Abstract
Background
Automated phenotyping technologies are continually advancing the breeding process. However, collecting various secondary traits throughout the growing season and processing massive amounts of data still take great efforts and time. Selecting a minimum number of secondary traits that have the maximum predictive power has the potential to reduce phenotyping efforts. The objective of this study was to select principal features extracted from UAV imagery and critical growth stages that contributed the most in explaining winter wheat grain yield. Five dates of multispectral images and seven dates of RGB images were collected by a UAV system during the spring growing season in 2018. Two classes of features (variables), totaling to 172 variables, were extracted for each plot from the vegetation index and plant height maps, including pixel statistics and dynamic growth rates. A parametric algorithm, LASSO regression (the least angle and shrinkage selection operator), and a non-parametric algorithm, random forest, were applied for variable selection. The regression coefficients estimated by LASSO and the permutation importance scores provided by random forest were used to determine the ten most important variables influencing grain yield from each algorithm.
Results
Both selection algorithms assigned the highest importance score to the variables related with plant height around the grain filling stage. Some vegetation indices related variables were also selected by the algorithms mainly at earlier to mid growth stages and during the senescence. Compared with the yield prediction using all 172 variables derived from measured phenotypes, using the selected variables performed comparable or even better. We also noticed that the prediction accuracy on the adapted NE lines (r = 0.58–0.81) was higher than the other lines (r = 0.21–0.59) included in this study with different genetic backgrounds.
Conclusions
With the ultra-high resolution plot imagery obtained by the UAS-based phenotyping we are now able to derive more features, such as the variation of plant height or vegetation indices within a plot other than just an averaged number, that are potentially very useful for the breeding purpose. However, too many features or variables can be derived in this way. The promising results from this study suggests that the selected set from those variables can have comparable prediction accuracies on the grain yield prediction than the full set of them but possibly resulting in a better allocation of efforts and resources on phenotypic data collection and processing.
Keywords: Unmanned aerial vehicle, Phenotyping, Yield prediction, LASSO, Random forest, Ridge regression, SVM
Background
As one of the leading sources for food production, wheat has the highest hectare over the world [1, 2]. Boosting grain yield to feed the ever growing world population is one of the major focuses in wheat breeding [3]. Recent developments in high-throughput field-based plant phenotyping have spawned various studies in wheat breeding programs, including using sensor-derived secondary traits to predict the primary trait of interest, such as yield. Accurate prediction of the primary trait can improve accuracy of genotypic selection, thus shortening breeding cycles and save costs. Two major sensing platforms have been widely used to measure the secondary traits in field: the ground-based and the aerial-based sensor platforms. Ground-based platforms provide large sensor payloads and throughputs. For example, a multi-sensor cart was developed for soybean and wheat breeding [4], mounted with ultrasonic sensor, NDVI sensor, thermal infrared radiometer, spectrometer, RGB sensor, as well as other ancillary sensors. Similar platforms include ‘phenocart’ [5], mobile ‘PhenoTrac’ [6, 7], and tractor-based semi-automatic system [8]. As for aerial-based platforms, the unmanned aerial vehicle (UAV) is gaining increased attention due to ease of operation, high spatial resolution, and quick coverage [9–12]. Typical sensors equipped on UAV in agricultural applications are RGB cameras [13], multispectral cameras [14], thermal cameras [15], and hyperspectral sensors [16].
It is of a great interest to use UAV-derived phenotypic traits for yield prediction. For winter wheat, the grain yield is usually estimated by vegetation indices [14, 17, 18] or morphological traits derived from aerial imagery at single growth stage [19]. For example, by deriving normalized difference vegetation index (NDVI) from UAV imagery on different growth stages, the highest correlation coefficient (r) of 0.91 was found between NDVI and final yield around flowering time [14]. In addition to looking at single growth-stage, researchers also attempted to exploit extra predictive power by integrating phenotypic traits from multiple growth-stages. In the study of Du & Noguchi [13], five RGB indices accumulated over eight flights were used as variables in the stepwise regression model, and the best model with four indices was selected (r = 0.69 on validation set). Additionally, Haghighattalab et al. [10] input multi-temporal phenotypic traits into principal component regression (PCR) and geographically weighted (GW) model to estimate wheat yield. The GW model considered the spatial relationship among acquired images, which performed better on grain yield prediction than PCR (r increased from 0.26 to 0.74 under the drought environment, and from 0.24 to 0.46 under irrigated environment).
Despite the promising findings on yield prediction with remotely sensed phenotypic traits throughout the growing season [20, 21], collecting and processing multi-temporal traits is still time-consuming and computationally expensive. For example, in this study, data collection on the winter wheat during spring growing season started from mid-April to mid-June in 2018 on a weekly basis. The collected imagery data after each flight took approximately 30 gigabytes of storage (around 9000 multispectral images and 1000 RGB images, over the 1.2 hectare field). Currently, processing such large dataset in a short time is still complicated. If several key UAV-derived phenotypic traits or growth stages for grain yield are available, data collection and processing efforts could be streamlined. The predictive model will also be simplified, allowing a better understanding of the predictive power of individual traits.
To determine critical phenotypic traits or growth stages, variable selection algorithms can be performed to reserve principal predictors, which in this case are features extracted from UAV imagery, based on the predictive powers of individual predictors on the response variable, which can be grain yield for example [22]. In this way, further processing can be narrowed down to those selected principal variables with reduced computational complexity, improved data analysis efficiency, and better data understanding [23, 24]. In this study, we adopted two common variable selection algorithms: LASSO regression and random forest. LASSO was firstly proposed by Tibshirani [25]. It adds penalty into parameter estimation to shrink the near-zero regression coefficients to zero, thus removing them out of the selection result. Random forest [26] aggregates hundreds of individual decision trees to achieve a better trade-off between bias and variance [27, 28]. It is a ranking-based nonparametric selection algorithm [28, 29], providing importance measurement for each variable. Both LASSO and random forest are feasible when the number of variables is greater than the number of observations [30, 31].
Only a few studies investigated the principal variable selection on UAV-derived phenotypic traits for wheat grain yield prediction [10, 13]. Furthermore, more features could be extracted from the ultra-high spatial resolution UAV images rather than common averaged statistical descriptions at the plot level (e.g., mean vegetation indices of each plot) [32–38]. In addition, it is meaningful to examine the predictive power of dynamic features from the multi-temporal UAV data, such as growth rate. To this end, the objective of this study was to select principal phenotypic variables that contribute most in explaining the grain yield in winter wheat, to potentially reduce the efforts in field phenotyping data collection and the subsequent data processing. Two specific objectives were:
To maximize the feature/variable extractions from the UAV-derived vegetation index (VI) and plant height maps including pixel statistics (e.g., mean, median) and dynamic growth rate.
To perform principal variable selection on extracted variables, and to evaluate predictive power for grain yield using the selected principal variables.
Clarification of terminology in this study:
Primary trait: grain yield;
Secondary traits: plant height, spectral reflectance;
UAV derived maps: plant height, NDVI, NDRE, and GNDVI map;
Features (variables) extracted from individual plots in UAV derived maps: trimmed mean, median, mode, 95th percentile, standard deviation, contrast, correlation, energy, and homogeneity.
Methods
Field layout
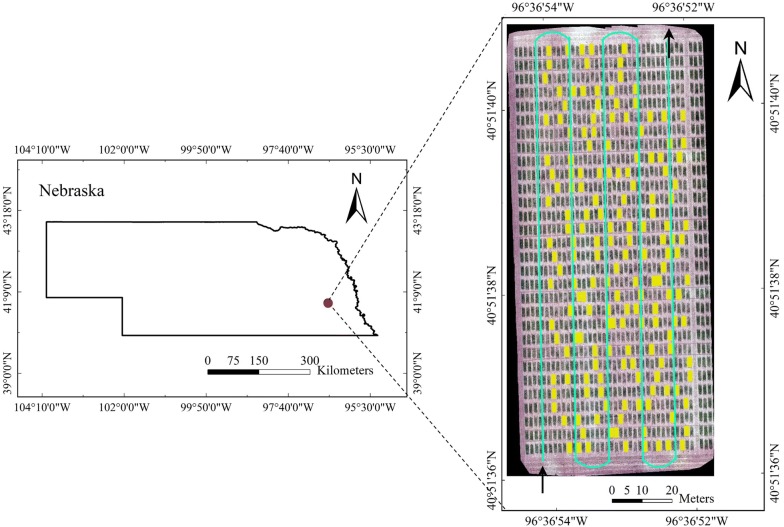
The studied field was located in Lincoln, Nebraska, USA (N 40.8581, W 96.6157), where winter wheat was grown during the growing season from the end of October, 2017 to early July, 2018. As part of a larger augmented design for yield trial, ten check lines with 17 replications, in total 170 plots, were used in this study (Fig. 1). The ten checks include three Nebraska (NE) lines (Freeman, Robidoux, Ruth), three Texas (TX) lines (TAM 304, TAM 113, TAM 114), two Westbred (WB) lines (WB Cedar, WB Grainfield), one Oklahoma line (Gallagher), and one Syngenta line (SY Wolf). The remaining plots in this trial were reserved proprietary lines at the time of this study. The checks were grown in plots of five rows of 3.0 m length and with 0.23 m spacing between the rows. Each check plot was planted with 35 grams of seeds, with a seeding rate of approximately 1,000,000 seeds per acre. Grain yield was measured in all five rows of each plot in early July, using a Zurn 150 Combine harvester (Zurn, Schöntal-Westernhausen, Germany) with a weigh system on the combine [39].
Fig. 1.
Field location and layout. The field was located in eastern Nebraska, USA (left). Cyan line indicates the flight path, and yellow rectangles indicate the studied 170 plots (right). The map was generated using images collected on April 27, 2018
UAV system and flight missions
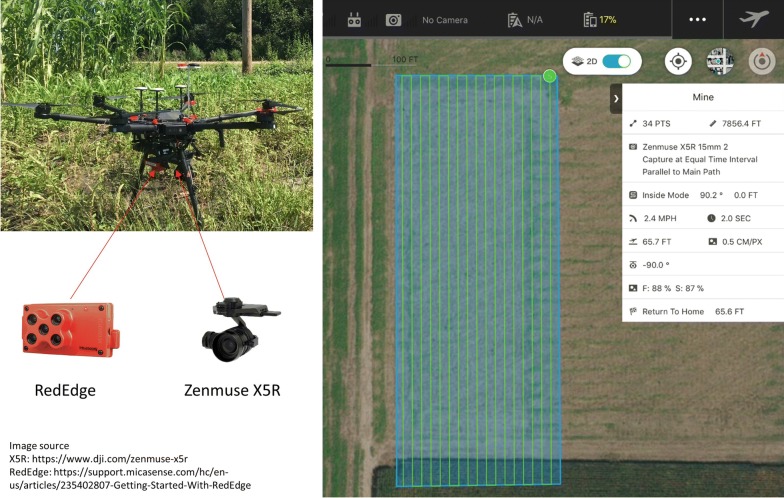
The UAV system used in this study consisted of a DJI Matrice 600 Pro multi-rotor platform (DJI, Shenzhen, China), a Zenmuse X5R RGB camera (DJI, Shenzhen, China), and a five-band multispectral camera RedEdge (Micasense, Seattle, USA) (Fig. 2). Each RGB image has an effective pixel size of 4608 by 3456, and each multispectral image has an effective pixel size of 1280 by 960. The multispectral camera also comes with a standard calibration panel for radiometric calibration, which was imaged on the ground right before or after each flight.
Fig. 2.
The UAV system (left) and the flight parameter settings on the DJI GS Pro application (right)
Seven RGB image sets and five multispectral image sets were acquired from mid-April to mid-June in 2018, and the corresponding growth stages are listed in Table 1. After several test flights, the flight altitude was set as 20 meters (65.7 FT in Fig. 2) above ground, and the forward and sideward overlaps were set as 88% and 87%, respectively. These flight parameters were set on the DJI GS pro application, as showed in Fig. 2 (right). The corresponding ground sampling distance (GSD) was 0.5 cm/pixel for the RGB image and 1.35 cm/pixel for the multispectral image. In order to do the geometric calibration, 21 ground control points (GCPs) using black and white cross-centered wooden boards were evenly placed over the field. GPS information of these GCPs was measured by a survey grade GNSS RTK GPS receiver (Topcon Positioning Systems, Inc., Tokyo, Japan), with centimeter accuracy in the X and Y directions, and centimeter accuracy in the Z direction. Three sources of data were used for the radiometric calibration: the data collected on the ground right before and after each flight over a MicaSense’s Calibrated Reflectance Panel, and the ambient light conditions in each of the five bands in a real time throughout the flight using a MicaSense Downwelling Light Sensor mounted on top of the UAV facing up towards the sky. The reflectance of the calibrated panel was 0.57, 0.57, 0.57, 0.56, and 0.53 in the blue, green, red, red edge, and near-infrared bands, respectively. In addition, standard calibration tarps were set up during data collections in the field with reflectance in 0.03, 0.22, and 0.48, respectively, to provide another source of information for radiometric calibration.
Table 1.
Seven data collections over the spring season of 2018
| Date | Acquired image type | Day of year (DOY) | Growth stage |
|---|---|---|---|
| April 22 | RGB | 111 | Tillering stage: Feekes 3 |
| April 27 | RGB and Multispectral | 116 | Green-up stage: Feekes 5 |
| May 7 | RGB and Multispectral | 126 | Jointing stage: Feekes 6 |
| May 15 | RGB | 134 | Flag leaf stage: Feekes 8 |
| May 21 | RGB and Multispectral | 140 | Boot stage: Feekes 9 |
| June 1 | RGB and Multispectral | 151 | Grain filling: Feekes 10.5.3 |
| June 18 | RGB and Multispectral | 168 | Physiological maturity: Feekes 11 |
Orthomosaicking and geometric and radiometric calibrations
Raw multispectral and RGB images were mosaicked or stitched using Pix4Dmapper (Pix4D, Lausanne, Switzerland), to further generate vegetation index (VI) and plant height maps [40]. Geometric calibration was performed during the orthomosaicking process in Pix4Dmapper using the GCPs setup at the data collection. Radiometric calibration included two steps. First one was the automatic radiometric calibration performed by Pix4Dmapper during the orthomosaicking process using the calibration data collected over the calibrated panel along with the ambient light changes collected during the flight from the downwelling light sensor. The output five-band maps from Pix4D were 16-bit GeoTIFFs, with pixel digital number ranging from 0 to 65,535. Pixel digital number in each map was further calibrated and converted to reflectance ratios ranging from 0 to 1, using the standard calibration tarps.
Vegetation index and plant height maps generation
Targeted plots were delineated with equal size and specific ID in ArcMap for following information extractions (Fig. 1). The plant height maps were calculated as the difference between the digital surface model (DSM) and the digital terrain model (DTM). The DSM was created automatically in Pix4Dmapper, representing the elevation of the canopy surface. A DTM map represents the elevation of the soil surface. In this study, the DTM was created by interpolating segmented soil pixels. Specifically, the RGB map from the earliest flight that had the highest proportion of bare soil exposure was transferred into the CIELAB color space. The histogram of the A channel in this color space is generally considered as a Gaussian-mixture model of vegetation pixels and soil pixels [41], thus being useful in segmenting soil and vegetation pixels. According to the threshold calculation method described in Liu et al. [42], a mask with only soil pixels was created. From this mask, thousands of soil points were randomly sampled to create DTM using Kriging interpolation in ArcMap 10.5.1 (Esri Inc. CA, USA).
Three classical VIs were calculated from the 5-band multispectral maps in RStudio 1.0.153 (RStudio, Inc. Boston, USA): NDVI, green NDVI (GNDVI), and normalized difference red edge (NDRE) (Eqs. 1–3). These traits are highly correlated with leaf chlorophyll contents and canopy structures, therefore, they are widely used for yield predictions [12, 14, 17, 18].
| 1 |
| 2 |
| 3 |
where R stands for the reflectance of the spectral band indicated in the subscript.
Variables extracted from VI and plant height maps
Two classes of variables were extracted in this study: pixel statistics and dynamic growth rate. For the pixel statistics, in addition to commonly extracted statistics (e.g., mean or median), each plot was further transferred into a texture feature matrix, i.e., gray-level co-occurrence matrix (GLCM), to derive statistical texture variables (contrast, correlation, energy, and homogeneity). The GLCM is a feature extraction method, allowing the extraction of second-order statistical texture variables [43]. The ‘second-order’ means that GLCM only considers the relationship between two pixels. Specifically, from un-transferred VI map, trimmed mean (mean value after trimming top and bottom 10% values), median (equals to 50th percentile), mode, and standard deviation were derived for each plot; similarly from un-transferred plant height map, trimmed mean, median, 95th percentile, and standard deviation were derived for each plot. After transferring each map into GLCM, another four statistical texture variables were calculated: contrast, correlation, energy, and homogeneity. Contrast represents the local gray level variations in an image; high contrast indicates the existence of any edges, noise, or wrinkled texture. Correlation measures the linear dependency of specified pixel pairs. Energy, also known as angular second moment, sums up the squared elements in GLCM. As for homogeneity, it is also called inverse difference moment and stands for the local homogeneity; high value represents the uniform local gray level.
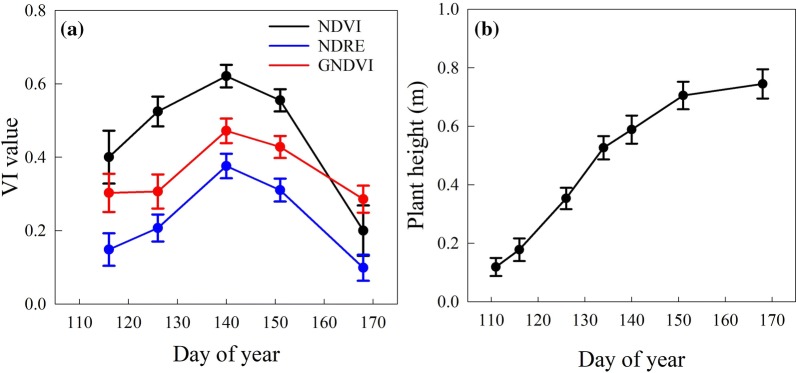
The second class of variable was dynamic growth rate, defined as the slope between two successive measurements along the season. In this study, dynamic growth rates were calculated for individual plots using the trimmed means of NDVI, NDRE, GNDVI, and plant height, respectively. An example was illustrated using NDVI dynamic curve in Fig. 3. With five NDVI measurements along the season, four growth rates were calculated with a negative number for the last one. The negative growth rate indicates the senescence process. As for plant height, with seven time-points in the dynamic curve (Fig. 5b), six growth rates were calculated.
Fig. 3.
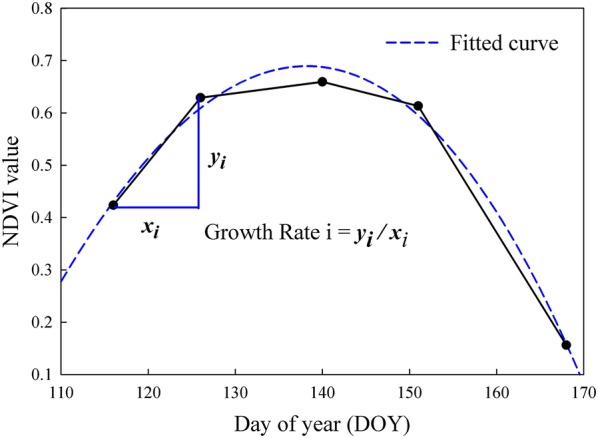
Dynamic growth rate calculation based on the dynamic curve of NDVI
Fig. 5.
Growth dynamic curves of VIs (a) and plant height (b) over the spring growing season in 2018. Error bars represent standard deviation among the 170 plots
Among the first class of variables, a significantly strong correlation (r = 0.99) was found between the trimmed mean and the median value. Therefore, the trimmed mean value was only used to calculate the dynamic growth rate and was not input into variable selection algorithms. Summing up two classes of variables, there were 172 variables for each experimental plot, as summarized in Table 2.
Table 2.
Summary of the 172 variables in each plot extracted from VI and plant height maps
| UAV derived map | Number of variables | |
|---|---|---|
| Pixel statistics | Dynamic growth rate | |
| Plant height | 7 features × 7 dates = 49* | 6 |
| NDVI | 7 features × 5 dates = 35** | 4 |
| NDRE | 7 features × 5 dates = 35** | 4 |
| GNDVI | 7 features × 5 dates = 35** | 4 |
| Total number of variables | 172 | |
* Median, 95th percentile, standard deviation, contrast, correlation, energy, and homogeneity
** Median, mode, standard deviation, contrast, correlation, energy, and homogeneity
Principal variable selection for grain yield estimation
The extracted 172 variables were treated as candidates in two variable selection algorithms to explain grain yield. Normalization was conducted before each selection procedure. The main parameter tuned in LASSO was lambda, a shrinkage penalty term. It was tuned through 10-fold cross-validation, with mean squared error (MSE) as the loss function. Lambda shrank some variable coefficients to zero while retaining non-zero variables. In addition to variable selection, LASSO also estimates the regression coefficients for selected variables. Since all variables were normalized, variables with higher absolute coefficient could be considered as contributing more in explaining grain yield. Therefore, the absolute regression coefficient was used as the ‘importance score’ for the variable selected by LASSO.
Random forest ranks variables according to the permutation importance of each variable. Generally, if a variable X is important for the dependent variable Y, permuting the order of X will break the correlation link between X and Y, thus increasing prediction error (MSE) [27]. In other words, the higher the increase in MSE after permuting variable X, the more critical the variable X is. Therefore, the increase in MSE (%IncMSE) was chose as the ‘importance score’ in random forest selection. Parameter tuned for the random forest in this study was the number of trees to grow, and the number of variables randomly sampled as candidates at each split. Both were optimized by grid search and were set as 1500 and 2 separately.
Compared to random forest, LASSO is more sensitive to multicollinearity among variables [44]. When there is a group of correlated variables, LASSO would arbitrarily select one variable from this group at each random run, thus resulting in inconsistent selections [45]. To alleviate this effect, each algorithm was set to run 30 times with different random seeds. Afterward, each variable had two lists of importance scores, with a length of 30, from LASSO and random forest. Ten variables with the highest averaged importance scores were chosen for LASSO and random forest, respectively.
To further evaluate the ability of grain yield prediction using the selected variable sets versus the original variables, ridge regression and Gaussian kernel-based support vector machine (SVM, non-parametric) were applied. Ridge regression is a parametric prediction algorithm. It is capable of addressing the collinearity issue that was not handled by multiple linear regression [46–48]. SVM can be used to solve classification and regression problems by constructing a hyperplane with maximized margins for separation in a high-dimensional space. The use of Gaussian radial basis function allows the SVM model to address nonlinearity data [49]. For each model, 80% observations were used as training data and 20% observations were treated as testing data. The predicting performance of each model using the selected variable sets was compared with the performance using all 172 variables and the correlation coefficients (r) and the root mean squared errors (RMSE) were shown. In addition, performance on lines grouped by their genetic background were also investigated, i.e., the NE lines, TX lines, WB lines, and two other lines (OK and SY line).
Results and discussion
Growth dynamics in terms of VI and plant height
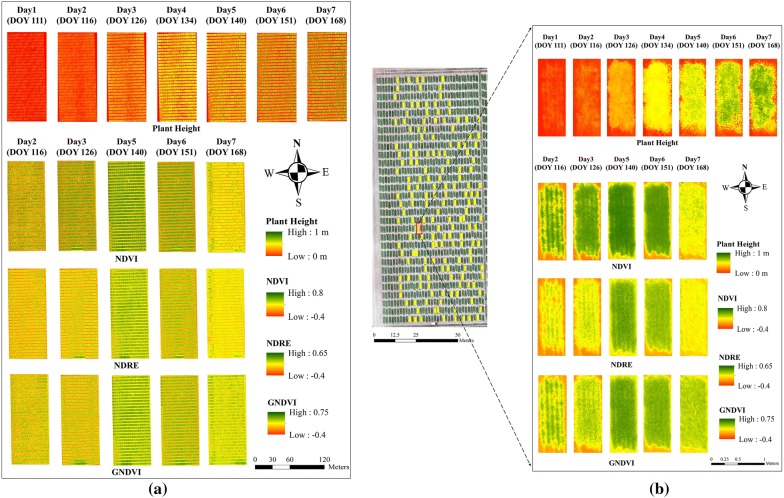
One application of the multi-temporal UAV data was to track the growth trend of winter wheat, using UAV-derived maps and dynamic curves. The multi-temporal maps tracking seasonal growth of the whole field was provided in Fig. 4a. Additionally, to visualize the growth trend of specific plot, one plot was randomly selected as the example and was presented in terms of plant height and VIs (Fig. 4b). Greener pixel of plant height means taller wheat plant, while greener pixel of VI indicates greater wheat vigor. As expected, plant height increased over the spring growing season; whereas the VI value peaked and had the greatest vigor on the 140 DOY. Among three VIs, a significant saturation issue could be found in NDVI around 140 DOY, which agrees with most other studies [50, 51] that NDVI tends to saturate with dense canopy cover.
Fig. 4.
Multi-temporal maps of plant height and three vegetation indices. DOY represents ‘day of year’ for each date, corresponding to x-axis in Fig. 5. a Whole field maps. b An example plot selected randomly (field map used images collected on May 7, 2019)
The dynamic curves provide a quantitative way to describe growth trend, using a trimmed mean of plant height or VI (Fig. 5). The NDVI, GNDVI, and NDRE followed a similar growth trend that reached a peak at the 140 DOY, with a significant drop after the 151 DOY (Fig. 5a). The dynamic curve also exhibited different growth rates over the season. For example, NDVI had almost equal growth rates between the 116 and 140 DOY, whereas NDRE and GNDVI had a smaller growth rate between the 116 and 126 DOY than that between the 126 and 140 DOY. In Fig. 5b, the plant height dynamic curve showed an increasing trend along seven data collections.
Both maps and dynamic curves exhibited similar winter wheat growth trends (Figs. 4 and 5): VI reached peak value around May 21 (boot stage) and plant height increased until June 18 (physiological maturity). The significantly low VI values on the last data collection were due to leaf yellowing at the end of the growth season. The seasonal changes of these VIs were similar to previous research [8, 52]. Similarly, the growth trend of plant height was also found in related research [36, 53], followed the pattern of the sigmoid curve. It was also noticeable that VI curves started to drop around May 21, whereas the growth rate of plant height decreased significantly after June 1. Considering the manually recorded flowering date (from end-May to early-June), it is possible that these changes, i.e., VI vigor starting to drop and height growth rate beginning to decrease, were synchronized with flowering.
Variable selection by LASSO and random forest
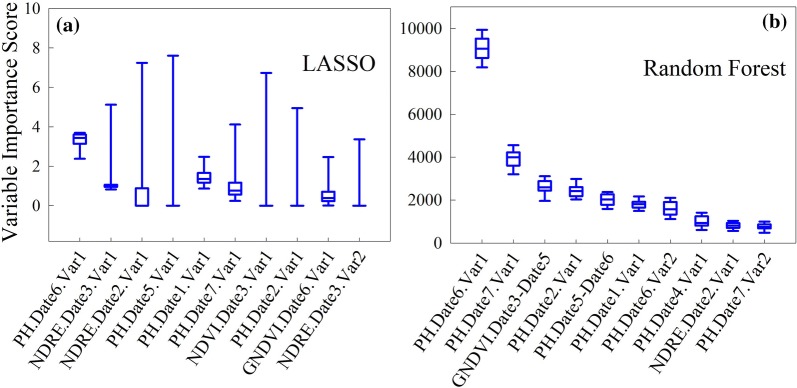
LASSO and random forest selected top ten variables were ranked in Table 3, according to averaged importance score. Each row represents one variable, with details of specific variable name, the corresponded map and on which times of data collection that the variable was derived from. Besides, an abbreviation was given for each selected variable and shown in x-axis of Fig. 6, with the purpose of presenting summarized variable importance scores. Two common variables selected by both algorithms were: PH.Date6.Var1 and NDRE.Date2.Var1.
Table 3.
Ten variables selected by LASSO and random forest, respectively
| Rank | Feature | Originated map | Date | Abbreviation |
|---|---|---|---|---|
| LASSO selected variables | ||||
| 1 | Median | Plant height | 6th | PH.Date6.Var1 |
| 2 | Contrast | NDRE | 3rd | NDRE.Date3.Var1 |
| 3 | Standard deviation | NDRE | 2nd | NDRE.Date2.Var1 |
| 4 | 95th percentile | Plant height | 5th | PH.Date5.Var1 |
| 5 | 95th percentile | Plant height | 1st | PH.Date1.Var1 |
| 6 | Homogeneity | Plant height | 7th | PH.Date7.Var1 |
| 7 | Standard deviation | NDVI | 3rd | NDVI.Date3.Var1 |
| 8 | Standard deviation | Plant height | 2nd | PH.Date2.Var1 |
| 9 | Correlation | GNDVI | 6th | GNDVI.Date6.Var1 |
| 10 | Standard deviation | NDRE | 3rd | NDRE.Date3.Var2 |
| Random forest selected variables | ||||
| 1 | Median | Plant height | 6th | PH.Date6.Var1 |
| 2 | Median | Plant height | 7th | PH.Date7.Var1 |
| 3 | Second growth rate | GNDVI | 3rd and 5th | GNDVI. Date3-Date5 |
| 4 | 95th percentile | Plant height | 2nd | PH.Date2.Var1 |
| 5 | Fifth growth rate | Plant height | 5th and 6th | PH.Date5-Date6 |
| 6 | Correlation | Plant height | 1st | PH.Date1.Var1 |
| 7 | 95th percentile | Plant height | 6th | PH.Date6.Var2 |
| 8 | Median | Plant height | 4th | PH.Date4.Var1 |
| 9 | Standard deviation | NDRE | 2nd | NDRE.Date2.Var1 |
| 10 | 95th percentile | Plant height | 7th | PH.Date7.Var2 |
Common variables selected by both algorithms were given in italic
Fig. 6.
Variable importance scores of top 10 variables selected by LASSO and random forest
PH.Date6.Var1, corresponded to the median value of the canopy height measurements within a plot around the grain filling stage (Table 3), was ranked as the top one by both models. The grain filling stage has already been proved to be critical for assessing wheat grain yield in previous research [20, 54, 55]. Both models selected more plant height related variables over VI related ones in this case. Though results may change in another case, this still confirmed the importance of plant height in wheat grain yield prediction. In fact, the relationship between plant height and wheat yield has always been an interesting topic for breeders. Back in 1978, Law [56] found a positive relationship between plant height and wheat yield. Further, with wheat plant height measured over multiple growth stages, strong positive correlations were found between plant height and final grain yield [57]. What is new with the UAS-based phenotyping compared with the traditional methods is that we are now able to obtain ultra-high resolution plot imagery to derive more features, such as the variation of plant height or vegetation indices within a plot other than just an averaged number, that are potentially very useful in breeding.
The potential importance of plant height, however, does not necessary mean that the vegetation indices are not useful in explaining grain yield. Good performance of VI on grain yield prediction have already be demonstrated in many studies [58, 59]. NDRE.Date2.Var1, another common variable selected by LASSO and random forest, was ranked as top third variable by LASSO. However, different from PH.Date6.Var1 that was derived at later growth stage, NDRE.Date2.Var1 was calculated from earlier growth stages. Besides, most of the other VI related variables selected were derived on earlier dates. This finding potentially indicates the importance of VI variables derived at earlier growth stages. In addition, compare to the most commonly known NDVI, NDRE and GNDVI related variables seem to appear more frequently in the selection results. As mentioned above, NDVI tends to saturate earlier than NDRE and GNDVI, which possibly result in less NDVI variables being selected.
Figure 6 summarized the importance scores of selected variables from 30 random runs. It was observed that the selection results of random forest were more consistent than those of LASSO. As mentioned above, since LASSO has higher sensitivity to multicollinearity among variables than random forest [44], the inconsistent results of LASSO indicated a possible multicollinearity issue among variables. Running the algorithm for 30 times randomly was the method adopted in this study to alleviate influence from this issue. Another solution that could be considered in future work is, to cluster correlated variables into groups first and then do selection on representative variables from each group [60, 61].
Grain yield prediction using selected variable sets
With the two sets of selected principal variables, the grain yield was estimated using both ridge regression and SVM model with Gaussian radial basis kernel. Performances on testing data (20%) were reported in Table 4, with r and RMSE averaged from 20 random sets of testing data.
Table 4.
Performance of grain yield prediction on testing data, using variable sets determined from LASSO and random forest, as well as all available variables
| Variables | LASSO selected variables | Random forest selected variables | All 172 variables | |||
|---|---|---|---|---|---|---|
| Sample size | r* | RMSE* (g/plot) | r | RMSE (g/plot) | r | RMSE (g/plot) |
| (1) Predictions of SVM model with Gaussian radial basis kernel | ||||||
| All lines | 0.32 | 320.19 | 0.39 | 306.15 | 0.29 | 314.77 |
| NE lines | 0.58 | 326.97 | 0.77 | 254.66 | 0.72 | 284.08 |
| TX lines | 0.21 | 271.44 | 0.36 | 255.51 | 0.57 | 215.92 |
| WB lines | 0.28 | 271.88 | 0.41 | 236.82 | 0.25 | 264.53 |
| OK and SY lines | 0.39 | 201.45 | 0.45 | 191.06 | 0.36 | 193.31 |
| (2) Predictions of ridge regression model | ||||||
| All lines | 0.49 | 283.86 | 0.39 | 301.89 | 0.25 | 314.83 |
| NE lines | 0.73 | 272.72 | 0.81 | 225.45 | 0.73 | 295.92 |
| TX lines | 0.55 | 235.37 | 0.50 | 255.68 | 0.47 | 242.99 |
| WB lines | 0.40 | 247.57 | 0.42 | 246.21 | 0.22 | 266.25 |
| OK and SY lines | 0.59 | 163.69 | 0.54 | 164.90 | 0.58 | 169.29 |
* Values of r and RMSE were averaged from 20 random sets of testing data
Comparing among three different variable sets, random forest selected variable set with SVM model had a slightly higher prediction accuracies (r = 0.36–0.77) than the other two variable sets (r = 0.21–0.58 for the LASSO selected set, and r = 0.25–0.72 for all variables); whereas the LASSO selected variable set with ridge regression (r = 0.40–0.73) had relatively but not significantly better performance than the other two sets (r = 0.39–0.81 in random forest selected set, and r = 0.22–0.73 in all variables set). It is noticeable that both random forest and SVM are non-parametric algorithm, while both LASSO and ridge regression are parametric algorithm. A possible suggestion could be made is that, non-parametric prediction model could be adopted to match the non-parametric variable selection, and vice versa. Another finding by comparing the three variable sets is that, the overall performance of using two selected variable sets with greatly reduced number of variables was better than using whole 172 variables.
The selected variables actually performed better on individual grouped lines with different genetic background than pooling all ten lines together (Table 4). The highest prediction performance was achieved for the NE lines (r = 0.58–0.81). Except for the NE lines which were bred for NE environments, the rest of them were bred for their target environments. Use the WB Cedar line as an example. This is a relatively shorter line and flowers at least a week before the NE lines. Hence, the variables that were picked as being most important might be different for Cedar compared to NE lines. And the way that we used sampling dates as opposed to developmental stages in the study would also exaggerate the issue. The observations with all 10 checks could be confounded by the different genetic backgrounds and development patterns. When analyzed them separately, this confounding effect was avoided and that is probably why the results looked a lot better. Even for a single line, there existed much variation in the field. For example, the yield of Freeman ranges from 1057 g (35.2 bu/a) to 1813 g (60.4 bu/a)—variation of nearly 25 bushels per acre. The variation can be expected to be even larger among those un-adapted lines. To illustrate the information in Table 4, the predicted grain yield of a random training–testing set with the ridge regression model was plotted versus measured yield (Fig. 7). The results of pooled ten lines and individual group of lines were shown.
Fig. 7.
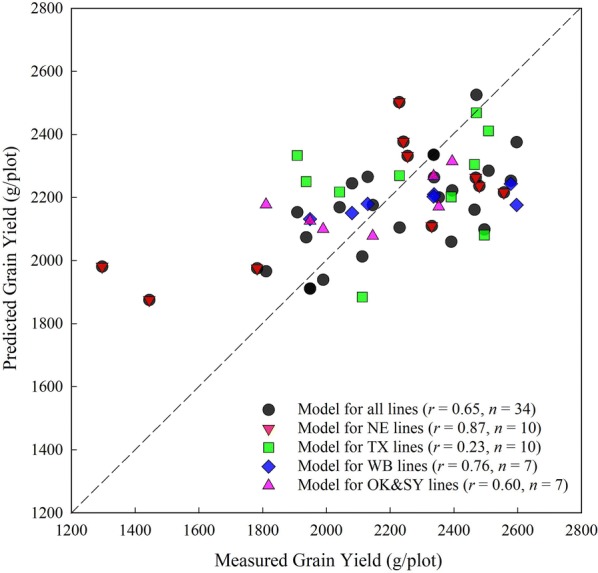
Relationship between measured and predicted grain yield of an example training–testing set for each group of lines. Dashed line is the 1:1 line. r is correlation coefficient; n is testing sample size
Conclusions
This study investigated the principal variable selection from features extracted from UAV-derived imagery for winter wheat grain yield prediction using LASSO and random forest algorithms. Selection results showed that plant height related variable derived at grain filling stage was ranked as the top one by both LASSO and random forest models; whereas temporal plant height and VIs are important throughout the season. Furthermore, the yield prediction using reduced variable sets selected by LASSO or random forest had comparable or even better accuracy than using all extracted variables. This indicated the possibility of a better allocation of efforts and resources on phenotypic data collection and processing by narrowing down the targeted secondary traits and growth stages. What can be noticed is that with the ultra-high resolution plot imagery obtained by the UAS-based phenotyping we are now able to derive more features, such as the variation of plant height or vegetation indices within a plot other than just an averaged number, that are potentially very useful for the breeding purpose. Further studies can be conducted to investigate the potential of genomic prediction by incorporating the selected key secondary traits measured by sensing systems into the genomic prediction models to increase the prediction accuracy. This study also serves as a preliminary study for future experiments, where the proposed variable selection methodology can be tested with more data collected in multiple years and locations to streamline the phenotyping to support the breeding process.
Acknowledgements
Not applicable.
Authors’ contributions
JL and YS formed the initial idea of this study. PSB, MB, NDG, HD, and VB provided the field and all the ground truth data. JL, AV, YS conducted the aerial data collections. JL performed the aerial data analysis. JL and YS wrote the manuscript. All authors read and approved the final manuscript.
Funding
This project is based on research that was supported by the Wheat Innovation Foundation fund from the Agricultural Research Division of the University of Nebraska-Lincoln, and the funding supported by the Nebraska Agricultural Experiment Station through the Hatch Act capacity funding program (Accession Number 1011130) from the USDA National Institute of Food and Agriculture.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors certified that they have no affiliations with or involvement in any organization or entity with any financial interest, or non-financial interest, in the subject matter or materials discussed in this manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jiating Li, Email: jiatingli@huskers.unl.edu.
Arun-Narenthiran Veeranampalayam-Sivakumar, Email: arun-narenthiran@huskers.unl.edu.
Madhav Bhatta, Email: bhattamadove@gmail.com.
Nicholas D. Garst, Email: nicholas.garst@huskers.unl.edu
Hannah Stoll, Email: h.donoho@gmail.com.
P. Stephen Baenziger, Email: pbaenziger1@unl.edu
Vikas Belamkar, Email: vikas.belamkar@unl.edu.
Reka Howard, Email: rekahoward@unl.edu.
Yufeng Ge, Email: yge2@unl.edu.
Yeyin Shi, Email: yshi18@unl.edu.
References
- 1.Belamkar V, Guttieri MJ, Hussain W, Jarquín D, El-basyoni I, Poland J, et al. Genomic selection in preliminary yield trials in a winter wheat breeding program. G3 Genes Genomes Genet. 2018;8(8):2735–2747. doi: 10.1534/g3.118.200415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Makino A. Photosynthesis, grain yield, and nitrogen utilization in rice and wheat. Plant Physiol. 2010;155(1):125–129. doi: 10.1104/pp.110.165076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ray DK, Mueller ND, West PC, Foley JA. Yield trends are insufficient to double global crop production by 2050. PLoS ONE. 2013;8(6):e66428. doi: 10.1371/journal.pone.0066428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai G, Ge Y, Hussain W, Baenziger PS, Graef G. A multi-sensor system for high throughput field phenotyping in soybean and wheat breeding. Comput Electron Agric. 2016;128:181–192. doi: 10.1016/j.compag.2016.08.021. [DOI] [Google Scholar]
- 5.Crain J, Reynolds M, Poland J. Utilizing high-throughput phenotypic data for improved phenotypic selection of stress-adaptive traits in wheat. Crop Sci. 2017;57(2):648–659. doi: 10.2135/cropsci2016.02.0135. [DOI] [Google Scholar]
- 6.Kipp S, Mistele B, Schmidhalter U. Identification of stay-green and early senescence phenotypes in high-yielding winter wheat, and their relationship to grain yield and grain protein concentration using high-throughput phenotyping techniques. Funct Plant Biol. 2014;41(3):227–235. doi: 10.1071/FP13221. [DOI] [PubMed] [Google Scholar]
- 7.Rischbeck P, Elsayed S, Mistele B, Barmeier G, Heil K, Schmidhalter U. Data fusion of spectral, thermal and canopy height parameters for improved yield prediction of drought stressed spring barley. Eur J Agron. 2016;78:44–59. doi: 10.1016/j.eja.2016.04.013. [DOI] [Google Scholar]
- 8.Comar A, Burger P, De Solan B, Baret F, Daumard F, Hanocq JF. A semi-automatic system for high throughput phenotyping wheat cultivars in-field conditions: description and first results. Funct Plant Biol. 2012;39(11):914–924. doi: 10.1071/FP12065. [DOI] [PubMed] [Google Scholar]
- 9.Geipel Jakob, Link Johanna, Wirwahn Jan, Claupein Wilhelm. A Programmable Aerial Multispectral Camera System for In-Season Crop Biomass and Nitrogen Content Estimation. Agriculture. 2016;6(1):4. doi: 10.3390/agriculture6010004. [DOI] [Google Scholar]
- 10.Haghighattalab A, Crain J, Mondal S, Rutkoski J, Singh RP, Poland J. Application of geographically weighted regression to improve grain yield prediction from unmanned aerial system imagery. Crop Sci. 2017;57(5):2478–2489. doi: 10.2135/cropsci2016.12.1016. [DOI] [Google Scholar]
- 11.Benincasa P, Antognelli S, Brunetti L, Fabbri CA, Natale A, Sartoretti V, et al. Reliability of NDVI derived by high resolution satellite and UAV compared to in-field methods for the evaluation of early crop N status and grain yield in Wheat. Exp Agric. 2018;54(4):604–622. doi: 10.1017/S0014479717000278. [DOI] [Google Scholar]
- 12.Guan Senlin, Fukami Koichiro, Matsunaka Hitoshi, Okami Midori, Tanaka Ryo, Nakano Hiroshi, Sakai Tetsufumi, Nakano Keiko, Ohdan Hideki, Takahashi Kimiyasu. Assessing Correlation of High-Resolution NDVI with Fertilizer Application Level and Yield of Rice and Wheat Crops using Small UAVs. Remote Sensing. 2019;11(2):112. doi: 10.3390/rs11020112. [DOI] [Google Scholar]
- 13.Du M, Noguchi N. Monitoring of wheat growth status and mapping of wheat yield’s within-field spatial variations using color images acquired from UAV-camera system. Remote Sens. 2017;9(3):289. doi: 10.3390/rs9030289. [DOI] [Google Scholar]
- 14.Duan T, Chapman SC, Guo Y, Zheng B. Dynamic monitoring of NDVI in wheat agronomy and breeding trials using an unmanned aerial vehicle. F Crop Res. 2017;210:71–80. doi: 10.1016/j.fcr.2017.05.025. [DOI] [Google Scholar]
- 15.Kefauver SC, Vicente R, Vergara-Díaz O, Fernandez-Gallego JA, Kerfal S, Lopez A, et al. Comparative UAV and field phenotyping to assess yield and nitrogen use efficiency in hybrid and conventional barley. Front Plant Sci. 2017;8:1–15. doi: 10.3389/fpls.2017.01733/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanning Martin, Kühling Insa, Trautz Dieter, Jarmer Thomas. High-Resolution UAV-Based Hyperspectral Imagery for LAI and Chlorophyll Estimations from Wheat for Yield Prediction. Remote Sensing. 2018;10(12):2000. doi: 10.3390/rs10122000. [DOI] [Google Scholar]
- 17.Kyratzis AC, Skarlatos DP, Menexes GC, Vamvakousis VF, Katsiotis A. Assessment of vegetation indices derived by UAV imagery for durum wheat phenotyping under a water limited and heat stressed mediterranean environment. Front Plant Sci. 2017;8:1–14. doi: 10.3389/fpls.2017.01114/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hassan MA, Yang M, Rasheed A, Yang G, Reynolds M, Xia X, et al. A rapid monitoring of NDVI across the wheat growth cycle for grain yield prediction using a multi-spectral UAV platform. Plant Sci. 2018 doi: 10.1016/j.plantsci.2018.10.022. [DOI] [PubMed] [Google Scholar]
- 19.Moravec D, Komárek J, Kumhálová J, Kroulík M, Prošek J, Klápště P. Digital elevation models as predictors of yield: comparison of an UAV and other elevation data sources. Agron Res. 2017;15(1):249–255. [Google Scholar]
- 20.Wang L, Tian Y, Yao X, Zhu Y, Cao W. Predicting grain yield and protein content in wheat by fusing multi-sensor and multi-temporal remote-sensing images. F Crop Res. 2014;164(1):178–188. doi: 10.1016/j.fcr.2014.05.001. [DOI] [Google Scholar]
- 21.Montesinos-López OA, Montesinos-López A, Crossa J, los Campos G, Alvarado G, Suchismita M, et al. Predicting grain yield using canopy hyperspectral reflectance in wheat breeding data. Plant Methods. 2017;13(1):1–23. doi: 10.1186/s13007-016-0152-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Degenhardt F, Seifert S, Szymczak S. Evaluation of variable selection methods for random forests and omics data sets. Brief Bioinform. 2017 doi: 10.1093/bib/bbx124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guyon I, Elisseeff A. An introduction to variable and feature selection. J Mach Learn Res. 2011;3:1157–1182. [Google Scholar]
- 24.Andersen CM, Bro R. Variable selection in regression-a tutorial. J Chemom. 2010;24(11–12):728–737. doi: 10.1002/cem.1360. [DOI] [Google Scholar]
- 25.Tibshirani R. Regression shrinkage and selection via the LASSO. J R Stat Soc Ser B. 1996;58(1):267–288. [Google Scholar]
- 26.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 27.Gregorutti B, Michel B, Saint-Pierre P. Correlation and variable importance in random forests. Stat Comput. 2017;27(3):659–678. doi: 10.1007/s11222-016-9646-1. [DOI] [Google Scholar]
- 28.Genuer R, Poggi JM, Tuleau-Malot C. Variable selection using random forests. Pattern Recognit Lett. 2010;31(14):2225–2236. doi: 10.1016/j.patrec.2010.03.014. [DOI] [Google Scholar]
- 29.Archer KJ, Kimes RV. Empirical characterization of random forest variable importance measures. Comput Stat Data Anal. 2008;52(4):2249–2260. doi: 10.1016/j.csda.2007.08.015. [DOI] [Google Scholar]
- 30.Leng C, Lin Y, Wahba G. A note on the LASSO and related procedures. Stat Sin. 2006;16:1273–1284. [Google Scholar]
- 31.Grömping U. Variable importance assessment in regression: linear regression versus random forest. Am Stat. 2009;63(4):308–319. doi: 10.1198/tast.2009.08199. [DOI] [Google Scholar]
- 32.Schirrmann M, Giebel A, Gleiniger F, Pflanz M, Lentschke J, Dammer KH. Monitoring agronomic parameters of winter wheat crops with low-cost UAV imagery. Remote Sens. 2016;8(9):706. doi: 10.3390/rs8090706. [DOI] [Google Scholar]
- 33.Shafian S, Rajan N, Schnell R, Bagavathiannan M, Valasek J, Shi Y, et al. Unmanned aerial systems-based remote sensing for monitoring sorghum growth and development. PLoS ONE. 2018;13:e0196605. doi: 10.1371/journal.pone.0196605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunt ER, Dean Hively W, Fujikawa SJ, Linden DS, Daughtry CST, McCarty GW. Acquisition of NIR-green-blue digital photographs from unmanned aircraft for crop monitoring. Remote Sens. 2010;2(1):290–305. doi: 10.3390/rs2010290. [DOI] [Google Scholar]
- 35.Holman FH, Riche AB, Michalski A, Castle M, Wooster MJ, Hawkesford MJ. High throughput field phenotyping of wheat plant height and growth rate in field plot trials using UAV based remote sensing. Remote Sens. 2016;8(12):1031. doi: 10.3390/rs8121031. [DOI] [Google Scholar]
- 36.Chu T, Chen R, Landivar JA, Maeda MM, Yang C, Starek MJ. Cotton growth modeling and assessment using unmanned aircraft system visual-band imagery. J Appl Remote Sens. 2016;10(3):036018. doi: 10.1117/1.JRS.10.036018. [DOI] [Google Scholar]
- 37.Bendig J, Bolten A, Bennertz S, Broscheit J, Eichfuss S, Bareth G. Estimating biomass of barley using crop surface models (CSMs) derived from UAV-based RGB imaging. Remote Sens. 2014;6(11):10395–10412. doi: 10.3390/rs61110395. [DOI] [Google Scholar]
- 38.Iqbal F, Lucieer A, Barry K, Wells R. Poppy crop height and capsule volume estimation from a single UAS flight. Remote Sens. 2017;9(7):24–27. doi: 10.3390/rs9070647. [DOI] [Google Scholar]
- 39.Bhatta M, Eskridge KM, Rose DJ, Santra DK, Baenziger PS, Regassa T. Seeding rate, genotype, and topdressed nitrogen effects on yield and agronomic characteristics of winter wheat. Crop Sci. 2017;57(2):951–963. doi: 10.2135/cropsci2016.02.0103. [DOI] [Google Scholar]
- 40.Li J, Shi Y, Veeranampalayam-Sivakumar A-N, Schachtman DP. Elucidating sorghum biomass, nitrogen and chlorophyll contents with spectral and morphological traits derived from unmanned aircraft system. Front Plant Sci. 2018;9:1–12. doi: 10.3389/fpls.2018.01406/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li L, Mu X, Macfarlane C, Song W, Chen J, Yan K, et al. A half-Gaussian fitting method for estimating fractional vegetation cover of corn crops using unmanned aerial vehicle images. Agric For Meteorol. 2018;262:379–390. doi: 10.1016/j.agrformet.2018.07.028. [DOI] [Google Scholar]
- 42.Liu Y, Mu X, Wang H, Yan G. A novel method for extracting green fractional vegetation cover from digital images. J Veg Sci. 2012;23(3):406–418. doi: 10.1111/j.1654-1103.2011.01373.x. [DOI] [Google Scholar]
- 43.Mohanaiah P, Sathyanarayana P, Gurukumar L. Image texture feature extraction using GLCM approach. Int J Sci Res Publ. 2013;3(1):2250–3153. http://www.ijsrp.org.
- 44.Wang L, Zhou X, Zhu X, Dong Z, Guo W. Estimation of biomass in wheat using random forest regression algorithm and remote sensing data. Crop J. 2016;4(3):212–219. doi: 10.1016/j.cj.2016.01.008. [DOI] [Google Scholar]
- 45.Lu F, Petkova E. A comparative study of variable selection methods in the context of developing psychiatric screening instruments. Stat Med. 2014;33(3):401–421. doi: 10.1002/sim.5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McDonald GC. Ridge regression. Wiley Interdiscip Rev Comput Stat. 2009;1(1):93–100. doi: 10.1002/wics.14. [DOI] [Google Scholar]
- 47.de Vlaming R, Groenen PJF. The current and future use of ridge regression for prediction in quantitative genetics. Biomed Res Int. 2015;2015:1–18. doi: 10.1155/2015/143712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orhan H, Eyduran E, Tatliyer A, Saygici H. Prediction of egg weight from egg quality characteristics via ridge regression and regression tree methods. Rev Bras Zootec. 2016;45(7):380–385. doi: 10.1590/S1806-92902016000700004. [DOI] [Google Scholar]
- 49.Basak D, Pal S, Patranabis DC. Support vector regression. Neural Inf Process Lett Rev. 2007;11(10):203–224. [Google Scholar]
- 50.Mutanga O, Skidmore AK. Narrow band vegetation indices overcome the saturation problem in biomass estimation. Int J Remote Sens. 2004;25(19):3999–4014. doi: 10.1080/01431160310001654923. [DOI] [Google Scholar]
- 51.Gu Y, Wylie BK, Howard DM, Phuyal KP, Ji L. NDVI saturation adjustment : a new approach for improving cropland performance estimates in the Greater Platte River Basin, USA. Ecol Indic. 2013;30:1–6. doi: 10.1016/j.ecolind.2013.01.041. [DOI] [Google Scholar]
- 52.Kalubarme MH, Potdar MB, Manjunath KR, Mahey RK, Siddhu SS. Growth profile based crop yield models: a case study of large area wheat yield modelling and its extendibility using atmospheric corrected NOAA AVHRR data. Int J Remote Sens. 2003;24(10):2037–2054. doi: 10.1080/01431160210156018. [DOI] [Google Scholar]
- 53.Chang A, Jung J, Maeda MM, Landivar J. Crop height monitoring with digital imagery from Unmanned Aerial System (UAS) Comput Electron Agric. 2017;141:232–237. doi: 10.1016/j.compag.2017.07.008. [DOI] [Google Scholar]
- 54.Hassan MA, Yang M, Rasheed A, Jin X, Xia X, Xiao Y, et al. Time-series multispectral indices from unmanned aerial vehicle imagery reveal senescence rate in bread wheat. Remote Sens. 2018;10(6):809. doi: 10.3390/rs10060809. [DOI] [Google Scholar]
- 55.Bowman BC, Chen J, Zhang J, Wheeler J, Wang Y, Zhao W, et al. Evaluating grain yield in spring wheat with canopy spectral reflectance. Crop Sci. 2015;55(5):1881–1890. doi: 10.2135/cropsci2014.08.0533. [DOI] [Google Scholar]
- 56.Law CN, Snape JW, Worland AJ. The genetical relationship between height and yield in wheat. Heredity. 1978;40:133–151. doi: 10.1038/hdy.1978.13. [DOI] [Google Scholar]
- 57.Girma K, Martin KL, Anderson RH, Arnall DB, Brixey KD, Casillas MA, et al. Mid-season prediction of wheat-grain yield potential using plant, soil, and sensor measurements. J Plant Nutr. 2006;29(5):873–897. doi: 10.1080/01904160600649187. [DOI] [Google Scholar]
- 58.Labus MP, Nielsen GA, Lawrence RL, Engel R, Long DS. Wheat yield estimates using multi-temporal NDVI satellite imagery. Int J Remote Sens. 2002;23(20):4169–4180. doi: 10.1080/01431160110107653. [DOI] [Google Scholar]
- 59.Moges SM, Raun WR, Mullen RW, Freeman KW, Johnson G V. Evaluation of Green, Red, and Near Infrared Bands for Predicting Winter Wheat Biomass, Nitrogen Uptake, and Final Grain Yield. 2006;4167.
- 60.Bühlmann P, Rütimann P, van de Geer S, Zhang CH. Correlated variables in regression: clustering and sparse estimation. J Stat Plan Inference. 2013;143(11):1835–1858. doi: 10.1016/j.jspi.2013.05.019. [DOI] [Google Scholar]
- 61.Bondell HD, Reich BJ. Simultaneous regression shrinkage, variable selection, and supervised clustering of predictors with OSCAR. Biometrics. 2008;64(1):115–123. doi: 10.1111/j.1541-0420.2007.00843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.