Abstract
Background and objectives
Lymph node metastasis is a key factor in predicting and determining the prognosis of patients with colorectal cancer (CRC). Sodium channels are highly expressed in a variety of tumors and are closely related to tumor development, metastasis, and invasion. We investigated the relationship between the expressions of different subtypes of Nav channels and lymph node metastasis of CRC.
Methods
Real-time PCR (RT-qPCR) was carried out to measure the expressions of different sodium channel subtypes, chemokine receptors (CCR2, CCR4, CCR7), and lymphocyte infiltration-related biomarkers (CD3e, CD8a, IL-2RA) in CRC tissues from 97 patients. The expressions of Nav1.5 and Nav1.6 in surgically isolated lymph nodes were detected by immunohistochemistry. Correlation analysis between expressions of different genes and lymph node metastasis was performed by two-tailed t test.
Results
Nav1.1 and Nav1.6 were highly expressed in CRC tissues and positively correlated with CRC lymph node metastasis. Nav1.6 was also highly expressed in metastatic lymph nodes. Further analysis showed that the high expression of Nav1.6 was closely related to the one of CCR2\CCR4 in tumor lymph node metastasis.
Conclusions
These results suggested that Nav1.6 might be a novel marker for CRC lymph node metastasis.
Keywords: Colorectal cancer, Sodium channels, Lymph nodes
Background
Colorectal cancer (CRC) is the second leading cause of tumor-related death and the third most common malignant tumor worldwide, only secondary to lung cancer and breast cancer [1, 2]. The main cause of death from CRC is distant metastasis of the tumor. The distant metastasis of CRC leads to its advanced stage with low remission and survival rates in patients. Lymph node metastasis is the most common metastatic pathway of CRC [3] and is also a key factor in predicting and determining the prognosis of patients [4]. O’Connell et al. reviewed clinical and pathological data of more than 100,000 patients with CRC [2]. Results showed that the 5-year stage-specific survivals were 93.2% for stage I, 84.7% for stage IIa, 72.2% for stage IIb, 83.4% for stage IIIa, 64.1% for stage IIIb, 44.3% for stage IIIc, and 8.1% for stage IV [2] (AJCC sixth edition system). Leonard L et al.’s statistics showed that the 5-year survival rate was 65.4% in patients with lymph node metastasis and 94.4% in patients without lymph node metastasis [5]. Therefore, the pathological examination of lymph nodes in CRC patients is crucial for post-operative treatment and patient prognosis [4].
The early diagnosis of tumor lymph node metastasis depends on the detection of specific markers of lymphangiogenesis. VEGFR3 is a relatively specific receptor expressed on lymphatic endothelial cells and involved in the signal transduction of activation of lymphatic endothelial cells [6]. However, VEGFR3 is also expressed on porous capillaries in endocrine organs as well as on monocytes and megakaryocytes. The other two lymphatic endothelial growth factors, VEGF-C and VEGF-D, promote lymphangiogenesis by binding to their receptor VEGFR-3 and induce anastomoses between lymphatic vessels and lymphatic vessels [6]. Other potential lymph node metastasis markers such as lymphatic vessel endothelial hyaluronan receptor 1 (LYVE-1) expressed on lymphatic vessel endothelium is considered to be a specific marker of lymphatic vessels. LYVE-1 was overexpressed in colon tumors compared with in unaffected colon tissues [7]. However, these lymphatic vessel markers only indicate the lymphatic hyperplasia of the tumor rather than directly indicate lymph node metastasis, and thereby cannot be used as a clinical diagnostic indicator to assess patient prognosis. As a result, it is essential to search for more specific markers of lymph node metastasis.
Voltage-gated Na+ channels (VGSCs) are macro-molecular protein complexes embedded in cell membranes, which are composed of a channel-forming alpha subunit and one or more smaller beta subunits [8]. The VGSC α subunit family contains nine members, Nav1.1–Nav1.9, encoded by the SCN1A–SCN11A genes [8]. VGSCs are abundantly expressed in excitable cells such as neurons and cardiomyocytes, which are responsible for the generation of action potentials and the transmission of neural signals [8]. However, VGSCs have been shown to be expressed in tumor cells from a variety of cancers, such as breast cancer, cervical cancer, colon cancer, melanoma, neuroblastoma, non-small cell lung cancer, ovarian cancer, and prostate cancer [9]. Abnormally high expression of VGSC alpha subunits in cancer cells promotes migration and invasion of tumor cells [9]. Nav1.1, Nav1.2, Nav1.3, Nav1.4, and Nav1.9 are expressed in ovarian cancer, non-small cell lung cancer, and prostate cancer [10]. The Nav1.7 alpha subunit promotes gastric cancer progression through MET transcriptional regulator-mediated upregulation of sodium-hydrogen antiporter 1 [10]. Other studies have shown that the expression of the Nav1.5 alpha subunit is closely related to a poor prognosis in breast cancer [11], non-small cell lung cancer [12], ovarian cancer [13], and prostate cancer [14]. The Nav1.5 alpha subunit is also a key regulator of the transcriptional regulatory network of genes that control CRC cell invasion [15]. The Nav1.5 alpha subunit controls colon cancer metastasis via regulation of Wnt signaling, cell migration, ectodermal development, steroid metabolism, and cell cycle-dependent protein expression [15]. Other studies have shown that Nav1.5 upregulates CRC-inducible gene expression through the MAPK signaling pathway, therefore promoting colon cancer metastasis [16].
Nav1.6 is expressed in breast cancer, cervical cancer, lymphoma, melanoma, mesothelioma, non-small cell lung cancer, prostate cancer, and small cell lung cancer [9]. Furthermore, the over-expression of Nav1.6 can promote the metastasis and invasion of cervical cancer [9, 17]. Similarly, our study found that Nav1.6 was highly expressed in CRC tissues and positively correlated with lymph node metastasis. Additionally, in the surgically removed lymph nodes, we found that Nav1.6, not Nav1.5, was highly expressed in metastatic lymph nodes.
Lymph node metastasis requires tumor cells to flow in or settle in the marginal sinus of the lymph nodes. Tumor-associated macrophages (TAMs) or M2 macrophages play an important role in the development, metastasis, and prognosis of a variety of malignant, metastatic tumors [18]. For example, in local lymph node metastasis of oral squamous cell carcinoma, M2 macrophage infiltration into the marginal sinus of the lymph node increased with malignancy [19]. Chemokine receptor 2 (CCR2)-positive monocytes and macrophages are classified as TAMs, both of which are regulated by the CCR2/CCL2 axis [20]. Another chemokine receptor CCR4 is an important chemokine receptor that regulates immune homeostasis and is thought to be involved in the progression of hematological malignancies [21]. CCR4 expression is positively correlated with HER2 expression, tumor recurrence, and lymph node, lung, and bone metastasis of breast cancer [21]. Autocrine and paracrine loops between cancer cells and macrophages promote lymph node metastasis via CCR4/CCL22 axis in head and neck squamous cell carcinoma [22]. Our result indicated that CCR2 and CCR4 were also highly expressed in CRC tissues and positively correlated with lymph node metastasis and over-expression of Nav1.6. This study suggests that Nav1.6 could be used as a potential biomarker for lymph node metastasis in patients with CRC. And the CCL2-CCR2 and CCL22-CCR4 axis may be the mechanism of high expression of Nav1.6 promoting lymph node metastasis of CRC.
Materials and methods
Biological samples
The enrolled cases in this study were patients with tumor lesions larger than 2 cm, who were diagnosed as stage II–III colon cancer (AJCC eighth edition system) without radiotherapy and chemotherapy. After laparoscopic surgery in CRC patients, the acquired specimens were sampled immediately in vitro. A 0.5 × 0.5-cm piece of CRC tumor tissue from the intestinal lumen surface was isolated as the tumor sample for further study. Meanwhile, 0.5 × 0.5 cm of the intestinal wall tissue (mainly mucosa and submucosa) 5 cm far from the edge of the tumor was isolated as a normal tissue control. All acutely isolated clinical samples were stored in a − 86 °C freezer within 2 min. The remaining tumor tissue was immediately preserved in 4% formalin for subsequent HE and immunochemistry (IHC) staining.
Real-time quantitative PCR
The total RNA from tumor and normal tissues from 97 CRC patients were prepared by using TRIzol reagent (Invitrogen). RNA (1 μg) was reverse-transcribed into cDNA using the Maxima First Strand cDNA Synthesis Kit according to the standard manufacturer’s protocol (Fermentas, USA). Real-time quantitative PCR (RT-qPCR) was performed by using the 2× Maxima SYBR Green/ROX qPCR Master Mix kit with ABI 7500 according to the standard manufacturer’s protocol (Fermentas, USA). Data were analyzed using the ΔΔCT relative quantification method. Primer sequences and corresponding genes are shown in Table 1.
Table 1.
Quantitative PCR primers used in this study. Designed with NCBI primer blast. Product length, 100–200 bp; annealing temperatures, 57–60 °C
| Gene | Genebank no. | Sequence |
|---|---|---|
| hNav1.1 | NM_001165963.1 |
ACATTACGTGCTGCTGGGAA CCAAGGTGGCCTGATTCTGT |
| hNav1.2 | NM_001040142.1 |
TGACAGAGTTTGTGGACCTGG GCGCAAACACGCTTAGACAG |
| hNav1.3 | NM_001081676.1 | TTCAGTTTCAGAGGTCGGGC TTTCTGTGGTGGTGCCGTTA |
| hNav1.4 | NM_000334.4 | CGGCTGGTGTGTCATACTGT TAGGCCAACAGAATGCCCTG |
| hNav1.5 | NM_000335.4 | TTTGTGGACCTGGGCAATGT GGCAGAAGACTGTGAGGACC |
| hNav1.6 | NM_001177984.2 | GGAAGCCTGTCTTGGTGTGT TTTGCGAGAGCCCCTTTGAT |
| hNav1.7 | NM_002977.3 |
GACGCAGCAGTAACATCAGC TTGATTGGTCGTGCCCTCTG |
| hNav1.8 | NM_001293306.2 | TCTCTGTGCCCATTGCTGAG CAGCTGCTCCTGTCCTTTGG |
| hNav1.9 | NM_001287223.1 | GTCTGGGAAAGACCAGCCTC CAGTGCTCTCTGCCTTTGGA |
| hCCR-2 | NM_001123041.2 | AGTCAACTGGACCAAGCCAC TGTGAAAAAGGCTTCTGAACTTCT |
| hCCR-3 | NM_001837.3 | CAGGAGTGGTGACGCCTAAG ACCAGCTTCTCATCTGCTGAA |
| hCCR-4 | NM_005508.4 | AAAGCAAGCTGCTTCTGGTTG AGGAAGAGCTCCCCAAATGC |
| hCCR-7 | NM_001301714.1 | CATGGACCTGGGTATGCCTG AAAGTTCCGCACGTCCTTCT |
| hBcl-2 | NM_000633.2 | TGAACTGGGGGAGGATTGTG CGTACAGTTCCACAAAGGCA |
| hTGFbetaR1 | NM_001130916.2 | TCCAACTACTGGTTTACCATTGC TTCTTCTCCCCGCCACTTTC |
| hCD3e | NM_000733.3 | GGCCTGAATCAGAGACGCAT TAGCCCAGGAAACAGGGAGT |
| hP53 | NM_000546.5 | ACCTATGGAAACTACTTCCTGAAA CTGGCATTCTGGGAGCTTCA |
| hCD8a | NM_001145873 | TGAGTTCTCTTCTCCCCTACGA GCCGTACTGAGGCAGAAACG |
| hIL2RA | NM_000417.2 | CAGTGCGTCCAGGGATACAG GACGAGGCAGGAAGTCTCAC |
| hIFNGR | NM_000416.2 | CAGCATGGCTCTCCTCTTTCTC TAGTTGGTGTAGGCACTGAGG |
| hGAPDH | NM_001256799.2 | ACATCAAGAAGGTGGTGAAGCA GTCAAAGGTGGAGGAGTGGGT |
Immunochemistry staining
Specimens were cut into 4-μm sections and adhered to glass slides. For HE staining, slides were deparaffinized and hydrated to water and then stained in hematoxylin solution, then differentiated sections with acid alcohol for two times and washed sections with ammonia solution. After being washed in a slow running tap water and stained in eosin, slides were dehydrated and mounted for imaging. The nucleus showed blue and the cytoplasm showed red. For immunochemistry staining, an anti-Nav1.5 antibody (ab56240 Abcam) and an anti-Nav1.6 antibody (ab6516 Abcam) were used to detect the expression of Nav1.5 and Nav1.6 in the lymph node, respectively. After the retrieval of antigens and an endogenous peroxidase block, slides were incubated with primary antibody overnight at 4 °C. After the incubation of the secondary antibody, add a freshly prepared DAB chromogenic reagent to mark the tissue and counterstain in the nucleus. Then dehydrated slides in gradient ethanol and then washed slides in xylene and mount with resin mounting medium. The nucleus stained with hematoxylin is blue. The positive cells developed by DAB reagent have a brown-yellow nucleus. For the negative control, samples were analyzed using a phosphate-buffered saline instead of the primary antibodies.
Data analysis
Statistical significance was analyzed using SPSS 20 software. Multiple comparisons were performed using a two-tailed t test. Differences were considered significant when P < 0.05. The asterisks in Figs. 2, 3, and 4 indicate significant differences (*P < 0.05, **P < 0.01, and ***P < 0.001).
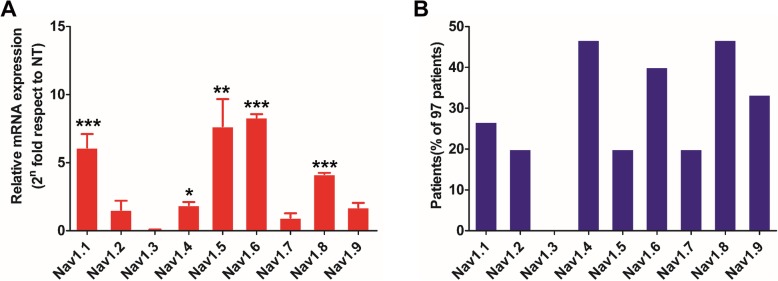
Fig. 2.
Analysis of mRNA expression of voltage-gated Na+ channel (VGSC) subtypes in colorectal cancer (CRC) tissues. Differences between the VGSC expression levels were expressed as fold change relative to normal tissue using the 2–∆∆CT method. a Relative mRNA expression of VGSC subtypes in 97 CRC tissues. b Proportion of patients with high expression levels of different subtypes of VGSCs in 97 CRC patient samples
Fig. 3.
Immunohistochemistry for Nav1.5 and Nav1.6 in surgically isolated lymph nodes. a, b Nav1.5 is lowly expressed in both non-metastatic and metastatic lymph nodes (magnification 10 × 40). Nav1.6 is lowly expressed in non-metastatic lymph nodes (c) but highly expressed in metastatic lymph nodes (d) (magnification, 10 × 40). e Comparison of the average optical density of Nav1.5- and Nav1.6-positive regions in metastatic lymph nodes (data from five patients). At least 3200-fold fields of view were randomly selected for each slice in each group for screenshots. Image-Pro Plus 6.0 software was used to select the same brown color as the unified standard for judging all photos. Each photo was analyzed to obtain the cumulative optical density (IOD) of each photo and the area of the tissue (AREA) and obtain the average optical density value (average optical, AO value), AO = IOD/AREA, the greater the AO value, the higher the positive expression level. f, g Expression of Nav1.5 and Nav1.6 in different regions of lymph node. The lower left area of the figure was the non-tumor metastatic region and the right area was metastatic region. There was no significant difference in the expression of Nav1.5 between these two regions while Nav1.6 is lowly expressed in the non-metastatic region but highly expressed in the metastatic region
Fig. 4.
Association of known markers of lymph node metastasis and infiltration with Nav1.6. a, b Quantitative PCR analysis of mRNA expression levels for lymphocyte infiltration- and lymph node metastasis-related molecular biomarkers in 97 CRC tissues. c Proportion of patients with a high expression of lymphocyte infiltration- and lymph node metastasis-related molecular biomarkers in 97 CRC patients. d Two-tailed t test analysis of the correlation between the upregulation of CCR2 and CCR4 and lymph node metastasis in CRC tissues. The independent sample t test in SPSS20.0 was used to determine whether nav1.6 was highly expressed as a grouping variable and CCR2\CCR4 as a test variable. When the sig value of the Levene test is greater than 0.05, the T value and the P value are obtained by assuming the variance is equal. When the sig value of the Levene test is less than 0.05, the T value and the P value are obtained by assuming that the variances are not equal
Results
CRC sampling and lymph node metastasis detection
Figure 1a shows the tissue removed via surgery of colon cancer radical resection, including the intestine approximately 25 cm from the anus, the corresponding total mesorectal and sigmoid mesenteric tissue, the inferior mesenteric artery, and the surrounding lymph node tissue. The tumor was round with an ulcerated diameter of approximately 1.5 cm, and the lower edge was 1 cm from the dentate line. A 10 × 10-fold hematoxylin and eosin (HE) staining map indicated that the cellular structure and cell size of a non-metastatic lymph node were normal. In the metastatic lymph node, the cancer tissue infiltrated the metastatic lymph node and arranged in a glandular tube (Fig. 1b, c). Figure 1d is a collective statistical analysis of lymph node metastasis from CRC patients. We collected 97 first-episode patients with CRC without preoperative chemo-radiotherapy. Among the 97 patients aging from 34 to 81 years old, with a male to female ratio of 61:36, the stage II/III ratio is 57:40, with/without an intravascular tumor thrombus ratio of 48:49, and with/without a perineural invasion ratio of 34:63. The total number of detected lymph nodes was 25.2 ± 9.8, of which 41.2% were lymph node metastasis and the total number of positive lymph nodes was 1.9 ± 3.3.
Fig. 1.
CRC sampling and lymph node metastasis detection. a The tissue removed via surgery for CRC radical resection, including the intestine approximately 25 cm from the anus, the corresponding total mesorectal and sigmoid mesenteric tissue, the inferior mesenteric artery, and the surrounding lymph node tissue. The tumor was round with an ulcerated diameter of approximately 3.5–4 cm (marked with red arrow). b is a 10 × 10-fold hematoxylin and eosin (HE) staining map of a non-metastatic lymph node, and its cellular structure and cell size are not significantly abnormal. c represents an HE staining map of a metastatic lymph node. d is a collective statistical analysis of lymph node metastasis and clinicopathological parameters from 97 CRC patients. For colon cancer, the resection range includes colon-centered, 10-cm upper and lower colon tissue and the corresponding mesenteric and mesenteric root tissues; for rectal cancer, the resected tissue includes tumor-centered proximal 10 cm, down 2–5 cm (depending on the location of the tumor from the anus and whether the anus is retained) and the corresponding mesenteric and mesenteric root tissues. All lymph nodes in the tissue were searched postoperatively and their metastases were counted
Various sodium channel subtypes were highly expressed in human CRC tissues
From the cancer tissues of 97 CRC patients, the total RNA was extracted and the expression levels of each VGSC were analyzed using qPCR. The expression levels in the normal tissues of patients were taken as a reference control. The patients with tissue VGSC expression levels twice as high as those of normal tissue were included in this metric. The results showed that Nav1.1, Nav1.2, Nav1.4, Nav1.5, Nav1.6, Nav1.8, and Nav1.9 were highly expressed in CRC tissues (Fig. 2a). In particular, the expression levels of Nav1.5, Nav1.6, and Nav1.8 increased more than eight times compared to normal tissues (Fig. 2a). Furthermore, patients with a high expression of Nav1.4, Nav1.6, and Nav1.8 accounted for more than 30% of the total number of CRC patients (Fig. 2b).
Over-expression of Nav1.6 positively correlated with lymph node metastasis in CRC
The correlation between the upregulation of VGSCs by each subtype and lymph node metastasis was further analyzed using the t test. Nav1.1 and Nav1.6 expression levels were significantly higher in CRC tissues with lymph node metastasis than those without metastasis, whereas other elevated expression levels of VGSC subtypes did not show significant correlation with CRC lymph node metastasis (Table 2). The immunochemistry staining result indicated that the non-metastatic lymph nodes are structurally intact and the lymphocytes are evenly distributed (Fig. 3a, c). In metastatic lymph nodes, multiple metastatic adenocarcinoma tissues were found (Fig. 3b, d). Nav1.5 was expressed at low levels in the interstitial tissues of non-metastatic lymph nodes and in the cytoplasm of metastatic lymph node glands (Fig. 3a, b). In contrast, Nav1.6 is highly expressed in the metastatic lymph nodes of the glandular epithelial cytoplasm (Fig. 3c, d). The analysis of the mean optical density values of the Nav1.5- and Nav1.6-positive regions in metastatic lymph nodes revealed that the expression of Nav1.6 was much higher than that of Nav1.5 (Fig. 3e). In the lymph node, there was a significant structural difference between the metastatic region and non-metastatic region. Although there was no significant difference in the expression of Nav1.5 between these two regions, the expression of Nav1.6 in the metastatic region was much higher than that in the non-metastatic region (Fig. 3f, g).
Table 2.
Two-tailed t test analysis of the correlation between the upregulation of different sodium channel subtypes and lymph node metastasis in CRC tissues
| Nav channel | Lymph node metastasis CRC | Non-lymph node metastasis CRC | T | P |
|---|---|---|---|---|
| Nav 1.1 | 3.19 ± 2.91 | 2.05 ± 1.96 | 2.304 | 0.023 |
| Nav 1.2 | 2.07 ± 2.37 | 1.97 ± 1.96 | 0.213 | 0.832 |
| Nav 1.3 | 0.34 ± 0.45 | 0.41 ± 0.52 | − 0.682 | 0.497 |
| Nav 1.4 | 3.08 ± 2.90 | 2.23 ± 2.21 | 1.630 | 0.106 |
| Nav 1.5 | 1.64 ± 1.34 | 1.17 ± 1.186 | 1.811 | 0.073 |
| Nav 1.6 | 3.63 ± 3.21 | 2.13 ± 1.96 | 2.534 | 0.014 |
| Nav 1.7 | 1.80 ± 1.76 | 2.13 ± 2.48 | − 0.683 | 0.496 |
| Nav 1.8 | 3.11 ± 2.72 | 2.59 ± 1.94 | 1.098 | 0.275 |
| Nav 1.9 | 1.66 ± 1.48 | 1.86 ± 1.70 | − 0.603 | 0.548 |
The independent sample t test in SPSS20.0 was used to determine whether lymph node metastasis was a grouping variable, and the expression of the sodium ion channel subtype was a test variable. When the sig value of the Levene test is greater than 0.05, the T value and the P value are obtained by assuming the variance is equal. When the sig value of the Levene test is less than 0.05, the T value and the P value are obtained by assuming that the variances are not equal. An obtained P value < 0.05 is indicated in italic
Since the presence of lymphovascular invasion is also a key parameter predicting patients’ prognosis in various types of cancer, we further analyzed the correlation between the upregulation of different Nav channels and lymphovascular invasion. The two-tailed t test analysis results suggested that the over-expression of Nav1.6 and Nav1.7 is positively related to lymphovascular invasion in CRC (Table 3).
Table 3.
Two-tailed t test analysis of the correlation between the upregulation of different sodium channel subtypes and lymphovascular invasion in CRC tissues
| Nav subtype | Lymphovascular invasion | Non-lymphovascular invasion | T | P |
|---|---|---|---|---|
| Nav1.1 | 2.70 ± 2.90 | 2.31 ± 1.98 | 0.791 | 0.431 |
| Nav1.2 | 1.58 ± 1.38 | 2.32 ± 2.48 | − 1.874 | 0.064 |
| Nav1.3 | 0.40 ± 0.51 | 0.37 ± 0.49 | 0.232 | 0.817 |
| Nav1.4 | 2.79 ± 2.84 | 2.36 ± 2.25 | 0.831 | 0.408 |
| Nav1.5 | 1.41 ± 1.38 | 1.30 ± 1.18 | 0.399 | 0.691 |
| Nav1.6 | 3.83 ± 3.15 | 1.86 ± 1.68 | 3.639 | 0.001 |
| Nav1.7 | 2.60 ± 2.58 | 1.57 ± 1.85 | 2.287 | 0.024 |
| Nav1.8 | 3.15 ± 2.68 | 2.50 ± 1.88 | 1.405 | 0.163 |
| Nav1.9 | 1.41 ± 1.30 | 2.06 ± 1.78 | − 1.959 | 0.053 |
The independent sample t test in SPSS20.0 was used to determine whether lymph node metastasis was a grouping variable, and the expression of the sodium ion channel subtype was a test variable. When the sig value of the Levene test is greater than 0.05, the T value and the P value are obtained by assuming the variance is equal. When the sig value of the Levene test is less than 0.05, the T value and the P value are obtained by assuming that the variances are not equal. An obtained P value of < 0.05 is indicated in italic
Over-expression of Nav1.6 positively correlated with the expression of CCR2 and CCR4
The biomarkers of lymphatic invasion and lymph node metastasis in clinical specimens were analyzed using RT-qPCR. The results showed that the chemokine receptor biomarkers CCR2, CCR4, and CCR7 were highly expressed in lymph node metastatic tissues compared with normal tissues (Fig. 4a). In addition, lymphocyte infiltration-related biomarkers CD3e, CD8a, and IL-2RA were highly expressed in CRC tissues (Fig. 4b). Further analysis found that the proportion of patients with high expression of CCR2, CCR4, and CCR7 in CRC patients was over 20%, whereas those with high expression of IL-2RA was as high as 47% (Fig. 4c). Patients with high expression of CD3e and CD8a were less than 20% (Fig. 4c). However, the further two-tailed t test analysis suggested that only the over-expression of CCR2 and CCR4 shows significant correlation with the high expression of Nav1.6 (Fig. 4d).
Over-expression of CCR2 and CCR4 positively correlated with lymph node metastasis in CRC
Based on the above RT-qPCR quantification results in combination with the patients’ diagnosis and treatment information, the correlation between lymph node metastasis and lymphocyte invasion and its relationship with the upregulation of metastasis-related molecular biomarkers were further analyzed using t tests. The results illustrated higher hCCR-2 and hCCR-4 expression in tumor tissues with lymph node metastasis than in those without lymph node metastasis (Table 4). This result suggested that high hCCR-2 and hCCR-4 expressions were significantly associated with lymph node metastasis in CRC.
Table 4.
Analysis of the correlation between the upregulation of lymph node metastasis-related molecular markers and lymph node metastasis in colorectal cancer (CRC) tissues
| Gene | Lymph node metastasis CRC | Non-Lymph node metastasis CRC | T | P |
|---|---|---|---|---|
| hCCR2 | 4.21 ± 3.67 | 2.24 ± 2.12 | 2.949 | 0.005 |
| hTGFbetaR | 0.94 ± 0.54 | 0.91 ± 0.55 | 0.215 | 0.830 |
| hCD3e | 2.65 ± 1.52 | 2.52 ± 2.12 | 0.304 | 0.762 |
| hCCR4 | 3.40 ± 3.50 | 2.03 ± 1.85 | 2.168 | 0.035 |
| hP53 | 2.23 ± 1.5 2 | 2.33 ± 1.59 | − 0.305 | 0.761 |
| hCCR7 | 6.21 ± 5.56 | 6.44 ± 6.65 | − 0.172 | 0.864 |
| hBcl2 | 2.41 ± 1.45 | 2.14 ± 1.50 | 0.883 | 0.380 |
| h1L2Ra | 6.45 ± 5.13 | 4.68 ± 3.65 | 1.973 | 0.051 |
| hCD8a | 11.55 ± 8.95 | 11.21 ± 12.49 | 0.144 | 0.886 |
Two-tailed t test in SPSS20.0 was used to determine whether lymph node metastasis was a group variable, and the expression of each immune factor was a test variable. When the sig value of the Levene test is greater than 0.05, the T value and the P value are obtained by assuming the variance is equal. When the sig value of the Levene test is less than 0.05, the T value and the P value are obtained by assuming that the variances are not equal. P < 0.05 is marked in italic
Discussion
The pathological examination of lymph nodes in patients with CRC is critical for post-operative treatment and prognosis prediction [4]. Early diagnosis of tumor lymph node metastasis relies on the use of specific sensitive lymphatic biomarkers and advanced techniques to make an accurate diagnosis of lymph angiogenesis and lymph node metastasis. However, the existing lymphatic metastasis markers are mostly lymphatic endothelial markers, which are not suitable for clinical diagnosis and evaluation of patient prognosis.
Abnormal expression of various sodium channel alpha subunits is closely related to tumor invasion and metastasis, but the relationship between sodium channel over-expression and tumor lymph node metastasis is still unclear [9]. Previous studies have shown that the Nav1.5 alpha subunit plays a key role in the transcriptional regulatory network that controls colon cancer cell invasion [15]. Our results showed that Nav1.1, Nav1.2, Nav1.4, Nav1.5, Nav1.6, Nav1.8, and Nav1.9 were highly expressed in CRC tissues. However, only a high expression of Nav1.1 and Nav1.6 rather than Nav1.5 was positively correlated with lymph node metastasis. Further immunohistochemistry shows that Nav1.5 and Nav1.6 were both lowly expressed in non-metastatic lymph nodes, while Nav1.6 was highly expressed in metastatic lymph nodes. These results suggested that a high expression of Nav1.1 and Nav1.6 may promote lymph node metastasis in colorectal cancer. Nav1.1 is mainly expressed in the central nervous system, especially the brain, whereas Nav1.6 is mainly expressed in the peripheral nervous system and epithelial cells, especially in the intestinal nerve center and intestinal epithelial cells [8]. Therefore, Nav1.6 is more suitable for in-depth study as a potential CRC lymph node metastasis marker.
CCR2, CCR4, and their respective ligand-mediated pathways, CCR2\CCL2 axis and CCR4\CCL22 axis, are considered to be important mechanisms of lymph node metastasis [21, 23]. In this study, we found that CCR2 and CCR4 were highly expressed in Nav1.6 high-expression CRC tissues and were positively correlated with lymph node metastasis. CCR2\CCL2 axis and CCR4\CCL22 axis may involve in the over-expression of Nav1.6 promoting lymph node metastasis. However, whether the upregulation of Nav1.6 expression promotes lymph node metastasis through CCR2\CCL2 axis and CCR4\CCL22 axis needs further study.
Tumor-infiltrating lymphocytes (TILs) are predictors of lymph node metastasis in many cancers such as early gastric cancers, melanoma, and rectal cancers [24–26]. High TILs were significantly associated with an absence of lymph node metastasis [24]. A high frequency of CD8-positive lymphocyte infiltration correlates with the lack of lymph node involvement in early rectal cancer [26]. Among the 97 patients, the proportion of patients with a high frequency of CD8-positive lymphocyte infiltration was about 15%. This result indicated that these patients with a high frequency of CD8-positive lymphocyte infiltration may have a less probability of lymph node metastasis in the future.
Conclusion
Our study suggests that Nav1.6 may be a potential biomarker for lymph node metastasis in patients with CRC.
Acknowledgements
First and foremost, we would like to show our deepest gratitude to all the doctors and nurses in the Department of Gastrointestinal Surgery, Quzhou People’s Hospital, for the collection of clinical samples. In addition, we thank the cooperation and support of participant patients with colorectal cancer.
Authors’ contributions
Z-HC and W-GL conceived and designed the experiments. YL, SL, JX, and XM performed the experiments. YL analyzed the data and wrote the paper. Authorship must be limited to those who have contributed substantially to the work reported. All authors read and approved the final manuscript.
Funding
None
Availability of data and materials
The datasets during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The study was done after an agreement from the local ethics committee (Quzhou People’s Hospital ethics committee) and with the patients’ informed consent.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shuiquan Lin, Yangbo Lv, Jianguang Xu and Zhenhong Chen contributed equally to this work.
Contributor Information
Zhenhong Chen, Phone: 86-0570-3055179, Email: qzczh@126.com.
Wuguang Lu, Phone: 86-25-52362182, Email: luwuguang@163.com.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst. 2004;96(19):1420–1425. doi: 10.1093/jnci/djh275. [DOI] [PubMed] [Google Scholar]
- 3.Ulintz PJ, Greenson JK, Wu R, Fearon ER, Hardiman KM. Lymph node metastases in colon cancer are polyclonal. Clin Cancer Res. 2018;24(9):2214–2224. doi: 10.1158/1078-0432.CCR-17-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin M, Frankel WL. Lymph node metastasis in colorectal cancer. Surg Oncol Clin N Am. 2018;27(2):401–412. doi: 10.1016/j.soc.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 5.Gunderson LL, Jessup JM, Sargent DJ, Greene FL, Stewart AK. Revised TN categorization for colon cancer based on national survival outcomes data. J Clin Oncol. 2010;28(2):264–271. doi: 10.1200/JCO.2009.24.0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Yi H, Liu Z, et al. Association between VEGFR-3 expression and lymph node metastasis in non-small-cell lung cancer. Vol 9. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langenes V, Svensson H, Börjesson L, et al. Expression of the chemokine decoy receptor D6 is decreased in colon adenocarcinomas. Cancer Immunol Immunother. 2013;62(11):1687–1695. doi: 10.1007/s00262-013-1472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Lera RM, Kraus RL. Voltage-gated sodium channels: structure, function, pharmacology, and clinical indications. J Med Chem. 2015;58(18):7093–7118. doi: 10.1021/jm501981g. [DOI] [PubMed] [Google Scholar]
- 9.Brackenbury WJ. Voltage-gated sodium channels and metastatic disease. Channels. 2012;6(5):352–361. doi: 10.4161/chan.21910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia J, Huang N, Huang H, et al. Voltage-gated sodium channel Nav 1.7 promotes gastric cancer progression through MACC1-mediated upregulation of NHE1. Int J Cancer. 2016;139(11):2553–2569. doi: 10.1002/ijc.30381. [DOI] [PubMed] [Google Scholar]
- 11.Fraser SP, Diss JK, Chioni AM, et al. Voltage-gated sodium channel expression and potentiation of human breast cancer metastasis. Clin Cancer Res. 2005;11(15):5381–5389. doi: 10.1158/1078-0432.CCR-05-0327. [DOI] [PubMed] [Google Scholar]
- 12.Roger S, Rollin J, Barascu A, et al. Voltage-gated sodium channels potentiate the invasive capacities of human non-small-cell lung cancer cell lines. Int J Biochem Cell Biol. 2007;39(4):774–786. doi: 10.1016/j.biocel.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 13.Gao R, Shen Y, Cai J, Lei M, Wang Z. Expression of voltage-gated sodium channel alpha subunit in human ovarian cancer. Oncol Rep. 2010;23(5):1293–1299. doi: 10.3892/or_00000763. [DOI] [PubMed] [Google Scholar]
- 14.Diss JK, Archer SN, Hirano J, Fraser SP, Djamgoz MB. Expression profiles of voltage-gated Na(+) channel alpha-subunit genes in rat and human prostate cancer cell lines. Prostate. 2001;48(3):165–178. doi: 10.1002/pros.1095. [DOI] [PubMed] [Google Scholar]
- 15.House CD. Vaske CJ, Schwartz AM, et al. Voltage-gated Na+ channel SCN5A is a key regulator of a gene transcriptional network that controls colon cancer invasion. Cancer Res. 2010;70(17):6957–6967. doi: 10.1158/0008-5472.CAN-10-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.CD H, Wang BD, Ceniccola K, et al. Voltage-gated Na+ channel activity increases colon cancer transcriptional activity and invasion via persistent MAPK signaling. Sci Rep. 2015;5:11541. doi: 10.1038/srep11541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hernandez-Plata E, Ortiz CS, Marquina-Castillo B, et al. Overexpression of Na V 1.6 channels is associated with the invasion capacity of human cervical cancer †. Int J Cancer. 2012;130(9):2013–2023. doi: 10.1002/ijc.26210. [DOI] [PubMed] [Google Scholar]
- 18.Colegio OR, Chu N-Q, Szabo AL, et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513:559. doi: 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mori K, Hiroi M, Shimada J, Ohmori Y. Infiltration of m2 tumor-associated macrophages in oral squamous cell carcinoma correlates with tumor malignancy. Cancers. 2011;3(4):3726–3739. doi: 10.3390/cancers3043726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J, Lv X, Chen J, et al. CCL2-CCR2 axis promotes metastasis of nasopharyngeal carcinoma by activating ERK1/2-MMP2/9 pathway. Oncotarget. 2015;7(13):15632–15647. doi: 10.18632/oncotarget.6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olkhanud PB, Baatar D, Bodogai M, et al. Breast cancer lung metastasis requires expression of chemokine receptor CCR4 and regulatory T cells. Cancer Res. 2009;69(14):5996–6004. doi: 10.1158/0008-5472.CAN-08-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsujikawa T, Yaguchi T, Ohmura G, et al. Autocrine and paracrine loops between cancer cells and macrophages promote lymph node metastasis via CCR4/CCL22 in head and neck squamous cell carcinoma. Int J Cancer. 2013;132(12):2755–2766. doi: 10.1002/ijc.27966. [DOI] [PubMed] [Google Scholar]
- 23.Fujita S, Ikeda T. The CCL2-CCR2 axis in lymph node metastasis from oral squamous cell carcinoma: an immunohistochemical study. J Oral Maxillofac Surg. 2017;75(4):742–749. doi: 10.1016/j.joms.2016.09.052. [DOI] [PubMed] [Google Scholar]
- 24.Kim JY, Kim CH, Lee Y, Lee JH, Chae YS. Tumour infiltrating lymphocytes are predictors of lymph node metastasis in early gastric cancers. Pathology. 2017;49(6):589–595. doi: 10.1016/j.pathol.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Diem S, Hasan Ali O, Ackermann CJ, et al. Tumor infiltrating lymphocytes in lymph node metastases of stage III melanoma correspond to response and survival in nine patients treated with ipilimumab at the time of stage IV disease. Cancer Immunol. 2018;67(1):39–45. doi: 10.1007/s00262-017-2061-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Däster Silvio, Eppenberger-Castori Serenella, Hirt Christian, Zlobec Inti, Delko Tarik, Nebiker Christian A., Soysal Savas D., Amicarella Francesca, Iezzi Giandomenica, Sconocchia Giuseppe, Heberer Michael, Lugli Alessandro, Spagnoli Giulio C., Kettelhack Christoph, Terracciano Luigi, Oertli Daniel, von Holzen Urs, Tornillo Luigi, Droeser Raoul A. High Frequency of CD8 Positive Lymphocyte Infiltration Correlates with Lack of Lymph Node Involvement in Early Rectal Cancer. Disease Markers. 2014;2014:1–7. doi: 10.1155/2014/792183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets during and/or analyzed during the current study are available from the corresponding author on reasonable request.