Abstract
A defining feature of eukaryotic cells is the segregation of complex biochemical processes among different intracellular compartments. The protein targeting, translocation, and trafficking pathways that sustain compartmentalization must recognize a diverse range of clients via degenerate signals. This recognition is imperfect, resulting in polypeptides at incorrect cellular locations. Cells have evolved mechanisms to selectively recognize mislocalized proteins and triage them for degradation or rescue. These spatial quality control pathways maintain cellular protein homeostasis, become especially important during organelle stress, and might contribute to disease when they are impaired or overwhelmed.
PROTEIN LOCALIZATION AND MISLOCALIZATION
Nearly all proteins in a eukaryotic cell are synthesized by ribosomes that reside in the cytosol. Roughly 35% of these proteins are segregated to the endoplasmic reticulum (ER), ∼25% to the nucleus, ∼5% to the mitochondria (and chloroplasts in plant cells), and <1% to peroxisomes and lipid droplets (Juszkiewicz and Hegde 2018; UniProt Consortium 2018). Thus, two-thirds of nascent proteins need to be selectively targeted from the cytosol to subcellular destinations where they are typically translocated across or embedded into a membrane. Because correct localization is critical for a protein's function, cells invest extensive resources toward generating and maintaining this spatial organization.
The molecular basis of the cell's different targeting and translocation pathways vary widely (Wickner and Schekman 2005; Rapoport 2007; Hegde and Keenan 2011; Ma et al. 2011; Shi and Theg 2013; Wiedemann and Pfanner 2017). Nevertheless, a fundamental shared principle is that protein targeting involves the recognition of a targeting signal within the nascent protein by a targeting factor for the destination organelle. Different types of targeting signals specify different destinations, whereas the corresponding targeting factors either have receptors at the intended destination or are already located at the destination. Thus, the fidelity of cellular spatial organization relies critically on the specificity and efficiency by which signals are recognized by their cognate targeting factors.
As with any molecular recognition event, a degree of inefficiency is inevitable because of intrinsic limits on specific binding. Most protein targeting pathways face the additional problem that targeting signals are degenerate and not sequence-specific (von Heijne 1985, 1995). Furthermore, the targeting signals for different destinations can share a degree of similarity, risking inappropriate recognition. Thus, even under optimal conditions, protein targeting can fail for a proportion of nascent proteins. Because proteins in the wrong cellular compartment are at risk of inappropriate interactions, unregulated function, or aggregation, they must be promptly recognized and either degraded or rescued.
How does a cell recognize when a protein is out of place? At first glance, the answer seems straightforward: Proteins in the wrong environment are unlikely to fold or assemble properly, so the quality control pathways dedicated to misfolded proteins would also deal with mislocalized proteins. Although this is certainly true to some extent, cells have additionally evolved recognition factors that seem to be specialized for protein mislocalization. As discussed in detail below, these factors often have a similar specificity as protein targeting factors, but instead of engaging a receptor at the destination organelle, they are coupled to ubiquitination and degradation machinery. In this way, mislocalization can be recognized by the prolonged exposure of a targeting signal that should have been recognized by a targeting factor.
This review discusses the challenges to high-fidelity protein localization and the extent to which it fails under physiologic and pathologic conditions. Failure of protein compartmentalization is emerging as a substantial quality control burden on cells and several factors have recently been implicated in their recognition and disposal. The importance of these spatial quality control pathways for organismal homeostasis is highlighted by the pathologic consequences of their saturation or failure.
MOLECULAR RECOGNITION OF DEGENERATE SIGNALS
The targeting signals that mediate protein compartmentalization are almost all degenerate motifs characterized by certain biophysical features rather than specific amino acid sequences. The most specific signal is a carboxy-terminal tripeptide (Ser-Lys-Leu-COOH, with some variation) for peroxisomal targeting that is specifically recognized by Pex5 (Ma et al. 2011). The X-ray structure of this interaction reveals the basis of its specificity (Gatto et al. 2000). In contrast to this exception, however, all other targeting signals (including most peroxisomal proteins that use other signals) are largely sequence-independent. This imposes a major challenge to high-fidelity recognition because the binding site for the signal must necessarily be flexible to accommodate diverse sequences.
The signal for cotranslational protein targeting to the ER contains the widest diversity as a result of the 7000 proteins (in mammals) that use this pathway (Guna and Hegde 2018; UniProt Consortium 2018). The minimal basis for this signal is simply a stretch of ∼7–9 primarily hydrophobic helix-compatible residues (von Heijne 1985). This can be found in either cleavable signal peptides or a transmembrane domain (TMD). Both of these elements are highly variable in amino acid sequence, composition, length, and flanking regions. Yet, signal sequences and TMDs must be recognized as a class while excluding all other nascent protein sequences of proteins destined for non-ER locations.
Signals for cotranslational ER targeting are recognized by the 54 kDa subunit of the signal recognition particle (SRP) (Keenan et al. 2001). The availability of multiple X-ray structures of SRP54 without (Keenan et al. 1998; Clemons et al. 1999) and with a bound signal (Janda et al. 2010; Hainzl et al. 2011; Hainzl and Sauer-Eriksson 2015) shows it to contain a hydrophobic binding groove that is accommodating in two ways. First, the helical scaffold that forms the hydrophobic groove can be in multiple conformations. Second, residues that line the groove are enriched in methionines whose side chains are flexible. This explains how the groove can bind a wide variety of signals; however, it is unclear from solely structural considerations how specificity for only ER-destined cargo is achieved (Fig. 1).
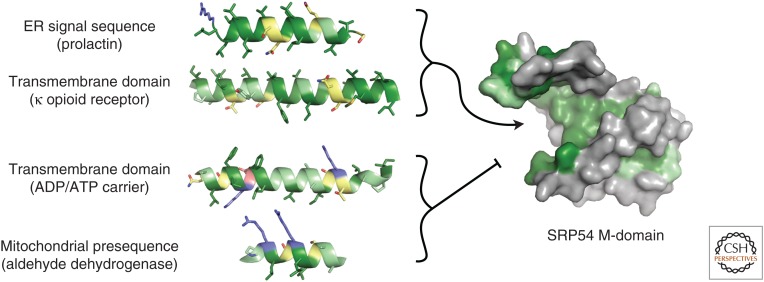
Figure 1.
The recognition problem for hydrophobic targeting sequences. Shown are examples of an endoplasmic reticulum (ER)-destined signal sequence (only the hydrophobic domain is depicted), the first transmembrane domains for proteins destined for the ER (opioid receptor) and mitochondria (ADP/ATP carrier), and a mitochondrial presequence (only the amphipathic helix is depicted). Despite many similarities among the four targeting sequences, the first two need to be recognized by the M-domain of SRP54, whereas the other two need to be avoided. The amino acids in the targeting sequences are colored as follows: acidic in pink, basic in light blue, highly hydrophobic in dark green, moderately hydrophobic in pale green, and polar in yellow. The substrate binding M-domain of SRP54 is taken from Protein Data Bank (PDB) code 3JAN, which was modeled from 1QB2, the X-ray structure of the human SRP54 M-domain (Clemons et al. 1999). Hydrophobic residues of SRP54 are colored green.
Part of the answer may lie in the observation that the hydrophobic groove appears to be autoinhibited by a carboxy-terminal amphipathic helix that might reduce promiscuous binding (Voorhees and Hegde 2015). Further contributions to fidelity arise from substrate-dependent modulation of SRP54's GTPase cycle and interaction with its receptor (Zhang et al. 2010). However, these mechanisms probably do not completely eliminate promiscuous recognition of TMDs in mitochondrial membrane proteins. Although the nascent polypeptide-associated complex (NAC) seems to aid in this discrimination (Gamerdinger et al. 2015), it is unclear how it functions in molecular terms.
A similar recognition problem exists for tail-anchored membrane proteins. These proteins have a single TMD close to the carboxyl terminus that serves as the targeting signal for either the ER, mitochondria, chloroplasts, or peroxisomes (Borgese and Fasana 2011; Hegde and Keenan 2011). The primary mode of recognition appears to be TMD hydrophobicity, with additional contributions from flanking charged residues (Kalbfleisch et al. 2007; Costello et al. 2017). Although the TMDs for ER-targeted tail-anchored proteins tend to be more hydrophobic than those destined for other organelles, the overlap is substantial (Guna and Hegde 2018). Furthermore, structures of the ER targeting factor Get3 bound to a TMD (Mateja et al. 2015) do not reveal an obvious basis on which all mitochondrial tail-anchored proteins could be rigorously excluded. As with cotranslational targeting, subtle kinetic differences in Get3 interactions with substrate and nonsubstrate TMDs may help increase fidelity (Rao et al. 2016), but would not eliminate errors altogether.
A number of observations indicate that targeting factors have broader intrinsic specificities than the clients for which they are intended. First, mitochondrial tail-anchored proteins can bind to the mammalian homolog of Get3 in vitro (Itakura et al. 2016) and their overexpression results in Get3-dependent ER targeting in yeast (Vitali et al. 2018). Conversely, deletion of components for the ER tail-anchored targeting pathway results in mitochondrial mislocalization (Schuldiner et al. 2008; Jonikas et al. 2009). Second, acute depletion of SRP allows at least some ER-destined proteins to be recognized by the mitochondrial import pathways, resulting in mistargeting (Costa et al. 2018). Third, depletion of NAC permits SRP to recognize cytosolic and mitochondrial proteins in vitro (Wiedmann et al. 1994) and target them to the ER in vivo (Gamerdinger et al. 2015). These types of experiments illustrate that when one targeting factor is missing or saturated, other targeting factors that ordinarily are not engaged now bind promiscuously. This suggests that targeting factors for different destinations have overlapping specificities, and that part of their overall fidelity comes from competition for substrates. The corollary to this idea is that such overlap necessarily results in some degree of intrinsic inefficiency and mistargeting even under normal circumstances.
Measurements of protein segregation efficiency to the ER via the SRP pathway suggest a failure rate of 5%–30% in cultured cells using heterologous expression (Rane et al. 2004; Levine et al. 2005; Kang et al. 2006; Emerman et al. 2010). Although this artificial situation may exaggerate the failure rate, studies of prion protein (PrP) translocation in transgenic mice suggest that even in the appropriate cell type with modest overexpression, the failure rate for PrP's targeting signal is 3%–5% (Rane et al. 2010). In unpublished observations, we have found that attaching a signal sequence to the biotin ligase BirA is insufficient to fully exclude its activity from the cytosol. Although this population of mislocalized BirA (or any other protein) is not obvious using green fluorescent protein (GFP)-tagging or biochemical fractionation, the far more sensitive enzymatic assay of target modification readily reveals it (Emerman et al. 2010). Thus, ER targeting has limits on its fidelity resulting in constitutive low-level mislocalization (Fig. 2). As discussed in detail later, mislocalized proteins have multiple potential fates, including proteasomal degradation, aggregation, and mistargeting to another organelle. Although less well studied, the targeting pathways to other destinations are similarly challenged and prone to at least some rate of failure.
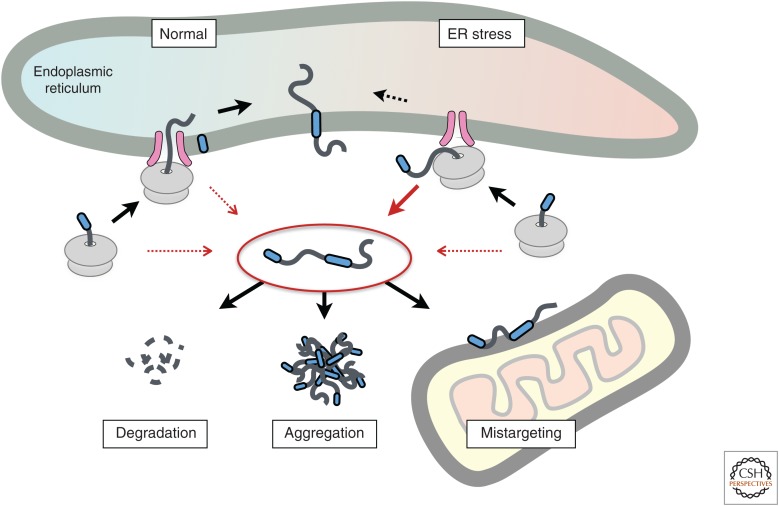
Figure 2.
Multiple fates for an endoplasmic reticulum (ER)-destined mislocalized protein. Protein import into the ER can fail, resulting in a mislocalized protein in the cytosol (inside the red oval in the center). This failure occurs constitutively at a low level under normal conditions (red arrows, left side) but can be enhanced for some proteins during acute ER stress (red arrows, right side). The mislocalized protein can be degraded, aggregate with itself or other proteins, or mistarget to another organelle.
Why has evolution tolerated this apparent inefficiency? The most plausible answer is that targeting elements are often part of the final protein and integral to its folding and function. For example, >3000 membrane proteins in the human genome are targeted via a TMD that has functions beyond targeting. In multipass membrane proteins, the TMD used for targeting is also a critical structural element of the final protein; in single-pass membrane proteins, the TMD often plays key roles in protein–protein interactions. Thus, such functional constraints imposed by these posttargeting roles necessarily means that TMDs used for targeting are not only diverse, but sequence features that are often detrimental for optimal targeting are nonetheless conserved for other reasons.
Because the targeting of proteins to other organelles is also frequently mediated by sequences that are part of the final protein, diversity of targeting sequences, and hence a degree of variability in targeting efficiency, would seem to be both universal and inevitable. Given the large proportion of cellular proteins that rely on subcellular targeting during their biogenesis, mislocalization is therefore a constant threat to the generation and maintenance of spatial organization.
CAUSES AND CONSEQUENCES OF IMPAIRED ORGANELLE IMPORT
In addition to intrinsic failures of localization attributable to inefficiency, the failure rate is sometimes exaggerated owing to environmental perturbations. For example, during ER stress, import is attenuated (Fig. 2). This appears to be substrate-selective and is influenced by the nature of the targeting signal (Kang et al. 2006). Some signal peptides are refractory to stress-dependent attenuation, including that for the BiP, which is needed in the ER lumen to alleviate the stress. By replacing the signal sequence of a stress-sensitive protein (PrP) with the signal sequence of a stress-resistant protein, it was possible to enforce ER translocation even during acute stress. The aggregation of PrP in the ER lumen under these conditions, and its adverse consequence for cell viability, indicates that transient attenuation of translocation during stress is a protective response (Kang et al. 2006).
Mitochondrial stress has also been shown to impair protein import into the stressed mitochondrion, leading to several physiologic consequences (Fig. 3). First, the load of new proteins entering stressed mitochondria is reduced at the expense of generating mislocalized proteins that must be degraded by cytosolic proteasomes (Wright et al. 2001). Second, failure to import PINK1 into the inner membrane of stressed mitochondria results in its accumulation at the outer membrane, thus marking these organelles for initiation of mitophagy (Sekine and Youle 2018). Third, reduced protein import into mitochondria can initiate stress responses aimed at stress alleviation. These include attenuated import of the transcription factor ATFS-1 in Caenorhabditis elegans (Nargund et al. 2012), proteasome up-regulation (Wrobel et al. 2015), an altered translational landscape (Wang and Chen 2015), and activation of the transcription factor Pdr3 in yeast (Weidberg and Amon 2018). Identifying the mechanism(s) of sensing such stress responses and complete delineation of the signaling pathways that culminate in the observed responses are important goals.
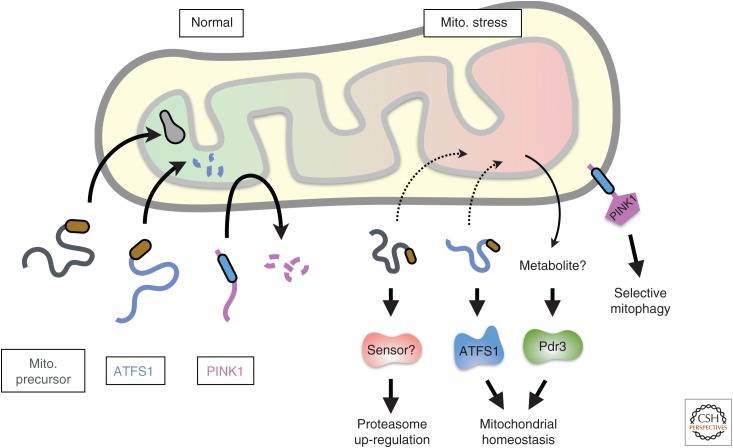
Figure 3.
Protein mislocalization and signaling during mito. (mitochondrial) stress. Depicted are the fates of several mitochondrial precursors during normal conditions (left) and acute stress (right). Failed import during acute stress results in (presumably misfolded) mitochondrial precursors in the cytosol that can trigger proteasomal up-regulation by a mechanism that is incompletely understood (Wrobel et al. 2015). In special cases, the nonimported precursor is not degraded selectively during mitochondrial stress and can signal downstream responses. This is the case for ATFS1, which can act as a transcription factor to up-regulate genes that improve mitochondrial homeostasis (Nargund et al. 2012). PINK1 is normally imported into the inner membrane where its cleavage permits retrotranslocation to the cytosol for degradation. During acute stress, PINK1 import to the inner membrane is aborted, allowing its insertion into the mitochondrial outer membrane where it initiates a series of events leading to mitochondrial autophagy (Sekine and Youle 2018). Pdr3 is also activated during certain types of mitochondrial stress (Weidberg and Amon 2018), although the signal for activation, perhaps a mitochondrial metabolite, is not known.
Although acute reduction in translocation is protective for both mitochondria and the ER, chronic protein mislocalization clearly has adverse consequences. Perhaps the most striking illustration of this comes from the rare human diseases caused by mutations in targeting signals (Arnold et al. 1990; Karaplis et al. 1995; Seppen et al. 1996; Cassanelli et al. 1998; Hussain et al. 2013; Guo et al. 2014). Such mutations typically have two consequences: reduced correctly produced protein and elevated mislocalized protein. In most cases, it appears to be the mislocalization that is particularly detrimental. This is supported by the finding that such mutations are autosomal dominant and cause more severe phenotypes than expected for haploinsufficiency in the gene.
In mice, it has been shown that even a partially inefficient signal sequence on PrP causes age-dependent neurodegeneration (Rane et al. 2008), whereas deletion of the signal produces a severe phenotype (Ma et al. 2002). Because PrP is nonessential and its ablation has little or no overt phenotype (Büeler et al. 1993), the consequences of signal-perturbed PrP expression is apparently attributable to its inappropriate residence in the cytosol (Norstrom et al. 2007). When considered with the observation that mislocalized insulin or parathyroid hormone also causes disruption to protein homeostasis and cell death (Arnold et al. 1990; Karaplis et al. 1995; Guo et al. 2014), it appears that protein mislocalization, even of a single highly expressed protein, is generally detrimental in vivo over time. The mechanistic basis of cell dysfunction caused by mislocalized proteins is not clear but may be related to their aggregation or inappropriate interactions with other proteins or organelles.
Consistent with this interpretation, disruption of protein targeting pathways in yeast or cultured mammalian cells results in cytosolic aggregation of nontargeted proteins (Schuldiner et al. 2008; Guna et al. 2018), mistargeting to other organelles (Schuldiner et al. 2008; Jonikas et al. 2009; Costa et al. 2018), induction of various stress responses (Jonikas et al. 2009; Brandman et al. 2012), and increased sensitivity to other perturbations of protein homeostasis pathways (Jonikas et al. 2009; Aviram et al. 2016). For example, perturbation of tail-anchored protein targeting pathways to the ER results in aggregation in the cytosol and detectable mistargeting to other organelles (Schuldiner et al. 2008; Jonikas et al. 2009; Guna et al. 2018). Similarly, depletion of NAC results in promiscuous SRP recognition of cytosolic and mitochondrial proteins, their mistargeting to the ER, aggregation of many proteins, and activation of ER stress pathways (Wiedmann et al. 1994; Gamerdinger et al. 2015). Finally, acute SRP depletion results in detectable mistargeting to mitochondria, impaired mitochondrial function, and subsequent mitochondrial fragmentation (Costa et al. 2018). Although all of these examples are gross exaggerations, they serve to illustrate the potential for cellular disruption posed by mislocalized proteins.
The fact that protein targeting has intrinsic limits on its fidelity and results in a small but constant flux of polypeptides mistargeted to the cytosol or another organelle implies that each compartment has mechanisms to recognize and eliminate mislocalized proteins. If such mechanisms did not exist, mislocalized proteins would accumulate over time and cause impaired cytosolic and organellar protein homeostasis similar to that observed when mislocalization is exaggerated. The factors that mediate spatial quality control of mistargeted proteins have only begun to emerge in the past several years.
CYTOSOLIC MONITORS OF PROTEIN MISLOCALIZATION
Genetic and biochemical strategies in yeast and mammalian systems, respectively, have begun to identify cytosolic monitors of protein mislocalization. One principle that has emerged from these studies is that targeting signals, which are often hydrophobic, are recognized by quality control factors that facilitate substrate ubiquitination and degradation by the proteasome (Fig. 4A). Several of these quality control factors with overlapping specificities are believed to patrol the cytosol for a range of potential targeting signals that would signify a mislocalized protein (Fig. 4B).
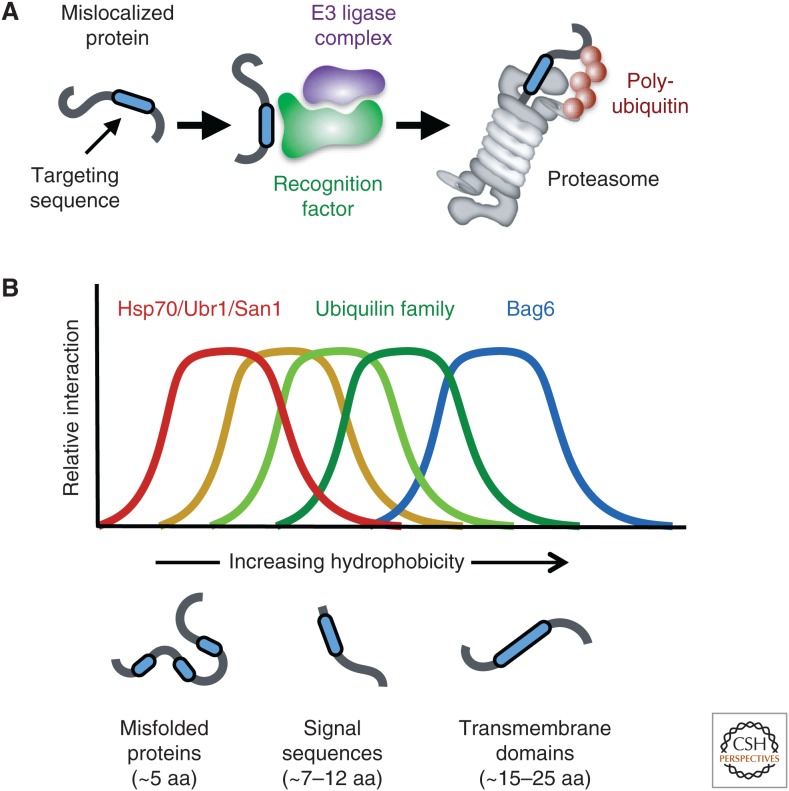
Figure 4.
Cytosolic recognition of mislocalized proteins. (A) Targeting signals (or transmembrane domains) can be recognized by dedicated cytosolic factors that recruit ubiquitination machinery to mediate substrate ubiquitination. These substrates are then degraded by proteasomes. (B) Schematic depiction of the concept that different cytosolic factors, all of which recognize exposed hydrophobicity, are tuned to different lengths and degrees of hydrophobic domains. Biochemical analyses suggest that there is substantial overlap in what is recognized.
Studies in yeast aimed at identifying cytosolic quality control factors primarily used ΔSS-CPY, a mutant carboxypeptidase Y (CPY) mislocalized to the cytosol because of deletion of its ER targeting signal (Medicherla et al. 2004). Genetic studies showed that ΔSS-CPY degradation requires the proteasome and the widely used ubiquitin-conjugating E2 enzymes Ubc4 and Ubc5 (Park et al. 2007). In addition, Ssa1 (an Hsp70 family chaperone) and its cochaperone Ydj1 also facilitated ΔSS-CPY degradation (Park et al. 2007). Based on solubility assays performed in the presence or absence of these factors, it appears that at least one function of chaperones in this pathway is to prevent ΔSS-CPY aggregation (or facilitate its resolubilization) to allow proteasome delivery.
In a second round of studies, the E3 ubiquitin ligases needed for ΔSS-CPY degradation were revealed to be the cytosolic ligase Ubr1 and the nuclear ligase San1 (Eisele and Wolf 2008; Heck et al. 2010). Importantly, the well-characterized role of Ubr1 in recognizing certain amino-terminal residues as part of the N-end rule degradation pathway was not the basis of its role in ΔSS-CPY degradation. Instead, subsequent studies have suggested that San1 (and probably Ubr1) recognize clients via exposed hydrophobic patches of ∼5 residues or more that typify misfolded proteins (Fredrickson et al. 2011).
In parallel with these studies on mislocalized protein substrates, misfolded cytosolic proteins were analyzed by similar strategies in yeast. This led to the finding of a similar set of factors, including chaperones (most notably Ssa1 and Ydj1) cooperating with Ubr1 and San1 E3 ligases (Prasad et al. 2010, 2018; Khosrow-Khavar et al. 2012; Guerriero et al. 2013; Scazzari et al. 2015; Maurer et al. 2016). Whereas additional factors were described for some substrates (such as Hsp90 and its cofactors [McClellan et al. 2005], Ubr2 [Nillegoda et al. 2010], Doa10 [Maurer et al. 2016], or other J-domain proteins [Summers et al. 2013; Prasad et al. 2018]), the overall principles for quality control of misfolded and mislocalized proteins seemed to be very similar. Thus, one mechanism of mislocalized protein quality control is apparently based on the simple fact that a protein in the wrong environment is not likely to fold correctly owing to the absence of suitable maturation factors and modifications.
The studies in yeast used model substrates lacking the signal sequence, an element that would be retained in bona fide mislocalized proteins caused by failed targeting. Furthermore, mislocalized membrane proteins, which would contain noninserted TMDs, were not examined. In contrast, more recent biochemical analyses of mislocalized proteins in a mammalian system included signal sequences and TMDs, a key difference from the yeast studies that led to the identification of different factors.
The study of mislocalized proteins in the mammalian system originates from in vitro studies of protein translocation into organelles (Sharma et al. 2010). A typical control reaction for protein targeting and translocation studies is a sample lacking the target organelle. Notably, secretory and membrane proteins synthesized in reticulocyte lysate lacking ER or mitochondria get ubiquitinated (Hessa et al. 2011; Itakura et al. 2016). This observation suggested that at least some quality control factors involved in degrading mislocalized proteins are present in the lysate, providing a tractable experimental system for their study.
Using this system, mislocalized ER-destined proteins were found to interact with Bag6, a protein whose depletion led to reduced mislocalized protein ubiquitination (Hessa et al. 2011). The E3 ligase RNF126 was later shown to associate with Bag6 and mediate mislocalized protein ubiquitination (Rodrigo-Brenni et al. 2014). Bag6 had previously been implicated in recognizing ubiquitinated aberrant proteins in the cytosol (Minami et al. 2010), but its substrate range and mechanism of action were unclear. Subsequent studies showed that Bag6 strongly favors long linear hydrophobic sequences such as ER targeting signals, TMDs, and carboxy-terminal GPI-anchoring signals (Mariappan et al. 2010; Hessa et al. 2011; Yamamoto et al. 2017). Removal of those elements from a mislocalized protein strongly impaired recognition by Bag6 even if the protein was misfolded. Because these sequences are removed or buried in the membrane upon successful targeting, it appears that their cytosolic exposure is a critical cue of targeting failure. Thus, the selectivity of Bag6 for mislocalized proteins arises from its specificity for targeting sequences.
How does Bag6 avoid competing or interfering with targeting factors? The answer appears to be twofold. First, the positioning of SRP54 at the mouth of the ribosome exit tunnel via tight ribosome–SRP interactions (Halic et al. 2004; Voorhees and Hegde 2015) probably prioritizes cotranslational SRP interaction with signal sequences and TMDs. Second, posttranslational priority for targeting factors or chaperones probably arises from a combination of their relative abundance, rate of binding, and intrinsic specificity. Thus, calmodulin, which is involved in posttranslational targeting of small secretory proteins and certain tail-anchored proteins (Shao and Hegde 2011; Guna et al. 2018), is more abundant than Bag6 and seems to prefer more moderately hydrophobic sequences relative to Bag6's preference for higher hydrophobicity. For other tail-anchored proteins, the on-rate of a factor called SGTA appears to afford a competitive advantage (Shao et al. 2017). SGTA can then hand over the tail-anchored protein to TRC40 (the mammalian homolog of Get3) for ER targeting (Wang et al. 2010; Mock et al. 2015; Shao et al. 2017).
Remarkably, the handover reaction from SGTA to TRC40 is bridged by Bag6 and its two interacting partners Ubl4A and TRC35 (Mock et al. 2015; Shao et al. 2017). If this handover fails to occur promptly, the substrate eventually dissociates from SGTA. Because this failure occurs in proximity to Bag6, it is positioned ideally to capture mistargeted tail-anchored proteins for subsequent degradation (Shao et al. 2017). Thus, in metazoans, a quality control factor specialized for mislocalized proteins is embedded within a targeting pathway to rapidly capture any failures.
In yeast, Bag6 apparently has been lost and the Ubl4A and TRC35 homologs (termed Get5 and Get4, respectively) interact directly with each other to form a bridge between Sgt2 (the SGTA homolog) and Get3 (Wang et al. 2010). It is possible that metazoan cells place a higher premium on eliminating mislocalized proteins than in yeast, where rapid cell division provides frequent opportunities to exclude aggregated proteins from the newly budded daughter cell (Liu et al. 2010).
Using very similar biochemical approaches, the Ubiquilin family of proteins was identified as TMD-binding factors involved in the quality control of mislocalized mitochondrial proteins (Itakura et al. 2016). Like Bag6, Ubiquilin family members bind hydrophobic elements such as TMDs and signal sequences (Itakura et al. 2016; Suzuki and Kawahara 2016; Whiteley et al. 2017). They then recruit a ubiquitin ligase (that remains to be identified) to facilitate degradation of their bound clients. As a class, Ubiquilins appear to prefer signals and TMDs of somewhat lower hydrophobicity than Bag6 (Itakura et al. 2016), although the overlap is substantial. In mammals, three of the four Ubiquilins are widely expressed in all tissues, as is Bag6. Thus, most mammalian cells appear to contain at least four quality control factors in the cytosol that favor long hydrophobic targets. The need for several such factors might reflect the fact that signals and TMDs are highly diverse in sequence and properties. An important goal is to define the substrate ranges of these factors and the extent to which they can compensate for each other.
It is noteworthy that Dsk2, the yeast homolog of the Ubiquilins, is not needed for degradation of ΔSS-CPY (Medicherla et al. 2004). As noted above, the absence of the signal sequence may have led to this result. It will therefore be of interest to determine whether in yeast, cytosolic factors with specificity for highly hydrophobic elements that characterize mislocalized proteins feature in their degradation separate from the pathways for misfolded cytosolic proteins. Based on the mammalian results, Dsk2 would seem to be an obvious candidate for such a factor.
Whereas the cytosolic factors for mislocalized proteins have thus far focused primarily on ER- and mitochondria-destined proteins, one might also expect analogous factors for mislocalized nuclear proteins. Although many nuclear proteins shuttle between the cytosol and nucleus, some are strictly nuclear. Examples include histones, certain components of the nuclear pore complex, the nuclear lamina proteins, and others. Whether or how these proteins are recognized in the cytosol remains to be investigated. One candidate is the recently identified hybrid E2/E3 enzyme UBE2O involved in degradation of ribosomal proteins that fail nuclear import (Nguyen et al. 2017; Yanagitani et al. 2017). Although the specificity of UBE2O remains to be fully explored, it appears to recognize domains that overlap with nuclear localization sequences in ribosomal proteins (Yanagitani et al. 2017). Whether it recognizes analogous sequences in proteins like histones or lamins remains unknown.
MEMBRANE MONITORS OF PROTEIN MISLOCALIZATION
In addition to cytosolic mislocalization caused by failed targeting, some proteins can also mistarget to the wrong membrane. Although the fate of these mistargeted proteins is poorly understood, it appears that there are mechanisms to selectively recognize and degrade them at both the ER and mitochondrial outer membrane (Fig. 5). In some cases, the mistargeted protein might also be rescued to the correct destination.
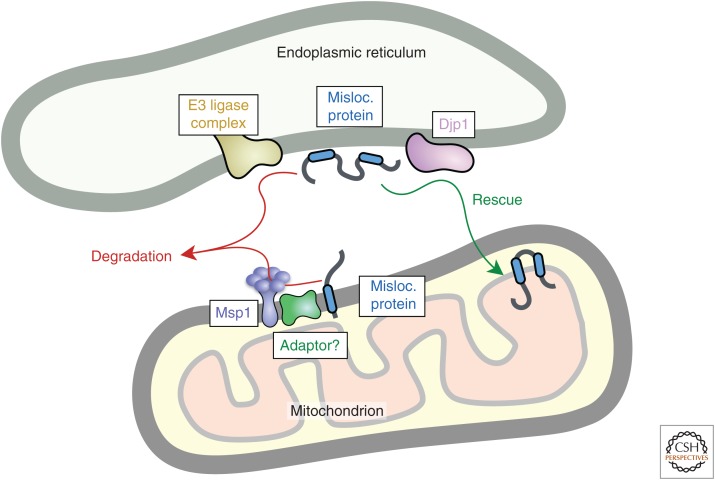
Figure 5.
Recognition of mistargeted proteins at organelle membranes. Mistargeted proteins are depicted at the endoplasmic reticulum (ER) and mitochondrion. At the ER, they might be recognized by E3 ubiquitin ligase complexes centered around TRC8 or MARCH6 (Doa10 in yeast) and routed for degradation in the cytosol (Metzger et al. 2008; Stefanovic-Barrett et al. 2018). At mitochondria, the AAA+ ATPase Msp1 (ATAD1 in mammals) can recognize mistargeted proteins, possibly with the aid of an adaptor, to mediate extraction into the cytosol (Chen et al. 2014; Okreglak and Walter 2014; Weir et al. 2017; Wohlever et al. 2017; Weidberg and Amon 2018). Whether ubiquitin ligases are involved and at what step they act to mediate Msp1-mediated degradation is not known. Mistargeted proteins can also be rescued, as exemplified by a pathway from the ER to mitochondria involving the chaperone Djp1 (Hansen et al. 2018). Misloc. protein, Mislocalized protein.
Studies in yeast and mammalian cells have identified the ATPase Msp1 (ATAD1 in humans) as a factor required for efficient clearance of mistargeted tail-anchored proteins from the outer mitochondrial membrane and peroxisomes (Chen et al. 2014; Okreglak and Walter 2014). Notably, ER-destined tail-anchored proteins are visible at these membranes in cells lacking Msp1/ATAD1, indicating that at least some degree of mistargeting occurs even under normal conditions.
Msp1 is anchored via a single amino-terminal TMD to the outer mitochondrial membrane and peroxisomes, with its ATPase domain facing the cytosol. The ATPase domain interacts with itself to homohexamerize (Wohlever et al. 2017), resulting in a central pore similar to other members of the AAA+ ATPase family (Olivares et al. 2018). Reconstitution studies have shown that purified Msp1 in liposomes is sufficient to drive extraction of a tail-anchored protein from the membrane to the cytosol in an ATP-dependent reaction (Wohlever et al. 2017). Interestingly, mutations to key residues within the Msp1 hexamer impair extraction, suggesting a model in which substrates are pulled through this pore similar to how other AAA+ ATPases use energy to apply force to their substrate proteins.
The substrate range of Msp1 and its mechanism of substrate recognition remain poorly understood. The primary clue comes from analysis of Pex15, a tail-anchored protein that is stable in peroxisomes, but degraded rapidly from the mitochondrial outer membrane (Weir et al. 2017). This observation seemed puzzling given that Msp1 is present in both membranes. The explanation lies in the fact that Pex15 forms a complex with the peroxisomal membrane protein Pex3, and this complex is refractory to Msp1-mediated degradation. Thus, Pex15 in the mitochondrial outer membrane lacks its binding partner, allowing orphaned Pex15 recognition. The region of Pex15 that is recognized and the specific factor(s) that mediate the recognition remain unclear. Because Msp1 has only one TMD, it is difficult to see how this could recognize the TMDs of tail-anchored proteins selectively. Thus, it is plausible that there are other factors that interact with Msp1 to provide specificity.
A potential adaptor for Msp1 is the protein Cis1. In yeast, it was observed that failed import into mitochondria results in induction of Cis1, which apparently bridges the interaction between Msp1 and the translocase of the outer mitochondrial membrane (Weidberg and Amon 2018). Because Cis1 (along with Msp1) are needed to survive conditions of import failure, it is believed that they participate in clearing mitochondrial translocase complexes of partially imported and trapped substrates to restore their availability. These observations suggest that Msp1 may have a broader client range than only tail-anchored proteins and suggest that its targeting to clients is mediated by adaptors such as Cis1 and potentially others that remain to be identified.
The converse problem of mistargeted proteins at the ER membrane also warrants clearance. Direct analysis of this problem is generally lacking, but some insight has come from the study of an artificial degron called CL1. Reporter proteins appended with this short amphipathic helix require the ER resident E3 ligase Doa10 for degradation in yeast (Metzger et al. 2008; Maurer et al. 2016). In the absence of Doa10-mediated degradation, the CL1-containing reporter is observed partially localized at the ER membrane. In mammalian cells, the resident ER ubiquitin ligases TRC8 and MARCH6 (a homolog of Doa10) are required for degradation of CL1-containing reporters (Stefanovic-Barrett et al. 2018). Because CL1 partially resembles a signal sequence or part of a TMD, it might represent a serendipitous model for protein mistargeting to the ER. If so, these results suggest that ubiquitin ligases implicated in ER-associated degradation are also involved in degrading mistargeted proteins.
Recently, it was shown that some mitochondrial membrane proteins mistargeted to the ER membrane are not degraded, but rather retrieved for successful import into mitochondria (Hansen et al. 2018). An ER-localized factor called Djp1 is required for this retrieval pathway. In the absence of Djp1, mitochondrial membrane proteins are promiscuously inserted into the ER, degraded, and/or aggregated. Whether analogous rescue pathways exist in other instances of mistargeting is unknown. In yeast cells, aggregates can be disaggregated with the help of Hsp104 (Doyle and Wickner 2009). The chaperone Sgt2, part of the tail-anchored insertion pathway (Hegde and Keenan 2011), can interact with Hsp104 (Wang et al. 2010). This suggests the possibility that mistargeted or aggregated tail-anchored proteins in yeast could be rescued for insertion.
All of the examples discussed thus far involve missegregation among different organelles. However, many organelles are subcompartmentalized, even within a single continuous membrane system. Because the protein compositions of different subcompartments are distinct, spatial quality control pathways may be needed to maintain this specialization. One notable example is represented by the Asi ubiquitin ligase complex located in the inner nuclear membrane. The inner membrane is continuous with the outer nuclear and ER membranes via nuclear pore complexes that provide a barrier to free diffusion. Nevertheless, proteins from the ER and outer membrane can sometimes be missegregated to the inner membrane. Degradation of these mislocalized proteins requires the Asi complex, which is believed to function analogously to ubiquitin ligase complexes that mediate ER-associated degradation (Foresti et al. 2014).
Many other continuous membrane systems are similarly specialized into spatially and functionally distinct domains. Yet other quality control systems probably exist to retain this specialization. For example, highly polarized cell types such as neurons have many plasma membrane regions with very specific protein compositions that must be tightly regulated for correct function. How stray proteins are eliminated from each subdomain is poorly understood, but likely to be of substantial biological importance.
MISLOCALIZATION IN DISEASE
As noted previously, the most obvious instances in which protein mislocalization features in disease are the rare cases of a mutated targeting sequence. These examples serve to highlight the potential toxicity of a mislocalized protein in physiologically relevant contexts (as opposed to overexpression in model systems). Given the detrimental consequences of even one protein being chronically mislocalized, it is worth considering the issue of whether and how the small amount of intrinsic protein mislocalization might influence pathogenesis more generally in human disease.
A prominent feature of numerous protein misfolding diseases is protein aggregates in the cytosol (Goedert 2015). Cultured cells containing cytosolic aggregates were observed to accumulate a cytosolic population of proteins destined for ER targeting (Chakrabarti et al. 2011). The basis for this accumulation was attributable to impaired degradation of the normally minor nontargeted population; hence, cytosolic accumulation could be minimized by maximizing the efficiency of the targeting signal. Notably, proteasome activity was normal in these cells, arguing for disruption of another component of the degradation pathway for mislocalized proteins.
Cytosolic aggregates of a polyglutamine-containing protein were found to be enriched in Ubiquilins (Doi et al. 2004; Wang and Monteiro 2007; Mori et al. 2012; Rutherford et al. 2013), perhaps for their disaggregation (Hjerpe et al. 2016). In cultured cells, such aggregates sequester so much Ubiquilin that the amount free in the cytosol is markedly depleted (Itakura et al. 2016). Because Ubiquilins play a role in eliminating mislocalized proteins, Ubiquilin sequestration in aggregates would allow accumulation of its substrates. Indeed, a mitochondrial membrane protein lacking its targeting sequence was found to be normally degraded in aggregate-lacking cells, but strongly stabilized in aggregate-containing cells (Itakura et al. 2016). Ubiquilins have been found sequestered in aggregates in various human diseases (Mori et al. 2012; Rutherford et al. 2013), suggesting that mislocalized proteins might accumulate in vivo similarly to these observations in cells. Conversely, overexpression of Ubiquilins is protective in a mouse model of polyglutamine protein aggregation (Safren et al. 2014), although the basis of this protection remains unclear.
In addition to protein aggregation, chronic organelle stress is a very common feature of many disease states, including several protein misfolding disorders (Yoshida 2007; Federico et al. 2012). Increased protein mislocalization may accompany organelle stress, imposing a higher demand on the respective clearance pathways. Thus, it is attractive to postulate that a combination of decreased import because of organelle stress and decreased clearance owing to sequestration of key factors may lead to accumulation of hydrophobic mislocalized proteins. Because these may themselves form aggregates or induce organelle stress, this process may be self-perpetuating once a threshold is reached. Indeed, such a process appears to be the explanation for why transient proteasome inhibition leads to runaway accumulation of nontranslocated PrP in the cytosol long after the proteasome inhibitor is removed (Ma and Lindquist 2002; Chakrabarti et al. 2011).
As the pathways for recognition, clearance, and rescue of mislocalized proteins become clearer, it is likely that partial loss-of-function mutations will feature in diseases of protein homeostasis. Indeed, mutations in Ubiquilin-2 (UBQLN2) are known to cause neurodegeneration in humans and in mice (Deng et al. 2011; Hjerpe et al. 2016; Le et al. 2016), although the pathogenic mechanism is unclear at this time. Furthermore, as better reporters for sensitively and specifically monitoring protein mislocalization are developed, one can investigate whether the spatial organization of the cellular proteome declines during various age-related diseases, or even as part of the normal aging process.
ACKNOWLEDGMENTS
We are grateful to past and present Hegde laboratory members for illuminating discussions. Work in the authors’ laboratory is supported by the UK Medical Research Council (UK MRC; MC_UP_A022_1007 to R.S.H.). E.Z. was supported by an MRC Career Development Fellowship and the Laboratory of Molecular Biology/AstraZeneca (LMB/AZ) Blue Sky Fund (BSF27).
Footnotes
Editors: Richard I. Morimoto, F. Ulrich Hartl, and Jeffery W. Kelly
Additional Perspectives on Protein Homeostasis available at www.cshperspectives.org
REFERENCES
- Arnold A, Horst SA, Gardella TJ, Baba H, Levine MA, Kronenberg HM. 1990. Mutation of the signal peptide-encoding region of the preproparathyroid hormone gene in familial isolated hypoparathyroidism. J Clin Invest 86: 1084–1087. 10.1172/JCI114811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviram N, Ast T, Costa EA, Arakel EC, Chuartzman SG, Jan CH, Haßdenteufel S, Dudek J, Jung M, Schorr S, et al. 2016. The SND proteins constitute an alternative targeting route to the endoplasmic reticulum. Nature 540: 134–138. 10.1038/nature20169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese N, Fasana E. 2011. Targeting pathways of C-tail-anchored proteins. Biochim Biophys Acta 1808: 937–946. 10.1016/j.bbamem.2010.07.010 [DOI] [PubMed] [Google Scholar]
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, et al. 2012. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151: 1042–1054. 10.1016/j.cell.2012.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büeler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, Weissmann C. 1993. Mice devoid of PrP are resistant to scrapie. Cell 73: 1339–1347. 10.1016/0092-8674(93)90360-3 [DOI] [PubMed] [Google Scholar]
- Cassanelli S, Bertolini S, Rolleri M, De Stefano F, Casarino L, Elicio N, Naselli A, Calandra S. 1998. A “de novo” point mutation of the low-density lipoprotein receptor gene in an Italian subject with primary hypercholesterolemia. Clin Genet 53: 391–395. 10.1111/j.1399-0004.1998.tb02752.x [DOI] [PubMed] [Google Scholar]
- Chakrabarti O, Rane NS, Hegde RS. 2011. Cytosolic aggregates perturb the degradation of nontranslocated secretory and membrane proteins. Mol Biol Cell 22: 1625–1637. 10.1091/mbc.e10-07-0638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Umanah GKE, Dephoure N, Andrabi SA, Gygi SP, Dawson TM, Dawson VL, Rutter J. 2014. Msp1/ATAD1 maintains mitochondrial function by facilitating the degradation of mislocalized tail-anchored proteins. EMBO J 33: 1548–1564. 10.15252/embj.201487943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemons WM Jr, Gowda K, Black SD, Zwieb C, Ramakrishnan V. 1999. Crystal structure of the conserved subdomain of human protein SRP54M at 2.1 Å resolution: Evidence for the mechanism of signal peptide binding. J Mol Biol 292: 697–705. 10.1006/jmbi.1999.3090 [DOI] [PubMed] [Google Scholar]
- Costa EA, Subramanian K, Nunnari J, Weissman JS. 2018. Defining the physiological role of SRP in protein-targeting efficiency and specificity. Science 359: 689–692. 10.1126/science.aar3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello JL, Castro IG, Camões F, Schrader TA, McNeall D, Yang J, Giannopoulou EA, Gomes S, Pogenberg V, Bonekamp NA, et al. 2017. Predicting the targeting of tail-anchored proteins to subcellular compartments in mammalian cells. J Cell Sci 130: 1675–1687. 10.1242/jcs.200204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, et al. 2011. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 477: 211–215. 10.1038/nature10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi H, Mitsui K, Kurosawa M, Machida Y, Kuroiwa Y, Nukina N. 2004. Identification of ubiquitin-interacting proteins in purified polyglutamine aggregates. FEBS Lett 571: 171–176. 10.1016/j.febslet.2004.06.077 [DOI] [PubMed] [Google Scholar]
- Doyle SM, Wickner S. 2009. Hsp104 and ClpB: Protein disaggregating machines. Trends Biochem Sci 34: 40–48. 10.1016/j.tibs.2008.09.010 [DOI] [PubMed] [Google Scholar]
- Eisele F, Wolf DH. 2008. Degradation of misfolded protein in the cytoplasm is mediated by the ubiquitin ligase Ubr1. FEBS Lett 582: 4143–4146. 10.1016/j.febslet.2008.11.015 [DOI] [PubMed] [Google Scholar]
- Emerman AB, Zhang ZR, Chakrabarti O, Hegde RS. 2010. Compartment-restricted biotinylation reveals novel features of prion protein metabolism in vivo. Mol Biol Cell 21: 4325–4337. 10.1091/mbc.e10-09-0742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federico A, Cardaioli E, Da Pozzo P, Formichi P, Gallus GN, Radi E. 2012. Mitochondria, oxidative stress and neurodegeneration. J Neurol Sci 322: 254–262. 10.1016/j.jns.2012.05.030 [DOI] [PubMed] [Google Scholar]
- Foresti O, Rodriguez-Vaello V, Funaya C, Carvalho P. 2014. Quality control of inner nuclear membrane proteins by the Asi complex. Science 346: 751–755. 10.1126/science.1255638 [DOI] [PubMed] [Google Scholar]
- Fredrickson EK, Rosenbaum JC, Locke MN, Milac TI, Gardner RG. 2011. Exposed hydrophobicity is a key determinant of nuclear quality control degradation. Mol Biol Cell 22: 2384–2395. 10.1091/mbc.e11-03-0256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamerdinger M, Hanebuth MA, Frickey T, Deuerling E. 2015. The principle of antagonism ensures protein targeting specificity at the endoplasmic reticulum. Science 348: 201–207. 10.1126/science.aaa5335 [DOI] [PubMed] [Google Scholar]
- Gatto GJ, Geisbrecht BV, Gould SJ, Berg JM. 2000. Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat Struct Biol 7: 1091–1095. 10.1038/81930 [DOI] [PubMed] [Google Scholar]
- Goedert M. 2015. Alzheimer's and Parkinson's diseases: The prion concept in relation to assembled Aβ, tau, and α-synuclein. Science 349: 1255555 10.1126/science.1255555 [DOI] [PubMed] [Google Scholar]
- Guerriero CJ, Weiberth KF, Brodsky JL. 2013. Hsp70 targets a cytoplasmic quality control substrate to the San1p ubiquitin ligase. J Biol Chem 288: 18506–18520. 10.1074/jbc.M113.475905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guna A, Hegde RS. 2018. Transmembrane domain recognition during membrane protein biogenesis and quality control. Curr Biol 28: R498–R511. 10.1016/j.cub.2018.02.004 [DOI] [PubMed] [Google Scholar]
- Guna A, Volkmar N, Christianson JC, Hegde RS. 2018. The ER membrane protein complex is a transmembrane domain insertase. Science 359: 470–473. 10.1126/science.aao3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Xiong Y, Witkowski P, Cui J, Wang LJ, Sun J, Lara-Lemus R, Haataja L, Hutchison K, Shan SO, et al. 2014. Inefficient translocation of preproinsulin contributes to pancreatic β cell failure and late-onset diabetes. J Biol Chem 289: 16290–16302. 10.1074/jbc.M114.562355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainzl T, Sauer-Eriksson AE. 2015. Signal-sequence induced conformational changes in the signal recognition particle. Nat Commun 6: 7163 10.1038/ncomms8163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainzl T, Huang S, Meriläinen G, Brännström K, Sauer-Eriksson AE. 2011. Structural basis of signal-sequence recognition by the signal recognition particle. Nat Struct Mol Biol 18: 389–391. 10.1038/nsmb.1994 [DOI] [PubMed] [Google Scholar]
- Halic M, Becker T, Pool MR, Spahn CMT, Grassucci RA, Frank J, Beckmann R. 2004. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427: 808–814. 10.1038/nature02342 [DOI] [PubMed] [Google Scholar]
- Hansen KG, Aviram N, Laborenz J, Bibi C, Meyer M, Spang A, Schuldiner M, Herrmann JM. 2018. An ER surface retrieval pathway safeguards the import of mitochondrial membrane proteins in yeast. Science 361: 1118–1122. 10.1126/science.aar8174 [DOI] [PubMed] [Google Scholar]
- Heck JW, Cheung SK, Hampton RY. 2010. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc Natl Acad Sci 107: 1106–1111. 10.1073/pnas.0910591107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde RS, Keenan RJ. 2011. Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nat Rev Mol Cell Biol 12: 787–798. 10.1038/nrm3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessa T, Sharma A, Mariappan M, Eshleman HD, Gutierrez E, Hegde RS. 2011. Protein targeting and degradation are coupled for elimination of mislocalized proteins. Nature 475: 394–397. 10.1038/nature10181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjerpe R, Bett JS, Keuss MJ, Solovyova A, McWilliams TG, Johnson C, Sahu I, Varghese J, Wood N, Wightman M, et al. 2016. UBQLN2 mediates autophagy-independent protein aggregate clearance by the proteasome. Cell 166: 935–949. 10.1016/j.cell.2016.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Mohd Ali J, Jalaludin MY, Harun F. 2013. Permanent neonatal diabetes due to a novel insulin signal peptide mutation. Pediatr Diabetes 14: 299–303. 10.1111/pedi.12011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Zavodszky E, Shao S, Wohlever ML, Keenan RJ, Hegde RS. 2016. Ubiquilins chaperone and triage mitochondrial membrane proteins for degradation. Mol Cell 63: 21–33. 10.1016/j.molcel.2016.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda CY, Li J, Oubridge C, Hernández H, Robinson CV, Nagai K. 2010. Recognition of a signal peptide by the signal recognition particle. Nature 465: 507–510. 10.1038/nature08870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, et al. 2009. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323: 1693–1697. 10.1126/science.1167983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S, Hegde RS. 2018. Quality control of orphaned proteins. Mol Cell 71: 443–457. 10.1016/j.molcel.2018.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalbfleisch T, Cambon A, Wattenberg BW. 2007. A bioinformatics approach to identifying tail-anchored proteins in the human genome. Traffic 8: 1687–1694. 10.1111/j.1600-0854.2007.00661.x [DOI] [PubMed] [Google Scholar]
- Kang SW, Rane NS, Kim SJ, Garrison JL, Taunton J, Hegde RS. 2006. Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell 127: 999–1013. 10.1016/j.cell.2006.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaplis AC, Lim SK, Baba H, Arnold A, Kronenberg HM. 1995. Inefficient membrane targeting, translocation, and proteolytic processing by signal peptidase of a mutant preproparathyroid hormone protein. J Biol Chem 270: 1629–1635. 10.1074/jbc.270.4.1629 [DOI] [PubMed] [Google Scholar]
- Keenan RJ, Freymann DM, Walter P, Stroud RM. 1998. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell 94: 181–191. 10.1016/S0092-8674(00)81418-X [DOI] [PubMed] [Google Scholar]
- Keenan RJ, Freymann DM, Stroud RM, Walter P. 2001. The signal recognition particle. Annu Rev Biochem 70: 755–775. 10.1146/annurev.biochem.70.1.755 [DOI] [PubMed] [Google Scholar]
- Khosrow-Khavar F, Fang NN, Ng AHM, Winget JM, Comyn SA, Mayor T. 2012. The yeast ubr1 ubiquitin ligase participates in a prominent pathway that targets cytosolic thermosensitive mutants for degradation. G3 (Bethesda) 2: 619–628. 10.1534/g3.111.001933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le NTT, Chang L, Kovlyagina I, Georgiou P, Safren N, Braunstein KE, Kvarta MD, Van Dyke AM, LeGates TA, Philips T, et al. 2016. Motor neuron disease, TDP-43 pathology, and memory deficits in mice expressing ALS–FTD-linked UBQLN2 mutations. Proc Natl Acad Sci 113: E7580–E7589. 10.1073/pnas.1608432113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine CG, Mitra D, Sharma A, Smith CL, Hegde RS. 2005. The efficiency of protein compartmentalization into the secretory pathway. Mol Biol Cell 16: 279–291. 10.1091/mbc.e04-06-0508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Larsson L, Caballero A, Hao X, Öling D, Grantham J, Nyström T. 2010. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell 140: 257–267. 10.1016/j.cell.2009.12.031 [DOI] [PubMed] [Google Scholar]
- Ma J, Lindquist S. 2002. Conversion of PrP to a self-perpetuating PrPSc-like conformation in the cytosol. Science 298: 1785–1788. 10.1126/science.1073619 [DOI] [PubMed] [Google Scholar]
- Ma J, Wollmann R, Lindquist S. 2002. Neurotoxicity and neurodegeneration when PrP accumulates in the cytosol. Science 298: 1781–1785. 10.1126/science.1073725 [DOI] [PubMed] [Google Scholar]
- Ma C, Agrawal G, Subramani S. 2011. Peroxisome assembly: Matrix and membrane protein biogenesis. J Cell Biol 193: 7–16. 10.1083/jcb.201010022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariappan M, Li X, Stefanovic S, Sharma A, Mateja A, Keenan RJ, Hegde RS. 2010. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature 466: 1120–1124. 10.1038/nature09296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateja A, Paduch M, Chang HY, Szydlowska A, Kossiakoff AA, Hegde RS, Keenan RJ. 2015. Structure of the Get3 targeting factor in complex with its membrane protein cargo. Science 347: 1152–1155. 10.1126/science.1261671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer MJ, Spear ED, Yu AT, Lee EJ, Shahzad S, Michaelis S. 2016. Degradation signals for ubiquitin-proteasome dependent cytosolic protein quality control (CytoQC) in yeast. G3 (Bethesda) 6: 1853–1866. 10.1534/g3.116.027953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan AJ, Scott MD, Frydman J. 2005. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell 121: 739–748. 10.1016/j.cell.2005.03.024 [DOI] [PubMed] [Google Scholar]
- Medicherla B, Kostova Z, Schaefer A, Wolf DH. 2004. A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep 5: 692–697. 10.1038/sj.embor.7400164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger MB, Maurer MJ, Dancy BM, Michaelis S. 2008. Degradation of a cytosolic protein requires endoplasmic reticulum-associated degradation machinery. J Biol Chem 283: 32302–32316. 10.1074/jbc.M806424200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami R, Hayakawa A, Kagawa H, Yanagi Y, Yokosawa H, Kawahara H. 2010. BAG-6 is essential for selective elimination of defective proteasomal substrates. J Cell Biol 190: 637–650. 10.1083/jcb.200908092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock JY, Chartron JW, Zaslaver M, Xu Y, Ye Y, Clemons WM. 2015. Bag6 complex contains a minimal tail-anchor-targeting module and a mock BAG domain. Proc Natl Acad Sci 112: 106–111. 10.1073/pnas.1402745112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori F, Tanji K, Odagiri S, Toyoshima Y, Yoshida M, Ikeda T, Sasaki H, Kakita A, Takahashi H, Wakabayashi K. 2012. Ubiquilin immunoreactivity in cytoplasmic and nuclear inclusions in synucleinopathies, polyglutamine diseases and intranuclear inclusion body disease. Acta Neuropathol 124: 149–151. 10.1007/s00401-012-0999-z [DOI] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. 2012. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 337: 587–590. 10.1126/science.1223560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AT, Prado MA, Schmidt PJ, Sendamarai AK, Wilson-Grady JT, Min M, Campagna DR, Tian G, Shi Y, Dederer V, et al. 2017. UBE2O remodels the proteome during terminal erythroid differentiation. Science 357: eaan0218 10.1126/science.aan0218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nillegoda NB, Theodoraki MA, Mandal AK, Mayo KJ, Ren HY, Sultana R, Wu K, Johnson J, Cyr DM, Caplan AJ. 2010. Ubr1 and Ubr2 function in a quality control pathway for degradation of unfolded cytosolic proteins. Mol Biol Cell 21: 2102–2116. 10.1091/mbc.e10-02-0098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norstrom EM, Ciaccio MF, Rassbach B, Wollmann R, Mastrianni JA. 2007. Cytosolic prion protein toxicity is independent of cellular prion protein expression and prion propagation. J Virol 81: 2831–2837. 10.1128/JVI.02157-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okreglak V, Walter P. 2014. The conserved AAA-ATPase Msp1 confers organelle specificity to tail-anchored proteins. Proc Natl Acad Sci 111: 8019–8024. 10.1073/pnas.1405755111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares AO, Baker TA, Sauer RT. 2018. Mechanical protein unfolding and degradation. Annu Rev Physiol 80: 413–429. 10.1146/annurev-physiol-021317-121303 [DOI] [PubMed] [Google Scholar]
- Park SH, Bolender N, Eisele F, Kostova Z, Takeuchi J, Coffino P, Wolf DH. 2007. The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol Biol Cell 18: 153–165. 10.1091/mbc.e06-04-0338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Kawaguchi S, Ng DTW. 2010. A nucleus-based quality control mechanism for cytosolic proteins. Mol Biol Cell 21: 2117–2127. 10.1091/mbc.e10-02-0111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Xu C, Ng DTW. 2018. Hsp40/70/110 chaperones adapt nuclear protein quality control to serve cytosolic clients. J Cell Biol 217: 2019–2032. 10.1083/jcb.201706091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane NS, Yonkovich JL, Hegde RS. 2004. Protection cytosolic prion protein toxicity by modulation of protein translocation. EMBO J 23: 4550–4559. 10.1038/sj.emboj.7600462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane NS, Kang SW, Chakrabarti O, Feigenbaum L, Hegde RS. 2008. Reduced translocation of nascent prion protein during ER stress contributes to neurodegeneration. Dev Cell 15: 359–370. 10.1016/j.devcel.2008.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane NS, Chakrabarti O, Feigenbaum L, Hegde RS. 2010. Signal sequence insufficiency contributes to neurodegeneration caused by transmembrane prion protein. J Cell Biol 188: 515–526. 10.1083/jcb.200911115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M, Okreglak V, Chio US, Cho H, Walter P, Shan SO. 2016. Multiple selection filters ensure accurate tail-anchored membrane protein targeting. eLife 5: e21301 10.7554/eLife.21301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport TA. 2007. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450: 663–669. 10.1038/nature06384 [DOI] [PubMed] [Google Scholar]
- Rodrigo-Brenni MC, Gutierrez E, Hegde RS. 2014. Cytosolic quality control of mislocalized proteins requires RNF126 recruitment to Bag6. Mol Cell 55: 227–237. 10.1016/j.molcel.2014.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford NJ, Lewis J, Clippinger AK, Thomas MA, Adamson J, Cruz PE, Cannon A, Xu G, Golde TE, Shaw G, et al. 2013. Unbiased screen reveals ubiquilin-1 and -2 highly associated with huntingtin inclusions. Brain Res 1524: 62–73. 10.1016/j.brainres.2013.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safren N, El Ayadi A, Chang L, Terrillion CE, Gould TD, Boehning DF, Monteiro MJ. 2014. Ubiquilin-1 overexpression increases the lifespan and delays accumulation of huntingtin aggregates in the R6/2 mouse model of Huntington's disease. PLoS ONE 9: e87513 10.1371/journal.pone.0087513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scazzari M, Amm I, Wolf DH. 2015. Quality control of a cytoplasmic protein complex: Chaperone motors and the ubiquitin-proteasome system govern the fate of orphan fatty acid synthase subunit Fas2 of yeast. J Biol Chem 290: 4677–4687. 10.1074/jbc.M114.596064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, Schmitt HD, Schwappach B, Weissman JS. 2008. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134: 634–645. 10.1016/j.cell.2008.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine S, Youle RJ. 2018. PINK1 import regulation; a fine system to convey mitochondrial stress to the cytosol. BMC Biol 16: 2 10.1186/s12915-017-0470-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppen J, Steenken E, Lindhout D, Bosma PJ, Elferink RP. 1996. A mutation which disrupts the hydrophobic core of the signal peptide of bilirubin UDP-glucuronosyltransferase, an endoplasmic reticulum membrane protein, causes Crigler-Najjar type II. FEBS Lett 390: 294–298. 10.1016/0014-5793(96)00677-1 [DOI] [PubMed] [Google Scholar]
- Shao S, Hegde RS. 2011. A calmodulin-dependent translocation pathway for small secretory proteins. Cell 147: 1576–1588. 10.1016/j.cell.2011.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, Rodrigo-Brenni MC, Kivlen MH, Hegde RS. 2017. Mechanistic basis for a molecular triage reaction. Science 355: 298–302. 10.1126/science.aah6130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Mariappan M, Appathurai S, Hegde RS. 2010. In vitro dissection of protein translocation into the mammalian endoplasmic reticulum. Methods Mol Biol 619: 339–363. 10.1007/978-1-60327-412-8_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LX, Theg SM. 2013. The chloroplast protein import system: From algae to trees. Biochim Biophys Acta 1833: 314–331. 10.1016/j.bbamcr.2012.10.002 [DOI] [PubMed] [Google Scholar]
- Stefanovic-Barrett S, Dickson AS, Burr SP, Williamson JC, Lobb IT, van den Boomen DJH, Lehner PJ, Nathan JA. 2018. MARCH6 and TRC8 facilitate the quality control of cytosolic and tail-anchored proteins. EMBO Rep 19: e45603 10.15252/embr.201745603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers DW, Wolfe KJ, Ren HY, Cyr DM. 2013. The type II Hsp40 Sis1 cooperates with Hsp70 and the E3 ligase Ubr1 to promote degradation of terminally misfolded cytosolic protein. PLoS ONE 8: e52099 10.1371/journal.pone.0052099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, Kawahara H. 2016. UBQLN4 recognizes mislocalized transmembrane domain proteins and targets these to proteasomal degradation. EMBO Rep 17: 842–857. 10.15252/embr.201541402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium T. 2018. UniProt: The universal protein knowledgebase. Nucleic Acids Res 46: 2699 10.1093/nar/gky092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali DG, Sinzel M, Bulthuis EP, Kolb A, Zabel S, Mehlhorn DG, Figueiredo Costa B, Farkas Á, Clancy A, Schuldiner M, et al. 2018. The GET pathway can increase the risk of mitochondrial outer membrane proteins to be mistargeted to the ER. J Cell Sci 131: jcs211110 10.1242/jcs.211110 [DOI] [PubMed] [Google Scholar]
- von Heijne G. 1985. Signal sequences. The limits of variation. J Mol Biol 184: 99–105. 10.1016/0022-2836(85)90046-4 [DOI] [PubMed] [Google Scholar]
- von Heijne G. 1995. Protein sorting signals: Simple peptides with complex functions. EXS 73: 67–76. [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Hegde RS. 2015. Structures of the scanning and engaged states of the mammalian SRP-ribosome complex. eLife 4: e07975 10.7554/eLife.07975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chen XJ. 2015. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature 524: 481–484. 10.1038/nature14859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Monteiro MJ. 2007. Ubiquilin interacts and enhances the degradation of expanded-polyglutamine proteins. Biochem Biophys Res Commun 360: 423–427. 10.1016/j.bbrc.2007.06.097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Brown EC, Mak G, Zhuang J, Denic V. 2010. A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol Cell 40: 159–171. 10.1016/j.molcel.2010.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Amon A. 2018. MitoCPR—A surveillance pathway that protects mitochondria in response to protein import stress. Science 360: eaan4146 10.1126/science.aan4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir NR, Kamber RA, Martenson JS, Denic V. 2017. The AAA protein Msp1 mediates clearance of excess tail-anchored proteins from the peroxisomal membrane. eLife 6: e28507 10.7554/eLife.28507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley AM, Prado MA, Peng I, Abbas AR, Haley B, Paulo JA, Reichelt M, Katakam A, Sagolla M, Modrusan Z, et al. 2017. Ubiquilin1 promotes antigen-receptor mediated proliferation by eliminating mislocalized mitochondrial proteins. eLife 6: e26435 10.7554/eLife.26435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W, Schekman R. 2005. Protein translocation across biological membranes. Science 310: 1452–1456. 10.1126/science.1113752 [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Pfanner N. 2017. Mitochondrial machineries for protein import and assembly. Annu Rev Biochem 86: 685–714. 10.1146/annurev-biochem-060815-014352 [DOI] [PubMed] [Google Scholar]
- Wiedmann B, Sakai H, Davis TA, Wiedmann M. 1994. A protein complex required for signal-sequence-specific sorting and translocation. Nature 370: 434–440. 10.1038/370434a0 [DOI] [PubMed] [Google Scholar]
- Wohlever ML, Mateja A, McGilvray PT, Day KJ, Keenan RJ. 2017. Msp1 is a membrane protein dislocase for tail-anchored proteins. Mol Cell 67: 194–202.e6. 10.1016/j.molcel.2017.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G, Terada K, Yano M, Sergeev I, Mori M. 2001. Oxidative stress inhibits the mitochondrial import of preproteins and leads to their degradation. Exp Cell Res 263: 107–117. 10.1006/excr.2000.5096 [DOI] [PubMed] [Google Scholar]
- Wrobel L, Topf U, Bragoszewski P, Wiese S, Sztolsztener ME, Oeljeklaus S, Varabyova A, Lirski M, Chroscicki P, Mroczek S, et al. 2015. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature 524: 485–488. 10.1038/nature14951 [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Hayashishita M, Minami S, Suzuki K, Hagiwara T, Noguchi A, Kawahara H. 2017. Elimination of a signal sequence-uncleaved form of defective HLA protein through Bag6. Sci Rep 7: 14545 10.1038/s41598-017-14975-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagitani K, Juszkiewicz S, Hegde RS. 2017. UBE2O is a quality control factor for orphans of multiprotein complexes. Science 357: 472–475. 10.1126/science.aan0178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H. 2007. ER stress and diseases. FEBS J 274: 630–658. 10.1111/j.1742-4658.2007.05639.x [DOI] [PubMed] [Google Scholar]
- Zhang X, Rashid R, Wang K, Shan SO. 2010. Sequential checkpoints govern substrate selection during cotranslational protein targeting. Science 328: 757–760. 10.1126/science.1186743 [DOI] [PMC free article] [PubMed] [Google Scholar]