Abstract
Ultra-hypermutation (>100 mutations/Mb) is rare in childhood cancer genomes and has been primarily reported in patients with constitutional mismatch repair deficiency (CMMRD) caused by biallelic germline mismatch repair (MMR) gene mutations. We report a 5-yr-old child with classic clinical features of CMMRD and an ultra-hypermutated medulloblastoma with retained MMR protein expression and absence of germline MMR mutations. Mutational signature analysis of tumor panel sequencing data revealed a canonical DNA polymerase-deficiency-associated signature, prompting further genetic testing that uncovered a germline POLE p.A456P missense variant, which has previously been reported as a recurrent somatic driver mutation in cancers. This represents the earliest known onset of malignancy in a patient with a germline mutation in the POLE proofreading polymerase. The clinical features in this child, virtually indistinguishable from those of CMMRD, suggest that polymerase-proofreading deficiency should be considered in the differential diagnosis of CMMRD patients with retained MMR function.
Keywords: cerebellar medulloblastoma
INTRODUCTION
DNA replication fidelity in humans is preserved via two distinct repair mechanisms: polymerase proofreading and the mismatch repair (MMR) system (Rayner et al. 2016). Proofreading of newly synthesized DNA strands during replication is performed by the 3′–5′ exonuclease activity of the Pol δ and Pol ε replicative polymerase catalytic subunits, POLD1 and POLE, respectively (Ganai and Johansson 2016; Rayner et al. 2016). The MMR pathway is a postreplicative error correction mechanism for mismatches evading polymerase proofreading and is primarily effected by MLH1, MSH2, MSH6, and PMS2 (Germano et al. 2018).
Although a functional deficiency in either pathway leads to a “mutator” cellular phenotype (Loeb 2016), the association between defective replication repair and cancer is better recognized for the MMR pathway, which is defective in two inherited cancer predisposition syndromes. Monoallelic heterozygous loss-of-function germline mutations in MMR genes cause Lynch syndrome, an autosomal dominant disorder (Lynch et al. 2015). In contrast, biallelic (compound heterozygous or homozygous) loss-of-function germline mutations in MMR genes result in constitutional MMR deficiency syndrome (CMMRD), which is inherited in an autosomal recessive pattern (Bakry et al. 2014; Wimmer et al. 2014).
Despite a shared pathogenesis, Lynch syndrome and CMMRD are distinct in inheritance pattern, epidemiology, and clinical features (Abedalthagafi 2018). CMMRD is characterized by a higher frequency of brain and hematological malignancies and a markedly earlier age of onset (median age at first tumor is <10 yr) of cancer (Tabori et al. 2017). In addition, there are nonmalignant dermatological manifestations including pigmented skin papules and café au lait macules reminiscent of neurofibromatosis type 1 as well as multiple pilomatricomas. These clinical features have been proposed as a scoring system for further diagnostic workup that includes tumor/normal MMR staining by immunohistochemistry and/or microsatellite instability (MSI) evaluation followed by genetic analysis (Bakry et al. 2014; Wimmer et al. 2014; Tabori et al. 2017). Tumors in CMMRD patients are invariably “hypermutated” (defined as >10 mutations/Mb) and often “ultra-hypermutated” (>100 mutations/Mb) with acquisition of secondary somatic POLE/POLD1 mutations (Shlien et al. 2015; Bouffet et al. 2016), and characteristic mutational signatures of MMR deficiency may be detected in the tumor genome (Alexandrov et al. 2013).
Recently, a highly penetrant dominant cancer predisposition syndrome was described in kindreds with colonic polyposis and/or carcinomas in germline carriers of POLE/POLD1 exonuclease domain mutations (EDMs) (Briggs and Tomlinson 2013; Palles et al. 2013). Initially termed polymerase proofreading-associated polyposis, emerging reports of extracolonic malignancies, including central nervous system (CNS) gliomas as well as pancreatic and endometrial adenocarcinomas (Rohlin et al. 2014; Hansen et al. 2015; Spier et al. 2015; Bellido et al. 2016) suggests that the POLE/POLD1 cancer predisposition syndrome is an evolving entity. POLE/POLD1 syndrome patients typically receive their first cancer diagnosis in adulthood (median 40 and 32.5 yr for POLE and POLD1, respectively), which is more similar to Lynch syndrome than CMMRD (Bellido et al. 2016). Tumors with POLE/POLD1 EDMs (Cancer Genome Atlas Network 2012; Church et al. 2013) are typically MMR-proficient and microsatellite stable (MSS) and are ultra-hypermutated with a characteristic genomic mutational signature (Alexandrov et al. 2013; Rayner et al. 2016; Campbell et al. 2017). Recurrent somatic polymerase EDMs are more commonly reported in POLE than POLD1, with certain hotspot codons (D275, P286, S297, V411, P436, A456, Y458, S459, and S461) associated with very high mutation burdens (Campbell et al. 2017). Germline EDMs in familial cases, however, are highly restricted to only a handful of recurrent missense changes including POLE L242V and in certain POLD1 codons (S478, L474, D316, and R409) (Bellido et al. 2016).
In this report, we present a 5-yr-old child with medulloblastoma and clinical features of CMMRD, whose ultra-hypermutated tumor was found to retain MMR expression and harbor a mutation signature of germline POLE deficiency. Genetic workup revealed an absence of germline MMR mutations and the discovery of a heterozygous germline POLE p.A456P mutation.
RESULTS
Clinical Presentation and Family History
The patient is a Latina female who presented at 5 yr of age with a several week progressive history of headache, nausea, vomiting, impaired balance, and frequent falling toward her right side. Initial physical examination revealed more than 100 hyperpigmented macules and papules over her entire body, including some with classic café au lait macule features (Fig. 1A,B). Maternal family history was negative for relatives with any early-onset cancers, and both maternal half-siblings are healthy without chronic medical conditions or hyperpigmented skin lesions. Paternal family and medical history were unavailable.
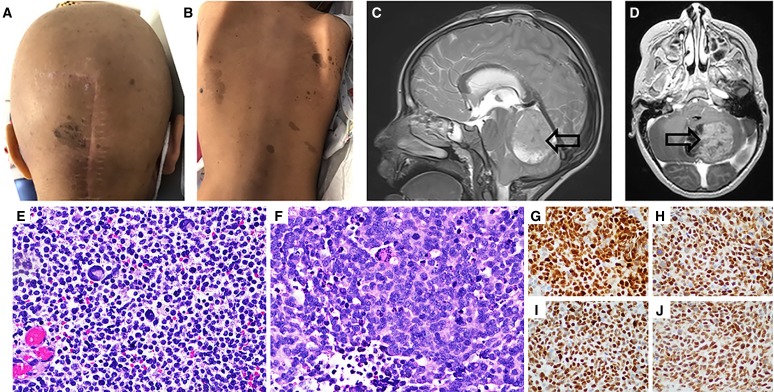
Figure 1.
Representative images of numerous hyperpigmented skin lesions on the head (A) and back (B). Representative sagittal (C) and axial (D) MRI images showing a posterior fossa mass. Representative low-power (E) and high-power (F) images of the anaplastic medulloblastoma specimen demonstrating small round cells, mitotic figures, and scattered multinucleated giant cells. Immunohistochemistry for MMR proteins (G–J) revealing retained expression of MLH1, MSH2, MSH6, and PMS2, respectively.
A head CT revealed a posterior fossa mass and obstructive hydrocephalus. Subsequent MRI confirmed the presence of a left cerebellar heterogeneously enhancing tumor with restricted diffusion, in addition to multifocal leptomeningeal metastatic disease along the cerebellum (Fig. 1C,D). The patient was additionally noted to have left-sided complex renal cyst, a right distal tibia osteochondroma, and three superficial hard palpable subcutaneous calcified nodules on her extremities, which were subsequently diagnosed as pilomatricomas on pathology examination.
A gross total resection of the primary posterior fossa tumor was performed. Pathologic examination revealed a large cell anaplastic medulloblastoma with several atypical mitotic figures, apoptosis, regional necrosis, and giant multinucleated cells (Fig. 1E,F). Additional medulloblastoma subclassification using fluorescence in situ hybridization probes for Chromosome 6q23 (monosomy 6), MYC, MYCN, EGFR, PTEN (monosomy 10), and immunohistochemistry (GLI1, YAP1, β-catenin, p53, FOXG1) revealed a non-WNT/non-SHH medulloblastoma without MYC/MYCN amplification. MMR immunohistochemistry of the tumor revealed staining for all four MMR proteins to be strongly retained in both tumor and normal cells (Fig. 1G–J).
Genomic Analyses
Given the suspected diagnosis of CMMRD, genetic analysis was performed on both germline and tumor specimens. Germline analysis of MMR pathway genes (MLH1, MSH2, MSH6, and PMS2) and other genes associated with childhood brain tumors (TP53, PTCH1, NF1, APC, etc.) (Table 1) using gene panels featuring variant and deletion/duplication analysis (including for EPCAM) was negative for any pathogenic or likely pathogenic variants or copy-number abnormalities.
Table 1.
Panel genes tested for pathogenic germline variants in the patient
| Panel | Genes |
|---|---|
| Hereditary high-risk CRC panel | APC, BMPR1A, EPCAM, MLH1, MSH2, MSH6, MUTYH, PMS2, PTEN, SMAD4, STK11, TP53 |
| Hereditary brain/CNS/PNS cancer panel (unique genes not on CRC panel) | ALK, ATM, MEN1, MRE11A, NBN, NF2, PALB2, PHOX2B, PTCH1, SUFU, VHL |
(CRC) Colorectal cancer, (CNS) central nervous system, (PNS) peripheral nervous system.
Tumor variant analysis was undertaken on the medulloblastoma specimen using a 124-gene next-generation sequencing (NGS) panel in a CLIA-certified laboratory (Supplemental Table S1A) revealing several loss-of-function variants in genes commonly mutated in medulloblastoma (Table 2), including pathogenic nonsense mutations in TP53 and APC that, as noted above, were absent in the germline of the patient in testing for familial cancer predisposition (Table 1), confirming a somatic origin for these mutations. The key finding, however, was that of an ultra-hypermutated tumor with a markedly increased tumor mutation burden (TMB) estimated at >150 mutations/Mb of the genome. Notably, 72/94 variants on the tumor panel report had no entries in reference population databases with at least 25% (18/72) being nonsense mutations.
Table 2.
Relevant tumor panel findings
| Gene | hg19 position | CDS change | AA change | Variant type | Origin | dbSNP | Variant allele fraction | COSMIC ID |
|---|---|---|---|---|---|---|---|---|
| APC | Chr 5:112175124 | c.3833C>A | p.Ser1278Ter | Nonsense | Somatic | 27.40% | COSM19313 | |
| KDM6A | Chr X:44950066 | c.3835C>T | p.Arg1279Ter | Nonsense | Somatic | rs863224886 | 50.00% | COSM28780 |
| NF2 | Chr 22:30070832 | c.1348G>T | p.Glu450Ter | Nonsense | Somatic | 49.60% | ||
| PTCH1 | Chr 9:98244436 | c.634G>T | p.Glu212Ter | Nonsense | Somatic | 43.70% | ||
| SUFU | Chr 10:104387001 | c.1365+1G>T | Splicing | Somatic | 46.30% | |||
| TP53 | Chr 17:7578212 | c.637C>T | p.Arg213Ter | Nonsense | Somatic | rs397516436 | 98.80% | COSM10654 |
(CDS) Coding sequence, (AA) amino acid.
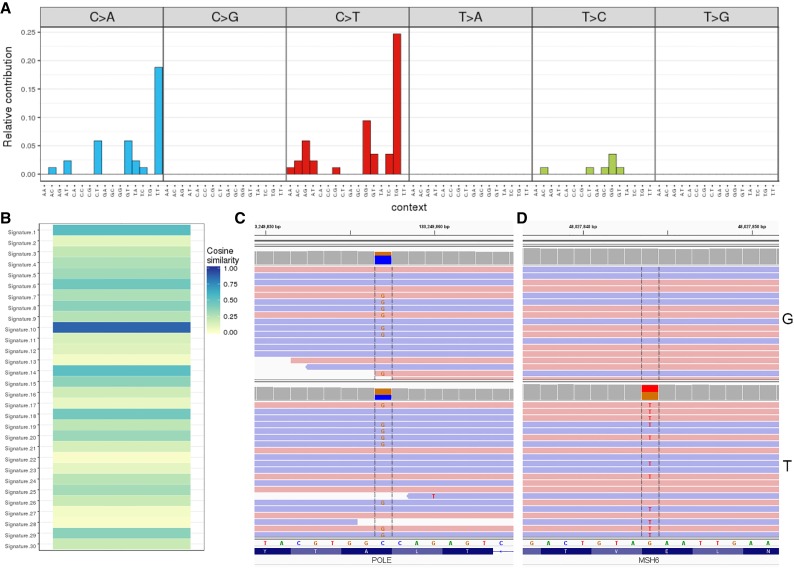
Given the profile of an MMR-proficient ultra-hypermutated tumor, the possibility of a polymerase proofreading error-related mechanism was entertained. Mutation signature analysis of all tumor point mutations and the surrounding trinucleotide context revealed a disproportionate frequency of C>A transversions (∼35%) and C>T transitions (∼50%), with the T[C>T]G (24%) and T[C>A]T (18%) signature together comprising >40% of the mutational burden (Fig. 2A). This mutational pattern is characteristic of COSMIC “signature 10” (Alexandrov et al. 2013; Rayner et al. 2016) and an unsupervised similarity analysis of the tumor mutational profile to all published signature patterns confirmed the highest cosine similarity of 0.87 (1 being identical) to that signature (Fig. 2B).
Figure 2.
Mutation signature analysis and identification of germline POLE mutation. Mutation signature profile of the tumor as represented on the 96-substitution classification (A) along with the trinucleotide context. Mutations and the contexts are plotted on the horizontal axis, and the vertical axis represents the proportional contribution of each pattern to the overall signature. A striking pattern of increased C>A [TCT] and C>T [TCG] trinucleotide contexts forming ∼40% of all changes is observed, which has been strongly associated with “signature 10,” and confirmed on mutational pattern similarity analysis against all 30 COSMIC signatures (B) showing closest match to “signature 10” (cosine similarity 0.87). The signature prediction of germline POLE alteration was corroborated by germline and tumor sequencing revealing a POLE p.A456P mutation (C) in the germline (top panel, “G”) and tumor (bottom panel, “T”). In contrast, a heterozygous MSH6 nonsense mutation was only seen in the tumor (D).
Because of the known association between signature 10 and germline POLE deficiency, a 981-gene comprehensive cancer panel (Supplemental Table S1B) containing POLE/POLD1 and MMR genes was used to sequence genomic DNA from germline and tumor specimens. The analysis revealed a heterozygous germline missense mutation in exon 14 of POLE: NM_006231.3(POLE):c.1366G>C (p.Ala456Pro) (Table 3; Fig. 2C). This variant, previously reported as a recurrent somatic POLE driver mutation and associated with a high mutation burden (Campbell et al. 2017), lies within the Exo III motif of the POLE exonuclease domain in close proximity to several recurrent somatic POLE mutations. The p.A456P mutation alters a highly conserved alanine residue that is predicted to affect DNA binding and lead to defective proofreading activity (Church et al. 2013). No additional somatic POLE mutation (or copy-number change and/or LOH) as a second hit was noted. Further genetic testing for POLE in the mother revealed that she did not harbor the germline p.A456P variant. Repeated attempts by the clinical team to reach the father for familial testing were unsuccessful; thus, a paternally inherited mutation cannot be ruled out.
Table 3.
Germline POLE and somatic MSH6 variants detected in the patient
| Gene | hg19 position | Depth at position (G, T) | CDS change | AA change | Variant type | Origin | Predicted effect | dbSNP | Geno | ClinVar ID |
|---|---|---|---|---|---|---|---|---|---|---|
| POLE | 12:133249857 | 51, 136 | c.1366G>C | p.A456P | Missense | G | Del | – | Het | SCV000825544.1 |
| MSH6 | 2:48027844 | 302, 1004 | c.2722G>T | p.Q908X | Nonsense | S | Del | – | Het |
(CDS) Coding sequence, (AA) amino acid, (G) germline, (S) somatic, (T) tumor, (Del) deleterious, (Het) heterozygous.
The comprehensive cancer panel testing of the tumor also revealed a nonsense mutation in exon 4 of MSH6 that was not present in the germline: NM_000179.2(MSH6):c.2722G>T (p.Glu908Ter) at 46.7% variant allele fraction (Fig. 2D). No second hit in MSH6 or other alterations in the MMR genes were detected, consistent with the retention of MMR staining in the cells (Fig. 1G–J). Both the germline POLE and somatic MSH6 variants were independently confirmed by direct Sanger sequencing. No other pathogenic alterations in any of the remaining polymerase genes were detected.
Clinical Course
Following clinical recovery from gross total resection of the primary tumor, the patient was treated with proton beam craniospinal irradiation (at 3600 cobalt Gray equivalent plus boost to tumor bed and nodules) followed by seven cycles of chemotherapy (including vincristine, cisplatin, cyclophosphamide, gemcitabine, and pemetrexed) on a high-risk arm of a multi-institutional phase 2 clinical trial for patients with newly diagnosed medulloblastoma. Her chemotherapy courses were complicated by malnutrition necessitating enteral caloric supplementation via nasogastric tube and recurrent episodes of Clostridium difficile colitis and methicillin-susceptible Staphylococcus aureus bacteremia. She otherwise tolerated her treatment without severe complication and is doing well without evidence of medulloblastoma recurrence or progression of leptomeningeal metastasis 4 mo after completion of treatment.
Clinical experts in pediatric cancer surveillance were consulted to devise a customized screening protocol including at least annual brain MRI imaging indefinitely given the need for surveillance for recurrence as well as new POLE polymerase cancer syndrome–related brain tumors. She additionally will soon be undergoing a baseline colonoscopy. The timing of subsequent serial evaluations will be based on the presence or absence of colon adenomas; if no adenomas are identified, a repeat colonoscopy can be delayed until the patient becomes a teenager. She is followed biannually by dermatology and will undergo annual gynecology exams on reaching adulthood.
DISCUSSION
This case illustrates several novel and striking aspects of the still-evolving POLE/POLD1 polymerase cancer syndrome as well as the utility of integrative tumor sequencing and mutation signature analysis in accurately predicting the underlying mutational mechanism in a tumor.
First, the patient presented with clinical features indistinguishable from that of CMMRD based on current diagnostic criteria proposed by the C4CMMRD consortium (Wimmer et al. 2014), including a pediatric-onset tumor (1 point), numerous café au lait macules (2 points), and multiple pilomatricomas (2 points), with a score of ≥3 suggested to prompt screening for MMR deficiency (MMR-D). Other investigators have recently reported a 14-yr-old boy with rectosigmoid carcinoma and CMMRD-like dermatological manifestations and a germline POLE missense mutation (p.V411L) (Wimmer et al. 2017), suggesting that the characteristic skin manifestations of CMMRD may be common to different DNA repair defect syndromes. Our results corroborate that and provide further evidence that a POLE/POLD1 deficiency should be considered in the differential of CMMRD patients, particularly if further testing excludes MMR deficiency.
Second, to our knowledge, at age 5 yr, this is the youngest cancer presentation reported in a patient with germline POLE/POLD1 cancer predisposition syndrome. Except for the 14-yr-old boy described by Wimmer et al. (2017) with a negative family history, to date, the median age at first cancer diagnosis in all known familial cases of POLE/POLD1 syndrome is reported to be in the fourth decade (earliest 27 yr) (Bellido et al. 2016). The presentation in early childhood is, therefore, highly unusual and contributed to further overlap with the clinical presentation of CMMRD.
Third, this is the first report of the POLE p.A456P mutation as a germline alteration. The p.A456P EDM is a known recurrent somatic hotspot mutation that has been previously reported in at least 32 tumors (Table 4) (COSM937319), including hyper/ultra-hypermutated tumors with mutational signatures characteristic of POLE proofreading deficiency (Shinbrot et al. 2014). Review of the literature reveals that the TMB, when reported, is invariably high in all tumors harboring the p.A456P mutation (Table 4). The alanine at codon 456 is highly conserved from humans to Drosophila, and structural analysis predicts mutation of this site to lead to altered DNA binding through distortion of the polymerase active site (Church et al. 2013). Taken together, the p.A456P EDM is considered a driver POLE mutation in human carcinogenesis (Campbell et al. 2017) and is the most likely cause of the phenotype in this patient. As important, the mutation was found to be absent in the germline of the asymptomatic mother, although paternal inheritance could not be ruled out.
Table 4.
Reported POLE p.A456P tumor mutations and associated mutation burden
| Tumor | Total cases | TMB-ultra | TMB-hyper | TMB-low | TMB-ND | Reference(s) |
|---|---|---|---|---|---|---|
| Colorectal carcinoma | 7 | 5 | 1 | - | 1 | Shinbrot et al. 2014; Stenzinger et al. 2014; Jansen et al. 2016; Campbell et al. 2017 |
| Endometrial carcinoma | 21 | 2 | 2 | - | 17 | Church et al. 2013; Kandoth et al. 2013; Billingsley et al. 2015; Talhouk et al. 2015; Espinosa et al. 2016; McConechy et al. 2016; Bellone et al. 2017; Campbell et al. 2017; Espinosa et al. 2017; Van Gool et al. 2018 |
| Glioblastoma multiforme | 1 | 1 | - | - | - | Hoadley et al. 2018 |
| Ovarian carcinoma | 2 | - | 1 | - | 1 | Campbell et al. 2017; Parra-Herran et al. 2017 |
| Cervical squamous cell carcinoma | 1 | - | 1 | - | - | Campbell et al. 2017 |
(TMB) Tumor mutation burden, (hyper) hypermutated (>10/Mb), (ultra) ultra-hypermutated (>100/Mb), (ND) not determined.
The combination of an unusually early age of onset with a novel germline occurrence of a recurrent somatic mutation raises the possibility of the p.A456P EDM leading to a more severe POLE proofreading defect. It is noteworthy that in a large screen of cancers for driver POLE EDMs (Campbell et al. 2017), both the p.A456P mutation and the p.V411L mutation reported previously in the 14-yr-old boy (Wimmer et al. 2017) were characterized as strong “mutators” because of a tight correlation with elevated TMB. In contrast, using functional assays and association with TMB, the only recurrent familial germline POLE mutation (p.L424V) was found to have a weak “mutator” phenotype (Shinbrot et al. 2014; Campbell et al. 2017), possibly correlating with a later age of onset seen in those individuals. Further reports of other POLE EDMs in the germline along with functional studies would be necessary to clarify the genotype–phenotype correlation between the severity of the POLE mutation and age of onset.
Ultra-hypermutation was first recognized in adult-onset MSS colon cancers that were found to be enriched for POLE and, less commonly, POLD1 EDMs (Cancer Genome Atlas Network 2012; Donehower et al. 2013; Palles et al. 2013). Childhood cancer genomes are typically characterized by a low TMB (Vogelstein et al. 2013; Schlesner and Eils 2015) with hyper/ultra-hypermutation seen in <1% of all pediatric cancers, primarily in the context of CMMRD (Gröbner et al. 2018). Tumor profiling using NGS panels to direct management of cancers is now common in clinical practice, and this report highlights the utility of tumor sequencing and analysis. Detection of an ultra-hypermutated tumor has potentially major therapeutic implications for this patient as a positive association between TMB and the response to therapeutic immune checkpoint inhibition, including improved progression-free survival and increased rates of objective response, presumed secondary to an increased number of neoantigens in hypermutated tumors (Rizvi et al. 2015; Bourdais et al. 2017; Le et al. 2017; Nebot-Bral et al. 2017), is now well-established. In the event of tumor recurrence in this patient, treatment with a PD-1 or PD-L1 inhibitor will be strongly considered. Multiple clinical trials of immune checkpoint inhibitors specifically enrolling patients with hypermutated tumors are currently available (Clinical Trials NCT02359565, NCT02992964).
In addition, the identification of ultra-hypermutation on tumor panel analysis in the absence of MMR-D (as evidenced by negative MMR genetic testing and retained MMR expression by IHC) for the first time raised the possibility of a germline POLE deficiency as the primary genetic defect, despite the unusually young age at presentation for that diagnosis. The tumor mutational signature analysis revealed an unambiguous “signature 10” pattern that is a “canonical” signature of POLE deficiency (Alexandrov et al. 2013; Campbell et al. 2017). This directed further evaluation of the tumor and germline specimens for POLE/POLD1 mutations revealing the deleterious POLE p.A456P mutation in the germline. This report corroborates the accuracy of mutational signature analysis of hypermutated tumors from tumor panel sequencing data in prospectively predicting the underlying genetic defect, as proposed recently in the form of a diagnostic algorithm for hypermutated childhood tumors (Campbell et al. 2017). The correlations reported here and elsewhere (Campbell et al. 2017) emphasize that germline testing for CMMRD and POLE/POLD1 inherited cancer syndrome may result from either clinical features or tumor mutation spectrum (when available). Notably, identification of the germline POLE defect in our patient along with the reported pediatric CRC case (Wimmer et al. 2017) has also led to modification in the follow-up care, as current cancer screening recommendations for patients with germline POLE mutations include colonoscopies every 1–2 yr and gastroduodenoscopies every 3 yr beginning at 18–25 yr old (Spier et al. 2015; Bellido et al. 2016; Rayner et al. 2016; Buchanan et al. 2018).
Finally, although both colonic and extracolonic malignancies, including CNS astrocytomas, have been previously reported in familial POLE/POLD1 mutation carriers, (Rohlin et al. 2014; Hansen et al. 2015; Spier et al. 2015; Bellido et al. 2016; Johanns et al. 2016; Esteban-Jurado et al. 2017), to our knowledge, this is the first report of a medulloblastoma as the presenting malignancy in a patient with germline POLE mutation. Except for the scattered giant multinucleated cells that have been previously reported in hypermutated glioblastomas with somatic POLE mutations (Erson-Omay et al. 2015), the histopathological characteristics of the anaplastic medulloblastoma were characteristic. Our report, therefore, extends both the age of diagnosis and the spectrum of tumors seen in POLE/POLD1 syndrome patients and suggests that the phenotype of this newly described cancer predisposition syndrome is still being defined.
METHODS
The study was performed after obtaining informed consent for enrollment on a Baylor College of Medicine Institutional Review Board-approved study.
Genomic DNA was extracted from blood and the medulloblastoma specimen utilizing the QIAamp DNA Mini Kit and the DNA FFPE Tissue Kit (QIAGEN), respectively. Germline DNA underwent clinical sequencing at two CLIA-certified reference laboratories: Baylor Genetics laboratory for the Hereditary High-risk CRC panel and the Hereditary Brain/CNS/PNS panel and at Invitae laboratories for POLE testing. Additional sequencing using the SeqCap EZ Human Oncology Panel (Roche Inc.) was performed at Texas Children's Hospital. Tumor DNA underwent clinical sequencing at the CLIA-certified Texas Children's Hospital laboratory using the Solid Tumor NGS panel as well as the Human Oncology Panel. A list of genes covered on the tumor panels is included in Supplemental Table S1.
Sequencing by the hereditary cancer panels at Baylor Genetics featured Illumina-based NGS variant analysis and MLPA/CGH-based deletion/duplication analysis of familial cancer genes (Table 1). For the human oncology panel and solid tumor panels, NGS libraries were generated using 50 ng of DNA and subsequently pooled in either 4-plex or 2-plex for hybridization capture with respective capture probe set. Captured pools were then sequenced on an Illumina MiSeq with 600V3 chemistry (2 × 150-bp read length). Coverage metrics for the sequenced samples are provided in Table 5. NextGENe (SoftGenetics) and Platypus (Rimmer et al. 2014) were utilized for variant calling, followed by annotation with VEP (McLaren et al. 2016). Mutation prediction algorithms (PolyPhen, SIFT, LRT, MutationTaster) were used using dbNSFP (Liu et al. 2011) to predict the biological effect of the mutation.
Table 5.
Sequencing metrics on the human oncology panel for the tumor and germline specimens
| Sample | Total reads | Percent aligned reads | Percent reads on target | Average coverage |
|---|---|---|---|---|
| Germline | 7,096,416 | 94.29% | 68.95% | 181.92 |
| Tumor | 20,774,903 | 95.49% | 77.56% | 581.33 |
Mutation signature analysis and visualization were generated using the Mutational Patterns Bioconductor package in R (Blokzijl et al. 2018). The package was utilized to generate the 96-substitution classification profile (Alexandrov et al. 2013) with all substitutions converted to the pyrimidine of the Watson–Crick base pair and representing the immediate trinucleotide context (5′ × 3′) of the mutated base. The optimal nonnegative linear combination of COSMIC signatures was then fitted to the substitution classification profile, followed by calculation of the cosine similarity between the tumor mutation signature in this patient and the COSMIC signatures to determine the mutation signature classes that best fit the signature of the patient.
Tumor mutation burden was calculated from the solid tumor mutation panel (∼1 Mb capture size) using the total number of nonsynonymous variants in the coding region of the panel adjusted for the panel size.
ADDITIONAL INFORMATION
Data Deposition and Access
The POLE variant can be found in ClinVar (https://www.ncbi.nnlm.nih.gov/clinvar/) under accession number SCV000825544.1. The MSH6 variant was submitted to ClinVar and can be found under accession number SCV000965671.1. Tumor and germline targeted sequencing data from the human oncology panel will be deposited to the National Center for Biotechnology Information (NCBI) Database of Genotypes and Phenotypes (dbGaP; http://www.ncbi.nlm.nih.gov/gap).
Ethics Statement
Informed consent for enrollment on study H-27233 “Molecular Characterization of Cancer Genomes” approved by the Institutional Review Board of Baylor College of Medicine was obtained by a study physician.
Author Contributions
Clinical care was provided by H.L., S.S., S.P.R., M.C., A.M.A., and S.E.P. Genomic data analysis and interpretation were performed by S.S., J.R., H.V., F.Y.L., K.E.F., D.W.P., S.E.P., and A.R. H.L. and A.R. wrote the initial draft of the manuscript. All authors contributed to reviewing and editing the final draft.
Funding
This study was partly funded by the Gillson Longenbaugh Foundation and Cookies for Kids Cancer (D.W.P.).
Competing Interest Statement
The authors have declared no competing interest.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Abedalthagafi M. 2018. Constitutional mismatch repair-deficiency: current problems and emerging therapeutic strategies. Oncotarget 9: 35458–35469. 10.18632/oncotarget.26249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale AL, et al. 2013. Signatures of mutational processes in human cancer. Nature 500: 415–421. 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakry D, Aronson M, Durno C, Rimawi H, Farah R, Alharbi QK, Alharbi M, Shamvil A, Ben-Shachar S, Mistry M, et al. 2014. Genetic and clinical determinants of constitutional mismatch repair deficiency syndrome: report from the constitutional mismatch repair deficiency consortium. Eur J Cancer 50: 987–996. 10.1016/j.ejca.2013.12.005 [DOI] [PubMed] [Google Scholar]
- Bellido F, Pineda M, Aiza G, Valdés-Mas R, Navarro M, Puente DA, Pons T, González S, Iglesias S, Darder E, et al. 2016. POLE and POLD1 mutations in 529 kindred with familial colorectal cancer and/or polyposis: review of reported cases and recommendations for genetic testing and surveillance. Genet Med 18: 325–332. 10.1038/gim.2015.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellone S, Bignotti E, Lonardi S, Ferrari F, Centritto F, Masserdotti A, Pettinella F, Black J, Menderes G, Altwerger G, et al. 2017. Polymerase ε (POLE) ultra-mutation in uterine tumors correlates with T lymphocyte infiltration and increased resistance to platinum-based chemotherapy in vitro. Gynecol Oncol 144: 146–152. 10.1016/j.ygyno.2016.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billingsley CC, Cohn DE, Mutch DG, Stephens JA, Suarez AA, Goodfellow PJ. 2015. Polymerase varepsilon (POLE) mutations in endometrial cancer: clinical outcomes and implications for Lynch syndrome testing. Cancer 121: 386–394. 10.1002/cncr.29046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokzijl F, Janssen R, van Boxtel R, Cuppen E. 2018. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med 10: 33 10.1186/s13073-018-0539-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouffet E, Larouche V, Campbell BB, Merico D, de Borja R, Aronson M, Durno C, Krueger J, Cabric V, Ramaswamy V, et al. 2016. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J Clin Oncol 34: 2206–2211. 10.1200/JCO.2016.66.6552 [DOI] [PubMed] [Google Scholar]
- Bourdais R, Rousseau B, Pujals A, Boussion H, Joly C, Guillemin A, Baumgaertner I, Neuzillet C, Tournigand C. 2017. Polymerase proofreading domain mutations: new opportunities for immunotherapy in hypermutated colorectal cancer beyond MMR deficiency. Crit Rev Oncol Hematol 113: 242–248. 10.1016/j.critrevonc.2017.03.027 [DOI] [PubMed] [Google Scholar]
- Briggs S, Tomlinson I. 2013. Germline and somatic polymerase ε and δ mutations define a new class of hypermutated colorectal and endometrial cancers. J Pathol 230: 148–153. 10.1002/path.4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan DD, Stewart JR, Clendenning M, Rosty C, Mahmood K, Pope BJ, Jenkins MA, Hopper JL, Southey MC, Macrae FA, et al. 2018. Risk of colorectal cancer for carriers of a germ-line mutation in POLE or POLD1. Genet Med 20: 890–895. 10.1038/gim.2017.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BB, Light N, Fabrizio D, Zatzman M, Fuligni F, de Borja R, Davidson S, Edwards M, Elvin JA, Hodel KP, et al. 2017. Comprehensive analysis of hypermutation in human cancer. Cell 171: 1042–1056.e10. 10.1016/j.cell.2017.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. 2012. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487: 330–337. 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church DN, Briggs SEW, Palles C, Domingo E, Kearsey SJ, Grimes JM, Gorman M, Martin L, Howarth KM, Hodgson SV, et al. 2013. DNA polymerase ε and δ exonuclease domain mutations in endometrial cancer. Hum Mol Genet 22: 2820–2828. 10.1093/hmg/ddt131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower LA, Creighton CJ, Schultz N, Shinbrot E, Chang K, Gunaratne PH, Muzny D, Sander C, Hamilton SR, Gibbs RA, et al. 2013. MLH1-silenced and non-silenced subgroups of hypermutated colorectal carcinomas have distinct mutational landscapes. J Pathol 229: 99–110. 10.1002/path.4087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erson-Omay EZ, Çağlayan AO, Schultz N, Weinhold N, Omay SB, Özduman K, Köksal Y, Li J, Serin Harmanci A, Clark V, et al. 2015. Somatic POLE mutations cause an ultramutated giant cell high-grade glioma subtype with better prognosis. Neuro Oncol 17: 1356–1364. 10.1093/neuonc/nov027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa I, D'Angelo E, Palacios J, Prat J. 2016. Mixed and ambiguous endometrial carcinomas: a heterogenous group of tumors with different clinicopathologic and molecular genetic features. Am J Surg Pathol 40: 972–981. 10.1097/PAS.0000000000000640 [DOI] [PubMed] [Google Scholar]
- Espinosa I, Lee CH, D'Angelo E, Palacios J, Prat J. 2017. Undifferentiated and dedifferentiated endometrial carcinomas with POLE exonuclease domain mutations have a favorable prognosis. Am J Surg Pathol 41: 1121–1128. 10.1097/PAS.0000000000000873 [DOI] [PubMed] [Google Scholar]
- Esteban-Jurado C, Giménez-Zaragoza D, Muñoz J, Franch-Expósito S, Álvarez-Barona M, Ocaña T, Cuatrecasas M, Carballal S, López-Ceron M, Marti-Solano M, et al. 2017. POLE and POLD1 screening in 155 patients with multiple polyps and early-onset colorectal cancer. Oncotarget 8: 26732–26743. 10.18632/oncotarget.15810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganai RA, Johansson E. 2016. DNA replication—a matter of fidelity. Mol Cell 62: 745–755. 10.1016/j.molcel.2016.05.003 [DOI] [PubMed] [Google Scholar]
- Germano G, Amirouchene-Angelozzi N, Rospo G, Bardelli A. 2018. The clinical impact of the genomic landscape of mismatch repair-deficient cancers. Cancer Discov 8: 1518–1528. 10.1158/2159-8290.CD-18-0150 [DOI] [PubMed] [Google Scholar]
- Gröbner SN, Worst BC, Weischenfeldt J, Buchhalter I, Kleinheinz K, Rudneva VA, Johann PD, Balasubramanian GP, Segura-Wang M, Brabetz S, et al. 2018. The landscape of genomic alterations across childhood cancers. Nature 555: 321–327. 10.1038/nature25480 [DOI] [PubMed] [Google Scholar]
- Hansen MF, Johansen J, Bjørnevoll I, Sylvander AE, Steinsbekk KS, Sætrom P, Sandvik AK, Drabløs F, Sjursen W. 2015. A novel POLE mutation associated with cancers of colon, pancreas, ovaries and small intestine. Fam Cancer 14: 437–448. 10.1007/s10689-015-9803-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V, et al. 2018. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 173: 291–304.e6. 10.1016/j.cell.2018.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen AML, van Wezel T, van den Akker BEWM, Ventayol Garcia M, Ruano D, Tops CMJ, Wagner A, Letteboer TGW, Gómez-García EB, Devilee P, et al. 2016. Combined mismatch repair and POLE/POLD1 defects explain unresolved suspected Lynch syndrome cancers. Eur J Hum Genet 24: 1089–1092. 10.1038/ejhg.2015.252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanns TM, Miller CA, Dorward IG, Tsien C, Chang E, Perry A, Uppaluri R, Ferguson C, Schmidt RE, Dahiya S, et al. 2016. Immunogenomics of hypermutated glioblastoma: a patient with germline POLE deficiency treated with checkpoint blockade immunotherapy. Cancer Discov 6: 1230–1236. 10.1158/2159-8290.CD-16-0575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. 2013. Mutational landscape and significance across 12 major cancer types. Nature 502: 333–339. 10.1038/nature12634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, et al. 2017. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357: 409–413. 10.1126/science.aan6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jian X, Boerwinkle E. 2011. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat 32: 894–899. 10.1002/humu.21517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb LA. 2016. Human cancers express a mutator phenotype: hypothesis, origin, and consequences. Cancer Res 76: 2057–2059. 10.1158/0008-5472.CAN-16-0794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch HT, Snyder CL, Shaw TG, Heinen CD, Hitchins MP. 2015. Milestones of lynch syndrome: 1895–2015. Nat Rev Cancer 15: 181–194. 10.1038/nrc3878 [DOI] [PubMed] [Google Scholar]
- McConechy MK, Talhouk A, Leung S, Chiu D, Yang W, Senz J, Reha-Krantz LJ, Lee C-H, Huntsman DG, Gilks CB, et al. 2016. Endometrial carcinomas with POLE exonuclease domain mutations have a favorable prognosis. Clin Cancer Res 22: 2865–2873. 10.1158/1078-0432.CCR-15-2233 [DOI] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, Flicek P, Cunningham F. 2016. The Ensembl Variant Effect Predictor. Genome Biol 17: 122 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebot-Bral L, Brandao D, Verlingue L, Rouleau E, Caron O, Despras E, El-Dakdouki Y, Champiat S, Aoufouchi S, Leary A, et al. 2017. Hypermutated tumours in the era of immunotherapy: the paradigm of personalised medicine. Eur J Cancer 84: 290–303. 10.1016/j.ejca.2017.07.026 [DOI] [PubMed] [Google Scholar]
- Palles C, Cazier JB, Howarth KM, Domingo E, Jones AM, Broderick P, Kemp Z, Spain SL, Guarino E, Salguero I, et al. 2013. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet 45: 136–144. 10.1038/ng.2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra-Herran C, Lerner-Ellis J, Xu B, Khalouei S, Bassiouny D, Cesari M, Ismiil N, Nofech-Mozes S. 2017. Molecular-based classification algorithm for endometrial carcinoma categorizes ovarian endometrioid carcinoma into prognostically significant groups. Mod Pathol 30: 1748–1759. 10.1038/modpathol.2017.81 [DOI] [PubMed] [Google Scholar]
- Rayner E, van Gool IC, Palles C, Kearsey SE, Bosse T, Tomlinson I, Church DN. 2016. A panoply of errors: polymerase proofreading domain mutations in cancer. Nat Rev Cancer 16: 71–81. 10.1038/nrc.2015.12 [DOI] [PubMed] [Google Scholar]
- Rimmer A, Phan H, Mathieson I, Iqbal Z, Twigg SRF, Consortium WGS, Wilkie AOM, McVean G, Lunter G. 2014. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet 46: 912–918. 10.1038/ng.3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al. 2015. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science 348: 124–128. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlin A, Zagoras T, Nilsson S, Lundstam ULF, Wahlström J, Hultén L, Martinsson T, Karlsson GB, Nordling M. 2014. A mutation in POLE predisposing to a multi-tumour phenotype. Int J Oncol 45: 771.1. 10.3892/ijo.2014.2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesner M, Eils R. 2015. Hypermutation takes the driver's seat. Genome Med 7: 31 10.1186/s13073-015-0159-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinbrot E, Henninger EE, Weinhold N, Covington KR, Göksenin AY, Schultz N, Chao H, Doddapaneni H, Muzny DM, Gibbs RA, et al. 2014. Exonuclease mutations in DNA polymerase ε reveal replication strand specific mutation patterns and human origins of replication. Genome Res 24: 1740–1750. 10.1101/gr.174789.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlien A, Campbell BB, de Borja R, Alexandrov LB, Merico D, Wedge D, Van Loo P, Tarpey PS, Coupland P, Behjati S, et al. 2015. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat Genet 47: 257–262. 10.1038/ng.3202 [DOI] [PubMed] [Google Scholar]
- Spier I, Holzapfel S, Altmüller J, Zhao B, Horpaopan S, Vogt S, Chen S, Morak M, Raeder S, Kayser K, et al. 2015. Frequency and phenotypic spectrum of germline mutations in POLE and seven other polymerase genes in 266 patients with colorectal adenomas and carcinomas. Int J Cancer 137: 320–331. 10.1002/ijc.29396 [DOI] [PubMed] [Google Scholar]
- Stenzinger A, Pfarr N, Endris V, Penzel R, Jansen L, Wolf T, Herpel E, Warth A, Klauschen F, Kloor M, et al. 2014. Mutations in POLE and survival of colorectal cancer patients—link to disease stage and treatment. Cancer Med 3: 1527–1538. 10.1002/cam4.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabori U, Hansford JR, Achatz MI, Kratz CP, Plon SE, Frebourg T, Brugières L. 2017. Clinical management and tumor surveillance recommendations of inherited mismatch repair deficiency in childhood. Clin Cancer Res 23: e32–e37. 10.1158/1078-0432.CCR-17-0574 [DOI] [PubMed] [Google Scholar]
- Talhouk A, McConechy MK, Leung S, Li-Chang HH, Kwon JS, Melnyk N, Yang W, Senz J, Boyd N, Karnezis AN, et al. 2015. A clinically applicable molecular-based classification for endometrial cancers. Br J Cancer 113: 299–310. 10.1038/bjc.2015.190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gool IC, Ubachs JEH, Stelloo E, de Kroon CD, Goeman JJ, Smit VTHBM, Creutzberg CL, Bosse T. 2018. Blinded histopathological characterisation of POLE exonuclease domain-mutant endometrial cancers: sheep in wolf's clothing. Histopathology 72: 248–258. 10.1111/his.13338 [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr., Kinzler KW. 2013. Cancer genome landscapes. Science 339: 1546–1558. 10.1126/science.1235122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer K, Kratz CP, Vasen HFA, Caron O, Colas C, Entz-Werle N, Gerdes AM, Goldberg Y, Ilencikova D, Muleris M, et al. 2014. Diagnostic criteria for constitutional mismatch repair deficiency syndrome: suggestions of the European consortium ‘care for CMMRD’ (C4CMMRD). J Med Genet 51: 355–365. 10.1136/jmedgenet-2014-102284 [DOI] [PubMed] [Google Scholar]
- Wimmer K, Beilken A, Nustede R, Ripperger T, Lamottke B, Ure B, Steinmann D, Reineke-Plaass T, Lehmann U, Zschocke J, et al. 2017. A novel germline POLE mutation causes an early onset cancer prone syndrome mimicking constitutional mismatch repair deficiency. Fam Cancer 16: 67–71. 10.1007/s10689-016-9925-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The POLE variant can be found in ClinVar (https://www.ncbi.nnlm.nih.gov/clinvar/) under accession number SCV000825544.1. The MSH6 variant was submitted to ClinVar and can be found under accession number SCV000965671.1. Tumor and germline targeted sequencing data from the human oncology panel will be deposited to the National Center for Biotechnology Information (NCBI) Database of Genotypes and Phenotypes (dbGaP; http://www.ncbi.nlm.nih.gov/gap).