Abstract
Genome sequencing was performed on matched normal and tumor tissue from a 6.5-yr-old boy with a diagnosis of recurrent medulloblastoma. A pathogenic heterozygous c.432+1G>A canonical splice donor site variant in GNAS was detected on analysis of blood DNA. Analysis of tumor DNA showed the same splice variant along with copy-neutral loss of heterozygosity on Chromosome 20 encompassing GNAS, consistent with predicted biallelic loss of GNAS in the tumor specimen. This case strengthens the evidence implicating GNAS as a tumor-suppressor gene in medulloblastoma and highlights a scenario in which therapeutics targeting the cAMP pathway may be of great utility.
Keywords: cerebellar medulloblastoma
CASE PRESENTATION
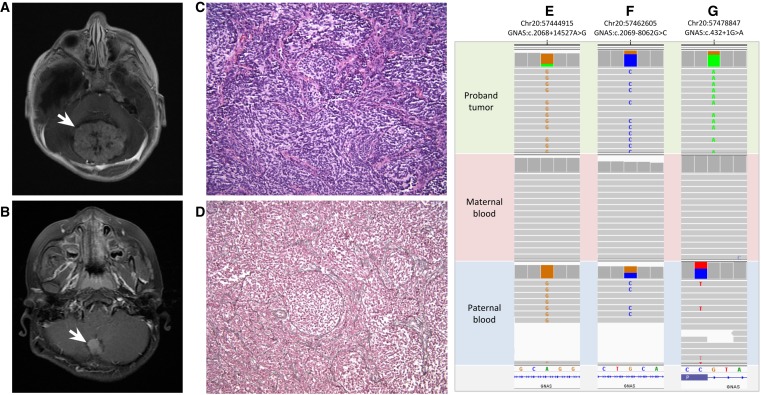
The patient is a 6.5-yr-old boy who was referred for research genome sequencing because of a diagnosis of recurrent medulloblastoma. His initial presentation at 22 mo was prompted by a several-week history of loss of developmental milestones, persistent head tilt, and decreased responsiveness. Past medical history was notable for delivery by cesarean section at 28 wk for oligohydramnios and breech presentation, and severe feeding difficulties in early infancy necessitating fundoplication and gastrostomy-tube placement. Pertinent findings on exam at presentation included macrocephaly, listlessness, truncal ataxia, and episodic apnea. Magnetic resonance imaging (MRI) of the brain showed a 5.5-cm mass in the midline posterior fossa (Fig. 1A), obstructive hydrocephalus, and mild tonsillar herniation. The patient underwent gross total resection of the tumor. Histopathologic analysis of the resected specimen was consistent with nodular/desmoplastic medulloblastoma. Immunohistochemical staining performed on a clinical basis and transcriptomic profiling performed on a research basis were both consistent with sonic hedgehog (SHH) subtype medulloblastoma. MRI of the spine and cerebrospinal fluid cytology demonstrated no evidence of leptomeningeal spread of disease. He was treated with induction, consolidation, and maintenance chemotherapy per study protocol (SJYC07) without radiation therapy. He remained disease-free until age 6.5 yr, when surveillance MRI showed a local recurrence in the original resection cavity (Fig. 1B) without evidence of dissemination. The patient underwent gross total resection in which pathology was again consistent with nodular/desmoplastic medulloblastoma (Fig. 1C,D). He was treated with craniospinal proton radiotherapy. Concurrently, he was enrolled in the Rady Children's Institute for Genomic Medicine (RCIGM) Biorepository Study (IRB #160468) to facilitate genomic analysis of matched tumor and normal tissue samples. The results of that analysis are discussed here. The patient was subsequently treated with high-dose craniospinal proton radiation therapy, had a complete response, and remained without central nervous system (CNS) disease 18 mo after completion of therapy.
Figure 1.
Representative MRI images of the midline posterior fossa medulloblastoma at (A) initial diagnosis and (B) recurrence. (C) H&E stain of the recurrent tumor (200× magnification) showing densely cellular proliferation of “small blue cells” with nodules showing neural differentiation with pale fibrillary background. (D) Reticulin stain of recurrent tumor (200× magnification) showing reticulin deposition within internodular areas, outlining reticulin-negative nodules. (E–G) IGV data showing the de novo c.432+1G>A pathogenic variant in GNAS is present on the paternally inherited allele. GNAS SNVs detected on analysis of proband tumor (top), maternal blood (middle), and paternal blood (bottom). The allele fraction of the paternally inherited SNVs in E and F (NM_080425) in the proband's tumor is similar to the allele fraction of the pathogenic splice variant (G) (NM_000516), indicating that the de novo splice variant has likely occurred on the paternal allele.
TECHNICAL ANALYSIS AND METHODS
Samples of blood and tumor were collected for genomic analysis. Whole-genome sequencing (WGS) was performed on DNA extracted from blood to assess for the presence of informative constitutional variants. Concurrently, WGS was performed on DNA extracted from tumor tissue at the time of recurrence to assess for functionally relevant somatic mutations. Attempts to isolate DNA from FFPE tumor blocks from the initial tumor resection failed to yield any usable product. WGS was performed as follows: PCR-free libraries of DNA fragments were constructed using the Illumina PCR-free Library Preparation kit according to manufacturer's recommendations. Paired-end sequencing (2 × 150 bp) was performed with Illumina chemistry on a HiSeq 4000 sequencer to a mean depth of coverage of 45× (blood) and 94× (tumor). Resulting sequences were aligned to the human genome reference (hg19) using the Burrows–Wheeler Aligner (BWA) and variants identified with the Genome Analysis Tool Kit (GATK). Somatic variants were identified using the Varscan, Vardict, and Strelka software packages. CONTRA analysis was applied to tumor sequence data to call copy-number variants (CNVs). Variants identified on analysis of tumor DNA that were absent from blood DNA were considered somatic mutations. All clinically reported single-nucleotide variants were orthogonally confirmed by Sanger sequencing, and all clinically reported CNVs were orthogonally confirmed by SNP array analysis (Illumina Infinium CytoSNP- 850K BeadChip v1.1).
VARIANT INTERPRETATION
Two potentially clinically significant single nucleotide variants were detected by germline and tumor WGS and are summarized in Table 1. Results of tumor WGS CNV analysis and tumor SNP microarray are summarized in Table 2. WGS performed on DNA extracted from the patient's blood sample detected a de novo heterozygous pathogenic c.432+1G>A variant in GNAS. This variant affects the canonical splice donor site of intron 5 and thus is predicted to alter splicing and result in loss of normal protein function. This variant had been previously reported in the literature as a de novo change in an individual with a diagnosis of pseudopseudohypoparathyroidism (Wilson et al. 1994). It is absent from the ExAC and gnomAD population databases and thus is presumed to be rare. A second maternally inherited heterozygous variant of uncertain clinical significance was detected in DICER1. This variant affects a highly conserved amino acid and is absent from the ExAC and gnomAD population databases. However, it has not been previously reported in the literature as a deleterious change and as such the clinical implications of this variant remain uncertain.
Table 1.
Single-nucleotide variants detected by genome sequencing of germline and tumor
| Gene | Transcript | Coordinates (hg19) | Nucleotide | Protein | Classification | Coverage (blood) | AF (blood) | Coverage (tumor) | AF (tumor) |
|---|---|---|---|---|---|---|---|---|---|
| DICER1 | NM_030621 | Chr 14:95574409 | c.2458A>G | p.T820A | VUS | 45× | 42% | 111× | 43% |
| GNAS | NM_000516 | Chr 20:57478847 | c.432+1G>A | p.? | Pathogenic | 47× | 57% | 121× | 78% |
(VUS) Variant of uncertain significance, (AF) allele fraction.
Table 2.
Structural variants detected by genome sequencing of tumor
| ISCN arr[GRCh37] | Event | Size (bp) | Cancer-related genes |
|---|---|---|---|
| 1p32.1p22.3 (60,489,417_86,741,437) × 1 | Loss | 26,252,021 | JAK1, FUBP1, BCL10 |
| 1p21.2p12 (102,000,184_120,532,214) × 1 | Loss | 18,532,031 | RBM15, TRIM33, NRAS, FAM46C, NOTCH2 |
| (7) × 3 | Gain | 159,138,663 | Many, e.g., CDK6 |
| 13q14.2q21.1 (49,392,118_55,549,433) × 1 | Loss | 6,157,316 | |
| 20q11.21qter (30,137,047_63,025,520) × 2 hmz | Copy-neutral LOH | 32,888,474 | ASXL1, MAFB, TOP1, SDC4, GNAS, SS18L1 |
(LOH) Loss of heterozygosity.
Both the GNAS and DICER1 variants were also detected by WGS of the tumor sample (Table 1). Based on the allelic fraction of the GNAS variant, the estimated tumor purity of the sample was 50%. WGS and SNP microarray analysis of tumor additionally showed trisomy 7 and several copy-number variants including multiple deletions involving regions on Chromosomes 1p and 13q (Table 2). Copy-neutral loss of heterozygosity of Chromosome 20q11.21qter encompassing GNAS (Table 2) was also detected, consistent with biallelic loss of GNAS in the tumor specimen. No focal amplifications of known oncogenes or deletions of known tumor-suppressor genes were present.
SUMMARY
GNAS resides in a complex imprinted locus from which multiple transcripts are generated, the best characterized of which encodes Gsα (MIM: 139320) (Bastepe and Jüppner 2005). Gsα is the alpha subunit of the stimulatory guanine nucleotide-binding protein, a key mediator of the cAMP signal transduction pathway (Mantovani et al. 2016). Biallelic expression of GNAS occurs in most tissues (Haldeman-Englert et al. 2017). However, predominant expression of the maternal allele has been demonstrated in certain tissues (e.g., thyroid epithelial cells) (Haldeman-Englert et al. 1993; Bastepe and Jüppner 2005) and likely explains the parent-of-origin specificity of phenotypes resulting from mutations in GNAS. Inactivating variants affecting the maternal GNAS allele cause pseudohypoparathyroidism, which is characterized by multihormone resistance with an Albright hereditary osteodystrophy (AHO) phenotype (short stature, round face, subcutaneous ossifications, brachydactyly, and intrauterine growth restriction) (Haldeman-Englert et al. 1993). Inactivating mutations on the paternal GNAS allele cause pseudopseudohypoparathyroidism, which presents as AHO without endocrine dysfunction (Mantovani et al. 2016). Paternal GNAS mutations have also been implicated in additional disorders including progressive osseous heteroplasia and osteoma cutis (Haldeman-Englert et al. 2017).
The detection of a pathogenic loss-of-function variant in GNAS on analysis of this patient's blood (Table 1) is consistent with a diagnosis of a constitutional GNAS-associated disorder. However, because the mutation occurred as a de novo change, the parent of origin of the mutated allele could not be determined on the basis of the germline sequence data alone. In addition, there were no informative single-nucleotide polymorphisms (SNPs) in close proximity to the de novo event that could be used to phase the pathogenic mutation. However, analysis of the tumor sample was informative and permitted phasing of the de novo variant to the paternal allele. WGS of the tumor showed the c.432+1G>A variant in GNAS at an allele fraction of 78%. This finding, in conjunction with copy-neutral loss of heterozygosity involving 20q11.21qter encompassing GNAS, indicated that the normal GNAS allele had likely been lost in part or all of the tumor specimen. Additional remote SNPs in the GNAS gene were analyzed and overrepresentation of the paternal allele to a degree consistent with the allele fraction of the pathogenic splice variant was evident (Fig. 1E–G), suggesting that somatic loss of the maternal GNAS allele and duplication of the paternal GNAS allele had occurred in the tumor. Thus, the c.432+1G>A variant could be phased to the paternal allele, indicating that this patient is at risk of developing the pseudopseudohypoparathyroidism, progressive osseous heteroplasia, and/or osteoma cutis phenotypes.
The pathogenic splice variant in GNAS was also observed in the patient's medulloblastoma sample. Together with copy-neutral loss of heterozygosity encompassing the GNAS gene, this finding is consistent with presumed loss of normal GNAS expression in tumor tissue. Much is known about the molecular drivers of pediatric medulloblastoma, which have been divided into four major subgroups on the basis of transcriptomic profiling: WNT, SHH, Group 3, and Group 4 (Taylor et al. 2012). The WNT and SHH subtypes are named for key signaling pathways implicated in medulloblastoma tumorigenesis. Less is known about the biological underpinnings of the latter two groups, which consequently have generic names (Taylor et al. 2012). Histopathologic analysis of the tumor in this patient was consistent with desmoplastic/nodular medulloblastoma, a subgroup typically associated with SHH type tumors (Northcott et al. 2012). Previous studies of medulloblastoma genomes have shown that the SHH subgroup is characterized by more regions of chromosome gain and/or loss relative to WNT medulloblastomas (Northcott et al. 2012). In addition, driver mutations in a specific set of genes involved in SHH signaling (e.g., PTCH1) are commonly found as somatic events (Northcott et al. 2012).
Germline mutations in PTCH1 and SUFU, both negative regulators of the SHH pathway (Northcott et al. 2012), have been linked with a predisposition to medulloblastoma development (Smith et al. 2014). Gsα, which is encoded by GNAS, promotes production of cAMP (Mantovani et al. 2016), which in turn represses SHH signaling through activation of cAMP-dependent protein kinase A (Makinodan and Marneros 2012; He et al. 2014). Although GNAS modulates SHH signaling in this way (Regard et al. 2013), inactivating variants in GNAS have not been tightly correlated with an increased risk of medulloblastoma or other malignant tumor formation (Mantovani et al. 2018). A recent report, however, describes a child with osteoma cutis and medulloblastoma who was found on biopsy of multiple affected tissues to have a homozygous nonsense variant in GNAS (Huh et al. 2014). The patient died from complications of surgery before a blood sample could be obtained for germline variant analysis, but it is reasonable to assume the patient was at least heterozygous for the inactivating GNAS variant. Furthermore, in a study of two independent cohorts of patients with SHH-driven medulloblastomas, reduced GNAS expression levels were closely correlated with unfavorable survival outcomes (He et al. 2014), suggesting a role for GNAS in medulloblastoma formation and/or progression.
In mice, loss of Gnas in neural stem/progenitor cells causes tumors that recapitulate the location and histologic features of human SHH-driven medulloblastomas (He et al. 2014). In tumor cells derived from mice with loss of Gnas, a significant decrease in intracellular cAMP levels was noted. This would be expected to reduce PKA-mediated repression of the Shh pathway, thereby contributing to hyperactivation of Shh signaling and tumor formation. Intriguingly, treatment of affected mice with rolipram, a phosphodiesterase type 4 inhibitor that blocks cAMP breakdown, led to reduced tumor volumes, decreased tumor cell proliferation, and significant improvements in life expectancy in treated relative to untreated mice (He et al. 2014). Although the clinical application of rolipram specifically is limited by its propensity to cause gastrointestinal toxicity (Prickaerts et al. 2017), these data underscore the potential utility of cAMP agonists for the treatment of GNAS-inactivated SHH-driven medulloblastomas (He et al. 2014).
The pathogenic splice variant in GNAS detected in this patient was found as a constitutional mutation, whereas the copy-neutral LOH was restricted to the tumor, indicating the latter event occurred as a somatic change. In contrast to the predicted loss-of-function effect of the variants identified in this patient, putative gain-of-function variants in GNAS are among the most commonly detected somatic mutations in cancer. Predominantly missense variants in GNAS have been detected as somatic events in pituitary tumors, colorectal cancer, pancreatic cancer, and other neoplasms (O'Hayre et al. 2013). Thus, GNAS may play a dual role in cancer development, acting either as a tumor suppressor or an oncogenic driver in a mutation- and possibly tumor-dependent fashion.
In summary, we describe a patient with recurrent nodular/desmoplastic medulloblastoma who was found on WGS to have a de novo pathogenic germline variant in GNAS. To our knowledge, this is the second report of medulloblastoma occurring in a patient with a constitutional GNAS variant but the first comprehensive genetic analysis incorporating germline and tumor WGS with single-nucleotide and structural variant detection. Together with a compelling mouse model (He et al. 2014), this report adds to the evidence implicating GNAS as a tumor-suppressor gene in SHH-driven medulloblastoma. With studies in mice showing the potential utility of cAMP-augmenting agents in GNAS-inactivated tumors, an investigation of the efficacy of such drugs in patients, particularly those with recurrent or refractory disease, is warranted.
ADDITIONAL INFORMATION
Database Deposition and Access
The reported variant in GNAS has been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession number SCV000623565.1. The reported variant in DICER1 has been submitted to ClinVar and can be found under accession number SCV000987202. Raw sequencing data was submitted to dbGaP (https://www.ncbi.nlm.nih.gov/gap/), accession numbers pending.
Ethics Statement
Written informed consent for genome sequencing and for publication was obtained from all study subjects. The study was approved by Institutional Review Board of the University of California at San Diego under protocol # 160468.
Acknowledgments
We are grateful to the patient and his family for participating in this study. We also thank the research and laboratory staff at Rady Children's Institute for Genomic Medicine.
Author Contributions
All authors contributed to scientific discussion and manuscript review.
Funding
Funding support was provided by Rady Children's Institute for Genomic Medicine, the Joseph Clayes III Charitable Trust, and by the National Cancer Institute (R01 CA159859 to R.W.-R.).
Competing Interest Statement
The authors have declared no competing interest.
REFERENCES
- Bastepe M, Jüppner H. 2005. GNAS locus and pseudohypoparathyroidism. Horm Res 63: 65–74. 10.1007/978-0-387-77576-0_3 [DOI] [PubMed] [Google Scholar]
- Haldeman-Englert CR, Hurst AC, and Levine MA. 2017. Disorders of GNAS inactivation. In GeneReviews® (ed. Adam MP, et al. ). University of Washington, Seattle; 1993–2019. Available from http://www.ncbi.nlm.nih.gov/books/NBK459117/ [Google Scholar]
- He X, Zhang L, Chen Y, Remke M, Shih D, Lu F, Wang H, Deng Y, Yu Y, Xia Y, et al. 2014. The G protein α subunit Gαs is a tumor suppressor in Sonic hedgehog–driven medulloblastoma. Nat Med 20: 1035–1042. 10.1038/nm.3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh JY, Kwon MJ, Seo KY, Kim MK, Chae KY, Kim SH, Ki CS, Yoon MS, Kim DH. 2014. Novel nonsense GNAS mutation in a 14-month-old boy with plate-like osteoma cutis and medulloblastoma. J Dermatol 41: 319–321. 10.1111/1346-8138.12284 [DOI] [PubMed] [Google Scholar]
- Makinodan E, Marneros AG. 2012. Protein kinase A activation inhibits oncogenic Sonic hedgehog signalling and suppresses basal cell carcinoma of the skin. Exp Dermatol 21: 847–852. 10.1111/exd.12016 [DOI] [PubMed] [Google Scholar]
- Mantovani G, Spada A, Elli FM. 2016. Pseudohypoparathyroidism and Gsα-cAMP-linked disorders: current view and open issues. Nat Rev Endocrinol 12: 347–356. 10.1038/nrendo.2016.52 [DOI] [PubMed] [Google Scholar]
- Mantovani G, Bastepe M, Monk D, de Sanctis L, Thiele S, Usardi A, Ahmed SF, Bufo R, Choplin T, De Filippo G, et al. 2018. Diagnosis and management of pseudohypoparathyroidism and related disorders: first International Consensus Statement. Nat Rev Endocrinol 14: 476–500. 10.1038/s41574-018-0042-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Jones DTW, Kool M, Robinson GW, Gilbertson RJ, Cho YJ, Pomeroy SL, Korshunov A, Lichter P, Taylor MD, et al. 2012. Medulloblastomics: the end of the beginning. Nat Rev Cancer 12: 818–834. 10.1038/nrc3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hayre M, Vazquez-Prado J, Kufareva I, Stawiski EW, Handel TM, Seshagiri S, Silvio Gutkind J. 2013. The emerging mutational landscape of G-proteins and G-protein coupled receptors in cancer. Nat Rev Cancer 13: 412–424. 10.1038/nrc3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prickaerts J, Heckman PRA, Blokland A. 2017. Investigational phosphodiesterase inhibitors in phase I and phase II clinical trials for Alzheimer's disease. Expert Opin Invest Drugs 26: 1033–1048. 10.1080/13543784.2017.1364360 [DOI] [PubMed] [Google Scholar]
- Regard JB, Malhotra D, Gvozdenovic-Jeremic J, Josey M, Chen M, Weinstein LS, Lu J, Shore EM, Kaplan FS, Yang Y. 2013. Activation of Hedgehog signaling by loss of GNAS causes heterotopic ossification. Nat Med 19: 1505–1512. 10.1038/nm.3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MJ, Beetz C, Williams SG, Bhaskar SS, O'Sullivan J, Anderson B, Daly SB, Urquhart JE, Bholah Z, et al. 2014. Germline mutations in SUFU cause Gorlin syndrome-associated childhood medulloblastoma and redefine the risk associated with PTCH1 mutations. J Clin Oncol 32: 4155–4161. 10.1200/JCO.2014.58.2569 [DOI] [PubMed] [Google Scholar]
- Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, Eberhart CG, Parsons DW, Rukowski S, Gajjar A, et al. 2012. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol 123: 465–472. 10.1007/s00401-011-0922-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson LC, Oude Luttikhuis ME, Clayton PT, Fraser WD, Trembath RC. 1994. Parental origin of Gs alpha gene mutations in Albright's hereditary osteodystrophy. J Med Genet 31: 835–839. 10.1136/jmg.31.11.835 [DOI] [PMC free article] [PubMed] [Google Scholar]