Abstract
Substantial progress has been made in recent years toward deciphering the molecular and genetic underpinnings of the pubertal process. The availability of powerful new methods to interrogate the human genome has led to the identification of genes that are essential for puberty to occur. Evidence has also emerged suggesting that the initiation of puberty requires the coordinated activity of gene sets organized into functional networks. At a cellular level, it is currently thought that loss of transsynaptic inhibition, accompanied by an increase in excitatory inputs, results in the pubertal activation of GnRH release. This concept notwithstanding, a mechanism of epigenetic repression targeting genes required for the pubertal activation of GnRH neurons was recently identified as a core component of the molecular machinery underlying the central restraint of puberty. In this chapter we will discuss the potential contribution of various mechanisms of epigenetic regulation to the hypothalamic control of female puberty.
Keywords: GnRH neurons, Kisspeptin neurons, Female puberty, Chromatin modifications, DNA methylation, Transcriptional repression, Transcriptional activation, Epigenetic regulators, microRNAs, Long noncoding RNAs
1. Introduction
It is well-established that the initiation of mammalian puberty requires an increased secretory activity of a handful of hypothalamic neurosecretory neurons that produce the decapeptide gonadotropin-releasing hormone (GnRH). Because GnRH neurons are able to produce and release GnRH long before puberty, it is also clear that they are neither the ultimate responsible for the initiation of puberty nor constitute – under normal conditions – a significant obstacle for the pubertal process to be initiated earlier [reviewed in Ojeda and Skinner (2006)]. Instead, the secretory activity of GnRH neurons depends on trans-synaptic and glial inputs provided by different neurotransmitters, neuromodulators and cell–cell signaling molecules, derived from either neuronal subsets or glial cells functionally connected to GnRH neurons [reviewed in Ojeda and Skinner (2006), Terasawa and Fernandez (2001), Plant and Witchel (2006)]. While the trans-synaptic input can be either excitatory or inhibitory, the glial input is almost invariably excitatory (Prevot, 2002).
The unquestionable complexity of the cellular systems regulating GnRH neuron activity poses two important questions: what is the impact that genes expressed in such diverse cell populations may have on the initiation of puberty, and what are the mechanisms providing dynamic coordination to genetic networks that – operating within this diversity of cellular phenotypes – contribute to the central control of the pubertal process. The availability of new tools to explore the human genome has facilitated the identification of several genes that are essential for puberty to take place. They include GNRHR, which is necessary for pituitary gonadotrophs to respond to GnRH because it encodes the GnRH receptor (Bedecarrats and Kaiser, 2007), LEP, the gene encoding leptin, a cytokinine produced by adipocytes (Strobel et al., 1998) that is essential not only for the regulation of energy homeostasis, but also for the initiation of puberty (Ahima et al., 2000; Elias, 2012), and LEPR (encoding the leptin receptor) (Clement et al., 1998). Mutations affecting genes that have a primary role in regulating hypothalamic GnRH release include mutations in KISS1R (encoding the kisspeptin receptor) (Seminara et al., 2003; de Roux et al., 2003), KiSS1 (encoding kisspeptins) (Lapatto et al., 2007; Topaloglu et al., 2012), TAC3 (encoding neurokinin B, NKB), and TAC3R (encoding the NKB receptor) (Topaloglu et al., 2008). Others genes are required for GnRH neuron migration [reviewed in Sykiotis et al. (2010)]. More recently, two mutations causing premature puberty, instead of pubertal failure, have been described. One of these mutations results in the constitutive activation of KISS1R (Teles et al., 2008); the other appears to involve loss of an inhibitory input, because it involves inactivating mutations of MKRN3, which encodes a protein likely involved in the inhibitory control of puberty (Abreu et al., 2013). Despite the importance of this information, the fact that known gene mutations affecting puberty account for only a small percentage (less than 2%) of individuals with pubertal disorders, and the demonstration that sequence variations in more than 40 genes are associated with an early age at menarche (Ong et al., 2009; Perry et al., 2009; Sulem et al., 2009; He et al., 2009; Elks et al., 2010; Cousminer et al., 2013; Tanikawa et al., 2013), suggest that puberty is not an event triggered by a single gene. Instead, it appears to involve a diversity of genes, which – based on studies in animal models – have been postulated to be organized into functionally modules wired into larger gene networks (Lomniczi et al., 2013; Ojeda et al., 2006).
Even if the notion of many genes contributing to the pubertal process is accepted at face value, gene diversity does not explain how inherited, permanent changes in DNA sequence can regulate gene expression dynamically, while also imposing an encompassing level of coordination and transcriptional plasticity to gene sets controlling female reproductive development. In this article we will develop the concept that a biological regulatory system able to perform these functions is epigenetics – i.e., those heritable changes in gene expression that occur without changing the primary nucleotide sequence of a gene (Wolffe and Matzke, 1999; Herman and Baylin, 2003). Epigenetic mechanisms can not only provide gene-specific gatekeeper functions (Garcia-Bassets et al., 2007), but are also endowed with an unsuspected degree of plasticity able to transiently change gene expression within hours (Miller and Sweatt, 2007), and even minutes (Kangaspeska et al., 2008; Metivier et al., 2008). It is now clear that epigenetic information is also essential for a variety of neural functions, including memory formation (Miller and Sweatt, 2007), recovery of learning and memory (Fischer et al., 2007), dendritic development (Wu et al., 2007), neuronal and behavioral plasticity (Kumar et al., 2005), estrogen-induced gene expression (Perillo et al., 2008; Subramanian et al., 2008), glial–neuronal interactions (Shen et al., 2008), circadian rhythms (Nakahata et al., 2008; Bellet and Sassone-Corsi, 2010), and sexual differentiation of the brain (McCarthy et al., 2009; Semaan et al., 2012).
2. Neuronal circuits controlling LH release at puberty
As indicated earlier, the transysnaptic control of GnRH neurons is dual, that is, effected by counteracting excitatory and inhibitory inputs. A substantial fraction of the excitatory transsynaptic input to GnRH neurons is provided by glutamatergic neurons (Ojeda and Skinner, 2006; Plant and Witchel, 2006), but a more powerful – and anatomically discrete – neuronal system stimulating GnRH release is provided by hypothalamic neurons that secrete a set of four biologically active peptides known as kisspeptins (Oakley et al., 2009; d’Anglemont et al., 2010). These peptides result from proteolytic processing of a kisspeptin precursor that is the product of the KISS1/Kiss1 gene (Ohtaki et al., 2001; Kotani et al., 2001). All kiss-peptins are potent stimulators of GnRH release (Oakley et al., 2009; Shahab et al., 2005). The critical importance of these peptides for puberty was demonstrated 10 years ago by studies in humans showing that loss of function of GPR54/KISS1R, the gene encoding the kisspeptin receptor, results in pubertal failure (Seminara et al., 2003; de Roux et al., 2003).
Opposing this excitatory influence, there are three main neuronal subsets providing inhibitory transsynaptic regulation to GnRH neurons (Fig. 1): opiatergic [reviewed in Terasawa and Fernandez (2001)], RFamide-related peptide (RFRP)-containing neurons (Tsutsui et al., 2010), and GABAergic neurons (Terasawa and Fernandez, 2001; Herbison and Moenter, 2011).
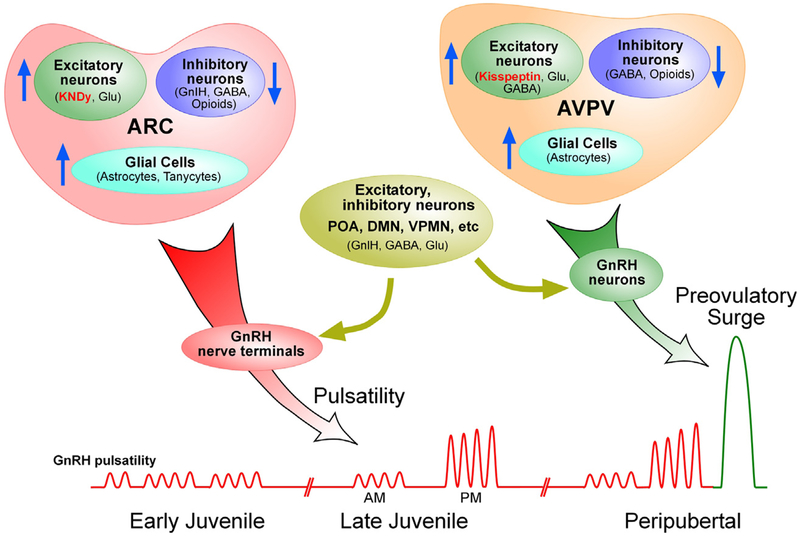
Fig. 1. The hypothalamic control of pulsatile and surge LH release.
The hypothalamic control of puberty involves excitatory and inhibitory transsynaptic inputs to GnRH neurons, in addition to facilitatory glia-to-neuron signaling. According to this concept, the initiation of puberty involves a shift from a predominantly inhibitory (shown by downward arrows) to an excitatory mode of control (upward arrows). This shift results in diurnal activation of pulsatile GnRH release, which leads to increased LH pulsatility, the first endocrine manifestation of puberty. The change in pulsatile GnRH release results from activation of excitatory networks (neuronal and glial) operating in the ARC of the hypothalamus, with KNDy neurons playing a central role. The neuronal and glial systems involved appear to predominantly target GnRH nerve terminals at the median eminence. The preovulatory surge of gonadotropins is a later event at puberty and is triggered by activation of AVPV kisspeptin neurons responding to an elevation in circulating estrogen levels. The potential involvement of other excitatory neurons, such as those that use glutamate (Glu) and GABA acting via GABAA receptors for neurotransmission, is also indicated. However not all the excitatory or inhibitory systems regulating pulsatile GnRH release are located in the ARC or AVPV. Additional inhibitory neurons, such as those releasing GnIH are located in the dorsomedial nucleus (DMN), and groups of excitatory/inhibitory neurons are located in the medial preoptic area (POA), medial amygdala and ventral premammillary nucleus (VPMN).
Opiatergic neurons inhibit GnRH neuronal activity by releasing different peptides that bind to specific cell membrane receptors (Kordon et al., 1994) located both on GnRH neurons (Dudas and Merchenthaler, 2006) and on neurons controlling GnRH secretion (Ojeda and Skinner, 2006; Terasawa and Fernandez, 2001). A prominent example of this latter type of interaction is found in the ARC. In this region of the hypothalamus, kisspeptin neurons produce the opioid peptide dynorphin, which inhibits GnRH secretion at least in part by repressing kisspeptin release via a paracrine/autocrine type of interaction (Navarro et al., 2009).
RFRP is the mammalian ortholog of the peptide gonadotropin-inhibiting hormone (GnIH), first described in birds (Ebling and Luckman, 2008). RFRP neurons use the peptides RFRP1 and RFRP3 for transsynaptic communication. Both peptides are recognized by a high-affinity receptor termed GPR147 or NPFFR1 (Tsutsui et al., 2010; Hinuma et al., 2000), and a low-affinity receptor termed GPR74 or NPFFR2 (Fukusumi et al., 2006). GPR147 is expressed in GnRH neurons (Ducret et al., 2009; Poling et al., 2012), suggesting that RFRP neurons can act directly on GnRH neurons to inhibit GnRH secretion.
The actions of GABAergic neurons on the GnRH neuronal network are more complex. GABA inhibits GnRH secretion via both GABAA and GABAB receptors expressed on neurons connected to the GnRH neuronal network (Ojeda and Skinner, 2006; Terasawa and Fernandez, 2001; Liu and Herbison, 2011), and via GABAB receptors located on GnRH neurons (Liu and Herbison, 2011). Despite these inhibitory actions, GABAergic neurons also excite GnRH neurons directly via activation of GABAA receptors (Herbison and Moenter, 2011).
Finally, not only neurons but also glial cells contribute to the hypothalamic control of puberty (Fig. 1) [reviewed in Prevot (2002), Lomniczi and Ojeda (2009)]. A series of studies have shown that astrocytes and ependymoglial cells lining the ventro-lateral surface of the third ventricle (known as tanycytes) facilitate GnRH secretion by both releasing growth factors and small molecules (such as ATP and prostaglandin E2, PGE2) and via cell–cell adhesive interactions (Prevot, 2002; Lomniczi and Ojeda, 2009; Clasadonte et al., 2011). While the former way of communication is exerted by diffusible factors, glial-GnRH neuron adhesive communication requires direct cell–cell contact and involves adhesion molecules with unique structural features that allow them to set in motion bidirectional intracellular signaling (Lomniczi and Ojeda, 2009). These molecules include sialylated neural cell adhesion molecule NCAM (PSA-NCAM) Parkash and Kaur, 2005; Perera et al., 1993, Synaptic Cell Adhesion Molecule 1 (SynCAM1) Sandau et al., 2011a,b, and Receptor-like Protein Tyrosine Phosphatase-β (RPTPβ) Parent et al., 2007.
2.1. Neuronal circuits required for pulsatile LH release
More than 40 years ago Boyar et al. (1972) introduced the now widely accepted concept that an increase in pulsatile LH release is the first endocrine manifestation of the initiation of puberty. This study, performed in humans, demonstrated that the amplitude of LH pulses detected in the blood stream increases at night during the juvenile-early puberty transition. Subsequent studies showed that a diurnal increase in pulsatile LH heralds the onset of puberty in other mammalian species, including rodents, sheep and nonhuman primates [reviewed in Ojeda and Skinner (2006)]. In rodents, the pubertal increase in LH pulsatility takes place in the afternoon, and at the end of late juvenile development, i.e., around postnatal day (PND) 28–30 Urbanski and Ojeda, 1985 (Fig. 1). A primary transsynaptic mechanism underlying pulsatile GnRH release is thought to be the synchronized activity of a subset of neurons located in the arcuate nucleus (ARC) of the hypothalamus, called KNDy neurons (Navarro et al., 2011; Lehman et al., 2010) (Fig. 1). They received this name because they produce kisspeptin, NKB and dynorphin (Navarro et al., 2011; Wakabayashi et al., 2010). KNDy neurons release NKB, which acts on other KNDy neurons via specific receptors to stimulate kisspeptin release (Navarro et al., 2011; Wakabayashi et al., 2010). NKB and kisspeptin are released periodically, and this oscillatory behavior is determined by a phase-delayed inhibitory feedback of dynorphin on NKB release (Navarro et al., 2011; Wakabayashi et al., 2010). Direct evidence for a role of KNDy neurons in the genesis of pulsatile LH release in rodents was recently provided (Beale et al., 2014). In addition to KNDy neurons, it is also likely that pulsatile GnRH release is regulated (directly or indirectly) by glutamatergic, GABAergic, opioid and RFRP neurons (Fig. 1).
2.2. Neuronal circuits required for the preovulatory LH surge
In addition to KNDy neurons of the ARC (Shahab et al., 2005; Clarkson et al., 2009; Gottsch et al., 2004), there is another population of kisspeptin neurons located in the anteroventral periventricular nucleus (AVPV) of rodents Clarkson et al., 2009; Gottsch et al., 2004 and the rostral periventricular area of both humans and rodents (Clarkson et al., 2009; Gottsch et al., 2004; Hrabovszky et al., 2010). A similar distribution has been observed in the ovine brain (Pompolo et al., 2006). These neurons contain neither dynorphin nor NKB and do not contribute to the control of pulsatile GnRH release. Instead, AVPV neurons are required for the pre-ovulatory surge of gonadotropins (Pinilla et al., 2012)( Fig. 1). Accordingly, they do not appear to be involved in the initiation of female puberty, because the gonadotropin surge occurs only after the pubertal process is well under way. As in the case of ARC-dependent pulsatile GnRH release, the surge mode of GnRH release may be regulated by additional excitatory neuronal systems (glutamatergic neurons binding to kainate and NMDA receptors and GABA neurons operating via GABAA receptors located on GnRH neurons) and by inhibitory neurons, including GABA neurons operating via GABAB receptors, opioid neurons operating via multiple receptors, and RFRP neurons using GPR147 receptors for transsynaptic inhibition (Fig. 1).
3. Gene networks and the neuroendocrine control of puberty
The concept of a diversity of genes affecting the time of puberty implies that they may be functionally organized into networks able to generate a coordinated output of biological signals. One such network operating in the peripubertal hypothalamus of rats and monkeys was postulated to contain genes that despite of having diverse cellular functions, were earlier identified as being involved in tumor suppression/tumor formation (Roth et al., 2007). In the hypothalamus, these genes, (referred to as Tumor Related Genes, TRGs), are predicted to be organized into a network structure containing “central” nodes that reside at the heart of the network, and a host of more peripherally located subordinate genes that are controlled transcriptionally by the central nodes. Although subordinate genes can act both as conduits of network output and inlets for incoming information, central nodes can also be directly influenced by outside inputs [reviewed in Lomniczi et al. (2013)]. The TRG network has five putative central nodes (CDP/CUTL1/CUX1, MAF, p53, YY1, and USF2), also called hubs. They are not only strongly connected to each other but also to additional upper-echelon genes (OCT2, TTF1, and EAP1) postulated to be involved in the transcriptional regulation of the pubertal process (Ojeda et al., 1999; Mastronardi et al., 2006; Heger et al., 2007), and that are themselves TRGs (Lomniczi et al., 2013).
One of these upper echelon genes (EAP1) and one predicted subordinate gene (KiSS1), deserve special mention, because their predicted regulation by central TRG nodes has been experimentally verified (Mueller et al., 2011, 2012). KISS1 was previously known as a metastasis suppressor gene (Steeg et al., 2003), and EAP1 was recently shown to be part of a transcriptional repressive complex that modulates apoptosis in breast cancer (Yeung et al., 2011). SynCAM1 (previously known as tumor suppressor of lung cancer, TSLC1) is another interesting TRG because of its pivotal role in the developmental control of glia-GnRH neuron adhesive communication, and in the mechanism by which astrocytic erbB4 receptors facilitate GnRH release and control normal female reproductive function (Sandau et al., 2011a,b).
More recently another puberty transcriptional gene network was reported in cattle (Fortes et al., 2011). Although the central nodes of this network are different from the TRG central nodes, they appear to regulate similar target genes, such as NELL2, NRG1 (which encodes an erbB4 ligand), and genes encoding adhesive molecules involved in cell–cell communication (such as SynCAM1). Interestingly, one of the transcription factors most heavily connected (top 10%) in the bovine puberty gene network is Mll3, a trithorax gene that appears to be involved in the epigenetic regulation of puberty (see Section 5 below). By doing a connectivity analysis of the core components of each network we noticed that seven of the most heavily connected TFs in the bovine network are also first neighbors of the TRG central nodes, with MLL3 connected to CUX1/CUTL1, EAP1 and YY1 (Fig. 2A).
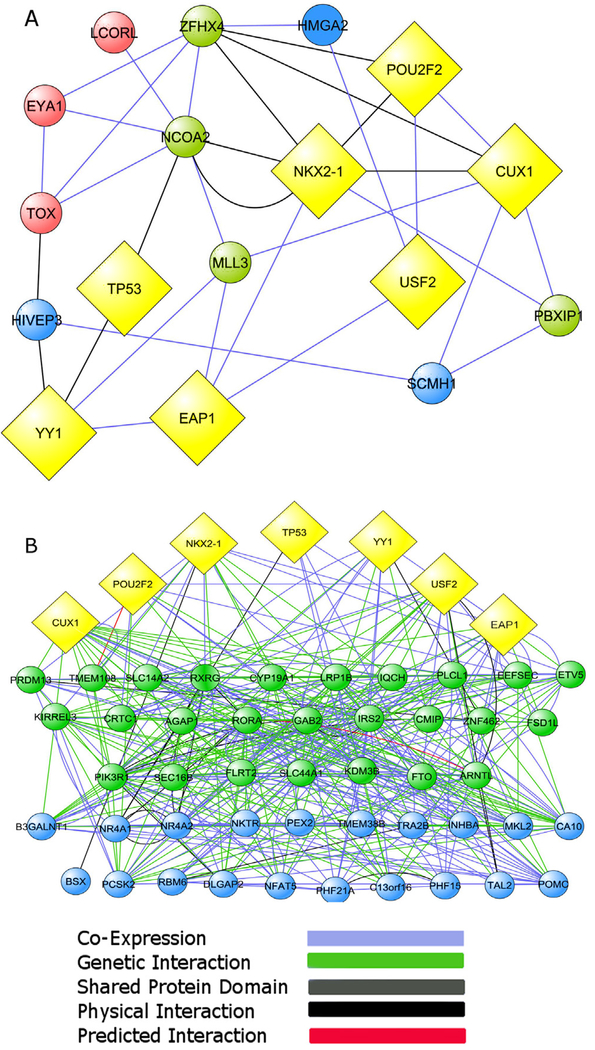
Fig. 2. Network connectivity of the most interconnected bovine puberty genes and human menarche-related genes to the central nodes of a TRG network derived from rats and nonhuman primates.
(A) The ten most connected genes of a bovine puberty gene network (Fortes et al., 2011) are first neighbors of the TRG central nodes (depicted as yellow diamonds). (B) Genes identified by GWAS as associated to the age of human menarche (Ong et al., 2009; Perry et al., 2009; Sulem et al., 2009; He et al., 2009; Elks et al., 2010; Cousminer et al., 2013; Tanikawa et al., 2013) are also highly connected to both the five original TRG central nodes (Roth et al., 2007 and to the upper-echelon transcriptional regulators TTF1/NKX2.1 and EAP1/IRF2BPL (yellow diamonds). Bovine puberty genes and menarche-related genes connected to multiple TRGs are depicted as green circles. Genes connected to at least one TRGs are shown as blue circles. Genes not connected to any TRG are represented as red circles. In both cases the connectivity is via co-expression (blue), predicted interaction (red), shared protein domain (gray), physical interactions (black), and genetic interaction (green) indicated by the GeneMANIA network construction algorithm. The thickness of each line indicates the strength of the evidence supporting that type of interaction in the Gene MANIA database. The distribution of nodes in B does not reflect a hierarchical distribution; instead it intends to emphasize the different degrees of connectivity that exists between central TRGs and menarche-related genes.
The draft model of these networks is bound to be modified as new information becomes available and as more gene interactions are elucidated. For instance, the cohort of genes shown to be associated with an earlier onset of menarche in humans (Ong et al., 2009; Perry et al., 2009; Sulem et al., 2009; He et al., 2009; Elks et al., 2010) is now known to be connected to EAP1 and TTF1 (Cukier et al., 2013). Moreover, a recent genome wide association study (GWAS) in a Japanese population showed that a SNP near the TTF1 locus (most commonly known as Nkx2.1) is significantly associated to early menarche (Tanikawa et al., 2013). Another recent study revealed five additional loci linked to pubertal timing (Cousminer et al., 2013). Even more interesting, analysis of the potential relationship that the five TRG central nodes, in addition to TTF1 and EAP1, may have with menarche-related gene using the GeneMANIA (Warde-Farley et al., 2010) network inference software and literature database indicates that the TRG central nodes are connected to as many as 22 of the menarche-related genes through co-expression or genetic interaction relationships (Fig. 2B). Moreover, 26 menarche related genes are connected to two or more TRG central nodes. Many of the genes that are connected to multiple TRG genes are themselves potential regulators of gene expression, such as KDM3B (also known as JMJD1B), a histone demethylase involved in removal of the repressive H3K9me2 and H3K9me1 histone marks (Kim et al., 2012; Krishnan et al., 2011). Overall, these results indicate the existence of significant interrelationships in regulation and expression between TRG central nodes, bovine puberty genes, and menarche-related genes discovered through GWAS studies.
In addition to the transcriptional repressors operating with the TRG network (YY1, EAP1, CUX1), there is a post-transcriptional repressor system that may contribute to controlling the timing of puberty. A central node of this system is LIN28b, which encodes an RNA binding protein that inhibits the maturation of let7 miRNAs (Lehrbach et al., 2009; Heo et al., 2009; Hagan et al., 2009), a family of microRNAs with tumor suppressor activity (Chang et al., 2009). The potential contribution of LIN28b to the regulation of puberty was suggested by the finding that a single nucleotide polymorphism near the LIN28B gene in human chromosome 6(q21) is associated with earlier puberty and shorter stature in girls (Ong et al., 2009; Perry et al., 2009; Sulem et al., 2009). In rats, the expression of Lin28b has been shown to decrease in the hypothalamus during prepubertal development in both males and females, a change that coincides with an increase in the abundance of let7a and let7b, two well-known LIN28b targets (Sangiao-Alvarellos et al., 2013). These opposite changes suggest that as the repressive effect of LIN28b on let7a and let7b maturation subsides, degradation of let7 target genes increases due to increased availability of these miRNAs. Should a predominance of these target genes be inhibitory to puberty, the net outcome would be expected to be an advancement of the timing of puberty.
4. Modes of epigenetic regulation
There are two well-established, and one emerging mechanism of epigenetic control. The former include: (a) chemical modifications of the DNA via DNA methylation and hydroxymethylation, and (b) modifications of chromatin structure caused by posttranslational modifications (PTMs) of histone proteins that, wrapped around by two superhelical turns of DNA, make up the nucleosome, the core unit of chromatin. The third and most recently unveiled mechanism of epigenetic control is exerted by non-coding RNAs, which provide epigenetic information as either microRNAs (miRNAs) or as long intergenic noncoding RNAs (lincRNAs).
4.1. DNA methylation
DNA methylation and hydroxymethylation are two covalent modifications of cytosine residues that mostly targets the dinucleotide sequence CpG (Jaenisch and Bird, 2003; Bjornsson et al., 2004). DNA methylation consists in the addition of a methyl group to position 5 of cytosine, resulting in the formation of 5-methylcytosine (5-mC) (Fig. 3A). Oxidation of 5-mC by the TET family of dio-oxygenase enzymes yields 5-hydroxymethylcytosine (5hmC) Tahiliani et al., 2009; Koh et al., 2011. In general, increased DNA methylation (5-mC) is associated with gene repression, and hypomethylation (less 5-mC, more 5-hmC) is associated with transcriptional activation. The balance of 5-mC and 5-hmC at a given genomic region depends on the activity of DNA methyltransferases that generate 5-mC, and the TET enzymes that catalyze the conversion of 5-mC to 5-hmC (Fig. 3A). The methyltransferases involved are DNA methyltransferase1 (DNMT1), which maintains basal levels of DNA methylation, and DNMT3a and DNMT3b responsible for de novo methylation of both unmethylated and hemimethylated DNA (Jaenisch and Bird, 2003). Although 5-mC and 5-hmC co-exist throughout the genome, their relative abundance varies in different regions. Thus, 5hmC has been found to be associated with euchromatin (i.e., chromatin in the open state) and enriched in promoter regions of active genes, whereas 5-mC displays an opposite pattern (Ficz et al., 2011).
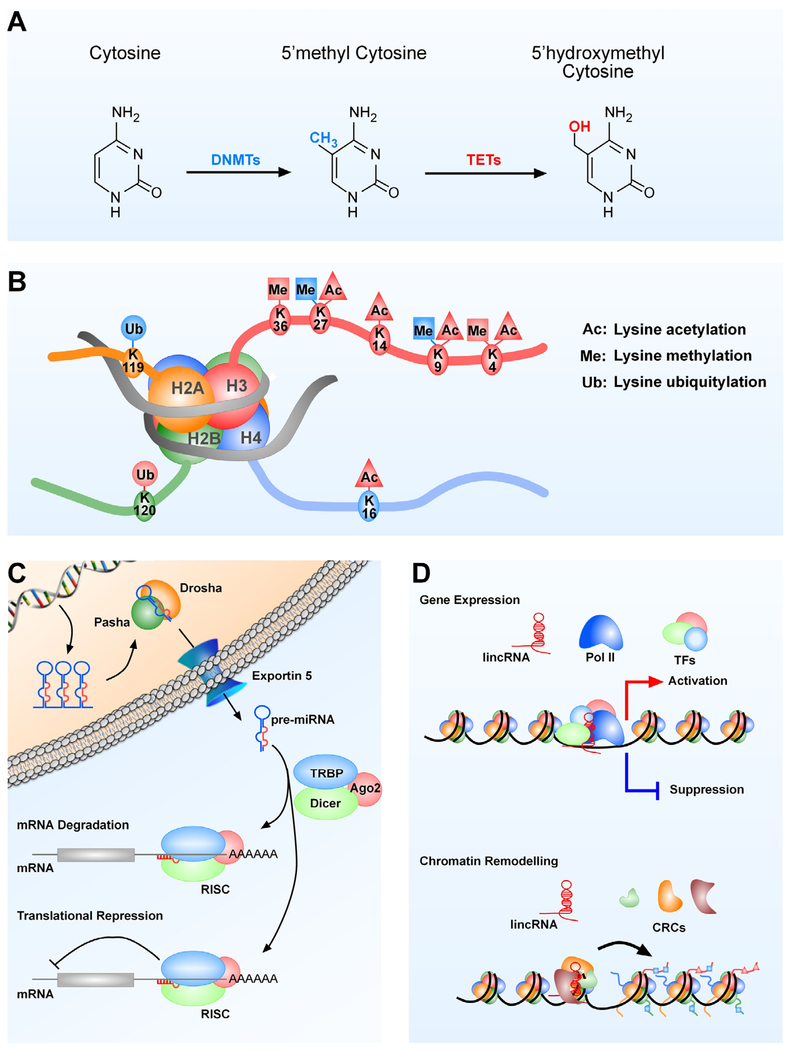
Fig. 3. Modes of epigenetic regulation.
(A) DNA methylation. Methylation of cytosine at position 5 is carried out by DNMTs (DNMT1, DNMT3a and DNMT3b), and inactivation of this methyl group by hydroxymethylation is carried out by the TET enzymes. Names in blue color indicate repression and red color indicates activation of gene expression. (B) Histone PTMs. Only the PTMs catalyzed by the PcG and TrxG complexes (methylation, ubiquitination) or associated (acetylation) with TrxG-dependent PTMs are shown. Histone PTM in blue = PTM associated with gene repression; histone PTM in red = PTM associated with gene activation. (C) miRNAs. The pathway leading to miRNA production is outlined and the fact that miRNAs silence mRNA expression by inducing either mRNA degradation or translational repression is emphasized. (D) Long intergenic noncoding RNAs. Two mechanisms of lincRNA action are depicted. In one of them, lincRNAs modify gene expression by serving as landing pads for transcription factors that either repress or activate transcription. The other mechanism consists of lincRNAs directing the organization of chromatin states to specific genomic regions involved in gene regulation. DNMTs = DNA methyltransferases, TETs = ten eleven translocation (dioxygenase) enzymes; H = histone; Pasha = nuclear protein that is part of the microprocessor complex required for miRNA processing. Pasha associates with the RNA III enzyme Drosha. Drosha = RNA III enzyme that cleaves pri-miRNA (the primary transcript of miRNAs) to precursor (pre)-miRNA, which contains a stem-loop structure; Dicer = endoribonuclease that cleaves pre-miRNA into 20–25 mer double-stranded miRNAs; TRBP = human immunodeficiency virus transactivating response RNA-binding protein; it recruits Dicer to Ago2 for miRNA processing; Ago2 = Argonaut 2, the catalytic component of RISC; RISC = RNA induced silencing complex; Pol II = RNA polymerase 2; TF = transcription factor; CRC = chromatin remodeling complex.
4.2. Histone post-translational modifications
Histone PTMs alter the N-terminus tails of the four histones (H2A, H2B, H3, and H4) that make up the protein core of nucleosomes (Kouzarides, 2007; Khorasanizadeh, 2004) (Fig. 3B). These modifications include acetylation, methylation, phosphorylation, ubiquitination, and sumoylation (Kouzarides, 2007; Khorasanizadeh, 2004). Acetylation by acetyltransferases (HATs), deacetylation by histone deacetylases (HDACs), and methylation by methyltransferases (HMTs) are perhaps the best well-characterized. As in the case of DNA methylation, some generalizations can be made: acetylation is associated with activation of transcription; deacetylation with gene silencing (Kouzarides, 2007; Khorasanizadeh, 2004). The functional consequences of the methylation status of histones are more complex. For instance, H3 methylation of lysine 9 and 27 (H3K9me and H3K27me) is usually seen in silenced genes (Wang et al., 2008; Ruthenburg et al., 2007) whereas trimethylation of histone 3 (H3) at lysine 4 (H3K4me3) is a feature of active transcription (Wang et al., 2008; Berger, 2007).
Importantly, changes in DNA methylation and histone modifications are not dissociated events as they usually work in sync to regulate gene expression (Cameron et al., 1999; Cedar and Bergman, 2009). For instance, H3K4me2/3 prevent DNA methylation (Cedar and Bergman, 2009; Ooi et al., 2007), and H3K9me facilitates DNMT3a and DNMTb recruitment to target genes (Cedar and Bergman, 2009). DNMTs do not work in isolation either; upon recruitment, they associate with HDACs bringing about gene silencing (Rountree et al., 2000; Robertson et al., 2000; Burgers et al., 2002). In addition to enzymes instituting these histone modifications, there is a similarly complex set of enzymes that reverse them (Kouzarides, 2007; Borrelli et al., 2008). Together, “writers” and “erasers” (Borrelli et al., 2008) allow epigenetic “readers” (such as PRC1, a subcomplex of the Polycomb group [PcG] of transcriptional repressors, see below) to regulate gene activity with a degree of flexibility and developmental plasticity not provided by DNA sequence. Because complex biological processes require integrative mechanisms of gene regulation displaying these attributes, it would appear logical to assume that the role of epigenetics in the regulation of puberty is substantial.
4.3. Noncoding RNAs
A series of recent studies have revealed that most of the human genome is transcribed into noncoding RNAs (ncRNAs), instead of protein-encoding messenger (m)RNAs Gruber and Zavolan, 2013; Batista and Chang, 2013. Non coding RNAs are divided into two main groups: small RNAs (sRNAs) and long noncoding RNAs (lncRNAs). While small RNAs are 20–30 nucleotides in length, lncRNAs are longer than 200 nucleotides (Batista and Chang, 2013; Filipowicz et al., 2008). There are three classes of sRNAs: microRNAs (miRNAs) Filipowicz et al., 2008, endo-small inhibitory RNAs (endo siRNAs) Okamura et al., 2008, and piwiRNAs (piRNAs) Kim, 2006, all of them involved in epigenetic silencing (Huang et al., 2014). Among them, the best known in terms of epigenetic regulation are miRNAs (Gruber and Zavolan, 2013) (Fig. 3C). They target a variety of mRNAs encoding proteins required for both DNA methylation and histone modifications (Gruber and Zavolan, 2013), including DNA methylation (DNMTs), DNA demethylation (TET1-3), histone 3 K27 trimethylation (EZH2), and histone deacetylation (HDAC1, 4, 6), among others (Gruber and Zavolan, 2013).
The contribution of lncRNAs to epigenetics is more complex. Although lncRNAs do not code for any protein, they are polyadenylated (Batista and Chang, 2013). Many of them are produced in gene-free regions of the genome; because of this, they are known as long intergenic noncoding RNAs (lincRNAs). LincRNAs bind to chromatin modifying complexes and guide these complexes to genomic regions involved in the control of gene expression (Spitale et al., 2011)( Fig. 3D). They appear to serve as scaffolds that bring together chromatin remodeling complexes targeting them to genomic regions involved in the control of gene expression (Batista and Chang, 2013; Spitale et al., 2011). Perhaps the most well-recognized lincRNA involved in epigenetic regulation is HOTAIR, which targets the silencing complexes PRC2 (a sub-group of the PcG complex of transcriptional repressors, see below) and LSD1-coREST complexes to the regulatory regions of downstream genes (Chu et al., 2011; Rinn et al., 2007). Because PRC2 catalyzes H3K27 trimethylation and LSD1-coREST demethylates H3K4me2, the net outcome of HOTAIR actions is gene silencing. Importantly, the very existence of lncRNAs is epigenetically determined as thousands of them have been identified in the genome by a chromatin signature consisting of trimethylation of histone 3 at lysine 4 (H3K4me3) at the promoter region of RNA polymerase II-transcribed genes accompanied by the presence of histone 3 trimethylated at lysine 36 (H3K36m3) along the transcribed region, resulting in a chromatin configuration known as the K4-K36 bivalency (Guttman et al., 2009).
4.4. Epigenetic regulation of distal enhancer regions
A hot new topic in epigenetic research is the existence of distal enhancers regions that regulate gene expression by looping from sites in the noncoding genome to either the promoter region of genes to regulate gene transcription, or to intronic sequences within the gene to regulate transcriptional elongation (Stadhouders et al., 2012). Distal enhancers are usually located thousands of base pairs away from the gene they regulate and their activity is defined by the presence of two different chromatin signatures. Active enhancers are marked by the simultaneous presence of histone 3 acetylated at lysine 27 (H3K27ac), histone 3 monomethylated at lysine 4 (H3K4me1), and polymerase II (Stadhouders et al., 2012; Calo and Wysocka, 2013). The “writers” of this epigenetic signature are proteins of the Trithorax group (TrxG) of epigenetic activators (Herz et al., 2012; Tie et al., 2014; Hu et al., 2013) (see below). Latent or “poised” enhancers lack polymerase II; they also contain H3K4me1, but instead of H3K27ac, they contain histone 3 trimethylated at lysine 27 (H3K27me3), a modification catalyzed by the PRC2 sub-complex of PcG proteins (see below) Herz et al., 2012; Tie et al., 2014; Hu et al., 2013.
It is becoming rapidly apparent that distal enhancers play a very important role in the regulation of gene transcription. They may not only be switched from latent to active by incoming stimuli (Ostuni et al., 2013), but can also be affected by common sequence variation. A striking example of distal enhancers influencing a phenotypic outcome was recently reported by Smemo et al. (2014), who demonstrated that intronic regions in the FTO locus known to be associated with risk of obesity in humans directly influence expression of the IRX3 homeobox gene through a chromatin looping mechanism. Evidence for the role of IRX3 in obesity was provided by the finding that Irx3-deficient mice exhibit significant loss of body weight and are resistant to a regime of high-fat diet (Smemo et al., 2014). Interestingly, FTO has also been shown to be associated with polymorphisms influencing age at menarche and appears to be coexpressed with TP53 and YY1 (Fig. 2). While the precise mechanism by which sequence variations, such as single nucleotide polymorphisms (SNPs) modify the transcriptional output of target genes is not clear in all cases, one compelling hypothesis suggests that many such variations alter the ability of transcription factors to bind to the enhancer, and therefore alter the ability of the affected enhancer to regulate gene expression (Smemo et al., 2014; Andersson et al., 2014).
There is good reason to believe that regulatory mechanisms underlying the neuroendocrine control of puberty involve input from distal enhancer regions. Recent evidence reported by the FANTOM 5 consortium indicates that SNPs in linkage with variants associated with phenotypic outcomes are enriched in regions identified as distal enhancers (Andersson et al., 2014). This analysis included several of the SNPs described as influencing the age at menarche (Elks et al., 2010).
5. An epigenetic mechanism of cellular memory: the counteracting role of Polycomb group (PcG) and Trithorax group (TrxG) proteins
It is now well established that PcG and TrxG proteins play a major, evolutionary conserved, role in the epigenetic control of gene expression. It is also clear that they have mutually antagonistic activities, and that their interplay at genomic regulatory regions provides the cell with a mitotically heritable “memory” of which genes are silenced and which are active throughout development (Schuettengruber et al., 2011; Diand Helin, 2013; Shilatifard, 2012).
5.1. The Polycomb group complex: an epigenetic silencer
In mammals the PcG system is composed of two repressive complexes (termed Polycomb Repressive Complexes, PRC1 and PRC2) (Fortes et al., 2011; Simon and Kingston, 2009) (Fig. 4). There are two types of PRC1 complexes (Schwartz and Pirrotta, 2013; Blackledge et al., 2014). Canonical PRC1 complexes contain a catalytic core consisting of one of two proteins, RING1 or RING2 (also known as RING1A and RING1B), which have E3 ubiquitin protein ligase activity, a combination of at least five Pc (Polycomb) proteins known as chromobox proteins (CBX2, CBX4, CBX6, CBX7 and CBX8), because they contain a conserved chromodomain (CBX) at their amino terminus, two Psc (posterior sex comb) proteins (BMI1, also known as PCGF4, for Polycomb group RING finger protein 4), and MEL18 (also known as PCGF2), in addition to three polyhomeotic-like proteins (PHC1-3). Although there are several mammalian CBX proteins (CBX2, 4, 6, 7, and 8) (Schwartz and Pirrotta, 2013), different CBX genes are expressed in different cells (Otte and Kwaks, 2003). Non canonical or variant PRC1 complexes lack CBX proteins; instead, RING1 or RING2 form a complex with either RYBP (RING1 and YY1 binding protein) or YAF2 (YY1-associated factor) and one of four PCGF proteins (PCGF1, 3, 5 or 6) (Gao et al., 2012). The complex containing RYBP was shown to be critical for PRC1-dependent mono-ubiquitylation of histone 2A at lysine 119 (Blackledge et al., 2014; Tavares et al., 2012). This modification (H2AK119ub1) appears to inhibit gene expression by both suppressing RNA polymerase activity in bivalent promoters and preventing H3K4 methylation (Di and Helin, 2013), a feature of activated genes (Wang et al., 2008; Berger, 2007; Guttman et al., 2009). Surprisingly, variant PRC1-dependent H2A ubiquitylation was found to recruit PRC2 and H3K27me3 to PcG target promoters (Blackledge et al., 2014).
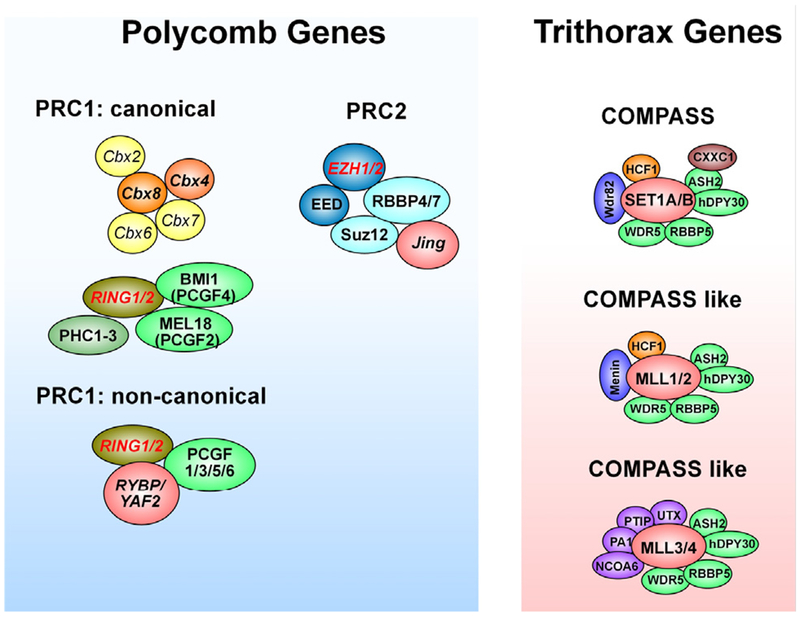
Fig. 4. Subunit composition of the PcG and COMPASS families.
The three major Polycomb repressive complexes (PRCs) are depicted. The PRC2 complex contains the histone methyltransferase enhancer of zeste homologue 1 (EZH1) or EZH2), which together with embryonic ectoderm development (EED) and suppressor of zeste 12 homolog (SUZ12) catalyzes the trimethylation of histone H3 at lysine K27 (H3K27me3). Multiple forms of the PRC1 complex exist. Canonical PRC1 complexes contain combinations of at least five Pc (Polycomb) proteins (known as chromobox proteins: CBX2, CBX4, CBX6, CBX7 and CBX8), two Psc (posterior sex comb) proteins (BMI1, also known as PCGF4 (polycomb group RING finger protein 4), MEL18 (also known as PCGF2) and one of two RING proteins, RING1 or RING 2 that provide the catalytic core to the complex because they have E3 ubiquitin ligase activity. In addition PRC1 complexes contain three polyhomeotic-like proteins (PHC1-3). Non canonical PRC1 complexes lack CBX proteins and contain instead a RING1 or RING2 protein that forms a complex with either RYBP (RING1 and YY1 binding protein) or YAF2 (YY1-associated factor) and one of four PCGF proteins different from BMI1 and MEL18 (PCGF1, 3, 5 or 6). The six known mammalian COMPASS complexes are also shown. Although all of them methylate H3K4 at lysine 4, different complexes are responsible for the mono, di or tri methylation of this amino acid (see text for details). Modified from Mohan et al. (2012).
The mammalian PRC2 complex contains a catalytic core consisting of one of two methyltransferases, enhancer of zeste 1 (EZH1) or EZH2, which trimethylate histone 3 at lysine 27. These subunits require two partners, suppressor of zeste (Suz12) and the WD40 domain protein EED, for catalytic activity (Di and Helin, 2013). In addition, PRC2 contains one of two histone binding proteins (RBBP4 or RBBP7) and the Zinc finger protein AEBP2 (Schwartz and Pirrotta, 2013).
5.2. The Trithorax group: an antagonist of epigenetic silencing
TrxG proteins counteract the effect of PcG proteins by implementing methylation of H3K4 (Schuettengruber et al., 2011; Shilatifard, 2012). In mammals, H3K4 methylation is mediated by six protein complexes termed COMPASS (Complex of Proteins associated to Set1) and COMPASS-like, because they are related to the original yeast SET1 methyltransferase (Schuettengruber et al., 2011; Shilatifard, 2012) (Fig. 4). Two of these complexes (SET1A and SET1B COMPASS) contain Drosophila SET1-related proteins; two (known as COMPASS-like) contain the proteins MLL1 or MLL2, which are related to Drosophila Trx (Trithorax), and the other two (also termed COMPASS-like) contain either MLL3 or MLL4, both of them related to Drosophila Trr (Trithorax-related). Recent studies have shown that SET1A/SET1B COMPASS mediates H3K4 trimethylation at promoters of active genes (Hu et al., 2013), MLL2 implements this PTM at bivalent promoters (Denissov et al., 2014; Wu et al., 2008), i.e. those promoters that are poised for activation (Bernstein et al., 2006), and MLL3/MLL4 catalyze monomethylation of H3K4 at enhancers sites (Hu et al., 2013).
6. Epigenetic regulation of early neuroendocrine reproductive development
6.1. Hypothalamic sexual differentiation
Estrogen-dependent sexual differentiation of the rodent preoptic area (POA) appears to be influenced by epigenetic modifications affecting either DNA methylation or the pattern of histone PTMs of the estrogen receptor alpha (ERα) gene (McCarthy and Nugent, 2013). While a portion of the ERα gene promoter was found to show a developmental pattern of DNA methylation that does not correlate with gene expression (Schwarz et al., 2010), DNA methylation of another portion of the promoter exhibited a good correlation (Kurian et al., 2010), suggesting that such changes are subtle and highly circumscribed to a limited number of CpG dinucleotides in specific segments of the ERα gene 5’flanking region. Histone PTMs appear to play a more decisive role than DNA methylation in the masculinization of the POA, as inhibition of HDAC activity targeted to this brain region of neonatal animals reduced adult male behavior (Matsuda et al., 2011). This study further demonstrated that HDAC2 and HDAC4, possibly associated with both the ERα and aromatase gene promoters, are required for male behavior. Whether other histone PTMs contribute to brain sexual differentiation is currently unknown.
A gene showing a striking sexually dimorphic expression in the AVPV of rodents is Kiss1 (Kauffman et al., 2009). Although Kiss1 expression is much greater in females than in males, DNA methylation of the Kiss1 promoter is greater in females than in males (Semaan et al., 2012). Though evidently counter intuitive, this difference may reflect the reported ability of DNA methylation to block the recruitment of transcriptional repressors under some conditions (Semaan et al., 2012).
6.2. GnRH neuronal maturation
The first evidence that GNRH expression may be under epigenetic control during development was provided by a study that used cultured GnRH neurons from nonhuman primates (Kurian et al., 2010). These investigators showed that methylation at 8 of 14 CpG sites in a region located about 2000 bp upstream from the GNRH transcription start site decreased in GnRH neurons coinciding with an increase in GnRH expression during in vitro embryonic development (Kurian et al., 2010). Because GnRH neurons derived from the nasal epithelium mature in vitro at about the same pace and time as GnRH neurons in vivo, these findings imply that demethylation of specific regions within the GNRH gene locus play a role in the in vivo activation of GNRH transcription. Importantly, this study opens the doors for additional investigation of the epigenetic mechanism regulating GnRH neuron biology not only during embryonic development, but also during the normal onset of puberty and under conditions that either advance or delay the pubertal process.
7. Epigenetic regulation of neuronal circuits controlling the onset of puberty
7.1. The kisspeptin gene and the preovulatory surge of gonadotropins
A role for E2 in the epigenetic control of AVPV kisspeptin neurons was recently demonstrated by Tomikawa and colleagues who showed that E2 increases acetylated H3 content at the Kiss1 promoter in the AVPV, but reduces acetylated H3 in the ARC (Tomikawa et al., 2012). Furthermore, E2 increased ERα binding to the Kiss1 promoter in the AVPV, but not the ARC. Tomikawa and colleagues did not detect changes in Kiss1 promoter DNA methylation in either the AVPV or ARC in response to estradiol, suggesting that DNA methylation may not be an epigenetic cue regulating Kiss1 promoter activity at the time of the preovulatory surge of gonadotropins. Lomniczi et al. (2013) made a similar observation when studying methylation of the Kiss1 promoter at the time of the pubertal increase in pulsatile LH secretion. Interestingly, there is an estrogen-responsive enhancer in the 3’-region of the Kiss1 gene, which operates in the AVPV, but not the ARC (Tomikawa et al., 2012), suggesting an epigenetic contribution to E2 positive feedback. Whether the inhibitory effect of estrogen on ARC Kiss1 expression also involves an epigenetic component remains to be elucidated.
7.2. The kisspeptin gene and the initiation of puberty
For years a prevailing view to explain the initiation of puberty assumed the existence of a pubertal “brake” (Grumbach and Styne, 1992). According to this notion, during the prepubertal period the secretory activity of GnRH neurons is under trans-synaptic inhibitory control. At puberty this inhibition would be lifted, resulting in increased GnRH release [reviewed in Terasawa (1999)]. A different, but not mutually exclusive, view is that puberty can only occur if there is activation of excitatory inputs (Ojeda, 1991). This latter concept was strongly supported by the demonstration that activation of kisspeptin neurons, which provide a significant fraction of the stimulatory inputs controlling GnRH neurons, is essential for puberty to occur [reviewed in Pinilla et al. (2012)]. Based on these and other observations the original concept has been refined to state that a concomitant decrease in inhibitory inputs and an increase in excitatory neurotransmission are the essential underpinnings of the pubertal process (Ojeda and Terasawa, 2002).
Notwithstanding the potential importance of this trans-synaptic interplay, very recent evidence suggests that the inhibitory and stimulatory control of puberty may not be exclusively provided by parallel cell–cell communication pathways impinging on GnRH neurons. Instead, a critical inhibitory/excitatory Yin-Yang mechanism regulating puberty appears to reside at a transcriptional level within neurons involved in stimulating GnRH release. The existence of a transcriptionally repressive mode controlling puberty-activating genes was initially suggested by the fact that some central nodes of the TRG network (YY1, EAP1, and the CUX isoform CUX1p120) can repress Kiss1 transcriptional activity (Heger et al., 2007; Mueller et al., 2011). Definitive proof was provided by the demonstration that the PcG silencing complex prevents the premature initiation of puberty by repressing the transcriptional activity of Kiss1 in KNDy neurons of the ARC (Lomniczi et al., 2013).
In this study the hypothalamus was interrogated via DNA arrays during the juvenile and peripubertal stages of female rat reproductive maturation. Three developmental phases were selected for inquiry: early juvenile (day 21); late juvenile (day 28), and the day of the first preovulatory surge of gonadotropins (days 30–36). Day 21 corresponds to the initiation of the juvenile period (Ojeda and Urbanski, 1994), day 28 to the beginning of puberty, and the first preovulatory surge of gonadotropins to the completion of puberty. Day 28 was considered as the approximated age when puberty is initiated in female rats, because it is at this time that a diurnal change in pulsatile LH release is first detected (Urbanski and Ojeda, 1985).
Because the Kiss1 and Tac2 genes are active within the same neurons, and their protein products are functionally connected (NKB stimulates kisspeptin release via TAC3R receptors) (Navarro et al., 2011; Wakabayashi et al., 2010), they have been postulated to be unique members of a class of “puberty-activating” genes (Lomniczi et al., 2013). Using the Kiss1 gene as a prototype, it was shown that two essential components of the PcG complex (Eed and Cbx7) are expressed in KNDy, and – importantly – that their encoded proteins are associated with the 5’ flanking region of the Kiss1 gene (Lomniczi et al., 2013). The results showed that at the end of juvenile development, methylation of the Eed and Cbx7 promoters increases in the ARC concurrent with decreased expression of both genes. These changes were accompanied by eviction of EED and CBX proteins from the Kiss1 promoter, and a notable change in the chromatin status of the promoter, which displayed increased levels of acetylated H3 (H3K9, 14ac) and trimethylated H3 (H3K4me3), two modifications associated with gene activation. As predicted by these changes, Kiss1 expression increased in the ARC at this time.
Unexpectedly, the content of H3K27me3, a histone modification catalyzed by PRC2 (Wang et al., 2008; Ruthenburg et al., 2007) decreased significantly later during peripubertal maturation, on the day of the first preovulatory surge of gonadotropins. This developmental pattern suggests that association of H3K27me3 and H3K4me3 to the Kiss1 promoter behaves as predicted by the hypothesis of bivalent domains (Bernstein et al., 2006), i.e., both marks co-exist in the regulatory region of genes “poised” for activation in response to incoming inputs (Bernstein et al., 2007).
The study of Lomniczi et al. (2013) also demonstrated that systemic administration of 5-azacytidine, an inhibitor of DNA methylation, prevented both the peripubertal decline in Cbx7 and Eed mRNA expression, and the eviction of CBX7 and EED from the Kiss1 promoter. In addition, it prevented the association of activating histone marks to the promoter, and the increase in Kiss1 expression that occur at end of juvenile development. These observations are consistent with the interpretation that if an increase in DNA methylation of PcG promoters is prevented, eviction of EED/CBX7 occupancy of the Kiss1 promoter fails to occur, and this diminishes the accessibility of activating histone marks to the promoter. The importance of PcG-mediated silencing for the timing of puberty was demonstrated by experiments in which EED was overexpressed in the ARC of early juvenile rats (via lentiviral-mediated delivery). In these animals, exogenous EED was recruited to the Kiss1 promoter, and Kiss1 expression was reduced as evidenced by the lower number of immunoreactive kisspeptin neurons in the ARC and a reduction in Kiss1 mRNA levels. Pulsatile GnRH release was blunted, puberty was delayed, and estrous cyclicity was disrupted. Importantly, the number of pups delivered by animals receiving EED in the ARC was markedly reduced suggesting that if the repressive influence of the PcG complex on KNDy neurons is maintained beyond juvenile days, fertility is compromised (Lomniczi et al., 2013).
The prepubertal increase in H3K4me3 abundance at the Kiss1 promoter is extremely interesting because it implies the recruitment of an activating complex concomitant to the loss of PcG inhibition. A likely candidate for this role is the TrxG complex because these proteins are known to antagonize PcG silencing by catalyzing H3K4 trimethylation and facilitating H3 acetylation (Shilatifard, 2012; Simon and Kingston, 2009). By doing so, the TrxG complex may provide the necessary trans-activational component to puberty-activating genes at the time when the inhibitory influence of repressive epigenetic information is waning. The potential importance of the TrxG complex in the control of puberty is suggested by the finding that inactivating mutations of CHD7, a chromatin remodeling protein that antagonizes PcG action by binding to H3K4me2 and H3K4me3 via its chromodomain, are associated with hypothalamic hypogonadism in humans (Bianco and Kaiser, 2009; Kim et al., 2008). Notwithstanding the potential importance of TrxG proteins as transcriptional activators of the pubertal process, the alternative contribution of other trans-activating protein complexes known to “read” the H3K4me3 mark needs consideration. A potential candidate for this role is the SAGA complex, which binds to the DNA of H3K4me2/3 containing promoters via the subunit Sgf29 (Vermeulen et al., 2010).
Another aspect made evident by the study of Lomniczi et al. is that the decrease in PcG expression that antedates the onset of puberty is not an estrogen (E2)-dependent phenomenon. Indeed, there are no canonical estrogen responsive elements (EREs) in PcG promoters. Moreover, E2 action is associated with gene activation instead of gene repression as evidenced by the ability of E2 to induce demethylation of DNA and loss of H3K9me2/3 from E2-target promoters (Kangaspeska et al., 2008; Metivier et al., 2008; Perillo et al., 2008). Although E2 may not be responsible for the dissociation of PcG proteins from the Kiss1 promoter in KNDy neurons, it appears to elicit epigenetic modifications affecting either other puberty-related genes or the Kiss1 gene itself expressed in AVPV kisspeptin neurons. This idea is supported by studies showing that E2 induces changes in H3 acetylation at the Kiss1 promoter in AVPV kisspeptin neurons (Tomikawa et al., 2012), triggers fluctuations in DNA methylation (Kangaspeska et al., 2008; Metivier et al., 2008), induces the formation of co-activating complexes containing histone acetyl-transferases and histone methyltransferases (Metivier et al., 2003), and enhances gene transcription by inducing demethylation of H3K9me2/3 (Garcia-Bassets et al., 2007; Perillo et al., 2008). Moreover, expression of ERα itself is regulated by DNA methylation (Issa et al., 1994; Westberry et al., 2008; Belinsky et al., 2002).
8. Environmental cues affecting the timing of puberty via epigenetic mechanisms
8.1. Nutrition
The influence of metabolic cues on female hypothalamic reproductive maturation was documented many years ago (Kennedy and Mitra, 1963; Frisch and Revelle, 1970; Frisch, 1980). Several metabolic signals have been postulated to play an important role in this process (Elias, 2012; Heger et al., 1999; Fernandez-Fernandez et al., 2006), and solid evidence has been provided that peripheral hormones exert these effects by modifying the activity of hypothalamic kisspeptin neurons. While leptin and IGF1 increase Kiss1 expression both directly and indirectly (Roa et al., 2010; Hiney et al., 2009), the gut-derived hormone, ghrelin, and the liver derived hormone FGF21, whose levels rise in fasting, repress the initiation of puberty (Tena-Sempere, 2008; Owen et al., 2013) by inhibiting Kiss1 mRNA expression in the ARC (Forbes et al., 2009) and AVPV (Owen et al., 2013), respectively.
There is a critical period during late gestation in humans and early postnatal life in rodents, in which the building blocks of energy homeostasis, including the hypothalamic regulation of food intake and energy balance, become established by a “developmental program” [reviewed in Remmers and Delemarre-van de Waal (2011), Gabory et al. (2011)]. If the availability of nutrients is increased or limited during this time, the program is irreversibly affected, resulting in persistent alterations in energy homeostasis and increased susceptibility to diabetes, cardiovascular and metabolic disease (Remmers and Delemarre-van de Waal, 2011; Grove et al., 2005).
Numerous reports (Kennedy and Mitra, 1963; Frisch and Revelle, 1970; Frisch et al., 1975; Ronnekleiv et al., 1978; Sloboda et al., 2009; Castellano et al., 2011; Wang et al., 2011) have also made clear that female puberty is delayed by early life nutritional challenge. A pivotal component of this developmental delay is the hypothalamic Kiss1 system. Kiss1 expression is reduced by early undernutrition (Castellano et al., 2005; True et al., 2011), and the connectivity of ARC kisspeptin neurons is impaired (Caron et al., 2012). Conversely, increasing nutrient availability during the critical period of nutritional programming advances puberty (Frisch et al., 1975; Castellano et al., 2011) and this advancement is associated with activation of the Kiss1 gene (Castellano et al., 2011).
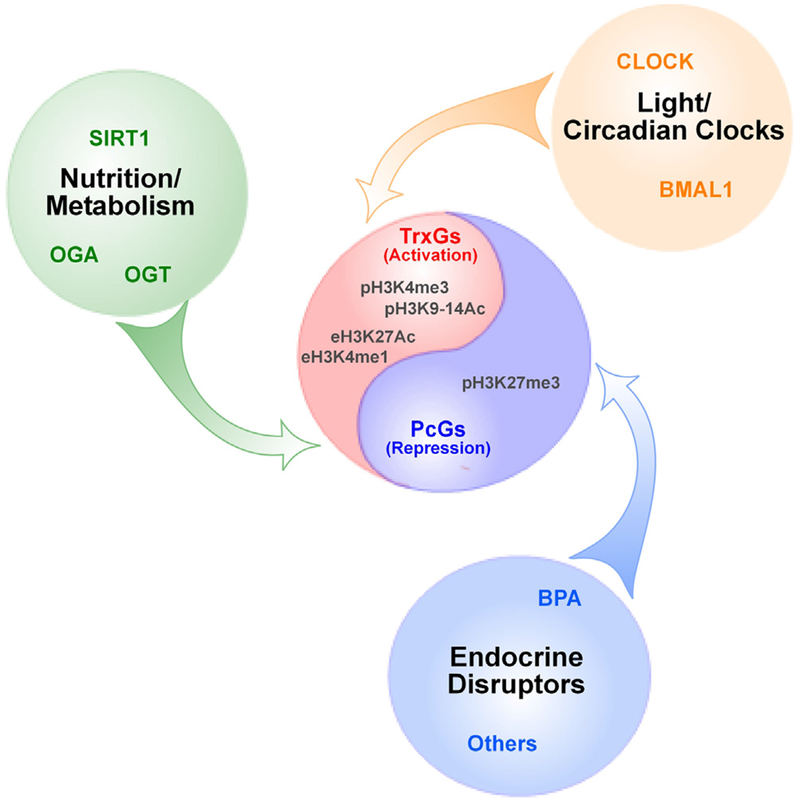
Epigenetics plays a central role in developmental programming (Bellet and Sassone-Corsi, 2010; Remmers and Delemarre-van de Waal, 2011; Gabory et al., 2011; Castellano et al., 2011), linking early-life alterations in energy balance to a variety of adult metabolic disorders (Remmers and Delemarre-van de Waal, 2011; Gabory et al., 2011; Vaquero and Reinberg, 2009; Choi and Friso, 2010). Among the major epigenetic modifications regulating transcriptional activity, chromatin remodeling stands out as a powerful means of epigenetic control underlying the mechanism by which external inputs modify gene expression in the brain and other tissues (Borrelli et al., 2008). Because cellular metabolites are used as a source of histone post-translational modifications (Cheung et al., 2000) – a central component of chromatin transitions – it would appear intuitively evident that changes in metabolic state affecting reproductive maturation would use epigenetic mechanisms to modify the expression of specific genes. After all, chromatin modifications are intimately involved in the regulation of energy homeostasis (Gabory et al., 2011; Vaquero and Reinberg, 2009). Surprisingly, however, little, if anything is known about the role of epi-genetics in conveying nutritional status information to the cellular networks involved in the hypothalamic control of GnRH secretion.
Two regulatory systems stand out as candidates to serve as epigenetic links between early alterations in nutritional input and the neuroendocrine control of puberty. One of these systems is represented by the sirtuins, a class of histone deacetylases (HDACs) Bellet and Sassone-Corsi, 2010; Vaquero and Reinberg, 2009; Shoba et al., 2009; Ruderman et al., 2010. The other is the hexosamine biosynthetic pathway (HBP).
8.1.1. The sirtuins
SIRT1 (one of the sirtuins) can silence gene expression by both promoting the synthesis of repressive histone marks and by forming inhibitory complexes with other transcriptional repressors (Vaquero and Reinberg, 2009; Ruderman et al., 2010; Mulligan et al., 2011; Vaquero et al., 2007). SIRT1 enzymatic activity depends on the availability of nicotinamide adenine dinucleotide (NAD+), which serves as a cofactor. Because of this, SIRT1 acts as a fuel-sensing molecule that allows the cell to respond to both reduction and increases in nutrient availability (Vaquero and Reinberg, 2009; Shoba et al., 2009; Ruderman et al., 2010). At the chromatin level, SIRT1 promotes the formation of facultative heterochromatin (Vaquero et al., 2007) (a tightly packed chromatin region that can be dynamically and reversibly restored to the more open conformation of euchromatin). SIRT1 changes chromatin structure by deacetylating histone 4 at lysine 16 (H4K16) and histone 3 at lysine 9 (H3K9ac) Vaquero and Reinberg, 2009, two histone marks associated with gene activation (Wang et al., 2008; Ruthenburg et al., 2007; Guttman et al., 2009). In addition, SIRT1 promotes methylation of histone 3 at lysine 9 (H3K9me3) Vaquero and Reinberg, 2009, a modification associated with gene silencing (Wang et al., 2008; Ruthenburg et al., 2007; Guttman et al., 2009; Schaefer et al., 2009). SIRT1 interacts with the PcG complex to silence genes (Furuyama et al., 2004; Chopra and Mishra, 2005; Hussain et al., 2009; Pruitt et al., 2006; O’Hagan et al., 2011), is activated in the hypothalamus by decreased nutrient availability (Cakir et al., 2009; Dietrich et al., 2010; Ramadori et al., 2008), and is abundant in the ARC (Cakir et al., 2009; Dietrich et al., 2010; Ramadori et al., 2008). These considerations suggest that SIRT1 either by itself or in collaboration with the PcG complex may serve as an epigenetic link between energy balance and reproductive function. Future studies are needed to test the validity of this notion.
8.1.2. The hexosamine biosynthetic pathway (HBP)
This pathway integrates the metabolism of amino acids, fat, carbohydrates and nucleotides (Love and Hanover, 2005; Butkinaree et al., 2010) via the synthesis of uridine diphosphate N-Acetylglucosamine (UDP-GlcNAc), a cytoplasmic sugar nucleotide synthesized from glucose, acetylCoA, glutamine and UTP (Marshall et al., 1991). The cytoplasmic levels of UDP-GlcNAc fluctuate in response to changes in nutrient influx into the cell, and represent the rate limiting step in the synthesis of β-D-N-acetylglucosamine (O-GlcNAc), a moiety that is added to and removed from target proteins via a process known as O-GlcNAc cycling. O-GlcNAc cycling requires two enzymes: N-acetylglucosamine transferase (OGT), which adds GlcNAc at Ser or Thr residues of target proteins, and O-GlcNAcase (OGA), which removes O-GlcNAc moieties from these sites. Both enzymes are highly conserved throughout evolution and encoded by single genes.
There is now indisputable evidence that O-GlcNAc cycling plays a critical role in maintaining chromatin structure and regulating gene transcription (Butkinaree et al., 2010; Hanover, 2010; Hanover et al., 2012). Because O-GlcNAcylation has been shown to contribute to both PcG-mediated repression (Love and Hanover, 2005; Hanover, 2010) and TrxG-mediated activation of gene transcription (Deplus et al., 2013; Fujiki et al., 2011, 2009) it appears plausible that early life nutrient excess may influence neuroendocrine reproductive development via a mechanism involving the balancing effect of a nutrient sensor (O-GlcNAcylation) on two opposing epigenetic forces of transcriptional regulation (PcG and TrxG). Studies addressing this issue should shed new light into the biological importance of this mechanism.
8.2. The circadian clock
8.2.1. Circadian rhythms
Several physiological functions contributing to body homeostasis, such as hormone secretion, body temperature, sleep-wake cycles, locomotor activity and feeding behavior, are regulated by an internal clock that controls their circadian (“circa diem”, about-a-day) rhythms of activity (Bellet and Sassone-Corsi, 2010; Masri and Sassone-Corsi, 2013). Intense efforts to unravel the molecular bases of these rhythms and the biological underpinnings of the process controlling them have shown that circadian rhythms are governed by the suprachiasmatic nucleus (SCN). Signals emanating from SCN neurons synchronize the circadian oscillations displayed by most cells in the body. These oscillations occur autonomously in the absence of external clues, because they are driven by peripheral clocks intrinsic to each cell type. However, the periodicity of peripheral clocks is not exactly 24 h and thus they need to be synchronized (entrained) by external cues (also called zeitgebers). This entrainment is provided by the SCN, which after detecting diurnal changes in luminosity conveys this information to a network of peripheral clocks (Etchegaray et al., 2003) to reset their activity daily, thereby preventing them to free-run out of phase (Bellet and Sassone-Corsi, 2010; Masri and Sassone-Corsi, 2013).
From the molecular standpoint, the clock machinery consists of a network of transcriptional-translational feedback loops initiated by the binding of CLOCK and BMAL1, two basic helix-loop-helix-PAS transcription factors, to DNA sequences termed E-boxes present in the regulatory regions of downstream clock-controlled genes (CCGs) Masri and Sassone-Corsi, 2013. The best characterized CCGs are the period genes Per1, Per2 and Per3, and the cryptochrome genes Cry1 and Cry2. The protein products of these genes form a heterodimeric repressive complex that inhibit CLOCK-BMAL1-mediated activation of Per and Cry genes, in addition to other CCGs. Protein degradation of the PER/CRY repressive complex maintains the amplitude of the oscillatory activation, and eventually diminishes the repressive effect to a point that allows the initiation of a new CLOCK-BMAL1 driven transcriptional cycle every 24 h (Bellet and Sassone-Corsi, 2010; Masri and Sassone-Corsi, 2013). DNA microarray analysis of various tissues has revealed that the abundance of 10–15% of all transcripts is subjected to circadian oscillation (Akhtar et al., 2002; Duffield et al., 2002). Many of these genes are directly targeted by CLOCK-BMAL1. Others encode transcription factors that regulate the expression of additional downstream CCGs. An example of this class is provided by TTF1, which is regulated by CLOCK-BMAL1 and in turn controls GnRH expression in the prepubertal hypothalamus (Matagne et al., 2012). That Kiss1 and Tac2 may be CCGs directly regulated by CLOCK-BMAL1 is suggested by the presence of canonical and non-canonical E-boxes in the regulatory region of both genes and the ability of the CLOCK-BMAL1 heterodimer to enhance Kiss1 transcription (unpublished results).
8.2.2. Chromatin remodeling and circadian biology
Almost 15 years ago Crosio et al. (2000) provided the first evidence that a chromatin PTM (phosphorylation of H3 at Ser10) plays a role in circadian gene expression. Subsequent studies expanded this finding by showing that histone PTMs at CCG promoters vary in a circadian fashion (Etchegaray et al., 2003; Doi et al., 2006; Naruse et al., 2004), and that both H3K4me3, a histone PTM associated with gene activation, and H3K27me3, a histone PTM associated with gene repression, occur in a circadian fashion (Ripperger and Schibler, 2006; Etchegaray et al., 2006). Whereas H3K4me3 abundance parallels CLOCK-BMAL1 binding (Ripperger and Schibler, 2006), that of H3K27me3 is required for implementation of the repressive phase of the circadian clock (Etchegaray et al., 2006). Perhaps the most revealing finding linking epigenetics to the circadian clock was the demonstration that CLOCK itself has HAT activity (Doi et al., 2006). This study demonstrated that CLOCK not only catalyzes the acetylation of H3 at lysine 9 and 14 (H3K9, 14ac) in a circadian manner, but even more importantly, it showed that this activity is essential for circadian gene expression (Doi et al., 2006). The HAT activity of CLOCK is counteracted by the HDAC activity of SIRT1 (Vaquero and Reinberg, 2009; Ruderman et al., 2010). Although neither CLOCK nor SIRT1 expression vary in a circadian fashion, the association of both into a complex recruited to CCG promoters is circadian (Nakahata et al., 2008). It thus appears that the biology of the circadian clock is inextricably linked to chromatin transitions occurring at CCG regulatory regions. Identifying the neuronal and glial networks where these mechanisms may operate to set in motion the circadian activation of the pubertal process is undoubtedly an exciting subject of future investigation.
8.2.3. Circadian clock function involves histone PTMs catalyzed by PcG and TrxG proteins
Ten years ago Etchegaray et al. (2003) provided the first evidence that transcriptional regulation of the core clock mechanism involves chromatin remodeling. These authors showed oscillatory acetylation of H3K9 and RNA polymerase II (Pol II) binding at CCG promoters. Importantly, they also showed that the mechanism by which CRY1 terminates CLOCK-BMAL1 transactivation of CCGs to reinitiate a circadian cycle involves a reduction in HAT activity, which by altering chromatin structure reduces the accessibility of CLOCK-BMAL1 heterodimers to their target E-box sequences. Because H3 acetylation is a PTM associated with TrxG-dependent H3K4me3, these early findings hinted at a potential role of TrxG complexes in circadian biology. That this is indeed the case was demonstrated several years later by the finding that MLL1, a TrxG protein (Fig. 4), is essential for circadian gene transcription. MLL1 implements cyclic H3K4 trimethylation, forms a complex with CLOCK and BMAL1, contributes to both H3 acetylation and to the cyclic recruitment of CLOCK-BMAL1 heterodimers to circadian promoters, and is required for circadian gene expression (Katada and Sassone-Corsi, 2010). It is therefore plausible that a mechanism involving these TrxG-clock gene interactions may contribute to the circadian activation of the pubertal process. Lack of Mll1 abolishes circadian changes in expression of circadian genes (Katada and Sassone-Corsi, 2010) and its recruitment to these promoters requires the presence of the CLOCK/BMAl1 complex (Katada and Sassone-Corsi, 2010).
PcG proteins counteract the effect of TrxG proteins by facilitating the repressive effect of CRY proteins on CLOCK-BMAL1 transcriptional activation (Etchegaray et al., 2006). In the absence of the PcG protein EZH2, circadian periodicity is disrupted (Etchegaray et al., 2006). Ezh2 encodes a PcG protein of the PRC2 subcomplex responsible for the tri-methylation of lysine 27 on histone 3 (H3K27me3). EZH2 co-precipitates with CLOCK/BMAL1 throughout the circadian cycle, bind to both the Perl and Per2 promoters coinciding with the presence of H3K27me3 at these sites, and is required for the transcriptional repressive activity of CRY1 (Etchegaray et al., 2006), a protein encoded by the clock controlled gene Cryl. This gene is a direct CLOCK/BMAL1 target that forms a heterodimeric complex with PER proteins to repress CLOCK/BMAL1-mediated gene activation, and thus is required for the oscillatory activity of the clock’s transcriptional cycle (Bellet and Sassone-Corsi, 2010). Loss of Ezh2 reduces the ability of CRY1 to repress CLOCK/BMAL1 mediated transcription, and disrupts the circadian cycle due to the inability of CRY proteins to inhibit transcription in the absence of H3K27me3 (Etchegaray et al., 2006).
These findings establish an essential role for both PcG and TrxG proteins in the maintenance of circadian rhythms, and support the existence of a similar mechanism underlying the ARC control of pulsatile GnRH release at puberty. It is, therefore, clear that by dynamically changing opposite chromatin states at specific promoters PcG and TrxG proteins play a fundamental role in establishing and maintaining circadian periodicity at puberty.
8.2.4. Circadian regulation of pulsatile and surge LH release at puberty
Studies in ovariectomized rats treated with estradiol have shown that both AVPV Kiss1 mRNA levels and the activity of AVPV kisspeptin neurons are increased by estradiol in a circadian-dependent manner (Robertson et al., 2009). This diurnal rhythmicity is not apparent in the absence of estrogen. Whether diurnal changes in Kiss1 (and/or Tac2/Dyn) expression occurs in KNDy neurons in association to the onset of puberty has never been explored, despite speculation that “rhythmic (ARC) Kiss1 expression and (kisspeptin) release may be responsible for the diurnal rhythms of LH secretion observed in (peripubertal) females (Tolson and Chappell, 2012). Because the pubertal increase in pulsatile LH release occurs in the presence of low – and unchanging – estrogen levels, it is likely that a circadian change in KNDy neuron activity is also estrogen-independent. This notion is supported by the increased activity of SCN neurons projecting to the ARC observed during the last part of the light cycle in the absence of changes in estrogen production (Saeb-Parsy and Dyball, 2003).
8.3. Endocrine disruptors
In recent years there has been an increasing awareness that man-made agents, such as pesticides, alcohol, asbestos, arsenic, heavy metals, air pollution, and a variety of agents structurally similar to steroid or amine hormones, can disrupt endocrine and neuroendocrine development by altering epigenetic regulatory mechanisms (Collotta et al., 2013; Christensen and Marsit, 2011; Fleisch et al., 2012; Knower et al., 2014). Of these agents, the latter group, collectively known as endocrine disruptor chemicals (EDCs), is probably the most studied [for recent reviews see [231,233].
Bisphenol A (BPA) is an EDC that requires special mention because of its widespread prevalence. BPA is used in in the manufacturing of a variety of consumer products, including plastic bottles, lining of metal cans for food and drinks, pacifiers, microwave wraps, water pipe linings, etc. The use of BPA is so pervasive that it has been detected in body fluids of more than 90% of the human population of the US (Calafat et al., 2005). BPA appears to alter development by increasing the methylation status of several genes (such as LAMP3, BRCA1, CCNA1 and others) (Qin et al., 2012), upregulating EZH2 expression (Doherty et al., 2010), and dysregulating miRNA expression (Tilghman et al., 2012). Though mostly obtained in breast cancer cells, these results suggest that man-made environmental toxins can affect cellular function by disrupting epigenetic regulation at various levels of control, including DNA methylation, histone PTMs and miRNA function.
Comparatively speaking much less is known about the effect of EDCs on the neuroendocrine control of puberty, although some evidence exists linking them to sexual precocity in humans (Parent et al., 2005, 2001). Evidently much remains to be done to obtain a more conclusive picture of the epigenetic processes affected by environmental toxins and the consequences that such actions might have on pubertal development. Although not discussed here, the contribution of environmentally induced trans-generational epigenetic inheritance (Guerrero-Bosagna and Skinner, 2012) to the timing of puberty remains as an exciting subject of investigation in need of attention.
9. Conclusions and perspectives
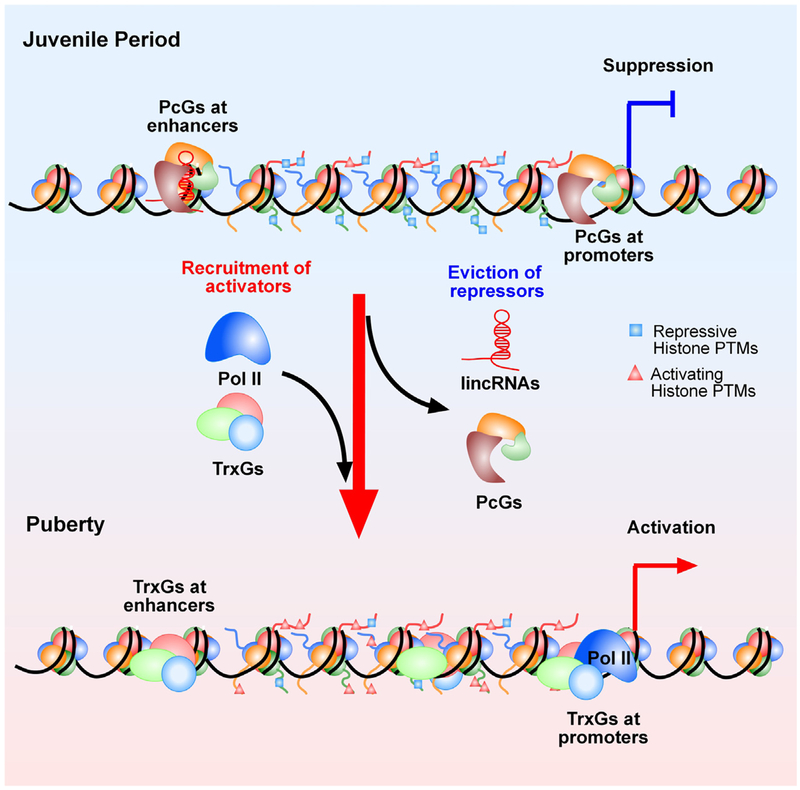
It is becoming increasing clear that epigenetic mechanisms play a significant role in the regulation of neuroendocrine reproductive development and the timing of puberty. Based on the information currently available, we speculate that the initiation of puberty requires a switch from transcriptional inhibition to transcriptional activation. We envision a scenario in which transcriptional repression is provided by epigenetic mechanism involving the PcG silencing complex, whereas transcriptional activation requires the contribution of TrxG proteins (Fig. 5). We further speculate that these interactions are integral components of a fundamental mechanism underlying the epigenetic control of puberty-activating genes, epitomized by the Kiss1 gene.
Fig. 5. Postulated epigenetic mechanisms controlling the onset of female puberty.
This model predicts the existence of an antagonistic (Yin-Yang) mechanism of transcriptional regulation underlying the developmental changes in expression of genes that facilitate pubertal development. According to this concept, the transcriptional activity of these genes (Kiss1, Tac2, Nell2, TTF1, others) is repressed during prepubertal development by silencing molecules, such as the PcG complex. PcG proteins catalyze the formation of a repressive chromatin structure characterized by an abundance of histone PTMs associated with gene silencing(such as H3K27me3). As puberty approaches, these “writers” of a repressive chromatin configuration are evicted from, and the content of histone repressive marks is reduced at, promoter regions controlling puberty-activating genes. Along with this change, writers of histone PTMs associated with transcriptional activation, such as H3K4me3 and H3K9, 14ac, are recruited to these regulatory regions resulting in enhanced gene expression. A strong candidate for this activational role is the TrxG activating complex, which antagonizes the silencing effect of PcG by both catalyzing the methylation of histone 3 at lysine 4 (H3K4me3, an activating histone mark) and binding to promoter DNA containing this mark. It is also envisioned that a similar relationship operates in distal enhancers regions controlling puberty-related genes. In this case, PcG deposition of the histone repressive mark H3K27me3, coupled to the presence of H3K4me1 and the absence of Pol II, define the presence of a latent enhancer. This inactive enhancer acquires an active configuration following the implementation of H3K27ac by the TrxG complex, and the recruitment of Pol II in the presence of H3K4me1 (also catalyzed by TrxG).
There is little doubt that we are just seeing the tip of the iceberg, and that much remains to be done to both identify the various epigenetic mechanism regulating puberty and the environmental stimuli that set them in motion. We speculate that a dynamic interplay of epigenetic repression and activation of gene transcription resides at the heart of the process by which different environmental stimuli, such as light, nutrition and endocrine disruptors, regulate pubertal development (Fig. 6). However, there is a plethora of questions that need to be addressed. For instance, what is the role of DNA methylation, histone PTMs and ncRNAs in the regulation of specific hypothalamic cell populations controlling puberty (e.g. kisspeptin, NKB, glutamatergic, and GABAergic neurons, in addition to astroglial cells and tanycytes)? Does the makeup of the epigenetic machinery differ from cell type to cell type? Is the composition and activity of transcriptional repressors and activators similar in different hypothalamic cell types controlling puberty? Do they use the same or different partners according to the chemical phenotype of the cell involved? Are different hypothalamic cell subsets affected differently by different environmental stimuli such as nutrition, the circadian clock, and environmental toxins? What are the epigenetic pathways used by such different environmental inputs to convey and direct information to the different components of the hypothalamic epigenome? What is the role played by microRNAs and lncRNAs in the regulation of hypothalamic cellular subsets involved in the control of puberty? Contemporary techniques for the isolation of specific cell populations coupled with genome-wide characterization of histone and DNA methylation landscapes may provide an effective tool to begin the daunting task of answering these questions. If, as one may suspect, different puberty-controlling cell populations in the hypothalamus have their own epigenetic landscape, it would be important to identify the factors that determine this cellular specificity and gain insights into the mechanism underlying the establishment of cell-specific epigenomes. There is little doubt that the emerging field of epigenetic of puberty is in its infancy, but it is also clear that, if the necessary effort and resources are provided, rapid maturation and coming of age of this exciting field of research may be closer than anticipated. Presumably too, these efforts will lead to the identification of epigenetic defects as an underlying cause of idiopathic precocious puberty and constitutional delayed puberty in humans.
Fig. 6.
Different external and internal environmental stimuli (nutrition/metabolism, light/circadian clocks, endocrine disruptors) are envisioned to affect the timing and progression of puberty by altering the counteractive activity of PcG/TrxG-mediated epigenetic machinery. According to this model the different stimuli depicted can affect the time of puberty by modifying the function of either the PcG or TrxG complex or both.
Acknowledgments
This research was supported by grant IOS IOS1121691 from the National Science Foundation (USA).
Abbreviations:
- 5hmC
5-hydroxymethylcytosine
- 5-mC
5-methylcytosine
- Ac
acetyl
- ARC
arcuate nucleus
- ASH2
absent, small, or homeotic-like 2
- AVPV
anteroventral periventricular nucleus
- BMAL1/ARNTL
brain and muscle ARNT-like 1/aryl hydrocarbon receptor nuclear translocator-like
- BMI1
B lymphoma Mo-MLV insertion region 1 homolog
- BPA
bisphenol A
- BRCA1
breast cancer 1
- CBX
chromobox
- CCCs
clock-controlled genes
- CCNA
cyclin A2
- CDP/CUX1/CUTL1
cut-like homeobox 1
- CHD7
chromodomain helicase DNA binding protein 7
- CLOCK
clock circadian regulator
- COMPASS
complex of proteins associated to Set1
- CRC
chromatin remodeling complex
- CRY
cryptochrome circadian clock
- CXXC1
CXXC finger protein 1
- DMN
dorso medial nucleus
- DNMTs
DNA methyltransferasess
- DYN
dynorphin
- E2
estrogen
- EAP1/IRF2BPL
enhanced at puberty/interferon regulatory factor 2 binding protein-like
- EDCs
endocrine disruptor chemicals
- EED
embryonic ectoderm development
- endo siRNAs
endo-small inhibitory RNAs
- ERa
estrogen receptor alpha
- EREs
estrogen responsive elements
- EZH1/2
enhancer of Zeste 1 or 2
- FGF21
fibroblast growth factor 21
- FTO
fat mass and obesity associated
- GABA
gamma-aminobutyric acid
- Glu
glutamate
- GnIH
gonadotropin-inhibiting hormone
- GnRH
gonadotropin-releasing hormone
- GNRHR
GnRH receptor
- GWAS
genome wide association study
- H
histone
- HATs
histone acetyltransferases
- HBP
hexosamine biosynthetic pathway
- HCF1
host cell factor C1
- HDACs
histone deacetylases
- hDPY30
human protein dumpy (Dpy)-30 homolog
- HMTs
histone methyltransferases
- IGF1
insulin-like growth factor 1
- IRX3
iroquois homeobox 3
- K
lysine
- KDM3B/ JMJD1B
lysine (K)-specific demethylase 3B
- KiSS1
gene encoding kisspeptins
- KISS1R/GPR54
gene encoding kisspeptin receptor/G-protein coupled receptor 54
- KNDy
neurons that produce kisspeptin, neurokinin B and dynorphin
- LAMP3
lysosomal-associated membrane protein 3
- LEP
leptin
- LEPR
leptin receptor
- LH
luteinizing hormone
- lincRNAs
long intergenic noncoding RNAs
- MAF
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog
- Me
methyl
- MEL18/PCGF2
polycomb group RING finger protein 2
- Menin
multiple endocrine neoplasia I
- miRNAs
microRNAs
- MKRN3
makorin ring finger protein 3
- MLL
myeloid/lymphoid or mixed-lineage leukemia
- NAD+
nicotinamide adenine dinucleotide
- NCOA6
nuclear receptor coactivator 6
- ncRNAs
noncoding RNAs
- NELL2
NEL-like 2
- NRG1
neuregulin 1
- OCT2/POU2F2
POU class 2 homeobox 2
- OGA
O-GlcNAcase
- O-GlcNAc
β-D-N-acetylglucosamine
- OGT
N-acetylglucosamine transferase
- p53/TP53
tumor suppressor protein p53
- PA1/PAGR1
PAXIP1 associated glutamate-rich protein 1
- PcG
polycomb group
- PCGFs
polycomb group RING finger proteins
- PER
period circadian clock
- piRNAs
piwiRNAs
- PHC
polyhomeotic-like protein
- PMN
pre mammillary nucleus
- POA
preoptic area
- Pol II
RNA polymerase II
- PRC
polycomb repressive complex
- PTIP/PAXIP1
PAX interacting (with transcription-activation domain) protein 1
- PTMs
posttranslational modifications
- RBBP4, 5, 7
retinoblastoma binding proteins 4, 5 and 7
- RFRP
RFamide-related peptide
- RING1/2
really interesting new gene finger domain protein 1 or 2
- RISC
RNA induced silencing complex
- RPTPβ
Receptor-like Protein Tyrosine Phosphatase-β
- RYBP
RING1 and YY1-bindingprotein
- SCN
suprachiasmatic nucleus
- SET1A/B
Su (var), enhancer of zeste, and trithorax] domain-containing A or B
- SIRT1
Sirtuin 1; SNP, single nucleotide polymorphism
- sRNAs
small RNAs
- SUZ12
suppressor of ZESTE12
- SynCAM1/TSLC1
Synaptic Cell Adhesion Molecule 1/Tumor Supressor of Lung Cancer 1
- TAC3/NKB
Tachykinin 3/Neurokinin B
- TAC3R
Tachykinin 3 receptor
- TET
Tet eleven translocation methylcytosine dioxygenase
- TFs
transcription factors
- TRGs
tumor related genes
- Trx
trithorax
- TrxG
trithorax group
- TTF1/Nkx2.1
thyroid transcription factor 1/NK2 homeobox 1
- Ub
ubiquitin
- UDP-GlcNAc
uridine diphosphate N-Acetylglucosamine
- USF2
upstream transcription factor 2
- UTX/KDM6A
ubiquitously transcribed x chromosome tetratricopeptide repeat protein/lysine (K)-specific demethylase 6A
- WDR5
WD (tryptophan-aspartic acid) repeat domain
- WDR82
WD (tryptophan-aspartic acid) repeat domain 82
- YAF2
YY1-associated factor 2
- YY1
YIN-YANG-1
- ZNF
zinc finger protein
References
- Abreu AP, Dauber A, Macedo DB, Noel SD, Brito VN, Gill JC, Cukier P, Thompson IR, Navarro VM, Gagliardi PC, Rodrigues T, Kochi C, Longui A, Beckers D, de ZF, Montenegro LR, Mendonca BB, Carroll RS, Hirschhorn JN, Latronico AC, Kaiser UB, 2013. Central precocious puberty caused by mutations in the imprinted gene MKRN3. N. Engl. J. Med 368, 2467–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahima RS, Saper CB, Flier JS, Elmquist JK, 2000. Leptin regulation of neuroendocrine systems. Front. Neuroendocrinol 21, 263–307. [DOI] [PubMed] [Google Scholar]
- Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP, 2002. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr. Biol 12, 540–550. [DOI] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, Ntini E, Arner E, Valen E, Li K, Schwarzfischer L, Glatz D, Raithel J, Lilje B, Rapin N, Bagger FO, Jorgensen M, Andersen PR, Bertin N, Rackham O, Burroughs AM, Baillie JK, Ishizu Y, Shimizu Y, Furuhata E, Maeda S, Negishi Y, Mungall CJ, Meehan TF, Lassmann T, Itoh M, Kawaji H, Kondo N, Kawai J, Lennartsson A, Daub O, Heutink P, Hume DA, Jensen TH, Suzuki H, Hayashizaki Y, Muller F, Forrest AR, Carninci P, Rehli M, Sandelin A, Kawaji H, Baillie JK, de Hoon MJ, Haberle V, Lassmann T, Kulakovskiy IV, Lizio M, Itoh M, Andersson R, Mungall CJ, Meehan TF, Schmeier S, Bertin N, Jorgensen M, Dimont E, Arner E, Schmid C, Schaefer U, Medvedeva YA, Plessy C, Vitezic M, Severin J, Semple CA, Ishizu Y, Young RS, Francescatto M, Alam I, Albanese D, Altschuler GM, Arakawa T, Archer JA, Arner P, Babina M, Rennie S, Balwierz PJ, Beckhouse AG, Pradhan-Bhatt S, Blake JA, Blumenthal A, Bodega B, Bonetti A, Briggs J, Brombacher F, Burroughs AM, Califano A, Cannistraci CV, Carbajo D, Chen Y, Chierici M, Ciani Y, Clevers HC, Dalla E, Davis CA, Detmar M, Diehl AD, Dohi T, Drablos F, Edge AS, Edinger M, Ekwall K, Endoh M, Enomoto H, Fagiolini M, Fairbairn L, Fang H, Farach-Carson MC, Faulkner GJ, Favorov AV, Fisher ME, Frith MC, Fujita R, Fukuda S, Furlanello C, Furuno M, Furusawa J, Geijtenbeek TB, Gibson AP, Gingeras T, Goldowitz D, Gough J, Guhl S, Guler R, Gustincich S, Ha TJ, Hamaguchi M, Hara M, Harbers M, Harshbarger J, Hasegawa A, Hasegawa Y, Hashimoto T, Herlyn M, Hitchens KJ, Ho Sui SJ, Hofmann OM, Hoof I, Hori F, Huminiecki L, Iida K, Ikawa T, Jankovic BR, Jia H, Joshi A, Jurman G, Kaczkowski B, Kai C, Kaida K, Kaiho A, Kajiyama K, Kanamori-Katayama M, Kasianov AS, Kasukawa T, Katayama S, Kato S, Kawaguchi S, Kawamoto H, Kawamura YI, Kawashima T, Kempfle JS, Kenna TJ, Kere J, Khachigian LM, Kitamura T, Klinken SP, Knox AJ, Kojima M, Kojima S, Kondo N, Koseki H, Koyasu S, Krampitz S, Kubosaki A, Kwon AT, Laros JF, Lee W, Lennartsson A, Li K, Lilje B, Lipovich L, Mackay-sim A, Manabe R, Mar JC, Marchand B, Mathelier A, Mejhert N, Meynert A, Mizuno Y, de Lima Morais DA, Morikawa H, Morimoto M, Moro K, Motakis E, Motohashi H, Mummery CL, Murata M, Nagao-Sato S, Nakachi Y, Nakahara F, Nakamura T, Nakamura Y, Nakazato K, van NE, Ninomiya N, Nishiyori H, Noma S, Nozaki T, Ogishima S, Ohkura N, Ohmiya H, Ohno H, Ohshima M, Okada-Hatakeyama M, Okazaki Y, Orlando V, Ovchinnikov DA, Pain A, Passier R, Patrikakis M, Persson H, Piazza S, Prendergast JG, Rackham OJ, Ramilowski JA, Rashid M, Ravasi T, Rizzu P, Roncador M, Roy S, Rye MB, Saijyo E, Sajantila A, Saka A, Sakaguchi S, Sakai M, Sato H, Satoh H, Savvi S, Saxena A, Schneider C, Schultes EA, Schulze-Tanzil GG, Schwegmann A, Sengstag T, Sheng G, Shimoji H, 2014. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Chang HY, 2013. Long noncoding RNAs: cellular address codes in development and disease. Cell 152, 1298–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale KE, Kinsey-Jones JS, Gardiner JV, Harrison EK, Thompson EL, Hu MH, Sleeth ML, Sam AH, Greenwood HC, McGavigan AK, Dhillo WS, Mora JM, Li XF, Franks S, Bloom SR, O’Byrne KT, Murphy KG, 2014. The physiological role of arcuate kisspeptin neurons in the control of reproductive function in female rats. Endocrinology 155, 1091–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedecarrats GY, Kaiser UB, 2007. Mutations in the human gonadotropin-releasing hormone receptor: insights into receptor biology and function. Semin. Reprod. Med 25, 368–378. [DOI] [PubMed] [Google Scholar]
- Belinsky SA, Snow SS, Nikula KJ, Finch GL, Tellez CS, Palmisano WA, 2002. Aberrant CpG island methylation of the p16(INK4a) and estrogen receptor genes in rat lung tumors induced by particulate carcinogens. Carcinogenesis 23, 335–339. [DOI] [PubMed] [Google Scholar]
- Bellet MM, Sassone-Corsi P, 2010. Mammalian circadian clock and metabolism – the epigenetic link. J. Cell Sci 123, 3837–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL, 2007. The complex language of chromatin regulation during transcription. Nature 447, 407–412. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES, 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES, 2007. The mammalian epigenome. Cell 128, 669–681. [DOI] [PubMed] [Google Scholar]
- Bianco SD, Kaiser UB, 2009. The genetic and molecular basis of idiopathic hypogonadotropic hypogonadism. Nat. Rev. Endocrinol 5, 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsson HT, Fallin MD, Feinberg AP, 2004. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 20, 350–358. [DOI] [PubMed] [Google Scholar]
- Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen LL, Ito S, Cooper S, Kondo K, Koseki Y, Ishikura T, Long HK, Sheahan TW, Brockdorff N, Kessler BM, Koseki H, Klose RJ, 2014. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 157, 1445–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P, 2008. Decoding the epigenetic language of neuronal plasticity. Neuron 60, 961–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyar R, Finkelstein J, Roffwarg H, Kapen S, Weitzman E, Hellman L, 1972. Synchronization of augmented luteinizing hormone secretion with sleep during puberty. N. Engl. J. Med 287, 582–586. [DOI] [PubMed] [Google Scholar]
- Burgers WA, Fuks F, Kouzarides T, 2002. DNA methyltransferases get connected to chromatin. Trends Genet. 18, 275–277. [DOI] [PubMed] [Google Scholar]
- Butkinaree C, Park K, Hart GW, 2010. O-linked beta-N-acetylglucosamine (O-GlcNAc): extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim. Biophys. Acta 1800, 96–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakir I, Perello M, Lansari O, Messier NJ, Vaslet CA, Nillni EA, 2009. Hypothalamic Sirt1 regulates food intake in a rodent model system. PLoS ONE 4, e8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calafat AM, Kuklenyik Z, Reidy JA, Caudill SP, Ekong J, Needham LL, 2005. Urinary concentrations of bisphenol A and 4-nonylphenol in a human reference population. Environ. Health Perspect 113, 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Wysocka J, 2013. Modification of enhancer chromatin: what, how, and why? Mol. Cell 49, 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB, 1999. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet 21, 103–107. [DOI] [PubMed] [Google Scholar]
- Caron E, Ciofi P, Prevot V, Bouret SG, 2012. Alteration in neonatal nutrition causes perturbations in hypothalamic neural circuits controlling reproductive function. J. Neurosci 32, 11486–11494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano JM, Navarro VM, Fernandez-Fernandez R, Nogueiras R, Tovar S, Roa J, Vazquez MJ, Vigo E, Casanueva FF, Aguilar E, Pinilla L, Dieguez C, Tena-Sempere M, 2005. Changes in hypothalamic KiSS-1 system and restoration of pubertal activation of the reproductive axis by kisspeptin in undernutrition. Endocrinology 146, 3917–3925. [DOI] [PubMed] [Google Scholar]
- Castellano JM, Bentsen AH, Sanchez-Garrido MA, Ruiz-Pino F, Romero M, Garcia-Galiano D, Aguilar E, Pinilla L, Dieguez C, Mikkelsen JD, Tena-Sempere M, 2011. Early metabolic programming of puberty onset: impact of changes in postnatal feeding and rearing conditions on the timing of puberty and development of the hypothalamic kisspeptin system. Endocrinology 152, 3396–3408. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y, 2009. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet 10, 295–304. [DOI] [PubMed] [Google Scholar]
- Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M, Jung J, Gao P, Dang CV, Beer MA, Thomas-Tikhonenko A, Mendell JT, 2009. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc. Natl. Acad. Sci. U.S.A 106, 3384–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P, Allis CD, Sassone-Corsi P, 2000. Signaling to chromatin through histone modifications. Cell 103, 263–271. [DOI] [PubMed] [Google Scholar]
- Choi SW, Friso S, 2010. Epigenetics: a new bridge between nutrition and health. Adv. Nutr. (Bethesda) 1, 8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra VS, Mishra RK, 2005. To SIR with Polycomb: linking silencing mechanisms. BioEssays 27, 119–121. [DOI] [PubMed] [Google Scholar]
- Christensen BC, Marsit CJ, 2011. Epigenomics in environmental health. Front. Genet 2, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY, 2011. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 44, 667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson J, d’Anglemont d.T.X., Colledge WH, Caraty A, Herbison AE, 2009. Distribution of kisspeptin neurones in the adult female mouse brain. J. Neuroendocrinol 21, 673–682. [DOI] [PubMed] [Google Scholar]
- Clasadonte J, Poulain P, Hanchate NK, Corfas G, Ojeda SR, Prevot V, 2011. Prostaglandin E2 release from astrocytes triggers gonadotropin-releasing hormone (GnRH) neuron firing via EP2 receptor activation. Proc. Natl. Acad. Sci. U.S.A 108, 16104–16109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement K, Vaisse C, Lahlou N, Cabrol S, Pelloux V, Cassuto D, Gourmelen M, Dina C, Chambaz J, Lacorte JM, Basdevant A, Bougneres P, LeBouc Y, Froguel P, Guy-Grand B, 1998. A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature 392, 398–401. [DOI] [PubMed] [Google Scholar]
- Collotta M, Bertazzi PA, Bollati V, 2013. Epigenetics and pesticides. Toxicology 307, 35–41. [DOI] [PubMed] [Google Scholar]
- Cousminer DL, Berry DJ, Timpson NJ, Ang W, Thiering E, Byrne EM, Taal R, Huikari V, Bradfield JP, Kerkhof M, Groen-Blokhuis MM, Kreiner-Moller E, Marinelli M, Holst C, Leinonen JT, Perry JR, Surakka I, Pietilainen O, Kettunen J, Anttila V, Kaakinen M, Sovio U, Pouta A, Das S, Lagou V, Power C, Prokopenko I, Evans DM, Kemp JP, St PB, Ring S, Palotie A, Kajantie E, Osmond C, Lehtimaki T, Viikari JS, Kahonen M, Warrington NM, Lye SJ, Palmer LJ, Tiesler CM, Flexeder C, Montgomery GW, Medland SE, Hofman A, Hakonarson H, Guxens M, Bartels M, Salomaa V, Murabito JM, Kaprio J, Sorensen TI, Ballester F, Bisgaard H, Boomsma DI, Koppelman GH, Grant SF, Jaddoe VW, Martin NG, Heinrich J, Pennell CE, Raitakari OT, Eriksson JG, Smith GD, Hypponen E, Jarvelin MR, McCarthy MI, Ripatti S, Widen E, 2013. Genome-wide association and longitudinal analyses reveal genetic loci linking pubertal height growth, pubertal timing and childhood adiposity. Hum. Mol. Genet 22, 2735–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosio C, Cermakian N, Allis CD, Sassone-Corsi P, 2000. Light induces chromatin modification in cells of the mammalian circadian clock. Nat. Neurosci 3, 1241–1247. [DOI] [PubMed] [Google Scholar]
- Cukier P, Wright H, Rulfs T, Silveira LF, Teles MG, Mendonca BB, Arnhold IJ, Heger S, Latronico AC, Ojeda SR, Brito VN, 2013. Molecular and gene network analysis of thyroid transcription factor 1 (TTF1) and enhanced at puberty (EAP1) genes in patients with GnRH-dependent pubertal disorders. Horm. Res. Paediatr 80, 257–266. [DOI] [PubMed] [Google Scholar]
- d’Anglemont d.T.X., Colledge WH, 2010. The role of kisspeptin signaling in reproduction. Physiology (Bethesda) 25, 207–217. [DOI] [PubMed] [Google Scholar]
- de Roux N, Genin E, Carel J-C, Matsuda F, Chaussain J-L, Milgrom E, 2003. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPr54. Proc. Natl. Acad. Sci. U.S.A 100, 10972–10976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissov S, Hofemeister H, Marks H, Kranz A, Ciotta G, Singh S, Anastassiadis K, Stunnenberg HG, Stewart AF, 2014. Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mill is redundant. Development 141, 526–537. [DOI] [PubMed] [Google Scholar]
- Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, Shih AH, Levine RL, Bernard O, Mercher T, Solary E, Urh M, Daniels DL, Fuks F, 2013. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 32, 645–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di CL, Helin K, 2013. Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol 20, 1147–1155. [DOI] [PubMed] [Google Scholar]
- Dietrich MO, Antunes C, Geliang G, Liu ZW, Borok E, Nie Y, Xu AW, Souza DO, Gao Q, Diano S, Gao XB, Horvath TL, 2010. Agrp neurons mediate Sirt1’s action on the melanocortin system and energy balance: roles for Sirt1 in neuronal firing and synaptic plasticity. J. Neurosci 30, 11815–11825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty LF, Bromer JG, Zhou Y, Aldad TS, Taylor HS, 2010. In utero exposure to diethylstilbestrol (DES) or bisphenol-A (BpA) increases EZH2 expression in the mammary gland: an epigenetic mechanism linking endocrine disruptors to breast cancer. Horm. Cancer 1, 146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Sassone-Corsi P, 2006. Circadian regulator CLOCK is a histone acetyltransferase. Cell 125, 497–508. [DOI] [PubMed] [Google Scholar]
- Ducret E, Anderson GM, Herbison AE, 2009. RFamide-related peptide-3, a mammalian gonadotropin-inhibitory hormone ortholog, regulates gonadotropin-releasing hormone neuron firing in the mouse. Endocrinology 150, 2799–2804. [DOI] [PubMed] [Google Scholar]
- Dudas B, Merchenthaler I, 2006. Three-dimensional representation of the neurotransmitter systems of the human hypothalamus: inputs of the gonadotrophin hormone-releasing hormone neuronal system. J. Neuroendocrinol 18, 79–95. [DOI] [PubMed] [Google Scholar]
- Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC, 2002. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr. Biol 12, 551–557. [DOI] [PubMed] [Google Scholar]
- Ebling FJ, Luckman SM, 2008. RFAmide-related peptide: another sexy peptide? Endocrinology 149, 899–901. [DOI] [PubMed] [Google Scholar]
- Elias CF, 2012. Leptin action in pubertal development: recent advances and unanswered questions. Trends Endocrinol. Metab 23, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elks CE, Perry JR, Sulem P, Chasman DI, Franceschini N, He C, Lunetta KL, Visser JA, Byrne EM, Cousminer DL, Gudbjartsson DF, Esko T, Feenstra B, Hottenga JJ, Koller DL, Kutalik Z, Lin P, Mangino M, Marongiu M, McArdle PF, Smith AV, Stolk L, van Wingerden SH, Zhao JH, Albrecht E, Corre T, Ingelsson E, Hayward C, Magnusson PK, Smith EN, Ulivi S, Warrington NM, Zgaga L, Alavere H, Amin N, Aspelund T, Bandinelli S, Barroso I, Berenson GS, Bergmann S, Blackburn H, Boerwinkle E, Buring JE, Busonero F, Campbell H, Chanock SJ, Chen W, Cornelis MC, Couper D, Coviello AD, d’Adamo P, de FU, de Geus EJ, Deloukas P, Doring A, Smith GD, Easton DF, Eiriksdottir G, Emilsson V, Eriksson J, Ferrucci L, Folsom AR, Foroud T, Garcia M, Gasparini P, Geller F, Gieger C, Gudnason V, Hall P, Hankinson SE, Ferreli L, Heath AC, Hernandez DG, Hofman A, Hu FB, Illig T, Jarvelin MR, Johnson AD, Karasik D, Khaw KT, Kiel DP, Kilpelainen TO, Kolcic I, Kraft P, Launer LJ, Laven JS, Li S, Liu J, Levy D, Martin NG, McArdle WL, Melbye M, Mooser V, Murray JC, Murray SS, Nalls MA, Navarro P, Nelis M, Ness AR, Northstone K, Oostra BA, Peacock M, Palmer LJ, Palotie A, Pare G, Parker AN, Pedersen NL, Peltonen L, Pennell CE, Pharoah P, Polasek O, Plump AS, Pouta A, Porcu E, Rafnar T, Rice JP, Ring SM, Rivadeneira F, Rudan I, Sala C, Salomaa V, Sanna S, Schlessinger D, Schork NJ, Scuteri A, Segre AV, Shuldiner AR, Soranzo N, Sovio U, Srinivasan SR, Strachan DP, Tammesoo ML, Tikkanen E, Toniolo D, Tsui K, Tryggvadottir L, Tyrer J, Uda M, van Dam RM, van Meurs JB, Vollenweider P, Waeber G, Wareham NJ, Waterworth DM, Weedon MN, Wichmann HE, Willemsen G, Wilson JF, Wright AF, Young L, Zhai G, Zhuang WV, Bierut LJ, Boomsma DI, Boyd HA, Crisponi L, Demerath EW, van Duijn CM, Econs MJ, Harris TB, Hunter DJ, Loos RJ, Metspalu A, Montgomery GW, Ridker PM, Spector TD, Streeten EA, Stefansson K, Thorsteinsdottir U, Uitterlinden AG, Widen E, Murabito JM, Ong KK, Murray A, 2010. Thirty new loci for age at menarche identified by a metaanalysis of genome-wide association studies. Nat. Genet, 42, 1077–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray JP, Lee C, Wade PA, Reppert SM, 2003. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421, 177–182. [DOI] [PubMed] [Google Scholar]
- Etchegaray JP, Yang X, DeBruyne JP, Peters AH, Weaver DR, Jenuwein T, Reppert SM, 2006. The polycomb group protein EZH2 is required for mammalian circadian clock function. J. Biol. Chem 281, 21209–21215. [DOI] [PubMed] [Google Scholar]
- Fernandez-Fernandez R, Martini AC, Navarro VM, Castellano JM, Dieguez C, Aguilar E, Pinilla L, Tena-Sempere M, 2006. Novel signals for the integration of energy balance and reproduction. Mol. Cell. Endocrinol 254–255, 127–132. [DOI] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W, 2011. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473, 398–402. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N, 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet 9, 102–114. [DOI] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH, 2007. Recovery of learning and memory is associated with chromatin remodelling. Nature 447, 178–182. [DOI] [PubMed] [Google Scholar]
- Fleisch AF, Wright RO, Baccarelli AA, 2012. Environmental epigenetics: a role in endocrine disease? J. Mol. Endocrinol 49, R61–R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes S, Li XF, Kinsey-Jones J, O’Byrne K, 2009. Effects of ghrelin on kisspeptin mRNA expression in the hypothalamic medial preoptic area and pulsatile luteinising hormone secretion in the female rat. Neurosci. Lett 460, 143–147. [DOI] [PubMed] [Google Scholar]
- Fortes MR, Reverter A, Nagaraj SH, Zhang Y, Jonsson NN, Barris W, Lehnert S, Boe-Hansen GB, Hawken RJ, 2011. A single nucleotide polymorphism-derived regulatory gene network underlying puberty in 2 tropical breeds of beef cattle. J. Anim. Sci 89, 1669–1683. [DOI] [PubMed] [Google Scholar]
- Frisch RE, 1980. Pubertal adipose tissue: is it necessary for normal sexual maturation? Evidence from the rat and human female. Fed. Proc 39, 2395–2400. [PubMed] [Google Scholar]
- Frisch RE, Revelle R, 1970. Height and weight at menarche and a hypothesis of critical body weight and adolescent events. Science 109, 397–399. [DOI] [PubMed] [Google Scholar]
- Frisch RE, Hegsted DM, Yoshinaga K, 1975. Bodyweight and food intake at early estrus of rats on a high-fat diet. Proc. Natl. Acad. Sci. U.S.A 72, 4172–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki R, Chikanishi T, Hashiba W, Ito H, Takada I, Roeder RG, Kitagawa H, Kato S, 2009. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature 459, 455–459. [DOI] [PubMed] [Google Scholar]
- Fujiki R, Hashiba W, Sekine H, Yokoyama A, Chikanishi T, Ito S, Imai Y, Kim J, He HH, Igarashi K, Kanno J, Ohtake F, Kitagawa H, Roeder RG, Brown M, Kato S, 2011. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature 480, JJ7–J60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukusumi S, Fujii R, Hinuma S, 2006. Recent advances in mammalian RFamide peptides: the discovery and functional analyses of PrRP, RFRPs and QRFP. Peptides 27, 1073–1086. [DOI] [PubMed] [Google Scholar]
- Furuyama T, Banerjee R, Breen TR, Harte PJ, 2004. SIR2 is required for polycomb silencing and is associated with an E(Z) histone methyltransferase complex. Curr. Biol 14, 1812–1821. [DOI] [PubMed] [Google Scholar]
- Gabory A, Attig L, Junien C, 2011. Developmental programming and epigenetics. Am. J. Clin. Nutr 94, 1943S–1952S. [DOI] [PubMed] [Google Scholar]
- Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D, 2012. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 45, 344–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS, Ju BG, Ohgi KA, Wang J, Escoubet-Lozach L, Rose DW, Glass CK, Fu XD, Rosenfeld MG, 2007. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell 128, 505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottsch ML, Cunningham MJ, Smith JT, Popa SM, Acohido BV, Crowley WF, Seminara S, Clifton DK, Steiner RA, 2004. A role for kisspeptins in the regulation of gonadotropin secretion in the mouse. Endocrinology 145, 4073–4077. [DOI] [PubMed] [Google Scholar]
- Grove KL, Grayson BE, Glavas MM, Xiao XQ, Smith MS, 2005. Development of metabolic systems. Physiol. Behav 86, 646–660. [DOI] [PubMed] [Google Scholar]
- Gruber AJ, Zavolan M, 2013. Modulation of epigenetic regulators and cell fate decisions by miRNAs. Epigenomics J, 671–683. [DOI] [PubMed] [Google Scholar]
- Grumbach MM, Styne DM, 1992. Puberty: ontogeny, neuroendocrinology, physiology, and disorders. In: Wilson JD, Foster DW (Eds.), Williams Textbook of Endocrinology, 8th ed W.B. Saunders, Co., Philadelphia, pp. 1139–1221. [Google Scholar]
- Guerrero-Bosagna C, Skinner MK, 2012. Environmentally induced epigenetic transgenerational inheritance of phenotype and disease. Mol. Cell. Endocrinol 3J4, 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES, 2009. Chromatin signature reveals over a thousand highly conserved large noncoding RNAs in mammals. Nature 4J8, 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI, 2009. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol 16, 1021–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanover JA, 2010. Epigenetics gets sweeter: O-GlcNAc joins the “histone code”. Chem. Biol 17, 1272–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanover JA, Krause MW, Love DC, 2012. Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nat. Rev. Mol. Cell Biol 13, 312–321. [DOI] [PubMed] [Google Scholar]
- He C, Kraft P, Chen C, Buring JE, Pare G, Hankinson SE, Chanock SJ, Ridker PM, Hunter DJ, Chasman DI, 2009. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat. Genet 41, 724–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger S, Partsch CJ, Peter M, Blum WF, Kiess W, Sippell WG, 1999. Serum leptin levels in patients with progressive central precocious puberty. Pediatr. Res 46, 71–75. [DOI] [PubMed] [Google Scholar]
- Heger S, Mastronardi C, Dissen GA, Lomniczi A, Cabrera R, Roth CL, Jung H, Galimi F, Sippell W, Ojeda SR, 2007. Enhanced at puberty 1 (EAP1) is a new transcriptional regulator of the female neuroendocrine reproductive axis. J. Clin. Invest 117, 2145–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN, 2009. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138, 696–708. [DOI] [PubMed] [Google Scholar]
- Herbison AE, Moenter SM, 2011. Depolarising and hyperpolarising actions of GABA(A) receptor activation on gonadotrophin-releasing hormone neurones: towards an emerging consensus. J. Neuroendocrinol 23, JJ7–J69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Baylin SB, 2003. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med 349, 2042–2054. [DOI] [PubMed] [Google Scholar]
- Herz HM, Mohan M, Garruss AS, Liang K, Takahashi YH, Mickey K, Voets O, Verrijzer CP, Shilatifard A 2012. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/MU4. Genes Dev. 26, 2604–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiney JK, Srivastava VK, Pine MD, Les DW, 2009. Insulin-like growth factor-I activates KiSS-1 gene expression in the brain of the prepubertal female rat. Endocrinology 150, 376–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinuma S, Shintani Y, Fukusumi S, Iijima N, Matsumoto Y, Hosoya M, Fujii R, Watanabe T, Kikuchi K, Terao Y, Yano T, Yamamoto T, Kawamata Y, Habata Y, Asada M, Kitada C, Kurokawa T, Onda H, Nishimura O, Tanaka M, Ibata Y, !Lost Data, 2000. New neuropeptides containing carboxy-terminal RFamide and their receptor in mammals. Nat. Cell Biol 2, 703–708. [DOI] [PubMed] [Google Scholar]
- Hrabovszky E, Ciofi P, Vida B, Horvath MC, Keller E, Caraty A, Bloom SR, Ghatei MA, Dhillo WS, Liposits Z, Kallo I, 2010. The kisspeptin system of the human hypothalamus: sexual dimorphism and relationship with gonadotropin-releasing hormone and neurokinin B neurons. Eur. J. Neurosci 31, 1984–1998. [DOI] [PubMed] [Google Scholar]
- Hu D, Gao X, Morgan MA, Herz HM, Smith ER, Shilatifard A, 2013. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol. Cell. Biol 33, 4745–4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Jiang C, Zhang R, 2014. Epigenetics: the language of the cell? Epigenomics 6, 73–88. [DOI] [PubMed] [Google Scholar]
- Hussain M, Rao M, Humphries AE, Hong JA, Liu F, Yang M, Caragacianu D, Schrump DS, 2009. Tobacco smoke induces polycomb-mediated repression of Dickkopf-1 in lung cancer cells. Cancer Res. 69, 3570–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB, 1994. Methylation of the oestrogen receptor CpG Island links ageing and neoplasia in human colon. Nat. Genet 7, 536–540. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A, 2003. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet 33 (Suppl.), 245–254. [DOI] [PubMed] [Google Scholar]
- Kangaspeska S, Stride B, Metivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G, 2008. Transient cyclical methylation of promoter DNA. Nature 452, 112–115. [DOI] [PubMed] [Google Scholar]
- Katada S, Sassone-Corsi P, 2010. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat. Struct. Mol. Biol 17, 1414–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman AS, Navarro VM, Kim J, Clifton DK, Steiner RA, 2009. Sex differences in the regulation of Kiss1/NKB neurons in juvenile mice: implications for the timing of puberty. Am. J. Physiol. Endocrinol. Metab 297, E1212–E1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy GC, Mitra J, 1963. Body weight and food intake as initiating factors for puberty in the rat. J. Physiol 166, 408–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorasanizadeh S, 2004. The nucleosome: from genomic organization to genomic regulation. Cell 116, 259–272. [DOI] [PubMed] [Google Scholar]
- Kim VN, 2006. Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes Dev. 20, 1993–1997. [DOI] [PubMed] [Google Scholar]
- Kim HG, Kurth I, Lan F, Meliciani I, Wenzel W, Eom SH, Kang GB, Rosenberger G, Tekin M, Ozata M, Bick DP, Sherins RJ, Walker SL, Shi Y, Gusella JF, Layman LC, 2008. Mutations in CHD7, encoding a chromatin-remodeling protein, cause idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am. J. Hum. Genet 83, 511–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Kim KB, Eom GH, Choe N, Kee HJ, Son HJ, Oh ST, Kim DW, Pak JH, Baek HJ, Kook H, Hahn Y, Kook H, Chakravarti D, Seo SB, 2012. KDM3B is the H3K9 demethylase involved in transcriptional activation of lmo2 in leukemia. Mol. Cell. Biol 32, 2917–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knower KC, To SQ, Leung YK, Ho SM, Clyne CD, 2014. Endocrine disruption of the epigenome: a breast cancer link. Endocr. Relat. Cancer 21, T33–T55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, Lahesmaa R, Orkin SH, Rodig SJ, Daley GQ, Rao A, 2011. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8, 200–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordon C, Drouva SV, Martinez de la Escalera G, Weiner RI, 1994. Role of classic and peptide neuromediators in the neuroendocrine regulation of luteinizing hormone and prolactin In: Knobil E, Neill JD (Eds.), The Physiology of Reproduction, second ed., vol. 1 Raven Press, New York, pp. 1621–1681. [Google Scholar]
- Kotani M, Detheux M, Vandenbogaerde A, Communi D, Vanderwinden JM, Le Poul E, Brezillon S, Tyldesley R, Suarez-Huerta N, Vandeput F, Blanpain C, Schiffmann SN, Vassart G, Parmentier M, 2001. The metastasis suppressor gene KiSS-1 encodes kisspeptins, the natural ligands of the orphan G protein-coupled receptor GPR54. J. Biol. Chem 276, 34631–34636. [DOI] [PubMed] [Google Scholar]
- Kouzarides T, 2007. Chromatin modifications and their function. Cell 128, 693–705. [DOI] [PubMed] [Google Scholar]
- Krishnan S, Horowitz S, Trievel RC, 2011. Structure and function of histone H3 lysine 9 methyltransferases and demethylases. ChemBioChem 12, 254–263. [DOI] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, Russo SJ, Laplant Q, Sasaki TS, Whistler KN, Neve RL, Self DW, Nestler J, 2005. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron 48, 303–314. [DOI] [PubMed] [Google Scholar]
- Kurian JR, Olesen KM, Auger AP, 2010. Sex differences in epigenetic regulation of the estrogen receptor-alpha promoter within the developing preoptic area. Endocrinology 151, 2297–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian JR, Keen KL, Terasawa E, 2010. Epigenetic changes coincide with in vitro primate GnRH neuronal maturation. Endocrinology 151, 5359–5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapatto R, Pallais JC, Zhang D, Chan YM, Mahan A, Cerrato F, Le WW, Hoffman GE, Seminara SB, 2007. Kiss1−/− mice exhibit more variable hypogonadism than Gpr54−/− mice. Endocrinology 148, 4927–4936. [DOI] [PubMed] [Google Scholar]
- Lehman MN, Coolen LM, Goodman RL, 2010. Minireview: kisspeptin/neurokinin B/dynorphin (KNDy) cells of the arcuate nucleus: a central node in the control of gonadotropin-releasing hormone secretion. Endocrinology 151, 3479–3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, Balasubramanian S, Miska EA, 2009. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat. Struct. Mol. Biol 16, 1016–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Herbison AE, 2011. Estrous cycle- and sex-dependent changes in pre- and postsynaptic GABAB control of GnRH neuron excitability. Endocrinology 152, 4856–4864. [DOI] [PubMed] [Google Scholar]
- Lomniczi A, Ojeda SR, 2009. A role for glial cells of the neuroendocrine brain in the central control of female sexual development In: Parpura V, Haydon P (Eds.), Astrocytes in (Patho)Physiology of the Nervous System. Springer, NY, pp. 487–511. [Google Scholar]
- Lomniczi A, Loche A, Castellano JM, Ronnekleiv OK, Bosh M, Kaidar G, Knoll JG, Wright H, Pfeifer GP, Ojeda SR, 2013. Epigenetic control of female puberty. Nat. Neurosci 16, 281–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomniczi A, Wright H, Castellano JM, Sonmez K, Ojeda SR, 2013. A system biology approach to identify regulatory pathways underlying the neuroendocrine control of female puberty in rats and nonhuman primates. Horm. Behav 64, 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love DC, Hanover JA, 2005. The hexosamine signaling pathway: deciphering the “O-GlcNAc code. Sci. STKE, re13. [DOI] [PubMed] [Google Scholar]
- Marshall S, Bacote V, Traxinger RR, 1991. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J. Biol. Chem 266, 4706–4712. [PubMed] [Google Scholar]
- Masri S, Sassone-Corsi P, 2013. The circadian clock: a framework linking metabolism, epigenetics and neuronal function. Nat. Rev. Neurosci 14, 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronardi C, Smiley GG, Raber J, Kusakabe T, Kawaguchi A, Matagne V, Dietzel A, Heger S, Mungenast AE, Cabrera R, Kimura S, Ojeda SR, 2006. Deletion of the Ttfl gene in differentiated neurons disrupts female reproduction without impairing basal ganglia function. J. Neurosci 26, 13167–13179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matagne V, Kim JG, Ryu BJ, Hur MK, Kim MS, Kim K, Park BS, Damante G, Smiley G, Lee BJ, Ojeda SR, 2012. Thyroid transcription factor 1, a homeodomain containing transcription factor, contributes to regulating periodic oscillations in GnRH gene expression. J. Neuroendocrinol 24, 916–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda KI, Mori H, Nugent BM, Pfaff DW, McCarthy MM, Kawata M, 2011. Histone deacetylation during brain development is essential for permanent masculinization of sexual behavior. Endocrinology 152, 2760–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MM, Nugent BM, 2013. Epigenetic contributions to hormonally-mediated sexual differentiation of the brain. J. Neuroendocrinol 25, 1133–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MM, Auger AP, Bale TL, De Vries GJ, Dunn GA, Forger NG, Murray EK, Nugent BM, Schwarz JM, Wilson ME, 2009. The epigenetics of sex differences in the brain. J. Neurosci 29, 12815–12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F, 2003. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115, 751–763. [DOI] [PubMed] [Google Scholar]
- Metivier R, Gallais R, Tiffoche C, Le PC, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, Benes V, Jeltsch A, Gannon F, Salbert G, 2008. Cyclical DNA methylation of a transcriptionally active promoter. Nature 452, 45–50. [DOI] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD, 2007. Covalent modification of DNA regulates memory formation. Neuron 53, 857–869. [DOI] [PubMed] [Google Scholar]
- Mohan M, Herz HM, Shilatifard A, 2012. SnapShot: histone lysine methylase complexes. Cell 149, 498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller JK, Dietzel A, Lomniczi A, Loche A, Tefs K, Kiess W, Danne T, Ojeda SR, Heger S, 2011. Transcriptional regulation of the human KiSS1 gene. Mol. Cell. Endocrinol 342, 8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller JK, Koch I, Lomniczi A, Loche A, Rulfs T, Castellano JM, Kiess W, Ojeda S, Heger S, 2012. Transcription of the human EAP1 gene is regulated by upstream components of a puberty-controlling tumor suppressor gene network. Mol. Cell. Endocrinol 351, 184–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan P, Yang F, Di SL, Ji JY, Ouyang J, Nishikawa JL, Toiber D, Kulkarni M, Wang Q, Najafi-Shoushtari SH, Mostoslavsky R, Gygi SP, Gill G, Dyson NJ, Naar AM, 2011. A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development. Mol. Cell 42, 689–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente P, Sassone-Corsi P, 2008. The NAD+dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente P, Sassone-Corsi P, 2008. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse Y, Oh-hashi K, Iijima N, Naruse M, Yoshioka H, Tanaka M, 2004. Circadian and light-induced transcription of clock gene Per1 depends on histone acetylation and deacetylation. Mol. Cell. Biol 24, 6278–6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro VM, Gottsch ML, Chavkin C, Okamura H, Clifton DK, Steiner RA, 2009. Regulation of gonadotropin-releasing hormone secretion by kisspeptin/dynorphin/neurokinin B neurons in the arcuate nucleus of the mouse. J. Neurosci 29, 11859–11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro VM, Gottsch ML, Wu M, Garcia-Galiano D, Hobbs SJ, Bosch MA, Pinilla L, Clifton DK, Dearth A, Ronnekleiv OK, Braun RE, Palmiter RD, Tena-Sempere M, Alreja M, Steiner RA, 2011. Regulation of NKB pathways and their roles in the control of Kiss1 neurons in the arcuate nucleus of the male mouse. Endocrinology 152, 4265–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley AE, Clifton DK, Steiner RA, 2009. Kisspeptin signaling in the brain. Endocr. Rev 30, 713–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hagan HM, Wang W, Sen S, Destefano SC, Lee SS, Zhang YW, Clements G, Cai Y, Van NL, Easwaran H, Casero RA, Sears CL, Baylin SB, 2011. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell 20, 606–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtaki T, Shintani Y, Honda S, Matsumoto H, Hori A, Kanehashi K, Terao Y, Kumano S, Takatsu Y, Masuda Y, Ishibashi Y, Watanabe T, Asada M, Yamada T, Suenaga M, Kitada C, Usuki S, Kurokawa T, Onda H, Nishimura O, Lost Data, 2001. Metastasis suppressor gene KiSS-1 encodes peptide ligand of a G-protein-coupled receptor. Nature 411, 613–617. [DOI] [PubMed] [Google Scholar]
- Ojeda SR, 1991. The mystery of mammalian puberty: how much more do we know? Perspect. Biol. Med 34, 365–383. [DOI] [PubMed] [Google Scholar]
- Ojeda SR, Skinner MK, 2006. Puberty in the rat In: Neill JD (Ed.), The Physiology of Reproduction, third ed Academic Press/Elsevier, San Diego, pp. 2061–2126. [Google Scholar]
- Ojeda SR, Terasawa E, 2002. Neuroendocrine regulation of puberty In: Pfaff D, Arnold A, Etgen A, Fahrbach S, Moss R, Rubin R (Eds.), Hormones, Brain and Behavior, vol. 4 Elsevier, New York, pp. 589–659. [Google Scholar]
- Ojeda SR, Urbanski HF, 1994. Puberty in the rat In: Knobil E, Neill JD (Eds.), The Physiology of Reproduction, second ed., vol. 2 Raven Press, New York, pp. 363–409. [Google Scholar]
- Ojeda SR, Hill J, Hill DF, Costa ME, Tapia V, Cornea A, Ma YJ, 1999. The Oct-2 POU-domain gene in the neuroendocrine brain: a transcriptional regulator of mammalian puberty. Endocrinology 140, 3774–3789. [DOI] [PubMed] [Google Scholar]
- Ojeda SR, Lomniczi A, Mastronardi C, Heger S, Roth C, Parent AS, Matagne V, Mungenast AE, 2006. Minireview: the neuroendocrine regulation of puberty: is the time ripe for a systems biology approach? Endocrinology 147, 1166–1174. [DOI] [PubMed] [Google Scholar]
- Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC, 2008. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 453, 803–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong KK, Elks CE, Li S, Zhao JH, Luan J, Andersen LB, Bingham SA, Brage S, Smith GD, Ekelund U, Gillson CJ, Glaser B, Golding J, Hardy R, Khaw KT, Kuh D, Luben R, Marcus M, McGeehin MA, Ness AR, Northstone K, Ring SM, Rubin C, Sims MA, Song K, Strachan DP, Vollenweider P, Waeber G, Waterworth DM, Wong A, Deloukas P, Barroso I, Mooser V, Loos RJ, Wareham NJ, 2009. Genetic variation in LIN28B is associated with the timing of puberty. Nat. Genet 41, 729–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH, 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448, 714–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G, 2013. Latent enhancers activated by stimulation in differentiated cells. Cell 152, 157–171. [DOI] [PubMed] [Google Scholar]
- Otte AP, Kwaks TH, 2003. Gene repression by Polycomb group protein complexes: a distinct complex for every occasion? Curr. Opin. Genet. Dev 13, 448–454. [DOI] [PubMed] [Google Scholar]
- Owen BM, Bookout AL, Ding X, Lin VY, Atkin SD, Gautron L, Kliewer SA, Mangelsdorf DJ, 2013. FGF21 contributes to neuroendocrine control of female reproduction. Nat. Med 19, 1153–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent AS, Krstevska-Konstantinova M, Matagne V, Gerard A, Lebrethon MC, Charlier C, Bourguignon J-P, 2001. Endocrine disrupter contribution to sexual precocity: suggestive detection of pesticide derivatives in patients immigrant to Belgium and stimulation of GnRH pulsatility in rat hypothalamus. Pediatr. Res 49, 139A.11158503 [Google Scholar]
- Parent AS, Rasier G, Gerard A, Heger S, Roth C, Mastronardi C, Jung H, Ojeda SR, Bourguignon JP, 2005. Early onset of puberty: tracking genetic and environmental factors. Horm. Res 64 (Suppl. 2), 41–47. [DOI] [PubMed] [Google Scholar]
- Parent AS, Mungenast AE, Lomniczi A, Sandau US, Peles E, Bosch MA, Ronnekleiv OK, Ojeda SR, 2007. A contactin-receptor-like protein tyrosine phosphatase beta complex mediates adhesive communication between astroglial cells and gonadotrophin-releasing hormone neurones. J. Neuroendocrinol 19, 847–859. [DOI] [PubMed] [Google Scholar]
- Parkash J, Kaur G, 2005. Neuronal-glial plasticity in gonadotropin-releasing hormone release in adult female rats: role of the polysialylated form of the neural cell adhesion molecule. J. Endocrinol 186, 397–409. [DOI] [PubMed] [Google Scholar]
- Perera AD, Lagenaur CF, Plant TM, 1993. Postnatal expression of polysialic acid-neural cell adhesion molecule in the hypothalamus of the male rhesus monkey (Macaca mulatta). Endocrinology 133, 2729–2735. [DOI] [PubMed] [Google Scholar]
- Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV, 2008. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science 319, 202–206. [DOI] [PubMed] [Google Scholar]
- Perry JR, Stolk L, Franceschini N, Lunetta KL, Zhai G, McArdle PF, Smith AV, Aspelund T, Bandinelli S, Boerwinkle E, Cherkas L, Eiriksdottir G, Estrada K, Ferrucci L, Folsom AR, Garcia M, Gudnason V, Hofman A, Karasik D, Kiel DP, Launer LJ, van MJ, Nalls MA, Rivadeneira F, Shuldiner AR, Singleton A, Soranzo N, Tanaka T, Visser JA, Weedon MN, Wilson SG, Zhuang V, Streeten EA, Harris TB, Murray A, Spector TD, Demerath EW, Uitterlinden, Murabito JM, 2009. Meta-analysis of genome-wide association data identifies two loci influencing age at menarche. Nat. Genet 41, 648–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinilla L, Aguilar E, Dieguez C, Millar RP, Tena-Sempere M, 2012. Kisspeptins and reproduction: physiological roles and regulatory mechanisms. Physiol. Rev 92, 1235–1316. [DOI] [PubMed] [Google Scholar]
- Plant TM, Witchel SF, 2006. Puberty in nonhuman primates and humans In: Neill JD (Ed.), The Physiology of Reproduction, third ed Academic Press/Elsevier, San Diego, pp. 2177–2230. [Google Scholar]
- Poling MC, Kim J, Dhamija S, Kauffman AS, 2012. Development, sex steroid regulation, and phenotypic characterization of RFamide-Related Peptide (Rfrp) gene expression and RFamide receptors in the mouse hypothalamus. Endocrinology 153, 1827–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompolo S, Pereira A, Estrada KM, Clarke IJ, 2006. Colocalization of kisspeptin and gonadotropin-releasing hormone in the ovine brain. Endocrinology 147, 804–810. [DOI] [PubMed] [Google Scholar]
- Prevot V, 2002. Glial-neuronal-endothelial interactions are involved in the control of GnRH secretion. J. Neuroendocrinol 14, 247–255. [DOI] [PubMed] [Google Scholar]
- Pruitt K, Zinn RL, Ohm JE, McGarvey KM, Kang SH, Watkins DN, Herman JG, Baylin SB, 2006. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2, e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin XY, Fukuda T, Yang L, Zaha H, Akanuma H, Zeng Q, Yoshinaga J, Sone H, 2012. Effects of bisphenol A exposure on the proliferation and senescence of normal human mammary epithelial cells. Cancer Biol. Ther 13, 296–306. [DOI] [PubMed] [Google Scholar]
- Ramadori G, Lee CE, Bookout AL, Lee S, Williams KW, Anderson J, Elmquist JK, Coppari R, 2008. Brain SIRT1: anatomical distribution and regulation by energy availability. J. Neurosci 28, 9989–9996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmers F, Delemarre-van de Waal HA, 2011. Developmental programming of energy balance and its hypothalamic regulation. Endocr. Rev 32, 272–311. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough H, Helms JA, Farnham PJ, Segal E, Chang HY, 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger JA, Schibler U, 2006. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet 38, 369–374. [DOI] [PubMed] [Google Scholar]
- Roa J, Garcia-Galiano D, Castellano JM, Gaytan F, Pinilla L, Tena-Sempere M, 2010. Metabolic control of puberty onset: new players, new mechanisms. Mol. Cell. Endocrinol 324, 87–94. [DOI] [PubMed] [Google Scholar]
- Robertson KD, it-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP, 2000. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet 25, 338–342. [DOI] [PubMed] [Google Scholar]
- Robertson JL, Clifton DK, de la Iglesia HO, Steiner RA, Kauffman AS, 2009. Circadian regulation of Kiss1 neurons: implications for timing the preovulatory gonadotropin-releasing hormone/luteinizing hormone surge. Endocrinology 150, 3664–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronnekleiv OK, Ojeda SR, McCann SM, 1978. Undernutrition, puberty and development of the estrogen positive feedback in the female rat. Biol. Reprod 19, 414–424. [DOI] [PubMed] [Google Scholar]
- Roth CL, Mastronardi C, Lomniczi A, Wright H, Cabrera R, Mungenast AE, Heger S, Jung H, Dubay C, Ojeda SR, 2007. Expression of a tumor-related gene network increases in the mammalian hypothalamus at the time of female puberty. Endocrinology 148, 5147–5161. [DOI] [PubMed] [Google Scholar]
- Rountree MR, Bachman KE, Baylin SB, 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet 25, 269–277. [DOI] [PubMed] [Google Scholar]
- Ruderman NB, Xu XJ, Nelson L, Cacicedo JM, Saha AK, Lan F, Ido Y, 2010. AMPK and SIRT1: a long-standing partnership? Am. J. Physiol. Endocrinol. Metab 298, E751–E760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD, 2007. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol 8, 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeb-Parsy K, Dyball RE, 2003. Defined cell groups in the rat suprachiasmatic nucleus have different day/night rhythms of single-unit activity in vivo. J. Biol. Rhythms 18, 26–42. [DOI] [PubMed] [Google Scholar]
- Sandau US, Mungenast AE, McCarthy J, Biederer T, Corfas G, Ojeda SR, 2011a. The synaptic cell adhesion molecule, SynCAM1, mediates astrocyte-to-astrocyte and astrocyte-to-GnRH neuron adhesiveness in the mouse hypothalamus. Endocrinology 152, 2353–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandau US, Mungenast AE, Alderman Z, Sardi SP, Fogel AI, Taylor B, Parent AS, Biederer T, Corfas G, Ojeda SR, 2011b. SynCAM1, a synaptic adhesion molecule, is expressed in astrocytes and contributes to erbB4 receptor-mediated control of female sexual development. Endocrinology 152, 2364–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangiao-Alvarellos S, Manfredi-Lozano M, Ruiz-Pino F, Navarro V, Sanchez-Garrido MA, Leon S, Dieguez C, Cordido F, Matagne V, Dissen GA, Ojeda SR, Pinilla L, Tena-Sempere M, 2013. Changes in hypothalamic expression of the Lin28/let-7 system and related microRNAs during postnatal maturation and after experimental manipulations of puberty. Endocrinology 154, 942–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, Sampath SC, Intrator A, Min A, Gertler TS, Surmeier DJ, Tarakhovsky A, Greengard P, 2009. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron 64, 678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Martinez AM, Iovino N, Cavalli G, 2011. Trithorax group proteins: switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol 12, 799–814. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V, 2013. A new world of polycombs: unexpected partnerships and emerging functions. Nat. Rev. Genet 14, 853–864. [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Nugent BM, McCarthy MM, 2010. Developmental and hormone-induced epigenetic changes to estrogen and progesterone receptor genes in brain are dynamic across the life span. Endocrinology 151, 4871–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semaan SJ, Dhamija S, Kim J, Ku EC, Kauffman AS, 2012. Assessment of epigenetic contributions to sexually-dimorphic kiss1 expression in the anteroventral periventricular nucleus of mice. Endocrinology 153, 1875–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS Jr., Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, Zahn D, Dixon J, Kaiser UB, Slaugenhaupt SA, Gusella JF, O’Rahilly S, Carlton MB, Crowley WF Jr., Aparicio SA, Colledge WH, 2003. The GPR54 gene as a regulator of puberty. N. Engl. J. Med 349, 1614–1627. [DOI] [PubMed] [Google Scholar]
- Shahab M, Mastronardi C, Seminara SB, Crowley WF, Ojeda SR, Plant TM, 2005. Increased hypothalamic GPR54 signaling: a potential mechanism for initiation of puberty in primates. Proc. Natl. Acad. Sci. U.S.A 102, 2129–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Sandoval J, Swiss VA, Li J, Dupree J, Franklin RJM, Casaccia-Bonnefil P, 2008. Age-dependent epigenetic control of differentiation inhibitors is critical for remyelination efficiency. Nat. Neurosci 11, 1024–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A, 2012. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem 81, 65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoba B, Lwin ZM, Ling LS, Bay BH, Yip GW, Kumar SD, 2009. Function of sirtuins in biological tissues. Anat. Rec. (Hoboken) 292, 536–543. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE, 2009. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol 10, 697–708. [DOI] [PubMed] [Google Scholar]
- Sloboda DM, Howie GJ, Pleasants A, Gluckman PD, Vickers MH, 2009. Pre- and postnatal nutritional histories influence reproductive maturation and ovarian function in the rat. PLoS ONE 4, e6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gomez-Marin C, Aneas, Credidio FL, Sobreira DR, Wasserman NF, Lee JH, Puviindran V, Tam D, Shen M, Son JE, Vakili NA, Sung HK, Naranjo S, Acemel RD, Manzanares M, Nagy A, Cox NJ, Hui CC, Gomez-Skarmeta JL, Nobrega MA, 2014. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507, 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale RC, Tsai MC, Chang HY, 2011. RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics 6, 539–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadhouders R, van den Heuvel A, Kolovos P, Jorna R, Leslie K, Grosveld F, Soler E, 2012. Transcription regulation by distal enhancers: who’s in the loop? Transcription 3, 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeg PS, Ouatas T, Halverson D, Palmieri D, Salerno M, 2003. Metastasis suppressor genes: basic biology and potential clinical use. Clin. Breast Cancer 4, 51–62. [DOI] [PubMed] [Google Scholar]
- Strobel A, Issad T, Camoin L, Ozata M, Strosberg AD, 1998. A leptin missense mutation associated with hypogonadism and morbid obesity. Nat. Genet 18, 213–215. [DOI] [PubMed] [Google Scholar]
- Subramanian K, Jia D, Kapoor-Vazirani P, Powell DR, Collins RE, Sharma D, Peng J, Cheng X, Vertino PM, 2008. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol. Cell 30, 336–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulem P, Gudbjartsson DF, Rafnar T, Holm H, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Alexandersen P, Feenstra B, Boyd HA, Aben KK, Verbeek AL, Roeleveld N, Jonasdottir A, Styrkarsdottir U, Steinthorsdottir V, Karason A, Stacey SN, Gudmundsson J, Jakobsdottir M, Thorleifsson G, Hardarson G, Gulcher J, Kong A, Kiemeney LA, Melbye M, Christiansen C, Tryggvadottir L, Thorsteinsdottir U, Stefansson K, 2009. Genome-wide association study identifies sequence variants on 6q21 associated with age at menarche. Nat. Genet 41, 734–738. [DOI] [PubMed] [Google Scholar]
- Sykiotis GP, Pitteloud N, Seminara SB, Kaiser UB, Crowley WF Jr., 2010. Deciphering genetic disease in the genomic era: the model of GnRH deficiency. Sci. Transl. Med 2, 32rv2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal, Iyer LM, Liu DR, Aravind L, Rao A, 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanikawa C, Okada Y, Takahashi A, Oda K, Kamatani N, Kubo M, Nakamura Y, Matsuda K, 2013. Genome wide association study of age at menarche in the Japanese population. PLoS ONE 8, e63821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, Wutz A, Vidal M, Elderkin S, Brockdorff N, 2012. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 148, 664–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teles MG, Bianco SD, Brito VN, Trarbach EB, Kuohung W, Xu S, Seminara SB, Mendonca BB, Kaiser UB, Latronico AC, 2008. A GPR54-activating mutation in a patient with central precocious puberty. N. Engl. J. Med 358, 709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena-Sempere M, 2008. Ghrelin as a pleotrophic modulator of gonadal function and reproduction. Nat. Clin. Pract. Endocrinol. Metab 4, 666–674. [DOI] [PubMed] [Google Scholar]
- Terasawa E, 1999. Hypothalamic control of the onset of puberty. Curr. Opin. Endocrinol. Diabetes 6, 44–49. [Google Scholar]
- Terasawa E, Fernandez DL, 2001. Neurobiological mechanisms of the onset of puberty in primates. Endocr. Rev 22, 111–151. [DOI] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Saiakhova AR, Howard B, Monteith KE, Scacheri PC, Cosgrove MS, Harte PJ, 2014. Trithorax monomethylates histone H3K4 and interacts directly with CBP to promote H3K27 acetylation and antagonize Polycomb silencing. Development 141, 1129–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilghman SL, Bratton MR, Segar HC, Martin EC, Rhodes LV, Li M, McLachlan JA, Wiese TE, Nephew KP, Burow ME, 2012. Endocrine disruptor regulation of microRNA expression in breast carcinoma cells. PLoS ONE 7, e32754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolson KP, Chappell PE, 2012. The changes they are a-timed: metabolism, endogenous clocks, and the timing of puberty. Front. Endocrinol. (Lausanne) 3, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomikawa J, Uenoyama Y, Ozawa M, Fukanuma T, Takase K, Goto T, Abe H, Ieda N, Minabe S, Deura C, Inoue N, Sanbo M, Tomita K, Hirabayashi M, Tanaka S, Imamura T, Okamura H, Maeda K, Tsukamura H, 2012. Epigenetic regulation of Kiss1 gene expression mediating estrogen-positive feedback action in the mouse brain. Proc. Natl. Acad. Sci. U.S.A 109, E1294–E1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topaloglu AK, Reimann F, Guclu M, Yalin AS, Kotan LD, Porter KM, Serin A, Mungan NO, Cook JR, Ozbek MN, Imamoglu S, Akalin NS, Yuksel B, O’Rahilly S, Semple RK, 2008. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for Neurokinin B in the central control of reproduction. Nat. Genet 41, 354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topaloglu AK, Tello JA, Kotan LD, Ozbek MN, Yilmaz MB, Erdogan S, Gurbuz F, Temiz F, Millar RP, Yuksel B, 2012. Inactivating KISS1 mutation and hypogonadotropic hypogonadism. N. Engl. J. Med 366, 629–635. [DOI] [PubMed] [Google Scholar]
- True C, Kirigiti MA, Kievit P, Grove KL, Smith MS, 2011. Leptin is not the critical signal for kisspeptin or luteinising hormone restoration during exit from negative energy balance. J. Neuroendocrinol 23, 1099–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui K, Bentley GE, Bedecarrats G, Osugi T, Ubuka T, Kriegsfeld LJ, 2010. Gonadotropin-inhibitory hormone (GnIH) and its control of central and peripheral reproductive function. Front. Neuroendocrinol 31, 284–295. [DOI] [PubMed] [Google Scholar]
- Urbanski HF, Ojeda SR, 1985. The juvenile-peripubertal transition period in the female rat: establishment of a diurnal pattern of pulsatile luteinizing hormone secretion. Endocrinology 117, 644–649. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Reinberg D, 2009. Calorie restriction and the exercise of chromatin. Genes Dev. 23, 1849–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D, 2007. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature 450, 440–444. [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, Mann M, 2010. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142, 967–980. [DOI] [PubMed] [Google Scholar]
- Wakabayashi Y, Nakada T, Murata K, Ohkura S, Mogi K, Navarro VM, Clifton DK, Mori Y, Tsukamura H, Maeda K, Steiner RA, Okamura H, 2010. Neurokinin B and dynorphin A in kisspeptin neurons of the arcuate nucleus participate in generation of periodic oscillation of neural activity driving pulsatile gonadotropin-releasing hormone secretion in the goat. J. Neurosci 30, 3124–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K, 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Gen 40, 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Moya N, Niessen S, Hoover H, Mihaylova MM, Shaw RJ, Yates JR III, Fischer WH, Thomas JB, Montminy M, 2011. A hormone-dependent module regulating energy balance. Cell 145, 596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q, 2010. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucl. Acids Res 38, W214–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westberry JM, Prewitt AK, Wilson ME, 2008. Epigenetic regulation of the estrogen receptor alpha promoter in the cerebral cortex following ischemia in male and female rats. Neuroscience 152, 982–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe AP, Matzke MA, 1999. Epigenetics: regulation through repression. Science 286, 481–486. [DOI] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, Crabtree GR, 2007. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron 56, 94–108. [DOI] [PubMed] [Google Scholar]
- Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A, 2008. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol. Cell. Biol 28, 7337–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung KT, Das S, Zhang J, Lomniczi A, Ojeda S, Xu CF, Neubert TA, Samuels HH, 2011. A novel transcription complex that selectively modulates apoptosis of breast cancer cells through regulation of FASTKD2. Mol. Cell. Biol 31, 2287–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]