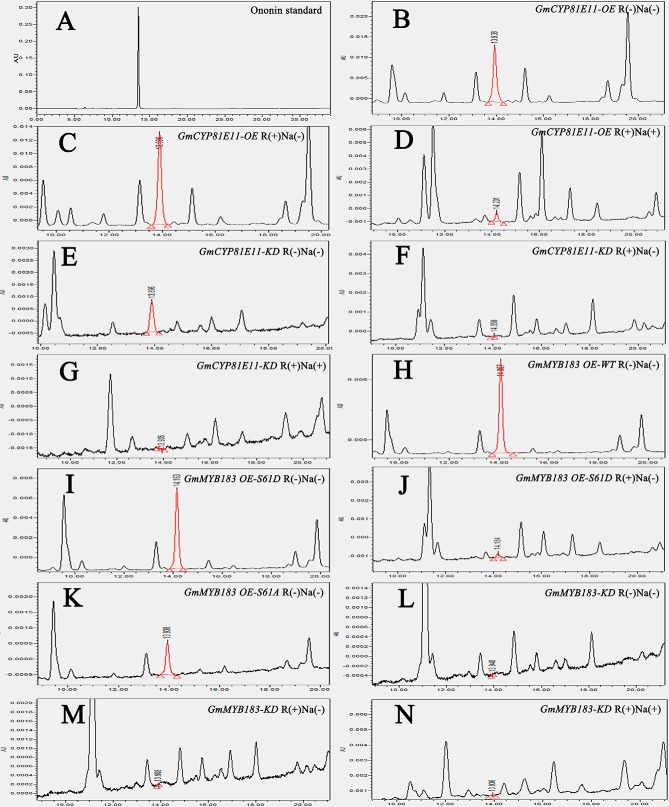
HPLC method for flavone quantification was carried out as previously described with minor modifications (10). Briefly, 25 mg roots were ground in liquid nitrogen, and extracted with 1.25 ml of pre-cooled methanol: acetic acid: water (9:1:10, v/v/v) at room temperature for 30 min. After 5 min of incubation, the supernatant of extract was filtered through a 0.22 μM membrane. Then, 10 μl of the purified extract was injected into a Waters HPLC system and separated by a 150 mm × 4.6 mm × 5 μm RP-Spherisorb-ODS2-C18 analytical column. The flavone compounds were monitored with a Waters 600 Controller and a PDA Detector between the wavelengths of 200 and 600 nm. The mobile phase A contained 10% acetonitrile and 0.1% trifluoroacetic acid in water (v/v/v), and mobile phase B contained 90% acetonitrile and 0.1% trifluoroacetic acid in water (v/v/v). The separation gradient was set as 10% to 40% mobile phase B for 30 min at a flow rate of 0.5 ml per minute, followed by 40% to 100% mobile phase B for 5 min. Then, the column was rinsed with 100% of the mobile phase B for 2 min. Peak identification was confirmed by both retention time against standards and UV absorption spectra. The flavone or flavone aglycone standard ononin was purchased from Sigma-Aldrich, St. Louis, MO, and the cyanidin 3-arabinoside chloride was purchased from Miragen, Boulder, Colorado.
Assay of Superoxide (O2·−) Scavenging Capacity Assay
To access the root's superoxide anion (O2·−) scavenging capacity, the guaiacol method was used as previously described (3). Briefly, 50 mg of root homogenate was suspended in 1 ml of 0.05 M phosphate buffer (pH 5.5) to extract antioxidant enzymes. After 10 min incubation, the mixture was centrifuged at 12,000 × g and 4 °C for 10 min. 0.5 ml supernatant (crude enzyme extract) was added into 2 ml; the reaction buffer consisted of 0.5 ml 0.05 M guaiacol (substrate, overdose), 1 ml 0.05 M phosphate buffer and 0.5 ml 2% hydrogen peroxide (H2O2). The increase in OD470 resulted from guaiacol oxidation in 4 min was recorded every 30 s until the reaction time reached 4 min.
Free Radical Scavenging Activity by ABTS+ Assay
To analyze the scavenging activity of soybean roots on cationic radical, the ABTS·+ decolorization assay was conducted as previously described (3). Briefly, 10 μl of extracts were mixed with 1.0 ml of ABTS·+ working solution, which contained 2.45 mM potassium persulphate and 7 mM ABTS (OD734 = 0.70 ± 0.02). The OD734 of the reaction mixture was measured after 30 min incubation at 30 °C.
Measurement of H2O2 Content
For analyzing the H2O2 content, 0.5 g soybean root was ground into fine powders in liquid nitrogen. The powders were suspended in 5 ml PBS buffer (pH 7.2) for 30 min at 4 °C. Then, the mixture was centrifuged at 12,000 rpm and 4 °C for 10 min. An aliquot contains 100 μl of suspension was analyzed using the H2O2 Quantitative Assay Kit (Water-compatible, C500069, Sangon Biotech, Shanghai, China) following the instructions of the manufacturer. Briefly, 100 μl of extract were mixed with 1.0 ml of Fe2+ working solution; the Fe2+could be oxidized into Fe3+ by H2O2 and generate an optical absorption at wavelength 560 nm. The OD560 of the reaction mixture was measured after 20 min incubation at 25 °C.
RESULTS
Phosphoprotein Profiles of Soybean Root Reveal Major Changes of Transcription Factors
Kinases, such as CCamK (45) and SOS2 (46), play key roles in rhizobia-legume symbiosis and plant salt tolerance. The phosphoproteomic technique provides an effective and comprehensive phosphoprotein profile for understanding the roles of these kinases in regulating these events in plants. In this study, quantitative phosphoproteomic approaches were used to find the key signaling components in soybean roots in response to rhizobia-inoculation and salt stress. The intensity of each phosphopeptide was normalized to the mean intensities of all phosphopeptides detected in all biological replicates, the change of intensity in log2 (Treated/Control) was calculated for each phosphopeptide (supplemental Table S2). The Student's t test (p values) was performed using the standard deviation of four biological replicates (supplemental Table S2).
In total, 4832 phosphorylated sites corresponding to 2288 phosphoproteins were identified. Among them, 4698 phosphopeptides were quantitatively analyzed (supplemental Tables S2 and S4). The annotations of these phosphoproteins were extracted from the protein database of green plants (http://www.matrixscience.com/). Among them, at least thirty-two unique phosphoproteins or their homologous have been reported to play key roles in regulating flavonoid synthesis or trafficking. These phosphoproteins include nine MYB (1), four WD40 repeat protein (47), seven bZip (48), five WRKY (49), and two bHLH (50), which are common transcription factors regulating the expression of flavonoid synthases genes, two V-type proton ATPase (51) and three ABC transporter C (52), which are usually involved in flavonoid trafficking. These results imply that, flavonoids might play important roles in root responses to salinity. Conceivably, phosphorylation of these proteins should play crucial roles in regulating flavonoid synthesis and/or trafficking.
The software MEME Suite and motif-X were used to extract overrepresented patterns from the 800 peptides which are differentially phosphorylated between the treated and control groups with a significant change (FDR ≤ 0.05 and p value ≤ 0.05). The phosphorylation intensities of differentially phosphorylated peptides from the treatment group (IpT) were compared with those from the control (IpC) group, and the phosphopeptides were divided into the Up group, when IpT > IpC, and the Down group when IpT < IpC. The Up group represented the peptides with higher phosphorylation level in the Rhizobia-inoculated (+) sample or Rhizobia-inoculated (+) and NaCl-treated (+) samples and the Down group represented peptides with lower phosphorylation levels as compared with that in the control.
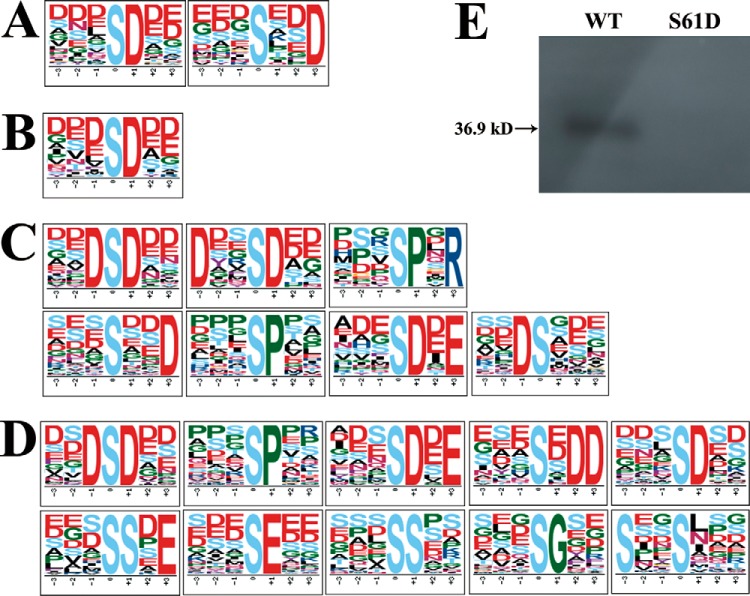
Two phosphopeptide motifs in the Up group (Fig. 1A) and one motif from the Down group were found to be enriched when the soybean root was inoculated with rhizobia (Fig. 1B). In addition, Ser was observed as the only centrally phosphorylated amino acid in both the Up and Down groups after rhizobia inoculation. The amino acid closest to the phosphorylated Ser was mostly Asp (Fig. 1A, 1B). These differentially regulated motifs were then searched for kinases that might target them as substrates using the tool and compendium of phosphorylation motifs curated in the human protein reference database (http://hprd.org/PhosphoMotif_finder). To cite an example, [sxxD] is known to be recognized by casein kinase-II (41, 53–56). The phosphorylated peptide DALAAGYAsADDAAPQNSGR in GmMYB173 (Q0PJH9) which contains the [sxxD] motif was found to be significantly enriched in the Up group (supplemental Table S2), indicating that phosphorylation of GmMYB173 is induced by the inoculation of rhizobia. On the other hand, phosphorylation of the [sxxD]-containing peptide DDAAGYAsADDAAPINSDK in GmMYB183 (Q0PJI2) was significantly suppressed after inoculation of the rhizobia.
Fig. 1.
Phosphorylation motifs enriched from differentially phosphorylated peptides after salt treatment. A, and B, are phosphorylation motifs extracted from the phosphopeptides in the Up group and Down group, respectively, when the soybean roots were inoculated with rhizobia. C, and D, show phosphorylation motifs extracted from the phosphopeptides in the Up and Down groups when the rhizobia-inoculated root was treated with NaCl. E, GmMYB183 could be a substrate of GmCK2α. Phosphorylation assay was carried out as described in the experimental procedures. WT and S61D: recombinant proteins of the N-terminal 168 aa of GmMYB183-S61D and GmMYB183-WT fused to a Trx- and a 6 x His-tags at its N terminus, and a 6 x His-tag at its C terminus. Data indicated that the Ser61 is the unique site in GmMYB183.
The MYB type TFs were reported to play pivotal roles in regulating the expression of enzymes involved in flavonoids syntheses (29, 33). Consistently we observed that all the salt responsive GmCHS, GmCHI and GmCYP contained several MYB-TF binding motifs in their promoters (1). Hence, the phosphorylation of TFs GmMYB173 and GmMYB183 could be important nodes connecting the upstream stress or rhizobia-triggered signals to the transcriptional control of the salt responsive genes. The functional significance of GmMYB173 has been covered in a separate study (1) and GmMYB183 will be the focus of the current research. Because the only phosphorylation peptide found in GmMYB183 contains a CK2 recognition motif (sxD), we expressed and purified the GmCK2α and confirmed that the GmMYB183 protein is a substrate of the soybean kinase GmCK2α (Fig. 1E).
When the rhizobia-inoculated root was treated with NaCl, we observed that seven phosphorylated motifs were enriched from the Up group (Fig. 1C) and ten motifs were enriched in the Down group (Fig. 1D). Ser was also observed as the only central phosphorylated amino acid residue in both groups. The amino acids closest to the phosphorylated Ser were most often Asp, Pro, Glu and Gly (Fig. 1C, 1D). Three phosphorylation motifs ([DsD], [sP], [sDxE]) were enriched from both Up and Down groups. Four phosphorylation motifs ([DxxsD], [sPxR], [sxxD], and [Ds]) were enriched from the Up group and seven phosphorylation motifs ([sxDD], [sD], [ssxE], [sE], [ss], [sG] and [sxxs]) were enriched from the Up group. These differentially phosphorylated motifs were then searched for the kinases that might use them as targets in the relevant databases. To cite a few examples, [sP] is recognized by ERK1 and ERK2 kinases, [sPxR] is the substrate of CDK1/2/4/6, [sDxE], [DxxsD], [sxDD], [ssxE] and [sxxs] are recognized by casein kinase-II (53–56).
Several transcription factors, including MYBs, bZIPs, pH-response factors, calmodulin-binding transcription factors (CAMTAs), ethylene-responsive factors (ERFs), WRKY factors, and GTE factors were found to be differentially phosphorylated (supplemental Tables S2 and S4). There were nine GmMYBs, five WRKYs, seven bZIPs, four CAMTAs and one pH-responsive factor which were among those quantitatively analyzed with one or more phosphorylated peptides. Interestingly, the phosphorylation of peptide DDAAGYAsADDAAPINSDK in GmMYB183 (Q0PJI2) was also found to be significantly decreased after Rhizobia inoculation (+) followed by NaCl(+) treatment, similar to the changes observed in the R(+)Na(-) treatment. In addition, the phosphorylation of peptide DALAAGYAsADDAAPQNSGR in GmMYB173 (Q0PJH9) was also up-regulated by R(+)Na(+) (supplemental Table S2). This implies that soybean GmMYB183S61p and GmMYB173S59p mediate responses triggered by rhizobia inoculation and salinity (supplemental Tables S2 and S4), where GmMYB183 seems to play a negative role and the GmMYB173 play a positive role in soybean responses to rhizobia inoculation and NaCl treatment.
It has been reported that MYB type TFs are involved in regulating the expression of enzymes involved in chalcone metabolism (29). Our previous research showed that all the salt responsive GmCHS, GmCHI and GmCYP contain several MYB-binding motifs in their promoters; GmMYB173 could bind to the promoter of GmCHS5 and regulate its transcriptional expression (1). Hence the phosphorylation of GmMYB173 and GmMYB183 might affect their binding efficiencies to these promoters and regulates the transcriptional expression of these chalcone synthesis genes. It appears that the rhizobia inoculation, although is not an ionic or osmotic stress, could triggers changes in the phosphorylation status of GmMYB173 and GmMYB183 like that induced by NaCl treatment, and prime or enhance the soybean's salt tolerance.
Metabolite Profiles of Soybean Root Reveal Prominent Changes in Flavonoid
Our previous research has revealed that rhizobia could enhance soybean's ability to adapt to saline soil, and changes in flavonoid metabolism is also associated with plant tolerance to salt stress (1, 3, 4). However, it is still unclear which flavonoid contributes to the soybean's tolerance to salinity under a certain treatment. Hence, metabolomics was used to discover the flavonoids involved in rhizobia-triggered tolerance to salinity. The metabolites were ionized via negatively and positively charge modes, and then subjected to MS analysis. Ten biological replicates of each sample were used to estimate the fold changes in metabolite concentrations between salt-treated or rhizobia-inoculated roots and control samples (supplemental Tables S5 and S6) using a label-free metabolomics approach (57). Negative-charge-mode and positive-charge-mode enabled us to detect 2075 and 4668 metabolites respectively, out of which 38 and 65 flavonoids were detected through both charge modes. The flavonoids with significant and reproducible differences (p ≤ 0.05) after treatments are presented in Table I.
Table I. Flavonoid content with significant changes under salt stress and rhizobia innoculation.
| Rhizobia(+) |
Rhizobia(+) & NaCl(+) |
||||
|---|---|---|---|---|---|
| Name | Fold change | p value | Name | Fold change | p value |
| *Lilaline | 2.8518 | 0.0000 | *Neoraunone | 36.5074 | 0.0000 |
| *Quercetin 3-(6¢′-acetylgalactoside) | 2.2393 | 0.0000 | *Robustone | 30.5624 | 0.0000 |
| *Diospyrin | 2.1141 | 0.0000 | Repenol | 18.0835 | 0.0000 |
| Kaempferol 3-(2¢′-hydroxypropionylglucoside)-4′-glucoside | 1.2795 | 0.0000 | *Luteolin 3′-methyl ether 7-glucuronosyl-(1→2)-glucuronide | 4.1935 | 0.0000 |
| *Kaempferol 3-sulfate-7-alpha-arabinopyranoside | 1.6116 | 0.0000 | *Kaempferol 3-sulfate-7-alpha-arabinopyranoside | 5.1385 | 0.0000 |
| Kaempferol 3-(4¢′-(E)-p-coumarylrobinobioside)-7-rhamnoside | 1.5148 | 0.0001 | Sophoracoumestan A | 11.2355 | 0.0000 |
| *Glabrone | 1.3232 | 0.0002 | Echinoisosophoranone | 6.2261 | 0.0000 |
| *Corylin | 1.3195 | 0.0003 | Quercetin 3,7,3′-tri-O-sulfate | 2.3300 | 0.0000 |
| *Neoraunone | 2.7213 | 0.0006 | *6-Hydroxykaempferol 3,5,7,4′-tetramethyl ether 6-rhamnoside | 5.2728 | 0.0000 |
| *Robustone | 2.4265 | 0.0007 | *Isochamaejasmin | 2.4987 | 0.0000 |
| Piscerythramine | 1.1913 | 0.0052 | *Naringin | 2.6854 | 0.0000 |
| *Cyanidin-3-arabinoside | 1.1090 | 0.0101 | *Cyanidin-3-arabinoside | 1.1752 | 0.0170 |
| *Isochamaejasmin | 1.2818 | 0.0065 | Quercimeritrin | 2.7887 | 0.0000 |
| *Bilobetin | 1.5005 | 0.0125 | Medicagol | 3.1639 | 0.0000 |
| *Luteolin 3′-methyl ether 7-glucuronosyl-(1→2)-glucuronide | 1.3764 | 0.0133 | Atovaquone | 4.2812 | 0.0000 |
| *6-Hydroxykaempferol 3,5,7,4′-tetramethyl ether 6-rhamnoside | 1.5204 | 0.0146 | Pongamoside A | 1.6110 | 0.0000 |
| *Naringin | 1.4490 | 0.0175 | Dalpanin | 2.9768 | 0.0000 |
| *Quercetin 3-(4¢′-malonylrhamnoside) | 1.4839 | 0.0293 | Syringetin 3-glucuronide | 1.4077 | 0.0002 |
| Naringin 6¢′-malonate | 1.3785 | 0.0333 | *Corylin | 2.5700 | 0.0004 |
| *Phenindione | 0.6657 | 0.0000 | Cyanidin 3-(6¢′-succinyl-glucoside) | 1.8170 | 0.0005 |
| *Daidzin | 0.6131 | 0.0000 | Karanjin | 1.3458 | 0.0005 |
| *Apigenin 7-(2¢′-acetyl-6¢′-methylglucuronide) | 0.5609 | 0.0000 | *Glabrone | 1.5755 | 0.0011 |
| *Macrocarposide | 0.6387 | 0.0000 | *Lilaline | 1.5627 | 0.0012 |
| *Malonylglycitin | 0.6107 | 0.0000 | Quercetin 3-alloside | 1.6699 | 0.0030 |
| Kaemferol | 0.7897 | 0.0001 | *Diospyrin | 1.3290 | 0.0059 |
| *Genistein | 0.7523 | 0.0019 | Irisflorentin | 1.3406 | 0.0059 |
| Atovaquone | 0.7041 | 0.0021 | Phellodensin E | 1.9397 | 0.0069 |
| Sophoracoumestan A | 0.3687 | 0.0024 | *Quercetin 3-(4¢′-malonylrhamnoside) | 1.9801 | 0.0096 |
| *Ononin | 0.8150 | 0.0003 | *Bilobetin | 2.1858 | 0.0098 |
| *Mirificin | 0.6717 | 0.0312 | 5′-Methoxybilobetin | 1.4243 | 0.0125 |
| *Cladrin 7-O-glucoside | 0.6532 | 0.0318 | Quercetin 3-(6¢′-methylglucuronide) | 1.7671 | 0.0295 |
| *Patuletin 3-(6¢′-(E)-feruloylglucoside) | 0.8286 | 0.0350 | *Quercetin 3-(6¢′-acetylgalactoside) | 1.1708 | 0.0297 |
| *Ikarisoside A | 0.8637 | 0.0387 | Naringenin | 0.0157 | 0.0000 |
| Kaempferol 3-(2¢′,3¢′-diacetyl-4¢′-p-coumaroylrhamnoside) | 0.7723 | 0.0461 | *Ononin | 0.8069 | 0.0007 |
| *Frutinone A | 0.7959 | 0.0484 | *Macrocarposide | 0.1317 | 0.0000 |
| Malvidin 3-glucoside-pyruvate | 0.1011 | 0.0000 | |||
| *Phenindione | 0.2988 | 0.0000 | |||
| *Malonylglycitin | 0.1268 | 0.0000 | |||
| Davallioside A | 0.4789 | 0.0000 | |||
| Isorhamnetin 3-rhamnoside | 0.3797 | 0.0000 | |||
| *Apigenin 7-(2¢′-acetyl-6¢′-methylglucuronide) | 0.2443 | 0.0000 | |||
| *Daidzin | 0.4528 | 0.0000 | |||
| Lupalbigenin | 0.3566 | 0.0000 | |||
| *Ikarisoside A | 0.6030 | 0.0000 | |||
| Kaempferol 3-(2¢′-hydroxypropionylglucoside)-4′-glucoside | 0.6986 | 0.0000 | |||
| Clitoriacetal | 0.3364 | 0.0000 | |||
| Daidzein | 0.2400 | 0.0000 | |||
| *Patuletin 3-(6¢′-(E)-feruloylglucoside) | 0.5913 | 0.0000 | |||
| Formononetin 7-O-(6¢′-acetylglcoside) | 0.5613 | 0.0000 | |||
| 6-Methoxykaempferol 3-rhamnoside-7-(4¢¢′-acetylrhamnoside) | 0.6455 | 0.0000 | |||
| Malonyldaidzin | 0.1228 | 0.0001 | |||
| Rhoifolin | 0.2778 | 0.0001 | |||
| Evodiamine | 0.6465 | 0.0001 | |||
| *Cladrin 7-O-glucoside | 0.3654 | 0.0003 | |||
| Kalbreclasine | 0.5762 | 0.0004 | |||
| Isoliquiritigenin 2′-glucosyl-(1→4)-rhamnoside | 0.5372 | 0.0006 | |||
| Ononin | 0.8069 | 0.0007 | |||
| Genistein 4′-O-glucoside | 0.6439 | 0.0007 | |||
| Malonylgenistin | 0.5811 | 0.0009 | |||
| Malonylgenistin | 0.6196 | 0.0010 | |||
| *Frutinone A | 0.6936 | 0.0013 | |||
| *Genistein | 0.7207 | 0.0019 | |||
| Glycitin | 0.3981 | 0.0024 | |||
| Silybin | 0.8227 | 0.0047 | |||
| Flavaprenin 7,4′-diglucoside | 0.8810 | 0.0060 | |||
| Kaempferol 3-(4¢′-(E)-p-coumarylrobinobioside)-7-rhamnoside | 0.8495 | 0.0128 | |||
| Formononetin | 0.8081 | 0.0153 | |||
| Neorauteen | 0.6721 | 0.0183 | |||
| *Mirificin | 0.7028 | 0.0356 | |||
| Delphinidin 3-(6¢′-malonylglucoside) | 0.6970 | 0.0421 | |||
*Metabolites found with similar changes after Rhizobia(+) and Rhizobia(+) & NaCl(+) treatments.
Inoculation of soybean roots by rhizobia resulted in significant increases in the endogenous levels of 19 flavonoids, including cyanidin-3-arabinoside, quercetin 3-(6′'-acetylgalactoside), luteolin 3′-methyl ether 7-glucuronosyl-(1->2)-glucuronide, and quercetin 3-(4′'-malonylrhamnoside) (Table I); and remarkable decreases in the concentrations of 16 flavonoids, such as ononin, genistein, and daidzin (Table I). When the rhizobia-inoculated roots were further treated with salt stress, the accumulations of 32 flavonoids were found to be significantly increased and 38 flavonoids were detected with a significant decrease. Interestingly, 27 out of the 36 differentially changed flavonoids after rhizobia inoculation were also found to have similar up-regulation or downregulation trends in the R(+)Na(+) treatment. This indicates that these flavonoids might play significant roles in both the rhizobia inoculation and salinity response pathways.
These observations promoted us to speculate whether branching in flavonoid metabolism pathways could significantly alter the symptoms of salt tolerance. As a precursor of all flavonoids, the accumulation of chalcone is essential for production of cyanidin derivatives which serve as potent ROS scavengers. The homologs of CHI and CYP that catalyze the conversion of chalcone to different types of derivatives (for example, cyanidin as a positive effector, and ononin as a negative effector) should play distinct roles in soybean responses to salinity.
GmMYB183 Regulates the Expression of GmCYP81E11 in Response to Salinity
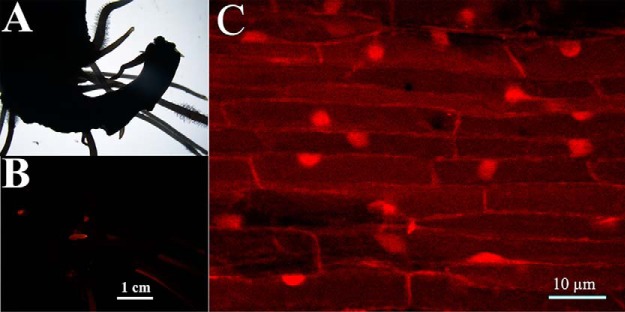
Because flavonoids are molecules involved in both soybean-rhizobia interaction and soybean salt tolerance, we expressed the phospho-mimetic/ablative mutants of all the 27 differentially phosphorylated transcription factors which are likely involved in regulation of flavonoid syntheses and evaluated the salt tolerance of the transgenic roots. This quick survey showed that only 3 transcription factors, GmMYB176, GmMYB173 and GmMYB183 are functionally related to salt tolerance of soybean, and the first two were reported elsewhere (1, 33). Hence the rest of the current study is mostly focused on GmMYB183. Although the Glyma06G187600 encoded protein (GmMYB183) is classified as a member of the MYB type transcription factor family based on its sequence (58), empirical data about the molecular and physiological functions of GmMYB183 are lacking. The coding sequence of GmMYB183 was tagged with RFP at its 3′ end and expressed in soybean roots (Fig. 2A, 2B) via A. rhizogenes-mediated transformation. Subcellular localization of GmMYB183-RFP was observed using a Zeiss LSM710 confocal microscope. Consistent with its function as a transcription factor, GmMYB183-RFP was found localized in the nucleus in soybean roots (Fig. 2C).
Fig. 2.
GmMYB183-RFP localizes in the nucleus of soybean root cells. The original roots of soybean seedling were removed and infected with A. rhizogenes carrying a GmMYB183-RFP expressing construct, transgenic roots with red fluorescence were selected using a Nikon SMZ1500 fluorescence stereoscope (A, B). The subcellular localization of the GmMYB183-RFP fusion protein was observed using a Zeiss LSM710 confocal microscope (C).
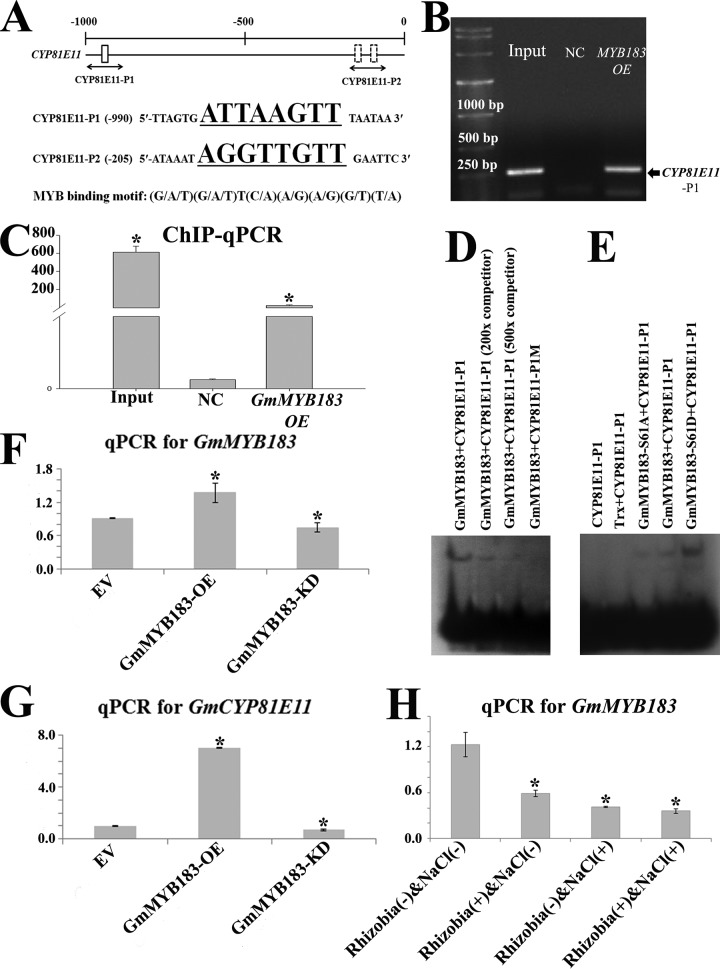
Glyma09g05440.1 encodes for a cytochrome P450 protein named GmCYP81E11, which was identified as a salt responsive gene (3). We searched for the 8 bp MYB recognition motif conforming to the (G/A/T)(G/A/T)T(C/A)(A/G)(A/G)(G/T)(T/A) format (53) in the promoter of GmCYP81E11, and found 2 copies in the −1kb region before the starting codon, an “ATTAAGTT” element located at −984 to −977 and an “AGGTTGTT” at −199 to −192 (Fig. 3A). The physical interaction between GmMYB183 and the promoter of GmCYP81E11 was tested using chromatin immuno-precipitation coupled with quantitative PCR (ChIP-qPCR) (Fig. 3B, 3C). A 35S::GmMYB183-RFP-HA plant expression construct or the empty vector (35S::RFP-HA) was transformed into soybean roots via A. rhizogenes-mediated transformation. The transgenic roots displaying red fluorescence were picked for the ChIP-PCR assay as previously described (44) using an anti-HA polyclonal antibody raised in rabbit. qPCR amplification using CYP81E11-P1 flanking primer pair (ChIPCYP81E11P1-F and ChIPCYP81E11P1-R in supplemental Table S3), a 222-fold enrichment in PCR amplification from ChIPed test sample (roots transformed with 35S::GmMYB183-RFP-HA) was observed when compared with that transformed with empty vector (35S::RFP-HA) (Fig. 3C). Because the PCR amplification from the ChIPed template DNA using CYP81E11-P2 flanking primer pair (ChIPCYP81E11P2-F and ChIPCYP81E11P2-R in supplemental Table S3) did not show any remarkable enrichment, we concluded that there is a physical interaction between GmMYB183 and the DNA section corresponding to the CYP81E11-P1 position in the promoter of GmCYP81E11.
Fig. 3.
GmMYB183 binds to a MYB-specific Cis-element present in the promoter of GmCYP81E11. A, Distribution of MYB binding motifs (G/A/T)(G/A/T)T(C/A)(A/G)(A/G)(G/T)(T/A) in the promoter of GmCYP81E11. Position of the probe (underlined sequence) used for ChIP-based binding assay is shown below the gene. Position Weight Matrix of MYB binding motifs was curated in the JASPAR database (http://jaspar.genereg.net/cgi-bin/jaspar_db.pl?rm=browse&db=core&tax_group=plants). B, and C, ChIP-qPCR assays indicates that GmMYB183 binds to the promoter sections containing the MYB-binding motifs in vivo. The empty vector was used as a negative control (NC). An asterisk indicates a significant difference (p ≤ 0.05) to the NC according to Student's t test. FC means fold change. D, and E, EMSA assays showed that GmMYB183 specially binds to the GmCYP81E11-P1 fragment from the GmCYP81E11 promoter (D), and phosphorylation at S61 of GmMYB183 enhances this interaction (E). GmCYP81E11-P1 (ttttATGTATTAGTGATTAAGTTTAATAACGTGA) or a mutated version with its ATTAAGTT core sequence changed to ATAAAGTT (GmCYP81E11-P1M) was labeled as a probe, and 200 or 500 folds of unlabeled double strand GmCYP81E115-P1 fragment was set as the competitor. F, and G, The transcription levels of GmMYB183 (F) and GmCYP81E11 (G) in soybean transgenic roots expressing empty vector (EV), GmMYB183-overexpression (GmMYB183-OE) and RNAi (GmMYB183-KD) constructs, respectively. Data presented are mean ± S.E. (n = 3). H, Transcription of GmMYB183 in soybean roots treated with R(-)Na(-), R(+)Na(-), R(-)Na(+) and R(+)Na(+). Data represent mean values ± S.E., each sample was analyzed with three biological replicates. An asterisk indicates a significant difference (p ≤ 0.05, Student's t test) between treatment [R(+)Na(-), R(-)Na(+) and R(+)Na(+)] and the control [R(-)Na(-)].
To further confirm the interaction between GmMYB183 and the promoter of GmCYP81E11, we performed EMSAs using double-stranded GmCYP81E11 probes containing a MYB motif (GmCYP81E11-P1 containing an ATTAAGTT) and a mutant (GmCYP81E11-P1M: ATAAAGTT) as described in the experimental procedures. The result showed that GmMYB183 binds to the CYP81E11-P1 probe in a sequence dependent manner (Fig. 3D). We also tested GmCYP81E11-P2 probes with EMSA, consistently no interactions were detected between them and the DNA binding domain of GmMYB183 (Fig. 3D). Because the S61 phosphorylation site is close to the DNA-binding domain (aa 74–130), we compared the interaction between GmCYP81E11-P1 probe and the WT, phospho-mimic (S61D) and phospho-ablative (S61A) mutant of GmMYB183, our results supported that phosphorylation at S61 significantly facilitate the binding of GmMYB183 to the promoter of GmCYP81E11 via the GmCYP81E11-P1 MYB-binding element (Fig. 3E).
To test whether GmMYB183 regulates the transcription of GmCYP81E11, we analyzed the transcription of GmCYP81E11 in GmMYB183-overexpression (pDL28-GmMYB183-OE/Ubq10::dsRed) and RNAi-mediated knock-down (pDL28-GmMYB183-KD) roots (Fig. 3F). The roots carrying the empty vector (pDL28/Ubq10::dsRed) were used as control. Our results showed that introduction of the GmMYB183-OE construct resulted in an over 2-fold increase in GmCYP81E11 expression as compared with roots transformed with the GmMYB183-KD construct (Fig. 3G), suggesting that GmMYB183 activates the expression of GmCYP81E11. We then compared the transcription of GmMYB183 before and after salt treatment and/or rhizobia-inoculation, significant difference was detected between every treatment [R(+)Na(-) or R(-)Na(+) or R(+)Na(+)] and the control [R(-)Na(-)] (Fig. 3H). These results demonstrated that both salt treatment and rhizobia-inoculation also suppress the expression of GmMYB183 (Fig. 3H).
GmCYP81E11 and GmMYB183 Affect Flavonoid Metabolism and Salt Tolerance in Soybean
Based on previous studies, it is reasonable to speculate that both the transcription factor GmMYB183 and its downstream target gene GmCYP81E11 could affect soybean tolerance to salinity. To further validate that both GmMYB183 and GmCYP81E11 contribute to salt tolerance by affecting flavonoid metabolism, composite soybean plants with gain-of-function and loss-of-function mutations of these two genes were generated and tested for their salt tolerance (Table II).
Table II. Ononin contents in OE or KD roots in response to salt stress.
| R(-)Na(-)* | R(+)Na(-) | R(+)Na(+) | |
|---|---|---|---|
| Ononin (μg/g fresh weight) | |||
| GmCYP81E11 | |||
| OE | 0.7221 ± 0.0070 | 0.5780 ± 0.0477 | 0.0609 ± 0.0033 |
| KD | 0.2413 ± 0.0173 | ND** | ND |
| GmMYB183 | |||
| OE-WT | 0.2278 ± 0.0039 | ND | ND |
| OE-S61D | 0.2800 ± 0.0049 | 0.0236 ± 0.0052 | ND |
| OE-S61A | 0.1044 ± 0.0048 | ND | ND |
| KD | ND | ND | ND |
*R(-)Na(-): partially cited from Reference 1;
**ND: no detectable.
Because it has been generally accepted as a negative effector, the flavonoid ononin, a product up-regulated by GmCYP81E11, was analyzed in all the tested transgenic roots (Fig. 4 and Table II). Contents of ononin were found to be positively correlated with the expression levels of GmCYP81E11 and GmMYB183 (Table II); in addition, it is also found to be negatively affected by rhizobia and/or NaCl treatments because the contents of ononin were significantly decreased in both R(+)Na(-) and R(+)Na(+) treated samples (Table II). Furthermore, our data showed that transgenic roots expressing GmCYP81E11-OE had the highest level of ononin when not treated (0.7221 ± 0.0070 μg/g fresh weight, R(-)Na(-)); it were moderately decreased when treated with rhizobia only (0.5780 ± 0.0477 μg/g fresh weight, R(+)Na(-)) and were further decreased when treated with both rhizobia and salt (0.0609 ± 0.0033 μg/g fresh weight, R(+)Na(+)).
Fig. 4.
HPLC-based analysis of ononin contents in transgenic roots of soybean. A, The chromatographic spectrum observed from ononin standard separation. B, to (N) The chromatographic spectrum observed from HPLC separation of transgenic GmCYP81E11-OE R(-)Na(-) (B), GmCYP81E11-OE R(+)Na(-) (C), GmCYP81E11-OE R(+)Na(+) (D), GmCYP81E11-KD R(-)Na(-) (E), GmCYP81E11-KD R(+)Na(-) (F), GmCYP81E11-KD R(+)Na(+) (G), GmMYB183 OE-WT R(-)Na(-) (H), GmMYB183 OE-S61D R(-)Na(-) (I), GmMYB183 OE-S61D R(+)Na(-) (J), GmMYB183 OE-S61A R(-)Na(-) (K), GmMYB183-KD R(-)Na(-) (L), GmMYB183-KD R(+)Na(-) (M), GmMYB183-KD R(+)Na(+) (N) roots.
To test the relevance of phosphorylation at Ser-61 of GmMYB183 to flavonoid metabolism, we generated the following plant expressing constructs driven by 35S promoter: (1) the full-length cds of GmMYB183 (GmMYB183 OE-WT), (2) phosphomimic mutant of GmMYB183 (GmMYB183 OE-S61D), (3) phospho-ablative mutant of GmMYB183 (GmMYB183 OE-S61A) and (4) RNAi-mediated knockdown construct of GmMYB183 (GmMYB183 KD). Interestingly, introduction of GmMYB183 KD into the transgenic roots resulted in no detectable content of ononin, whereas the GmMYB183 OE-S61A, GmMYB183 OE-WT, GmMYB183 OE-S61D constructs resulted in 0.1044 ± 0.0048, 0.2278 ± 0.0039, and 0.2800 ± 0.0049 μg/g fresh weight (Table II). This indicates that the S61 phosphorylation in GmMYB183 enhances the accumulation of ononin.
GmCYP81E11 and GmMYB183 are Involved in ROS Elimination in Soybean
The antioxidant property of plant tissue is generally accepted to correlate with plant tolerance to salinity. The antioxidant properties in terms of H2O2 and ABTS●+ [2, 2′-azinobis (3-ethylbenzothiazoline 6-sulfonate)] radical scavenging capacities in soybean root were analyzed as previously described (3). To test the relevance of ROS elimination capacity to flavonoid metabolism, all the transgenic roots mentioned above whose flavonoid content had been evaluated were also analyzed for their ROS elimination capacity. Our results revealed that the ROS elimination capacities in these tested roots were arranged in a trend contrary to that of endogenous ononin levels (Table III). Generally, the H2O2 and ABTS●+ scavenging capacities in all transgenic roots were found to be stimulated by salt stress. Elevated expression of both GmMYB183 and GmCYP81E11 negatively affected ROS scavenging capacities in the soybean transgenic roots. For example, the H2O2 elimination capacity was measured to be 1.5750 ± 0.0424 U*g−1*min−1 in GmCYP81E11-OE for transformed roots, remarkably lower than 2.1800 ± 0.0283 U*g−1*min−1 measured in the GmCYP81E11-KD transformed roots when treated with R(+)Na(+) (Table III). In addition, the GmMYB183 KD construct resulted in the highest capacities for eliminating both H2O2 and ABTS●+; whereas the GmMYB183 OE-S61D construct displayed the lowest ROS scavenging capacity (Table III).
Table III. ROS elimination capacity of OE or KD roots.
| R(-)Na(-) | R(+)Na(-) | R(+)Na(+) | |
|---|---|---|---|
| H2O2 (U*g−1*min−1) | |||
| GmCYP81E11 | |||
| OE | 1.1800 ± 0.0057 | 1.2833 ± 0.0424 | 1.5750 ± 0.0424 |
| KD | 1.5388 ± 0.0159 | 1.6757 ± 0.0384 | 2.1800 ± 0.0283 |
| GmMYB183 | |||
| OE-WT | 1.1867 ± 0.0094 | 1.3300 ± 0.0236 | 1.4100 ± 0.0000 |
| OE-S61D | 0.9586 ± 0.0303 | 1.0988 ± 0.0018 | 1.3043 ± 0.0020 |
| OE-S61A | 1.2588 ± 0.0548 | 2.0638 ± 0.0513 | 2.2700 ± 0.0255 |
| KD | 1.9600 ± 0.0672 | 2.5520 ± 0.0453 | 2.8267 ± 0.0094 |
| ABTS (10 μmol*g−1) | |||
| GmCYP81E11 | |||
| OE | 0.2923 ± 0.0132 | 0.5063 ± 0.0127 | 0.5563 ± 0.0118 |
| KD | 0.3658 ± 0.0028 | 0.5722 ± 0.0292 | 0.6116 ± 0.0140 |
| GmMYB183 | |||
| OE-WT | 0.2514 ± 0.0361 | 0.3232 ± 0.0187 | 0.3880 ± 0.0212 |
| OE-S61D | 0.1546 ± 0.0274 | 0.3155 ± 0.0071 | 0.3245 ± 0.0105 |
| OE-S61A | 0.3563 ± 0.0374 | 0.4946 ± 0.0150 | 0.5426 ± 0.0112 |
| KD | 0.4158 ± 0.0150 | 0.5334 ± 0.0299 | 0.6254 ± 0.0113 |
Phosphorylation of GmMYB183 at S61 Represses Salt Tolerance of Transgenic Soybean Roots
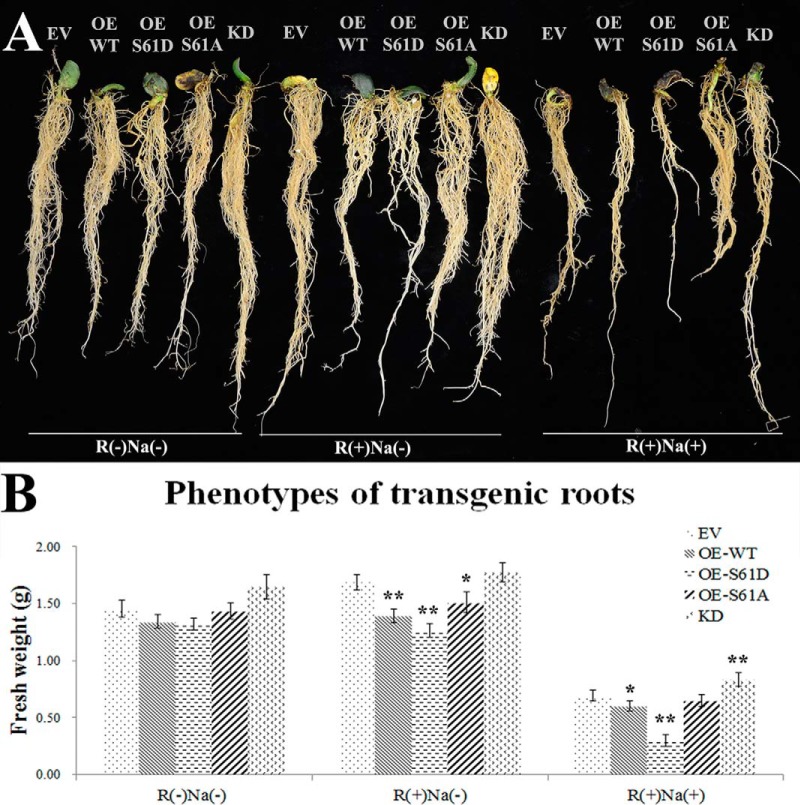
To further understand the contribution of GmMYB183 phosphorylation at its Ser61 on regulating the salt tolerance of soybean, fresh weights of transgenic roots were analyzed in this research. Consistent with the abovementioned ROS scavenging capacities, roots expressing GmMYB183S61D (OE-S61D) showed lower tolerance than those transformed with GmMYB183S61A (OE-S61A) and GmMYB183 (OE-WT) constructs (Fig. 5A and 5B). Furthermore, the transgenic roots whose GmMYB183 expression is knocked-down (KD) displayed the highest tolerance. These results indicate that phosphorylation of GmMYB183 plays a negative role in soybean salt tolerance (Fig. 5A and 5B).
Fig. 5.
Effects of salt treatment on transgenic roots of composite soybean plants. Composite soybean plants were generated by infection with A. rhizogenes strain K599 carrying empty vector (EV), overexpression constructs of GmMYB183 (OE-WT), GmMYB183S61D (OE-S61D), GmMYB183S61A (OE-S61A) and RNAi-mediated knockdown construct of GmMYB183 (KD). Transgenic roots were confirmed by DsRed fluorescence as described in the materials and methods section. One-week old composite plants were treated in 1/4 fold Fahräeus medium without (control) or with 200 mM NaCl [NaCl (+)] for 4 weeks. A, Plants are typical representatives of three repeats of salt treatments. B, Fresh weights of transgenic roots: data expressed are means ± s.d. of root samples from three plants and the experiments are repeated three times with similar results; an asterisk indicates a significant difference from the control (EV) based on p ≤ 0.05 of student's t test;double asterisks indicates a significant difference from the control (EV) based on p ≤ 0.01 of student's t test.
We further evaluated the salt tolerance of the whole composite plants although their shoots remain to be wild-type. Our results indicated similar correlations exist between their tolerance performance and the corresponding transgenes expressed in their roots. For example, composite shoots with root expressing GmMYB183S61D (OE-S61D) showed lower tolerance than those transformed with GmMYB183S61A (OE-S61A) and GmMYB183 (OE-WT) constructs (supplemental Fig. S1). Furthermore, the composite shoots with transgenic roots in which GmMYB183 expression is knocked-down (KD) displayed the highest growth under salinity. Consistently, the composite plants with root over-expressing GmCYP81E11 (OE-GmCYP81E11) showed lower tolerance than those composites with transgenic roots in which GmCYP81E11 expression is knocked-down (KD-GmCYP81E11) in roots (supplemental Fig. S2).
Salt Treatment and Rhizobia Inoculation Triggered Different Accumulation of H2O2 In Soybean Roots
By looking for phosphorylation events similarly triggered by both rhizobia inoculation and salt treatments, we found that GmMYB183 is a critical player contributes to the rhizobia potentiated soybean tolerance to salt stress. In the meantime, one could also notice that rhizobia inoculation of soybean roots triggers effects similar to those induced by NaCl treatment including decrease in GmMYB183 phosphorylation, inhibition of GmCYP81E11 gene expression, decrease of ononin accumulation, and enhancement of ROS scavenging capacities in plants (Fig. 3, Table II, III). However, caution is recommended to conclude that the effect of NaCl treatment is better than the rhizobia inoculation in inducing these responses because the time scale of rhizobia inoculation and salt treatment in these experiments were not comparable. Whereas the time for salt treatment is 24 h, the time for rhizobia inoculation is over a couple of weeks. It is noteworthy that inoculation of rhizobia also had some opposite or alleviating effects on NaCl stress. Taking the phosphor-proteome as an example, the up-regulated phosphorylated peptides in the roots inoculated with rhizobia [(R(+) Na(-)] (98 peptides listed in supplemental Table S2 in this research) are much less than those in roots treated with NaCl [R(-)Na(+)] (412 peptides listed in supplemental Table S5, in Ref 1 where the salt treatment setup is the same as that in this research). Moreover, the rhizobia and NaCl treatments indeed showed opposite effects on many phosphorylated peptides as documented in the two abovementioned supplemental tables, for example, the phosphor-peptide IVLPPELPPEIPDDEVEVSDDDLQFVK in the A0A0B2P066 protein (Nucleolar complex protein 3 like).
Because the accumulation of ROS stimulated by salt stress has been known to trigger oxidative damages in plant cells (5, 6), we compared the time course of ROS accumulation in soybean roots within 96 h of salt treatment and rhizobia inoculation (supplemental Fig. S3). Consistently we saw NaCl treatments triggered rapid accumulation of H2O2 within the first 24 h post treatment (hpt). The H2O2 level was kept at a relatively high levels with slight decrease throughout the rest of the 96 h testing period in the NaCl single treatment group [R(-)Na(+)], but dropped much more quickly after reach the pinnacle at 24 hpts in the NaCl treated roots pre-inoculated with rhizobia for 4 weeks [R(+)Na(+)]. On the other hand, rhizobia inoculation [R(+)Na(-)] triggered a rapid H2O2 accumulations comparable to those induced by NaCl treatments[R(-)Na(+) and R(+)Na(+)] during the first 6 h of inoculation, since then it start to decease steadily and restored to the untreated level at 96 h post inoculation. Although confirming that salt stress [R(-)Na(+)] hurts plant cells with excessive and long lasting accumulation of ROS, these results showed that inoculation of rhizobia [R(+)Na(-)] only trigger a short burst of H2O2 in soybean roots immediately after inoculation which serves as an effective signaling event potentiating protective responses apparently upstream the actions of GmMYB183 and GmCYP81E11, a scenario as happen in the [R(+)Na(+)] double treatment group.
DISCUSSION
Flavonoids in leguminous plants have been well characterized for their roles in plant-microbe interactions; they are the key signals that determine the host-symbiont specificity between rhizobia and their host legumes (9). A wide variety of flavonoids have been shown to induce the expression of nod-genes of rhizobia (9); quercetin from alfalfa was reported to stimulate spore germination and hyphal branching of arbuscular mycorrhizal fungi (AMF) (59, 60). In addition, flavonoids were also found to be involved in the AMF-induced defense reactions in alfalfa, and it has been suggested that this AMF-trigged change of flavonoid profiles could initiate a general plant defense response against a broad range of stresses including salinity (61). However, the mechanism of how these microbes enhance plant's salt tolerance is far from clear.
We previous demonstrated that rhizobia could trigger the transcriptional expression of MYB genes and strengthen the soybean roots' ROS scavenging capabilities (4). Furthermore, some MYB transcription factors were revealed to transcriptionally activate genes encoding for biosynthesis of flavonoids under salt stress (1, 62, 63). Although the anti-oxidative property of flavonoids is not novel (64), its physiological impacts have not been adequately addressed at the whole plant level. The chemical basis of the anti-oxidative property of flavonoids has been attributed to the hydroxy groups in their structures. The dihydroxy B-ring flavonoids were proven to have higher efficiencies in reducing ROS as compared with monohydroxy B-ring flavonoids (1, 63–65). Consistent with these findings, the dihydroxy B-ring (e.g. quercetin) and dihydroxy B-ring-substituted flavonoids, such as quercetin 3-(6′'-methylglucuronide), quercetin 3-(2G-(E)-p-coumaroylrutinoside), quercetin 3-(6′'-acetylgalactoside), and quercetin 3-alloside were found to be significantly up-regulated in soybean roots after salt stress in our current metabolomic study (Table I). In contrast, the monohydroxy B-ring flavonoids, such as ononin, daidzin, genistein, kaempferol 3-sulfate-7-alpha-arabinopyranoside, and apigenin 7-(2″-acetyl-6″-methylglucuronide) were shown to be significantly downregulated.
The flavonoid derivatives, primarily formed through ortho-dihydroxy B-ring glycosylation, were shown to donate electrons or hydrogen atoms in anti-oxidative reactions (66), and quercetin glycosides, which dominate the flavonoid profile in soybean roots (Table I), are believed to be superior electron donors compared with other glycosides (43, 67, 68). Legume flavonoids are diversified in their structure as well as physiological functions (9). For example, formononetin was an essential intermediate of leguminous phytoalexins with methoxy groups and plays a critical role in legume-rhizobium symbiosis (21, 63). In addition, daidzein and genistein are precursors of 2′-hydroxydaidzein and 2′-hydroxygenistein, respectively (69); the two products both have 2′, 3′-hydroxy (dihydroxy) in the B-ring and are accepted to have higher radical-scavenging efficiency than their corresponding precursors. The monohydroxy B-ring flavonoids, such as apigenin and 7, 4′-dihydroxyflavone, are well known as signaling molecules mediating plant responses to pathogen attack (8, 70). When excess ROS is induced by salinity, soybean roots are reprogrammed to produce flavonoids with better radical-scavenging efficiency. In other words, the quercetin derivatives could assume positive roles in soybean tolerance to salt stress, whereas the ononin, daidzin, genistein, kaempferol and their derivatives have negative contributions to soybean's resistance to oxidative stress.
As the flavonoids vary in antioxidant efficiencies, their differential syntheses may imply distinct requirements to help plants survive salt stress. Flavonoids are mainly synthesized via enzymes involved in the shikimate-phenylpropanoid pathway, such as CYP. CYPs are heme-containing proteins which catalyze a wide range of oxidative reactions (71, 72). Lam et al. (2014) demonstrated that cytochrome P450 93G2 (CYP93G2) functions as a flavanone (monohydroxy B-ring flavonoids) 2-hydroxylase (F2H) that supplies 2-hydroxyflavanones for C-glycosylation in rice (Oryza sativa) (73). In addition, the soybean proteins CYP81E11, CYP81E12 and CYP81E18 catalyze daidzein and genistein to dihydroxy B-ring flavonoids 2′-hydroxydaidzein and 2′-hydroxygenistein, respectively (69). Therefore, these enzymes should have the potential to enhance soybean tolerance to oxidative stresses. The licorice CYP81E1 was also shown to be isoflavone 2′-hydroxylase (I2′H) (74) that catalyzes hydroxylation of isoflavones at C-2′ of the B-rings (75).
Expressions of enzymes in flavonoid synthesis are controlled largely by MYB-type TFs (32, 76–79). The N-terminal DNA-binding domain of MYB TFs bind to a conserved octamer (G/A/T)(G/A/T)T(C/A)(A/G)(A/G)(G/T)(T/A) in promoters of their downstream genes. Based on the number of imperfect helix-turn-helix motifs (one, two, three, or four) in their MYB domain, MYB proteins are categorized into four sub-families, including MYB1R, MYBR2R3, MYB3R and 4R-MYB; most plant MYB proteins are the R2R3 type (37, 80–81). MYB transcription factors constitute a big family involved in various physiological responses. In the present study, the phosphorylation of GmMYB173 and GmMYB183 were found to be stimulated and repressed respectively by salt stress and rhizobia inoculation (supplemental Table S2).
MYB proteins vary in binding capacities to different cis-elements; therefore, a MYB factor may have preference in recognizing and regulating a particular set of downstream genes (82). Accumulated results imply that GmMYB173 and GmMYB183 proteins may affect stress tolerance through regulation of downstream genes, GmCHS5 and GmCYP81E11, respectively (1 and Fig. 3). In the current study, we found that overexpression of GmMYB183 stimulated the expression of the GmCYP81E11 and the knockdown of GmMYB183 roots repressed the expression of the GmCYP81E11. Consistent with the GmCYP81E11 OE roots, the GmMYB183 OE roots also demonstrated increased ononin contents. Likewise, the GmMYB173 induces the accumulation of cyanidin 3-arabinoside chloride (C3A) via up-regulation of GmCHS5 (1). Compared with C3A, the ononin is a 'low efficient' ROS scavenger compared with C3A (63–66); hence GmMYB173 and GmCHS5 are positive effectors of physiological response to salt stress, whereas the GmMYB183 and GmCYP81E11 are negative regulators in the ROS scavenging process.
The GmMYB183 protein contains a Ser residue at position 61 (Ser-61), which is conserved among its homologous proteins in plants, including those from Arachis ipaensis, Medicago truncatula, Glycine max, Cajanus cajan, Phaseolus vulgaris, Vigna angularis, Prunus persica, Malus domestica, Morus notabilis and Citrus clementine (Fig. 6). Additionally, this Ser residue is followed by a [xDD], forming a common casein kinase-II target motif (53–55). The physiological functions of MYB type TFs are known to be significantly affected by the post-translational phosphorylation. Li et al. (79, 83) demonstrated that the 14–3-3 proteins regulated the intracellular localization of the transcriptional activator GmMYB176 and affected the isoflavonoid synthesis in soybean. In this research, replacement of Ser-61 with Asp-61 (a phosphorylation mimic mutation) in GmMYB183 was found to be positively correlated with ononin accumulation in soybean roots (Fig. 4 and Table II).
Fig. 6.
The phosphorylated peptide GYAsxDDA in GmMYB183 protein is highly conserved in several plants. The peptide GYAsxDDA was found in homologous proteins of GmMYB183 in several plants including Arachis ipaensis, Medicago truncatula, Glycine max, Cajanus cajan, Phaseolus vulgaris, Vigna angularis, Prunus persica, Malus domestica, Morus notabilis and Citrus clementine.
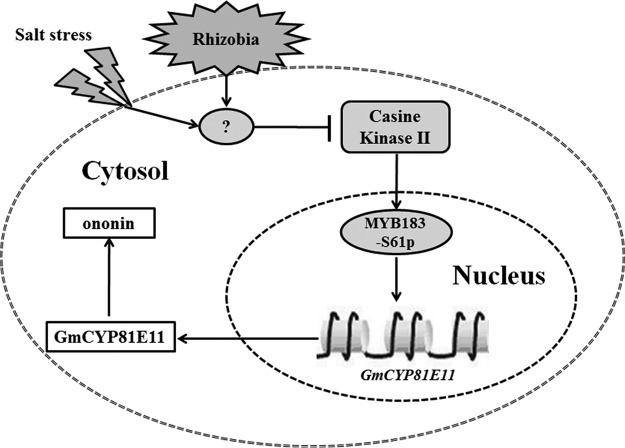
In summary, rhizobia symbiosis helps the soybean plant to cope with the detrimental effects of high soil salinity mostly because of its effect in enhancing the ROS scavenging activities, although the underlying mechanism is poorly understood. In this study, we showed that the rhizobia-inoculation, obviously not a salt stress, was found to stimulate and repress the phosphorylation of GmMYB173 and GmMYB183, two transcription factors control the expression of GmCHS5 and GmCYP81E11, enzymes involved in flavonoid syntheses, thereby help soybean plants to adapt to salt stress. It is noteworthy that monohydroxy B-ring flavonoid derivatives, such as ononin, are produced through the involvement of GmCYP81E11, and might act as an effector in other physiological processes other than protecting cells from oxidative stress. Moreover, the synthesis of ononin derivatives depletes a large amount of chalcone and adversely affects the accumulation of dihydroxy B-ring flavonoids. Our results showed that the expression of GmCYP81E11 is directly activated by GmMYB183 (Fig. 3); and the “GmMYB183-GmCYP81E11-monohydroxy B-ring flavonoids” pathway seems to play a negative role in soybean's tolerance to salt-induced oxidative stress. This pathway could be activated by pre-inoculation of rhizobia at germination stage; then the potentiated soybean seedlings may gain significantly higher tolerance to saline soil when they are transplanted (Fig. 7).
Fig. 7.
A hypothetical model for transcription factors GmMYB183 in regulating soybean's responses to salinity. After the perception of salinity or rhizobia inoculation signal, the phosphorylation of GmMYB183 was inhibited. Furthermore, the expression of its downstream gene GmCYP81E11 was downregulated and the GmCYP81E11-induced monohydroxy B-ring flavonoid (ononin) declined. Finally, the adjusted flavonoids appropriately reduced the ROS for enhancing the soybean's tolerance to salinity.
Accession Numbers
Accession numbers for the proteins mentioned in this article: GmMYB173 (Glyma17G094400): Q0PJH9; GmMYB183 (Glyma06G187600): Q0PJI2; GmCHS5 (Glyma01g43880.1): P48406; and GmYP81E11 (Glyma09g05440.1): Q2LAL0.
DATA AVAILABILITY
The mass spectrometry raw data are available via ProteomeXchange with identifier PXD006600.
Supplementary Material
Acknowledgments
We thank Prof. Maojun Xu and Miss Zhifang Jiang (from Hangzhou Normal University) for their kind assistance during the HPLC analysis of flavonoid contents.
Footnotes
* This work was supported by the grants 31970286 and 31301053 from the National Science Foundation of China, LY17C020004 from the Natural Science Foundation of Zhejiang Province, 20170432B01 and 20180432B11 from the Hangzhou Science and Technology Bureau, PF14002004014, PD11002002018001, 2016XJSGWXM27 and 2016XJSGWXM32 from Hangzhou Normal University.
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- ROS
- reactive oxygen species
- hpt
- hours post treatment
- CHS
- chalcone synthase
- CHI
- chalcone isomerase
- CYP
- cytochrome P450 monooxygenase.
REFERENCES
- 1. Pi E., Zhu C., Fan W., Huang Y., Qu L., Li Y., Zhao Q., Qiu L., Wang H., Poovaiah B. W., and Du L. (2018) Quantitative phosphoproteomic and metabonomic analyses reveal GmMYB173 optimizes flavonoid metabolism in soybean to survive salt stress. Mol. Cell. Proteomics 17, 1209–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lu Y. H., Lam H. M., Pi E. X., Zhan Q. L., Tsai S. N., Wang C. M., Kwan Y. W., and Ngai S. M. (2013) Comparative metabolomics in Glycine max and Glycine soja under salt stress to reveal the phenotypes of their offspring. J. Agr. Food Chem. 61, 8711–8721 [DOI] [PubMed] [Google Scholar]
- 3. Pi E. X., Qu L. Q., Hu J. W., Huang Y. Y., Qiu L. J., Lu H., Jiang B., Liu C., Peng T. T., Zhao Y., Wang H. Z., Tsai S. N., Ngai S. M., and Du L. Q. (2016) Mechanisms of soybean roots' tolerances to salinity revealed by proteomic and phosphoproteomic comparisons between two cultivars. Mol. Cell. Proteomics 15, 266–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Qu L. Q., Huang Y. Y., Zhu C. M., Zeng H. Q., Shen C. J., Liu C., Zhao Y., and Pi E. X. (2016) Rhizobia-inoculation enhances the soybean's tolerance to salt stress. Plant Soil 400, 209–222 [Google Scholar]
- 5. Affenzeller M. J., Darehshouri A., Andosch A., Lutz C., and Lutz-Meindl U. (2009) Salt stress-induced cell death in the unicellular green alga Micrasterias denticulata. J. Exp. Bot. 60, 939–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fahnenstich H., Scarpeci T. E., Valle E. M., Flugge U. I., and Maurino V. G. (2008) Generation of hydrogen peroxide in chloroplasts of Arabidopsis overexpressing glycolate oxidase as an inducible system to study oxidative stress. Plant Physiol. 148, 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu T., Pi E. X., Tsai S. N., Lam H. M., Sun S. M., Kwan Y. W., and Ngai S. M. (2011) GmPHD5 acts as an important regulator for crosstalk between histone H3K4 di-methylation and H3K14 acetylation in response to salinity stress in soybean. BMC Plant Biol. 11, 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. MartensYan J. H., Wang B. A., Jiang Y. N., Cheng L. J., and Wu T. L. (2014) GmFNSII-controlled soybean flavone metabolism responds to abiotic stresses and regulates plant salt tolerance. Plant Cell Physiol. 55, 74–86 [DOI] [PubMed] [Google Scholar]
- 9. Aoki T., Akashi T., and Ayabe S. (2000) Flavonoids of leguminous plants: structure, biological activity, and biosynthesis. J. Plant Res. 113, 475–488 [Google Scholar]
- 10. Jiang Y. N., Wang B. A., Li H., Yao L. M., and Wu T. L. (2010) Flavonoid production is effectively regulated by RNA1 interference of two flavone synthase genes from Glycine max. J. Plant Biol. 53, 425–432 [Google Scholar]
- 11. Shen X., Zhou Y., Duan L., Li Z., Eneji A. E., and Li J. (2010) Silicon effects on photosynthesis and antioxidant parameters of soybean seedlings under drought and ultraviolet-B radiation. J. Plant Physiol. 167, 1248–1252 [DOI] [PubMed] [Google Scholar]
- 12. Bandurska H., Niedziela J., and Chadzinikolau T. (2013) Separate and combined responses to water deficit and UV-B radiation. Plant Sci. 213, 98–105 [DOI] [PubMed] [Google Scholar]
- 13. Soledad O. N., Florencia M. M., Laura F. M., Raul D. G., Balbina A. A., and Pia O. F. (2015) Potassium phosphite increases tolerance to UV-B in potato. Plant Physiol. Bioch. 88, 1–8 [DOI] [PubMed] [Google Scholar]
- 14. Graham T. L., Graham M. Y., Subramanian S., and Yu O. (2007) RNAi silencing of genes for elicitation or biosynthesis of 5-deoxyisoflavonoids suppresses race-specific resistance and hypersensitive cell death in Phytophthora sojae infected tissues. Plant Physiol. 144, 728–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jia L. G., Sheng Z. W., Xu W. F., Li Y. X., Liu Y. G., Xia Y. J., and Zhang J. H. (2012) Modulation of anti-oxidation ability by proanthocyanidins during germination of Arabidopsis thaliana seeds. Mol. Plant 5, 472–481 [DOI] [PubMed] [Google Scholar]
- 16. Sakao K., Fujii M., and Hou D. X. (2009) Acetyl derivate of quercetin increases the sensitivity of human leukemia cells toward apoptosis. BioFactors 35, 399–405 [DOI] [PubMed] [Google Scholar]
- 17. Yang X., Kang S. M., Jeon B. T., Kim Y. D., Ha J. H., Kim Y. T., and Jeon Y. J. (2011) Isolation and identification of an antioxidant flavonoid compound from citrus-processing by-product. J. Sci. Food Agric. 91, 1925–1927 [DOI] [PubMed] [Google Scholar]
- 18. Chobot V., Huber C., Trettenhahn G., and Hadacek F. (2009) (±)-catechin: chemical weapon, antioxidant, or stress regulator? J. Chem. Ecol. 35, 980–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim H. J., Suh H. J., Kim J. H., Park S., Joo Y. C., and Kim J. S. (2010) Antioxidant activity of glyceollins derived from soybean elicited with Aspergillus sojae. J. Agric. Food Chem. 58, 11633–11638 [DOI] [PubMed] [Google Scholar]
- 20. Tirichine L., Imaizumi-Anraku H., Yoshida S., Murakami Y., Madsen L. H., Miwa H., Nakagawa T., Sandal N., Albrektsen A. S., Kawaguchi M., Downie A., Sato S., Tabata S., Kouchi H., Parniske M., Kawasaki S., and Stougaard J. (2006) Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441, 1153–1156 [DOI] [PubMed] [Google Scholar]
- 21. Zhang J., Subramanian S., Stacey G., and Yu O. (2009) Flavones and flavonols play distinct critical roles during nodulation of Medicago truncatula by Sinorhizobium meliloti. Plant J. 57, 171–183 [DOI] [PubMed] [Google Scholar]
- 22. Chen S. L., Hawighorst P., Sun J., and Polle A. (2014) Salt tolerance in populus: significance of stress signaling networks, mycorrhization, and soil amendments for cellular and whole-plant nutrition. Environ. Exp. Bot. 107, 113–124 [Google Scholar]
- 23. Miransari M., and Smith D. L. (2009) Alleviating salt stress on soybean (Glycine max (L.) Merr.) - Bradyrhizobium japonicum symbiosis, using signal molecule genistein. Eur. J. Soil Biol. 45, 146–152 [Google Scholar]
- 24. Ruiz-Lozano J. M., Porcel R., Azcon C., and Aroca R. (2012) Regulation by arbuscular mycorrhizae of the integrated physiological response to salinity in plants: new challenges in physiological and molecular studies. J. Exp. Bot. 63, 4033–4044 [DOI] [PubMed] [Google Scholar]
- 25. Bianco C., and Defez R. (2009) Medicago truncatula improves salt tolerance when nodulated by an indole-3-acetic acid-overproducing Sinorhizobium meliloti strain. J. Exp. Bot. 60, 3097–3107 [DOI] [PubMed] [Google Scholar]
- 26. Koes R., Verweij W., and Quattrocchio F. (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10, 236–242 [DOI] [PubMed] [Google Scholar]
- 27. Ferreyra M. L. F., Rius S. P., and Casati P. (2012) Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 3, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ishida J. K., Wakatake T., Yoshida S., Takebayashi Y., Kasahara H., Wafula E., dePamphilis C. W., Namba S., and Shirasu K. (2016) Local auxin biosynthesis mediated by a YUCCA flavin monooxygenase regulates haustorium development in the parasitic plant Phtheirospermum japonicum. Plant Cell 28, 1795–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liao Y., Zou H. F., Wang H. W., Zhang W. K., Ma B., Zhang J. S., and Chen S. Y. (2008) Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res. 18, 1047–1060 [DOI] [PubMed] [Google Scholar]
- 30. Broun P. (2005) Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr. Opin. Plant Biol. 8, 272–279 [DOI] [PubMed] [Google Scholar]
- 31. Schwinn K. E., Boase M. R., Bradley J. M., Lewis D. H., Deroles S. C., Martin C. R., and Davies K. M. (2014) MYB and bHLH transcription factor transgenes increase anthocyanin pigmentation in petunia and lisianthus plants, and the petunia phenotypes are strongly enhanced under field conditions. Front. Plant Sci. 5, 603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. de VettenLotkowska M. E., Tohge T., Fernie A. R., Xue G. P., Balazadeh S., and Mueller-Roeber B. (2015) The arabidopsis transcription factor MYB112 promotes anthocyanin formation during salinity and under high light stress. Plant Physiol. 169, 1862–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yi J. X., Derynck M. R., Li X. Y., Telmer P., Marsolais F., and Dhaubhadel S. (2010) A single-repeat MYB transcription factor, GmMYB176, regulates CHS8 gene expression and affects isoflavonoid biosynthesis in soybean. Plant J. 62, 1019–1034 [DOI] [PubMed] [Google Scholar]
- 34. Borevitz J. O., Xia Y., Blount J., Dixon R. A., and Lamb C. (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12, 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gonzalez A., Zhao M., Leavitt J. M., and Lloyd A. M. (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827 [DOI] [PubMed] [Google Scholar]
- 36. Yuan Y., Qi L. J., Yang J., Wu C., Liu Y. J., and Huang L. Q. (2015) A Scutellaria baicalensis R2R3-MYB gene, SbMYB8, regulates flavonoid biosynthesis and improves drought stress tolerance in transgenic tobacco. Plant Cell Tiss. Org. 120, 961–972 [Google Scholar]
- 37. Su L. T., Wang Y., Liu D. Q., Li X. W., Zhai Y., Sun X., Li X. Y., Liu Y. J., Li J. W., and Wang Q. Y. (2015) The soybean gene, GmMYBJ2, encodes a R2R3-type transcription factor involved in drought stress tolerance in Arabidopsis thaliana. Acta Physiol. Plant. 37, 138 [Google Scholar]
- 38. Bassel G. W., Gaudinier A., Brady S. M., Hennig L., Rhee S. Y., and De Smet I. (2012) Systems analysis of plant functional, transcriptional, physical interaction, and metabolic networks. Plant Cell 24, 3859–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Albert R. (2007) Network inference, analysis, and modeling in systems biology. Plant Cell 19, 3327–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Niehaus T. D., Nguyen T. N., Gidda S. K., ElBadawi-Sidhu M., Lambrecht J. A., McCarty D. R., Downs D. M., Cooper A. J., Fiehn O., Mullen R. T., and Hanson A. D. (2014) Arabidopsis and maize RidA proteins preempt reactive enamine/imine damage to branched-chain amino acid biosynthesis in plastids. Plant Cell 26, 3010–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schwartz D., and Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 42. Du L., Ali G. S., Simons K. A., Hou J., Yang T., Reddy A. S., and Poovaiah B. W. (2009) Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457, 1154–1158 [DOI] [PubMed] [Google Scholar]
- 43. Chen F., Dong G., Wu L., Wang F., Yang X., Ma X., Wang H., Wu J., Zhang Y., Wang H., Qian Q., and Yu Y. (2016) A nucleus-encoded chloroplast protein YL1 is involved in chloroplast development and efficient biogenesis of chloroplast ATP synthase in rice. Sci. Rep-UK 6, 32295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schmidt R., Mieulet D., Hubberten H. M., Obata T., Hoefgen R., Fernie A. R., Fisahn J., Segundo B. S., Guiderdoni E., Schippers J. H. M., and Mueller-Roeber B. (2013) SALT-RESPONSIVE ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell 25, 2115–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Routray P., Miller J. B., Du L., Oldroyd G., and Poovaiah B. W. (2013) Phosphorylation of S344 in the calmodulin-binding domain negatively affects CCaMK function during bacterial and fungal symbioses. Plant J. 76, 287–296 [DOI] [PubMed] [Google Scholar]
- 46. Liu J., Ishitani M., Halfter U., Kim C.-S., and Zhu J.-K. (2000) The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. U.S.A. 97, 3730–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Walker A. R., Davison P. A., Bolognesi-Winfield A. C., James C. M., Srinivasan N., Blundell T. L., Esch J. J., Marks M. D., and Gray J. C. (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11, 1337–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stracke R., Favory J. J., Gruber H., Bartelniewoehner L., Bartels S., Binkert M., Funk M., Weisshaar B., and Ulm R. (2010) The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 33, 88–103 [DOI] [PubMed] [Google Scholar]
- 49. Verweij W., Spelt C. E., Bliek M., de Vries M., Wit N., Faraco M., Koes R., and Quattrocchio F. M. (2016) Functionally similar WRKY proteins regulate vacuolar acidification in Petunia and hair development in Arabidopsis. Plant Cell 28, 786–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xie X. B., Li S., Zhang R. F., Zhao J., Chen Y. C., Zhao Q., Yao Y. X., You C. X., Zhang X. S., and Hao Y. J. (2012) The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 35, 1884–1897 [DOI] [PubMed] [Google Scholar]
- 51. Faraco M., Spelt C., Bliek M., Verweij W., Hoshino A., Espen L., Prinsi B., Jaarsma R., Tarhan E., de Boer A. H., Di Sansebastiano G. P., Koes R., and Quattrocchio F. M. (2014) Hyperacidification of vacuoles by the combined action of two different P-ATPases in the tonoplast determines flower color. Cell Rep. 6, 32–43 [DOI] [PubMed] [Google Scholar]
- 52. Francisco R. M., Regalado A., Ageorges A., Burla B. J., Bassin B., Eisenach C., Zarrouk O., Vialet S., Marlin T., Chaves M. M., Martinoia E., and Nagy R. (2013) ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-glucosides. Plant Cell 25, 1840–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Broin P. O., Smith T. J., and Golden A. A. J. (2015) Alignment-free clustering of transcription factor binding motifs using a genetic-k-medoids approach. BMC Bioinformatics 16, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Matsuura H., Ishibashi Y., Shinmyo A., Kanaya S., and Kato K. (2010) Genome-wide analyses of early translational responses to elevated temperature and high salinity in Arabidopsis thaliana. Plant Cell Physiol. 51, 448–462 [DOI] [PubMed] [Google Scholar]
- 55. Prasad T. S. K., Goel R., Kandasamy K., Keerthikumar S., Kumar S., Mathivanan S., Telikicherla D., Raju R., Shafreen B., Venugopal A., Balakrishnan L., Marimuthu A., Banerjee S., Somanathan D. S., Sebastian A., Rani S., Ray S., Kishore C. J. H., Kanth S., Ahmed M., Kashyap M. K., Mohmood R., Ramachandra Y. L., Krishna V., Rahiman B. A., Mohan S., Ranganathan P., Ramabadran S., Chaerkady R., and Pandey A. (2009) Human protein reference database-2009 update. Nucleic Acids Res. 37, D767–D772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lv D. W., Subburaj S., Cao M., Yan X., Li X. H., Appels R., Sun D. F., Ma W. J., and Yan Y. M. (2014) Proteome and phosphoproteome characterization reveals new response and defense mechanisms of Brachypodium distachyon leaves under salt stress. Mol. Cell. Proteomics 13, 632–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Masclaux-Daubresse C., Clement G., Anne P., Routaboul J. M., Guiboileau A., Soulay F., Shirasu K., and Yoshimoto K. (2014) Stitching together the multiple dimensions of autophagy using metabolomics and transcriptomics reveals impacts on metabolism, development, and plant responses to the environment in Arabidopsis. Plant Cell 26, 1857–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Du H., Yang S. S., Liang Z., Feng B. R., Liu L., Huang Y. B., and Tang Y. X. (2012) Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 12, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tsai S. M., and Phillips D. A. (1991) Flavonoids released naturally from alfalfa promote development of symbiotic glomus spores in vitro. Appl. Environ. Microb. 57, 1485–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Becard G., Douds D. D., and Pfeffer P. E. (1992) Extensive in vitro hyphal growth of vesicular-Arbuscular Mycorrhizal fungi in the presence of CO2 and flavonols. Appl. Environ. Microb. 58, 821–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Volpin H., Elkind Y., Okon Y., and Kapulnik Y. (1994) A vesicular Arbuscular mycorrhizal fungus (Glomus intraradix) induces a defense response in alfalfa roots. Plant Physiol. 104, 683–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nakabayashi R., Yonekura-Sakakibara K., Urano K., Suzuki M., Yamada Y., Nishizawa T., Matsuda F., Kojima M., Sakakibara H., Shinozaki K., Michael A. J., Tohge T., Yamazaki M., and Saito K. (2014) Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 77, 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Agati G., Azzarello E., Pollastri S., and Tattini M. (2012) Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 196, 67–76 [DOI] [PubMed] [Google Scholar]
- 64. Hideo Yamasaki Yasuko Sakihama and Ikehara N. (1997) Flavonoid-peroxidase reaction as a detoxification mechanism of plant cells against H2O2. Plant Physiol. Bioch. 115, 1405–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Brown J. E., Khodr H., Hider R. C., and Rice-Evans C. A. (1998) Structural dependence of flavonoid interactions with Cu2+ ions: implications for their antioxidant properties. Biochem. J. 330, 1173–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hernandez I., Alegre L., Van Breusegem F., and Munne-Bosch S. (2009) How relevant are flavonoids as antioxidants in plants? Trends Plant Sci. 14, 125–132 [DOI] [PubMed] [Google Scholar]
- 67. Agati G., Biricolti S., Guidi L., Ferrini F., Fini A., and Tattini M. (2011) The biosynthesis of flavonoids is enhanced similarly by UV radiation and root zone salinity in L. vulgare leaves. J. Plant Physiol. 168, 204–221 [DOI] [PubMed] [Google Scholar]
- 68. Agati G., Matteini P., Goti A., and Tattini M. (2007) Chloroplast-located flavonoids can scavenge singlet oxygen. New Phytol. 174, 77–89 [DOI] [PubMed] [Google Scholar]
- 69. Uchida K., Akashi T., and Aoki T. (2015) Functional expression of cytochrome P450 in Escherichia coli: An approach to functional analysis of uncharacterized enzymes for flavonoid biosynthesis. Plant Biotechnol-Nar 32, 205–213 [Google Scholar]
- 70. Martens S., and Mithofer A. (2005) Flavones and flavone synthases. Phytochemistry 66, 2399–2407 [DOI] [PubMed] [Google Scholar]
- 71. Carelli M., Biazzi E., Panara F., Tava A., Scaramelli L., Porceddu A., Graham N., Odoardi M., Piano E., Arcioni S., May S., Scotti C., and Calderini O. (2011) Medicago truncatula CYP716A12 is a multifunctional oxidase involved in the biosynthesis of Hemolytic Saponins. Plant Cell 23, 3070–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Seki H., Sawai S., Ohyama K., Mizutani M., Ohnishi T., Sudo H., Fukushima E. O., Akashi T., Aoki T., Saito K., and Muranaka T. (2011) Triterpene functional genomics in licorice for identification of CYP72A154 involved in the biosynthesis of glycyrrhizin. Plant Cell 23, 4112–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lam P. Y., Zhu F. Y., Chan W. L., Liu H. J., and Lo C. (2014) Cytochrome P450 93G1 is a flavone synthase II that channels flavanones to the biosynthesis of tricin O-linked conjugates in rice. Plant Physiol. 165, 1315–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Akashi T., Aoki T., and Ayabe S. (1998) Identification of a cytochrome P450 cDNA encoding (2S)-flavanone 2-hydroxylase of licorice (Glycyrrhiza echinata L., Fabaceae) which represents licodione synthase and flavone synthase II. FEBS Lett. 431, 287–290 [DOI] [PubMed] [Google Scholar]
- 75. Kessmann H., Choudhary A. D., and Dixon R. A. (1990) Stress responses in alfalfa (Medicago sativa L.) III. Induction of medicarpin and cytochrome P450 enzyme activities in elicitor-treated cell suspension cultures and protoplasts. Plant Cell Rep. 9, 38–41 [DOI] [PubMed] [Google Scholar]
- 76. Morse A. M., Whetten R. W., Dubos C., and Campbell M. M. (2009) Post-translational modification of an R2R3-MYB transcription factor by a MAP Kinase during xylem development. New Phytol. 183, 1001–1013 [DOI] [PubMed] [Google Scholar]
- 77. Zhang J. A., Subramanian S., Zhang Y. S., and Yu O. (2007) Flavone synthases from Medicago truncatula are flavanone-2-hydroxylases and are important for nodulation. Plant Physiol. 144, 741–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yan J., Wang B., Zhong Y., Yao L., Cheng L., and Wu T. (2015) The soybean R2R3 MYB transcription factor GmMYB100 negatively regulates plant flavonoid biosynthesis. Plant Mol. Biol. 89, 35–48 [DOI] [PubMed] [Google Scholar]
- 79. Li X., and Dhaubhadel S. (2012) 14–3-3 proteins act as scaffolds for GmMYB62 and GmMYB176 and regulate their intracellular localization in soybean. Plant Signaling & Behavior 7, 965–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Stracke R., Werber M., and Weisshaar B. (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456 [DOI] [PubMed] [Google Scholar]
- 81. Li X. W., Li J. W., Zhai Y., Zhao Y., Zhao X., Zhang H. J., Su L. T., Wang Y., and Wang Q. Y. (2013) A R2R3-MYB transcription factor, GmMYB12B2, affects the expression levels of flavonoid biosynthesis genes encoding key enzymes in transgenic Arabidopsis plants. Gene 532, 72–79 [DOI] [PubMed] [Google Scholar]
- 82. Romero I., Fuertes A., Benito M. J., Malpica J. M., Leyva A., and Paz-Ares J. (1998) More than 80 R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J. 14, 273–284 [DOI] [PubMed] [Google Scholar]
- 83. Li X. Y., Chen L., and Dhaubhadel S. (2012) 14–3-3 proteins regulate the intracellular localization of the transcriptional activator GmMYB176 and affect isoflavonoid synthesis in soybean. Plant J. 71, 239–250 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry raw data are available via ProteomeXchange with identifier PXD006600.