Abstract
The continuous refreshment of the proteome is critical to maintain protein homeostasis and to adapt cells to changing conditions. Thus, de novo protein biogenesis by ribosomes is vitally important to every cellular system. This process is delicate and error-prone and requires, besides cytosolic chaperones, the guidance by a specialized set of molecular chaperones that bind transiently to the translation machinery and the nascent protein to support early folding events and to regulate cotranslational protein transport. These chaperones include the bacterial trigger factor (TF), the archaeal and eukaryotic nascent polypeptide-associated complex (NAC), and the eukaryotic ribosome-associated complex (RAC). This review focuses on the structures, functions, and substrates of these ribosome-associated chaperones and highlights the most recent findings about their potential mechanisms of action.
The life of every protein, irrespective of its function or origin, starts by its messenger RNA (mRNA)-templated translation on ribosomes. Upon synthesis by ribosomes, the emerging polypeptide chains directly start their folding program into the unique three-dimensional (3D) structure to become biologically active. However, about one-third of newly made proteins are cotranslationally transported to another destination before folding, for example, to the endoplasmic reticulum (ER). The de novo folding and transport of proteins is problematic because hydrophobic residues of unfolded polypeptide chains are accessible, which enhances the probability that the newly synthesized proteins follow an unproductive off pathway leading to misfolding and aggregation or premature degradation (Deuerling and Bukau 2004; Hartl et al. 2011; Balchin et al. 2016). Moreover, a large fraction of new proteins are cotranslationally modified, for example, by amino-terminal acetylation or/and cleavage of the amino-terminal methionine. Thus, different factors act cotranslationally on nascent chains assumingly in a highly specific and coordinated manner, both temporally and spatially, to ensure the functionality of the translatome (Kramer et al. 2009; Gamerdinger 2016). To accomplish productive folding and transport and to prevent off pathways, newly synthesized polypeptides interact with ribosome-associated chaperones that prevent inappropriate inter- and intramolecular interactions and thus promote transport and/or folding into the native state. Ribosome-associated chaperones are found in every cell but differ significantly among the different kingdoms of life with regard to their structure and mechanism of action. Whereas prokaryotes have only one ribosome-associated chaperone, which is called trigger factor (TF), eukaryotes use two different TF-unrelated chaperone systems at the ribosome (Fig. 1), the conserved heterodimeric nascent polypeptide-associated complex (NAC) and a specialized Hsp70/Hsp40 dimeric chaperone system called ribosome-associated complex (RAC). RAC acts as a cofactor for an additional ribosome-attached Hsp70 partner in yeast (called Ssb) or a cytosolic Hsp70 in higher eukaryotes (Deuerling and Bukau 2004; Hartl et al. 2011; Preissler and Deuerling 2012). Recent analyses of the nascent interactome of these chaperones suggest that they act on almost every new protein synthesized with only a few exceptions (see below). A subset of newly synthesized proteins bind to other cytosolic chaperones, for example, Hsp70 or Hsp60 family members, later during synthesis or after the release from ribosomes for further assistance of their de novo folding program (for review, see Buskiewicz et al. 2004; Hartl et al. 2011; Preissler and Deuerling 2012).
Figure 1.
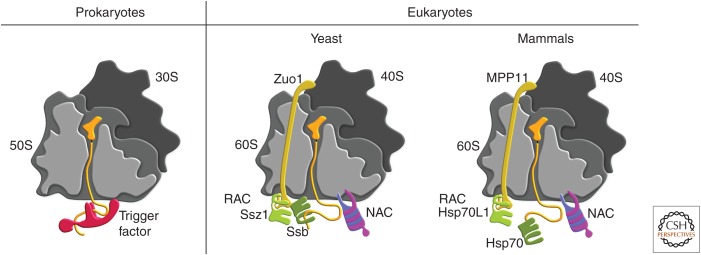
Ribosome-binding chaperones. The concept of ribosome-associated chaperones that assist de novo protein folding is conserved in prokaryotes and eukaryotes, albeit realized by different types of chaperones. The 30S or 40S ribosomal subunit is schematically drawn in dark gray and the 50S or 60S subunit in middle and light gray, indicating that the 50S/60S is sliced in half to illustrate the interior with the ribosomal tunnel. The nascent polypeptide (orange) attached to a transfer RNA (tRNA) in the P-site migrates through the tunnel and interacts with chaperones when it leaves the ribosome at the exit site. In prokaryotes (left), a single chaperone called trigger factor ([TF], red) binds transiently to the ribosome to welcome nascent polypeptides. In eukaryotes (right), two ribosome-associated chaperone systems exist: the heterodimeric nascent polypeptide-associated complex ([NAC], shown in pink and purple) and the ribosome-associated complex ([RAC], shown in yellow and light green), which consists of Ssz (light green) and Zuotin (Zuo, yellow) in yeast and the Zuo-homolog MPP11 (yellow) and its complex partner Hsp70L1 (light green) in mammals, respectively. Restricted to fungi, the Hsp70 Ssb (dark green) additionally binds to ribosomes and acts on nascent polypeptides, whereas in mammals RAC (MPP11/Hsp70L1) recruit a cytosolic Hsp70 (dark green) to bind to nascent polypeptides.
THE RIBOSOME AS PARTNER IN CRIME
The ribosome is a large ribonucleoprotein particle of ∼2.4 MDa in bacteria and ∼4 MDa in eukaryotes (Steitz 2008; Jenner et al. 2012; Klinge et al. 2012). It consists of two subunits in all organisms, a small subunit (30S in bacteria, 40S in eukaryotes) and a large subunit (50S in bacteria, 60S in eukaryotes). Four functional features are intrinsic to ribosomes (Fig. 2A): The first feature is the decoding center, which ensures selection of the correct aminoacyl-transfer RNA (tRNA) specified by the codon in the mRNA. The decoding center lies in the small subunit and recognizes the geometry of codon–anticodon base pairing and sterically discriminates against mismatches (Schmeing and Ramakrishnan 2009). The second feature represents the peptidyl-transferase center (PTC), the active site of the ribosome where peptide bond formation occurs. The PTC is located in a cleft within the subunit interface within the large ribosomal subunit. A third feature is the ribosomal tunnel inside the large subunit. With a length of 80–100 Å and a diameter of ∼10 Å at its narrowest point and ∼20 Å at its widest point it connects the PTC with the ribosome exit site (Wilson and Beckmann 2011). The tunnel wall is predominantly composed of ribosomal RNA (rRNA) and only few ribosomal proteins. The ribosomal proteins uL4 and uL22 contribute to the tunnel wall and form a 10 Å narrow constriction ∼30 Å from the PTC. The high content of rRNA gives the tunnel an overall electronegative potential. Once thought of as a merely passive conduit, the tunnel is now acknowledged to play an important role in protein biogenesis, for example, in context of arrest sequences, which trigger PTC inactivation as well as forming a secluded environment in which first folding events of the nascent polypeptide take place, mostly the formation of α-helical elements (Wilson and Beckmann 2011). The factor-binding platform centered around the tunnel exit represents the fourth functional feature of the ribosome (Fig. 2B). It comprises several ribosomal proteins and rRNA elements and constitutes the binding sites for various ribosome-associated factors that act on nascent polypeptides including chaperones, processing enzymes (such as N-acetyltransferases [NATs]) and targeting factors (e.g., signal recognition particle [SRP]).
Figure 2.
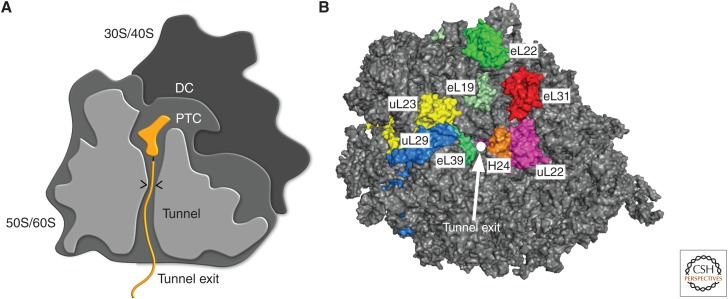
Functional features of a ribosome. (A) Schematic depiction of a ribosome. The decoding center (DC) is located in the small ribosomal subunit (30S or 40S). Peptidyl-transferase center (PTC), ribosomal tunnel (tunnel), and the tunnel exit are located in the large ribosomal subunit (50S or 60S). A peptidyl-transfer RNA (tRNA) in the PTC with nascent chain (orange) is included to illustrate the path of a nascent chain. The constriction site within the ribosomal tunnel is indicated by the two arrowheads. (B) Top view on ribosome tunnel exit. Surface rendering of the yeast 60S subunit (gray) with ribosomal proteins implicated in cofactor binding around the tunnel exit (white circle) highlighted. eL19 (pale green), eL22 (green), uL22 (magenta), uL29 (marine), eL31 (red), eL39 (lime green). Helix 24 (H24) of the 25S ribosomal RNA (rRNA) is depicted in orange.
The tunnel exit has a diameter of ∼20 Å and is composed of rRNA and four conserved ribosomal proteins, uL22, uL23, uL24, and uL29 (Fig. 2B). In addition, kingdom-specific ribosomal proteins are also present at the exit site (e.g., eL19, eL31, eL39 in archea and eukaryotes; bL17 and bL32 only in bacteria). Some of these ribosomal proteins have been shown to serve as binding sites for ribosome-associated factors. In particular, uL23 associates with multiple nascent chain-processing factors, including TF, NAC, SRP, and Sec61, and was therefore dubbed the “universal ribosome docking site” (Kramer et al. 2002; Pool et al. 2002; Wegrzyn et al. 2006). From a multitude of cross-linking and cryo-electron microscopy (cryo-EM) studies, however, a more complex picture emerged, in which additional ribosomal proteins, such as eL19, eL22, uL22, uL29, eL31, and eL39 also represent crucial contact sites for ribosome-associated factors (Pool et al. 2002; Peisker et al. 2008; Polevoda et al. 2008; Pech et al. 2010; Leidig et al. 2013; Zhang et al. 2014; Gumiero et al. 2016; Lee et al. 2016). In addition, ribosome-associated factors also contact rRNA elements (Leidig et al. 2013; Zhang et al. 2014; Gumiero et al. 2016; Lee et al. 2016). These interactions are typically based on electrostatic interactions between positively charged stretches within the associated factors and negatively charged RNA. In general, ribosome association of individual factors is highly dynamic and in cases in which factors compete for the same or close nearby binding site(s), binding of factors is often mutually exclusive. Notably, the order in which factors associate with the ribosome and bind the nascent chain is primarily determined by the chemical attributes of the emerging nascent chain itself.
It is likely that several factors can adopt different conformations on the ribosome and have more than one binding site on the ribosome depending on their functional state. For example, for the ribosome-attached Hsp70 Ssb in yeast, different conformations and contacts with the ribosome have been suggested for the ATP-bound open and ADP-bound closed conformation (Gumiero et al. 2016). Clearly, we are only at the beginning of a detailed mechanistic understanding and appreciation of the intricate dynamic processes taking place at the ribosome exit site and within the ribosomal tunnel.
STRUCTURES, FUNCTIONS, AND MECHANISMS OF ACTION OF RIBOSOME-ASSOCIATED CHAPERONES
Bacterial Trigger Factor
TF is a highly abundant chaperone found in bacteria and chloroplasts but not in the cytosol of eukaryotes. It associates via a ribosome-binding motif in its ND (Fig. 3) transiently in a 1:1 ratio with ribosomes mainly via interaction with the ribosomal protein uL23 at the tunnel exit (Kramer et al. 2002; Ferbitz et al. 2004). First evidence that TF acts as a chaperone for nascent polypeptides was provided by experiments showing that combined loss of TF and the Hsp70 chaperone DnaK provoked synergistic defects in de novo protein folding resulting in global protein aggregation and decreased viability of Escherichia coli cells (Deuerling et al. 1999; Teter et al. 1999).
Figure 3.
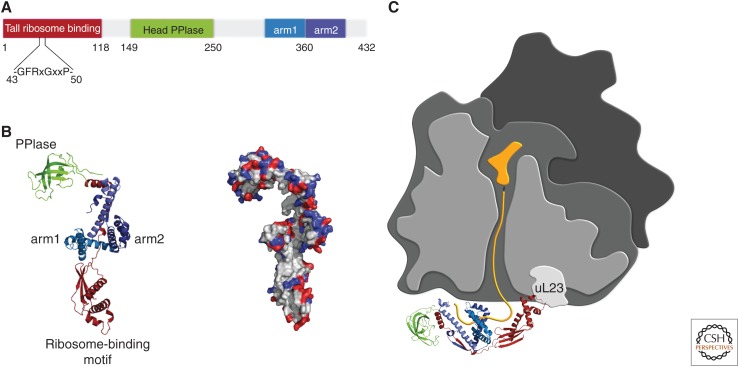
Escherichia coli trigger factor (TF). (A) Schematic representation of the linear domain organization of TF. The ribosome-binding domain (“tail”) with the ribosome-binding motif (residues 43–50) is shown in red, the PPIase “head” in green and “arm” 1 and “arm” 2 in light blue and dark blue, respectively. (B) TF adopts an extended three-dimensional fold. (Left) Ribbon diagram of the TF fold. Color code is similar to A. In positions of the ribosome-binding motif (residues 43–50), the arms and the PPIase are indicated. (Right) Surface charge distribution of TF in the same orientation as in the ribbon diagram. Positively and negatively charged residues are shown in blue and red, respectively. (C) Structural model of TF bound to the ribosome (different shades of gray as in Fig. 1). The main contact between TF (colors as in A) involves the ribosome-binding motif in the N domain and the ribosomal protein uL23 (light gray). The nascent polypeptide (yellow) migrates into the TF cradle during synthesis. All TF structures were prepared using PyMOL (DeLano Scientific LLC) based on Ferbitz et al. (2004).
TF has a unique 3D conformation. It is an extended dragon-like structure with a central body including two protruding arms, a head, and a tail region (Fig. 3). Bound to the ribosome as monomer by its amino-terminal tail, TF leans over the ribosomal exit tunnel thereby exposing its large interior surface speckled with multiple hydrophobic patches toward the exiting nascent chain (Fig. 3B,C; Ferbitz et al. 2004). Thus, TF is ideally positioned to capture emerging chains. Based on the crystal structure, TF can accommodate entire protein domains or even small proteins (up to a length of 130 aa) in its central cavity. Ribosome-bound TF can prevent premature and incorrect folding of proteins during synthesis. For example, TF retards cotranslational folding of recombinantly expressed firefly luciferase in E. coli cells, thereby enhancing the total yield of active luciferase (Agashe et al. 2004; Kaiser et al. 2006). More recent data suggest that TF can reshape and improve the folding pathway of a protein by protecting partially folded intermediates (Singhal et al. 2015; Wruck et al. 2018). Even more intriguing, TF can reverse premature folding by facilitating unfolding of preformed structures in nascent polypeptides, which allows the nascent peptide to reenter the productive folding path (Hoffmann et al. 2012; Mashaghi et al. 2013).
A more detailed understanding of the TF mechanism of action was provided by a recent study of Saio and colleagues using sophisticated nuclear magnetic resonance (NMR) techniques (Gamerdinger and Deuerling 2014; Saio et al. 2014). These investigators determined the structure and dynamics of purified TF interacting with unfolded model substrates in solution. TF forms a binding scaffold with up to four distinct substrate-binding sites distributed along TFs’ inner surface (Fig. 3B) with a variable order of binding and occupancy. The binding sites are composed of nonpolar residues that can form numerous hydrophobic pockets to bind to hydrophobic peptide stretches of 6–10 residues in substrate proteins. Additionally, polar residues proximate to the hydrophobic-binding sites can be used to form hydrogen bonds with the substrate probably to enhance affinity and navigate binding. Importantly, these binding sites show a flexible local architecture and the engagement of individual residues within the binding sites is variable depending on the substrate segment bound to it. This high degree of plasticity of TFs’ binding surfaces explains how TF can serve such a promiscuous and large pool of nascent substrates (Gamerdinger and Deuerling 2014; Saio et al. 2014). Using up all of its binding sites, TF can directly bind up to 50 amino-acid residues of a substrate. The hydrophobic peptide stretches bound by TF are separated by linker regions in between that remain unbound and may even loop outward of the central cavity. Perhaps, multiple TF molecules can bind simultaneously to a nascent substrate, which enables TF to retain also large polypeptides in an unfolded state and protect them from aggregation by shielding their exposed hydrophobic regions. This interaction mode of TF may even exert the force driven by the binding energy to unfold misfolded peptide segments of low thermodynamic stability.
More than 300 different aggregation-prone protein species were found in TF- and DnaK-deficient E. coli cells. The identified proteins are involved in many different cellular processes, range in size from 16 kDa to 140 kDa, and are specifically enriched for large (>40 kDa) multidomain proteins (Deuerling et al. 2003). Selective ribosome profiling (SeRP) provided the first global analysis of the nascent interactome of TF (Oh et al. 2011). This technique combines affinity purification of ribosome–TF complexes and subsequent identification of the mRNA segment that is being read by TF-bound ribosomes (Becker et al. 2013). This elegant approach revealed new fundamental features of the cotranslational activity of TF. TF recruitment to translating ribosomes is delayed in vivo until nascent peptides reach a length of ∼100 aa. Assuming that 30–40 aa of the nascent chains are buried in the ribosomal tunnel, TF must bind to emerging peptides only once they have exposed at least 60–70 aa outside the ribosome. This initial exclusion of TF, which only correlates with the length of the polypeptides, provides a time window to allow processing enzymes to access nascent proteins (Oh et al. 2011). These enzymes are required, for example, to remove the formyl-moiety and the initiator methionine from the amino termini of nascent polypeptide chains. The second important finding from the ribosome profiling study is that TF interacts with all newly synthesized proteins, except for those that are inserted into the cytoplasmic membrane via the SRP targeting pathway. In contrast, nascent outer membrane β-barrel proteins (Omps) were among the strongest TF interactors during their synthesis on ribosomes. This suggests that the chaperone activity of E. coli TF is particularly important for keeping Omps in a translocation-competent conformation, so that they can be efficiently exported by the Sec machinery (Oh et al. 2011). It should be mentioned that this finding is in agreement with pioneering studies by Wickner and coworkers, who in 1987 initially identified E. coli TF in a reconstituted in vitro translocation experiment, as a cytosolic component that maintains proOmpA in a transport-competent conformation for delivery into inside-out membrane vesicles (Crooke and Wickner 1987).
Eukaryotic Ribosome-Associated Systems
The Nascent Polypeptide-Associated Complex
The NAC is an evolutionarily conserved eukaryotic heterodimeric protein complex composed of a α- and β-subunit, referred to as αNAC and βNAC (Fig. 4A) (Wiedmann et al. 1994). Both subunits contain a homologous NAC domain, which dimerize by forming a β-barrel-like structure with a hydrophobic core (Fig. 4A,B) (Liu et al. 2010; Wang et al. 2010). Besides the dimerization domain, structural information is also available for the carboxyl terminus of αNAC, which forms a compact three-helix-bundle motif characteristic for ubiquitin-associated domains (UBAs) (Fig. 4B) (Spreter et al. 2005). The remaining parts of NAC including the NDs of both subunits and the carboxy-terminal domain of βNAC have not been structurally resolved yet, and many regions in these domains are predicted to be intrinsically disordered, suggesting that overall NAC shows high flexibility (Pech et al. 2010; Martin et al. 2018).
Figure 4.
The nascent polypeptide-associated complex (NAC) in eukaryotes. (A) Schematic representation of the linear domain organization of human αNAC and βNAC. Both subunits contain a homologous NAC dimerization domain. A conserved positively charged ribosome-binding motif (RRKKK) is located in the amino-terminal domain of βNAC. A conserved ubiquitin-associated domain (UBA) is located at the very carboxyl terminus of αNAC. (B) Crystal structure of the human NAC dimerization domain, which forms a compact β-barrel-like structure (blue, βNAC; violet, αNAC, PDB: 3MCB). The structure of the UBA domain (from archaeal NAC, PDB: 1TR8) consists of a typical three-helix-bundle. Gray dashed lines indicate parts of NAC that are not structurally resolved yet. (C) Surface rendering of yeast 60S (gray) and 40S (wheat) subunits with potential major contact points of NAC near the tunnel exit revealed by cross-linking experiments. βNAC cross-links to both uL23 and eL31, whereas αNAC cross-links to uL29.
NAC is abundantly expressed in equimolar concentration relative to ribosomes and reversibly binds in a 1:1 stoichiometry to ribosomes in close proximity to the ribosomal tunnel exit (Raue et al. 2007; Preissler and Deuerling 2012). Cross-linking data suggest that both subunits can interact with nascent chains (Wiedmann et al. 1994), while specifically the amino-terminal region of βNAC is critical for ribosome binding (Wegrzyn et al. 2006; Pech et al. 2010). Studies in yeast showed that deletion of the first 11 amino-terminal residues or mutation of a conserved positively charged motif in the center (RRKxxKK) abolishes ribosome binding, suggesting that the amino-terminal part of βNAC makes the main ribosomal contact (Wegrzyn et al. 2006; Pech et al. 2010). Cross-linking data suggest that βNAC contacts the ribosome via the ribosomal protein uL23 (Wegrzyn et al. 2006); however, other studies also suggest an interaction with eL31 (Pech et al. 2010; Zhang et al. 2012; Nyathi and Pool 2015). Both ribosomal proteins are located next to the nascent peptide tunnel exit but on opposite sides (Fig. 4C), indicating some flexibility of NAC on ribosomes. In addition, also the α-subunit may contribute to ribosome binding as a recent study showed a cross-link between αNAC and the ribosomal protein uL29, a neighboring protein of uL23 (Fig. 4C) (Nyathi and Pool 2015). However, apart from these cross-linking data no further structural information is available yet and the exact positioning and structure of NAC at the ribosomal tunnel exit remains obscure.
The abundance and the position at the ribosomal tunnel exit indicate a central role for NAC in guarding de novo protein synthesis. The deletion of NAC leads to embryonic lethality in worms, flies, and mice, demonstrating a fundamental role of NAC in the protein homeostasis network (Deng and Behringer 1995; Markesich et al. 2000; Bloss et al. 2003). Despite its essential role, the in vivo function of NAC remained enigmatic for a long time and only recently important functional insights were obtained in Caenorhabditis elegans showing a primary function of NAC in regulating the cotranslational protein transport to the ER (Gamerdinger et al. 2015). In the absence of NAC, translating ribosomes unspecifically associate with the Sec61 translocon in the ER membrane leading to the mislocalization of nascent substrates to the ER. To prevent incorrect ribosome–Sec61 interactions, ribosome binding of NAC was found to be essential, indicating that NAC functions as a regulatory device blocking a high-affinity Sec61-binding site on ribosomes near the tunnel exit. This hypothesis is based on the fact that ribosomes per se show a very high intrinsic affinity to the Sec61 complex in the low nanomolar range independent of whether or not a signal sequence is exposed (Borgese et al. 1974; Jungnickel and Rapoport 1995). Thus, to maintain ER targeting specificity the binding of ribosomes to Sec61 must be negatively regulated by NAC. This is in agreement with a previous in vitro study showing that purified NAC prevents unspecific binding of ribosomes to ER-derived membranes (Moller et al. 1998). The activity of NAC opposes that of the SRP, which promotes the binding of correct, signal sequence-containing, ribosome-nascent chain complexes (RNCs) to the ER translocon (Keenan et al. 2001). This antagonistic “sort and countersort” interplay between NAC and SRP is essential to enhance accuracy and efficiency of the cellular protein-sorting machinery (Gamerdinger et al. 2015). The pivotal physiological role of NAC in ER protein transport is underscored by the fact that NAC deficiency leads to strong induction of the unfolded protein response (UPR) in the ER and accompanying induction of cell apoptosis, as shown in C. elegans, zebrafish, and human cells (Hotokezaka et al. 2009; Arsenovic et al. 2012; Murayama et al. 2015). Moreover, in addition to controlling the inherent Sec61–ribosome-binding affinity, NAC may be also important to regulate the binding specificity of SRP to translating ribosomes. A previous in vitro study indicated that NAC is required to prevent SRP to bind to signal sequenceless RNCs, indicating that NAC and SRP use overlapping binding sites on ribosomes and NAC inhibits the low-affinity binding of SRP to nonsecretory RNCs (Wiedmann et al. 1994). This finding was partially reproduced in vivo in yeast, showing that NAC modulates SRP-binding specificity to some degree (del Alamo et al. 2011). However, the NAC–SRP interplay remains obscure and is controversially discussed, as other in vitro studies did not find evidence for an altered SRP-binding specificity (Neuhof et al. 1998; Raden and Gilmore 1998), and, most importantly, because overall SRP-directed ER membrane targeting of RNCs seems not to be affected in vivo in the absence of NAC (del Alamo et al. 2011; Gamerdinger et al. 2015).
In addition to its function as a negative regulator of ER protein transport, several lines of evidence also suggest a function of NAC as an ATP-independent molecular chaperone. Cross-linking data indicate a direct binding of NAC to nascent chains and NAC deletion in yeast and human cells leads to an increased ubiquitylation of nascent polypeptides suggesting that NAC directly binds to nascent substrates to protect them from premature degradation (Wiedmann et al. 1994; Duttler et al. 2013; Wang et al. 2013). Furthermore, NAC is required to promote growth of yeast cells treated with the proline analog azetidine-2-carboxylic acid (AZC), which is known to corrupt the folding of newly synthesized proteins (Duttler et al. 2013). NAC deletion also exacerbates widespread protein aggregation in yeast cells lacking the major, ATP-driven cotranslational chaperone system, the RAC–Ssb system (Koplin et al. 2010). Altogether, these data support a role for NAC as an early acting molecular chaperone assisting the cotranslational folding of nascent chains. The principle that ATP-independent ribosome-associated chaperones have a crucial function in the folding of many newly synthesized proteins is well established for TF in bacteria, as outlined above. TF protects nascent chains against premature aggregation and degradation by using several flexible binding sites to shield hydrophobic peptide stretches in substrate proteins. However, whether NAC acts similar to the holdase chaperone TF is speculative. Apart from cross-linking data suggesting that both NAC subunits interact with nascent chains, little is known about NAC's substrate-binding specificity and even the particular substrate-binding site(s) of αNAC and βNAC have not been mapped so far. A recent cross-link-mass spectrometry study indicated that NAC predominantly interacts with substrates via the flexible amino-terminal regions of αNAC and βNAC; however, whether a crucial chaperone domain is located in these domains is yet unknown (Martin et al. 2018). Studies in yeast showed that NAC can associate with practically every ribosome-attached nascent polypeptide of a cell indicating that NAC may serve a very large pool of nascent substrates (del Alamo et al. 2011). In contrast to other organisms, yeast cells express two different βNAC subunits, βNAC and β′NAC, the latter expressed in lower amounts (Ott et al. 2015). Interestingly, the cotranslational substrate pool greatly differed between the two NAC species that exist in yeast. NAC containing β′NAC preferentially associates with proteins showing high intrinsic disorder and lower hydrophobicity. Conversely, NAC containing the β-subunit binds to longer polypeptides with high hydrophobicity and lower intrinsic disorder (del Alamo et al. 2011). These differences indicate that each NAC subunit recognizes specific physicochemical properties of the nascent polypeptide. However, β′NAC is only present in yeast, suggesting that its specific function is either not required or adopted by canonical NAC or other proteins in higher eukaryotes.
Further evidence also suggests a function for NAC in protein transport to mitochondria. Knockdown of NAC induces the mitochondrial UPR in C. elegans and causes mitochondrial dysfunction in human cells (Hotokezaka et al. 2009; Gamerdinger et al. 2015). Moreover, in yeast, NAC was found to enhance the efficiency of protein import to mitochondria (George et al. 1998; Fünfschilling and Rospert 1999). In this respect, NAC promotes the interaction of ribosomes with the mitochondrial surface, suggesting that NAC stimulates import of mitochondrial proteins in a cotranslational manner (George et al. 2002). A recent study in yeast showed that cotranslational protein transport to mitochondria is more widespread than previously thought (Williams et al. 2014). The targeting process of RNCs exposing a mitochondrial targeting sequence (MTS) to the translocase (TOM complex) of the organelle is not yet established. However, NAC could play a major role therein as it binds to the mitochondrial outer membrane protein OM14 in yeast (Lesnik et al. 2014). Through this interaction, translating ribosomes get localized to mitochondria and both NAC and OM14 are required to enhance protein import efficiency. Consistent with a mitochondrial targeting function, β′NAC was found to associate preferentially with ribosomes translating mitochondrial precursors in yeast (del Alamo et al. 2011). Moreover, mitochondrial proteins in particular get mistargeted and mislocalized to the ER on NAC depletion in C. elegans (Gamerdinger et al. 2015). In sum, these findings support a function of NAC in cotranslational protein transport to mitochondria. However, the question arises how NAC, which broadly binds to RNCs, can exert discriminative targeting activity toward RNCs, exposing an MTS. Moreover, OM14 is not conserved and a potential receptor for NAC on mitochondria in higher organisms has not been identified yet.
The Ribosome-Associated Complex
Eukaryotes feature a second conserved ribosome-associated chaperone system that is involved in initial protein folding, targeting, and prevention of aggregation of newly synthesized proteins, the RAC. RAC acts together with a ribosome-bound Hsp70 (heat-shock 70 kDa protein) in yeast (Ssb) or recruits cytosolic Hsp70 to nascent polypeptides in mammals (Fig. 1B,C) (Preissler and Deuerling 2012; Zhang et al. 2017). RAC is a stable heterodimeric complex of an Hsp40 protein (Zuo1 in yeast, ZRF1/MPP11 in humans) and a degenerated ATPase-inactive Hsp70 protein (Ssz1 in yeast, Hsp70L1 in humans) (Fig. 5A,B) (Gautschi et al. 2001, 2002; Huang et al. 2005; Conz et al. 2007). Most of our knowledge of the function and structure of the RAC–Hsp70 system is derived from studies in the baker's yeast Saccharomyces cerevisiae. RAC stimulates the ATPase activity of Ssb in yeast and thereby enhances the affinity of this Hsp70 for unfolded polypeptides ultimately assisting de novo protein folding (Preissler and Deuerling 2012; Zhang et al. 2017). RAC itself seems not to directly contact the nascent chain (Gautschi et al. 2002; Conz et al. 2007), but nonetheless it plays an important role in coordinating the substrate-binding specificity of Ssb in yeast (Koplin et al. 2010; Willmund et al. 2013; Döring et al. 2017). A functional cooperation between components of the yeast Hsp40/Hsp70–chaperone triad was first revealed by genetic analyses. Cells lacking either Ssb or RAC or both display a similar phenotype, which includes sensitivity to high salt concentrations, low temperature, and translation inhibitory drugs such as aminoglycosides (Nelson et al. 1992; Yan et al. 1998; Gautschi et al. 2002; Hundley et al. 2002). The first hint to a role of the RAC–Ssb system in folding of nascent chains at the ribosome came from cross-linking experiments establishing a RAC-dependent interaction of Ssb with short nascent chains (Pfund et al. 1998; Gautschi et al. 2002; Hundley et al. 2002). An involvement of RAC–Ssb in de novo protein folding was further corroborated by the fact that the aminoglycoside sensitivity of racΔ ssbΔ yeast cells can be partially suppressed by expression of the prokaryotic ribosome-associated chaperone TF (Rauch et al. 2005). In addition, cells devoid of Ssb showcase an increased aggregation of newly synthesized proteins, as well as of ribosomal proteins and ribosome biogenesis factors (Koplin et al. 2010; Willmund et al. 2013). These findings indicate that a loss of Ssb not only negatively impacts on folding of cytosolic proteins but also on ribosome biogenesis. Interestingly, the RAC–Hsp70 system is also important for maintaining translational fidelity (Rakwalska and Rospert 2004; Muldoon-Jacobs and Dinman 2006; Lee et al. 2016; Zhang et al. 2017).
Figure 5.
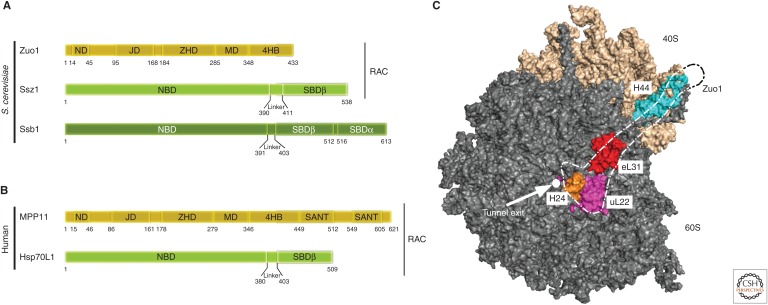
The ribosome-associated Hsp70/40 system in eukaryotes. (A) The yeast ribosome-associated chaperone system consists of the Hsp40 Zuo1, and the Hsp70s Ssb and Ssz1. Zuo1 and Ssz1 form the stable heterodimeric ribosome-associated complex (RAC). Zuo1 binds to the Ssz1 nucleotide-binding (NBD) and substrate-binding domain β (SBDβ) via its amino-terminal domain (ND). The J domain (JD) of Zuo1 is required for stimulation of the ATPase activity of its Hsp70 partner Ssb. The Zou1 homology domain (ZHD), the highly charged middle domain (MD), and the four-helix bundle (4HB) are involved in ribosome binding. Ssz1 and Ssb contain an amino-terminal NBD and a carboxy-terminal SBD. Ssz1 contains an incomplete SBD, which consists of only the SBDβ moiety, whereas the SBD of Ssb is complete and consists of SBDα and SBDβ. Autonomous ribosome binding of Ssb is mostly mediated via a positively charged stretch at the carboxyl terminus. RAC is conserved in mammals while Ssb homologs are absent (and a soluble Hsp70 is used instead). (B) The human RAC complex is formed by the Hsp40 MPP11 and Hsp70L1. MPP11 contains two SANT domains at its carboxyl terminus. (C) Surface rendering of yeast 60S (gray) and 40S (wheat) subunits with major contact points with Zuo1 highlighted. The dotted empty outline coarsely traces the shape of Zuo1. Exit tunnel (white circle), uL22 (magenta), uL31 (red), helix 24 (H24) of the 25S ribosomal RNA (rRNA) in 60S subunit (orange), and helix 44 (H44) of expansion segment 12 of 18S rRNA in 40S subunit (cyan). S. cerevisiae, Saccharomyces cerevisiae.
Recent structural work significantly expanded our understanding of the fungal RAC–Ssb system on the ribosome (Leidig et al. 2013; Zhang et al. 2014, 2017; Lee et al. 2016). Ribosome binding of the RAC complex is mediated solely by the Zuo1 subunit, which contacts the ribosomal protein eL31 close to the tunnel exit via a conserved charged region (Yan et al. 1998; Peisker et al. 2008). Moreover, recent cryo-EM studies unveiled a much more intricate interaction between Zuo1 and the ribosome. Zuo1 not only contacts the 60S subunit but also the 40S subunit and spans ∼190 Å across the subunits (Fig. 5A,C) (Zhang et al. 2014; Lee et al. 2016). While the central Zuo1 homology domain (ZHD) binds the 60S subunit (Leidig et al. 2013; Zhang et al. 2014; Kaschner et al. 2015; Lee et al. 2016), a four-helix bundle (4HB) at the carboxyl terminus of Zuo1 binds to the 40S subunit, and the middle domain (MD) forms a hinge between the two domains (Zhang et al. 2014; Lee et al. 2016).
The unique binding mode of RAC to translating ribosomes has several mechanistic implications. A communication between the ribosomal tunnel and chaperone activity at the tunnel exit has been anticipated given that Zuo1 contacts helix 24 (H24) of the 25S rRNA in the 60S subunit, and H24 in turn is in contact with ribosomal protein uL22, which extends into the ribosomal tunnel and makes contact with nascent chain segments at the constriction site. Conceivably, the nature of the nascent chain in the tunnel is sensed at the constriction site by uL22 and the information might be relayed via H24 and the ZHD to the Zuo1 J domain, which may influence J domain positioning with respect to Ssb and hence ultimately regulate Ssb activity at the tunnel exit (Lee et al. 2016; Zhang et al. 2017). Moreover, Zuo1 interacts via a positively charged surface with helix 44 (H44) of extension segment 12 (ES12) of 18S rRNA in the 40S subunit (Zhang et al. 2014; Lee et al. 2016). Because the extended H44 forms the core of the decoding center (DC) and mutations within H44 affect translational fidelity (Lee et al. 2016), binding of Zuo1 to the tip of H44 might explain how RAC influences translational fidelity (Lee et al. 2016; Zhang et al. 2017).
An unusual attribute of RAC is the stable interaction of the Hsp40 Zuo1 and the degenerated Hsp70 Ssz1, because Hsp40–Hsp70 interactions are normally transient (Mayer and Kityk 2015). First biochemical insights into the stable interaction of the two proteins came from hydrogen–deuterium exchange experiments, which showed that the Zuo1 amino-terminal domain (ND) binds stably to Ssz1, making direct contacts with both the substrate-binding domain (SBD) and the nucleotide-binding domain (NBD) of Ssz1 (Fiaux et al. 2010). The recent crystal structure of full-length Ssz1 in complex with the Zuo1 ND (both from the thermophilic ascomycete Chaetomium thermophilum) fully confirmed these findings and further revealed unique features of the interaction of the two proteins (Weyer et al. 2017; Zhang et al. 2017). In contrast to canonical Hsp70s, the linker between NBD and SBD is longer in Ssz1, and is detached from ATP-bound NBD allowing Zuo1 ND to bind this site in trans instead. In addition, parts of the Zuo1 ND complement the β-sandwich of Ssz1 SBDβ thereby mimicking SBDβ of canonical ADP-bound Hsp70s (despite having ATP bound). Overall, the conformation of Ssz1 in the RAC heterodimer looks like a hybrid between ADP- and ATP-bound Hsp70 (Weyer et al. 2017).
Ssz1 represents in several aspects an atypical Hsp70: First, it binds ATP but is not able to hydrolyze it (at least at a detectable rate), and ATP binding is not strictly required for its function (Huang et al. 2005; Conz et al. 2007). Second, it lacks the lid domain of the SBD and no binding to a substrate could be detected (Hundley et al. 2002; Huang et al. 2005). An Ssz1 truncation variant lacking the entire SBD fully complements growth defects of an ssz1Δ strain (Hundley et al. 2002; Conz et al. 2007). Zuo1 can only efficiently stimulate ATP hydrolysis by Ssb in the presence of Ssz1 (Huang et al. 2005). Consequently, it was proposed that Ssz1's predominant function is to facilitate Zuo1's ability to function as a J protein partner of Ssb on the ribosome (Huang et al. 2005). It was also speculated that Ssz1 might fulfill regulatory functions by introducing structural rearrangements within in the Zuo1 J domain, which might strengthen the contact to Ssb (Fiaux et al. 2010), and that Ssz1 might play a role in recruitment of substrates by Ssb (Leidig et al. 2013). Regardless, despite the immense progress on the structural site, the function of the unique RAC SBDβ conformation, as well as the mechanistic role of Ssz1 remain hitherto largely enigmatic.
In yeast, two Ssb homologs exist, Ssb1 and Ssb2, which differ only in four amino acids (here, collectively referred to as Ssb). The structure of Ssb is similar to canonical Hsp70 proteins, except that it contains in addition a nuclear export sequence (NES) at its carboxyl terminus, which likely facilitates shuttling between the nucleus and cytosol (Shulga et al. 1999). As for canonical Hsp70s, the Ssb reaction cycle is driven by cochaperones (Preissler and Deuerling 2012; Mayer and Kityk 2015). ATP hydrolysis, which results in tight substrate binding, is stimulated by RAC (Gautschi et al. 2002; Hundley et al. 2002), and nucleotide exchange factors for Ssb are Sse1, Sln1, and Fes1 (Peisker et al. 2010). The highly abundant Ssb interacts independent of RAC with ribosomes, and in wild-type cells ∼50% of Ssb is found associated with ribosomes (Nelson et al. 1992; Yan et al. 1998; Peisker et al. 2010). A positively charged region close to the end of the carboxy-terminal SBDα is essential for ribosome binding (Gumiero et al. 2016; Hanebuth et al. 2016). Likely, this region mediates ribosome association of Ssb via electrostatic interactions with expansion segment ES24 and/or ES41 within the 25S rRNA (Gumiero et al. 2016), consistent with a salt-sensitive binding of Ssb to vacant ribosomes (Pfund et al. 1998). In contrast, Ssb binding to ribosomes exposing a nascent chain (RNCs) is resistant to high salt concentrations, presumably because of hydrophobic Ssb–nascent chain interactions (Powers and Walter 1996; Pfund et al. 1998; Raue et al. 2007). A second positively charged region within the Ssb SBDβ in addition contributes to ribosome binding (Hanebuth et al. 2016). Cross-linking experiments revealed that Ssb contacts ribosomal proteins uL29, eL39, and eL19 in close proximity of the tunnel exit (Gumiero et al. 2016). Together with the crystal structure of full-length C. thermophilum Ssb (in the ATP-bound state), a picture emerges in which an exact positioning of Ssb on the ribosome close to the exit tunnel is critical for its function (Gumiero et al. 2016; Hanebuth et al. 2016). The positioning of Ssb on the ribosome is modulated by RAC (as well as the nucleotide-status of Ssb itself) indicating dynamic changes of Ssb on the ribosome on ATP hydrolysis (Gumiero et al. 2016). Intriguingly, in the presence of RAC, autonomous ribosome binding of Ssb is not essential for protein folding, suggesting a RAC-mediated interaction of Ssb with RNCs (Gumiero et al. 2016; Hanebuth et al. 2016). This is mostly consistent with the situation in organisms other than fungi, in which no dedicated autonomously ribosome-anchored Hsp70 exists and RAC collaborates with a cytosolic Hsp70 instead (Fig. 1; Nelson et al. 1992; Gautschi et al. 2002).
Two recent studies investigated the cotranslational substrates of Ssb on a global scale (Willmund et al. 2013; Döring et al. 2017). In the first study by Willmund and colleagues, ribosome–Ssb complexes were isolated and the ribosome-associated mRNA analyzed using genome-wide microarrays. This study revealed that the nascent chain substrate pool of Ssb is very broad, encompassing ∼80% of cytosolic and nuclear proteins. General features of the Ssb-bound proteins were the presence of larger domains and a generally large size. Ssb substrates were also enriched for subunits of oligomeric complexes (e.g., TRiC/CCT, proteasome, and ribosomal particle), which in most cases do not represent large proteins, but usually interact with several other subunits within the complex. Further features common to most Ssb substrates were a general low translation rate, and an enrichment of β-sheets, and short linear hydrophobic elements. Together, these data suggest that Ssb assists cotranslational folding of larger proteins harboring complicated structures or that require binding partners for their stability (Willmund et al. 2013).
In the second analysis, SeRP (Becker et al. 2013) was used to analyze Ssb interaction with nascent chains on a proteome-wide scale at near-codon resolution (Döring et al. 2017). Consistent with Willmund et al. (2013), Ssb bound to ∼80% of cytosolic and nuclear proteins in the SeRP study. Furthermore, and in dissent with Willmund et al. (2013), ∼80% of nascent mitochondrial proteins and more than 40% of ER-targeted proteins were found associated with Ssb, suggesting a function of Ssb in targeting/translocation of these proteins (Döring et al. 2017). Because Ssb was found to bind nascent mitochondrial proteins preferentially at lengths of 100 residues, it has been speculated that Ssb might increase the targeting efficiency of these proteins by preventing premature folding and misfolding (Döring et al. 2017). Such a scenario is supported by the fact that a subset of nuclear-encoded mitochondrial proteins aggregated in the absence of Ssb (Willmund et al. 2013). The SeRP study further showed that Ssb binds positively charged, degenerate sequences close to the ribosomal surface, when the substrate recognition motif extends ∼5 aa from the tunnel exit, whereby disordered regions seem to be disfavored. Ssb nascent chain binding close to the tunnel exit is in good agreement with a wealth of previous biochemical and structural data (Pfund et al. 1998; Gautschi et al. 2002; Hundley et al. 2002; Gumiero et al. 2016). Notably, Ssb engages most substrates by multiple binding-release cycles (Döring et al. 2017). As Ssb binds preferentially segments that will be surface exposed or form the hydrophobic core of the folded protein, Ssb likely assists domain-wise folding by retarding premature/unproductive folding (Döring et al. 2017).
Challenging the common view that SRP is the first and only cytosolic interactor of nascent chains of the cotranslational translocation pathway before docking to the translocon, at least for a subset of SRP-targeted nascent chains, Ssb engagement before SRP binding has been observed (Döring et al. 2017). Whether this implies a handover of nascent chains from Ssb–RAC to SRP and/or the existence of an alternative targeting route to the ER membrane is currently unknown (Döring et al. 2017).
An influence of RAC on the substrate specificity of Ssb was observed in both studies (Willmund et al. 2013; Döring et al. 2017). The absence of RAC severely impaired Ssb binding to emerging recognition motifs and delayed RNC engagement of Ssb (Döring et al. 2017). In addition, deletion of RAC was found to relax the specificity of Ssb (Willmund et al. 2013). Intriguingly, Ssb binding to RNCs coincides with an increased translation speed, which can be attributed to both mRNA features as well as nascent chain features (Döring et al. 2017). It has been speculated that faster translation could reduce the number of ribosomes required to maintain protein synthesis and may therefore represent a strategy to economize protein synthesis (Döring et al. 2017). Either way, this finding highlights the close coordination of protein synthesis and cotranslational protein folding.
Whereas Ssb is unique to fungi, homologs of RAC are present in mammals as well (Hundley et al. 2005; Otto et al. 2005). This indicates that the presence of Hsp70/40 chaperones on ribosomes is common in the eukaryotic world. Zuo1 homologs in mammals (MPP11) contain two additional SANT domains at their carboxyl terminus (Hundley et al. 2005; Otto et al. 2005; Chen et al. 2014). SANT domains are normally involved in DNA binding and chromatin remodeling (Boyer et al. 2004), their exact role in MPP11 however is currently unknown. Knockdown of human MPP11 in HeLa cells results in growth defects and sensitivity toward drugs similar to what was observed for yeast cells lacking RAC (Jaiswal et al. 2011), suggesting that RAC fulfills similar functions in yeast as well as in metazoans. However, because higher eukaryotes lack a dedicated ribosome-anchored Hsp70-like Ssb, it was suggested that cytosolic Hsp70s act as functional partners for RAC in higher eukaryotes (Jaiswal et al. 2011). Interestingly, although Zuo homologs are found in all eukaryotic species in which the whole genome is sequenced, not all of them also reveal an Ssz-like Hsp70 (e.g., there is no Ssz homolog found in plants), suggesting that the RAC system may have a more versatile setting than the yeast and mammalian versions characterized so far.
INTERPLAY WITH OTHER RIBOSOME-ASSOCIATED FACTORS: SRP, PDF, MAP, AND NAT
The ribosome-associated chaperone systems presented in this review are integral and essential parts of a greater cotranslational proteostasis machinery that controls the quality and localization of newly synthesized proteins. Numerous other cytosolic protein biogenesis factors, including peptide deformylase (PDF), methionine aminopeptidases (MAPs), NATs, and the SRP also bind in close proximity to the ribosome exit site, and many of these factors use partially overlapping binding surfaces on ribosomes. An emerging question is how these factors gain regulated access to nascent polypeptides and whether they compete and/or collaborate at the ribosome exit site (Gamerdinger and Deuerling 2014; Gloge et al. 2014).
The only tunnel exit-binding factors that are at least as abundant as ribosomes are TF in bacteria and NAC in eukaryotes (Merz et al. 2006; Raue et al. 2007). These major cotranslational systems are supposed to play an important role in orchestrating nascent chain-processing events at the ribosome exit site. A molecular interplay between TF and SRP is well documented in vitro. TF generally prevents SRP binding to ribosomes, except to those RNCs presenting signal sequences (Bornemann et al. 2014). This RNC prefilter function by TF could explain why a small amount of SRP (ratio of SRP to ribosome about 1:100) is sufficient for effective targeting of membrane proteins (Jensen and Pedersen 1994). Moreover, TF also enhances the specificity of SRP-dependent protein targeting. Both factors can bind simultaneously to signal sequence-containing RNCs, and TF regulates SRP function at multiple targeting steps, including initial binding, targeting of RNC to the membrane via SRP–SRP receptor (FtsY) assembly, and removal of SRP from RNCs exposing nascent chains exceeding a critical length that compromises cotranslational translocation (Buskiewicz et al. 2004; Bornemann et al. 2014; Ariosa et al. 2015). Together, these activities enhance the fidelity of substrate selection by SRP and promote the efficiency of membrane protein transport in the cell. In contrast to SRP, TF seems not to modulate directly PDF and MAP binding to translating ribosomes, indicating that these enzymes can process their nascent substrates before or in parallel with TF binding (Bornemann et al. 2014).
The regulatory function of TF in cotranslational protein transport partially resembles that of NAC in eukaryotes. As outlined above, NAC is critical for accurate substrate selection by the Sec61 translocon, but also modulates the binding specificity of SRP (del Alamo et al. 2011; Gamerdinger et al. 2015). The presence of a not-yet-exposed signal anchor sequence inside the ribosomal tunnel increases the affinity of SRP for ribosomes, and the early binding of SRP to those RNCs depends on NAC (Berndt et al. 2009; Zhang et al. 2012). This early SRP recruitment is likely mediated by subtle structural alterations of the ribosome and NAC at the exit site such that SRP binding is favored. Moreover, in the absence of NAC, SRP partially binds to nonsecretory RNCs thereby perturbing the accurate substrate selection of MAP1 by blocking its ribosomal access (del Alamo et al. 2011; Nyathi and Pool 2015). Thus, although NAC and MAP1 can bind simultaneously to ribosomes and do not directly modulate each other's binding properties (Nyathi and Pool 2015), uncontrolled binding of a third factor can cause unforeseen problems in protein biogenesis. This example shows the delicateness of the molecular interplay at the tunnel exit and highlights the dynamic binding and release of ribosome-associated protein biogenesis factors through a concerted action. However, we are only beginning to understand the complex interdependency of these factors and how this affects the specific and timely targeting of nascent chains into the correct protein biogenesis pathway.
CONCLUDING REMARKS
The myriad of recently accumulated data on the in vivo nascent substrates, structures, and mechanisms of ribosome-associated chaperones from prokaryotic and eukaryotic kingdoms suggests that they are versatile and vital elements of the chaperone network crucial to control the quality and transport of newly synthesized proteins. In addition, not discussed here at all, there is culminating evidence that these chaperones may display additional functions off the ribosome, for example, TF and RAC–Ssb have been suggested to promote the assembly of ribosomal particles and NAC was found to enhance the refolding of aggregated luciferase in C. elegans (Martinez-Hackert and Hendrickson 2009; Koplin et al. 2010; Kirstein-Miles et al. 2013). The future challenge will be to dissect their individual roles during protein biogenesis and gain a deeper structural and mechanistic understanding of their ribosomal and nonribosomal activities.
ACKNOWLEDGMENTS
We apologize that we could not discuss all aspects of de novo folding and chaperone functions in depth. Moreover, we apologize to all our colleagues whose research was not or only very briefly discussed or not cited. We thank Dr. Christina Schlatter for assistance in figure preparation and Sandra Fries for proofreading the manuscript. M.G., S.G.K., and E.D. are supported by the Deutsche Forschungsgemeinschaft (DFG)-funded collaborative research center SFB969 on the “Chemical and Biological Principles of Cellular Proteostasis.”
Footnotes
Editors: Richard I. Morimoto, F. Ulrich Hartl, and Jeffery W. Kelly
Additional Perspectives on Protein Homeostasis available at www.cshperspectives.org
REFERENCES
- Agashe VR, Guha S, Chang HC, Genevaux P, Hayer-Hartl M, Stemp M, Georgopoulos C, Hartl FU, Barral JM. 2004. Function of trigger factor and DnaK in multidomain protein folding: Increase in yield at the expense of folding speed. Cell 117: 199–209. 10.1016/S0092-8674(04)00299-5 [DOI] [PubMed] [Google Scholar]
- Ariosa A, Lee JH, Wang S, Saraogi I, Shan SO. 2015. Regulation by a chaperone improves substrate selectivity during cotranslational protein targeting. Proc Natl Acad Sci 112: E3169–E3178. 10.1073/pnas.1422594112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsenovic PT, Maldonado AT, Colleluori VD, Bloss TA. 2012. Depletion of the C. elegans NAC engages the unfolded protein response, resulting in increased chaperone expression and apoptosis. PLoS ONE 7: e44038 10.1371/journal.pone.0044038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balchin D, Hayer-Hartl M, Hartl FU. 2016. In vivo aspects of protein folding and quality control. Science 353: aac4354 10.1126/science.aac4354 [DOI] [PubMed] [Google Scholar]
- Becker AH, Oh E, Weissman JS, Kramer G, Bukau B. 2013. Selective ribosome profiling as a tool for studying the interaction of chaperones and targeting factors with nascent polypeptide chains and ribosomes. Nat Protoc 8: 2212–2239. 10.1038/nprot.2013.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt U, Oellerer S, Zhang Y, Johnson AE, Rospert S. 2009. A signal-anchor sequence stimulates signal recognition particle binding to ribosomes from inside the exit tunnel. Proc Natl Acad Sci 106: 1398–1403. 10.1073/pnas.0808584106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloss TA, Witze ES, Rothman JH. 2003. Suppression of CED-3-independent apoptosis by mitochondrial βNAC in Caenorhabditis elegans. Nature 424: 1066–1071. 10.1038/nature01920 [DOI] [PubMed] [Google Scholar]
- Borgese N, Mok W, Kreibich G, Sabatini DD. 1974. Ribosomal–membrane interaction: In vitro binding of ribosomes to microsomal membranes. J Mol Biol 88: 559–580. 10.1016/0022-2836(74)90408-2 [DOI] [PubMed] [Google Scholar]
- Bornemann T, Holtkamp W, Wintermeyer W. 2014. Interplay between trigger factor and other protein biogenesis factors on the ribosome. Nat Commun 5: 4180 10.1038/ncomms5180 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Latek RR, Peterson CL. 2004. The SANT domain: A unique histone-tail-binding module? Nat Rev Mol Cell Biol 5: 158–163. 10.1038/nrm1314 [DOI] [PubMed] [Google Scholar]
- Buskiewicz I, Deuerling E, Gu SQ, Jockel J, Rodnina MV, Bukau B, Wintermeyer W. 2004. Trigger factor binds to ribosome-signal-recognition particle (SRP) complexes and is excluded by binding of the SRP receptor. Proc Natl Acad Sci 101: 7902–7906. 10.1073/pnas.0402231101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DH, Huang Y, Liu C, Ruan Y, Shen WH. 2014. Functional conservation and divergence of J-domain-containing ZUO1/ZRF orthologs throughout evolution. Planta 239: 1159–1173. 10.1007/s00425-014-2058-6 [DOI] [PubMed] [Google Scholar]
- Conz C, Otto H, Peisker K, Gautschi M, Wolfle T, Mayer MP, Rospert S. 2007. Functional characterization of the atypical Hsp70 subunit of yeast ribosome-associated complex. J Biol Chem 282: 33977–33984. 10.1074/jbc.M706737200 [DOI] [PubMed] [Google Scholar]
- Crooke E, Wickner W. 1987. Trigger factor: a soluble protein that folds pro-OmpA into a membrane-assembly-competent form. Proc Natl Acad Sci 84: 5216–5220. 10.1073/pnas.84.15.5216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Alamo M, Hogan DJ, Pechmann S, Albanese V, Brown PO, Frydman J. 2011. Defining the specificity of cotranslationally acting chaperones by systematic analysis of mRNAs associated with ribosome-nascent chain complexes. PLoS Biol 9: e1001100 10.1371/journal.pbio.1001100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng JM, Behringer RR. 1995. An insertional mutation in the BTF3 transcription factor gene leads to an early postimplantation lethality in mice. Transgenic Res 4: 264–269. 10.1007/BF01969120 [DOI] [PubMed] [Google Scholar]
- Deuerling E, Bukau B. 2004. Chaperone-assisted folding of newly synthesized proteins in the cytosol. Crit Rev Biochem Mol Biol 39: 261–277. 10.1080/10409230490892496 [DOI] [PubMed] [Google Scholar]
- Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B. 1999. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400: 693–696. 10.1038/23301 [DOI] [PubMed] [Google Scholar]
- Deuerling E, Patzelt H, Vorderwülbecke S, Rauch T, Kramer G, Schaffitzel E, Mogk A, Schulze-Specking A, Langen H, Bukau B. 2003. Trigger factor and DnaK possess overlapping substrate pools and binding specificities. Mol Microbiol 47: 1317–1328. 10.1046/j.1365-2958.2003.03370.x [DOI] [PubMed] [Google Scholar]
- Döring K, Ahmed N, Riemer T, Suresh HG, Vainshtein Y, Habich M, Riemer J, Mayer MP, O'Brien EP, Kramer G, et al. 2017. Profiling Ssb-nascent chain interactions reveals principles of Hsp70-assisted folding. Cell 170: 298–311.e20. 10.1016/j.cell.2017.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duttler S, Pechmann S, Frydman J. 2013. Principles of cotranslational ubiquitination and quality control at the ribosome. Mol Cell 50: 379–393. 10.1016/j.molcel.2013.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferbitz L, Maier T, Patzelt H, Bukau B, Deuerling E, Ban N. 2004. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature 431: 590–596. 10.1038/nature02899 [DOI] [PubMed] [Google Scholar]
- Fiaux J, Horst J, Scior A, Preissler S, Koplin A, Bukau B, Deuerling E. 2010. Structural analysis of the ribosome-associated complex (RAC) reveals an unusual Hsp70/Hsp40 interaction. J Biol Chem 285: 3227–3234. 10.1074/jbc.M109.075804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fünfschilling U, Rospert S. 1999. Nascent polypeptide-associated complex stimulates protein import into yeast mitochondria. Mol Biol Cell 10: 3289–3299. 10.1091/mbc.10.10.3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamerdinger M. 2016. Protein quality control at the ribosome: focus on RAC, NAC and RQC. Essays Biochem 60: 203–212. 10.1042/EBC20160011 [DOI] [PubMed] [Google Scholar]
- Gamerdinger M, Deuerling E. 2014. Biochemistry. Trigger factor flexibility. Science 344: 590–591. 10.1126/science.1254064 [DOI] [PubMed] [Google Scholar]
- Gamerdinger M, Hanebuth MA, Frickey T, Deuerling E. 2015. The principle of antagonism ensures protein targeting specificity at the endoplasmic reticulum. Science 348: 201–207. 10.1126/science.aaa5335 [DOI] [PubMed] [Google Scholar]
- Gautschi M, Lilie H, Funfschilling U, Mun A, Ross S, Lithgow T, Rucknagel P, Rospert S. 2001. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc Natl Acad Sci 98: 3762–3767. 10.1073/pnas.071057198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi M, Mun A, Ross S, Rospert S. 2002. A functional chaperone triad on the yeast ribosome. Proc Natl Acad Sci 99: 4209–4214. 10.1073/pnas.062048599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George R, Beddoe T, Landl K, Lithgow T. 1998. The yeast nascent polypeptide-associated complex initiates protein targeting to mitochondria in vivo. Proc Natl Acad Sci 95: 2296–2301. 10.1073/pnas.95.5.2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George R, Walsh P, Beddoe T, Lithgow T. 2002. The nascent polypeptide-associated complex (NAC) promotes interaction of ribosomes with the mitochondrial surface in vivo. FEBS Lett 516: 213–216. 10.1016/S0014-5793(02)02528-0 [DOI] [PubMed] [Google Scholar]
- Gloge F, Becker AH, Kramer G, Bukau B. 2014. Co-translational mechanisms of protein maturation. Curr Opin Struct Biol 24: 24–33. 10.1016/j.sbi.2013.11.004 [DOI] [PubMed] [Google Scholar]
- Gumiero A, Conz C, Gese GV, Zhang Y, Weyer FA, Lapouge K, Kappes J, von Plehwe U, Schermann G, Fitzke E, et al. 2016. Interaction of the cotranslational Hsp70 Ssb with ribosomal proteins and rRNA depends on its lid domain. Nat Commun 7: 13563 10.1038/ncomms13563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanebuth MA, Kityk R, Fries SJ, Jain A, Kriel A, Albanese V, Frickey T, Peter C, Mayer MP, Frydman J, et al. 2016. Multivalent contacts of the Hsp70 Ssb contribute to its architecture on ribosomes and nascent chain interaction. Nat Commun 7: 13695 10.1038/ncomms13695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. 2011. Molecular chaperones in protein folding and proteostasis. Nature 475: 324–332. 10.1038/nature10317 [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Becker AH, Zachmann-Brand B, Deuerling E, Bukau B, Kramer G. 2012. Concerted action of the ribosome and the associated chaperone trigger factor confines nascent polypeptide folding. Mol Cell 48: 63–74. 10.1016/j.molcel.2012.07.018 [DOI] [PubMed] [Google Scholar]
- Hotokezaka Y, van Leyen K, Lo EH, Beatrix B, Katayama I, Jin G, Nakamura T. 2009. αNAC depletion as an initiator of ER stress-induced apoptosis in hypoxia. Cell Death Differ 16: 1505–1514. 10.1038/cdd.2009.90 [DOI] [PubMed] [Google Scholar]
- Huang P, Gautschi M, Walter W, Rospert S, Craig EA. 2005. The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nat Struct Mol Biol 12: 497–504. 10.1038/nsmb942 [DOI] [PubMed] [Google Scholar]
- Hundley H, Eisenman H, Walter W, Evans T, Hotokezaka Y, Wiedmann M, Craig E. 2002. The in vivo function of the ribosome-associated Hsp70, Ssz1, does not require its putative peptide-binding domain. Proc Natl Acad Sci 99: 4203–4208. 10.1073/pnas.062048399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundley HA, Walter W, Bairstow S, Craig EA. 2005. Human Mpp11 J protein: Ribosome-tethered molecular chaperones are ubiquitous. Science 308: 1032–1034. 10.1126/science.1109247 [DOI] [PubMed] [Google Scholar]
- Jaiswal H, Conz C, Otto H, Wolfle T, Fitzke E, Mayer MP, Rospert S. 2011. The chaperone network connected to human ribosome-associated complex. Mol Cell Biol 31: 1160–1173. 10.1128/MCB.00986-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner L, Melnikov S, Garreau de Loubresse N, Ben-Shem A, Iskakova M, Urzhumtsev A, Meskauskas A, Dinman J, Yusupova G, Yusupov M. 2012. Crystal structure of the 80S yeast ribosome. Curr Opin Struct Biol 22: 759–767. 10.1016/j.sbi.2012.07.013 [DOI] [PubMed] [Google Scholar]
- Jensen CG, Pedersen S. 1994. Concentrations of 4.5S RNA and Ffh protein in Escherichia coli: The stability of Ffh protein is dependent on the concentration of 4.5S RNA. J Bacteriol 176: 7148–7154. 10.1128/jb.176.23.7148-7154.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungnickel B, Rapoport TA. 1995. A posttargeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell 82: 261–270. 10.1016/0092-8674(95)90313-5 [DOI] [PubMed] [Google Scholar]
- Kaiser CM, Chang HC, Agashe VR, Lakshmipathy SK, Etchells SA, Hayer-Hartl M, Hartl FU, Barral JM. 2006. Real-time observation of trigger factor function on translating ribosomes. Nature 444: 455–460. 10.1038/nature05225 [DOI] [PubMed] [Google Scholar]
- Kaschner LA, Sharma R, Shrestha OK, Meyer AE, Craig EA. 2015. A conserved domain important for association of eukaryotic J-protein co-chaperones Jjj1 and Zuo1 with the ribosome. Biochim Biophys Acta 1853: 1035–1045. 10.1016/j.bbamcr.2015.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan RJ, Freymann DM, Stroud RM, Walter P. 2001. The signal recognition particle. Annu Rev Biochem 70: 755–775. 10.1146/annurev.biochem.70.1.755 [DOI] [PubMed] [Google Scholar]
- Kirstein-Miles J, Scior A, Deuerling E, Morimoto RI. 2013. The nascent polypeptide-associated complex is a key regulator of proteostasis. EMBO J 32: 1451–1468. 10.1038/emboj.2013.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge S, Voigts-Hoffmann F, Leibundgut M, Ban N. 2012. Atomic structures of the eukaryotic ribosome. Trends Biochem Sci 37: 189–198. 10.1016/j.tibs.2012.02.007 [DOI] [PubMed] [Google Scholar]
- Koplin A, Preissler S, Ilina Y, Koch M, Scior A, Erhardt M, Deuerling E. 2010. A dual function for chaperones SSB–RAC and the NAC nascent polypeptide-associated complex on ribosomes. J Cell Biol 189: 57–68. 10.1083/jcb.200910074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer G, Rauch T, Rist W, Vorderwülbecke S, Patzelt H, Schulze-Specking A, Ban N, Deuerling E, Bukau B. 2002. L23 protein functions as a chaperone docking site on the ribosome. Nature 419: 171–174. 10.1038/nature01047 [DOI] [PubMed] [Google Scholar]
- Kramer G, Boehringer D, Ban N, Bukau B. 2009. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol 16: 589–597. 10.1038/nsmb.1614 [DOI] [PubMed] [Google Scholar]
- Lee K, Sharma R, Shrestha OK, Bingman CA, Craig EA. 2016. Dual interaction of the Hsp70 J-protein cochaperone Zuotin with the 40S and 60S ribosomal subunits. Nat Struct Mol Biol 23: 1003–1010. 10.1038/nsmb.3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidig C, Bange G, Kopp J, Amlacher S, Aravind A, Wickles S, Witte G, Hurt E, Beckmann R, Sinning I. 2013. Structural characterization of a eukaryotic chaperone—The ribosome-associated complex. Nat Struct Mol Biol 20: 23–28. 10.1038/nsmb.2447 [DOI] [PubMed] [Google Scholar]
- Lesnik C, Cohen Y, Atir-Lande A, Schuldiner M, Arava Y. 2014. OM14 is a mitochondrial receptor for cytosolic ribosomes that supports co-translational import into mitochondria. Nat Commun 5: 5711 10.1038/ncomms6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Hu Y, Li X, Niu L, Teng M. 2010. The crystal structure of the human nascent polypeptide-associated complex domain reveals a nucleic acid-binding region on the NACA subunit. Biochemistry 49: 2890–2896. 10.1021/bi902050p [DOI] [PubMed] [Google Scholar]
- Markesich DC, Gajewski KM, Nazimiec ME, Beckingham K. 2000. bicaudal encodes the Drosophila β NAC homolog, a component of the ribosomal translational machinery. Development 127: 559–572. [DOI] [PubMed] [Google Scholar]
- Martin EM, Jackson MP, Gamerdinger M, Gense K, Karamonos TK, Humes JR, Deuerling E, Ashcroft AE, Radford SE. 2018. Conformational flexibility within the nascent polypeptide-associated complex enables its interactions with structurally diverse client proteins. J Biol Chem 293: 8554–8568. 10.1074/jbc.RA117.001568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Hackert E, Hendrickson WA. 2009. Promiscuous substrate recognition in folding and assembly activities of the trigger factor chaperone. Cell 138: 923–934. 10.1016/j.cell.2009.07.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashaghi A, Kramer G, Bechtluft P, Zachmann-Brand B, Driessen AJ, Bukau B, Tans SJ. 2013. Reshaping of the conformational search of a protein by the chaperone trigger factor. Nature 500: 98–101. 10.1038/nature12293 [DOI] [PubMed] [Google Scholar]
- Mayer MP, Kityk R. 2015. Insights into the molecular mechanism of allostery in Hsp70s. Front Mol Biosci 2: 58 10.3389/fmolb.2015.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz F, Hoffmann A, Rutkowska A, Zachmann-Brand B, Bukau B, Deuerling E. 2006. The C-terminal domain of Escherichia coli trigger factor represents the central module of its chaperone activity. J Biol Chem 281: 31963–31971. 10.1074/jbc.M605164200 [DOI] [PubMed] [Google Scholar]
- Moller I, Jung M, Beatrix B, Levy R, Kreibich G, Zimmermann R, Wiedmann M, Lauring B. 1998. A general mechanism for regulation of access to the translocon: Competition for a membrane attachment site on ribosomes. Proc Natl Acad Sci 95: 13425–13430. 10.1073/pnas.95.23.13425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muldoon-Jacobs KL, Dinman JD. 2006. Specific effects of ribosome-tethered molecular chaperones on programmed–1 ribosomal frameshifting. Eukaryot Cell 5: 762–770. 10.1128/EC.5.4.762-770.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama E, Sarris M, Redd M, Le Guyader D, Vivier C, Horsley W, Trede N, Herbomel P. 2015. NACA deficiency reveals the crucial role of somite-derived stromal cells in haematopoietic niche formation. Nat Commun 6: 8375 10.1038/ncomms9375 [DOI] [PubMed] [Google Scholar]
- Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA. 1992. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 71: 97–105. 10.1016/0092-8674(92)90269-I [DOI] [PubMed] [Google Scholar]
- Neuhof A, Rolls MM, Jungnickel B, Kalies KU, Rapoport TA. 1998. Binding of signal recognition particle gives ribosome/nascent chain complexes a competitive advantage in endoplasmic reticulum membrane interaction. Mol Biol Cell 9: 103–115. 10.1091/mbc.9.1.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyathi Y, Pool MR. 2015. Analysis of the interplay of protein biogenesis factors at the ribosome exit site reveals new role for NAC. J Cell Biol 210: 287–301. 10.1083/jcb.201410086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Becker AH, Sandikci A, Huber D, Chaba R, Gloge F, Nichols RJ, Typas A, Gross CA, Kramer G, et al. 2011. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell 147: 1295–1308. 10.1016/j.cell.2011.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott AK, Locher L, Koch M, Deuerling E. 2015. Functional dissection of the nascent polypeptide-associated complex in Saccharomyces cerevisiae. PLoS ONE 10: e0143457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto H, Conz C, Maier P, Wolfle T, Suzuki CK, Jeno P, Rucknagel P, Stahl J, Rospert S. 2005. The chaperones MPP11 and Hsp70L1 form the mammalian ribosome-associated complex. Proc Natl Acad Sci 102: 10064–10069. 10.1073/pnas.0504400102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pech M, Spreter T, Beckmann R, Beatrix B. 2010. Dual binding mode of the nascent polypeptide-associated complex reveals a novel universal adapter site on the ribosome. J Biol Chem 285: 19679–19687. 10.1074/jbc.M109.092536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisker K, Braun D, Wolfle T, Hentschel J, Funfschilling U, Fischer G, Sickmann A, Rospert S. 2008. Ribosome-associated complex binds to ribosomes in close proximity of Rpl31 at the exit of the polypeptide tunnel in yeast. Mol Biol Cell 19: 5279–5288. 10.1091/mbc.e08-06-0661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisker K, Chiabudini M, Rospert S. 2010. The ribosome-bound Hsp70 homolog Ssb of Saccharomyces cerevisiae. Biochim Biophys Acta 1803: 662–672. 10.1016/j.bbamcr.2010.03.005 [DOI] [PubMed] [Google Scholar]
- Pfund C, Lopez-Hoyo N, Ziegelhoffer T, Schilke BA, Lopez-Buesa P, Walter WA, Wiedmann M, Craig EA. 1998. The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. EMBO J 17: 3981–3989. 10.1093/emboj/17.14.3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polevoda B, Brown S, Cardillo TS, Rigby S, Sherman F. 2008. Yeast Nα-terminal acetyltransferases are associated with ribosomes. J Cell Biochem 103: 492–508. 10.1002/jcb.21418 [DOI] [PubMed] [Google Scholar]
- Pool MR, Stumm J, Fulga TA, Sinning I, Dobberstein B. 2002. Distinct modes of signal recognition particle interaction with the ribosome. Science 297: 1345–1348. 10.1126/science.1072366 [DOI] [PubMed] [Google Scholar]
- Powers T, Walter P. 1996. The nascent polypeptide-associated complex modulates interactions between the signal recognition particle and the ribosome. Curr Biol 6: 331–338. 10.1016/S0960-9822(02)00484-0 [DOI] [PubMed] [Google Scholar]
- Preissler S, Deuerling E. 2012. Ribosome-associated chaperones as key players in proteostasis. Trends Biochem Sci 37: 274–283. 10.1016/j.tibs.2012.03.002 [DOI] [PubMed] [Google Scholar]
- Raden D, Gilmore R. 1998. Signal recognition particle-dependent targeting of ribosomes to the rough endoplasmic reticulum in the absence and presence of the nascent polypeptide-associated complex. Mol Biol Cell 9: 117–130. 10.1091/mbc.9.1.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakwalska M, Rospert S. 2004. The ribosome-bound chaperones RAC and Ssb1/2p are required for accurate translation in Saccharomyces cerevisiae. Mol Cell Biol 24: 9186–9197. 10.1128/MCB.24.20.9186-9197.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch T, Hundley HA, Pfund C, Wegrzyn RD, Walter W, Kramer G, Kim SY, Craig EA, Deuerling E. 2005. Dissecting functional similarities of ribosome-associated chaperones from Saccharomyces cerevisiae and Escherichia coli. Mol Microbiol 57: 357–365. 10.1111/j.1365-2958.2005.04690.x [DOI] [PubMed] [Google Scholar]
- Raue U, Oellerer S, Rospert S. 2007. Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. J Biol Chem 282: 7809–7816. 10.1074/jbc.M611436200 [DOI] [PubMed] [Google Scholar]
- Saio T, Guan X, Rossi P, Economou A, Kalodimos CG. 2014. Structural basis for protein antiaggregation activity of the trigger factor chaperone. Science 344: 1250494 10.1126/science.1250494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing TM, Ramakrishnan V. 2009. What recent ribosome structures have revealed about the mechanism of translation. Nature 461: 1234–1242. 10.1038/nature08403 [DOI] [PubMed] [Google Scholar]
- Shulga N, James P, Craig EA, Goldfarb DS. 1999. A nuclear export signal prevents Saccharomyces cerevisiae Hsp70 Ssb1p from stimulating nuclear localization signal-directed nuclear transport. J Biol Chem 274: 16501–16507. 10.1074/jbc.274.23.16501 [DOI] [PubMed] [Google Scholar]
- Singhal K, Vreede J, Mashaghi A, Tans SJ, Bolhuis PG. 2015. The trigger factor chaperone encapsulates and stabilizes partial folds of substrate proteins. PLoS Comput Biol 11: e1004444 10.1371/journal.pcbi.1004444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreter T, Pech M, Beatrix B. 2005. The crystal structure of archaeal nascent polypeptide-associated complex (NAC) reveals a unique fold and the presence of a ubiquitin-associated domain. J Biol Chem 280: 15849–15854. 10.1074/jbc.M500160200 [DOI] [PubMed] [Google Scholar]
- Steitz TA. 2008. A structural understanding of the dynamic ribosome machine. Nat Rev Mol Cell Biol 9: 242–253. 10.1038/nrm2352 [DOI] [PubMed] [Google Scholar]
- Teter SA, Houry WA, Ang D, Tradler T, Rockabrand D, Fischer G, Blum P, Georgopoulos C, Hartl FU. 1999. Polypeptide flux through bacterial Hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell 97: 755–765. 10.1016/S0092-8674(00)80787-4 [DOI] [PubMed] [Google Scholar]
- Wang L, Zhang W, Wang L, Zhang XC, Li X, Rao Z. 2010. Crystal structures of NAC domains of human nascent polypeptide-associated complex (NAC) and its αNAC subunit. Protein Cell 1: 406–416. 10.1007/s13238-010-0049-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Durfee LA, Huibregtse JM. 2013. A cotranslational ubiquitination pathway for quality control of misfolded proteins. Mol Cell 50: 368–378. 10.1016/j.molcel.2013.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn RD, Hofmann D, Merz F, Nikolay R, Rauch T, Graf C, Deuerling E. 2006. A conserved motif is prerequisite for the interaction of NAC with ribosomal protein L23 and nascent chains. J Biol Chem 281: 2847–2857. 10.1074/jbc.M511420200 [DOI] [PubMed] [Google Scholar]
- Weyer FA, Gumiero A, Gesé GV, Lapouge K, Sinning I. 2017. Structural insights into a unique Hsp70-Hsp40 interaction in the eukaryotic ribosome-associated complex. Nat Struct Mol Biol 24: 144–151. 10.1038/nsmb.3349 [DOI] [PubMed] [Google Scholar]
- Wiedmann B, Sakai H, Davis TA, Wiedmann M. 1994. A protein complex required for signal-sequence-specific sorting and translocation. Nature 370: 434–440. 10.1038/370434a0 [DOI] [PubMed] [Google Scholar]
- Williams CC, Jan CH, Weissman JS. 2014. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science 346: 748–751. 10.1126/science.1257522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmund F, del Alamo M, Pechmann S, Chen T, Albanese V, Dammer EB, Peng J, Frydman J. 2013. The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell 152: 196–209. 10.1016/j.cell.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DN, Beckmann R. 2011. The ribosomal tunnel as a functional environment for nascent polypeptide folding and translational stalling. Curr Opin Struct Biol 21: 274–282. 10.1016/j.sbi.2011.01.007 [DOI] [PubMed] [Google Scholar]
- Wruck F, Avellaneda MJ, Koers EJ, Minde DP, Mayer MP, Kramer G, Mashaghi A, Tans SJ. 2018. Protein folding mediated by trigger factor and Hsp70: New insights from single-molecule approaches. J Mol Biol 430: 438–449. 10.1016/j.jmb.2017.09.004 [DOI] [PubMed] [Google Scholar]
- Yan W, Schilke B, Pfund C, Walter W, Kim S, Craig EA. 1998. Zuotin, a ribosome-associated DnaJ molecular chaperone. EMBO J 17: 4809–4817. 10.1093/emboj/17.16.4809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Berndt U, Gölz H, Tais A, Oellerer S, Wölfle T, Fitzke E, Rospert S. 2012. NAC functions as a modulator of SRP during the early steps of protein targeting to the endoplasmic reticulum. Mol Biol Cell 23: 3027–3040. 10.1091/mbc.e12-02-0112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ma C, Yuan Y, Zhu J, Li N, Chen C, Wu S, Yu L, Lei J, Gao N. 2014. Structural basis for interaction of a cotranslational chaperone with the eukaryotic ribosome. Nat Struct Mol Biol 21: 1042–1046. 10.1038/nsmb.2908 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sinning I, Rospert S. 2017. Two chaperones locked in an embrace: Structure and function of the ribosome-associated complex RAC. Nat Struct Mol Biol 24: 611–619. 10.1038/nsmb.3435 [DOI] [PubMed] [Google Scholar]