Abstract
Radiation-induced lung injury is a major dose-limiting toxicity that occurs due to thoracic radiotherapy. Metabolomics is a powerful quantitative measurement of low-molecular-weight metabolites in response to environmental disturbances. However, the metabolomic profiles of radiation-induced lung injury have not been reported yet. In this study, male Sprague-Dawley rats were subjected to a single dose of 10 or 20 Gy irradiation to the right lung. One week after radiation, the obvious morphological alteration of lung tissues after radiation was observed by hematoxylin and eosin staining through a transmission electron microscope. We then analyzed the metabolites and related pathways of radiation-induced lung injury by gas chromatography–mass spectrometry, and a total of 453 metabolites were identified. Compared to the nonirradiated left lung, 19 metabolites (8 upregulated and 11 downregulated) showed a significant difference in 10 Gy irradiated lung tissues, including mucic acid, methyl-β-d-galactopyranoside, quinoline-4-carboxylic acid, and pyridoxine. There were 31 differential metabolites (16 upregulated and 15 downregulated) between 20 Gy irradiated and nonirradiated lung tissues, including taurine, piperine, 1,2,4-benzenetriol, and lactamide. The Kyoto Encyclopedia of Genes and Genomes–based pathway analysis enriched 32 metabolic pathways between the irradiated and nonirradiated lung tissues, including pyrimidine metabolism, ATP-binding cassette transporters, aminoacyl-tRNA biosynthesis, and β-alanine metabolism. Among the dysregulated metabolites, we found that taurine promoted clonogenic survival and reduced radiation-induced necrosis in human embryonic lung fibroblast (HELF) cells. This study provides evidence indicating that radiation induces metabolic alterations of the lung. These findings significantly advance our understanding of the pathophysiology of radiation-induced lung injury from the perspective of metabolism.
Keywords: radiation, radiation-induced lung injury, metabolomics, taurine
Introduction
Despite technological advances in radiotherapy, radiation-induced toxicity in previously healthy tissues remains a serious concern. The current radiotherapy techniques expose both healthy tissues and tumors to a wide range of dose sizes and fractionations, with a substantial amount of healthy tissue being potentially irradiated.1,2 In addition, radiological incidents, accidental exposure, and nuclear terrorism have the potential to expose a large number of humans to ionizing radiation.3-5 The lungs are major organs targeted by radiation in cancer radiotherapy of the thoracic region.6-8 Pulmonary complications following exposure to ionizing radiation include acute lung injury (ALI) and pneumonitis, which can progress to fibrosis.9-12 Acute lung injury is a severe respiratory disease. To date, no medical interventions have been proven effective in improving the protection and the outcome of ALI.13 Therefore, radiation-induced lung injury has long been considered a treatment-limiting factor for patients requiring thoracic radiation and a major obstacle to the successful treatment of thorax-associated tumors.14 In the early stages of radiation-induced lung injury (1-30 days after radiation), type I epithelial cells are damaged, macrophages increase, type II epithelial cells are activated, alveolar septal capillary endothelium is injured and proliferate, platelets and erythrocytes aggregate, and microthrombi form. In the advanced stages (60-180 days), type II epithelial cells proliferate abnormally, fibroblasts (collagen) proliferate, vascular cells atrophy and decrease, and alveolar fibrosis develops.15 It is important to find therapeutic targets to prevent radiation-induced lung injury. However, the exact mechanisms of radiation-induced lung injury are not clear, and there is no reliable early biomarker for this disease.
Metabolomics captures a snapshot of the complex interactions between genetic alterations, enzymatic activity, and metabolic reactions, providing information on molecular changes during disease progression. Therefore, metabolomics represents a powerful quantitative measurement of the low-molecular-weight metabolites of an organism at a specified time in specific environmental conditions.16 Because metabolomics can identify and quantify hundreds, and potentially thousands, of small molecules, it greatly enhances our ability to characterize patterns of metabolites correlating with a disease. In addition, it is possible to obtain many metabolic fingerprints to show the progression of the disease from a single patient’s samples over time without the need for the time-consuming techniques associated with genomics and proteomics. Therefore, quantitative metabolomics provides a powerful new tool for the development of biomarkers for numerous diseases, particularly those with a rapid onset and high mortality rate, such as ALI. Radiation exposure triggers a complex network of gene expression that may impact metabolic processes and alter the levels of metabolites.17,18 Such metabolites have a potential capacity as biomarkers of radiation damage. A range of metabolite-detecting technologies are available for characterizing small chemicals in patient samples. These approaches include gas chromatography–mass spectrometry (GC/MS), liquid chromatography–mass spectrometry, and nuclear magnetic resonance (NMR).19
The metabolic profiles of radiation-induced lung injury have not been reported. The purpose of this study is to explore changes in metabolites in irradiated lung tissues to reveal new biomarkers and potential molecular mechanisms for their progression by GC/MS in rat models.
Materials and Methods
All chemicals used in this study were of the highest purity and analytical grade. Taurine was purchased from Sigma-Aldrich (St. Louis, Missouri). Piperine was purchased from Aladdin (Shanghai, China).
Animals and Treatments
The protocols for experiments involving animals were approved by the Animal Experimentation Ethics Committee at Soochow University (Suzhou, China). Male Sprague-Dawley (SD) rats (4 weeks of age) were purchased from the Shanghai SLAC Laboratory Animal Ltd (Shanghai, China). These animals were housed in a pathogen-free environment at the Medical School facility of Soochow University. Sprague-Dawley rats were randomly divided into the control group (mock irradiated) and 10 and 20 Gy irradiation groups (n = 6). After the rats were anesthetized, the radiation site (right lung) was located by computed tomography–guided precise positioning system (Figure 1A). A single dose of 10 or 20 Gy irradiation was administered to the right lung at a dose rate of 200 cGy/min with 6-MeV X-ray radiation (Clinac 2100EX, Varian Medical Systems, Inc, Palo Alto, California). Lung tissue samples were collected 1 week after irradiation.
Figure 1.
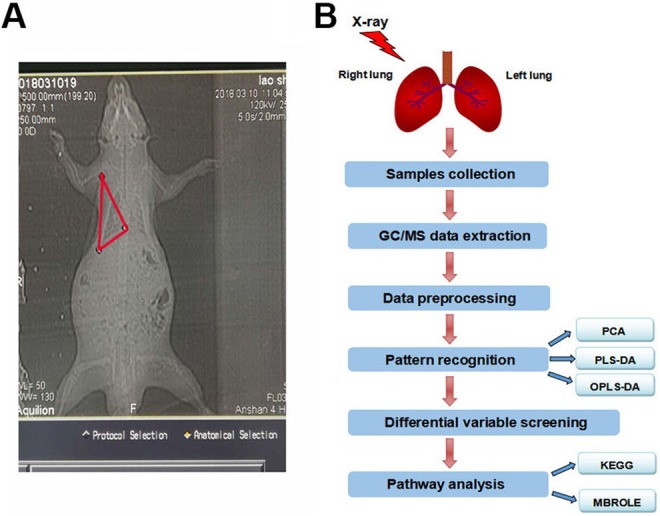
The procedures for metabolomics analysis. A, CT image of the right lung in a rat. The selected area was exposed to 10 or 20 Gy X-ray irradiation. B, The procedures for metabolomics analysis using GC/MS. CT indicates computed tomography; GC/MS, gas chromatography–mass spectrometry.
Hematoxylin and Eosin Staining
The lung tissues were fixed in 10% neutral buffered formalin and embedded in paraffin. Three-micrometer paraffin sections were deparaffinized and heat-treated with citrate buffer of pH 6.0 for 7 minutes following an epitope retrieval protocol. The sections of rat lung tissues were stained with hematoxylin and eosin.
Transmission Electron Microscopy
The lung tissues were fixed for 2 hours with 2.5% glutaraldehyde in a 0.05 M sodium cacodylate buffer at a pH of 7.2 at 25°C, followed by 2 hours in 2% OsO4 in a 0.1 M sodium cacodylate buffer and 18 hours in 1% aqueous uranyl acetate. After dehydration through an ethanol series, the specimens embedded in Epon 812 and ultrathin sections were collected on copper grids. After being stained with uranyl acetate and lead citrate, the sections were examined using a Tecnai G2 spirit BioTwin transmission electron microscope (FEI Company, Hillsboro, Oregon).
Sample Preparation for Metabolomic Analysis
All chemicals and solvents were analytical or HPLC grade. Methanol, pyridine, n-hexane, methoxylamine hydrochloride (97%), and BSTFA with 1% trimethylchlorosilane (TMCS) were purchased from CNW Technologies GmbH (Düsseldorf, Germany). Trichloromethane was obtained from Sinopharm Chemical Reagent Co, Ltd (Shanghai, China). l-2-Chlorophenylalanine was obtained from Shanghai Hengchuang Biotechnology Co, Ltd (Shanghai, China).
A 30 mg accurately weighed sample was transferred to a 1.5 mL Eppendorf tube. Two small steel balls were added to the tube. Next, 20 μL of internal standard (2-chloro-l-phenylalanine in methanol, 0.3 mg/mL) and 600 μL of the extraction solvent with methanol/water (4/1, v/v) were added to each sample. Samples were stored at −80°C for 2 minutes and then ground at 60 Hz for 2 minutes. Then, 120 μL of chloroform was added to the samples, and the samples were vigorously vortexed, followed by a 10 minutes ultrasound-associated extraction at room temperature, then stored at 4°C for 10 minutes. The samples were centrifuged at 12 000 rpm for 10 minutes at 4°C. The QC sample was prepared by mixing aliquots of all samples to produce a pooled sample. An aliquot of the 400-μL supernatant was transferred to a glass sampling vial for vacuum drying at 25°C. In addition, 80 μL of methoxylamine hydrochloride (dissolved in pyridine, 15 mg/mL) was added. The resultant mixture was vortexed vigorously for 2 minutes and incubated at 37°C for 90 minutes. Next, 80 μL of BSTFA (with 1% TMCS) and 20 μL n-hexane were added to the mixture, which was vortexed vigorously for 2 minutes and then derivatized at 70°C for 60 minutes. The samples were placed at 25°C for 30 minutes before the GC/MS analysis (Figure 1B).
Gas Chromatography/Mass Spectrometry Data Extraction
The lung samples were analyzed on an Agilent 7890B gas chromatography system coupled to an Agilent 5977A MSD system (Agilent Technologies Inc, Folsom, California). A DB-5MS fused-silica capillary column (30 m × 0.25 mm × 0.25 μm; Agilent J&W Scientific, Folsom, California) was utilized to separate the derivatives. Helium (>99.999%) was used as the carrier gas at a constant flow rate of 1 mL/min through the column. The injector temperature was maintained at 260°C. The injection volume was 1 μL by splitless mode, and the solvent delay time was set to 5 minutes. The initial oven temperature was 60°C, ramped to 125°C at a rate of 8°C/min, to 210°C at a rate of 5°C/min, to 270°C at a rate of 10°C/min, to 305°C at a rate of 20°C/min, and finally held at 305°C for 5 minutes. The temperature of the MS quadrupole and ion source (electron impact) was set to 150°C and 230°C, respectively. The collision energy was 70 eV. Mass spectrometric data were acquired in a full-scan mode (m/z 50-500). The QCs were injected at regular intervals (every 12 samples) throughout the analytical run to provide a set of data from which repeatability could be assessed.
Data Processing
Metabolites were annotated through the Fiehn or NIST database. After alignment with the Statistic Compare component, the “raw data array” (.cvs) was obtained from the raw data with 3-dimensional data sets including sample information, peak names (or retention time and m/z), and peak intensities. There were 1388 peaks detected from all samples. In the “data array,” all internal standards and any known pseudo positive peaks (caused by background noise, column bleed, or the BSTFA derivatization procedure) were removed. The data were normalized to the total peak area of each sample and multiplied by 10 000, and the peaks from the same metabolites were combined.
Pattern Recognition
The GC/MS data were processed with Simca-P 13.0 (Umetrics AB, Umea, Sweden) to perform the principal component analysis (PCA), partial least squares discriminant analysis (PLS-DA), and orthogonal partial least squares discrimination analysis (OPLS-DA) to discriminate the metabolic differences between the groups of the study. The Hoteling T2 region, shown as an ellipse in score plots of the models, defines the 95% confidence interval of the modeled variation. The variable importance in the projection (VIP) ranks the overall contribution of each variable to the OPLS-DA model, and those variables with VIP > 1 are considered relevant for group discrimination.20 In this study, the default 7-round cross-validation was applied with one-seventh of the samples being excluded from the mathematical model in each round to prevent overfitting.
Selection of Differential Metabolites
The differential metabolites were selected on the basis of the combination of a statistically significant threshold of VIP values obtained from the OPLS-DA model and P values from a 2-tailed Student t test on the normalized peak areas from different groups, where metabolites with VIP values larger than 1.0 and P values less than .05 were considered as differential metabolites.21
Pathway Analysis
The pathway enrichment analysis of differential metabolites aids in understanding the mechanisms of metabolic pathway changes in differential samples. Common pathway analysis is based on the pathway analysis of the Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathway.22 We mapped the differential metabolites to the KEGG database to obtain the enrichment results of their metabolic pathways.
Through the Metabolite Biological Role, the KEGG ID of differential metabolites was used for the pathway enrichment analysis to obtain metabolic pathway enrichment results. Values of P < .05 was significant, and the smaller the P value, the more significant the difference in the metabolic pathway, and the −log(P-value) was calculated. The differential metabolic pathway is displayed by the KEGG pathway mapper function, and the differential metabolites are visualized according to the upregulation and downregulation information. The small circles in the metabolic pathway map represent metabolites. The red-labeled metabolite in the pathway map is the experimentally detected upregulated metabolite, and the blue is the downregulated metabolite.23,24
Cell Culture and Irradiation
Human embryonic lung fibroblast (HELF) cells were maintained in Dulbecco’s modified Eagle medium. Cells were grown at 37°C in 5% CO2 incubators. Cells were exposed to different dosages of ionizing radiation using an X-ray linear accelerator (Rad Source, Suwanee, Georgia) at a fixed dose rate of 1.15 Gy/min.
Cell Viability Assay
Cell viability was determined using an MTT Cell Proliferation and Cytotoxicity Assay Kit (Beyotime, Nantong, China). Human embryonic lung fibroblast cells (5 × 103 cells/well) were seeded into a 96-well culture plate in 100 μL of cell culture medium and allowed to adhere overnight. Cells were then treated with taurine, piperine, or vehicle (phosphate-buffered saline or DMSO) for the desired time and were exposed to 10 Gy of ionizing radiation using an X-ray linear accelerator (Rad Source). The optical absorption value was measured using a microplate reader at 490 nm. For the last 4 hours of culture, MTT was added at a final concentration of 5 mg/mL. Then, plates were centrifuged at 1000 rpm for 5 minutes, the supernatant was discarded, and formazan crystals were solubilized in 0.15 mL DMSO.
Clonogenic Assay
Cells were seeded on 6-well plates in specified numbers in triplicate. Twenty-four hours later, cells were treated with taurine, piperine, or its vehicle for 6 hours before exposure to 2 Gy of ionizing radiation. After incubation for an additional 10 or 14 days, the cells were fixed with methanol and stained with crystal violet. Colonies of at least 50 cells were counted.
Low-Density Lipoprotein-Release Assay
To further measure the extent of cellular damage, the low-density lipoprotein (LDH) activity was also tested. After incubation, the culture supernatants were collected. The activity of LDH was detected at 600 nm according to the LDH assay kit (Beyotime). The LDH activity is measured by a 2-step reaction. At the first step, LDH catalyzes the reduction of NAD+ to NADH coupled with the oxidation of lactate acid to pyruvic acid. At the second step, diaphorase as a coupling enzyme uses NADH to catalyze the reduction of tetrazolium INT to formazan.
Measurement of Cell Apoptosis
The cells were treated with taurine, piperine, or its vehicle for 2 hours before receiving irradiation. Apoptosis was measured using a 7-AAD/Annexin-V double-staining apoptosis kit (BD Biosciences, Franklin Lakes, New Jersey) and flow cytometry (BD Biosciences, California). The Annexin-V+/7-AAD− cells indicated early apoptosis, and the Annexin-V+/7-AAD+ cells indicated late apoptosis. The percentages of both types of cells were counted.
Statistical Analysis
Results were expressed as the mean ± standard error of the mean of at least 3 independent experiments. Significance was calculated using 1-way analysis of variance. Statistical analyses were performed using SPSS software. Value of P < .05 was considered statistically significant.
Results
Morphological Alteration of the Lung Tissues After Radiation
The lung tissues of the control group had a normal tissue structure; no pathological changes such as acute or chronic inflammation were observed. The structure of the alveolus and alveolar septum were normal, no pulmonary veins were congested, capillary walls were intact, and bronchial lumens were not inflammatory. Seven days after radiation, lesions mainly showed exudation in the lung tissues of the 2 irradiated groups. In addition, the lesions showed a slight thickening of the alveolar septum, pulmonary interstitial and pulmonary capillary congestion, edema, and some alveolus with fluid (Figure 2A). The above pathological changes in the 20 Gy irradiation group were more obvious than those in the 10 Gy irradiation group (Figure 2B).
Figure 2.
Morphological alteration of irradiated lung tissues. The representative hematoxylin and eosin (H&E) staining of lung tissues and corresponding nonirradiated lung tissues of irradiated rats. A, After 7 days of irradiation, in the lung tissues of the 10 Gy irradiated group, the lesions mainly showed exudation. B, After 7 days of irradiation, the 20 Gy irradiated group showed a slight thickening of the alveolar septum, pulmonary interstitial and pulmonary capillary congestion, edema, and some alveolus with fluid. The transmission electron micrograph of the lung tissues. C, After 7 days of irradiation, the local alveolar type I epithelial cells were significantly swollen in the 10 Gy irradiation group. Some broken organelles were identified in the alveolar cavity. D, In the 20 Gy irradiation group after 7 days of irradiation, the lamellar bodies of alveolar type II epithelial cells began to empty. The nuclear division occurred in the alveolar type II epithelial cells.
As shown in Figure 2C, 7 days after irradiation, the local alveolar type I epithelial cells were highly swollen in the 10 Gy irradiation group, and the cytoplasm had become pale. The number of alveolar type II epithelial cells increased, and some cells were found to be slightly swollen. In addition, it was found that there were swollen vacuoles in the cytoplasm and broken organelles in the alveolar cavity (Figure 2C). In the 20 Gy irradiation group after 7 days of irradiation, the lamellar bodies of alveolar type II epithelial cells began to be found empty, and some organelles were found to be broken off. Alveolar wall vasodilatation and hyperemia occurred. Furthermore, we identified platelet aggregation in the alveolar capillaries and nuclear division in the alveolar type II epithelial cells (Figure 2D).
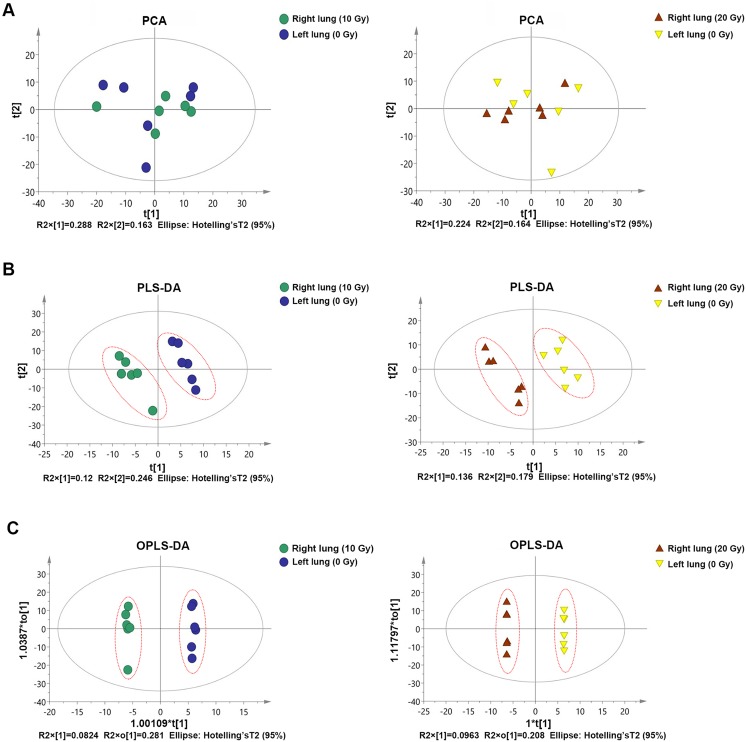
Pattern Recognition
First, we performed a PCA of the sample. It was found that there was a significant degree of separation between the 2 groups of samples in the PCA score map, reflecting a large difference in metabolism between the 2 groups of samples (Figure 3A). Therefore, the PCA model can be used to explain the metabolic differences between the 2 groups of samples more reliably. The 2 groups also showed significant differences in the PLS-DA score map, so the PLS-DA model can explain and predict the differences between the 2 groups of samples (Figure 3B). Finally, the model can eliminate noise information that is not related to classification (such as control and treatment) and can obtain more reliable metabolite information, which causes significant differences between the 2 groups of samples. We used OPLS-DA to filter out signals that were not related to model classification to obtain the OPLS-DA model. It was also found that the 2 groups of samples had significant differences in the OPLS-DA score map (Figure 3C).
Figure 3.
The score maps of various analysis patterns. A, The degree of separation between the 2 groups of samples in the PCA score maps. B, The degree of separation between the 2 groups of samples in the PLS-DA score maps. C, The degree of separation between the 2 groups of samples in the OPLS-DA score maps. OPLS-DA indicates orthogonal partial least squares discrimination analysis; PCA, principal component analysis; PLS-DA, partial least squares discriminant analysis.
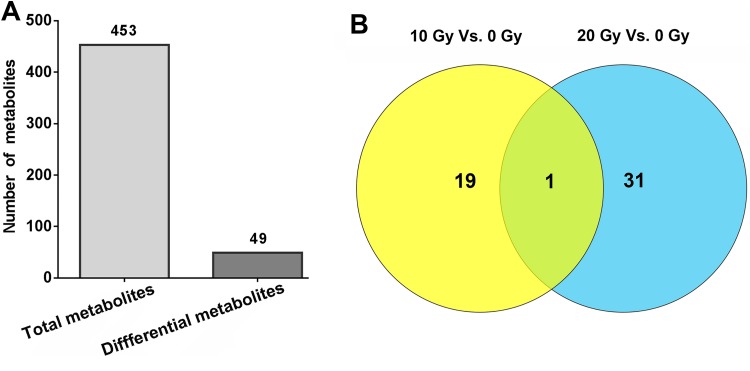
Analysis of the Lung Tissue Metabolites
A total of 453 metabolites were detected in lung tissues by GC/MS (Figure 4A). Compared with the control group, there were 49 differential metabolites with significance in the lung tissues of the 2 irradiated groups (Figure 4A and B). Among the 19 metabolites differentially present between the 10 Gy irradiation and the control group, 8 metabolites were upregulated and 11 metabolites were downregulated. The preferential metabolites in the 10 Gy irradiated rat lung included mucic acid, methyl-β-d-galactopyranoside, quinoline-4-carboxylic acid, and pyridoxine (Table 1). There were 31 differential metabolites between 20 Gy irradiation and the control group (16 upregulated and 15 downregulated), including taurine, piperine, 1,2,4-benzenetriol, and lactamide (Table 2). Cytidine was the only common differential metabolite that was downregulated in both 10 and 20 Gy radiation groups (Figure 4B, Tables 1 and 2). Cytidine is a component of RNA and is a nucleoside molecule formed by the attachment of cytosine to the ribose ring.
Figure 4.
Identified and dysregulated metabolites by GC/MS. A, The total metabolites and total differential metabolites in rat lung tissues. The total number of metabolites detected was much more than the total number of differential metabolites. B, Comparison of the differential metabolites between different dose groups. There was only 1 identical differential metabolite in the 10 and 20 Gy groups. GC/MS indicates gas chromatography–mass spectrometry.
Table 1.
Dysregulated Metabolites in the Lung Tissues (10 Gy Group vs 0 Gy Group).
| Number | Metabolites | Quantitative Mass | VIP | P Value | Fold-Change |
|---|---|---|---|---|---|
| 1 | Mucic acid | 447 | 1.65 | 1.42E-02 | 2.32 |
| 2 | Methyl-β-d-galactopyranoside | 204 | 1.19 | 4.15E-02 | 1.64 |
| 3 | Quinoline-4-carboxylic acid | 128 | 2.21 | 2.56E-02 | 1.56 |
| 4 | Pyridoxine | 280 | 2.13 | 2.51E-02 | 1.48 |
| 5 | N-Ethylmaleamic acid | 306 | 2.42 | 5.33E-03 | 1.46 |
| 6 | Cellobiose | 205 | 2.26 | 2.24E-02 | 1.30 |
| 7 | 2-Ketovaleric acid | 89 | 2.04 | 4.60E-02 | 1.24 |
| 8 | Thymidine | 170 | 2.11 | 4.97E-02 | 1.23 |
| 9 | O-Phosphonothreonine | 71 | 2.16 | 3.54E-02 | 0.90 |
| 10 | Toluenesulfonic acid | 229 | 2.23 | 2.46E-02 | 0.88 |
| 11 | Fructose-2,6-biphosphate degr prod | 270 | 2.30 | 2.00E-02 | 0.80 |
| 12 | Leucine | 104 | 2.68 | 3.42E-03 | 0.76 |
| 13 | 2-Mercaptoethanesulfonic acid | 181 | 2.08 | 3.68E-02 | 0.68 |
| 14 | Guanosine-5′-monophosphate | 169 | 1.36 | 4.89E-02 | 0.56 |
| 15 | N-(3-Aminopropyl)-morpholine | 237 | 1.90 | 4.44E-02 | 0.49 |
| 16 | Cytidine | 223 | 2.95 | 2.14E-04 | 0.44 |
| 17 | 2-Hydroxybutanoic acid | 131 | 1.48 | 2.62E-02 | 0.41 |
| 18 | 6-Hydroxynicotinic acid | 256 | 1.65 | 3.40E-03 | 0.24 |
| 19 | Carbobenzyloxy-l-leucine degr3 | 122 | 2.49 | 1.14E-02 | 2.79E-06 |
Abbreviation: VIP, variable importance in the projection.
Table 2.
Dysregulated Metabolites in the Lung Tissues (20 Gy Group vs 0 Gy Group).
| Number | Metabolites | Quantitative Mass | VIP | P Value | Fold-Change |
|---|---|---|---|---|---|
| 1 | Piperine | 172 | 1.73 | 2.09E-02 | 14.59 |
| 2 | 1,2,4-Benzenetriol | 342 | 2.14 | 1.92E-02 | 5.03 |
| 3 | Lactamide | 357 | 1.90 | 2.83E-02 | 2.26 |
| 4 | Guanine | 226 | 1.16 | 3.06E-02 | 1.81 |
| 5 | Histidine | 254 | 1.95 | 4.85E-02 | 1.34 |
| 6 | 4-Vinylphenol dimer | 192 | 2.33 | 6.81E-03 | 1.33 |
| 7 | Glutamine | 156 | 2.09 | 1.59E-02 | 1.32 |
| 8 | 4-Aminobutyric acid | 174 | 2.03 | 3.57E-02 | 1.31 |
| 9 | Serine | 306 | 2.26 | 1.42E-02 | 1.28 |
| 10 | Norleucine | 158 | 2.05 | 3.42E-02 | 1.27 |
| 11 | Methionine | 176 | 2.00 | 4.11E-02 | 1.25 |
| 12 | Valine | 144 | 2.02 | 3.83E-02 | 1.20 |
| 13 | Proline | 142 | 2.07 | 2.86E-02 | 1.17 |
| 14 | Threonine | 292 | 2.12 | 2.21E-02 | 1.17 |
| 15 | N-Acetyl-β-d-mannosamine | 319 | 1.96 | 3.36E-02 | 1.15 |
| 16 | Aspartic acid | 306 | 2.20 | 1.33E-02 | 1.14 |
| 17 | Palatinitol | 218 | 1.86 | 4.65E-02 | 0.92 |
| 18 | 5-Aminovaleric acid | 174 | 1.99 | 3.15E-02 | 0.91 |
| 19 | Malonamide | 343 | 2.14 | 1.90E-02 | 0.91 |
| 20 | Octadecanol | 327 | 1.96 | 3.60E-02 | 0.89 |
| 21 | Abietic acid | 154 | 1.87 | 4.17E-02 | 0.89 |
| 22 | 2-Monopalmitin | 218 | 2.14 | 1.82E-02 | 0.88 |
| 23 | Guanidinosuccinic acid | 328 | 1.96 | 3.67E-02 | 0.85 |
| 24 | 1-Monopalmitin | 401 | 2.30 | 7.77E-03 | 0.84 |
| 25 | Monostearin | 203 | 2.36 | 5.76E-03 | 0.83 |
| 26 | Adenosine 5-monophosphate | 315 | 2.12 | 1.95E-02 | 0.82 |
| 27 | 2-Hydroxyvaleric acid | 248 | 1.76 | 3.88E-02 | 0.72 |
| 28 | Cytidine | 223 | 2.61 | 1.95E-03 | 0.71 |
| 29 | Taurine | 326 | 1.53 | 4.86E-02 | 0.67 |
| 30 | Orotic acid | 254 | 1.54 | 3.89E-02 | 0.58 |
| 31 | Spermine | 144 | 1.94 | 3.88E-02 | 0.49 |
Abbreviation: VIP, variable importance in the projection.
Pathway Analysis of the Lung Tissues in the Irradiated Lungs of Rats
After analyzing all the metabolic pathways that were associated with the dysregulated metabolites, we found that there were 10 enriched metabolic pathways in the 10 Gy irradiation group (Figure 5A) and 29 enriched metabolic pathways in the 20 Gy irradiation group (Figure 5B). After comparing all the metabolic pathways of the 2 radiation dose groups, we found that there were 7 common metabolic pathways: pyrimidine metabolism; propanoate metabolism; nicotinate and nicotinamide metabolism; aminoacyl-tRNA biosynthesis; ATP-binding cassette transporters; valine, leucine, and isoleucine degradation; and valine, leucine, and isoleucine biosynthesis.
Figure 5.
Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of the dysregulated metabolites in irradiated lung tissues. A, Ten metabolic pathways were found to be enriched by differential metabolites in the 10 Gy irradiated group. B, Twenty-nine metabolic pathways were found to be enriched by differential metabolites in the 20 Gy irradiated group.
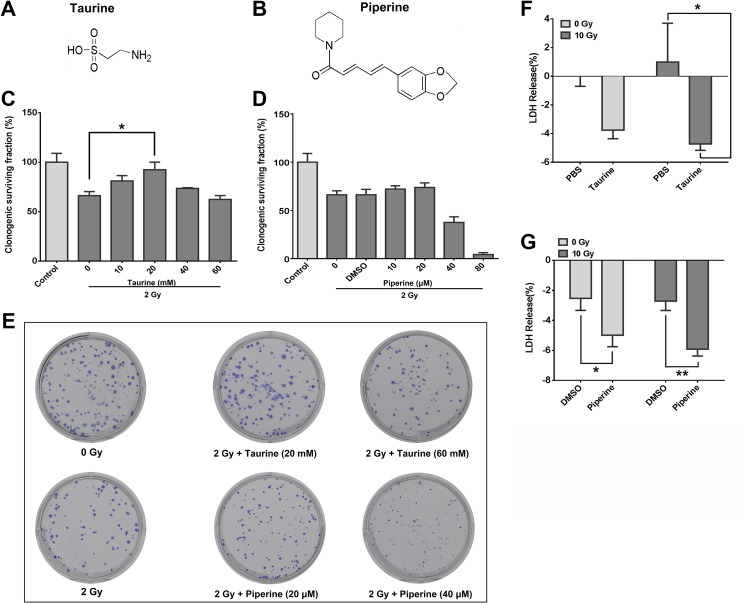
Protective Effect of Taurine and Piperine on HELF Cells
Taurine (Figure 6A) and piperine (Figure 6B) were further investigated in this study to explore the effect of dysregulated metabolites on the radiosensitivity of lung fibroblasts, as they have been associated with cell radiosensitivity.25,26 Human embryonic lung fibroblast cells were incubated with various concentrations of taurine and piperine for 24 or 48 hours, and cell viability was determined using an MTT assay. As shown in Supplementary Figure 1, 10 to 60 mM taurine and 10 to 80 μM piperine were used in the following experiments. Human embryonic lung fibroblast cells were treated with taurine and piperine for 6 hours before being exposed to 2 Gy ionizing radiation, to test their effect on cell radiosensitivity. The results of the clonogenic assay showed that pretreatment with 20 mM of taurine significantly promoted the survival of HELF cells (Figure 6C and E). Pretreatment with less than 20 μM piperine did not modulate cell radiosensitivity, whereas 40 and 80 μM piperine sensitized HELF cells to radiation (Figure 6D and E). The LDH-release assays and apoptosis analyses were used to test the effects of taurine and piperine on radiation-induced cell death. The results of the LDH-release assay showed that the HELF cells treated with taurine and piperine for 6 hours before radiation had a significant protective effect against radiation-induced cell necrosis (Figure 6F and G). However, the cells pretreated with taurine or piperine before radiation did not show a significant influence on the apoptotic rate of HELF cells (Supplementary Figure 2).
Figure 6.
The effects of taurine and piperine on the radiosensitivity of HELF cells. A, The chemical structure of taurine. B, The chemical structure of piperine. C, The effects of 2 Gy ionizing radiation with or without taurine (10-60 mM) pretreatment on cell proliferation. *P < .05. D, The effects of 2 Gy ionizing radiation with or without piperine (10-80 μM) pretreatment on cell proliferation. E, Representative images of cell plates of the clonogenic assay. F, The effect of taurine on the low-density lipoprotein (LDH) release of HELF cells. HELF cells were treated with taurine (10 mM) 6 hours before being exposed to 10 Gy ionizing radiation. G, The effect of piperine on the LDH release of HELF cells. HELF cells were treated with piperine (20 μM) 6 hours before being exposed to 10 Gy ionizing radiation. HELF indicates human embryonic lung fibroblast.
Discussion
Radiation-induced lung injury is an extraordinarily complex lung-associated disorder characterized by a neutrophilic inflammatory response and associated with increased pulmonary vascular permeability. In recent years, metabolomics has become an important and valuable tool in the life sciences, which can analyze the changing metabolite levels in biological systems in response to biological stimuli or perturbations.16-19 As the final downstream products of gene expression, metabolites are directly linked to phenotypes and reflect cellular activities at the functional level.27 Ionizing radiation has been reported to modulate the metabolism of cells and tissues.28-31 For example, Wu et al29 identified 12 differentially expressed metabolites in the irradiated mouse melanoma cell line B16 by NMR-based untargeted metabolic profiling. After whole liver irradiation, a total of 407 metabolites were identified in irradiated liver tissues.30 Ghosh et al31 reported a distinct dose- and time-dependent metabolic response to gastrointestinal tissue injury induced by ionizing radiation. In this study, we analyzed the metabolites in radiation-induced lung injury by GC/MS and identified a total of 453 metabolites. Compared to the nonirradiated left lung, 19 metabolites showed significant differences in the 10 Gy irradiation lung, whereas 31 differential metabolites were identified in the 20 Gy irradiated lung tissues. Interestingly, metabolic dysregulation in 10 and 20 Gy irradiated lung tissues differs a lot. We postulated that 10 and 20 Gy radiation resulted in different damages in nuclear DNA, proteins, lipids, and carbohydrates, which lead to different metabolic response to survive. Golla et al32 found 67 metabolic biomarkers in radiosensitive tissues and biofluids (serum and urine) in response to 6 Gy radiation, which indicated perturbations in pathways, including DNA methylation, energy, nucleic acid, amino acid, glutathione, and bile acid metabolism. Therefore, through previous studies and our research, it can be concluded that radiation-induced metabolic changes may be tissue specific.
Among the dysregulated metabolites, we focused on taurine and piperine. Taurine is a simple sulfur-containing amino acid and has several beneficial physiological actions, including antioxidation, detoxification, calcium homeostasis, cell membrane stabilization, and neuromodulation. We found that taurine was downregulated in 20 Gy irradiated lung tissues of rats. Yang et al reported that taurine is protective against radiation-induced reproductive system dysfunction. Oral administration of taurine ameliorated radiation-induced brain damage by scavenging reactive oxygen species and decreasing apoptosis.33 In our clonogenic assay, 10, 20, and 40 mM taurine increased clonogenic survival and reduced the radiation-induced death of HELF cells, in which the beneficial effect of 20 mM of taurine was the strongest. Moreover, HELF cells treated with 20 mM of taurine exhibited highest proliferative ability. These results are consistent with previous reports and indicate that taurine may be used as a radioprotector for radiogenic lung injury. Piperine, as a plant alkaloid, is an important constituent present in black pepper (Piper nigrum).34 The accumulation of piperine is associated with cell apoptosis.35,36 Piperine sensitizes cancer cells to radiation by the augmentation of apoptosis.37 In this study, we found that the piperine content was increased in 20 Gy irradiated lung tissues. A further functional study showed that higher concentrations of piperine reduced the clonogenic survival of HELF cells after radiation. However, 20 μM of piperine reduced radiation-induced necrosis. The effect of piperine on the progression of radiation-induced lung injury may warrant further in vivo studies using animal models.
In conclusion, this study provides evidence that irradiation induces metabolic alterations in the irradiated lungs of rats. In particular, taurine and piperine are involved in the radiosensitivity and radiation-induced death of lung fibroblasts. These findings significantly advance our understanding about the pathophysiology of radiation-induced lung injury from the perspective of metabolism.
Supplemental Material
Supplemental Material, Supplementary_Figure_and_Figure_legend for Metabolomic Analysis of Radiation-Induced Lung Injury in Rats: The Potential Radioprotective Role of Taurine by Yiying Gao, Xugang Li, Jingjing Gao, Zhen Zhang, Yang Feng, Jihua Nie, Wei Zhu, Shuyu Zhang and Jianping Cao in Dose-Response
Footnotes
Authors’ Note: Yiying Gao, Xugang Li, and Jingjing Gao contributed equally to this work.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work is supported by the National Natural Science Foundation of China (81803166, 81673100, 31770911, and 81872552) and the Social Development Program of Jiangsu Province (BE2017634 and BE2017652).
ORCID iD: Shuyu Zhang  https://orcid.org/0000-0003-1419-3635
https://orcid.org/0000-0003-1419-3635
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Aznar MC, Duane FK, Darby SC, Wang Z, Taylor CW. Exposure of the lungs in breast cancer radiotherapy: a systematic review of lung doses published 2010–2015. Radiother Oncol. 2018;126(1):148–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Klein D, Steens J, Wiesemann A, et al. Mesenchymal stem cell therapy protects lungs from radiation-induced endothelial cell loss by restoring superoxide dismutase 1 expression. Antioxid Redox Signal. 2017;26(11):563–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. DiCarlo AL, Maher C, Hick JL, et al. Radiation injury after a nuclear detonation: medical consequences and the need for scarce resources allocation. Disaster Med Public Health Prep. 2011;5(suppl 1):32–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Waselenko JK, Macvittie TJ, Blakely WF, et al. Medical management of the acute radiation syndrome: recommendations of the strategic national stockpile radiation working group. Ann Intern Med. 2004;140(12):1037–1051. [DOI] [PubMed] [Google Scholar]
- 5. Laiakis EC, Mak TD, Anizan S, et al. Development of a metabolomic radiation signature in urine from patients undergoing total body irradiation. Radiat Res. 2014;181(4):350–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yu J, Che J, Liu L, Yang F, Zhu X, Cao B. Tetrahydropalmatine attenuates irradiation induced lung injuries in rats. Life Sci. 2016;153:74–81. [DOI] [PubMed] [Google Scholar]
- 7. Mishra MV, Louie AV, Gondi V, Slotman B. The evolving role of radiotherapy in the management of small cell lung cancer. J Thorac Dis. 2018;10(suppl 21):2545–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ghobadi G, Wiegman EM, Langendijk JA, Widder J, Coppes RP, van Luijk P. A new CT-based method to quantify radiation-induced lung damage in patients. Radiother Oncol. 2015;117(1):4–8. [DOI] [PubMed] [Google Scholar]
- 9. Giridhar P, Mallick S, Rath GK, Julka PK. Radiation induced lung injury: prediction, assessment and management. Asian Pac J Cancer Prev. 2015;16(7):2613–2617. [DOI] [PubMed] [Google Scholar]
- 10. Palma DA, van Sörnsen de Koste JR, Verbakel WF, Senan S. A new approach to quantifying lung damage after stereotactic body radiation therapy. Acta Oncol. 2011;50(4):509–517. [DOI] [PubMed] [Google Scholar]
- 11. Ghafoori P, Marks LB, Vujaskovic Z, Kelsey CR. Radiation-induced lung injury. Assessment, management, and prevention. Oncology. 2008;22 (1):52–53. [PubMed] [Google Scholar]
- 12. Ghosh SN, Wu Q, Mäder M, et al. Vascular injury following whole thoracic X-ray irradiation in the rat. Int J Radiat Oncol Biol Phys. 2009; 74 (1):192–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xiong Z, Wang Y, Lang L, et al. Tissue metabolomic profiling to reveal the therapeutic mechanism of reduning injection on LPS-induced acute lung injury rats. RSC Adv. 2018;8(18):10023–10031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Malaviya R, Gow AJ, Francis M, Abramova EV, Laskin JD, Laskin DL. Radiation-induced lung injury and inflammation in mice: role of inducible nitric oxide synthase and surfactant protein D. Toxicol Sci. 2015;144(1):27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. S N SG, Raviraj R, Nagarajan D, Zhao W. Radiation-induced lung injury: impact on macrophage dysregulation and lipid alteration—a review. Immunopharmacol Immunotoxicol. 2019;41(3):370–379. [DOI] [PubMed] [Google Scholar]
- 16. Chen Y, Ma Z, Min L, et al. Biomarker identification and pathway analysis by serum metabolomics of lung cancer. Biomed Res Int. 2015;2015(7):183624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xie L, Zhou J, Zhang S, et al. Integrating microRNA and mRNA expression profiles in response to radiation-induced injury in rat lung. Radiat Oncol. 2014;9(1):111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rube CE, Uthe D, Schmid KW, et al. Dose-dependent induction of transforming growth factor beta (TGF-beta) in the lung tissue of fibrosis-prone mice after thoracic irradiation. Int J Radiat Oncol Biol Phys. 2000;47(4):1033–1042. [DOI] [PubMed] [Google Scholar]
- 19. Lacy P. Metabolomics of sepsis-induced acute lung injury: a new approach for biomarkers. Am J Physiol Lung Cell Mol Physiol. 2011;300(1):1–3. [DOI] [PubMed] [Google Scholar]
- 20. Koo I, Wei X, Zhang X. Analysis of metabolomic profiling data acquired on GC-MS. Meth Enzymol. 2014;543:315–324. [DOI] [PubMed] [Google Scholar]
- 21. Wang Y, Xu L, Shen H, et al. Metabolomic analysis with GC-MS to reveal potential metabolites and biological pathways involved in Pb & Cd stress response of radish roots. Sci Rep. 2015;5:18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42(database issue):D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chagoyen M, Pazos F. MBRole: enrichment analysis of metabolomic data. Bioinformatics. 2011;27 (5):730–731. [DOI] [PubMed] [Google Scholar]
- 24. Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(database issue):D109–D114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang W, Huang J, Xiao B, et al. Taurine protects mouse spermatocytes from ionizing radiation-induced damage through activation of Nrf2/HO-1 signaling. Cell Physiol Biochem. 2017;44(4):1629–1639. [DOI] [PubMed] [Google Scholar]
- 26. Ren C, Liang Z. Piperine alleviates lipopolysaccharide-induced inflammatory injury by down-regulating microRNA-127 in murine chondrogenic ATDC5 cells. Biomed Pharmacother. 2018;103:947–954. [DOI] [PubMed] [Google Scholar]
- 27. Cui L, Zheng D, Lee YH, et al. Metabolomics investigation reveals metabolite mediators associated with acute lung injury and repair in a murine model of influenza pneumonia. Sci Rep. 2016; 6:26076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roh C. Metabolomics in radiation-induced biological dosimetry: a mini-review and a polyamine study. Biomolecules. 2018;8(2):E34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu L, Hu Z, Huang Y, et al. Radiation changes the metabolic profiling of melanoma cell line B16. PLoS One. 2016;11(9):e0162917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kurland IJ, Broin PÓ, Golden A, et al. Integrative metabolic signatures for hepatic radiation injury. PLoS One. 2015;10(6):e0124795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ghosh SP, Singh R, Chakraborty K, et al. Metabolomic changes in gastrointestinal tissues after whole body radiation in a murine model. Mol Biosyst. 2013;9(4):723–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Golla S, Golla JP, Krausz KW, et al. Metabolomic analysis of mice exposed to gamma radiation reveals a systemic understanding of total-body exposure. Radiat Res. 2017;187(5):612–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. El-Maraghi EF, Abdel-Fattah KI, Soliman SM, El-Sayed WM. Taurine provides a time-dependent amelioration of the brain damage induced by γ-irradiation in rats. J Hazard Mater. 2018;359:40–46. [DOI] [PubMed] [Google Scholar]
- 34. Verma A, Kushwaha HN, Srivastava AK, et al. Piperine attenuates UV-R induced cell damage in human keratinocytes via NF-kB, Bax/Bcl-2 pathway: an application for photoprotection. J Photochem Photobiol B Biol. 2017;172:139–148. [DOI] [PubMed] [Google Scholar]
- 35. Greenshields AL, Doucette CD, Sutton KM, et al. Piperine inhibits the growth and motility of triple-negative breast cancer cells. Cancer Lett. 2015;357(1):129–140. [DOI] [PubMed] [Google Scholar]
- 36. Ba Y, Malhotra A. Potential of piperine in modulation of voltage gated K+ current and its influences on cell cycle arrest and apoptosis in human prostate cancer cells. Eur Rev Med Pharmacol Sci. 2018;22(24):8999–9011. [DOI] [PubMed] [Google Scholar]
- 37. Tak JK, Lee JH, Park JW. Resveratrol and piperine enhance radiosensitivity of tumor cells. BMB Rep. 2012;45(4):242–246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, Supplementary_Figure_and_Figure_legend for Metabolomic Analysis of Radiation-Induced Lung Injury in Rats: The Potential Radioprotective Role of Taurine by Yiying Gao, Xugang Li, Jingjing Gao, Zhen Zhang, Yang Feng, Jihua Nie, Wei Zhu, Shuyu Zhang and Jianping Cao in Dose-Response