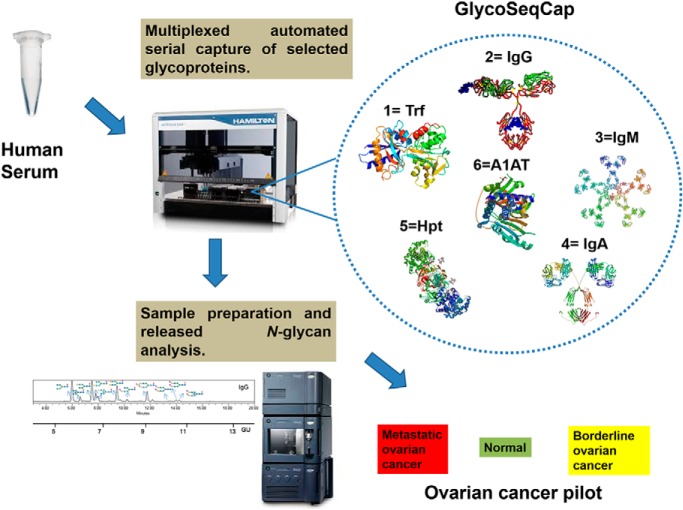
On a fully automated liquid handling station, the N-glycomes of antibodies IgG, IgM, and IgA and acute phase proteins transferrin, haptoglobin and alpha-1-antitrypsin from human serum have been purified and structurally characterized using UPLC, exoglycosidase digestions and LC-MS. The glycoprofiles of the resultant AQC labelled N-glycans from each glycoprotein can be used to quantitatively compare the structural differences between healthy, borderline and metastatic ovarian cancer. Statistical tools and discrimination models are employed to generate a diagnostic tool to discriminate between different stages of ovarian cancer. Future applications of the technology termed “GlycoSeqCap” include an extension to other inflammatory diseases.
Keywords: glycomics, glycoproteins, glycosylation, ovarian cancer, antibodies, plasma or serum analysis, acute phase proteins
Graphical Abstract
Highlights
Quantitative high-throughput glycoanalytical technology as a diagnostic tool for ovarian cancer detection.
Multiplexed approach harnessing N-glycan data for six glycoproteins from a single biological sample.
Detailed characterization of human serum N-glycans from antibodies IgG, IgM and IgA and acute phase proteins transferrin, haptoglobin and alpha-1-antitrypsin.
Structural differences in antibody and acute phase protein glycosylation for mechanistic insights.
Abstract
The direct association of the genome, transcriptome, metabolome, lipidome and proteome with the serum glycome has revealed systems of interconnected cellular pathways. The exact roles of individual glycoproteomes in the context of disease have yet to be elucidated. In a move toward personalized medicine, it is now becoming critical to understand disease pathogenesis, and the traits, stages, phenotypes and molecular features that accompany it, as the disruption of a whole system. To this end, we have developed an innovative technology on an automated platform, “GlycoSeqCap,” which combines N-glycosylation data from six glycoproteins using a single source of human serum. Specifically, we multiplexed and optimized a successive serial capture and glycoanalysis of six purified glycoproteins, immunoglobulin G (IgG), immunoglobulin M (IgM), immunoglobulin A (IgA), transferrin (Trf), haptoglobin (Hpt) and alpha-1-antitrypsin (A1AT), from 50 μl of human serum. We provide the most comprehensive and in-depth glycan analysis of individual glycoproteins in a single source of human serum to date. To demonstrate the technological application in the context of a disease model, we performed a pilot study in an ovarian cancer cohort (n = 34) using discrimination and classification analyses to identify aberrant glycosylation. In our sample cohort, we exhibit improved selectivity and specificity over the currently used biomarker for ovarian cancer, CA125, for early stage ovarian cancer. This technology will establish a new state-of-the-art strategy for the characterization of individual serum glycoproteomes as a diagnostic and monitoring tool which represents a major step toward understanding the changes that take place during disease.
Glycosylation is the most common and complex type of post-translational modification and glycoscience is rightly recognized as a current frontier. Over 1% of total human genome codes for ∼300 functional glycogenes known to exist in humans (1). Post-translational modifications (PTMs)1 of a protein are critical late events in protein biosynthesis and glycosylation is one of the most important. It reflects the genome of the individual person and also the many other influences on the cellular pathways. In fact, the glycoproteome can be thought of as the final readout of the genome that confers function on the gene products. A range of human genetic disorders, including congenital disorders of glycosylation (2, 3), MODY type diabetes (4), galactosemia (5) and muscular dystrophies (6) have been directly linked to or shown to involve faulty glycosylation. Cancer associated aberrant gene regulation also results in alterations of glycan structures and has been well studied both in the serum glycome and specific serum glycoproteins (7–9). In addition, chronic inflammatory diseases display altered glycosylation (10).
Immunoglobulin G (IgG) N-glycome is well characterized using liquid chromatography and mass spectrometry methods, (11–13) but N-glycosylation analysis of other glycoproteins has been studied to a far lesser extent. For example, N-glycosylation analysis of acute phase proteins, such as transferrin (Trf), alpha-1-antitrypsin (A1AT) and haptoglobin (Hpt), has been conducted in our laboratory (8, 14) using complex methodologies such as isoelectric focusing (IEF) or 2D-gel electrophoresis for glycoprotein separation. In addition, N-glycosylation of serum immunoglobulin M (IgM) (15) and immunoglobulin A (IgA) (16) have been characterized previously using normal phase high performance liquid chromatography (NP-HPLC) a decade ago with only a proportion of the glycans identified, a limitation of the technology at the time. More recently, IgA site specific glycosylation (N- and O-glycopeptides from IgA1) was elucidated in serum of patients with rheumatoid arthritis (RA) during pregnancy using matrix-assisted laser/desorption ionization Fourier transform ion cyclotron resonance (MALDI-FTICR) mass spectrometry (17). Of note, the team additionally performed released N-glycan analysis and could identify major N-glycans that were not identified in the characterized IgA1 N-glycopeptide fractions. As such, a detailed structural characterization of serum IgA N-glycome analysis remains to be undertaken. In a separate study, protein-specific differential glycosylation of immunoglobulins (IgG, IgM, and IgA) were studied in serum of ovarian cancer patients (18) using multiple reaction monitoring on a triple quadrupole mass spectrometer. This technology presents similar limitations as to the total structural elucidation of the N-glycome, but is highly revealing, whereby the authors hypothesize that within the total serum glycome profile, which is largely dominated by the highest abundance proteins (such as antibodies IgG, IgM, and IgA or acute phase proteins Trf, Hpt, A1AT) that protein- and site-specific glycosylation profiles will be likely to provide further insights into protein specific alterations in glycosylation of the glycans related to ovarian cancer as well as serve as more specific biomarkers for ovarian cancer than current tools (18).
Although some advances have been made in the sample purification and target glycoprotein enrichment (19), there is a growing necessity for the development of more robust, consistent, sensitive and versatile methods that can be automated and applied to personalized medicine approaches. To address this need, we optimized and automated a glycoanalytical technology to capture and glycoprofile six abundant individual glycoproteins by serial extraction-IgG, IgM, IgA, Trf, Hpt, and A1AT. We performed a detailed analysis of the N-glycomes from these six glycoproteins using 50 μl of pooled normal human serum (NHS). Then we applied this technology to an ovarian cancer patient cohort, consisting of 7 healthy controls, 6 borderline and 21 metastatic ovarian cancer patients and tested its potential to be used as a diagnostic tool for earlier detection of ovarian cancer. This study is a follow-up of our previously reported sample preparation and chromatography technologies for glycan analysis (11, 20, 21) and biomarker discovery.
EXPERIMENTAL PROCEDURES
Serum Samples
Normal human serum (NHS) samples were used as described previously (20). Briefly human adult serum sample from healthy male and female blood donors were pooled and used as a source of glycoproteins for subsequent glycan analysis (courtesy of the U.K. Blood Transfusion Service). Serum samples from 7 healthy women (hereafter called normal) and 27 ovarian cancer patients classified as either metastatic (n = 21) or borderline (n = 6) were used for the GlycoSeqCap technology (supplemental Tables S9) following ethical approval and obtaining informed consent. After allowing the blood to clot for 30–60 min, serum was obtained by centrifugation at 2000 × g for 10 min and stored at −80 °C until analysis.
Chemicals and Reagents
All chemical reagents and solvents were purchased from Merck KGaA (Darmstadt, Germany). Pre-packed Protein G (PTH93–20-02) 300 μl tip columns obtained from Phynexus (San Jose, CA). Albumin (Cat 191297005), IgM (Cat# 289005), IgA(Cat# 190288005), Trf (Cat# 191306005) and A1AT (Cat# 191287005) affinity matrix resins were obtained from ThermoFisher CaptureSelect (Waltham, MA) CaptureSelectTM and Hpt (Cat# 291.2950.05) obtained from BAC BV, Capture Select. Exoglycosidase and PNGase F enzymes were obtained from NEB (Ipswich, MA): PNGase F (P0709L), Bovine Kidney Fucosidase (BKF, P0748S), and other exoglycosidases Arthrobacter urefaciens sialidase (ABS, PZGK80090), Bovine Testes Galactosidase (BTG, PZGKX-5013), N-Acetyl Hexosaminidase (GUH, PZGK80050), and Jack Bean Mannosidase (JBM, GKX-5003) obtained from Europa (Prozyme, Hayward, CA). Solid-supported Ultralink hydrazide (Cat# 11809410) and Fermentas PageRuler Pre-stained Protein Ladder (Cat# 11832124) was obtained from ThermoFisher Scientific. The following plates were used;1 μm 96-well (PALL Acroprep Advance 350, VWR (Radnor, PA) 518-0026), 96-well 2 ml collection plate (X50 MICROPLATE, Fisher, 11511963), 96-well assay (Greiner, 450 μl, Cruinn (Dublin, Ireland) 731-1372), 384-well ultrafiltration (Acroprep, 10KDa, PALL 5077), 384-well PCR (Fisher, Armadillo AB-2384/O, orange, 30 μl, VWR 732 1578), and a 384-well storage/collection (Corning, 240 μl VWR 736 0202 3347). Reagents were placed on the robotic platform in a 12-well reagent trough (VWR 732–1390). Samples were prepared on a Hamilton robotics StarLet liquid-handling platform. The instrument is equipped with eight software-controlled pipettes, a vacuum manifold, and an automated heater shaker. Samples were analyzed on a Waters Acquity H-class UPLC instrument. The following workflow was implemented as robotic program on Hamilton Robotics Venus One software.
Custom Phytip Glycoprotein Affinity Matrix Resin Tip Column Preparation
Each specific anti-protein and anti-glycoprotein matrix was packed in Phytips by PhyNexus Inc. Briefly, 20 μl each of Albumin, IgG, IgM, IgA, Trf, and A1AT affinity matrix resins purchased from Life Technologies CaptureSelectTM and Hpt purchased from BAC BV were packed individually in a 300 μl Hamilton tips.
Phytip Glycoprotein Affinity Purification
Glycoproteins from 50 μl of whole serum sample were captured in the following sequence: Albumin, Trf, IgG, IgM, IgA, Hpt, and A1AT on the automated liquid handling station (Hamilton Starlet) in a 96-well format using the custom Phytips (Fig. 1). In this method Phytip equilibration, capture, wash, and elution refers to cycling a solution through the resin bed for a fixed number of aspirations and dispense steps termed cycles at defined flow rates. In a sequential fashion Albumin and the glycoproteins were captured from serum or from serum depleted of the preceding glycoproteins. For example, in the case of Trf, the Albumin depleted serum was used for the affinity chromatography. In the case of IgG affinity, the serum was depleted of both Albumin and Trf before IgG purification.
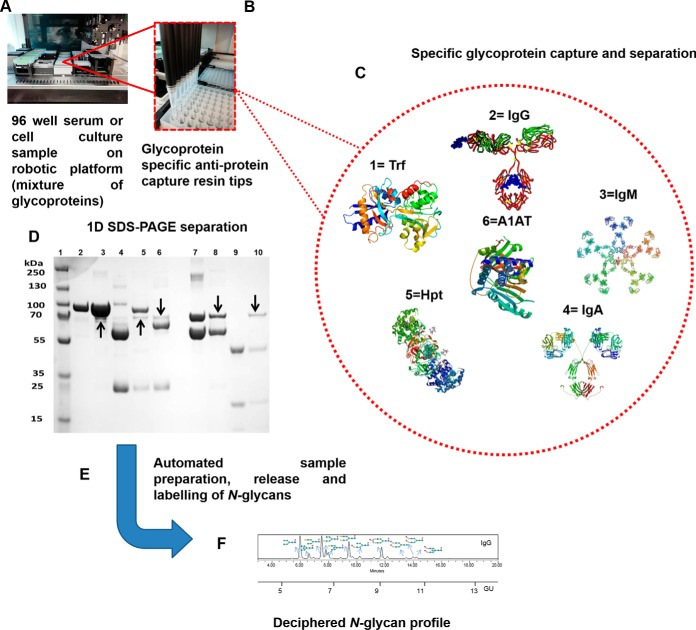
Fig. 1.
Multiplexed automated serial capture of glycoprotein N-glycoprofiling. 96-well format robotic platform (A), specific anti-glycoprotein capture resin packed in PhyNexus phytip (B), serial capture of selected glycoproteins 1.Trf, 2.IgG, 3.IgM, 4.IgA, 5.Hpt, 6.A1AT (C), 1D SDS-PAGE separation of multiplexed automated capture of six selected glycoproteins from pooled human serum using PhyNexus Phytips. Lane 1: Protein Marker, Lane 2: Trf Standard, Lane 3: Bound Trf, Lane 4: Bound IgG, Lane 5: Bound IgM, Lane 6: Bound IgA, Lane 7: A1AT Standard, Lane 8: Bound A1AT, Lane 9: Hpt Standard, Lane 10: Bound Hpt. The protein bands marked with arrows are traces of albumin protein (non-glycosylated) (D), automated glycoprotein sample preparation, PNGaseF release and aminoquinoline carbamate (AQC) labeling of N-glycans (E) and finally ultra-high performance liquid chromatography (UPLC) separation and glycan structural analysis (F).
Albumin and the glycoproteins were captured using Phytips from serum/depleted serum in 96-well Greiner plates (50 μl per well, 100 μl mixing volume, 6 cycles, 20 μl/s) using pre-equilibrated Phytips (200 μl per well, 0.1 m sodium phosphate buffer, 0.15 m NaCl, pH 7.4, 3 cycles, 5 μl/s). The albumin/glycoprotein in Phytip resin were washed three times in 96-well collection plates, each containing binding buffer (170 μl per well mixing volume, 0.1 m sodium phosphate buffer, 0.15 m NaCl, pH 7.4, 6 cycles, 5 μl/s) followed by treatment with washing buffer (170 μl per well mixing volume, 1% NaCl + 1g/L NaN3,, 4 cycles, 5 μl/s). The albumin/glycoproteins were eluted (80 μl per well, 0.2 m Glycine-HCl buffer, pH 2.5, 3 cycles, 4 μl/s) into three Greiner plates. Neutralization buffer (10 μl per well, 1 m Tris-HCl buffer, pH 9.0) was added to two Greiner plates and the samples were pooled together for two Greiner plates (180 μl resulting solution per well, mean concentrations 0.22, 0.90, 0.85, 0.12, 0.81, and 0.79 mg/ml respectively for IgG, IgM, IgA, Trf, Hpt, and A1AT).
1D SDS-PAGE
Affinity purified glycoproteins were reduced and loaded on SDS-PAGE gels (NuPAGE® Novex 4–12% Bis-Tris) and separated in a XCell SureLock™ Mini-Cell (Invitrogen, Carlsbad, CA) for 90min at 120V using a MES running buffer. 5 μg of total protein was loaded in each lane of the gel. Once electrophoretic separation was completed, proteins were visualized by staining with Coomassie blue staining solution followed by destaining with multiple changes of deionized water (Fig. 1).
Automated Glycoprotein Denaturation and N-Glycan Release
As described previously (22), briefly N-glycan analysis was performed using a pooled serum sample from 100 apparently healthy male and female adult blood donors (U.K. Blood Transfusion Service). The antibodies IgG, IgA, IgM and the acute phase proteins Trf, Hpt, and A1AT were purified from the pool using the respective affinity resin (Life Technologies) packed in Phynexus Phytips as previously described (in above section). On an automated platform, the glycoprotein samples were dispensed into two 384-well ultrafiltration plates (maximum loading: 60 μg protein, 10kDa). Ultra-filtration was performed by centrifugation (3700 × g, 30 min, room temperature) or on a vacuum manifold (> 25 in Hg vacuum, 30min). Denaturation buffer (25 μl per well, 100 mm sodium bicarbonate, 50 mm dithiothreitol (DTT), 0.1% sodium dodecyl sulfate (SDS)) was dispensed into 2 × 384-well ultrafiltration plates. After 10min incubation at room temperature, the samples were mixed 10 times (mixing volume: 15 μl, flow rate: 10 μl/s) and transferred to a 384-well PCR plate (ThermoFisher). The plate was placed into a robotic incubation chamber at 95 °C for 10 min. The plate was removed from the incubator and equilibrated to room temperature for 10 min. 1 m iodoacetamide (IAA), 10 μl, was dispensed into each well of the ultrafiltration plate and the samples (25 μl) were transferred back into the 384-well PCR plate. The samples were mixed 5 times (mixing volume 20 μl, flow rate, 10 μl/s). After 10 min incubation at room temperature, the ultrafiltration plate was stacked onto a 240 μl collection plate (Corning block) and centrifuged (3700 × g, 30 min, room temperature) and the supernatant was removed. Next, 10 μl of a 25 mm sodium bi-carbonate solution was dispensed into each well and the ultra-filtration plate was centrifuged (3700 × g, 30 min, room temperature) and the supernatant was removed.
The deglycosylation mix, 12 μl (0.4 μl of PNGase F (2.5U/ml), 25 mm sodium bicarbonate) was dispensed into each well and the plate was then covered with a lid and incubated on a robotic orbital shaker (shaking orbit: 2 mm, shaking speed: 700 rpm, temperature: 38 °C, incubation time: 30 min). The ultrafiltration plate was stacked onto a 30 μl/well PCR plate (Armadillo) and centrifuged (3700 × g, 10 min, room temperature). Finally, 10 μl of 25 m sodium bicarbonate solution was dispensed into each well of the ultrafiltration plate (still stacked onto the PCR plate) and the assembly was centrifuged (3700 × g, 10 min, room temperature). The released N-glycans in the PCR plate were subsequently fluorescently labeled. For glycan labeling, 5 μl of glycan sample (PCR plate) was transferred to a 95 μl Corning block and 11.6 μl of AQC (3 mg/ml MeCN) was added. Three microliters of this crude mixture was directly injected into the UPLC system. Alternatively, after the PNGase F release, glycans can be frozen and are stable at −20 °C for labeling later.
Ultra-Performance Liquid Chromatography (UPLC)
As previously described (22), briefly the separation of AQC-derivatized N-glycans was carried out by UPLC with fluorescence detection on a Waters ACQUITY UPLC H-Class instrument consisting of a binary solvent manager, sample manager, and fluorescence detector under the control of Empower 3 software (Waters, Milford, MA). The HILIC separations were performed using a Waters Ethylene Bridged Hybrid (BEH) Glycan column (150 × 2.1 mm i.d., 1.7 μm particles) with 50 mm ammonium formate (pH 4.4) as solvent A and MeCN as solvent B. The column was fitted with an ACQUITY in-line 0.2 μm filter. The separation was performed using a linear gradient of 70–53% MCN 0.56 ml/min in 16.5 min for IgG N-glycan separation. An injection volume of 3 μl prepared in 70% v/v MeCN was used throughout. Samples were maintained at 5 °C before injection, and the separation temperature was 40 °C. The FLD excitation/emission wavelength were λex = 245 nm and λem = 395 nm, respectively. The system was calibrated using an external standard of hydrolyzed and 2-AB-labeled glucose oligomers to create a dextran ladder, as described previously (11). A fifth-order polynomial distribution curve was fitted to the dextran ladder to assign glucose unit (GU) values from retention times (using Empower software from Waters).
Liquid Chromatography-Mass Spectrometry (LC-MS)
Online coupled fluorescence (FLR)-mass spectrometry detection was performed using a Waters Xevo G2 QTof with Acquity® UPLC (Waters Corporation, Milford, MA) and BEH Glycan column (2.1 × 150 mm, 1.7 μm particle size). For MS acquisition data the instrument was operated in positive-sensitivity mode with a capillary voltage of 3kV. The ion source block and nitrogen desolvation gas temperatures were set at 120 °C and 350 °C, respectively. The desolvation gas was set to a flow rate of 800 L/h. The cone voltage was maintained at 40V. Full-scan data for glycans were acquired over m/z range of 300 to 2000. Data collection and processing were controlled by MassLynx 4.1 software. The fluorescence detector settings were as follows: λexcitation: 245 nm, λemission: 395 nm; data rate was 10pts/second and a PMT gain = 20. Sample injection volume was 10 μl (75% MeCN). The flow rate was 0.400 ml/min (unless specified) and column temperature was maintained at 60 °C; solvent A was 50 mm ammonium formate (pH 4.4) and solvent B was MeCN. A 60min linear gradient was used and was as follows: 25–46% A for 35 min, 46–80% A for 8 min (flow rate at 0.2 ml/min), 80–25% A for 27 min. To avoid contamination of system, flow was sent to waste for the first 1.2 min and after 55 min.
Exoglycosidase Digestions
The AQC labeled N-glycans from the glycoproteins (IgG, IgM, IgA, Trf, Hpt, and A1AT) purified from healthy human serum were treated with exoglycosidases according to the literature procedure (23). For enzymes ABS (Arthrobacter urefaciens sialidase), BTG (Bovine testes beta-galactosidase), GUH (β-N-(1–2,3,4,6) Acetylglucosaminidase S), BKF (Bovine kidney alpha-fucosidase) and JBM (α1–2,3,6 Mannosidase J, Jack Bean): 1, 2, 2, 4, and 4 μl of each was used per digestion to give the final concentrations of 0.5, 1, 8, 3200 and 60U/ml respectively. All digestions were carried out in a final volume of 10 μl, at 50 mm NaOAc pH 5.5 for 24 h–96 h.
Computational Procedures
Statistical analysis: Variables age, menopause status, logCA125, C-reactive protein levels (CRP), protein titer and glycan peak areas between patients and controls were compared using a Tukey honest significant difference (HSD) test with ANOVA. Peak areas are compositional data (presented as a % of the total area under the graph) and therefore the constant-sum constraint (CSC) occurs. The CSC means individual variables do not vary independently, violating common assumptions on which standard statistical analyses are performed. This was avoided by performing a log transform, log(Peaki)/(100-Peaki), on all peak areas (24). To calculate the p values the data were controlled for age confounder by creating a linear regression model (ANCOVA). Correction for multiple testing was performed using Benjamini-Hochberg procedure with a 10% false positive rate.
Missing Values
There was one individual with missing age, menopause status, CA125, CRP and protein status. One individual had a missing menopause status. The small number of missing values were imputed using the K nearest neighbor approach where missing attributes were predicted. This is an acceptable procedure when the number of missing values are small (25).
Classification Model
To classify individuals into normal, borderline or metastatic neural networks without hidden layers were trained with combinations of age, menopause status, logCA125, CRP, protein and glycan peak relative abundances. A one against all binary classification was performed (e.g. normal versus borderline+metastatic). In order to test the performance of the models the data used to optimize parameters (train set) was split and a 10-fold cross validation (90% of data for training and 10% of data to test the model repeated 10 times) was performed. Discriminative power was calculated by Area under the Receiver Operating Characteristic (ROC) curve (AUC), sensitivity (SEN) and specificity (SPE).
Clustering
Principal component analysis (PCA) was performed using variables peaks areas, age, logCA125, CRP, protein titer, and menopause status. For hierarchical clustering of the glycomic profiles, GP % areas were normalized to the range [0, 1] and a hierarchy of clusters were built using the Ward algorithm (26). Spearman correlations were used to calculate the similarity among GP, age, logCA125, CRP and protein titer. Therefore, correlated variables appear close together in the hieratical dendrogram produced by the clustering.
RESULTS AND DISCUSSION
GlycoSeqCap-Automated N-glycan Analysis Platform Using Serial Capture of Individual Serum Glycoproteins
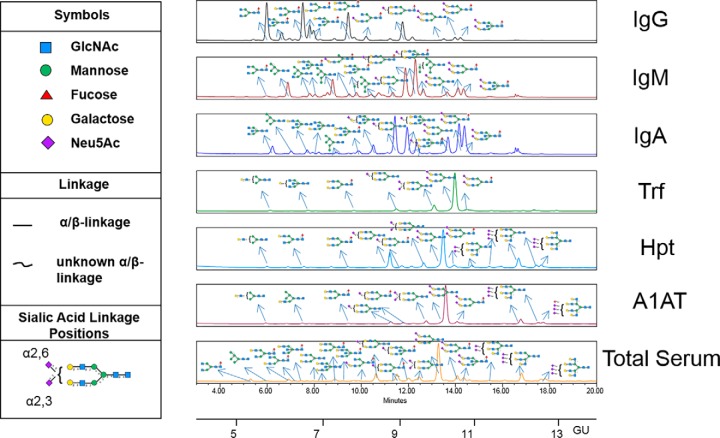
We developed a technology termed “GlycoSeqCap” involving the serial capture and subsequent N-glycoprofiling of individual glycoproteins from human serum. The multiplexed automated serial capture of six abundant serum glycoproteins (the immunoglobulins, IgG, IgM, IgA, and the acute phase proteins, transferrin (Trf), alpha-1-antitrypsin (A1AT), haptoglobin (Hpt)), and N-glycan release from pooled human serum was optimized using a 96 well format on a robotic platform (Fig. 1). Specific anti-glycoprotein capture resins (containing antibodies against specific glycoproteins) were packed in a series of PhyNexus phytips and each serum sample was passed successively through each resin in a sequential order Trf, IgG, IgM, IgA, Hpt, and A1AT. The purified proteins, >98% pure as determined by 1D-SDS page, were then subjected to PNGaseF release, ultrafiltration and aminoquinoline carbamate (AQC) fluorescent labeling of the N-glycan pools using the Hamilton Starlet robotic system using a previously published protocol (Fig. 1) (11). These pools were subjected to hydrophilic ultra-high-performance liquid chromatography (HILIC-UPLC) followed by exoglycosidase array glycan analysis according to the established procedures in our laboratory (23). The UPLC chromatograms for the fluorescently labeled released N-glycans of affinity purified glycoproteins (IgG, IgM, IgA, Trf, Hpt, and A1AT) from human serum and the corresponding released N-glycans from total human serum (21) are presented with annotations for the major N-glycans (Fig. 2).
Fig. 2.
UPLC-HILIC-FLD chromatograms of AQC labeled N-glycans released from human serum for affinity purified glycoproteins IgG, IgM and IgA, Trf, Hpt, A1AT and 2-AB labeled N-glycans released from total serum. Only major glycans are annotated. All major N-glycans identified within the total serum UPLC chromatogram are present in the individual glycoprotein chromatograms which showcases that serum glycosylation is largely dominated by the highest abundance proteins (IgG, IgM and IgA, Trf, Hpt and A1AT). SNFG glycan nomenclature is used throughout.
Advantages and Limitations of GlycoSeqCap Technology
This technological advancement provides N-glycosylation information for IgG, IgM, IgA, Trf, Hpt, and A1AT and serves as a template for many other glycoproteins of interest which may find use as a personalized medicine tool for future applications. A major advantage of the developed workflow is the ability to have multiplexed glycomics information from a single clinical source. In addition, the Phytips are reusable and show good reproducibility over three consecutive runs and after 1 month of storage. However, one limitation regarding the extension of the technology to include other glycoproteins is the requirement for an affinity resin with high selectivity and specificity for the targeted glycoprotein (e.g. antibodies or lectins) and in certain cases high amounts of biomaterial (in this case serum) may be required for lower abundant glycoproteins. In addition, the analyst must be cognizant of the purity of the targeted glycoprotein before enzymatic release of the N-glycans. For example, a recent publication cautions the users to account for the ubiquitous presence of varying levels of other contaminating plasma glycoproteins (27). In our study, we were careful to assess the purity of the glycoproteins by SDS-PAGE (Fig. 1) and systematically altered the number of capture, binding and wash steps to maximize purity before PNGase F release of N-glycans. As in the case of any affinity purification, it is possible that small traces of other glycoproteins have contributed to the N-glycan structures identified, albeit in tiny proportions.
The glycosylation data for six abundant glycoproteins in normal human serum aligns nicely with the total glycan pool identified for human serum previously (Fig. 2) (21, 28). All major glycans from human serum are identified in one/more of the individual glycoproteins that were characterized. In a large part, this identification on an individual glycoprotein level was enabled on such a small amount of serum (50 μl) because of the use of a fluorescent label AQC, which shows a twenty fold increase in fluorescent detection compared with the traditional label, 2-AB for released N-glycans which we have previously described for human IgG previously (11, 23). Various high-throughput large-scale studies have been conducted on a serum N-glycome level since 2009 (29–31). These studies have yielded significant contributions to understanding the role of glycosylation in many diseases but cannot pinpoint the exact glycosylation processing pathways involved. On an individual glycoprotein level, large scale studies have focused on IgG N-glycome only to date, (32) leaving plenty of scope for future investigations on an individual glycoprotein level. In addition, it may also be possible to link significant alterations from the serum N-glycome to a specific glycoprotein by extrapolation using the major glycans identified in this work.
Profiling and Detailed N-glycan Analysis of Serum IgG, IgA, and IgM
In order to execute glycoprofiling to compare glycosylation of two distinct populations (such as serum IgG N-glycosylation from a normal population compared with an ovarian cancer cohort) it is important to generate reproducible profiles and fully characterize the N-glycomes. To this end, we have undertaken the most comprehensive study of released N-glycans for individual glycoproteins in human serum to date. UPLC chromatograms (for each individual glycoprotein) were integrated and split into glycan peaks (GPs) and each GP typically accounts for one or more glycans.
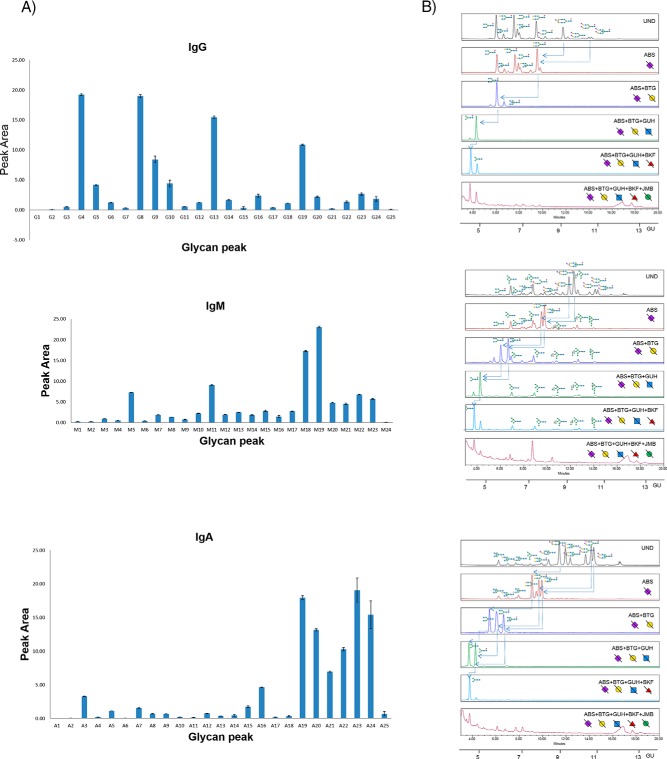
The serum IgG N-glycome UPLC chromatogram was split into 25 GPs (adapted from a literature procedure using 23 GPs11) to account for N-glycans prominent in IgG human serum from patients with ovarian cancer. The 25 IgG GP areas (G1-G25) were plotted for five technical replicates over three different days to provide standard error bars on the integrated peak areas (Fig. 3A, supplemental Table S7). Most of the GPs (21/25) are strongly reproducible for the technical replicates with coefficient of variance (CV) values for each GP below 25% with the exceptions of GPs G1, G15 and G25. These GPs cumulatively account for a small amount (0.35%) of the total peak area of IgG N-glycome in the technical replicates. Characterization proceeded as follows: the preliminary structures, assigned from glucose unit (GU) values (obtained by matching elution positions to a standard dextran hydrolysate curve and using Glycobase software (data now migrated to Glycostore (https://www.glycostore.org/) (33)), were confirmed by exoglycosidase array digestions (Fig. 3B) and intact mass using electrospray (ESI-LC) liquid chromatography mass spectrometry (supplemental Table S1). They agree with literature (11, 32) (with the addition of a high mannose species (M7) not previously identified) and are presented in supplemental Table S6.
Fig. 3.
Serial capture and N-glycoprofiling of human serum IgG, IgM and IgA, purified from human serum. The 25 IgG glycan peak areas (G1-G25), 24 IgM peak areas (M1–24) and 25 IgA glycan peak areas (A1-A25) plotted for five technical replicates over three different days (3A). The standard error shown as error bars. Sequencing of AQC labeled IgG, IgM and IgA N-glycans visualized by UPLC-HILIC chromatograms using exoglycosidase enzymes with glucose units (GU) to facilitate glycan identification (3B). Digestion of AQC labeled IgG, IgM and IgA N-glycans with addition of sialidase (ABS), galactosidase (BTG), hexosaminidase (GUH), fucosidase (BKF) and mannosidase (JBM) in the following order ABS, ABS+BTG, ABS+BTG+GUH, ABS+BTG+GUH+BKF and ABS+BTG+GUH+BKF+JBM. For IgG, arrows indicate the cleavage of sugar residues for selected peaks: major glycans FA2G2S1 and FA2G2S2. For IgM, arrows indicate the cleavage of sugar residues for selected peaks: major glycans FA2G2S1 and FA2BG2S1. For IgA, arrows indicate the cleavage of sugar residues for selected peaks: major glycans A2G2S1, FA2G2S2 and FA2BG2S2. SNFG nomenclature is used for glycan representation.
The equivalent data for serum IgM and IgA N-glycomes were split into 24 (M1-M24) and 25 (A1-A25) GPs respectively and are shown in Fig. 3 (reproducibility data in supplemental Table S7). For IgM, all GPs (M1-M24) show good reproducibility for the technical replicates (below 25%). Similarly, IgA shows good reproducibility for all major GPs (A2-A25, below 25%) but exhibits slightly higher CVs for the first and last GPs (A1, A25) accounting for 0.92% of the total IgG N-glycome. Structural assignments of IgM and IgA N-glycans were characterized in a similar manner to serum IgG using GU values and LC-MS (supplemental Table S2 and S3) and are presented in supplemental Table S6. For serum IgM, structural assignment of N-glycans were in agreement with the previously reported literature values (15) with some additional glycans identified in our analysis (13 newly characterized compositions out of a total of 53) including some of the M5A1 Series (M5A1, M5A1G1 and M5A1G1S(6)1), FA1 Series (FA1, FA1G1 and FA1G1S1), A2 Series (A2G1S(6)1 and A2G2S(6,6)2) and the A2B Series (A2BG1S(6)1). For IgA, structural assignment of N-glycans agreed with the literature reports previously described (16, 17, 34). As in the case of IgM, additional glycans were identified (16/55) in our in-depth analysis not previously described in the literature including some monoantennary and biantennary species (A1, A2[6]G1, A2[3]G1, FA2), hybrid species (M4A1, M4A1G1, M4A1G1S(6)1, M4A1BG1, M5A1G1, M5A1G1S(6)1), A3 Series (A3, A3G2, A3G2S(6)1, A3G3S2, FA3) and M10.
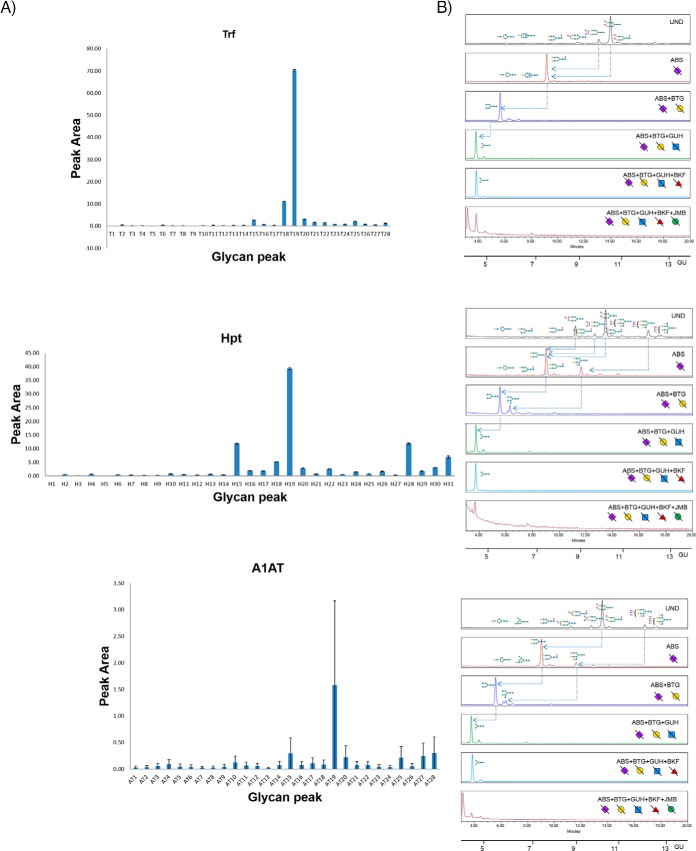
Profiling and Detailed N-glycan Analysis of Serum Trf, Hpt, and A1AT
The acute phase proteins, Trf, Hpt, and A1AT were purified from human serum by serial extraction as described earlier and the released N-glycans labeled with AQC analyzed by UPLC-HILIC. The 28 Trf (T1-T28), 31 Hpt (H1-H31), and 28 A1AT (AT1-AT28) glycan peak areas were plotted for at least five technical replicates over three different days to provide the standard error bars (Fig. 4A, 4B and reproducibility data in supplemental Table S7). All GPs in Trf, Hpt show good reproducibility (CV below 25%) except GPs T1 and T5 (total glycan peak area of 0.03%) in Trf and H5 (glycan peak area of 0.02%) in Hpt, accounting for very minor constituents of the total N-glycome in the technical replicates. The reproducibility of A1AT N-glycome is the lowest (being the last glycoprotein to be purified in the sequence) with only 19/28 glycan peaks exhibiting good reproducible CV values below 25%. The remainder (AT1, AT3–5, AT8–10, AT12, AT14) accounts for a total of 2.67% of the total A1AT N-glycome, a relatively small proportion of the total N-glycome.
Fig. 4.
Serial capture and N-glycoprofiling of human serum Trf, Hpt and A1AT, purified from human serum. The 28 Trf glycan peak areas (T1-T28), 31 Hpt glycan peak areas (H1-H31) and 28 A1AT glycan peak areas (AT1-AT28) were plotted for five technical replicates over three different days (4A). The standard error shown as error bars. Sequencing of AQC labeled Trf, Hpt and A1AT N-glycans visualized by UPLC-HILIC chromatograms using exoglycosidase enzymes with glucose units (GU) to facilitate glycan identification (4B). Digestion of AQC labeled Trf, Hpt and A1AT N-glycans with addition of sialidase (ABS), galactosidase (BTG), hexosaminidase (GUH), fucosidase (BKF) and mannosidase (JBM) in the following order ABS, ABS+BTG, ABS+BTG+GUH, ABS+BTG+GUH+BKF and ABS+BTG+GUH+BKF+JBM. In Trf, arrows indicate the cleavage of sugar residues for selected peaks: major glycans A2G2S(3,6)2 and A2G2S2(6,6)2. In Hpt, arrows indicate the cleavage of sugar residues for selected peaks: major glycans A3G3S3, A2G2S2 and A2G2S1. In A1AT, arrows indicate the cleavage of sugar residues for selected peaks: major glycans A3G3S3 and A2G2S2.SNFG nomenclature is used for glycan representation.
The AQC labeled Trf, Hpt and A1AT (Fig. 4) N-glycan pools were sequenced using exoglycosidase arrays and the data combined with glucose unit (GU) values and LC-MS to facilitate glycan identification. As for the immunoglobulins, many newly identified N-glycans are presented as well as agreement with the major glycans already presented in the literature for Trf (8),Hpt (8, 35), and A1AT (8, 36). The list of newly identified glycans are highlighted with a Δ symbol in supplemental Tables S6. For Trf, 28 out of 48 glycan compositions are identified for the first time in this publication. Most significantly, we identify the FA3G3 series not identified for Trf previously which accounts for almost 4% of the total N-glycome. Similarly, for Hpt and A1AT, 32/47 and 28/44 glycan compositions are identified respectively for the first time. For Hpt, the FA3 series is significant accounting for approx. 3% of the total N-glycome and the newly identified glycan A3F1G3S3 accounts for approx. 2% of the peak area.
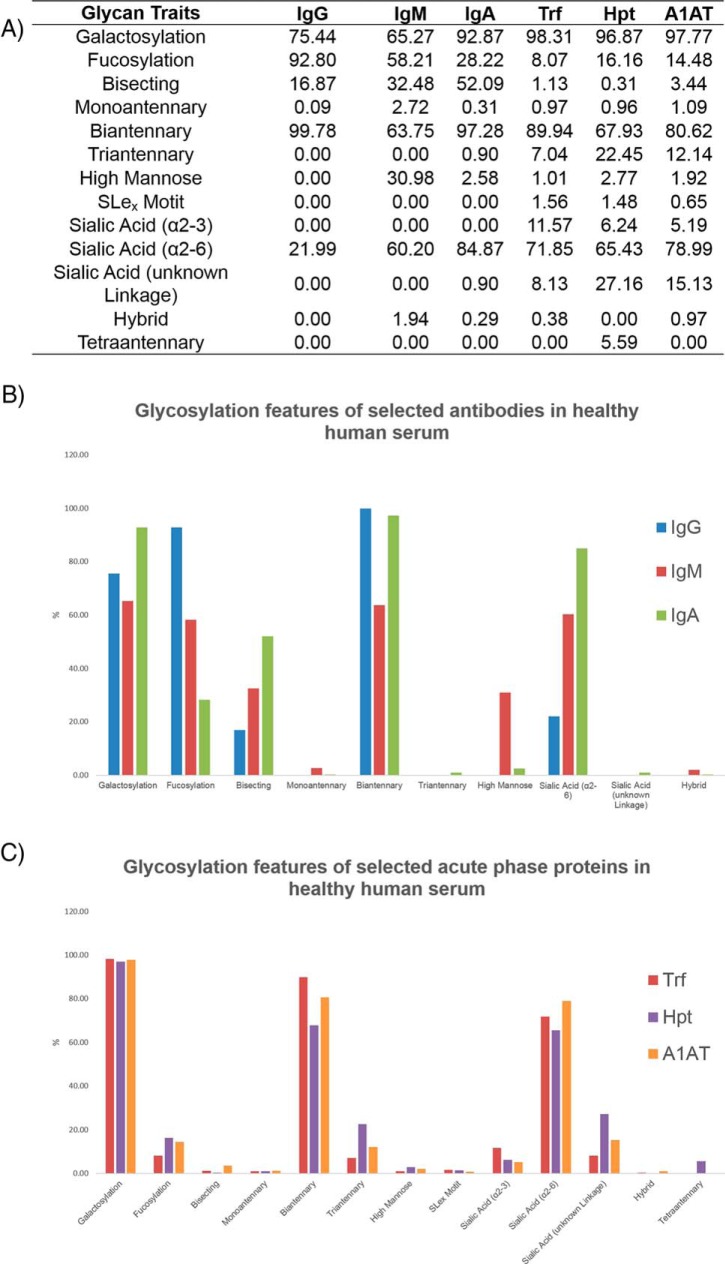
Comparison of Glycosylation Between Antibody Classes: IgG, IgM, and IgA
We probed the glycosylation of selected antibodies to gain an insight into the structure-function relationship of antibody classes. One important point to consider is that serum glycoproteins are a combination of active components and waste products and this may complicate interpretations. Structural similarities and differences in antibody glycosylation of IgG, IgM and IgA are presented for pooled normal human serum (Fig. 5A, 5B). The derived glycosylation traits were generated from UPLC data (supplemental Tables S6) and calculated from the summation of individual GPs for each glycoprotein (supplemental Table S8). Striking variation in glycosylation traits such as galactosylation, fucosylation, sialylation, bisecting GlcNAcs (glycans with bisecting N-acetylglucosamine residues), high mannose, hybrid or the degree of branching (monoantennary, biantennary, triantennary) can be observed. Overall, all antibodies exhibit a high proportion of biantennary structures (>60%) with little/no monoantennary/triantennary structures, as documented previously in the literature (15, 32, 34). Additionally, a high degree of galactosylation is observed for all three glycoproteins (>60%). Only α2,6-linked sialic acids (containing the Neu5Ac form) and no corresponding α2,3-linked sialic acids are identified for IgG or IgM which is consistent with literature reports (15, 32). Additionally, only minor species contained α2,3-linked sialic acids in IgA, as identified previously (17). Increased sialylation was observed from IgG to IgM to IgA (22%, 60%, and 85% respectively). IgG contains the highest proportion of core fucosylation (93%) relative to IgM or IgA (58 and 28% respectively) and no outer arm fucose were identified for the serum antibody series. This observation is consistent with other reports for serum antibodies (15, 17, 32). IgA contains the highest abundances of bisecting GlcNAcs (52%) and galactose residues (93%) and a very small proportion of triantennary structures (0.9%) not observed for the other antibodies. IgM exhibits the lowest amount of biantennary structures (64%) and galactosylation (65%) but significantly higher amounts of high mannose structures (31%).
Fig. 5.
Glycosylation traits for selected antibodies (IgG, IgM, IgA) and acute phase proteins (Trf, Hpt, A1AT) for healthy human serum are presented (5A). The values are represented as a % of the total glycosylation and are calculated from data provided in supplemental Table S15. Glycosylation features of antibodies (IgG, IgM and IgA) and acute phase proteins (Trf, Hpt, A1AT) are presented as control charts (5B and 5C).
Acute Phase Protein Glycosylation Traits: Trf, Hpt, and A1AT
Unlike the selected antibodies which share structural similarities in terms of their protein structure, the selected acute phase proteins (Trf, Hpt and A1AT) have less commonality and exhibit more varied glycosylation traits. These structural similarities and differences are presented for pooled normal human serum (Fig. 5A, 5C). As for the antibodies, a high degree of galactosylation was observed (95%) for selected acute phase proteins but relatively low amounts of total fucosylation (<20%) were identified, accounting for a combination of core fucose and the outer arm fucose residues (not identified in the selected serum antibodies in this study). Sialyl Lewisx (SLex) motifs (Neu5Acα2–3Galβ1–4[Fucα1–3]GlcNAcβ), which contain outer arm fucose residues, are identified in human serum acute phase proteins, albeit in small proportions. Notably, as in the case for the antibody glycosylation, α2,6-linked sialic acids (65%) are dominant over the corresponding α2,3-linked sialic acids (<10%) of the annotated sialic acids. In previous studies of acute phase proteins, many sialic acid linkage types were left ambiguous (8, 35), as such we provide the most extensive breakdown to date for acute phase protein N-glycosylation. A larger proportion of higher branched structures are observed such as triantennary structures (10%) and the presence of tetraantennary glycans are observed. Hpt exhibits the highest relative amount of both triantennary (22%) and tetraantennary (6%) structures compared with Trf or A1AT.
Glycosylation Traits for Functional Studies
The quantification of glycosylation traits for a series of glycoproteins is presented here for the first time from a single source of human serum (Fig. 5). One can envisage this information as being a starting point for functional studies to investigate the specific role of glycosylation in our immune system and has the added advantage over traditional approaches in glycoanalytics to explore the effect of more than one glycoprotein in a single experiment. For example, little is known about how the most abundant antibody isotypes in human serum (IgG, IgM, and IgA) compete for antigens but using quantified glycan traits, combined with antibody-antigen assays could provide a clearer picture for the role of glycosylation in this context. From our glycosylation traits alone, it is impossible to predict biological function, but it may provide a basis for further investigations. For example, taking into consideration functional examples of afucosylation of IgG1 increasing antibody-dependent cellular cytotoxicity (ADCC) (37, 38), and looking at levels of antibody fucosylation in human serum, the striking difference between IgG (93%) and IgM or IgA core fucosylation (58 and 28% respectively) may hint that serum IgA recruit effector cells for ADCC more efficiently compared with IgG or IgM. Importantly, it must be considered that we have measured only in human serum samples and these data does not necessarily reflect on a tissue/cell specific antibody glycosylation level. In addition to core fucosylation, we reported no outer arm fucose for the antibody classes. Noteworthy, outer arm fucosylation has been reported for secretory component (SC) N-glycans of IgA, but none for the corresponding J or H chain N-glycans (34). It is known that serum IgA is predominantly monomeric and does not contain the J chain and the SC and as such we report no conflicting findings in this study. Another interesting differentiation between the glycosylation traits of the antibody isotypes is the high abundance of sialylation for IgA (85%) relative to IgM (60%) and IgG (22%). We propose that serum IgA uses electrostatic interactions and steric obstruction/adhesion as the main mechanistic mode for fighting infection but it is not clear if this sialylation is employed for antigen binding/effector function purposes. A more in-depth analysis for Fc and Fab N-glycosylation of serum IgA would shed more light in this regard. Lastly for the antibody class, we reported the highest proportion of high mannose structures (31%) for serum IgM. This proportion of high mannose was higher than the value of 23% previously reported for serum IgM and the corresponding amount of galactosylation was lower (65%) compared with the literature value of 84% (15). We suggest that advances in modern technology have allowed for a more accurate and precise assignment of N-glycans in our study- however it cannot be ignored that the human serum populations used in the two studies may have been different.
In contrast with immunoglobulins, which are mainly produced by B-cells, major plasma glycoproteins including Trf, Hpt and A1AT originate from hepatocytes, which express only very low levels of the FUT8 fucosyltransferase and thus contain a low percentage of core fucosylated glycans (32). As anticipated, we observed lower levels of core fucose for the acute phase proteins compared with the antibody classes in our study (Fig. 5). Another interesting point relates to the increased complexity of the acute phase protein N-glycosylation compared with immunoglobulin classes (Fig. 5), with tetraantennary glycans present in these glycoproteins not isolated from the antibody classes. We cannot find a biological rationale for the reasons as to why the N-glycans are more branched in these acute phase proteins, but it may be related to their inflammatory properties. Also, these glycoproteins contain SLex motifs. SLex are known to play a vital role in cell-cell recognition and other processes and are known to be overexpressed in cancer cells (39). As such, this feature is particularly important in the context of our ovarian cancer cohort.
Glycoprofiling and Statistical Significance of Selected Glycoproteins in Ovarian Cancer
Having comprehensively assigned N-glycans and calculated derived glycosylation traits for the selected glycoproteins IgG, IgM, IgA, Trf, Hpt, and A1AT in normal human serum, we glycoprofiled a cohort of patients with ovarian cancer to exploit the quantifiable N-glycosylation alterations for (1) detection of ovarian cancer and (2) to differentiate between stage of the disease in an effort to provide a clinical tool for early diagnosis in the future.
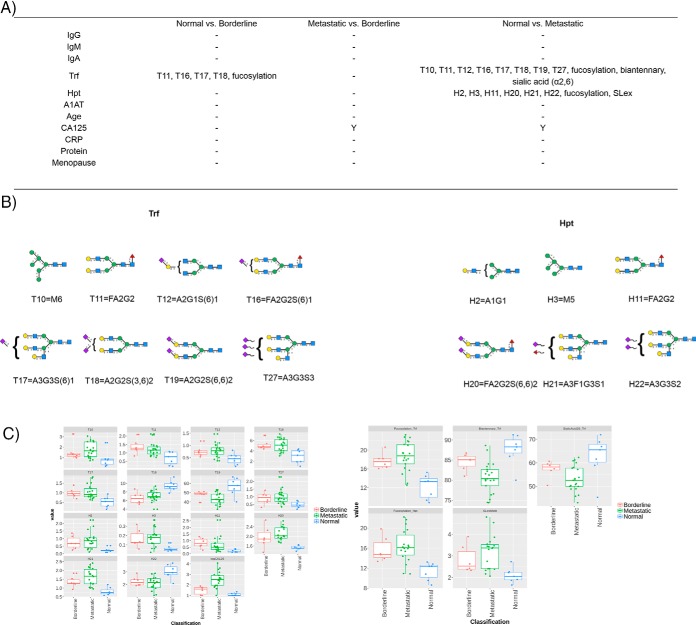
The N-glycome of selected proteins (IgG, IgM, IgA, Trf, Hpt, and A1AT) from 7 normal controls and 27 ovarian cancer patients (either classified as borderline or metastatic presented in biological manifest in supplemental Table S9) as a test group were analyzed resulting in a total of 204 processed glycoprofiles. The profiles were split into glycan peaks (GPs) according to the individual glycoproteins (e.g. G1–G25 for IgG) and the values are presented as a proportion of total % peak area in supplemental Tables S10–S15 for glycoproteins IgG, IgM, IgA, Trf, Hpt, and A1AT respectively. The derived glycosylation traits were also calculated and were derived as described earlier (supplemental Table S8). Controlling for age, the statistical significance (Tukey honest significant difference (HSD) with ANOVA, normally distributed (data not shown)) was measured for all GPs and glycosylation traits for each selected glycoprotein and are presented in supplemental Tables S16–S21, as well as for the clinical variables (supplemental Table S22) between normal versus borderline, metastatic versus borderline, normal versus metastatic clinical samples. The p values were corrected for multiple testing error, using a 5% false discovery rate (FDR) approach proposed by Benjamini-Hochberg (40). An adjusted p < 0.05 was considered statistically significant. Significant GPs, glycosylation traits and clinical parameters for the patients are presented in Fig. 6 for the glycoprotein series. Boxplots (Fig. 6C) and the major glycan (Fig. 6B) for the statistically significant GPs/glycosylation traits are also presented.
Fig. 6.
Statistically significant GPs and glycosylation traits for each glycoproteins and clinical parameters (Y = yes) with p values <0. 05 (corrected for multiple testing error using a 5% FDR proposed by Benjamini-Hochberg (40)) for ovarian cancer cohort of normal versus borderline, metastatic versus borderline and normal versus metastatic (6A). The major glycan for the each statistically significant GP is presented (6B). Boxplots are presented for the statistically significant GPs, glycosylation traits and logCA125 for borderline (red), metastatic (green) and normal (blue) samples (6C).
From the glycoproteins selected, Trf shows the best discrimination between normal patients (n = 7) and borderline (n = 6) or metastatic patients (n = 21). Most notably, Trf glycosylation can distinguish between normal and borderline samples using four individual GPs (T11, T16, T17 and T18), whereby CA125, the gold standard for ovarian cancer detection does not show the same statistical significant separation. Fucosylation also stands out as a feature that may be exploited for this discrimination. The GPs/glycosylation features of Trf cannot differentiate between metastatic and borderline samples in this cohort however, whereas CA125 does show a significance.
Several studies have presented evidence implicating Trf in ovarian cancer biology (41). In addition, alterations in Trf glycosylation in the context of cancer and other inflammatory diseases have been reviewed in the literature (36, 42). Taken together, Trf sialylation is often altered in disease states and is no exception in this study, whereby sialic acid (α2,6) is altered in the metastatic cancer cohort compared with normal controls. Because desialylated Trf has faster clearance, it may be an evolutionary tactic of bacteria/pathogens to contribute to oncogenesis. What is more striking in our study is the alterations of Trf fucosylation. Trf fucosylation has been shown to be dysregulated in diseases such as classical galactosemia (43) but to the best of our knowledge has not been reported in the context of ovarian cancer to date. Future studies are warranted to investigate further.
In addition, haptoglobin glycosylation is significantly altered in metastatic cancer (n = 18) compared with normal patients (n = 7) in this cohort by six distinct GPs (H2, H3, H11, H20, H21, and H22) as well as glycosylation features fucosylation and SLex motif. These data is consistent with literature reports (44), whereby the main glycosylation alterations of Hpt in cancer appear to be the presence of aberrantly fucosylated and sialylated structures as well as increased branching.
Discrimination and Cluster Analysis in Ovarian Cancer
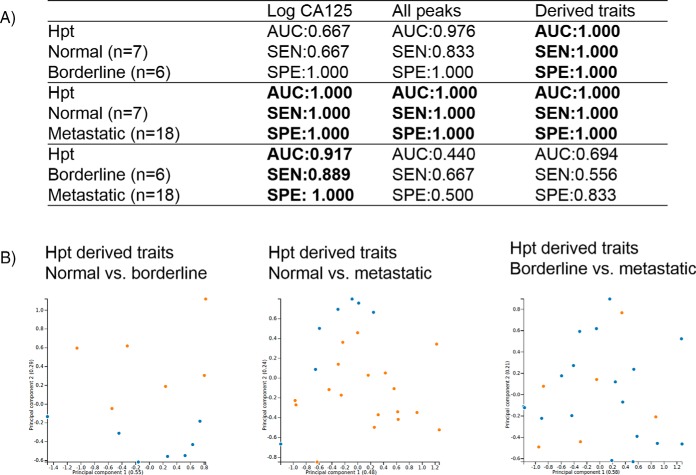
A discrimination analysis was explored for the probabilistic classification of healthy (normal) versus ovarian cancer samples using the glycosylation data and comparing to CA125 values for the clinical cohort. CA125 antigen is a high molecular weight glycoprotein, which is expressed by a large proportion of epithelial ovarian cancers and is currently regarded as the golden standard for ovarian cancer diagnosis, despite its poor sensitivity and specificity. It is only raised in ∼50% of stage 1 epithelial ovarian cancers and in 75–90% of patients with advanced disease and false positive results have been noted in many medical disorders, both malignant and benign (45). The area under the ROC curve (AUC), sensitivity (SEN) and specificity (SPE) were calculated and associated with normal versus borderline, metastatic versus borderline and normal versus metastatic clinical samples (supplemental Table S23, S24, and S25 respectively). The individual models performed well with regards to distinguishing normal from borderline patients, with the most promising result using derived traits from Hpt (AUC = 1.000, SEN = 1.000 and SPE = 1.000) (Fig. 7A). This finding, if validated could have major ramifications for early detection of ovarian cancer using Hpt glycosylation. Similarly, good discrimination was observed for normal versus metastatic patients. Again, Hpt showed perfect discrimination for all peaks, derived traits and combinations thereof. However, CA125 also allows perfect discrimination so this negates the glycome data in this instance. No glycosylation feature could discriminate between borderline and metastatic patients and superior results were observed for CA125 in this case.
Fig. 7.
Discrimination performance for noteworthy glycosylation traits (Hpt) for ovarian cancer cohort of normal versus borderline, normal versus metastatic and borderline versus metastatic. Linear regression model including AUC, SEN and SPE (7A), cluster analysis using PCA using Hpt derived traits as input for normal (blue, n = 7) versus borderline (orange, n = 6) separation, normal (blue, n = 7) versus metastatic (orange, n = 18) and borderline (orange, n = 6) versus metastatic blue, n = 18) respectively the following (7B) to highlight part B. In brackets the variance of the principal component.
Cluster analysis was performed using principal component analysis (PCA) and hierarchical clustering. PCA results are provided in supplemental Figs. S7–S12, and hierarchical clustering is presented in supplemental Figs. S13–S18 for the respective glycoproteins IgG, IgM, IgA, Trf, Hpt, and A1AT for individual glycan peaks. No clear discrimination was observed between normal, borderline or metastatic samples for PCA analysis on individual glycan peaks. For hierarchical clustering, no major clustering was observed for glycoproteins IgG, IgM, or A1AT but clustering was observed for IgA, Trf, and Hpt with clear clustering of metastatic and normal samples respectively with the most pronounced affect for Hpt. Taking glycosylation traits into consideration, Hpt again outperforms the other glycoproteins and PCA plots are shown in Fig. 7B showing a clear separation for normal and borderline samples and normal and metastatic samples but no separation between borderline versus metastatic samples, consistent with the AUC, SEN, and SPE observed for this dataset.
Glycoanalytical Diagnostic Tools for Ovarian Cancer
The availability of diagnostic tools for ovarian cancer remains somewhat elusive. CA125 is currently the best diagnostic tool for ovarian cancer on the market but is not reliable for diagnosing early stage ovarian cancer (46, 47). Early stage detection and subsequent treatment of ovarian cancer is an attractive approach to reduce morbidity from ovarian cancer. In this study, we corroborate these literature findings and show that CA125 cannot differentiate between borderline (n = 6) and normal (n = 7) samples (Fig. 6). For IgG, IgA, IgM, Hpt, and A1AT, again we cannot see any significant alterations between these classes but find instead that Trf glycosylation can discriminate using GPs T11, T16, T17, and T18 (Fig. 6) in our small clinical cohort. If these findings could be replicated and validated, Trf glycosylation could be exploited as an early detection tool for ovarian cancer. To probe the potential role of Trf glycosylation in ovarian cancer on a very basic level, we measured the up-regulation/down-regulation of the relative abundance of N-glycans for the statistically significant GPs (Fig. 6B) and hypothesize that the dominant glycosylation features may be significant in the progression of the disease. Two of the major glycans that are up-regulated in the borderline samples (n = 6) contain core fucose- FA2G2 (T11) and FA2G2S(6)1 (T16) whereas there is a corresponding decrease in afucosylated structure A2G2S(3,6)2 (T18). Taking the classification and clustering data into consideration Trf glycosylation does not provide superior differentiation between classes compared with the other glycoproteins- no clear PCA differentiation is observed (supplemental Fig. S10) but the hierarchical clustering does show its promise for resolving power between the groups (supplemental Fig. S16).
With respect to clustering and classification analysis, the best performer is Hpt glycosylation (Fig. 7) which displays clear groupings in the PCA analysis. In addition, regarding the statistical findings for Hpt glycosylation, there is a notable increase in two core fucosylated species FA2G2 (H11) and FA2G2S(6,6)2 (H20) in metastatic cancer patients (n = 18) compared with normal controls (n = 7) and a decrease in the afucosylated complex glycan A3G3S2 (H22). Taken collectively, these data suggests that core fucosylation (the FA2 Series) is significantly up-regulated in this ovarian cancer cohort in both Trf and Hpt. Importantly, FA2 was previously found to be significantly altered (increased) in sera of ovarian cancer patients using an independent N-glycoanalytical technology (46). We propose these changes reflect either differences in the expression levels of the α(1–6)-fucosyltransferase (FUT8) or donor substrate (GDP-fucose) in the medial-Golgi in ovarian cancer specific cells.
As discussed above, Trf glycosylation is the most significantly altered in our study and Hpt shows the greatest promise with respect to cluster analyses from the selected acute phase proteins and antibodies in the discrimination of ovarian cancer stages. Both glycoproteins warrant further functional studies in the future. Remarkably, very few investigations are described in the literature with respect to the possible role of Trf in ovarian cancer, despite its use as part of a commercial product, a multivariate index assay called OVA1 (approved by the FDA in 2016) which incorporates five serum biomarkers into a malignancy risk score of 0–10 using a proprietary algorithm and is recommended by the American College of Obstetricians and Gynecologists (48) as a tool to aid in evaluating women with adnexal masses. There is precedence of altered expression of Hpt glycosylation in ovarian cancer whereby Turner and colleagues found enhanced expression of branching, especially triantennary glycans in patients with ovarian cancer (49). As such Trf and Hpt present as interesting targets for ovarian cancer treatment. Regarding the remaining glycoproteins, IgG, IgM and IgA glycosylation were assessed previously in the context of ovarian cancer from late stage patients only-the results indicated that combining IgG glycosylation profiles with CA125 could improve accuracy of epithelial ovarian cancer prediction (18), in contrast to our findings. In a separate study, IgG galactosylation was used to assist a differential diagnosis of ovarian cancer with CA125 (50). A very early study investigated A1AT glycosylation in the context of ovarian cancer (51). The authors concluded that fucosylation was a glycosylation feature that became elevated in the presence of tumor growth but remained low in remission and during chemotherapy. This study does not reflect the utility of A1AT fucosylation in the selected cohort of patients.
CONCLUSION
This study presents the development of an elegant glycoanalytical platform for the detailed characterization and investigation of a sequence of glycoprotein N-glycosylation: antibodies IgG, IgM and IgA and acute phase proteins Trf, Hpt and A1AT. In the course of this analysis, besides achieving a detailed glycoprofile for each glycoprotein through the identification and profiling of more than a hundred glycan structures, we were able to identify novel glycan motifs and traits that have not been outlined to date. Its utility in the context of ovarian cancer is highlighted-Trf and Hpt glycosylation can be exploited as biomarker tools for ovarian cancer and may play a cardinal role. More importantly, this comprehensive and reproducible glycoprofiling technology could provide significant insights and serve as a baseline for the identification of biomarkers and their regulation in a series of diseases.
Data Availability
The mass spectrometry data in this study are freely available through the repository MassIVE (https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=a66ade995ac8431e80f6e27f11c55674) with the dataset identifier MSV000083877.
Supplementary Material
Acknowledgments
We thank the Leeds Multidisciplinary Research Tissue Bank for samples. We acknowledge Thermo Scientific for the donation of anti-haptoglobin capture resins.
Footnotes
* This work was supported by the EU FP7 programs High Glycan and GlycoBioM (278535, 259869). R.S. acknowledges funding from the Science Foundation Ireland Starting Investigator Research grant (SFI SIRG) under Grant Number 13/SIRG/2164. I.W. acknowledges funding from HighGlycoART: under Grant Number 1335G00086.
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- PTMs
- post-translational modifications
- IgG
- immunoglobulin G
- Trf
- transferrin
- A1AT
- alpha1antitrypsin
- Hpt
- haptoglobin
- IEF
- isoelectric focusing
- IgM
- immunoglobulin M
- IgA
- immunoglobulin A
- RA
- rheumatoid arthritis
- NP
- normal phase
- CSC
- constant-sum constraint.
REFERENCES
- 1. Taniguchi N. (2006) From glycobiology to systems glycobiology: international network with Japanese scientists through consortia. IUBMB Life 58, 269–272 [DOI] [PubMed] [Google Scholar]
- 2. Butler M., Quelhas D., Critchley A. J., Carchon H., Hebestreit H. F., Hibbert R. G., Vilarinho L., Teles E., Matthijs G., Schollen E., Argibay P., Harvey D. J., Dwek R. A., Jaeken J., and Rudd P. M. (2003) Detailed glycan analysis of serum glycoproteins of patients with congenital disorders of glycosylation indicates the specific defective glycan processing step and provides an insight into pathogenesis. Glycobiology 13, 601–622 [DOI] [PubMed] [Google Scholar]
- 3. Grunewald S., Matthijs G., and Jaeken J. (2002) Congenital disorders of glycosylation: a review. Pediatr Res 52, 618–624 [DOI] [PubMed] [Google Scholar]
- 4. Thanabalasingham G. Huffman J. E., Kattla J. J., Novokmet M., Rudan I., Gloyn A. L., Hayward C., Adamczyk B., Reynolds R. M., Muzinic A., Hassanali N., Pucic M., Bennett A. J., Essafi A., Polasek O., Mughal S. A., Redzic I., Primorac D., Zgaga L., Kolcic I., Hansen T., Gasperikova D., Tjora E., Strachan M. W., Nielsen T., Stanik J., Klimes I., Pedersen O. B., Njølstad P. R., Wild S. H., Gyllensten U., Gornik O., Wilson J. F., Hastie N. D., Campbell H., McCarthy M. I., Rudd P. M., Owen K. R., Lauc G., and Wright A. F. (2013) Mutations in HNF1A result in marked alterations of plasma glycan profile. Diabetes 62, 1329–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Colhoun H. O. Treacy E. P., MacMahon M., Rudd P. M., Fitzgibbon M., O'Flaherty R., Stepien K. M. (2018) Validation of an automated UPLCIgG N-glycan analytical method applicable to Classical Galactosaemia. Ann. Clin. Biochem. 55, 593–603 [DOI] [PubMed] [Google Scholar]
- 6. Freeze H. H., Eklund E. A., Ng B. G., and Patterson M. C. (2015) Neurological aspects of human glycosylation disorders. Annu. Rev. Neurosci. 38, 105–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Saldova R., Struwe W. B., Wynne K., Elia G., Duffy M. J., and Rudd P. M. (2013) Exploring the glycosylation of serum CA125. Int JMol Sci 14, 15636–15654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sarrats A., Saldova R., Pla E., Fort E., Harvey D. J., Struwe W. B., de Llorens R., Rudd P. M., and Peracaula R. (2010) Glycosylation of liver acute-phase proteins in pancreatic cancer and chronic pancreatitis. Proteomics Clin Appl 4, 432–448 [DOI] [PubMed] [Google Scholar]
- 9. Pinho S. S., and Reis C. A. (2015) Glycosylation in cancer: mechanisms and clinical implications. Nat. Rev. Cancer 15, 540–555 [DOI] [PubMed] [Google Scholar]
- 10. Marino K., Saldova R., Adamczyk B., and Rudd P. M. (2011) Changes in serum N-glycosylation profiles: functional significance and potential for diagnostics. Spr. Carb. Ch. 37, 57–93 [Google Scholar]
- 11. Stockmann H., Duke R. M., Millan Martin S., and Rudd P. M. (2015) Ultrahigh throughput, ultrafiltration-based n-glycomics platform for ultraperformance liquid chromatography (ULTRA(3)). Anal Chem 87, 8316–8322 [DOI] [PubMed] [Google Scholar]
- 12. O'Flaherty R., Trbojevic-Akmacic I., Greville G., Rudd P. M., and Lauc G. (2018) The sweet spot for biologics: recent advances in characterization of biotherapeutic glycoproteins. Expert Rev. Proteomics 15, 13–29 [DOI] [PubMed] [Google Scholar]
- 13. Lauc G., Pezer M., Rudan I., and Campbell H. (2016) Mechanisms of disease: The human N-glycome. Bba-Gen Subjects 1860, 1574–1582 [DOI] [PubMed] [Google Scholar]
- 14. McCarthy C., Saldova R., O'Brien M. E., Bergin D. A., Carroll T. P., Keenan J., Meleady P., Henry M., Clynes M., Rudd P. M., Reeves E. P., and McElvaney N. G. (2014) Increased outer arm and core fucose residues on the N-glycans of mutated alpha-1 antitrypsin protein from alpha-1 antitrypsin deficient individuals. JProteome Res 13, 596–605 [DOI] [PubMed] [Google Scholar]
- 15. Arnold J. N., Wormald M. R., Suter D. M., Radcliffe C. M., Harvey D. J., Dwek R. A., Rudd P. M., and Sim R. B. (2005) Human serum IgM glycosylation: identification of glycoforms that can bind to mannan-binding lectin. Biol, J., Chem. 280, 29080–29087 [DOI] [PubMed] [Google Scholar]
- 16. Mattu T. S., Pleass R. J., Willis A. C., Kilian M., Wormald M. R., Lellouch A. C., Rudd P. M., Woof J. M., and Dwek R. A. (1998) The glycosylation and structure of human serum IgA1, Fab and Fc regions and the role of N-glycosylation on Fcalpha receptor interactions. Biol, J., Chem. 273, 2260–2272 [DOI] [PubMed] [Google Scholar]
- 17. Bondt A., Nicolardi S., Jansen B. C., Stavenhagen K., Blank D., Kammeijer G. S., Kozak R. P., Fernandes D. L., Hensbergen P. J., Hazes J. M., van der Burgt Y. E., Dolhain R. J., and Wuhrer M. (2016) Longitudinal monitoring of immunoglobulin A glycosylation during pregnancy by simultaneous MALDI-FTICR-MS analysis of N- and O-glycopeptides. Sci. Rep. 6, 27955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ruhaak L. R., Kim K., Stroble C., Taylor S. L., Hong Q., Miyamoto S., Lebrilla C. B., and Leiserowitz G. (2016) Protein-specific differential glycosylation of immunoglobulins in serum of ovarian cancer patients. JProteome Res 15, 1002–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhu R., Zacharias L., Wooding K. M., Peng W., and Mechref Y. (2017) Glycoprotein enrichment analytical techniques: advantages and disadvantages. Methods Enzymol. 585, 397–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stockmann H., Adamczyk B., Hayes J., and Rudd P. M. (2013) Automated, high-throughput IgG-antibody glycoprofiling platform. Anal. Chem. 85, 8841–8849 [DOI] [PubMed] [Google Scholar]
- 21. Stockmann H., O'Flaherty R., Adamczyk B., Saldova R., and Rudd P. M. (2015) Automated, high-throughput serum glycoprofiling platform. Integr. Biol. 7, 1026–1032 [DOI] [PubMed] [Google Scholar]
- 22. Stockmann H., Duke R. M., Millan Martin S., and Rudd P. M. (2015) Ultrahigh throughput, ultrafiltratoin-based n-glycomics platform for ultraperformance liquid chromatography (ULTRA(3)). Anal. Chem. 18, 8316–8322 [DOI] [PubMed] [Google Scholar]
- 23. O'Flaherty R. Harbison A. M., Hanley P. J., Taron C. H., Fadda E., and Rudd P. M. (2017) Aminoquinoline fluorescent labels obstruct efficient removal of N-glycan core alpha(1–6) fucose by bovine kidney alpha-l-fucosidase (BKF). J. Proteome Res. 16, 4237–4243 [DOI] [PubMed] [Google Scholar]
- 24. Aitchison J. (1986) The statistical analysis of compositional data. Chapman & Hall, Ltd., University of Hong Kong, China [Google Scholar]
- 25. Crookston N. L., and Finley A. O. (2008) yaImpute: An R package for kNN imputation. J. Stat. Softw 23, 1–16 [Google Scholar]
- 26. Blashfield R. K. (1976) Mixture model tests of cluster-analysis - accuracy of 4 agglomerative hierarchical methods. Psychol. Bull. 83, 377–388 [Google Scholar]
- 27. Lauc G., Vuckovic F., Bondt A., Pezer M., and Wuhrer M. (2018) Trace N-glycans including sulphated species may originate from various plasma glycoproteins and not necessarily IgG. Nat. Commun. 9, 2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saldova R., Asadi Shehni A., Haakensen V. D., Steinfeld I., Hilliard M., Kifer I., Helland A., Yakhini Z., Børresen-Dale A. L., and Rudd P. M. (2014) Association of N-glycosylation with breast carcinoma and systemic features using high-resolution quantitative UPLC. J. Proteome Res. 13, 2314–2327 [DOI] [PubMed] [Google Scholar]
- 29. Knezevic A., Polasek O., Gornik O., Rudan I., Campbell H., Hayward C., Wright A., Kolcic I., O'Donoghue N., Bones J., Rudd P. M., and Lauc G. (2009) Variability heritability and environmental determinants of human plasma N-glycome. J. Proteome Res. 8, 694–701 [DOI] [PubMed] [Google Scholar]
- 30. Knezevic A., Gornik O., Polasek O., Pucic M., Redzic I., Novokmet M., Rudd P. M., Wright A. F., Campbell H., Rudan I., and Lauc G. (2010) Effects of aging, body mass index, plasma lipid profiles, and smoking on human plasma N-glycans. Glycobiology 20, 959–969 [DOI] [PubMed] [Google Scholar]
- 31. Lauc G., et al. (2010) Genomics meets glycomics - the first GWAS study of human N-glycome identifies HNF1 alpha as a master regulator of plasma protein fucosylation. Plos Genet. 6, doi:ARTN e1001256 10.1371/journal.pgen.1001256 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pučić M., Knežević A., Vidič J., Adamczyk B., Novokmet M., Polašek O., Gornik O, Supraha-Goreta S., Wormald M. R., Redzić I., Campbell H., Wright A., Hastie N. D., Wilson J. F., Rudan I., Wuhrer M., Rudd P. M., Josić D., and Lauc G. (2011). High Throughput Isolation and Glycosylation Analysis of IgG-Variability and Heritability of the IgG Glycome in Three Isolated Human Populations. Mol Cell Proteomics. 10, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhao S., Walsh I., Abrahams J. L., Royle L., Nguyen-Khuong T., Spencer D., Fernandes D. L., Packer N. H., Rudd P. M., and Campbell M. P. (2018) GlycoStore: a database of retention properties for glycan analysis. Bioinformatics 34, 3231–3232 [DOI] [PubMed] [Google Scholar]
- 34. Royle L., Roos A., Harvey D. J., Wormald M. R., van Gijlswijk-Janssen D., Redwan el R. M., Wilson I. A., Daha M. R., Dwek R. A., and Rudd P. M. (2003) Secretory IgA N- and O-glycans provide a link between the innate and adaptive immune systems. Biol. J. Chem. 278, 20140–20153 [DOI] [PubMed] [Google Scholar]
- 35. Fujimura T., Shinohara Y., Tissot B., Pang P. C., Kurogochi M., Saito S., Arai Y., Sadilek M., Murayama K., Dell A., Nishimura S., and Hakomori S. I. (2008) Glycosylation status of haptoglobin in sera of patients with prostate cancer vs. benign prostate disease or normal subjects. Int. J. Cancer 122, 39–49 [DOI] [PubMed] [Google Scholar]
- 36. McCarthy C., Saldova R., Wormald M.R., Rudd P.M., McElvaney N.G., and Reeves E. P. (2014) The role and importance of glycosylation of acute phase proteins with focus on alpha-1 antitrypsin in acute and chronic inflammatory conditions. J. Proteome Res. 13, 3131–3143 [DOI] [PubMed] [Google Scholar]
- 37. Liu SD., Chalouni C., Young J. C., Junttila T. T., Sliwkowski M. X., and Lowe J. B. (2015) Afucosylated antibodies increase activation of FcgammaRIIIa-dependent signaling components to intensify processes promoting ADCC. Cancer Immunol. Res. 3, 173–183 [DOI] [PubMed] [Google Scholar]
- 38. Pereira N. A., Chan K. F., Lin P. C., and Song Z. (2018) The “less-is-more” in therapeutic antibodies: Afucosylated anti-cancer antibodies with enhanced antibody-dependent cellular cytotoxicity. MAbs 10, 693–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liang J. X., Liang Y., and Gao W. (2016) Clinicopathological and prognostic significance of sialyl Lewis X overexpression in patients with cancer: a meta-analysis. Onco Targets Ther. 9, 3113–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Benjamini Y., and Hochberg Y. (1995) Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 [Google Scholar]
- 41. Rockfield S., Raffel J., Mehta R., Rehman N., and Nanjundan M. (2017) Iron overload and altered iron metabolism in ovarian cancer. Biol. Chem. 398, 995–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gornik O., and Lauc G. (2008) Glycosylation of serum proteins in inflammatory diseases. Dis. Markers 25, 267–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sturiale L., Barone R., Fiumara A., Perez M., Zaffanello M., Sorge G., Pavone L., Tortorelli S., O'Brien J. F., Jaeken J, and Garozzo D. (2005) Hypoglycosylation with increased fucosylation and branching of serum transferrin N-glycans in untreated galactosemia. Glycobiology 15, 1268–1276 [DOI] [PubMed] [Google Scholar]
- 44. Zhang S., Shang S., Li W., Qin X., and Liu Y. (2016) Insights on N-glycosylation of human haptoglobin and its association with cancers. Glycobiology 26, 684–692 [DOI] [PubMed] [Google Scholar]
- 45. Moss E. L., Hollingworth J., and Reynolds T. M. (2005) The role of CA125 in clinical practice. J. Clin. Pathol. 58, 308–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Saldova R., Royle L., Radcliffe C. M., Abd Hamid U. M., Evans R., Arnold J. N., Banks R. E., Hutson R., Harvey D. J., Antrobus R., Petrescu S. M., Dwek R. A., and Rudd P. M. (2007) Ovarian cancer is associated with changes in glycosylation in both acute-phase proteins and IgG. Glycobiology 17, 1344–1356 [DOI] [PubMed] [Google Scholar]
- 47. Scholler N., and Urban N. (2007) CA125 in ovarian cancer. Biomark. Med. 1, 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. American College of, O, and Gynecologists' Committee on Practice (2016) Practice Bulletin No B-G174: Evaluation and management of adnexal masses. Obstet. Gynecol. 128, e210–e226 [DOI] [PubMed] [Google Scholar]
- 49. Turner G. A., Goodarzi M. T., and Thompson S. (1995) Glycosylation of alpha-1-proteinase inhibitor and haptoglobin in ovarian cancer: evidence for two different mechanisms. Glycoconj. J. 12, 211–218 [DOI] [PubMed] [Google Scholar]
- 50. Qian Y., Wang Y., Zhang X., Zhou L., Zhang Z., Xu J., Ruan Y., Ren S., Xu C., and Gu J. (2013) Quantitative analysis of serum IgG galactosylation assists differential diagnosis of ovarian cancer. J. Proteome Res. 12, 4046–4055 [DOI] [PubMed] [Google Scholar]
- 51. Thompson S., Guthrie D., and Turner G. A. (1988) Fucosylated forms of alpha-1-antitrypsin that predict unresponsiveness to chemotherapy in ovarian-cancer. Brit. J. Cancer 58, 589–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry data in this study are freely available through the repository MassIVE (https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=a66ade995ac8431e80f6e27f11c55674) with the dataset identifier MSV000083877.