Abstract
Background: Emerging technologies such as smartphones and wearable sensors have enabled the paradigm shift to new patient-centered healthcare, together with recent mobile health (mHealth) app development. One such promising healthcare app is incision monitoring based on patient-taken incision images. In this review, challenges and potential solution strategies are investigated for surgical site infection (SSI) detection and evaluation using surgical site images taken at home.
Methods: Potential image quality issues, feature extraction, and surgical site image analysis challenges are discussed. Recent image analysis and machine learning solutions are reviewed to extract meaningful representations as image markers for incision monitoring. Discussions on opportunities and challenges of applying these methods to derive accurate SSI prediction are provided.
Conclusions: Interactive image acquisition as well as customized image analysis and machine learning methods for SSI monitoring will play critical roles in developing sustainable mHealth apps to achieve the expected outcomes of patient-taken incision images for effective out-of-clinic patient-centered healthcare with substantially reduced cost.
Keywords: surgical site infection, wound healing, wound management
The U.S. healthcare sector is transitioning from reactive care to proactive care, with the emphasis shifting to preventive measures and early interventions. Patients are increasingly empowered in this endeavor, both as a goal and as a result. Empowered patients may actively seek ways to engage in their healthcare using emerging technologies such as smartphones and wearable sensors. Recent studies have shown that patients are already using camera phones to e-mail and text incision photos to their providers [1], with access often prompted by providers. As a response to growing technology use among patients, a number of smartphone apps have been developed, such as the mPOWEr app (mobile Post-Operative Wound Evaluator; https://mpowercare.org), that enable patients to monitor their surgical sites for signs and symptoms of surgical site infection (SSI) at home and transmit photographs and self-reported incision and clinical observations to physicians. This generates promising new types of data that may address many challenging SSI problems, whereas the enormous scale of the data and the novelty also bring exciting intellectual challenges requiring close collaboration between medical professionals, statisticians, and computer scientists.
Increasingly available image data, together with other new types of data, naturally inspire the adoption of data-driven methods, such as machine learning, to extract information and make use of information-rich but statistically complex data. However, classic machine learning algorithms often have difficulty extracting semantic features directly from raw data. This phenomenon, commonly known as the semantic gap [2], requires assistance from domain knowledge for hand-crafted feature representations, on which machine learning models operate more effectively. In contrast, more recent deep learning approaches derive semantically meaningful representations, through construction of a hierarchy of features to represent a sophisticated concept. Deep learning requires less hand-engineered features and expert knowledge, and has recently achieved tremendous success in visual object recognition [3–6], face recognition and verification [7,8], object detection [9–12], image restoration and enhancement [13–18], clustering [19], emotion recognition [20], aesthetics and style recognition [21–24], scene understanding [25,26], speech recognition [27], machine translation [28], image synthesis [29], and even playing Go [30] and poker [31].
This promising progress has also motivated the widespread usage of deep learning in the medical image fields. For example, UNet [32] was first applied successfully in medical image segmentation. Esteva et al. [33] trained an end-to-end deep network and achieved dermatologist-level performance in classifying skin cancer. Moreover, deep learning is also applied successfully to many other tasks in the medical image context such as knee cartilage segmentation [34], diabetic retinopathy detection [35–37], lymph node detection [38–41], pulmonary nodule detection [42–44], brain lesion segmentation [45–48], and Alzheimer's disease classification [49–53]. A comprehensive survey can be found in Litjens et al. [54].
Our goal is to borrow strengths and translate the successes achieved those domains in incision image analysis for SSI, and further reinforce those domains with new methods developed to tackling new challenges in SSI. In this article, we propose a roadmap for developing incision image algorithms for automatic SSI detection and evaluation. Challenges persist, ranging from limited photo quality and uncontrolled imaging variations (e.g., light and angle), to the enormous heterogeneity of patients that calls for personalization in our algorithms [55]. We introduce both novelty and challenges in using incision images for SSI detection and evaluation and provide an overview of recent and related developments in computer vision, medical imaging processing, and analysis. We discuss a roadmap that could lead us to a systematic development of computational algorithms to detect and track SSI risk accurately using incision images captured by smartphones in a variety of conditions by a heterogeneous population.
Challenges in User-Generated Incision Images
As the National Patient Safety Agency has recently reported, 11% of serious medical events leading to mortality or substantial morbidity incidents are a function of unrecognized progression of disease [56,57]. This is particularly true for SSI because its natural history is still largely unknown. Whereas incision image data (and other types of user-generated data) offer great promise in helping clinicians capture disease progression with unprecedented and fine-grained resolution, it is challenging to extract risk-predictive patterns that correlate with the underlying disease progression because of challenges we discuss in the following subsections.
Quality issues in image acquisition
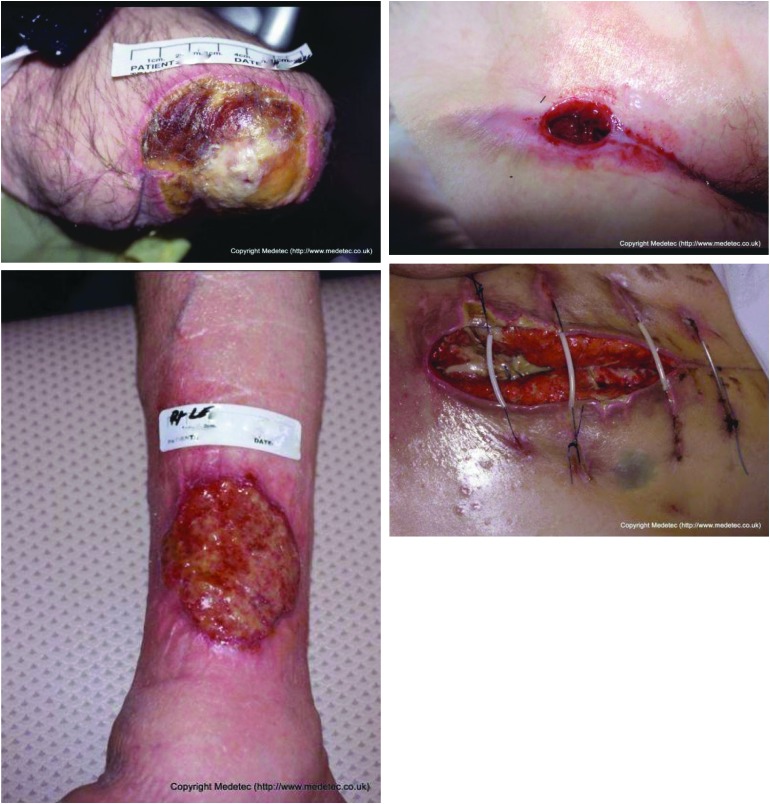
To extract clinically meaningful features from the user-generated incision images, we have to overcome the challenges presented by images taken by patients or family members who do not have clinical backgrounds and are taken using different types of devices in a variety of naturalistic environments. These challenges have not been addressed adequately in the existing literature. For example, images may be taken under different lighting conditions, and the positioning and size of the incision in the image may change between different images of the same incision (Fig. 1). Frequently, obstructing objects are included in incision images.
FIG. 1.
Example of the challenges faced characterizing surgical site images, including poor lighting conditions, obstructing objects such as hair, stitches, and thin films, as well as different camera angles and positioning. Color image is available online.
Several methods have been proposed to alleviate the issues above, including using apertures to ensure consistent lighting conditions, using transparent films over the incision, and using color or size fiduciaries such as a common object or uniform color template. One goal might be to develop automated guidance for patient–photographers, such as the image guidance provided in check-deposit banking applications, which may prompt the user to move closer or further away to optimize size and focus, or that may give warning of inadequate contrast or lighting. However, even with automated guidance, different healthcare during guidance will produce images with different styles [58]. Depending on whether these image differences are accounted for, these solutions may weaken the monitoring framework. In Shenoy et al. [59], the light condition is consistent in the incision images and incision is always centered. Under these conditions, their network achieved a high F1 score and this indicates the promising performance of available approaches after the quality issue is solved.
Feature extraction
It is apparent that image data provides an unprecedentedly rich source of data for SSI research, although this information may be masked by the wide variations in image acquisition. Important features extracted from images of the surgical site might include incision size, granularity, color, and morphology [60]. Image processing and analysis algorithms have been developed to extract these features directly from incision images. In addition to these traditional image analysis algorithms focusing on predefined image features, recent deep learning algorithms can automatically find feature representations that would be useful to monitor SSI, allowing for the discovery of new image features indicative of SSI progression [61].
Traditional teaching is that redness (erythema) adjacent to the incision indicates SSI, however, work by Sanger et al. [62] in characterizing the predictive value of provider incision observations of hospitalized patients did not show erythema to be predictive of infection, suggesting the importance of further investigation. There is no universal definition of imaged erythema, which would need to incorporate different skin tones, possibly individual patient responses to infection/inflammation, and imaging condition variability. Systematic characterization would be needed to understand the relation between SSI and either erythema as an extracted image feature, or as a deep learning feature representation of incision images.
Incision segmentation
To overcome the enormous variations in image acquisition and lay a reliable foundation for effective feature extraction, incision segmentation (spatial identification of the incision within the image) is a critical tool. An immediate goal of incision segmentation is to extract the size information of the incision area for SSI evaluation [63]. Segmentation can also be used to remove complex background distractions. Algorithms for incision segmentation have mostly been developed using incision images captured by skilled professionals with relatively well-controlled experiments, i.e., with consistent use of image acquisition device and procedure on a selected cohort. Early work involved applying a region growing method and automatic selection of the best channel [64] and developing active contour model in which the minimax principle was used adaptively to regularize the contour according to the local conditions in the incision image [65]. However, this method often contained many parameters that required manual adjustment for different images. Moreover, as we target user-generated incision images that come from amateur imaging devices (e.g., hand-held mobile phones), we expect image quality to be both decreased and variable [55]. In addition to the diverse incision characteristics, the imprecise definition of incision boundaries also complicates the problem. There are typically transition regions between incision and normal skin, but there is no clear consensus on image criteria to identify an incision boundary.
Recent Developments in Computer Vision and Medical Imaging Analysis
Deep learning
A basic neural network is composed of a set of perceptrons (artificial neurons), each of which maps inputs to output values with a simple activation function. Taking image classification as an example [3], a deep learning-based image classification system represents an object by gradually extracting edges, textures, and structures, from lower to middle-level hidden layers, which becomes more and more associated with the target semantic concept as the model grows deeper. Driven by the emergence of big data and hardware acceleration, the intricacy of data can be extracted with higher and more abstract level representation from raw inputs, gaining more power for deep learning to solve even traditionally intractable problems.
Among recent deep neural network architectures, convolutional neural networks (CNNs) and recurrent neural networks (RNNs) are the two main streams, differing in their connectivity patterns. Convolutional neural networks deploy convolution operations on hidden layers for weight sharing and parameter reduction. Convolutional neural networks can extract local information from grid-like input data and have mainly shown successes in computer vision and image processing, with many popular instances such as LeNet [66], AlexNet [3], VGG [67], GoogLeNet [68], and ResNet [69]. Recurrent neural networks are dedicated to processing sequential input data with variable length. Recurrent neural networks produce an output at each time step. The hidden neuron at each time step is calculated based on input data and hidden neurons at previous time step. To avoid vanishing/exploding gradients of RNNs in long-term dependency, long short-term memory (LSTM) [70] and gated recurrent unit (GRU) [71] with controllable gates are used widely in practical applications.
Recent deep networks have been shown to accomplish many tasks with substantial performance improvements over traditional image processing methods. For example, UNet [32], SegNet [72], ReSeg [73], MaskRCNN [74], and PSPNet [75] have achieved considerable performance progress in image segmentation. ResNet [69], GoogleNet [67], InceptionV3 [76], VGG [77], and NASNet [78] have achieved exceptional performance in image classification. Deep models can also be enhanced with particular robustness to real-world image degradations such as low resolution and noise [4,5,79], and therefore becoming more applicable to non-ideal quality photos from mobile devices. Readers with further interest are referred to the comprehensive deep learning textbook [80].
Feature extraction from incision images
Feature extraction methods for incision images have relied on identifying interpretable features indicative of incision progression. For example, a prevalent system widely used in incision assessment is the red-yellow-black system to identify granulation, slough, and necrotic tissue types [81]. Most traditional feature extraction methods have relied on first segmenting the incision into these distinct classes and then proceeding to extract features from these classes separately. For example, Mukherjee et al. [82] extracted features such as mean, standard deviation, skewness, kurtosis, and local contrast from 15 different color spaces. Using these features, a support vector machine (SVM) was used to segment the incision images into the red-yellow-black system for tissue type identification.
Many studies have indicated that color features are more useful for incision tissue classification compared with textural features [83–85], showing the effectiveness of the red-yellow-black system. However, textural features could further refine the incision assessment. Kolesnik and Fexa [86] studied the robustness of SVM models that only used color features, in comparison with SVM models that used both color and textural features for incision segmentation. Their study indicated that the textural features reduced the average magnitude of segmentation error compared to using only color features.
Moreover, color correction of the incision images has been shown to make feature extraction more robust. A study done by Wannous et al. [85] decomposed color correction into two distinct problems by obtaining a consistent color response via adjusting camera settings first and then determining the relation between the device-dependent color data and the device-independent color data. They conquered the second problem by placing a small Macbeth pattern in the camera field and achieved an improvement in accuracy from 68% to 76%.
Incision segmentation
As mentioned earlier, incision segmentation would be a critical tool to facilitate and enhance feature extraction from incision images. Particularly, machine learning-based methods [87] for incision segmentation have been found particularly promising to achieve full-automation and self-adaption. Early works include the use of SVM classifier, e.g., as in Kolesnik and Fexa [84], by treating incision segmentation as a binary classification task. Kolesnik and Fexa [86] further evaluated the robustness of SVM for incision segmentation. Their research indicates that it is not stable for new incision images and therefore not feasible for an automatic system. In addition, neural networks, Bayesian classifiers, and random forest decision trees were also utilized [88,89]. However, these methods relied highly on the choice of hand-crafted features that were created based on prior knowledge, which could only explore a limited amount of the image information.
The recent advance of deep learning brought tremendous developments to incision image processing, although many of them were not developed for user-generated incision image data. Since Long et al. [90] proposed fully convolutional networks (FCN), which extended the successful deep learning classification framework to segmentation task by replacing the fully connected layers with convolutional layers, many incision segmentation models were developed based on it. These models could be trained end-to-end and achieve superior performance. An encoder–decoder was utilized for the segmentation of incision by Lu et al. [91] and Wang et al. [92]. The WoundSeg, with higher performance and efficiency, was proposed by Liu et al. [63]. The architecture of the WoundSeg is shown in Figure 2. It modifies the FCN [90] structure by adding a skip connection from previous feature map with higher resolution for making the segmentation result finer. They also leverage data augmentation and post-processing, which together improve the accuracy to 98.12%. However, their dataset is taken by a professional in a hospital environment and the quality of images is ensured.
FIG 2.
The architecture of WoundSeg [63]. Color image is available online.
Because all these methods used supervised learning that required a lot of expert labeled data, it is an expensive approach if the labeling cost is considerable. Many studies also explored unsupervised methods such as clustering in incision segmentation. Yadav et al. [93] compared k-means and fuzzy c-means on Dr and Db color channels. The spectral approach based on the affinity matrix was explored in Dhane et al. [94]. Dhane et al. [55] further proposed fuzzy spectral clustering (FSC) that constructed similarity matrix with gray-based fuzzy similarity measure using spatial knowledge of an image. How scalable and practical these methods for analyzing user-generated incision images to tackle SSI problems demands many further studies and validations.
Discussion
With the rapid development of machine learning and medical image analysis, many developing techniques can potentially be adopted to address the existing challenges. In this section, we discuss some of the possibilities.
Learning-based image processing and enhancement
As we have elaborated earlier, incision images taken by amateur users are, in general, of low quality compared with professional skin scans. Importantly, they would display huge variations of lighting conditions, e.g., because of shadows or over-/underexposures, which will greatly jeopardize both feature extraction and incision region segmentation. Many deep learning based enhancement algorithms, which are trained to regress low-quality images to enhanced versions, could potentially be used. In other words, it is to map those low-quality images into high-quality incision image templates created by incision image data collected by professionals such that the low-quality image could be automatically calibrated, de-noised, enhanced, and imputed. Although this is a promising idea, more challenges arise from the fact that in the field of incision image analysis, there currently does not exist, nor will it be easy to collect, a large set of paired low-quality/enhanced images for training such models.
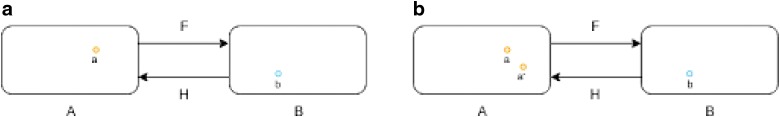
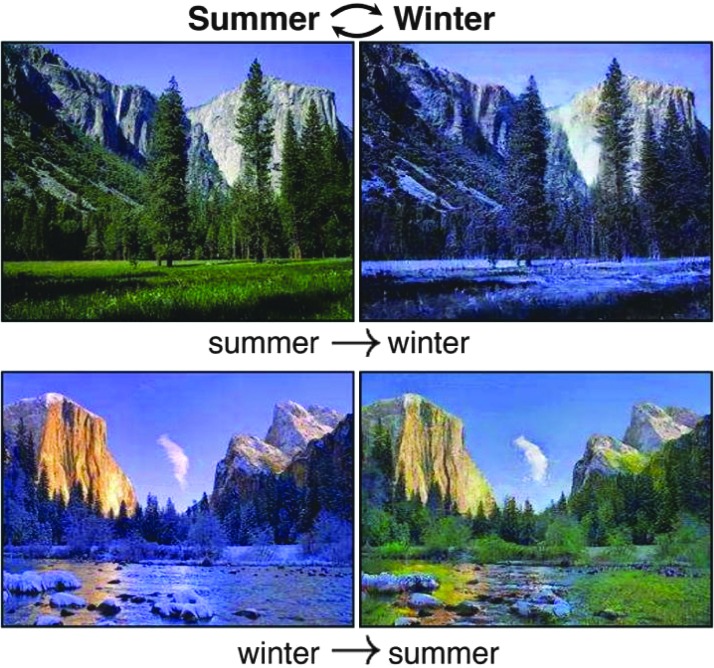
A tentative solution is to treat this as an image domain translation problem, in which domain is formed by a set of images with the same pattern. For example, landscape pictures taken in summer and winter can be treated as from two domains. The summer pictures are from domain A and the winter pictures are from domain B as shown in Figure 3(a). For each summer picture a from domain A, there exists a corresponding winter picture b in domain B, and thus, a can be transformed to b through a function F. The function F is called as domain translation function and the function H is the domain translation function from B to A. A translation demonstration between them is shown in Figure 4. Whereas in many applications, the reality is that there is no paired sample from domain A and B, we could resort to optimization solutions in a statistical framework. For example, as shown in Zhu et al. [95], a novel consistency loss was developed to measure the distance of the sample a and a 0 = H(F(a)) as shown in Figure 3(b). For good domain translation functions F and H, the distance between a and a 0 should be small. Therefore, with unpaired data from two domains, we can optimize the translation functions through minimizing the consistency loss. Similar techniques were lately applied to natural image enhancement applications [96], and we expect the idea to also be helpful for incision image enhancement.
FIG 3.
(a) Summer picture and (b) corresponding winter picture while F and H are translation functions. (b) Consistency loss measures the distance between a and a 0, in which a 0 = H(F(a)). Color image is available online.
FIG 4.
The domain translation between summer and winter pictures [95]. Color image is available online.
Interactive image capture
An important question is whether smartphone-based systems have any disadvantage in image capture quality compared with traditional medical workstations. Kumar et al. [97] showed that teleophthalmology images taken by a smartphone had a near-identical quality to images taken by a standard medical workstation. This suggests that images captured using smartphones do not suffer from any major quality issues compared with those taken using medical workstations, as long as sufficient quality control is put in place.
A solution that can potentially increase the quality of SSI images acquisition is to interactively guide the user when taking the SSI images. Indeed, a system that aids patients and medical personnel in adjusting/finding correct lighting conditions, exposure, and incision location could solve most of the image quality issues we highlighted previously. Such a system not only needs to have sufficient quality control mechanisms to obtain accurate SSI incision features, but also needs to be user-friendly. Recent medical image capture systems have utilized mobile phone-based applications to facilitate image capture, such as in teleophthalmology, clinical microscopy, and diabetic wound treatment [97–99]. Such interactive image capture methods have many notable advantages. Agu et al. [99] noted that the use of smartphones for medical image capture had the benefits of easy deployment as smartphone applications are easily developed and installed, and accessibility as smartphones are conveniently available to any person. In addition, new hardware can be timely leveraged to help SSI monitoring because smartphone hardware is upgraded frequently.
For the case of SSI image capture, existing incision assessment systems have relied on using peripheral or ancillary devices to control the lighting and incision positions. Moreover, thin films that overlay graph paper with mesh grids are typically used when measuring incision size [100–102]. An interactive incision image capturing system would need to have these mechanisms in place to ensure proper image quality.
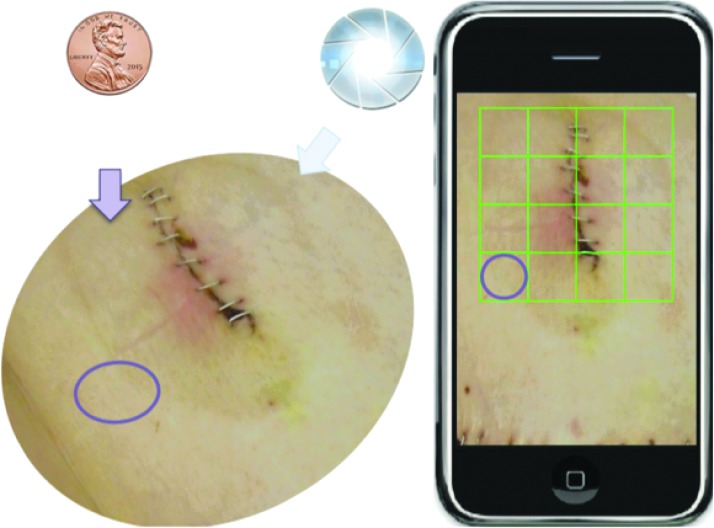
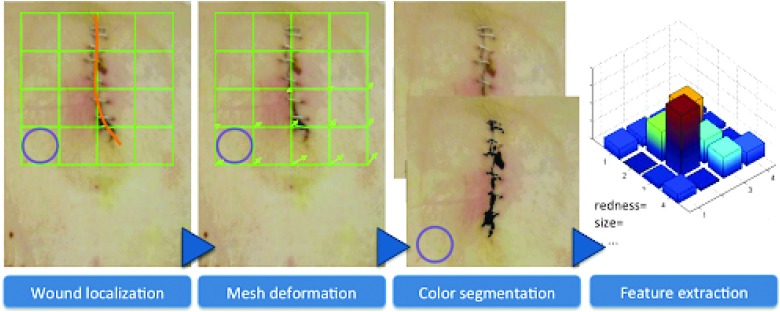
Such an interactive system can be shown in Figures 5 and 6. As shown in Figure 5, a coin is placed near the incision image as a size indicator, while a virtual mesh grid is used to replace the graphpaper thin film traditionally used. After the picture is taken, the incision is localized as shown in Figure 6. The user is asked to draw the contour of the incision region to allow a deformation of the mesh grid to normalize the incision region size. With this normalized image, color segmentation can be conducted, and interpretable feature of the incision can be extracted. Lighting conditions can also be normalized, and features can be readily extracted from the SSI images for effective and reliable SSI monitoring.
FIG 5.
Interactive procedure for image capture using mobile phones. Color image is available online.
FIG 6.
Envisioned interactive image capture system. Color image is available online.
Incision image segmentation and assessment
As mentioned earlier, challenges in segmenting and assessing incision images using deep learning center around three main issues: the labeled data volume, the segmentation label quality, and the interpretability and domain knowledge integration.
To overcome the first challenge, we resort to data augmentation, a widely utilized tool in deep learning to artificially increase the labeled data volume and diversity, without extra data collection efforts [3]. The common means relies on identifying label preserving transformations, i.e., variations known to exist in real data but not affecting semantic annotations. Moreover, models trained with such augmented data will also gain invariance to the selected types of variations. For example, one can alter the lighting conditions as well as color tones of labeled incision images for robust feature learning to varying lighting and skin color. For example, following Wang et al. [21], we can apply γ correction to the luminance channel with random γ values; a gradient in illumination could be further added to simulate an oriented light source. The lighting augmentation will not affect either image or pixel-level annotations.
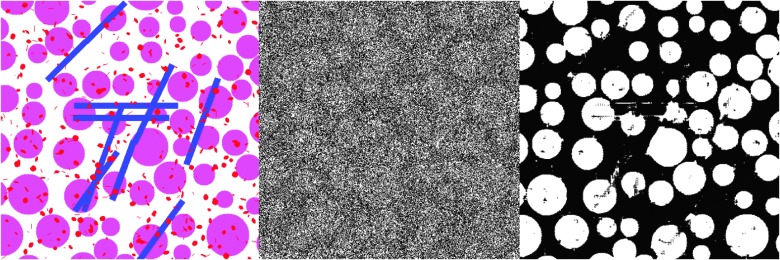
The second challenge of segmentation label quality results from the fact that pixel-wise annotations are often generated using semi-automatic methods such as the watershed algorithm; hence there could be incorrectly labeled pixels, making the supervision information for the segmentation task “noisy.” Motivated by recent success of training deep models with noisy labels [103,104], we could adopt the bootstrap strategy of Reed et al. [103] and introduce a noise layer into the deep image segmentation model as done by Sukhbaatar et al. [104] when we use noisy segmentation supervision. Our preliminary result [107] has shown promising potential of this approach, i.e., we have modified the U-net with a noise layer, which can take noisy segmentation results (Fig. 7, middle) for training and segment synthetic images (Fig. 7, left). Figure 7 (right) shows that such a modification can achieve promising segmentation results.
FIG 7.
Deep image segmentation with noisy labels: (left) synthetic testing image example; (middle) simulated noisy segmented image; (right) image segmentation by modified U-net using the simulated noisy segmentation. Color image is available online.
Eventually, another crucial capability for deep learning models to address SSI is how to make their predictive results interpretable to human. In this regard, many recent works from the interpretable deep learning fields show promise. For example, sensitivity analysis [108] tried to explain a prediction based on the model's locally evaluated gradient (partial derivative). Matching similar image parts could also be a promising method as done by Chen et al. [61]. We refer the readers to a literature review by Chakraborty et al. [107]. Different from existing incision image assessment methods that were either rule-based [100,108,109] or data-driven [63], we advocate to draw the complementary power of knowledge-based feature design and data-driven feature learning to maximize the information extraction from the incision images with robust performances.
Acknowledgments
This work was supported by U.S. Centers for Disease Control and Prevention (CDC) award #200-2016-91803 through the Safety and Healthcare Epidemiology Prevention Research Development (SHEPheRD) Program, which is managed by the Division of Healthcare Quality Promotion. The content is solely the responsibility of the authors and does not represent the official views of the CDC.
Author Disclosure Statement
All authors report no competing financial interests exist.
References
- 1. Gunter R, Chouinard S, Fernandes-Taylor S, et al. Current use of telemedicine for post-discharge surgical care: A systematic review. Am Coll Surg 2016;222:915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhao R, Grosky WI. Narrowing the semantic gap—Improved text-based web document retrieval using visual features. IEEE Trans Multimedia 2002;4:189–200 [Google Scholar]
- 3. Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. In: Bartlett P, Pereira F, Burges CJC, et al. (eds): Advances in Neural Information Processing Systems 25. (NIPS 2012). 2012:1097–1105 [Google Scholar]
- 4. Wang Z, Chang S, Yang Y, et al. Studying very low resolution recognition using deep networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2016:4792–4800 [Google Scholar]
- 5. Liu D, Cheng B, Wang Z, et al. Enhance visual recognition under adverse conditions via deep networks. IEEE Trans Image Proc (in press) [DOI] [PubMed] [Google Scholar]
- 6. Wu Z, Wang Z, Wang Z, Jin H. Towards privacy-preserving visual recognition via adversarial training: A pilot study. In: Proceedings of the European Conference on Computer Vision (ECCV) 2018:606–624 [Google Scholar]
- 7. Bodla N, Zheng J, Xu H, et al. Deep heterogeneous feature fusion for template-based face recognition. In: 2017 IEEE Winter Conference on Applications of Computer Vision (WACV) March 24, 2017 IEEE 2017:586–595 [Google Scholar]
- 8. Ranjan R, Bansal A, Xu H, et al. Crystal loss and quality pooling for unconstrained face verification and recognition. arXiv preprint arXiv:1804.01159. April 3, 2018
- 9. Ren S, He K, Girshick R, Sun J. Faster R-CNN: Towards real-time object detection with region proposal networks. In: Cortes C, Lawrence ND, Lee DD, et al. (eds): Advances in Neural Information Processing Systems 28. ACM. 2015:91–99 [Google Scholar]
- 10. Yu J, Jiang Y, Wang Z, et al. Unitbox: An advanced object detection network. In: Proceedings of the 2016 ACM on Multimedia Conference ACM 2016:516–520 [Google Scholar]
- 11. Gao J, Wang Q, Yuan Y. Embedding structured contour and location prior in Siamesed fully convolutional networks for road detection. In: 2017 IEEE International Conference on Robotics and Automation (ICRA) IEEE 2017:219–224 [Google Scholar]
- 12. Xu H, Lv X, Wang X, Ren Z, Bodla N, Chellappa R. Deep regionlets for object detection. In: Proceedings of the European Conference on Computer Vision (ECCV) 2018:798–814 [Google Scholar]
- 13. Timofte R, Agustsson E, Van Gool L, et al. Ntire 2017 challenge on single image super-resolution: Methods and results. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW) IEEE 2017:1110–1121 [Google Scholar]
- 14. Li B, Peng X, Wang Z, et al. Aod-net: All-in-one dehazing network. In: Proceedings of the IEEE International Conference on Computer Vision 2017:4770–4778 [Google Scholar]
- 15. Li B, Peng X, Wang Z, Xu J, Feng D. An all-in-one network for dehazing and beyond. arXiv preprint arXiv:170706543. 2017
- 16. Li B, Peng X, Wang Z, et al. End-to-end united video dehazing and detection. arXiv preprint arXiv:170903919. 2017
- 17. Liu D, Wen B, Jiao J, et al. Connecting image denoising and high-level vision tasks via deep learning. arXiv preprint arXiv:180901826. 2018 [DOI] [PubMed]
- 18. Prabhu R, Yu X, Wang Z, et al. U-finger: Multi-scale dilated convolutional network for fingerprint image denoising and inpainting. arXiv preprint arXiv:180710993. 2018
- 19. Wang Z, Chang S, Zhou J, et al. Learning a task-specific deep architecture for clustering. In: Proceedings of the 2016 SIAM International Conference on Data Mining, June 30, 2016. Society for Industrial and Applied Mathematics, 2016:360–377 [Google Scholar]
- 20. Cheng B, Wang Z, Zhang Z, et al. Robust emotion recognition from low quality and low bit rate video: A deep learning approach. arXiv preprint arXiv:170903126. 2017
- 21. Wang Z, Yang J, Jin H, et al. Deepfont: Identify your font from an image. In: Proceedings of the 23rd ACM International Conference on Multimedia ACM 2015:451–459 [Google Scholar]
- 22. Wang Z, Yang J, Jin H, et al. Real-world font recognition using deep network and domain adaptation. arXiv preprint arXiv:150400028. 2015
- 23. Wang Z, Chang S, Dolcos F, et al. Brain-inspired deep networks for image aesthetics assessment. arXiv preprint arXiv:160104155. 2016
- 24. Huang TS, Brandt J, Agarwala A, et al. Deep learning for font recognition and retrieval. In: Roopaei M, Rad P. (eds): Applied Cloud Deep Semantic Recognition: Advanced Anomaly Detection. Boca Raton, FL: Taylor & Francis Group, 2018:109–130 [Google Scholar]
- 25. Farabet C, Couprie C, Najman L, LeCun Y. Learning hierarchical features for scene labeling. IEEE Trans Pattern Anal Machine Intell 2013;35:1915–1929 [DOI] [PubMed] [Google Scholar]
- 26. Wang Q, Gao J, Yuan Y. A joint convolutional neural networks and context transfer for street scenes labeling. IEEE Trans Intell Transp Syst 2017;19:1457–1470 [Google Scholar]
- 27. Saon G, Kuo HKJ, Rennie S, Picheny M. The IBM 2015 English conversational telephone speech recognition system. arXiv preprint arXiv:150505899. 2015
- 28. Sutskever I, Vinyals O, Le QV. Sequence to sequence learning with neural networks. In: Advances in Neural Information Processing Systems 27 2014:3104–3112 [Google Scholar]
- 29. Goodfellow I, Pouget-Abadie J, Mirza M, et al. Generative adversarial nets. In: Advances in Neural Information Processing Systems 27 2014:2672–2680 [Google Scholar]
- 30. Silver D, Huang A, Maddison CJ, et al. Mastering the game of Go with deep neural networks and tree search. Nature 2016;529:484–489 [DOI] [PubMed] [Google Scholar]
- 31. Moravčík M, Schmid M, Burch N, et al. Deepstack: Expert-level artificial intelligence in no-limit poker. arXiv preprint arXiv:170101724. 2017 [DOI] [PubMed]
- 32. Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. In: International Conference on Medical image Computing and Computer-Assisted Intervention Springer 2015:234–241 [Google Scholar]
- 33. Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Prasoon A, Petersen K, Igel C, et al. Deep feature learning for knee cartilage segmentation using a triplanar convolutional neural network. In: International Conference on Medical Image Computing and Computer-Assisted Intervention Springer 2013:246–253 [DOI] [PubMed] [Google Scholar]
- 35. Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016;316:2402–2410 [DOI] [PubMed] [Google Scholar]
- 36. Gargeya R, Leng T. Automated identification of diabetic retinopathy using deep learning. Ophthalmology 2017;124:962–969 [DOI] [PubMed] [Google Scholar]
- 37. Abràmoff MD, Lou Y, Erginay A, et al. Improved automated detection of diabetic retinopathy on a publicly available dataset through integration of deep learning. Invest Ophthalmol Vis Sci 2016;57:5200–5206 [DOI] [PubMed] [Google Scholar]
- 38. Roth HR, Lu L, Seff A, et al. A new 2.5 D representation for lymph node detection using random sets of deep convolutional neural network observations. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer: 2014:520–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roth HR, Lu L, Liu J, et al. Improving computer-aided detection using convolutional neural networks and random view aggregation. IEEE Trans Med Imaging 2016;35:1170–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shin HC, Roth HR, Gao M, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging 2016;35:1285–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Litjens G, Sánchez CI, Timofeeva N, et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci Rep 2016;6:26286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hua KL, Hsu CH, Hidayati SC, et al. Computer-aided classification of lung nodules on computed tomography images via deep learning technique. Onco Targets Therapy 2015;8:2015–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Setio AAA, Ciompi F, Litjens G, et al. Pulmonary nodule detection in CT images: False positive reduction using multiview convolutional networks. IEEE Trans Med Imaging 2016;35:1160–1169 [DOI] [PubMed] [Google Scholar]
- 44. Ciompi F, Chung K, Van Riel SJ, et al. Towards automatic pulmonary nodule management in lung cancer screening with deep learning. Sci Rep 2017;7:46479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kamnitsas K, Ledig C, Newcombe VF, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal 2017;36:61–78 [DOI] [PubMed] [Google Scholar]
- 46. Kamnitsas K, Baumgartner C, Ledig C, et al. Unsupervised domain adaptation in brain lesion segmentation with adversarial networks. In: International Conference on Information Processing in Medical Imaging Springer 2017:597–609 [Google Scholar]
- 47. Ghafoorian M, Mehrtash A, Kapur T, Transfer learning for domain adaptation in MRI: Application in brain lesion segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention Springer 2017:516–524 [Google Scholar]
- 48. Chen H, Dou Q, Yu L, et al. VoxResNet: Deep voxelwise residual networks for brain segmentation from 3D MR images. NeuroImage 2018;170:446–455 [DOI] [PubMed] [Google Scholar]
- 49. Suk HI, Shen D. Deep learning-based feature representation for AD/MCI classification. In: International Conference on Medical Image Computing and Computer-Assisted Intervention Springer 2013:583–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hosseini-Asl E, Gimel'farb G, El-Baz A. Alzheimer's disease diagnostics by a deeply supervised adaptable 3D convolutional network. arXiv preprint arXiv:160700556. 2016 [DOI] [PubMed]
- 51. Payan A, Montana G. Predicting Alzheimer's disease: A neuroimaging study with 3D convolutional neural networks. arXiv preprint arXiv:150202506. 2015
- 52. Ortiz A, Munilla J, Gorriz JM, Ramirez J. Ensembles of deep learning architectures for the early diagnosis of the Alzheimer's disease. Int J Neural Syst 2016;26:1650025. [DOI] [PubMed] [Google Scholar]
- 53. Sarraf S, Tofighi G. Classification of Alzheimer's disease using fMRI data and deep learning convolutional neural networks. arXiv preprint arXiv:160308631. 2016
- 54. Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60–88 [DOI] [PubMed] [Google Scholar]
- 55. Dhane DM, Maity M, Mungle T, et al. Fuzzy spectral clustering for automated delineation of chronic wound region using digital images. Comput Biol Med 2017;89:551–560 [DOI] [PubMed] [Google Scholar]
- 56. Beck A, Tetruashvili L. Safer Care for the Acutely Ill Patient: Learning from Serious Incidents. National Patient Safety Agency. 2013
- 57. Gaynes RP, Culver DH, Horan TC, et al. Surgical site infection (SSI) rates in the United States, 1992–1998: The National Nosocomial Infections Surveillance System basic SSI risk index. Clin Infect Dis 2001;33(Suppl 2):S69–S77 [DOI] [PubMed] [Google Scholar]
- 58. Tartari E, Weterings V, Gastmeier P, et al. Patient engagement with surgical site infection prevention: An expert panel perspective. Antimicrob Resist Infect Control 2017;6:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shenoy VN, Foster E, Aalami L, et al. Deepwound: Automated postoperative wound assessment and surgical site surveillance through convolutional neural networks. In: 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) IEEE 2018:1017–1021 [Google Scholar]
- 60. van Walraven C, Musselman R. The Surgical Site Infection Risk Score (SSIRS): A model to predict the risk of surgical site infections. PLoS One 2013;8:e67167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chen C, Li O, Barnett A, et al. This looks like that: Deep learning for interpretable image recognition. arXiv preprint arXiv:180610574. 2018
- 62. Sanger PC, van Ramshorst GH, Mercan E, et al. A prognostic model of surgical site infection using daily clinical wound assessment. J Am Coll Surg 2016;223:259-270.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Liu X, Wang C, Li F, et al. A framework of wound segmentation based on deep convolutional networks. In: 2017 10th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISPBMEI) IEEE 2017:1–7 [Google Scholar]
- 64. Perez AA, Gonzaga A, Alves JM. Segmentation and analysis of leg ulcers color images. In: Proceedings International Workshop on Medical Imaging and Augmented Reality IEEE 2001:262–266 [Google Scholar]
- 65. Jones TD, Plassmann P. An active contour model for measuring the area of leg ulcers. IEEE Trans Med Imaging 2000;19:1202–1210 [DOI] [PubMed] [Google Scholar]
- 66. LeCun Y, et al. LeNet-5, convolutional neural networks. http://yann lecun com/exdb/lenet 2015. (Last accessed July26, 2019).
- 67. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:14091556. 2014
- 68. Szegedy C, Liu W, Jia Y, et al. Going deeper with convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2015:1–9 [Google Scholar]
- 69. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Rrecognition 2016:770–778 [Google Scholar]
- 70. Gers FA, Schmidhuber J, Cummins F. Learning to forget: Continual prediction with LSTM. Neural Comput 2000;12:2451–2471 [DOI] [PubMed] [Google Scholar]
- 71. Chung J, Gulcehre C, Cho K, Bengio Y. Empirical evaluation of gated recurrent neural networks on sequence modeling. arXiv preprint arXiv:14123555. 2014
- 72. Badrinarayanan V, Kendall A, Cipolla R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. arXiv preprint arXiv:151100561. 2015 [DOI] [PubMed]
- 73. Visin F, Ciccone M, Romero A, et al. Reseg: A recurrent neural network-based model for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops 2016:41–48 [Google Scholar]
- 74. He K, Gkioxari G, Dollár P, Girshick R. Mask R-CNN. In: 2017 IEEE International Conference on Computer Vision (ICCV) 2017:2980–2988 [Google Scholar]
- 75. Zhao H, Shi J, Qi X, et al. Pyramid scene parsing network. In: IEEE Conference on Computer Vision and Pattern Recognition (CVPR) IEEE 2017:2881–2890 [Google Scholar]
- 76. Szegedy C, Vanhoucke V, Ioffe S, et al. Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2016:2818–2826 [Google Scholar]
- 77. Szegedy C, Ioffe S, Vanhoucke V, Alemi AA. Inception-v4, inception-resnet and the impact of residual connections on learning. In: Thirty-First AAAI Conference on Artificial Intelligence February 12, 2017 [Google Scholar]
- 78. Zoph B, Vasudevan V, Shlens J, Le QV. Learning transferable architectures for scalable image recognition. arXiv preprint arXiv:170707012. 2017
- 79. Liu D, Wen B, Liu X, et al. When image denoising meets high-level vision tasks: A deep learning approach. arXiv preprint arXiv:170604284. 2017
- 80. Goodfellow I, Bengio Y, Courville A. Deep learning. Cambridge MA: MIT Press, 2016 [Google Scholar]
- 81. Krasner D. Wound care how to use the red-yellow-black system. Am J Nurs 1995;95:44–47 [PubMed] [Google Scholar]
- 82. Mukherjee R, Manohar DD, Das DK, et al. Automated tissue classification framework for reproducible chronic wound assessment. Biomed Res Int 2014;2014:851582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wang L, Pedersen PC, Agu E, et al. Area determination of diabetic foot ulcer images using a cascaded two-stage SVM-based classification. IEEE Trans Biomed Eng 2017;64:2098–2109 [DOI] [PubMed] [Google Scholar]
- 84. Kolesnik M, Fexa A. Segmentation of wounds in the combined color-texture feature space. Proc SPIE 5370, Medical Imaging 2004: Image Processing 2004:549–557 [Google Scholar]
- 85. Wannous H, Lucas Y, Treuillet S. Efficient SVM classifier based on color and texture region features for wound tissue images. Proc SPIE 6915, Medical Imaging 2008: Computer-Aided Diagnosis 2008:69152T [Google Scholar]
- 86. Kolesnik M, Fexa A. How robust is the SVM wound segmentation? In: Proceedings of the 7th Nordic Signal Processing Symposium-NORSIG 2006 IEEE 2006:50–53 [Google Scholar]
- 87. Zhao L, Li K, Wang M, et al. Automatic cytoplasm and nuclei segmentation for color cervical smear image using an efficient gapsearch MRF. Comput Biol Med 2016;71:46–56 [DOI] [PubMed] [Google Scholar]
- 88. Veredas F, Mesa H, Morente L. Binary tissue classification on wound images with neural networks and Bayesian classifiers. IEEE Trans Med Imaging 2010;29:410–427 [DOI] [PubMed] [Google Scholar]
- 89. Veredas FJ, Luque-Baena RM, Martín-Santos FJ, et al. Wound image evaluation with machine learning. Neurocomputing 2015;164:112–122 [Google Scholar]
- 90. Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. In: IEEE Conference on Computer Vision and Pattern Recognition 2015:3431–3440 [DOI] [PubMed] [Google Scholar]
- 91. Lu H, Li B, Zhu J, et al. Wound intensity correction and segmentation with convolutional neural networks. Concurr Comput 2017;29:e3927 [Google Scholar]
- 92. Wang C, Yan X, Smith M, et al. A unified framework for automatic wound segmentation and analysis with deep convolutional neural networks. In: 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) IEEE 2015:2415–2418 [DOI] [PubMed] [Google Scholar]
- 93. Yadav MK, Manohar DD, Mukherjee G, Chakraborty C. Segmentation of chronic wound areas by clustering techniques using selected color space. J Med Imaging Health Inform 2013;3:22–29 [Google Scholar]
- 94. Dhane DM, Krishna V, Achar A, et al. Spectral clustering for unsupervised segmentation of lower extremity wound beds using optical images. J Med Syst 2016;40:207. [DOI] [PubMed] [Google Scholar]
- 95. Zhu JY, Park T, Isola P, Efros AA. Unpaired imageto-image translation using cycle-consistent adversarial networks. In: Proceedings of the IEEE International Conference on Computer Vision 2017:2223–2232 [Google Scholar]
- 96. Zhu H, Chanderasekh V, Lim LLJH. Singe image rain removal with unpaired information: A differentiable programming perspective. 2019. https://www.aaai.org/ojs/index.php/AAAI/article/view/497/ (last accessed August8, 2019)
- 97. Kumar S, Wang EH, Pokabla MJ, Noecker RJ. Teleophthalmology assessment of diabetic retinopathy fundus images: Smartphone versus standard office computer workstation. Telemed J E Health 2012;18:158–162 [DOI] [PubMed] [Google Scholar]
- 98. Breslauer DN, Maamari RN, Switz NA, et al. Mobile phone based clinical microscopy for global health applications. PloS One 2009;4:e6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Agu E, Pedersen P, Strong D, et al. The smartphone as a medical device: Assessing enablers, benefits and challenges. In: 2013 IEEE International Workshop of Internet-of-Things Networking and Control (IoT-NC) IEEE 2013:48–52 [Google Scholar]
- 100. Nejati H, Pomponiu V, Do TT, et al. Smartphone and mobile image processing for assisted living: Health-monitoring apps powered by advanced mobile imaging algorithms. IEEE Signal Proc Mag 2016;33:30–48 [Google Scholar]
- 101. Poon TWK, Friesen MR. Algorithms for size and color detection of smartphone images of chronic wounds for healthcare applications. IEEE Access 2015;3:1799–1808 [Google Scholar]
- 102. Wang L, Pedersen PC, Strong DM, et al. Smartphone-based wound assessment system for patients with diabetes. IEEE Trans Biomed Eng 2015;62:477–488 [DOI] [PubMed] [Google Scholar]
- 103. Reed S, Lee H, Anguelov D, et al. Training deep neural networks on noisy labels with bootstrapping. arXiv preprint arXiv:1412.6596. December 20, 2014
- 104. Sukhbaatar S, Bruna J, Paluri M, et al. Training convolutional networks with noisy labels. arXiv preprint arXiv:1406.2080. June 9, 2014
- 105. Li W, Qian X, Ji J. Noise-tolerant deep learning for histopathological image segmentation. In: 2017 IEEE International Conference on Image Processing (ICIP) IEEE 2017:3075–3079 [Google Scholar]
- 106. Samek W, Wiegand T, Müller KR. Explainable artificial intelligence: Understanding, visualizing and interpreting deep learning models. arXiv preprint arXiv:170808296. 2017
- 107. Chakraborty S, Tomsett R, Raghavendra R, et al. Interpretability of deep learning models: A survey of results. In: 2017 IEEE SmartWorld, Ubiquitous Intelligence & Computing, Advanced & Trusted Computed, Scalable Computing & Communications, Cloud & Big Data Computing, Internet of People and Smart City Innovation (SmartWorld/SCALCOM/UIC/ATC/CBDCom/IOP/SCI) IEEE 2017:1–6 [Google Scholar]
- 108. Wang L, Pedersen P, Strong D, et al. Smartphone-based wound assessment system for patients with diabetes. IEEE Trans Biomed Eng 2015;62:477–488 [DOI] [PubMed] [Google Scholar]
- 109. Poon T, Friesen M. Algorithms for size and color detection of smartphone images of chronic wounds for healthcare applications. IEEE Access 2015;3:1799–1808 [Google Scholar]