Abstract
Background
Fecal carriage of extended-spectrum β-lactamase-producing Enterobacteriaceae (ESBL-PE) remains poorly documented in Africa. The objective of this study was to determine the prevalence of ESBL-PE fecal carriage in Chad.
Methods
In total, 200 fresh stool samples were collected from 100 healthy community volunteers and 100 hospitalized patients from January to March 2017. After screening using ESBL-selective agar plates and species identification by MALDI-TOF mass spectrometry, antibiotic susceptibility was tested using the disk diffusion method, and ESBL production confirmed with the double-disc synergy test. The different ESBL genes in potential ESBL-producing isolates were detected by PCR and double stranded DNA sequencing. Escherichia coli phylogenetic groups were determined using a PCR-based method.
Results
ESBL-PE fecal carriage prevalence was 44.5% (51% among hospitalized patients vs 38% among healthy volunteers; p < 0.05). ESBL-producing isolates were mostly Escherichia coli (64/89) and Klebsiella pneumoniae (16/89). PCR and sequencing showed that 98.8% (87/89) of ESBL-PE harbored blaCTX-M genes: blaCTX-M-15 in 94.25% (82/87) and blaCTX-M-14 in 5.75% (5/87). Phylogroup determination by quadruplex PCR indicated that ESBL-producing E. coli isolates belonged to group A (n = 17; 27%), C (n = 17; 27%), B2 (n = 9; 14%), B1 (n = 8; 13%), D (n = 8; 13%), E (n = 1; 1.6%), and F (n = 1; 1.6%). The ST131 clone was identified in 100% (9/9) of E. coli B2 strains.
Conclusions
The high fecal carriage rate of ESBL-PE associated with CTX-M-15 in hospital and community settings of Chad highlights the risk for resistance transmission between non-pathogenic and pathogenic bacteria.
Keywords: ESBL, Enterobacteriaceae, Fecal carriage, Chad
Introduction
Extended-Spectrum β-Lactamase-Producing Enterobacterial (ESBL-PE) have spread worldwide, and have become endemic in several countries since their first description in 1983 [1, 2]. Their diffusion is mainly attributed to ESBL-encoding genes that are often carried by mobile genetic elements, such as plasmids, that facilitate their dissemination [3]. Enterobacterial are commensal bacteria present in the intestinal tract of humans and various animals, [4] are an important reservoir of resistance genes, leading to ESBL-PE dissemination in communities. This might result in hospital and community infections if ESBL-encoding genes are acquired by pathogenic bacteria, or if commensal ESBL-PE become pathogenic [5, 6]. Indeed, colonization by ESBL-PE is one of the main risk factors for infection with antibiotic-resistant bacteria (ARB) [7]. These infections pose a great challenge due to the limited therapeutic options (including prolonged hospitalization) and the increased morbidity and mortality rates [8]. Therefore, they are particularly problematic in low-income countries [9].
Fecal carriage of ESBL-PE has been increasingly reported worldwide over the last decade. The highest ESBL-PE carriage prevalence has been described in Asia [10], whereas prevalence rates are lower in Europe and North America [11, 12]. Conversely, data on ESBL-PE fecal carriage in Sub-Saharan Africa are limited, and a few available studies reported a prevalence ranging from 6 (in Mauritania) to 66% (in Cameroon) in healthy volunteers and hospitalized patients [13, 14]. This small number of studies does not facilitate the identification of factors associated with this carriage in order to prevent and reduce the spread of ARB.
We recently reported a high prevalence of ESBL-PE containing the CTX-M-15 enzyme in clinical isolates from three major Chadian hospital [15] (48%). However, the rare studies on fecal carriage of ESBL-PE in Chad [16–19] and the absence of molecular data on ESBL-encoding genes limit our understanding of the genes and clones circulating among the Chadian population.
The aim of this study was to investigate the prevalence of ESBL-PE fecal carriage among hospitalized patients and healthy volunteers in Chad, and to identify the associated ESBL-encoding genes.
Materials and methods
Settings and bacterial isolates
Fresh stool samples were collected from 100 patients hospitalized for more than 48 h at the Mother and Child hospital, and from 100 healthy volunteers in the community (healthy volunteers were recruited from health staff and university students), from January to March 2017. This university teaching hospital is in N’Djamena, the capital city of Chad (1.5 million inhabitants), and is the reference mother-child hospital in Chad. It has a capacity of 261 beds (including an intensive care unit), with about 5000 admissions and 45,000 outpatients in 2016. People with diarrheal diseases were excluded from the study. This study was approved by the hospital ethics board and the Chadian Ministry of Public Health (No 676/PR/PM/MSP/SE/SG/DRGP/DRH/SGF/16). Informed written consent was obtained from all subjects and from at least one parent for each child before enrolment in the study. Only information about age, and sex was available.
ESBL-PE detection, bacterial identification, and antimicrobial susceptibility testing
Briefly, 0.5 g of each fresh stool sample was suspended in 5 ml of sterile saline solution (0.9%) and 100 ml aliquots were plated on ESBL agar plates (bioMérieux, Marcy-l’Etoile, France). Plates were examined after 24 and 48 h of incubation at 37 °C. Bacterial isolates were identified using biochemical tests and then confirmed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (Bruker Daltonics, Bremen, Germany). Antimicrobial susceptibility testing was performed using the disk diffusion method on Müller-Hinton agar and the clinical breakpoints recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines (Version 7.1) (http://www.eucast.org/clinical_breakpoints/). The following antibiotics were tested: amoxicillin, amoxicillin-clavulanic acid, ticarcillin, ticarcillin-clavulanic acid, piperacillin, piperacillin-tazobactam, temocilin, cephalexin, aztreonam, cefotaxim, ceftazidim, cefepime, cefoxitin, ertapenem, imipenem, gentamicin, tobramycin, netilmicin, amikacin, trimethoprim + sulfamethoxazole, nalidixic acid, ofloxacin, ciprofloxacin, levofloxacin, chloramphenicol, and fosfomycin. ESBL production was detected phenotypically with the double-disk synergy method [20]. In the case of high-level cephalosporinase production, the double-disk synergy test was performed using cloxacillin-supplemented medium (250 mg/L).
Molecular characterization of β-lactamase-encoding genes
DNA was extracted from each isolate using the boiling lysis method. Briefly, one single colony was transferred in 100 μL of distilled water, heated at 100 °C for 10 min, and then centrifuged. The presence of the most common ESBL-encoding genes, including blaCTX-M (CTX-M group 1, 2, 8, 9 and 25), blaTEM, blaSHV and blaOXA-like, was tested by multiplex PCR using the previously described primers and conditions [21]. DNA samples from reference blaCTX-M, blaTEM, blaSHV and blaOXA-like-positive strains were used as positive controls. PCR products were visualized by electrophoresis (100 V for 90 min) on 2% agarose gels containing ethidium bromide. A 100 bp DNA ladder (Promega, USA) was used as marker size. PCR products were sequenced bidirectionally on a 3100 ABI Prism Genetic Analyzer (Applied Biosystems). Sequencing data were analyzed online using the BLAST tool available at the National Center for Biotechnology Information web page (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Genotyping of ESBL-producing E. coli isolates
The phylogroup (A, B1, B2, C, D, E, and F) of each ESBL-producing E. coli isolate was determined using the quadruplex PCR-based method developed by Clermont et al. [22] For strains assigned to the B2 phylogenetic group, the presence of the E. coli sequence type (ST) 131 also was analyzed using a O25b-specific PCR method with pabB and trpA (control) allele-specific primers, as previously described [23].
Statistics
Statistical analyses were performed using the Epi Info software, version 3.5.3 (Centers for Disease Control and Prevention, Atlanta, GA, USA). Differences in the proportion of ESBL-producers between patient groups were assessed using the Chi-square test. A p-value < 0.05 was considered as statistically significant.
Results
Characteristics of participants and ESBL-PE prevalence
During the study period, 100 healthy volunteers and 100 hospitalized patients (n = 115 women and n = 85 men; age: 1 to 54 years) were screened. The overall frequency of ESBL-PE fecal carriage was 44.5%. ESBL-PE rate was significantly higher among hospitalized patients (51%) than healthy volunteers (51% vs 38%; p < 0.05, Chi-square test). Carriage rate was not different between sexes and in the three age groups (Table 1).
Table 1.
Demographic characteristics and prevalence of ESBL-PE carriage in the study population
| Variable | Hospitalized patients (n = 51) |
Healthy volunteers (n = 38) |
Total (n = 89) |
|---|---|---|---|
| Sex | |||
| Men, n (%) | 15 (29%) | 23 (60%) | 38 (43%) |
| Women n (%) | 36 (71%) | 15 (40%) | 51 (57%) |
| Age groups (years) | |||
| 1–11 n (%) | 18 (35%) | 3 (8%) | 21 (24%) |
| 12–24 n (%) | 13 (26%) | 9 (24%) | 22 (25%) |
| 25–54 n (%) | 20 (39%) | 26 (68%) | 46 (51%) |
| ESBL-PE carriers (%) | 51% | 38% | 44.5% |
Bacterial identification and antimicrobial susceptibility testing
The identified ESBL-PE strains included Escherichia coli (n = 64; n = 35 isolates from hospitalized patients and n = 29 from healthy volunteers), Klebsiella pneumoniae (n = 16; n = 11 isolates from hospitalized patients and n = 5 from healthy volunteers), Enterobacter cloacae (n = 6), Citrobacter freundii (n = 2), and Morganella morganii (n = 1) (Table 2).
Table 2.
Fecal carriage rate of ESBL-PE among hospitalized patients and healthy volunteers
| Strain | Hospitalized patients (n = 51) |
Healthy volunteers (n = 38) |
Total |
|---|---|---|---|
| Escherichia coli n (%) | 35 (69%) | 29 (76%) | 64 (72%) |
| Klebsiella pneumoniae n (%) | 11 (21%) | 5 (13%) | 16 (18%) |
| Enterobacter cloacae n (%) | 5 (10%) | 1 (3%) | 6 (7%) |
| Citrobacter freundii n (%) | 0 | 2 (5%) | 2 (2%) |
| Morganella morgani n (%) | 0 | 1 (3%) | 1 (1%) |
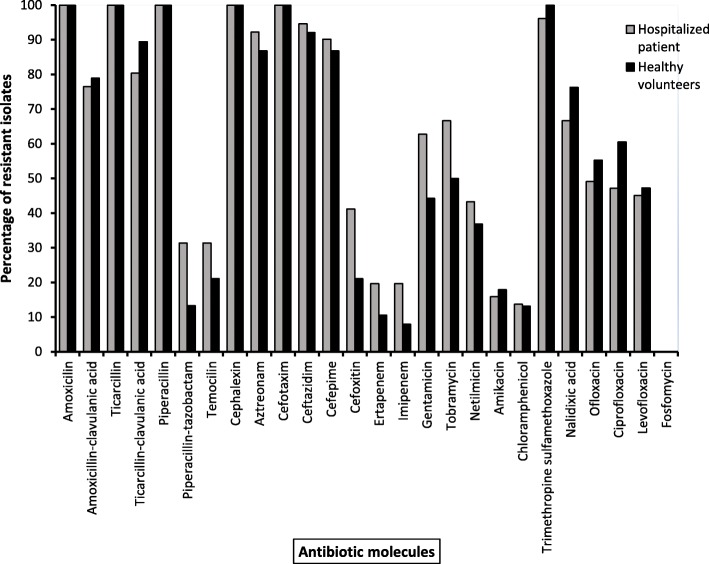
Antimicrobial susceptibility testing showed that few isolates were resistant to carbapenems (ertapenem and imipenem), particularly among hospitalized patients (Fig. 1). Resistance to aminoglycosides (gentamicin, tobramycin and netilmicin) was relatively high among hospitalized patients. Conversely, resistance to quinolones (nalidixic acid, ofloxacin, ciprofloxacin and levofloxacin) was higher among healthy volunteers. Most isolates were also resistant to sulfamethoxazole–trimethoprim and all isolates were susceptible to fosfomycin.
Fig. 1.
Percentage of antimicrobial resistance of ESBL-PE from healthy volunteers (n = 38) and hospitalized patients (n = 51) (X: antibiotic molecules, Y: percentage of resistant isolates). ESBL-PE, extended-spectrum ß-lactamase-producing Enterobacteriaceae
Characterization of β-lactamase-encoding genes
Molecular characterization of the ESBL-PE isolates revealed that they harbored different ß-lactamase-encoding genes (blaCTX-M, blaTEM and blaOXA). blaCTX-M genes were detected in 97.8% (87/89) of ESBL-PE isolates. The distribution of the different ß-lactamase-encoding genes in the different Enterobacteriaceae species from hospitalized patients and healthy volunteers is shown in Table 3.
Table 3.
ß-lactamase-encoding genes carried by the 89 ESBL-PE isolates from hospitalized patients (n = 51) and healthy volunteers (n = 38)
| Species | CTX-M-15 | CTX-M-14 | TEM | CTX-M-15/TEM | CTX-M-15/OXA-1 | CTX-M-14/OXA-1 | CTX-M-15/ TEM/OXA-1 |
|---|---|---|---|---|---|---|---|
| Hospitalized patients | |||||||
| Escherichia coli n = 35 | 14 (40%) | 0 | 0 | 1 (3%) | 10 (28%) | 1 (3%) | 9 (25%) |
| Klebsiella pneumoniae n = 11 | 3 (27%) | 0 | 0 | 0 | 8 (73%) | 0 | 0 |
| Enterobacter cloacae n = 5 | 1 (20%) | 0 | 1 (20%) | 0 | 2 (40%) | 1 (20%) | 0 |
| Healthy volunteers | |||||||
| Escherichia coli n = 29 | 13 (49%) | 2 (7%) | 0 | 1 (3%) | 13 (49%) | 0 | 0 |
| Klebsiella pneumoniae n = 5 | 3 (60%) | 1 (20%) | 0 | 0 | 1 (20%) | 0 | 0 |
| Enterobacter cloacae n = 1 | 1 (100%) | 0 | 0 | 0 | 0 | 0 | 0 |
| Citrobacter freundii n = 2 | 1 (50%) | 0 | 0 | 0 | 1 (50%) | 0 | 0 |
| Morganella morgana n = 1 | 0 | 0 | 1 (100%) | 0 | 0 | 0 | 0 |
| Total n = 89 | 36 (40%) | 3 (3%) | 2 (2%) | 2 (2%) | 35 (39%) | 2 (2%) | 9 (10%) |
Sequence analysis showed that among the 87 blaCTX-M-positive isolates, 82 contained blaCTX-M-15 and 5 carried blaCTX-M-14 alone or in combination with blaTEM and blaOXA (Table 3). The blaOXA-1 and blaTEM genes were detected in 51.7% (46/89) and 14.6% (13/89) of isolates, respectively. The blaCTX-M-15, blaCTX-M-14 and blaTEM genes were detected alone in 36, 3 and 2 isolates respectively, whereas blaCTX-M-15, blaOXA-1 and blaTEM were associated in 9 isolates. Moreover, 35 isolates coproduced blaCTX-M-15 and blaOXA-1, 2 harbored blaCTX-M-15 and blaTEM, and 2 expressed blaCTX-M-15 and blaOXA-1.
Phylogenetic grouping
The majority of the 64 ESBL-producing E. coli isolates belonged to the phylogenetic groups A 17/64 (27%) and C 17/64 (27%), followed by groups B2 9/64 (14%), B1 8/64 (13%), D 8/64 (13%), and groups E and F (one isolate/each), The phylogenetic group of three isolates could not be classified according to Clermont and al. method [22] Table 4. The pandemic E. coli ST131 clone was identified in 100% (9/9) of the B2 group isolates. The carriage rate of B2-ST131 E. coli isolates among hospitalized patients and healthy volunteers was 11.42% (4/35) and 17.24% (5/29), respectively. Isolates classified in group C and D were more frequent among hospitalized patients than heathy volunteers (12% vs 5, and 6% vs 2%, respectively).
Table 4.
Phylogenetic groups of 64 ESBL-producing E. coli isolates in hospitalized patients and healthy volunteers
| Phylogenetic groups | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| A | B1 | B2 | C | D | E | F | ND | ST131 | |
| Hospitalized patients n = 35 | 8 (23%) | 4 (11%) | 4 (11%) | 12 (34%) | 6 (17%) | 1 (3%) | 0 | 0 | 4 |
| Healthy volunteers n = 34 | 9 (26%) | 4 (12%) | 5 (15%) | 5 (15%) | 2 (6%) | 0 | 1 (3%) | 3 (9%) | 5 |
| Total n = 64 | 17 (27%) | 8 (13%) | 9 (14%) | 17 (27%) | 8 (13%) | 1 (1%) | 1 | 3 (4%) | 9 |
Discussion
Fecal carriage of ESBL-PE isolates is one of the main drivers for their dissemination in hospital and community settings worldwide, but has received little attention in Africa. Our study shows high rate (45%) of ESBL-PE fecal carriage in hospitalized patients and also healthy volunteers in Chad. Our results confirm the high intestinal carriage of ESBL-PE in Sub-Saharan Africa reported in Cameroon (54%) and Burkina Faso (32%) [24, 25]. Among ESBL-PE isolates, resistance was associated predominantly with CTX-M15 genes. ESBL-producing E. coli isolates belonged to different phylogroups (mainly to the commensal phylogenetic groups A and B1). Our findings confirm the widespread dissemination of ESBL-encoding genes among different Enterobacteriaceae species isolated in the community and also in hospital.
ESBL-PE carriage was significantly higher in hospitalized patients compared with healthy volunteers (51% vs. 38%, p < 0.05), as previously described in similar settings [25–28]. This result could be explained by higher antibiotic consumption that favors ARB selection and/or higher rate of in-hospital acquisition of ARB that then colonize the patient’s intestinal track [29–31]. Several factors can contribute to high ARB selection in low income countries, particularly poor drug quality or inadequate posology, long treatments, antibiotic misuse by health professionals, unskilled practitioners, auto-medication (antibiotics can be purchased without prescription), unhygienic conditions accounting for the spread of resistant bacteria, and inadequate surveillance programs [9, 32, 33]. Future studies should evaluate the specific risk factors contributing to the observed high ESBL-PE level in Chad.
In this study, most ESBL-PE isolates were multidrug-resistant (i.e., resistant to three or more classes of antimicrobials) [34]. Other antibiotics included mainly quinolones, aminoglycosides (except amikacin), and co-trimoxazole (trimethoprim/sulfamethoxazole), as previously reported in Burkina Faso and Gabon [25, 35]. This could be explained by the presence of multiple resistance genes on the same plasmid [28, 36]. For example, quinolones and co-trimoxazole are often used to treat some mild to moderate infections (e.g., urinary tract infections) caused by Enterobacteriaceae in various clinical and community settings without previous laboratory testing of the bacteria involved in the infection [37].
Our study shows that the blaCTX-M genes were the most common ß-lactamase-producing genes (98.8%) in the identified ESBL-PE isolates, with the predominance of blaCTX-M-15, in agreement with the reported worldwide spread of CTX-M-15 in community and hospital settings [3, 38]. Our finding also confirms the frequent association between blaCTX-M-15 and blaOXA-1 and/or blaTEM genes in ESBL-PE isolates, which reduces the therapeutic options for treatment with β-lactam antibiotics [25, 28]. Moreover, the presence of the blaCTX-M-14 gene, which is predominant in Asian countries [39] but very rare in Chad, could be due to migration from Chad to China for medical treatment and economic reasons.
Molecular analysis revealed that the ESBL-producing E. coli isolates belonged to several phylogenetic groups, mainly to the commensal phylogenetic groups A and B1, the virulent extra-intestinal phylogenetic groups B2 and D, and group C. This finding is consistent with previous studies that showed a high phylogenetic diversity among ESBL-producing E. coli, and supports the hypothesis of a frequent horizontal gene transfers between distant E. coli phylogroups [3]. Moreover, the human pandemic O25b-ST13 clone was identified in all ESBL-producing E. coli from phylogroup B2 (9/9). ST131 is the most prevalent clone among the group B2 E. coli and is known to colonize the human digestive tract of healthy subjects [40].
Overall, the high carriage frequency of different ESBL-producing Enterobacteriaceae and E. coli strains suggest that ESBL diffusion is not caused by the epidemic spread of a single resistant clone, as previously found for extra-intestinal E. coli belonging to group B2 [41, 42].
Conclusions
To our knowledge, this is the first study on the intestinal carriage of ESBL-PE in hospitalized patients and healthy community volunteers in Chad, and shows high carriage rate associated with the CTX-M-15 enzyme. The intestinal carriage of ESBL-PE is a significant challenge for public health, and highlights the urgent necessity to improve sanitation and implement antibiotic stewardship in African countries. Future studies should explore mechanisms involved in plasmid transfer and the determinants of the observed intestinal carriage.
Acknowledgments
We would like to thank IRD and CHU. We thank Elisabetta Andermarcher for assistance in preparing and editing the manuscript.
Authors’ contributions
OOM, CC, MH, JPH and GS Conceived and designed the experiments. OOM, ML, CS and SS Performed the experiments. OOM, ML, YD, AT, JB, AO, ALB, CC and GS Contributed to the writing of the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by a grant from Islamic Development Bank (IDB). IDB was not involved in the study design, and in data collection, analysis, interpretation, and in writing the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Ethics approval and consent to participate
This study was approved by the ethics board of each hospital and by the Chadian Ministry of Public Health (No 676/PR/PM/MSP/SE/SG/DRGP/DRH/ SGF/16). Informed written consent was obtained from all subjects and from at least one parent for each child before enrolment in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yann Dumont and Sylvain Godreuil contributed equally to this work.
References
- 1.Ghafourian S, Sadeghifard N, Soheili S, Sekawi Z. Extended Spectrum Beta-lactamases: definition, classification and epidemiology. Curr Issues Mol Biol. 2015;17:11–21. [PubMed] [Google Scholar]
- 2.Shaikh S, Fatima J, Shakil S, SMohdD R, Kamal MA. Antibiotic resistance and extended spectrum beta-lactamases: Types, epidemiology and treatment. Saudi J Biol Sci. 2015;22(1):90–101. doi: 10.1016/j.sjbs.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bevan ER, Jones AM, Hawkey PM. Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother. 2017;72(8):2145–2155. doi: 10.1093/jac/dkx146. [DOI] [PubMed] [Google Scholar]
- 4.Kariuki S, Hart CA. Global aspects of antimicrobial-resistant enteric bacteria. Curr Opin Infect Dis oct. 2001;14(5):579–586. doi: 10.1097/00001432-200110000-00012. [DOI] [PubMed] [Google Scholar]
- 5.Rolain J-M. Food and human gut as reservoirs of transferable antibiotic resistance encoding genes. Front Microbiol. 2013;4. 10.3389/fmicb.2013.00173. [DOI] [PMC free article] [PubMed]
- 6.Zhang H, Zhou Y, Guo S, Chang W. High prevalence and risk factors of fecal carriage of CTX-M type extended-spectrum beta-lactamase-producing Enterobacteriaceae from healthy rural residents of Taian, China. Front Microbiol. 2015;6. 10.3389/fmicb.2015.00239. [DOI] [PMC free article] [PubMed]
- 7.Thenmozhi S, Moorthy K, Sureshkumar BT, Suresh M. Antibiotic Resistance Mechanism of ESBL Producing Enterobacteriaceae in Clinical Field: A Review. Int J Pure App Biosci. 2014;2(3):207-26.
- 8.Lynch JP, Clark NM, Zhanel GG. Evolution of antimicrobial resistance among Enterobacteriaceae (focus on extended spectrum β-lactamases and carbapenemases) Expert Opin Pharmacother. 2013;14(2):199–210. doi: 10.1517/14656566.2013.763030. [DOI] [PubMed] [Google Scholar]
- 9.Ouedraogo AS, Jean Pierre H, Bañuls AL, Ouédraogo R, Godreuil S. Emergence and spread of antibiotic resistance in West Africa : contributing factors and threat assessment. Med Sante Trop. 2017;27(2):147–154. doi: 10.1684/mst.2017.0678. [DOI] [PubMed] [Google Scholar]
- 10.Hawkey PM. Multidrug-resistant Gram-negative bacteria: a product of globalization. J Hosp Infect. 2015;89(4):241–247. doi: 10.1016/j.jhin.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 11.Pilmis B, Cattoir V, Lecointe D, Limelette A, Grall I, Mizrahi A, et al. Carriage of ESBL-producing Enterobacteriaceae in French hospitals: the PORTABLSE study. J Hosp Infect. 2018;98(3):247–252. doi: 10.1016/j.jhin.2017.11.022. [DOI] [PubMed] [Google Scholar]
- 12.Islam S, Selvarangan R, Kanwar N, McHenry R, Chappell JD, Halasa N, et al. Intestinal Carriage of Third-Generation Cephalosporin-Resistant and Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae in Healthy US Children. J Pediatr Infect Dis Soc. 2018;7(3):234–240. doi: 10.1093/jpids/pix045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sallem RB, Slama KB, Estepa V, Cheikhna EO, Mohamed AM, Chairat S, et al. Detection of CTX-M-15-producing Escherichia coli isolates of lineages ST410-A, ST617-A and ST354-D in faecal samples of hospitalized patients in a Mauritanian hospital. J Chemother. 2015;27(2):114–116. doi: 10.1179/1973947814Y.0000000172. [DOI] [PubMed] [Google Scholar]
- 14.Djuikoue IC, Woerther P-L, Toukam M, Burdet C, Ruppé E, Gonsu KH, et al. Intestinal carriage of Extended Spectrum Beta-Lactamase producing E. coli in women with urinary tract infections, Cameroon. J Infect Dev Ctries. 2016;10(10):1135–1139. doi: 10.3855/jidc.7616. [DOI] [PubMed] [Google Scholar]
- 15.Ouchar Mahamat O, Lounnas M, Hide M, Dumont Y, Tidjani A, Kamougam K, et al. High prevalence and characterization of extended-spectrum ß-lactamase producing Enterobacteriaceae in Chadian hospitals. BMC Infect Dis. 2019;19(1):205. doi: 10.1186/s12879-019-3838-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yandaï FH, Zongo C, Moussa AM, Bessimbaye N, Tapsoba F, Savadogo A, et al. Prevalence and antimicrobial susceptibility of faecal carriage of extended-Spectrum β-lactamase (ESBL) producing Escherichia coli at the “Hôpital de la Mère et de l’Enfant” in N’Djamena. Chad Sci J Microbiol. 2014;3(2):25–31. [Google Scholar]
- 17.Ndoutamia G, Yandai FH, Nadlaou B. Antimicrobial resistance in extended spectrum-lactamases (ESBL)-producing Escherichia coli isolated from human urinary tract infections in Ndjamena. Chad Afr J Microbiol Res. 2015;9(11):776–780. doi: 10.5897/AJMR2014.7338. [DOI] [Google Scholar]
- 18.Nadlaou B, Abdelsalam T, Guelmbaye N. Prevalence Multi-Resistant Bacteria in Hospital N’djamena, Chad. Chemo Open Access. 2015;4:4.
- 19.Kengne M, Dounia AT, Nwobegahay JM. Bacteriological profile and antimicrobial susceptibility patterns of urine culture isolates from patients in Ndjamena, Chad. Pan Afr Med J. 2017;28:258. [DOI] [PMC free article] [PubMed]
- 20.Jarlier V, Nicolas MH, Fournier G, Philippon A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis août. 1988;10(4):867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- 21.Dallenne C, Da Costa A, Decré D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother mars. 2010;65(3):490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 22.Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep. 2013;5(1):58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 23.Clermont O, Dhanji H, Upton M, Gibreel T, Fox A, Boyd D, et al. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J Antimicrob Chemother août. 2009;64(2):274–277. doi: 10.1093/jac/dkp194. [DOI] [PubMed] [Google Scholar]
- 24.Lonchel Magoué C, Melin P, Gangoué-Piéboji J, Assoumou M-CO, Boreux R, De Mol P. Prevalence and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae in Ngaoundere. Cameroon Clin Microbiol Infect sept. 2013;19(9):E416–E420. doi: 10.1111/1469-0691.12239. [DOI] [PubMed] [Google Scholar]
- 25.Ouédraogo A-S, Sanou S, Kissou A, Poda A, Aberkane S, Bouzinbi N, et al. Fecal Carriage of Enterobacteriaceae Producing Extended-Spectrum Beta-Lactamases in Hospitalized Patients and Healthy Community Volunteers in Burkina Faso. Microb Drug Resist. 2016;23(1):63–70. doi: 10.1089/mdr.2015.0356. [DOI] [PubMed] [Google Scholar]
- 26.Ebrahimi F, Mózes J, Monostori J, Gorácz O, Fésűs A, Majoros L, et al. Comparison of rates of fecal colonization with extended-spectrum beta-lactamase-producing Enterobacteriaceae among patients in different wards, outpatients and medical students. Microbiol Immunol mai. 2016;60(5):285–294. doi: 10.1111/1348-0421.12373. [DOI] [PubMed] [Google Scholar]
- 27.Zahar J-R, Lesprit P, Ruckly S, Eden A, Hikombo H, Bernard L, et al. Predominance of healthcare-associated cases among episodes of community-onset bacteraemia due to extended-spectrum β-lactamase-producing Enterobacteriaceae. Int J Antimicrob Agents janv. 2017;49(1):67–73. doi: 10.1016/j.ijantimicag.2016.09.032. [DOI] [PubMed] [Google Scholar]
- 28.Babu R, Kumar A, Karim S, Warrier S, Nair SG, Singh SK, et al. Faecal carriage rate of extended-spectrum β-lactamase-producing Enterobacteriaceae in hospitalised patients and healthy asymptomatic individuals coming for health check-up. J Glob Antimicrob Resist. 2016;6:150–153. doi: 10.1016/j.jgar.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 29.Çakir Erdoğan D, Cömert F, Aktaş E, Köktürk F, Külah C. Fecal carriage of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella spp. in a Turkish community. Turk J Med Sci. 2017;47(1):172–179. doi: 10.3906/sag-1512-9. [DOI] [PubMed] [Google Scholar]
- 30.Rubio-Perez Ines, Martin-Perez Elena, Domingo Garcia Diego, Lopez-Brea Calvo Manuel, Larrañaga Barrera Eduardo. Extended-spectrum beta-lactamase-producing bacteria in a tertiary care hospital in Madrid: epidemiology, risk factors and antimicrobial susceptibility patterns. Emerging Health Threats Journal. 2012;5(1):11589. doi: 10.3402/ehtj.v5i0.11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu M, Fan Y, Wang M, Lu X. Characteristics of Extended-Spectrum β-Lactamases-Producing Escherichia coli in Fecal Samples of Inpatients of Beijing Tongren Hospital. Jpn J Infect Dis. 2017;70(3):290–294. doi: 10.7883/yoken.JJID.2016.023. [DOI] [PubMed] [Google Scholar]
- 32.Saravanan M, Ramachandran B, Barabadi H. The prevalence and drug resistance pattern of extended spectrum β–lactamases (ESBLs) producing Enterobacteriaceae in Africa. Microb Pathog. 2018;114:180–192. doi: 10.1016/j.micpath.2017.11.061. [DOI] [PubMed] [Google Scholar]
- 33.Lonchel CM, Meex C, Gangoué-Piéboji J, Boreux R, Assoumou M-CO, Melin P, et al. Proportion of extended-spectrum ß-lactamase-producing Enterobacteriaceae in community setting in Ngaoundere, Cameroon. BMC Infect Dis. 2012;12(1):53. doi: 10.1186/1471-2334-12-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Falagas ME, Karageorgopoulos DE. Pandrug resistance (PDR), extensive drug resistance (XDR), and multidrug resistance (MDR) among Gram-negative bacilli: need for international harmonization in terminology. Clin Infect Dis Off Publ Infect Dis Soc Am. 2008;46(7):1121–1122. doi: 10.1086/528867. [DOI] [PubMed] [Google Scholar]
- 35.Schaumburg F, Alabi A, Kokou C, Grobusch MP, Köck R, Kaba H, et al. High burden of extended-spectrum β-lactamase-producing Enterobacteriaceae in Gabon. J Antimicrob Chemother. 2013;68(9):2140–2143. doi: 10.1093/jac/dkt164. [DOI] [PubMed] [Google Scholar]
- 36.Morgan DJ, Okeke IN, Laxminarayan R, Perencevich EN, Weisenberg S. Non-prescription antimicrobial use worldwide: a systematic review. Lancet Infect Dis sept. 2011;11(9):692–701. doi: 10.1016/S1473-3099(11)70054-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kothari A, Sagar V. Antibiotic resistance in pathogens causing community-acquired urinary tract infections in India: a multicenter study. J Infect Dev Ctries. 2008;2(5):354–358. doi: 10.3855/jidc.196. [DOI] [PubMed] [Google Scholar]
- 38.Farra A, Frank T, Tondeur L, Bata P, Gody JC, Onambele M, et al. High rate of faecal carriage of extended-spectrum β-lactamase-producing Enterobacteriaceae in healthy children in Bangui, Central African Republic. Clin Microbiol Infect. 2016;22(10):891.e1–891.e4. doi: 10.1016/j.cmi.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Sidjabat HE, Paterson DL. Multidrug-resistant Escherichia coli in Asia: epidemiology and management. Expert Rev Anti-Infect Ther. 2015;13(5):575–591. doi: 10.1586/14787210.2015.1028365. [DOI] [PubMed] [Google Scholar]
- 40.Higa S, Sarassari R, Hamamoto K, Yakabi Y, Higa K, Koja Y, et al. Characterization of CTX-M type ESBL-producing Enterobacteriaceae isolated from asymptomatic healthy individuals who live in a community of the Okinawa prefecture. Japan J Infect Chemother Off J Jpn Soc Chemother avr. 2019;25(4):314–317. doi: 10.1016/j.jiac.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 41.Kõljalg S, Truusalu K, Stsepetova J, Pai K, Vainumäe I, Sepp E, et al. The Escherichia coli phylogenetic group B2 with integrons prevails in childhood recurrent urinary tract infections. APMIS Acta Pathol Microbiol Immunol Scand mai. 2014;122(5):452–458. doi: 10.1111/apm.12167. [DOI] [PubMed] [Google Scholar]
- 42.Basu S, Mukherjee SK, Hazra A, Mukherjee M. Molecular characterization of Uropathogenic Escherichia coli: Nalidixic acid and ciprofloxacin resistance, virulent factors and phylogenetic background. J Clin Diagn Res JCDR déc. 2013;7(12):2727–2731. doi: 10.7860/JCDR/2013/6613.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.