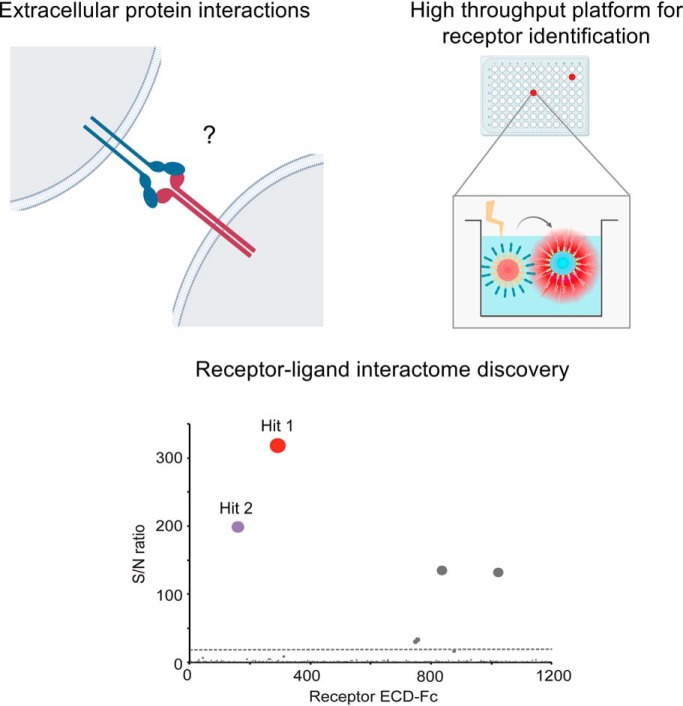
The Conditioned Media AlphaScreen represents a new platform for high throughput and sensitivity detection of interactions between plasma membrane proteins. The interleukin receptor IL20RA is identified as a binding partner for the orphan checkpoint inhibitor B7-H3. Further, a new functional interaction between the KIR2DL5 and the immune receptor PVR regulates natural killer cell cytolysis of tumor cells. This technology represents a versatile approach for receptor interactome discovery that provides insights into plasma membrane protein biology.
Keywords: assay development, automation, cancer therapeutics, cell-cell interactions, protein-protein interactions, secretome
Graphical Abstract
Highlights
New platform for high throughput detection of transient interactions between membrane proteins.
IL20RA is a receptor for the orphan checkpoint inhibitor B7-H3.
The natural killer cell protein KIR2DL5A binds the immune receptor PVR.
KIR2DL5 binding to PVR regulates natural killer cell cytotoxicity and inhibits tumor cell killing.
Elucidation of receptor interactomes to gain insights into extracellular protein biology.
Abstract
Receptors expressed on the plasma membrane and their interacting partners critically regulate cellular communication during homeostasis and disease, and as such represent main therapeutic targets. Despite its importance for drug development, receptor-ligand proteomics has remained a daunting field, in part because of the challenges associated to the study of membrane-expressed proteins. Here, to enable sensitive detection of receptor-ligand interactions in high throughput, we implement a new platform, the Conditioned Media AlphaScreen, for interrogation of a library consisting of most single transmembrane human proteins. Using this method to study key immune receptors, we identify and further validate the interleukin receptor IL20RA as the first binding partner for the checkpoint inhibitor B7-H3. Further, KIR2DL5, a natural killer cell protein that had remained orphan, is uncovered as a functional binding partner for the poliovirus receptor (PVR). This interaction is characterized using orthogonal assays, which demonstrate that PVR specifically engages KIR2DL5 on natural killer cells leading to inhibition of cytotoxicity. Altogether, these results reveal unappreciated links between protein families that may importantly influence receptor-driven functions during disease. Applicable to any target of interest, this technology represents a versatile and powerful approach for elucidation of receptor-ligand interactomes, which is essential to understand basic aspects of the biology of the plasma membrane proteins and ultimately inform the development of novel therapeutic strategies.
Cells respond to their environment through intricate signaling networks that are initiated and modulated by receptors present at the cell surface. As such, these proteins and their cognate ligands represent a main source of therapeutically relevant targets (1, 2). Notwithstanding the unprecedented success of drugs directed against cell surface proteins, the landscape of extracellular protein interactions (ePPIs)1 and molecular underpinnings of the receptor-ligand networks remain poorly characterized, thus challenging the development of new or improved therapeutics. This discrepancy is in part because of the complex biochemical nature of the cell surface proteins and thus limited compatibility of most available technologies for detection of ePPIs (3, 4). First, extracellular proteins often require functionally relevant post-translational modifications that are not added in widely used expression systems such as bacteria or baculoviruses, posing challenges for production as recombinant products. Secondly, membrane-tethered proteins are often expressed in low abundance, and in many cases establish weak, transient interactions that critically regulate multiple biological processes (5). These interactions can be easily disrupted under the harsh buffer conditions required to solubilize proteins present in membrane compartments, altogether making widely employed methods that require solubilization and washing steps, such as affinity purification/mass spectrometry (AP/MS), not optimal for detection of binding partners for membrane-expressed proteins (3, 4, 6). The more recently developed proximity-dependent biotinylation approaches coupled to MS overcome some of these limitations and have represented an important step forward toward elucidation of transient protein interaction networks (7, 8). Notwithstanding these advances, technical challenges remain, and extracellular protein interactions are significantly underrepresented in current databases. As such, the development of complementary methodologies to map receptor-ligand interactions is a key priority to advance basic research and drive drug development efforts.
The immune response against tumors is critically regulated by inhibitory receptors (also known as immune checkpoints or checkpoint inhibitors) such as CTLA4, TIM-3 or PD-1. These receptors have evolved to restrain T-cell responses and maintain homeostasis and immunological tolerance to protect the integrity of the healthy tissues. However, in the context of the tumor microenvironment, immunosuppressive signals triggered by these inhibitory receptors promote immune escape and growth of the tumor (9). In the last decade, antibody-mediated blockade of these inhibitory molecules has shown remarkable success in the clinics and has been established as a cornerstone in the treatment of cancer (9–11). Nevertheless, many inhibitory receptors are orphan and new interacting partners continue to be identified for relatively well-studied proteins, knowledge that is important to fully understand receptor functions and assess their therapeutic potential.
To advance research on the receptor-ligand interactions that regulate cell communication and immunity, we have developed a technology that enables single transmembrane (STM) receptor interactome discovery. To accomplish this, we took advantage of a recently established collection consisting of most human STM proteins, expressed as extracellular domain (ECD)-Fc fusions that are secreted to the conditioned media of human cells (12). To implement identification of ePPI using this collection of receptors, we have adapted the Amplified Luminescent Proximity Proteomics Homogenous Assay (13) (AlphaScreen) for small-scale and semi-automated detection of ePPIs. Altogether, here we present the Conditioned Media AlphaScreen, a new platform that combines the miniaturized AlphaScreen technology with our extensive STM receptor library expressed in human cells for high throughput and sensitivity identification of receptor-ligand interactions.
In this study, we employ this technology to investigate the landscape of interactions established by key inhibitory immune receptors. Expected binding partners characterized by a wide range of binding affinities were detected with significant signal to noise ratios, demonstrating the power of the method to capture challenging ePPIs. Importantly, these studies identified the interleukin-20 receptor subunit alpha (IL20RA) as a binding partner for B7-H3/CD276, a prominent immune checkpoint and emerging therapeutic target that had remained orphan. Next, to further demonstrate the applicability of the newly established platform, we explored the interactome of the poliovirus receptor (PVR/CD155), a central member of the PVR/nectin family that is highly expressed in tumors and a main regulator of the inhibitory receptor TIGIT (14, 15). Notably, in addition to the expected PVR interactors we identified a previously unrecognized binding partner, KIR2DL5, an orphan immune receptor expressed in natural killer (NK) cells (16, 17). This interaction was further characterized using orthogonal technologies that demonstrate a specific interaction between PVR and KIR2DL5, but not related family members. Moreover, our functional studies show that KIR2DL5 controls NK cytotoxicity in a PVR-dependent fashion, altogether uncovering a new association between the PVR/nectin and KIR families that may influence NK responses.
In summary, here we describe a versatile technology that can be used to study any extracellular target of interest, enabling elucidation of receptor interactomes with remarkable sensitivity and minimal background. Using this technology, we identify IL20RA as the first receptor discovered for the immune checkpoint B7-H3 and uncover the orphan receptor KIR2DL5 as a previously unappreciated modulator of NK cell functions via direct engagement of PVR. These findings suggest potential targets with therapeutic potential and provide unique insights into immune receptor biology.
EXPERIMENTAL PROCEDURES
Cell Lines and CRISPR/Cas9 Assays
The conditioned media library was prepared using Expi293F cells (Thermo-Fisher, Waltham, MA), a suspension cell line adapted from HEK293 cells. Cells were cultivated under the following conditions: 37 °C, 8% CO2, 80% humidity and 150 rpm agitation speed. Expi293 expression medium (Life Technologies, Waltham, MA) was used as the seed train and production media. COS7 cells, a monkey cell line derived from kidney fibroblasts, were used for transient expressions of selected receptors and cultured in DMEM media supplemented with 10% FBS, 2 mm l-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin, in a 37 °C humidified, 5% CO2 incubator. For competition assays with recombinant proteins or antibody, HEK293T cells purchased from ATCC (CRL-11268) were transiently transfected with PVR or CD226, expressed as full length-untagged constructs (obtained from Genentech collection). Transfections were performed using lipofectamine LTX reagent was used for cell transfection unless indicated, following manufacturer's instructions.
A-427 is an adherent human lung carcinoma cell line. The parental line (obtained from external vendor such as ATCC) was maintained at a dedicated internal cell line facility. Knockout of PVR/CD155 and CD112 expression in A-427 cells was performed by co-transfection with plasmids containing PVR/CD155- or CD112-targeted guide RNAs and Cas9 (Integrated DNA Technologies, Coralville, IA) using Lipofectamine LTX with PLUS Reagent (Life Technologies) in Opti-MEM (Life Technologies). Cells were expanded for 4 days, then single cell sorted for CD155/CD112 double negative cells to establish an A-427 CD155/CD112 double knockout line (referred to as A-427.DKO). Cells were cultured in RPMI 1640 media supplemented with 10% FBS, 2 mm l-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin, in a 37 °C humidified, 5% CO2 incubator.
Receptor Library Preparation and Small-scale Cell Transfection
The list of single transmembrane (STM) receptors was built by curation of public databases, followed by bioinformatics analysis for prediction of protein features, and finally by manual curation and review of published annotations. The boundaries of the ECD for each predicted extracellular protein were determined using publicly available algorithms, including Phobius, Signal P and TMHMM servers. Those proteins predicted to contain both a signal peptide as well as a single transmembrane region (according to both Phobius and TMHMM algorithms) were selected for the receptor library. The ECD of each protein was synthesized according to Uniprot sequences and cloned into a pRK5 vector (Genentech) containing a C-terminal Fc (hIgG1) tag. To maximize protein quality and incorporation of the relevant post-translational modifications, we used an automated platform for small-scale transfections in human cells, essentially as described (18). The prey library consisting of STM receptor ECD-Fc was prepared using micro-scale 1 ml transient transfections. Twenty-five KDa linear PEI was used as the transfection reagent, and conditioned media was harvested 7 days post-transfection. After centrifugation to remove any cell debris, supernatants were stored short-term at 4 °C.
Assay Optimization and Conditioned Media AlphaScreens Workflow for STM Interactome Discovery
Interactions between selected receptors and their binding partners, expressed as recombinant ECDs or in the conditioned media from transfected cells, were analyzed using the AlphaScreen technology (Perkin Elmer, Waltham, MA). The manufacturer's protocol was modified as described below for optimization of reagent consumption and increased throughput. The following beads were used: streptavidin-coated, donor beads (5 mg/ml, Perkin Elmer #6760002); Protein A-coated beads (5 mg/ml, Perkin Elmer #6760137R). First, donor and acceptor beads were titrated to measure robust detection of binding partners with optimal sample consumption. To do this, interactions between selected receptor ECDs, expressed as recombinant proteins, were tested by incubating different amounts of the donor beads (streptavidin-coated) with a range of molar concentrations of the biotinylated query protein. A single concentration of the relevant binding partners (as ECD-Fc proteins) was incubated with different amounts of acceptor beads (protein A-coated). As shown in the Results section, 0–16 μg of beads were tested, using equal amounts of donor and acceptor beads per reaction. Streptavidin-coated donor beads and protein A-coated acceptor beads were incubated with the query protein and binding partners for 1 h at room temperature, respectively. Following incubation, the beads were pelleted by centrifugation in a tabletop centrifuge at 25,000 rpm, and washed once with PBS containing 0.5% BSA. Next, donor and acceptor beads were incubated for 30 min at room temperature, followed by sample reading in a plate reader (Envision, Perkin Elmer). Plate reader settings were set according to manufacturer's instructions (excitation 680 nm, emission 520–620 nm). Incubations were performed in a final 50 μl volume using 384-well microplates. Four micrograms of beads/reaction were enough for identification of micromolar affinity receptor interactions with significant signal to noise ratio even when interacting partners were assayed a nanomolar concentrations. This concentration was used for subsequent assays.
Next, the assay was set up to screen for binding partners expressed directly in the conditioned media, without any further protein purification. Binding between the query protein (expressed as recombinant ECD) and the library of STM receptors expressed in the conditioned media was analyzed semi-automated procedures and liquid handling devices to increase data quality and throughput. The library of STM receptors was expressed in small scale using an automated cell transfection library previously developed in house (36). Seven days post-transfection, the conditioned media was harvested and spun down to eliminate any cell debris. The supernatants were stored at 4 °C until the day of the assay. The day of the assay, 100 μl of conditioned media were incubated with the donor beads, using 4 μg per well, in 96-deep-well plates. The conditioned media was then incubated with the beads for 1 h at room temperature on a plate shaker. Following incubation, the beads were pelleted using a microplate centrifuge (Beckman) and then washed with PBS-2% BSA. The bead pellets were resuspended and transferred to 384-well microplates (Optiplate-384, Perkin Elmer) using a Viaflo instrument Integra LifeSciences (Plainsboro Township, NJ). The biotinylated query protein was incubated with the acceptor beads for 1 h at room temperature, protected from light, washed once and resuspended in PBS-2% BSA. The biotinylated query protein was assayed at 60 nm concentration. Finally, the donor beads/query protein were dispensed to the 384-well plates containing the acceptor bead/STM receptor using a Tempest liquid handler (Formulatrix, Bedford, MA). The beads were then incubated for 30 min at room temperature and protected from light, and plates were red using an Envision plate reader (Perkin Elmer), using manufacturer's designed settings (excitation 680 nm; emission 520–620 nm).
Data Analysis for Hit Calling
Background was calculated by measuring average signal across all wells in each plate, and data was represented as signal over background (S/N) by dividing the alpha beads signal for each interaction by the background. Target-specific interactors have been empirically determined as hits characterized by S/N ∼20 or above, and thus this threshold was used for hit calling. Circle size in the plots represent S/N ratio for binding to each protein in the library. Data from unrelated query protein screens was compiled to identify promiscuous proteins in the library. These proteins were labeled as nonspecific binders and as such, were not considered as hits for the query receptors under study. Common nonspecific binders identified across multiple screens include: MXRA5, IGF2R, PCDHB9, and SORL1.
Recombinant Proteins and Biolayer Interferometry
All recombinant proteins used for the BLI studies, flow cytometry or AlphaScreens assays were purchased from R&D unless indicated. GDF15, B7-H3 and PVR recombinant ECDs, expressed as Avi-tagged proteins, were purified in house using standard affinity purification procedures. The proteins were biotinylated using the BirA enzyme following standard experimental procedures described elsewhere (19). His-tagged Ephrin B4, PD-1 and CD86 proteins used for the Conditioned Media AlphaScreens were purchased from R&D and biotinylated in vitro using the EZ-Link Sulfo-NHS-Biotin kit (Thermo Scientific). A biotin/protein 5:1 molar ratio was used.
Interactions between PVR and the relevant receptors were tested by biolayer interferometry, using an Octet Red system. Recombinant PVR was captured onto streptavidin-coated sensors and tested for binding to the protein analytes indicated in each case, assayed in PBS buffer. To test the interaction between PD-1 and podoplanin and their ligands PD-L1 and CLEC-2, respectively, PD-1 and podoplanin were expressed in the conditioned media of human cells as ECD-Fc proteins, as described, subsequently captured onto anti-human Fc sensors, and then analyzed for binding to PD-L1 and CLEC-2 expressed as recombinant his-tagged proteins assayed in PBS buffer. All data were analyzed using Forte Pall (Port Washington, NY) software v9.0.
Cell Surface Binding Assays
The indicated interleukin receptors or the KIR receptors or PVR binding partners were expressed on cells for analysis of B7-H3 or PVR binding to the cell surface, respectively. COS7 cells were transiently transfected with the selected binding partners, as indicated, and grown in glass-bottom microplates. DNAs encoding for the full-length receptors belong to a Genentech proprietary collection. After 48 h, the cells were incubated with recombinant B7-H3 or PVR to test binding to receptors expressed on the cell surface. Briefly, the cells were blocked with PBS containing 2% BSA, followed by incubation with soluble protein for 1 h at 4 °C. Following incubation, the cells were washed and subsequently fixed with 4% PFA. B7-H3 or PVR binding to the cell surface was detected using APC-conjugated streptavidin. Images were acquired using high content microscope (IN Cell 6000, Chicago, IL) and analyzed using the INCell Developer software to quantify signal intensity on the cell surface. Transfections were performed in duplicates and B7-H3 or PVR binding to the cells was represented as intersection plots.
Isolation of NK Cells and Generation of Lymphokine-Activated Killer (LAK) Cells
Purified NK cells were isolated from buffy coats drawn from normal healthy donors by negative selection performed using EasySep Human NK Cell Isolation Kit (StemCell Technologies, Vancouver, Canada), according to manufacturer's instructions. NK cells were cultured in complete RPMI media (RPMI 1640 supplemented with 10% FBS, 2 mm l-glutamine, 2 μm 2-ME, 1 mm sodium pyruvate, 100 U/ml penicillin and 100 μg/ml streptomycin) supplemented with 1000 U/ml recombinant human IL-2 (Peprotech, Rocky Hill, NJ), in a 37 °C humidified, 5% CO2 incubator.
KIR2DL5 Expression in NK Cells
All donor NK cells were determined to be KIR2DL5 negative by flow cytometry (data not shown). To express KIR2DL5 in LAK cells, IL-2 cultured NK cells were nucleofected with KIR2DL5 expression construct (catalogue number RG217119; OriGene Technologies, Rockville, MD) using the Amaxa Human NK Cell Nucleofector Kit (catalogue number VPA-1005; Lonza, Benicia, CA), according to manufacturer's instructions. Nucleofected cells were cultured as previously described and KIR2L5 expression was validated by flow cytometry 3 days following nucleofection.
Antibodies and Flow Cytometry
The following antibodies used for staining were purchased from BioLegend, San Diego, CA: PE-conjugated KIR2DL5 (clone UP-R1), APC- CD226 (clone 11A8), BV421-CD96 (clone NK92.39), BV605-TIGIT (clone A15153G), BV650-CD3 (clone OKT3), BV711-CD56 (clone 5.1H11), PE-human Fc (HP6017). Unconjugated anti-KIR2DL5A (clone UP-R1) was purchased from LSBio. LAK cell samples were acquired on LSRFortessa using CellQuest Pro v5.1.1. software (BD Biosciences, San Jose, CA) and data analysis performed using FlowJo v9.4.4 software (Tree Star, Inc., Ashland, OR). Cell sorting was performed on FACS Aria (BD Biosciences) to isolate KIR2DL5+ or KIR2DL5− LAK cells for killing assays. For single cell sorting of CD155/CD112 double-negative A-427 cells, cells were stained with PE-CD155 (clone TX24) and APC-CD112 (clone TX31). Samples were acquired on FACSCanto II using FACSDiva 8.0 software and data analysis performed using FlowJo v10 software (Tree Star, Inc.).
Competition Assays and KIR2DL5 Blocking Assays
KIR2DL5 binding to PVR in the presence of an anti-KIR2DL5 antibody was analyzed on cells transiently expressing PVR by flow cytometry. Recombinant KIR2DL5-Fc (at 50 nm concentration) was pre-incubated with 0–1.5 μm of anti-KIR2DL5 antibody (clone UP-R1) before incubation with PVR-expressing cells for 30 min at 4 °C. The cells were fixed for 10 min at room temperature with 4% PFA (ThermoFisher), and stained with PE-conjugated anti-human Fc for 30 min at 4 °C so as to detect the amount of KIR2DL5-Fc bound on the cells.
To test PVR binding to CD226 in the presence of other PVR binders, CD266 was transiently expressed on cells and binding studies were performed 48 h post-transfection. Biotinylated PVR (at 5 nm concentration) was pre-incubated with 0, 0.5 μm and 1 μm of KIR2DL5, TIGIT or CD226, expressed as recombinant ECD-Fc proteins, befure incubation with CD226-expressing cells for 30 min at 4 °C. APC-conjugated streptavidin was used for detection of PVR binding to the cell surface.
NK-mediated Cytotoxicity Assay
Real time-cell electronic sensing using the xCELLigence RTCA MP system (Biosciences) was performed to assess cytotoxicity. 1 × 105 A-427 WT or A-427.DKO target cells in 100 μl were plated to 96-well E-plate and cultured in the xCELLigence system installed in a 37 °C, 5% CO2 incubator. Optimal seeding conditions were established by monitoring Cell Index (CI), a value directly reflecting strength of cell adhesion and cell number as measured by electrical impedance values across the high-density electrode array coating the bottom of each well of the E-plate. After 4 h, 2 × 105 sorted KIR2DL5+ or KIR2DL5− LAK cells in 100 μl were added to appropriate wells, for an E:T ratio of 2:1, predetermined to provide an optimal window for detecting KIR2DL5-mediated inhibition of killing. For antibody blockade, LAK cells were pre-incubated with 20 μg/ml purified anti-KIR2DL5 (clone UP-R1, BioLegend), anti-CD226 Ab (clone DX11, BD Biosciences) or mouse IgG1, k, isotype control for 30 min on ice before addition to E-plate. The antibody was present throughout the entire duration of the killing assay. Negative control wells were target cells without effector cells. After addition of effector cells, CI was measured every 10 min in real time. CI at each time point was normalized against CI at the time of LAK cell addition, and cytotoxicity was calculated as % cytolysis = (normalized CIno effector − normalized Cieffector)/normalized CIno effector × 100.
Statistical Analysis
Statistical tests were conducted using paired two-tailed Student t test between the indicated groups.
RESULTS
Conditioned Media AlphaScreens for High Sensitivity Detection of Receptor-Ligand Interactions
STM receptors are modulators of physiologically and disease-relevant pathways and therefore represent prominent targets for biotherapeutic development. Thus, to enable receptor-ligand discovery for extracellular drug target candidates, we recently established an extensive library of human receptors, consisting of most STM proteins. To retain their binding properties while removing the insoluble transmembrane region, the ECDs of the receptors were expressed as Fc tag fusions for generation as secreted proteins in the conditioned media of transfected cells (12). In turn, the AlphaScreen method is a beads-based proximity method used to study diverse molecular interactions (20). To query PPI using this method, the binding partners of interest are immobilized on donor and acceptor beads, respectively. When the two proteins under study interact bringing the beads in proximity, a series of energy transfer reactions take place, generating a chemiluminescence signal that can be easily measured using a plate reader. Here, we sought to adapt this technology for screening of receptor-ligand interactions in combination with our human cell-produced receptor library, to enable evaluation of a query protein of interest for binding to most STM receptors in the human genome.
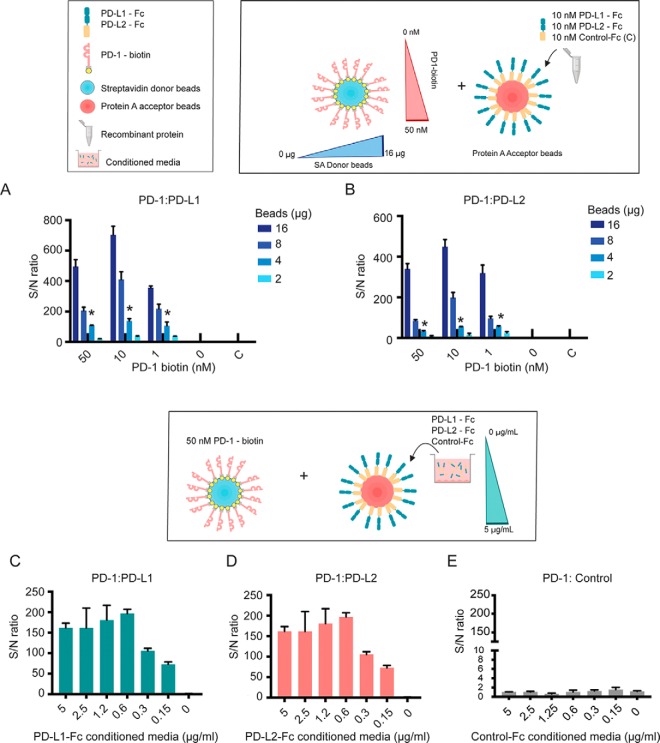
We hypothesized that immobilization of receptors and their ligands on the donor and acceptor beads would provide enhanced binding avidity, similarly to other multimerization strategies developed by our group and others, thus enabling detection of transient PPIs (21). Thus, we first sought to analyze the sensitivity of the AlphaScreen method to detect low affinity interactions between cell surface proteins, expressed as purified, recombinant ECDs. To do so, we studied the immune receptor PD-1, known to bind the cell surface receptors PD-L1/CD274 and PD-L2/PDCD1LG2 with low affinities characterized by micromolar KD (22). Recombinant PD-1 was biotinylated and captured on streptavidin-coated donor beads at different concentrations, whereas its binding partners PD-L1 and PD-L2, expressed as Fc-tagged proteins, were captured on acceptor beads. To evaluate and further optimize the AlphaScreen method for ePPI detection, PD-1 binding to its receptors was assayed using decreasing amounts of donor and acceptor beads and 384-well microplates for increased throughput (Fig. 1A). Notably, both PD-1/PD-L1 and PD-1/PD-L2 interactions were detected with significant signal to noise ratios and minimal background, even when the query protein PD-1 was present at nanomolar concentrations and low amounts of alpha beads were used (Fig. 1A and 1B). These results confirmed the feasibility of this approach to identify transient interactions between receptors.
Fig. 1.
Conditioned Media AlphaScreens for sensitive detection of transient receptor-ligand interactions. Biotinylated PD-1 protein was incubated with different amounts of streptavidin-coated donor beads (16 to 0 μg) at the molar concentrations indicated (50–0 nm). Binding to the known interacting partners A, PD-L1 or B, PD-L2 was analyzed using the procedure indicated Experimental procedures. PD-L1, PD-L2, or a control protein (labeled as “C”) expressed as recombinant ECD fused to a Fc tag, were captured on protein A-coated acceptor beads by incubating with different amounts of beads (16 to 0 μg). For these assays, PD-L1 and PD-L2 were tested at 10 nm concentration. Based on these results, 4 μg of beads were used for subsequent assays to minimize reagent consumption while allowing detection of low affinity PPIs with high signal/noise ratios. Plots show one representative assay out of two independent runs, assayed in triplicates. C–E, Biotinylated PD-1 (50 nm concentration) was captured on donor beads to analyze binding to acceptor beads that had been incubated with of conditioned media enriched in C, PD-L1, D, PD-L2 or E, a control receptor, expressed as ECD-Fc fusions. Fc-tagged receptors ECD in the conditioned media were titrated and diluted to the indicated concentrations (0–5 μg/ml) to assess sensitivity of the binding assays. Experiments shown are representative of at least two independent assays run in triplicates. Error bars indicate S.D.
Next, we evaluated the use of this method for detection of ePPI using conditioned media enriched in individual receptors, expressed as ECD-Fc soluble proteins. To do so, PD-1 binding partners were produced in human cells as indicated under Experimental Procedures, and assayed directly in the conditioned media. As described above, biotinylated PD-1 was immobilized on donor beads and then incubated with the acceptor beads that had been incubated with the conditioned media from PD-L1, PD-L2 or a control receptor ECD-Fc expressing cells (Fig. 1C). The receptor ECDs expressed in the conditioned media were titrated and incubated with the acceptor beads at different concentrations to evaluate sensitivity of detection. The expected interactions were readily detected when PD-L1 and PD-L2 were expressed in the conditioned media, with negligible background (Fig. 1E) and significant sensitivity, even when the receptor ECD-Fc were assayed at low concentrations (Fig. 1C and 1D). An important consideration when studying interactions between membrane proteins is the addition of relevant post-translational modifications, including proper folding and glycosylation. To further prove the quality of the conditioned media protein library, we next studied two interacting pairs known to depend on protein glycosylation, including PD-1/PD-L1 (23–25) and the cell surface protein podoplanin and CLEC-2, an interaction that has been shown to take place through O-glycosylated residues in podoplanin (26). Importantly, we detected specific binding when podoplanin and PD-1 were expressed in the conditioned media of human cells (supplemental Fig. S1), further demonstrating that interactions that require properly folded, glycosylated proteins are readily detected using the method described in this study.
Together, these results illustrated the sensitivity of the miniaturized AlphaScreen method for detection of transient ePPIs, including those that depend on receptor glycosylation, and demonstrated that these interactions can be detected when proteins are expressed in the conditioned media from human cell cultures.
Conditioned Media AlphaScreens for High Throughput Receptor Discovery
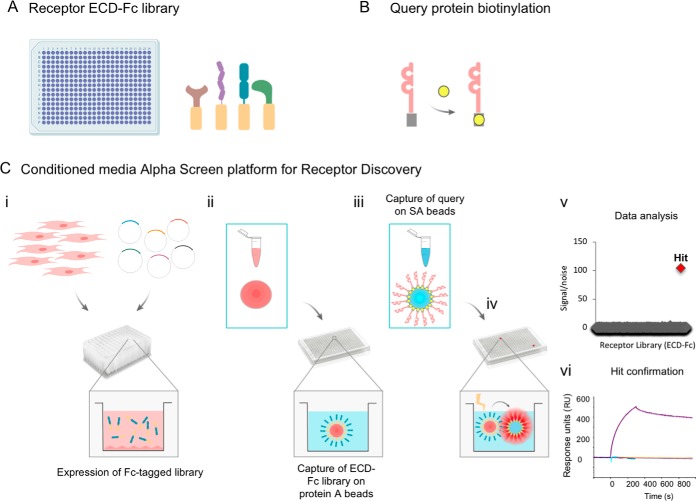
Having demonstrated the use of the method for robust detection of interactions using proteins expressed in the conditioned media, we then implemented a workflow to query the receptor library in high throughput (Fig. 2). To accomplish this, the library of STM receptors (Fig. 2A) was expressed using an automated platform optimized for small-scale transfection of human cells (Fig. 2C.i). The target of interest, expressed as a recombinant product, was biotinylated in vitro or expressed as an avi-tagged fusion protein for site-directed biotinylation (19) (Fig. 2B). After transfection and harvesting of the cell supernatants, the STM proteins enriched in the conditioned media were captured on protein A-coated acceptor beads (Fig. 2C.ii). Next, the STM protein-coated acceptor beads were pelleted and transferred to microplates for increased throughput and lower sample consumption. Concomitantly, the biotinylated query protein of interest was immobilized on streptavidin-coated donor beads (Fig. 2B and 2C.iii), followed by incubation with the library of STM receptors that had been previously captured on acceptor beads (Fig. 2C.iv). Upon excitation of the donor beads, the chemiluminescence signal was recorded using a plate reader and data was analyzed for hit calling and identification of binding partners (Fig. 2C.v). As described under Experimental Procedures, a semi-automated procedure was implemented to minimize sample manipulation. Finally, any hits of interest were confirmed using independent methodologies such as Biolayer Interferometry (BLI) (Fig. 2C.vi).
Fig. 2.
Automated Conditioned Media AlphaScreens workflow. A, A library consisting of ≈1,200 unique single transmembrane (STM) human receptors, expressed as extracellular domains fused to a Fc tag, was generated. B, The query protein under study is expressed as a recombinant protein for in vitro biotinylation, or expressed as an avi-tagged protein for site-specific biotinylation. C, Schematic representation of the Conditioned Media AlphaScreen platform for receptor interactome discovery: i, The STM receptor library is expressed using a high throughput platform for small-scale cell transfection. ii, Conditioned media enriched in individual receptor-Fc proteins is harvested and transferred to multi-well plates, following incubation with protein A-coated acceptor beads. iii, The biotinylated query protein is captured on streptavidin-coated donor beads and iv, incubated with the library of STM receptors, previously immobilized on acceptor beads. v, Query protein-receptor interactions are detected by reading the chemiluminescence signal, followed by data analysis for identification of interacting partners for the query protein under study. vi, Confirmation of binding partners of interest using additional methodologies such as biolayer interferometry.
Next, to test the functionality of this workflow, as a proof-of-concept we first used this platform to screen the immune checkpoint PD-1 for interactions with the collection of STM proteins. Recombinant PD-1 was biotinylated for capture on donor beads and incubated with the STM receptor library immobilized on acceptor beads, using the workflow described above. Importantly, these assays identified the expected interacting partners PD-L1 and PD-L2 as the top most hits, with low background and strong signal to noise ratio (Fig. 3A and supplemental Table S1). Specific proteins, including PCDHB9 or IGF2R, were identified as high scoring hits across independent screens for unrelated query proteins, indicating that these proteins are nonspecific binders and therefore should not be considered PD-1-specific hits (Fig. 3A and 3B, and not shown). To further evaluate the performance of the Conditioned Media Alpha beads methodology, we then screened the co-stimulatory receptor CD86 against the STM protein library. These assays identified the expected counter-receptors, CTLA4 and CD28, as the most prominent hits with significant signal to noise ratio, alongside a few additional high scoring hits, including PCDHB9 or SORL1, identified as nonspecific binders (Fig. 3B and supplemental Table S2). Finally, to further demonstrate the applicability of the newly established technology, two unrelated proteins were studied, including the secreted factor GDF15 and the receptor tyrosine kinase Ephrin B4. Importantly, these results demonstrated a unique interaction between GDF15 and its recently identified receptor, GFRAL (27) (supplemental Fig. S2A), and identified the Ephrin family members EphB1, EphB2 and EphB3 as binders for Ephrin B4 (supplemental Fig. S2B), as expected (28).
Fig. 3.
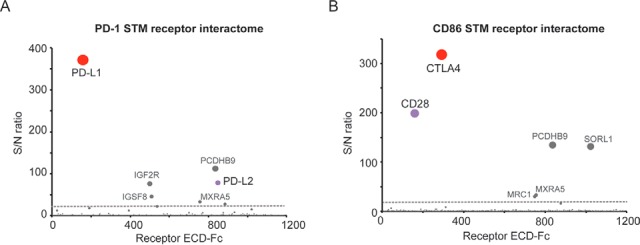
Conditioned Media AlphaScreen for high sensitivity elucidation of immune receptor interactomes. Recombinant PD-1 and CD86 proteins were biotinylated and analyzed for binding to the library of STM receptors following the experimental procedure described in the text. S/N ratio ≥ 20 was determined as experimentally-validated cut-off for hit calling (dotted gray line). A, PD-1 screens using the Conditioned Media AlphaScreen platform identify PD-L1 and PD-L2 and B, CD86 screening using the Conditioned Media AlphaScreen workflow allows identification of the binding partners CTLA4 and CD28 as top scoring hits. Hits labeled in gray color have been empirically determined to be nonspecific binders.
Altogether, these results demonstrate the applicability of the newly implemented platform to identify relevant receptor interactions with high sensitivity and specificity when a protein of interest is queried for binding to the extensive collection of STM receptors. These results can be achieved with low sample and reagent consumption and minimal requirements for purified protein.
IL20RA Is A Receptor for the Orphan Immune Checkpoint B7-H3
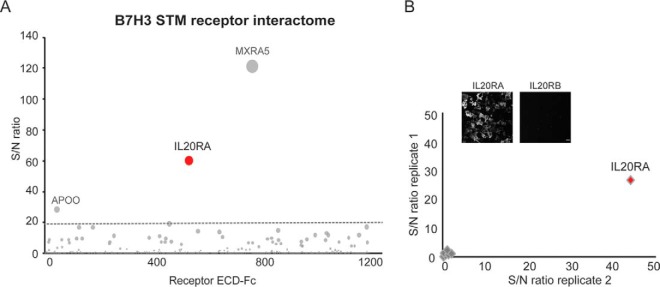
Next, we sought to study the interactome of the immune checkpoint B7-H3, a member of the B7 and CD28 families (29). In addition to acting as an inhibitory receptor that down-regulates T cell functions, B7-H3 is highly overexpressed in multiple tumors and has often been correlated with poor prognosis, positioning this receptor as an ideal candidate for the development on cancer immunotherapies. Notwithstanding its promise as a drug target, the receptor(s) for B7-H3 have remained unknown therefore limiting our understanding of its biological functions and challenging the development of therapeutics for B7-H3 modulation (30, 31). Thus, to enable elucidation of B7-H3 receptors and identification of potentially targetable interactions, B7-H3 was expressed as a recombinant protein engineered as an Avi-tagged ECD for screening using our receptor discovery platform. Surprisingly, IL20RA, known to act as a coreceptor for several members of the interleukin cytokine family, was identified as the only high scoring hit that specifically bound to B7-H3 (Fig. 4A and supplemental Table S3). MXRA5 was identified as a hit in screens for unrelated query proteins (Figs. 3, 5 and not shown) and was therefore labeled as a nonspecific binder.
Fig. 4.
The interleukin receptor IL20RA is a receptor for the immune checkpoint B7-H3. A, B7-H3 was expressed as a recombinant protein and screened using the Conditioned Media AlphaScreen technology. The B7-H3 STM receptor interactome identifies IL20RA as a top hit. Proteins labeled in gray color represent nonspecific binders. S/N ratio ≥ 20 was used as experimentally-determined cut-off for hit calling (dotted gray line). B, B7-H3 binding to IL20RA on the cell surface. Binding of recombinant biotinylated B7-H3 to cells expressing IL20RA or control receptors (IL20RA, IL20RB, IL17RB, IL10RA, IL10RB, LIFR, OSMR, and IL6R) was analyzed by immunofluorescence, using fluorophore-conjugated streptavidin for detection. Intersection plot shows B7-H3 binding to the surface of cells expressing the indicated receptors; transfections were performed in duplicates. Micrographs show images corresponding to B7-H3 binding to IL20RA- and IL20RB-expressing cells, as control. Scale bar = 50 μm.
Fig. 5.
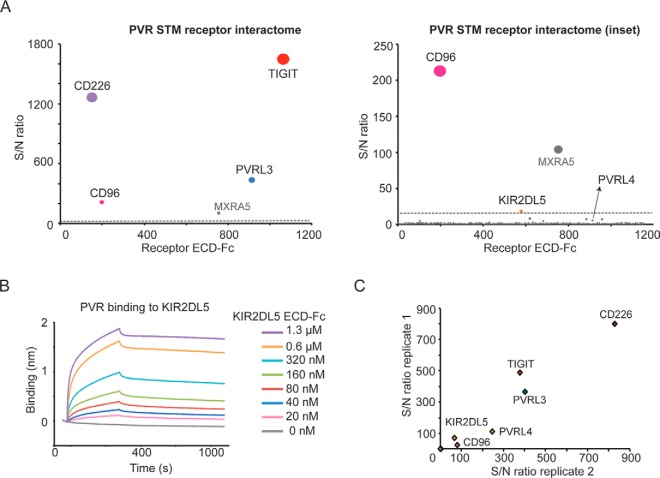
The orphan NK cell receptor KIR2DL5A is a novel binding partner for PVR. A, PVR screens using the Conditioned Media AlphaScreens platform identify known interacting partners as well as a previously unknown interaction with the receptor KIR2DL5A (inset). B, Analysis of PVR binding to KIR2DL5A, expressed as recombinant purified proteins, by BLI. Biotinylated PVR was immobilized on streptavidin sensors and tested for binding to KIR2DL5, as a soluble ECD-Fc tag analyte, at the indicated concentrations. C, PVR binding to receptors expressed on the cell surface. Individual KIR receptors (KIR2DL1, KIR2DL2, KIR2DL3, KIR2DL4, KIR2DL5A, KIR2DL5B, KIR2DS1, KIR2DS3, KIR2DS4, KIR2DS5, KIR3DL1, KIR3DL3) or known PVR binders (CD96, CD226, TIGIT, PVRL3, and PVRL4) were transiently expressed in cells, and binding of PVR to the cell surface was analyzed by immunofluorescence using fluorescent streptavidin for detection. Binding is represented as signal intensity/noise, and shown as intersection plots representing two independent transfections. Representative images for PVR binding to the cell surface are shown.
Next, to confirm and further study this interaction, we tested binding of recombinant B7-H3 to cells transiently expressing IL20RA or other interleukin receptors as controls. These assays confirmed the newly identified receptor for B7-H3, demonstrating a specific interaction with IL20RA expressed on the cell surface (Fig. 4B).
The Orphan Natural Killer Cell Receptor KIR2DL5A Is A New Binding Partner for PVR
PVR/CD155 is an immunoglobulin domain-containing protein that belongs to the nectin family, which also encompasses the inhibitory receptor and clinical target TIGIT (15). PVR is up-regulated in cancer and has been associated with enhanced tumor proliferation and migration (32–34). Although PVR functions have not been fully characterized, the receptor is thought to modulate T and NK cells responses through finely tuned interactions with its binding partners TIGIT, CD96 and CD226. As such, targeting these interactions has represented a major focus of immunotherapy research. To gain insights into the PVR/nectin family biology and identify interactions that could be relevant in the tumor microenvironment, we sought to characterize the PVR receptor interactome. To this end, PVR was expressed as an avi-tagged protein for site-specific biotinylation and analyzed for binding to the STM receptor library. These efforts identified the expected interacting partners, including CD226, TIGIT, CD96 and PVRL3 (Fig. 5A and supplemental Table S4). Of note, the NK cell receptor KIR2DL5 was also identified as a PVR binding partner in these screens, alongside PVRL4 that was detected as a low scoring hit (Fig. 5A, inset). To validate these findings, we studied the newly identified interaction between KIR2LD5 and PVR by BLI using purified recombinant proteins. These results confirmed PVR binding to KIR2DL5A (Fig. 5B) and not to other members of the family including the closely related member KIR2DL4 (supplemental Fig. S3). Next, to fully assess binding specificity for the KIR receptor family, we analyzed PVR binding to the cell surface of cells transiently expressing the KIR receptors. These results confirmed that PVR selectively binds to KIR2DL5A expressed on the cell surface (Fig. 5C), consistent with our receptor discovery results (Fig. 5A), and corroborated binding to PVRL4, a receptor highly related to the known PVR binder PVRL3, to our knowledge describing a new interaction within the PVR/PVRL4 family.
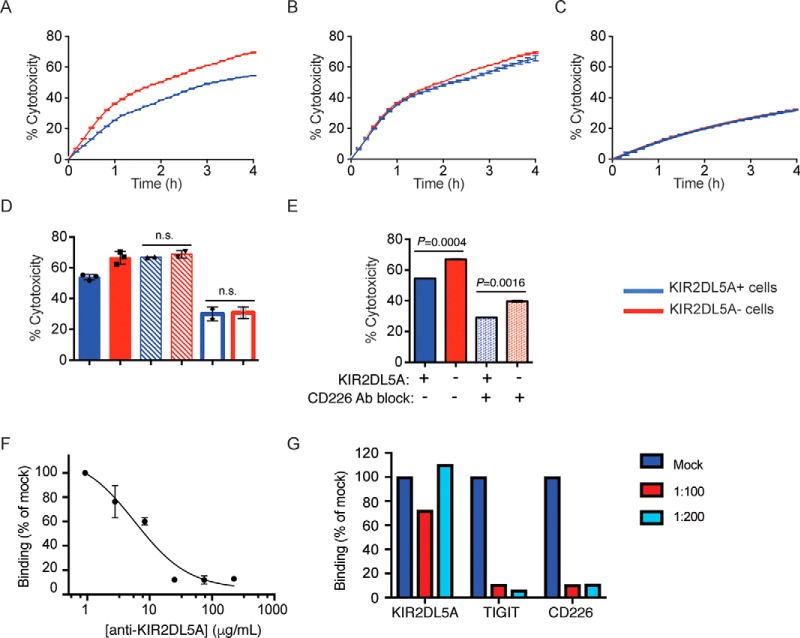
PVR Regulates NK Cytotoxicity Upon KIR2DL5A Engagement
KIR2DL5 is characterized by the presence of two immunoreceptor tyrosine-based inhibitory motifs (ITIM) in its cytosolic domain, and has been shown to play an inhibitory role in the human NK cell line NK92 (16). We then asked if PVR could impact cytotoxicity of human primary NK cells through direct targeting of KIR2DL5 on the cell surface. Real-time cell killing assays were used to assess the ability of PVR, expressed on tumor cells used as targets, to modulate cytolysis of KIR2DL5-expressing NK cells. NK cells isolated from normal healthy donors were cultured with IL-2 to generate LAK effector cells, as it is well established that LAK cells have higher killing activity than freshly isolated NK cells. Because none of the donor NK cells studied expressed detectable levels of endogenous KIR2DL5 in circulating NK cells (not shown), LAK cells were nucleofected with KIR2DL5, typically producing 15–30% KIR2DL5+ CD56+ NK cells (supplemental Fig. S4A). Real-time profiling of cytotoxicity against PVR-expressing A-427 target cells demonstrated that KIR2DL5− LAK cells efficiently recognized and killed the target cells, with ∼70% cytotoxicity after 4 h (Fig. 6A). Notably, KIR2DL5 expression in the NK cells resulted in reduced cytolytic activity (Fig. 6A and 6D), suggesting that KIR2DL5 delivers a negative signal that downmodulates NK activation. To determine whether the reduced effector function of KIR2DL5-expressing LAK cells was because of direct engagement by PVR expressed on the target cells, cytotoxicity was evaluated in the presence of an anti-KIR2DL5 antibody (16) that we found to potently block the PVR-KIR2DL5 interaction (Fig. 6F). The anti-KIR2DL5 antibody increased cytotoxicity by KIR2DL5+ LAK cells to the full capacity of the LAK cells, indicating that the inhibitory function of KIR2DL5 was blocked and further demonstrating the specificity of this interaction (Fig. 6B and 6F).
Fig. 6.
PVR modulates NK cell cytotoxicity in a KIR2DL5A-dependent manner. A, Sorted KIR2DL5+ (blue) or KIR2DL5− (red) LAK cells were used as effector cells against A-427 targets at a ratio of 2:1. Data are shown as % cytotoxicity determined from cell index values measured every 10 min over a 4 h period. Each time point is mean ± S.D. of duplicate wells. Real-time profiling of cytotoxicity shown is representative of three independent experiments. B, Sorted KIR2DL5+ (blue) or KIR2DL5− (red) LAK cells were pre-incubated with anti-KIR2DL5 (clone UP-R1) antibody before addition to A-427 target cells. Data shown is representative of two independent experiments. C, Sorted KIR2DL5+ (blue) or KIR2DL5− (red) LAK cells were tested against A-427(ΔPVR/ΔPVRL2) target cells. Data shown is representative of two independent experiments. D, Comparison of LAK-mediated killing at 4 h. Compiled data for KIR2DL5+ (blue) or KIR2DL5− (red) LAK cell killing against A-427 WT target cells (solid bars, n = 3), A-427 WT cytotoxicity in the presence of anti-KIR2DL5 antibody (hatched bars, n = 2), or killing of A-427.DKO targets (open bars, n = 2). E, Effect of CD226 antibody blockade. KIR2DL5+ (blue) or KIR2DL5− (red) LAK cell killing against A-427 WT in the absence (solid bars) or presence (dotted bars) of anti-CD226 antibody at 4 h. Data shown is representative of two independent experiments. F, Blocking of KIR2DL5A binding to PVR-expressing cells in the presence of an anti-KIR2DL5A antibody. KIR2DL5A binding to the cell surface is represented as mean fluorescence intensity (MFI) relative to binding in the absence of antibody. Data represents mean ± S.D. of duplicate wells. G, PVR binding to CD226-expressing cells in the presence of KIR2DL5A, TIGIT or CD226, expressed as recombinant proteins. Biotinylated PVR was incubated alone or in the presence of the PVR binders at the indicated molar ratios, and PVR bound to the surface was detected with fluorophore-conjugated streptavidin. Data shown is representative of two independent experiments.
CD226 is an important activating receptor for NK cells, which binds to PVR and the related family member PVRL2/CD112. We found that LAK cells expressed CD226, but not CD96 or TIGIT, additional PVR binders (supplemental Fig. S4C and S4D). Next, we generated A-427 cells lacking PVR and PVRL2 (A-427.DKO) using the CRISPR-cas9 technology, (supplemental Fig. S4B) an employed them as cellular targets in the cytotoxicity assays. Absence of PVR and PVRL2 significantly reduced sensitivity to killing by LAK cells, with only 30% cytotoxicity at the 4-hour time point (Fig. 6C and 6D). The reduction in killing of A-427.DKO targets as compared with A-427 WT cells may be attributable to the loss of activation signals provided by CD226 engagement of PVR and/or PVRL2 (15, 33). This effect was independent of KIR2DL5 expression on the NK cells, further demonstrating that KIR2DL5 functions in this context require engagement of PVR on the target cell (Fig. 6C and 6D). In keeping with this notion, addition of an anti-CD226 blocking antibody led to decreased cytotoxicity (Fig. 6E), confirming the role of CD226 in LAK cell effector function (Fig. 6E). To further study these interactions, we then performed competition studies between the relevant PVR binding partners, expressed on the cell surface. Interestingly, these assays showed that TIGIT effectively blocked PVR interaction with CD226 expressed on the cell surface (Fig. 6G), as previously described. On the contrary, KIR2DL5 did not compete PVR binding to CD226, suggesting an alternative mechanism of action that is consistent with the different immunoglobulin domain organization of KIR2DL5, relative to CD226 and TIGIT (Fig. 6G). Collectively, these functional assays demonstrate that the novel binding partner KIR2DL5 interacts with PVR to inhibit NK cell-mediated cytotoxicity, even in the presence of the activating receptor CD226 (Fig. 7).
Fig. 7.
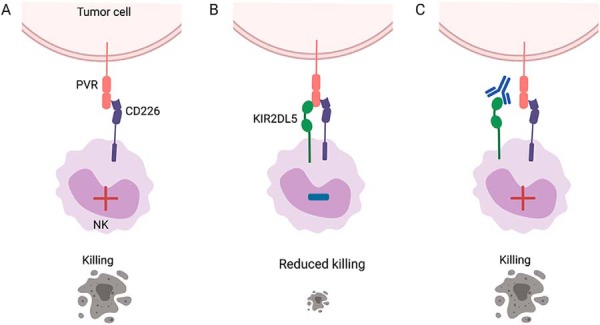
KIR2DL5 modulates NK cell effector function upon interaction with PVR on the cell surface. A, Recognition of PVR on the tumor cell by the activating receptor CD226 on the NK cell results in cell activation and killing. B, NK cell-expressed KIR2DL5 binds to PVR, even in the presence of CD226 co-expressed on the tumor cell, delivering an inhibitory signal that reduces NK-mediated cytolysis of the tumor cell. C, Antibody blockade of KIR2DL5 restores cytotoxic functions and killing of the tumor cell.
DISCUSSION
Cellular communication and signaling during homeostasis and disease are largely dictated by interactions between proteins expressed at the cell surface. Nevertheless, our understanding of the receptor-ligand interactions in the extracellular space is fragmentary and for many receptors, no binding partners have been identified, altogether posing significant challenges for the development of targeted therapeutic strategies. Antibody-mediated blockade of immune checkpoints such as PD-1 has represented an unparalleled step forward in the cancer immunotherapies (9, 11). However, many patients fail to respond to these therapies and only a subset exhibit durable responses, indicating that multiple non-redundant inhibitory pathways function independently to suppress anti-tumor immunity, possibly through interactions with unknown molecules in the tumor. This is well illustrated by inhibitory receptors such as B7-H3, TIM-3, or VISTA, emerging therapeutic targets currently under study in multiple exploratory and pre-clinical efforts that have nonetheless remained orphan (29, 35). Developing additional methods for sensitive and comprehensive elucidation of the ligands and thus signaling pathways for these molecules is key to understand basic aspects of receptor biology and improve antibody-based therapeutics, and it may also lead to identification of additional targets.
Here, we present the Conditioned Media AlphaScreen, a robust platform that combines the miniaturized AlphaScreen method for screening of a large collection of human receptors, implemented to enable high sensitivity and throughput identification of STM receptor interactomes for targets of interest. These interactions are studied using STM proteins expressed in the conditioned media of human cells, therefore avoiding any solubilization or stringent washing conditions that may otherwise disrupt relevant interactions. In-trans PPIs, which take place between two juxtaposed cells, are fundamental for cell communication and have dominated the landscape of therapeutically relevant targets such as TIGIT/PVR or PD-1/PD-L1, which are thought to mediate interaction between different immune cell types, or immune cells and tumor cells. These interactions involve clusters of receptors tightly arrayed on the plasma membrane and as such, are often characterized by KD in the micromolar range. As shown by analysis of the immune receptor interactomes using our platform, the Conditioned Media AlphaScreen is optimally suited for high sensitivity detection of in-trans interactions, which by nature are undetectable by most commonly used techniques that focus on in-cis interactions in a given cell type. In combination with the relevant collection of receptor ECDs, this technology allows unbiased identification of receptor-ligand interactions, thus not being restricted to expression of binding partners in specific cell types, such as primary cell populations, which can be challenging to study using other proteomics methods.
Given the size of the receptor library presented in this study, it is possible that a fraction of the STM receptors are not properly folded or lack some of the relevant posttranslational modifications when expressed as soluble proteins. Similarly, the presence of the Fc tag fused to the receptor ECD might interfere with certain interactions, leading to false negatives. Despite these important considerations, our results so far, which include data for unrelated receptors (PD-1, CD86, B7-H3, PVR, Ephrin B4) and the secreted factor GDF15, protein interactions that involve glycosylated residues such as podoplanin/CLEC-2, as well as several other targets from different protein families (over 10 additional query proteins screened by the time this manuscript was submitted) have shown significant sensitivity and minimal off-target effects, altogether demonstrating the suitability of this method for receptor interactome discovery. Notwithstanding the wide applicability of this platform to study query proteins characterized by different domain structures and biochemical properties, including receptors and secreted factors, the method in its current format is not optimal for detection of other interaction types such as those that involve multi-transmembrane receptors, which in most cases cannot be engineered for expression as soluble proteins. Similarly, the current methodology would not enable detection of interactions that involve heterodimers, those that require the transmembrane or intracellular regions for complex formation. Developing alternative methodologies, such as cell-based cDNA libraries or mass spectrometry-based approaches, will be essential to study these challenging proteins types. Despite the aforementioned limitations, the platform presented here serves as a powerful, complementary approach for STM receptor interactome discovery, and represents a unique workflow for elucidation of both in-trans and in-cis binary interactions characterized by a broad range of binding affinities.
The B7 family contains prominent immune receptors such as PD-L1, PD-L2 or CD80, known to interact with members of the CD28 family of co-stimulatory and co-inhibitory molecules. Interestingly, B7-H3 is significantly overexpressed in multiple cancers and a substantial number of studies have demonstrated important roles for B7-H3 beyond its functions in immune suppression, including epithelial-mesenchymal transition and increased tumor growth and metastasis (30, 31, 36, 37). The finding that B7-H3 interacts with IL20RA indicates a previously unknown connection between the B7 receptors and the IL20 cytokine family. These cytokines function through heterocomplexes on the cell surface, including IL-19, IL-20 and IL-24 that signal through a complex formed by IL20RA and IL20RB (38). Here, we identify and validate a binary interaction between IL20RA and B7-H3, suggesting that B7-H3 may regulate interleukin-induced signaling independently of IL20RB. IL20RA is mostly absent from hematopoietic lineages but highly expressed in skin where it plays various roles in inflammation and autoimmune disease (39–41). Interestingly, several studies have shown B7-H3 as a significant factor for melanoma progression and metastasis, and in fact, clinical trials to treat melanoma and carcinoma patients using anti-B7-H3 antibodies as monotherapy are currently ongoing (39, 42, 43). Although the functional outcome of this interaction has not been addressed, the identification of a receptor for B7-H3 represents an important leap forward that should inform therapeutic strategies, development of neutralizing antibodies. Further investigation of the crosstalk between the B7 receptors and IL20 cytokine family in the context of cancer are warranted.
Although most existing immunotherapeutic antibodies for treatment of cancer are thought to exert their blocking effects predominantly through T cells, NK cells also constitute promising targets for emerging immunotherapies (44, 45). The PVR/nectin receptors have emerged as important regulators of NK cell functions (15, 44). PVR is thought to modulate immune cell activation through complex protein interactions that involve the inhibitory receptors TIGIT and CD96, as well as the activating receptor CD226. TIGIT and CD96 are known to mediate several immunosuppressive functions that are counterbalanced by the competitive interaction with the activating receptor CD226 (44, 46, 47). Given the expression of the PVR/nectin proteins in tumors and their prominent role in immunosurveillance, modulation of these interactions represents major ongoing efforts for therapeutic development (14, 15). Interestingly, our results suggest that, unlike the well-defined PVR binder TIGIT, KIR2DL5A does not block the interaction with CD226, suggesting a different mechanism of action that should be addressed in future studies. Here we show that even in the presence of the potent activating receptor CD226, KIR2DL5 engages PVR leading to reduced NK cytotoxicity, thus suggesting that this interaction contributes to the balance of inhibitory and activating signals and may influence tumor cell killing.
The KIR receptor family plays a major role in regulating NK cell development, maturation and NK activation (48, 49) and is increasingly recognized as important modulators of autoimmune disease and infection. KIR2DL5 expression has been associated with faster progression in Alzheimer's disease patients, impaired responses to anti-viral therapies and increased susceptibility to viral infection (50–53), altogether pointing toward an important and yet poorly uncharacterized role for KIR2DL5 in immunity. Interestingly, the domain organization in the KIR2DL5 ECD more closely resembles that of KIR2DL4, an activating receptor that recognizes non-classical MHC class I, whereas the intracellular domain, characterized by one typical ITIM motif and one atypical ITIM, is only observed in a structurally unrelated member of the family, KIR3DL2 (16, 17). However, contrary to KIR2DL4, KIR3DL2 and most KIR receptors, KIR2DL5 do not appear to recognize any MHC alleles, and no other interacting partners have been reported. Altogether these observations suggest that KIR2DL5 has evolved to exert unique functions, possibly upon activation by ligands unrelated to MHC class I. Our findings that KIR2DL5 specifically interacts with PVR support this hypothesis and highlights a potential mechanism of action. Although KIR2DL5 has been shown to inhibit cytotoxicity of NK92 cells, the inhibitory mechanisms mediated by this receptor have not been explored in depth, primarily because of the lack of physiological ligands. Here, we show that KIR2DL5 plays an inhibitory function in NK cytolysis through direct interaction with PVR expressed on tumor cells. It is plausible that, similarly to TIGIT, KIR2DL5 also delivers a signal in the PVR-expressing cells triggering a functional response that should be addressed in future studies.
The PVR-KIR2DL5 interaction characterized in this work establishes a new association between the nectin and KIR receptor families, otherwise unrelated immune molecules. In fact, to our knowledge, this study is the first to reveal receptor-ligand interactions unrelated to MHC for a KIR receptor, suggesting functions for this family beyond MHC recognition. In healthy individuals, the expression of KIR2DL5 seems to be restricted to a low percentage of NK cells and to a lower extent CD8+ T cells (16). Nevertheless, whether KIR2DL5 expression is up-regulated in NK or T cells infiltrating the tumors remains to be addressed. Emerging preclinical evidence has shown promise in modulating the PVR/nectin family and understanding the dynamic interaction between these receptors will be crucial to further develop efficacious therapies. Notably, a recent phase 1 trial has shown a safety profile and potential for reinvigoration of NK cell responses upon administration of an anti-KIR2DL-1, -2 and -3 antibody for the treatment of solid tumors (45). Much remains to be understood about the molecular mechanisms by which the nectin and KIR families regulate immune function, and whether the crosstalk with the KIR family can be exploited to enhance immune responses. Notwithstanding these challenges, here we present a new workflow for extracellular interactome discovery and provide important insights into the biology of these receptor families that may ultimately inform therapeutic development.
Supplementary Material
Acknowledgments
We thank Genentech Reviewers for critically reading the manuscript. Thank you to Shaila Shrinagesh and Karen Bellici for their help compiling the receptor libraries. We are grateful to Hong Li and May Lin for providing the protein reagents, and to Lovejit Singh and Ya Naing Win for help with DNA reagents. Randy Yen and Daniel Tran have provided excellent technical assistance and support with automation procedures.
Footnotes
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- ePPI
- extracellular protein interactions
- AP/MS
- affinity purification/MS
- STM
- single transmembrane
- LAK
- lymphokine-activated killer.
REFERENCES
- 1. Hopkins A. L., and Groom C. R. (2002) The druggable genome. Nat. Rev. Drug Discov. 1, 727–730 [DOI] [PubMed] [Google Scholar]
- 2. Overington J. P., Al-Lazikani B., and Hopkins A. L. (2006) How many drug targets are there? Nat. Rev. Drug Discov. 5, 993–996 [DOI] [PubMed] [Google Scholar]
- 3. Wright G. J., Martin S., Bushell K. M., and Sollner C. (2010) High-throughput identification of transient extracellular protein interactions. Biochem. Soc. Trans. 38, 919–922 [DOI] [PubMed] [Google Scholar]
- 4. Martinez-Martin N. (2017) Technologies for proteome-wide discovery of extracellular host-pathogen interactions. J. Immunol. Res. 2017, 2197615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nooren I. M., and Thornton J. M. (2003) Diversity of protein-protein interactions. EMBO J. 22, 3486–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wright G. J. (2009) Signal initiation in biological systems: the properties and detection of transient extracellular protein interactions. Mol. Biosyst. 5, 1405–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gingras A. C., Abe K. T., and Raught B. (2019) Getting to know the neighborhood: using proximity-dependent biotinylation to characterize protein complexes and map organelles. Curr. Opin. Chem. Biol. 48, 44–54 [DOI] [PubMed] [Google Scholar]
- 8. Rees J. S., Li X. W., Perrett S., Lilley K. S., and Jackson A. P. (2015) Protein Neighbors and Proximity Proteomics. Mol. Cell. Proteomics 14, 2848–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen D. S., and Mellman I. (2013) Oncology meets immunology: the cancer-immunity cycle. Immunity 39, 1–10 [DOI] [PubMed] [Google Scholar]
- 10. Chen D. S., and Mellman I. (2017) Elements of cancer immunity and the cancer-immune set point. Nature 541, 321–330 [DOI] [PubMed] [Google Scholar]
- 11. Catakovic K., Klieser E., Neureiter D., and Geisberger R. (2018) T cell exhaustion: from pathophysiological basics to tumor immunotherapy. Cell Commun. Signal. 15, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martinez-Martin N., Marcandalli J., Huang C. S., Arthur C. P., Perotti M., Foglierini M., Ho H., Dosey A. M., Shriver S., Payandeh J., Leitner A., Lanzavecchia A., Perez L., and Ciferri C. (2018) An unbiased screen for human Cytomegalovirus identifies neuropilin-2 as a central viral receptor. Cell 174, 1158–1171 [DOI] [PubMed] [Google Scholar]
- 13. Taouji S., Dahan S., Bosse R., and Chevet E. (2009) Current screens based on the AlphaScreen technology for deciphering cell signalling pathways. Curr. Genomics 10, 93–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gao J., Zheng Q., Xin N., Wang W., and Zhao C. (2017) CD155, an onco-immunologic molecule in human tumors. Cancer Sci. 108, 1934–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Manieri N. A., Chiang E. Y., and Grogan J. L. (2017) TIGIT: A key inhibitor of the cancer immunity cycle. Trends Immunol. 38, 20–28 [DOI] [PubMed] [Google Scholar]
- 16. Estefania E., Flores R., Gomez-Lozano N., Aguilar H., Lopez-Botet M., and Vilches C. (2007) Human KIR2DL5 is an inhibitory receptor expressed on the surface of NK and T lymphocyte subsets. J. Immunol. 178, 4402–4410 [DOI] [PubMed] [Google Scholar]
- 17. Cisneros E., Moraru M., Gomez-Lozano N., Lopez-Botet M., and Vilches C. (2012) KIR2DL5: an orphan inhibitory receptor displaying complex patterns of polymorphism and expression. Front. Immunol. 3, 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bos A. B., Luan P., Duque J. N., Reilly D., Harms P. D., and Wong A. W. (2015) Optimization and automation of an end-to-end high throughput microscale transient protein production process. Biotechnol. Bioeng. 112, 1832–1842 [DOI] [PubMed] [Google Scholar]
- 19. Fairhead M., and Howarth M. (2015) Site-specific biotinylation of purified proteins using BirA. Methods Mol. Biol. 1266, 171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yasgar A., Jadhav A., Simeonov A., and Coussens N. P. (2016) AlphaScreen-based assays: ultra-high-throughput screening for small-molecule inhibitors of challenging enzymes and protein-protein interactions. Methods Mol. Biol. 1439, 77–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martinez-Martin N., Ramani S. R., Hackney J. A., Tom I., Wranik B. J., Chan M., Wu J., Paluch M. T., Takeda K., Hass P. E., Clark H., and Gonzalez L. C. (2016) The extracellular interactome of the human adenovirus family reveals diverse strategies for immunomodulation. Nat. Commun. 7, 11473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cheng X., Veverka V., Radhakrishnan A., Waters L. C., Muskett F. W., Morgan S. H., Huo J., Yu C., Evans E. J., Leslie A. J., Griffiths M., Stubberfield C., Griffin R., Henry A. J., Jansson A., Ladbury J. E., Ikemizu S., Carr M. D., and Davis S. J. (2015) Structure and interactions of the human programmed cell death 1 receptor. J. Biol. Chem. 288, 11771–11785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin D. Y., Tanaka Y., Iwasaki M., Gittis A. G., Su H. P., Mikami B., Okazaki T., Honjo T., Minato N., and Garboczi D. N. (2008) The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors. Proc. Natl. Acad. Sci. U.S.A. 105, 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lazar-Molnar E., Yan Q., Cao E., Ramagopal U., Nathenson S. G., and Almo S. C. (2008) Crystal structure of the complex between programmed death-1 (PD-1) and its ligand PD-L2. Proc. Natl. Acad. Sci. U.S.A. 105, 10483–10488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hsu J. M., Li C. W., Lai Y. J., and Hung M. C. (2018) Posttranslational modifications of PD-L1 and their applications in cancer therapy. Cancer Res. 78, 6349–6353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nagae M., Morita-Matsumoto K., Kato M., Kaneko M. K., Kato Y., and Yamaguchi Y. (2014) A platform of C-type lectin-like receptor CLEC-2 for binding O-glycosylated podoplanin and nonglycosylated rhodocytin. Structure 22, 1711–1721 [DOI] [PubMed] [Google Scholar]
- 27. Mullican S. E., Lin-Schmidt X., Chin C. N., Chavez J. A., Furman J. L., Armstrong A. A., Beck S. C., South V. J., Dinh T. Q., Cash-Mason T. D. Cavanaugh C. R., Nelson S., Huang C., Hunter M. J., and Rangwala S. M. (2007) GFRAL is the receptor for GDF15 and the ligand promotes weight loss in mice and nonhuman primates. Nat. Med. 23, 1150–1157 [DOI] [PubMed] [Google Scholar]
- 28. Coulthard M. G., Morgan M., Woodruff T. M., Arumugam T. V., Taylor S. M., Carpenter T. C., Lackmann M., and Boyd A. W. (2012) Eph/Ephrin signaling in injury and inflammation. Am. J. Pathol. 181, 1493–1503 [DOI] [PubMed] [Google Scholar]
- 29. Pardoll D. M. (2012) The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 12, 252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Castellanos J. R., Purvis I. J., Labak C. M., Guda M. R., Tsung A. J., Velpula K. K., and Asuthkar S. (2017) B7-H3 role in the immune landscape of cancer. Am J Clin Exp Immunol, 6, 66–75 [PMC free article] [PubMed] [Google Scholar]
- 31. Picarda E., Ohaegbulam K. C., and Zang X. (2016) Molecular pathways: targeting B7-H3 (CD276) for human cancer immunotherapy. Clin. Cancer Res. 22, 3425–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mahnke K., and Enk A. H. (2016) TIGIT-CD155 Interactions in melanoma: a novel co-inhibitory pathway with potential for clinical intervention. J. Invest. Dermatol. 136, 9–11 [DOI] [PubMed] [Google Scholar]
- 33. Chan C. J., Martinet L., Gilfillan S., Souza-Fonseca-Guimaraes F., Chow M. T., Town L., Ritchie D. S., Colonna M., Andrews D. M., and Smyth M. J. (2016) The receptors CD96 and CD226 oppose each other in the regulation of natural killer cell functions. Nat. Immunol. 15, 431–438 [DOI] [PubMed] [Google Scholar]
- 34. Nishiwada S., Sho M., Yasuda S., Shimada K., Yamato I., Akahori T., Kinoshita S., Nagai M., Konishi N., and Nakajima Y. (2015) Clinical significance of CD155 expression in human pancreatic cancer. Anticancer Res. 35, 2287–2297 [PubMed] [Google Scholar]
- 35. Chen L., and Flies D. B. (2013) Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat. Rev. Immunol. 13, 227–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Flem-Karlsen K., Fodstad O., Tan M., and Nunes-Xavier C. E. (2018) B7-H3 in cancer - beyond immune regulation. Trends Cancer 4, 401–404 [DOI] [PubMed] [Google Scholar]
- 37. Dong P., Xiong Y., Yue J., Hanley S. J. B., and Watari H. (2018) B7H3 as a promoter of metastasis and promising therapeutic target. Front. Oncol. 8, 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rutz S., Wang X., and Ouyang W. (2014) The IL-20 subfamily of cytokines–from host defense to tissue homeostasis. Nat. Rev. Immunol. 14, 783–795 [DOI] [PubMed] [Google Scholar]
- 39. Tekle C., Nygren M. K., Chen Y. W., Dybsjord I., Nesland J. M., Maelandsmo G. M., and Fodstad O. (2012) B7-H3 contributes to the metastatic capacity of melanoma cells by modulation of known metastasis-associated genes. Int. J. Cancer 130, 2282–2290 [DOI] [PubMed] [Google Scholar]
- 40. Wei C. C., Hsu Y. H., Li H. H., Wang Y. C., Hsieh M. Y., Chen W. Y., Hsing C. H., and Chang M. S. (2006) IL-20: biological functions and clinical implications. J. Biomed. Sci. 13, 601–612 [DOI] [PubMed] [Google Scholar]
- 41. Stenderup K., Rosada C., Worsaae A., Clausen J. T., and Norman Dam T. (2007) Interleukin-20 as a target in psoriasis treatment. Ann. N.Y. Acad. Sci. 1110, 368–381 [DOI] [PubMed] [Google Scholar]
- 42. Flem-Karlsen K., Tekle C., Andersson Y., Flatmark K., Fodstad O., and Nunes-Xavier C. E. (2017) Immunoregulatory protein B7-H3 promotes growth and decreases sensitivity to therapy in metastatic melanoma cells. Pigment Cell Melanoma Res. 30, 467–476 [DOI] [PubMed] [Google Scholar]
- 43. Wang J., Chong K. K., Nakamura Y., Nguyen L., Huang S. K., Kuo C., Zhang W., Yu H., Morton D. L., and Hoon D. S. (2013) B7-H3 associated with tumor progression and epigenetic regulatory activity in cutaneous melanoma. J. Invest. Dermatol. 133, 2050–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Martinet L., and Smyth M. J. (2015) Balancing natural killer cell activation through paired receptors. Nat. Rev. Immunol. 15, 243–254 [DOI] [PubMed] [Google Scholar]
- 45. Vey N., Karlin L., Sadot-Lebouvier S., Broussais F., Berton-Rigaud D., Rey J., Charbonnier A., Marie D., Andre P., Paturel C., Zerbib R., Bennouna J., Salles G., and Goncalves A. (2018) A phase 1 study of lirilumab (antibody against killer immunoglobulin-like receptor antibody KIR2D; IPH2102) in patients with solid tumors and hematologic malignancies. Oncotarget 9, 17675–17688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Blake S. J., Dougall W. C., Miles J. J., Teng M. W., and Smyth M. J. (2016) Molecular pathways: targeting CD96 and TIGIT for cancer immunotherapy. Clin. Cancer Res. 22, 5183–5188 [DOI] [PubMed] [Google Scholar]
- 47. Yu X., Harden K., Gonzalez L. C., Francesco M., Chiang E., Irving B., Tom I., Ivelja S., Refino C. J., Clark H., Eaton D., and Grogan J. L. (2009) The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat. Immunol. 10, 48–57 [DOI] [PubMed] [Google Scholar]
- 48. Cheent K., and Khakoo S. I. (2009) Natural killer cells: integrating diversity with function. Immunology. 126, 449–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Campbell K. S., and Purdy A. K. (2011) Structure/function of human killer cell immunoglobulin-like receptors: lessons from polymorphisms, evolution, crystal structures and mutations. Immunology 132, 315–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Carneiro V. L., Lemaire D. C., Bendicho M. T., Souza S. L., Cavalcante L. N., Angelo A. L., Freire S. M., Mendes C. M., Santana N., Lyra L. G., and Lyra A. C. (2010) Natural killer cell receptor and HLA-C gene polymorphisms among patients with hepatitis C: a comparison between sustained virological responders and non-responders. Liver Int. 30, 567–573 [DOI] [PubMed] [Google Scholar]
- 51. Tavis J. E., Donlin M. J., Aurora R., Fan X., and Di Bisceglie A. M. (2011) Prospects for personalizing antiviral therapy for hepatitis C virus with pharmacogenetics. Genome Med. 3, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chong M. S., Goh L. K., Lim W. S., Chan M., Tay L., Chen G., Feng L., Ng T. P., Tan C. H., and Lee T. S. (2013) Gene expression profiling of peripheral blood leukocytes shows consistent longitudinal downregulation of TOMM40 and upregulation of KIR2DL5A, PLOD1, and SLC2A8 among fast progressors in early Alzheimer's disease. J. Alzheimers Dis. 34, 399–405 [DOI] [PubMed] [Google Scholar]
- 53. Jones D. C., Peacock S., Hughes D., Traherne J. A., Allen R. L., Barnardo M. C., Friend P., Taylor C. J., Fuggle S., Trowsdale J., and Young N. T. (2014) Killer immunoglobulin-like receptor gene repertoire influences viral load of primary human cytomegalovirus infection in renal transplant patients. Genes Immun. 15, 562–568 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.