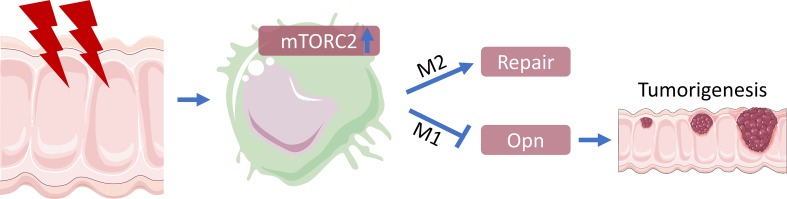
Abstract
The mechanistic target of rapamycin complex 2 (mTORC2) is a potentially novel and promising anticancer target due to its critical roles in proliferation, apoptosis, and metabolic reprogramming of cancer cells. However, the activity and function of mTORC2 in distinct cells within malignant tissue in vivo is insufficiently explored. Surprisingly, in primary human and mouse colorectal cancer (CRC) samples, mTORC2 signaling could not be detected in tumor cells. In contrast, only macrophages in tumor-adjacent areas showed mTORC2 activity, which was downregulated in stromal macrophages residing within human and mouse tumor tissues. Functionally, inhibition of mTORC2 by specific deletion of Rictor in macrophages stimulated tumorigenesis in a colitis-associated CRC mouse model. This phenotype was driven by a proinflammatory reprogramming of mTORC2-deficient macrophages that promoted colitis via the cytokine SPP1/osteopontin to stimulate tumor growth. In human CRC patients, high SPP1 levels and low mTORC2 activity in tumor-associated macrophages correlated with a worsened clinical prognosis. Treatment of mice with a second-generation mTOR inhibitor that inhibits mTORC2 and mTORC1 exacerbated experimental colorectal tumorigenesis in vivo. In conclusion, mTORC2 activity is confined to macrophages in CRC and limits tumorigenesis. These results suggest activation but not inhibition of mTORC2 as a therapeutic strategy for colitis-associated CRC.
Keywords: Gastroenterology, Immunology
Keywords: Colorectal cancer, Drug therapy, Innate immunity
A pro-inflammatory metabolic reprogramming of mTORC2-deficient macrophages promotes tumor growth in colorectal cancer.
Introduction
The mechanistic target of rapamycin (mTOR) is an evolutionary conserved serine/threonine kinase that exists in 2 complexes and regulates a broad set of basic cellular and metabolic processes (1). mTOR complex 1 (mTORC1) and mTORC2 have distinct functions for regulating growth, proliferation, and energy homeostasis at the cellular and organismal level (2–7). mTORC2 is defined by the rapamycin-insensitive companion of mTOR (Rictor) and directly phosphorylates Akt at serine 473 and PKCα at serine 657 to perform these functions (8). Due to mTOR’s manifold growth-promoting roles, mTORC1 and mTORC2 are implicated in pathological conditions, such as inflammation and cancer (2, 3, 9–12).
The gastrointestinal tract is the largest immune compartment of the human body (13). A dysregulation of gut homeostasis occurs in Crohn’s disease or ulcerative colitis that belongs to a group of inflammatory disorders of the intestine known as inflammatory bowel disease (IBD) (14). IBD is of unclear etiology and affects about 3.5–4 million people in Europe and the United States (15). Chronic inflammation of the colon in IBD can cause injury of the epithelial barrier and lead to an imbalanced gut homeostasis, which are driving forces for carcinogenesis (16). Hence, IBD predisposes to colorectal cancer (CRC), referred to as colitis-associated cancer (CAC), the third most common cancer, globally (17). CAC develops in 20% of patients diagnosed with IBD, and mortality rates are still over 50% in this patient group (18).
Myeloid immune cells such as monocytes and macrophages are an important part of the innate immune system. They are instrumental in controlling the barrier function in the small and the large intestine to maintain tissue homeostasis and prevent inflammation, as well as tumorigenesis (19, 20). The mTOR pathway controls many aspects of innate immune cell function, such as the production of pro- versus antiinflammatory mediators (5, 21). Myeloid mTORC1 has been associated with a protective role for intestinal homeostasis (22). Mice deficient in mTORC1 signaling in CD11c+ cells are more susceptible to dextran sodium sulfate–induced (DSS-induced) colitis via a mechanism that includes the production of insufficient amounts of IL-10 (22). Moreover, Rictor-deficient macrophages have alterations in their polarization potential (23–27). However, the role of myeloid mTORC2 during colitis and CAC remained elusive so far, but it is of importance due to the regained recognition of the immune system as controller and mediator of antitumor responses and therapies. Its role during inflammation-driven tumorigenesis is especially of interest, as new ATP-competitive inhibitors that block both mTORC1 and mTORC2 are currently being developed for the treatment of cancer (28, 29).
First-generation inhibitors such as rapamycin that mainly target mTORC1 showed only limited antitumor efficacy in clinical trials (30–32). This was attributed to a failure to efficiently inhibit mTORC2. The catalytic inhibitors that target both mTORC1 and mTORC2 showed greater potential in preclinical tumor models and are currently tested in various clinical trials (28, 29, 33–37). While mTORC1 or mTORC2 are essentially always active in cancer cells in vitro when cultured under normal cell culture conditions, the mTOR activation status of normal and malignant tissue cells in vivo is often unknown. Here, we addressed the heterogeneity of mTOR activation during CRC in mice and human patients. We propose that understanding the role of mTORC2 in vivo in different cellular compartments, such as the innate immune system, is necessary to develop a rationale for the clinical use of novel inhibitors targeting the mTOR complex.
Results
mTORC2 is downregulated in infiltrating macrophages in human and mouse CRC.
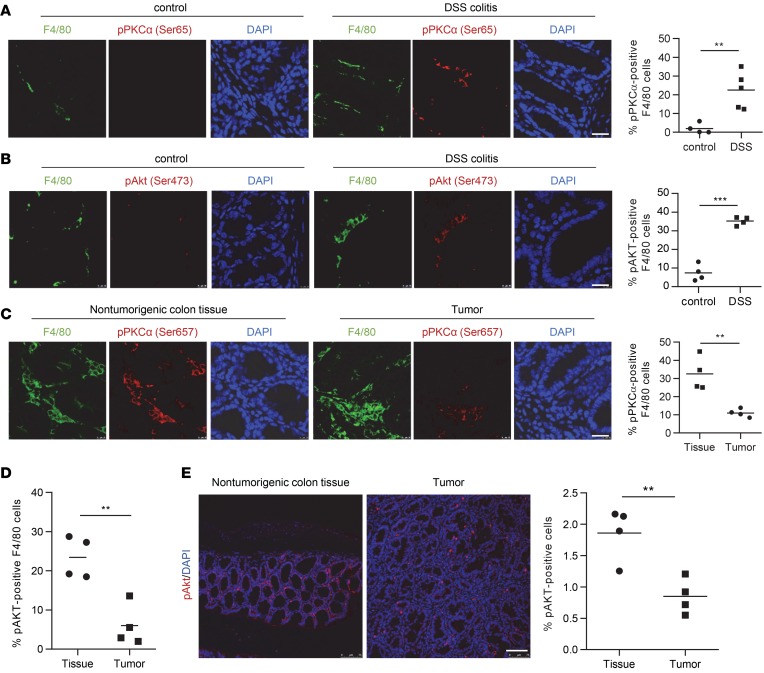
To investigate the activation status of mTORC2 of colon tissue cells in vivo, we profiled phosphorylation of the direct mTORC2 targets Akt at serine 473 and PKCα at serine 657 by immunofluorescence in mouse colonic tissue in the DSS-induced colitis model and the azoxymethane/DSS/CAC (AOM/DSS/CAC) model (38–40). Representative colitis and CAC examples are shown in Supplemental Figure 1 (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.124164DS1). Interestingly, activation of mTORC2 identified by phosphorylation of PKCα and Akt was largely confined to macrophages and increased during DSS-induced colitis (Figure 1, A and B). Moreover, we observed in the CAC model that, in nontumorigenic colon tissue, 30%–40% of F4/80 tissue macrophages stained positive for pPKCα, whereas — in the tumor stroma — only around 10% of the macrophages were positive for pPKCα (Figure 1C). Similarly, we noticed reduced pAKT+ tumor–associated F4/80 macrophages compared with nontumorigenic tissue (Figure 1D). Analysis of total pAKT+ cells yielded similar results (Figure 1E).
Figure 1. mTORC2 is downregulated in infiltrating macrophages in mouse colitis and CAC.
(A and B) IF double staining for F4/80 (green) and (A) pPKCα Ser657 (red) or (B) pAKT Ser473 (red) in mouse control and DSS colitis sections and quantitative number and means of pAKT-positive F4/80 macrophages. Scale bar: 20 µm. (C–E) IF for F4/80 (green) and pPKCα Ser657 (red) (C) or pAKT Ser473 (red) (D and E) in nontumorigenic mouse colon tissue and colorectal tumor sections, and quantitative number and means of pPKCα-positive F4/80 macrophages. Scale bar: 20 µm. A–C evaluated from 4 IF samples. P values were determined by unpaired 2-tailed Student’s t test. **P < 0.01, ***P < 0.001.
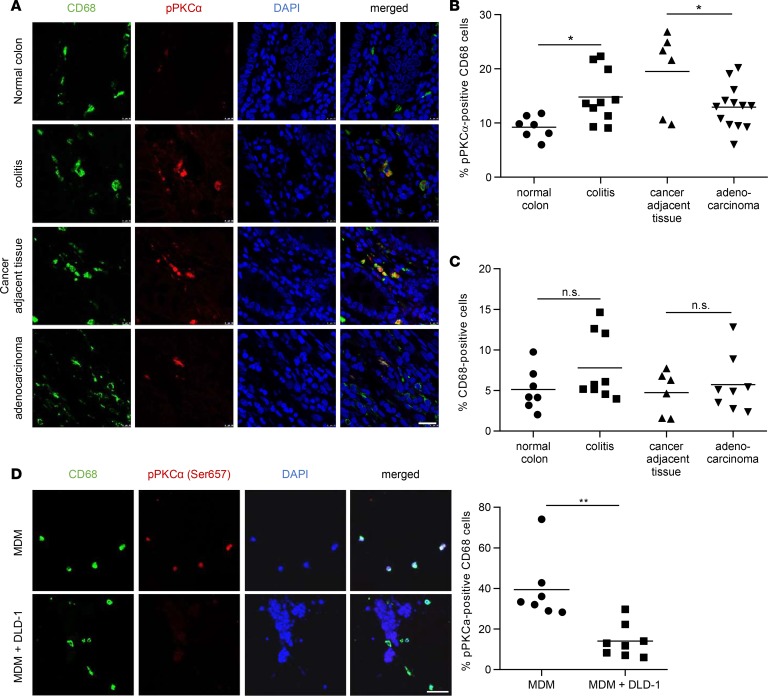
Next, we examined human colon tissue. Similar to the mouse, pPKCα staining was hardly detectable and confined to CD68+ macrophages in healthy colons (Figure 2A). In chronic colitis patients, colonic macrophages showed increased mTORC2 activation compared with healthy controls (Figure 2, A and B). In patients with CRC, stromal macrophages in nontumorigenic colonic tissue adjacent to the tumor site similarly had high mTORC2 activity (Figure 2, A and B). CD68+ tumor–associated macrophages, present within the tumor stroma, showed a significantly decreased staining for pPKCα when compared with stromal macrophages in nontumorigenic colonic tissue (Figure 2, A and B). Tumor or other stromal cells showed no detectable staining for pPKCα in any of the analyzed conditions. Absolute numbers of CD68+ cells did not significantly differ between the groups (Figure 2C). To corroborate these findings in vitro, we investigated pPKCα phosphorylation in a 3-dimensional (3-D) coculture model using human monocyte-derived macrophages (MDMs) and the human CRC cell line DLD-1. Similar to the in vivo situation, in the presence of tumor cells, reduced pPKCα signaling was observed in macrophages (Figure 2D). These results suggest that mTORC2 activity in mouse and humans is largely confined to macrophages during chronic colitis, as well as CAC, and that mTORC2 activation is reduced in tumor-associated macrophages compared with cancer adjacent tissue.
Figure 2. mTORC2 is downregulated in infiltrating macrophages in human CRC.
(A) IF for CD68 (green) and pPKCα (red) in human colorectal tissue sections. Scale bar: 20 μm. (B) Quantitative number and means of pPKCα+CD68+ macrophages. (C) Quantitative number and means of CD68+ macrophages. A–C were evaluated from at least 6 different IF samples. P values were determined by 1-way ANOVA and Bonferroni correction for pairwise comparisons. (D) IF double stain for CD68 (green) and pPKCα Ser657 (red) in collagen gels either with monocyte-derived macrophages (MDM) alone (upper panel) or with MDM cultured with DLD-1 cells (lower panel, nuclear stain only). Quantitative number and means of pPKCα+CD68+ macrophages, evaluated from at least 6 samples. Scale bar: 50 μm. P values were determined by unpaired 2-tailed Student’s t test. *P < 0.05; **P < 0.01.
mTORC2 in macrophages suppresses colorectal tumor formation.
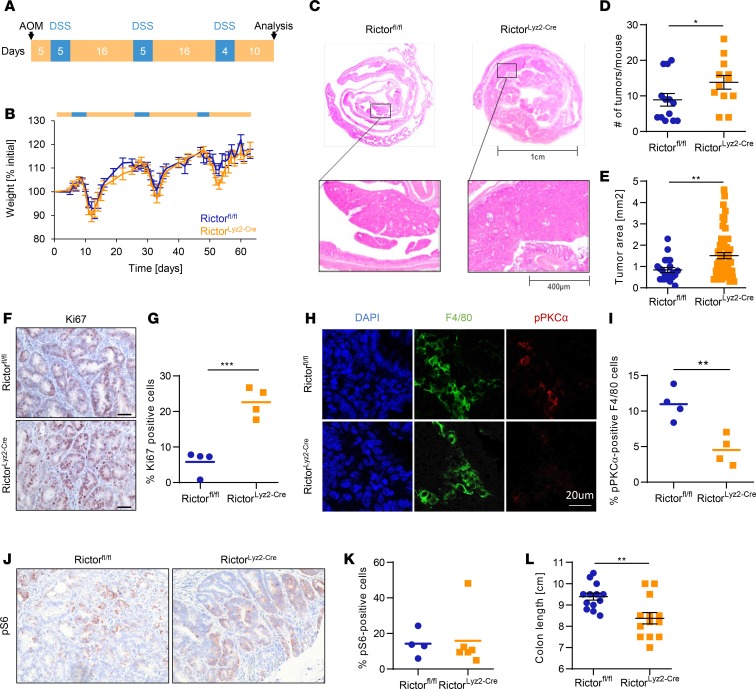
Since we observed a downregulation of mTORC2 activity in tumor-associated macrophages in human and mouse CAC, we wanted to further investigate the role of mTORC2 in macrophages in vivo. Therefore, we generated mice with a conditional deletion of Rictor in cells expressing the lysozyme M gene (RictorLyz2-Cre) and applied the AOM/DSS/CAC model (Figure 3A). We confirmed efficient recombination of the floxed allele in peritoneal and colonic macrophages but not in epithelial, dendritic, or T cells of the colon (Supplemental Figure 2). Diarrhea-mediated weight loss during the 3 DSS cycles was similar between RictorLyz2-Cre and control mice, although there was a nonsignificant trend to higher weight loss in the later DSS cycles in the RictorLyz2-Cre mice (Figure 3B). Interestingly, the number of tumors in the colon, which is the main readout of this model, was significantly higher in the RictorLyz2-Cre animals when analyzed 10 days after the last DSS cycle (Figure 3, C and D). In addition, the tumor area was also increased, suggesting that mTORC2 activity in myeloid cells limits tumorigenesis (Figure 3, C and E). The percent of Ki-67+ tumor cells was significantly higher in RictorLyz2-Cre mice (Figure 3, F and G). As expected, the low activity of mTORC2 in tumor-associated macrophages of Rictorfl/fl mice was further reduced in macrophages of RictorLyz2-Cre mice (Figure 3, H and I, and Supplemental Figure 3). Phosphorylation of pS6 as readout for mTORC1 activity indicated that mostly tumor cells and not stromal cells stained positive for pS6 (Figure 3, J and K). However, we observed a very uneven distribution of pS6 positivity, with many tumors not showing detectable mTORC1 activity (data not shown). The colon length was decreased in RictorLyz2-Cre mice, pointing toward enhanced colitis (41) in the mice (Figure 3L). These data, in total, indicate that mTORC2 signaling in macrophages negatively regulates colorectal tumorigenesis.
Figure 3. RictorLyz2-Cre mice have increased susceptibility for colorectal cancer.
(A) Scheme of the AOM/DSS-induced colitis–associated colorectal cancer model. To induce colorectal cancer, RictorLyz2-Cre and control (Rictorfl/fl) mice were challenged with AOM, followed by 3 cycles of 2% DSS in the drinking water. (B) Weight of Rictorfl/fl and RictorLyz2-Cre mice was monitored during the procedure (n = 12–13). (C) Representative H&E colon stainings of AOM/DSS-treated mice are shown. (D and E) Tumor number (D) and tumor area (E) are depicted. Data represent the mean ± SEM (n = 12–13). (F) IHC for Ki67 in colorectal tumor sections. Scale bar: 40 μm. (G) Quantitative number and means of Ki67+ cells. (H) IF for F4/80 (green) and pPKCα (red) in colorectal tumor sections. Scale bar: 20 μm. (I) Quantitative number and means for pPKCα+ F4/80 macrophages. (J) IHC for pS6 in colorectal tumor sections. Scale bar: 40 μm. (K) Quantitative number and means of pS6+ cells. (L) Colon length of tumor-bearing mice is shown. Data represent the mean ± SEM (n = 12–13). P values were determined by unpaired 2-tailed Student’s t test. *P < 0.05; **P < 0.01; *** P < 0.001.
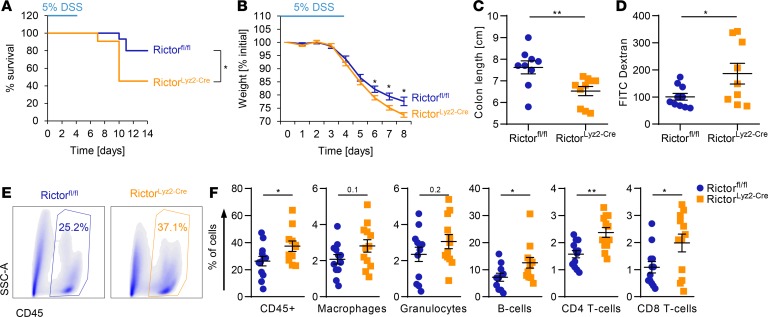
Rictor in macrophages prevents exacerbated DSS-induced colitis.
To examine mechanisms responsible for the observed enhanced CRC susceptibility of RictorLyz2-Cre mice, we analyzed early phases of CAC development when chronic inflammation acts as a major driver. We challenged mice with 5% DSS in the drinking water for 5 days. Survival analysis revealed a significantly increased lethality in the RictorLyz2-Cre mice compared with control mice (Rictorfl/fl) (Figure 4A). In addition, RictorLyz2-Cre mice lost more weight than Rictorfl/fl mice after the DSS cycle (Figure 4B). Moreover, the length of the colon was significantly shorter in the RictorLyz2-Cre mice on day 8 (Figure 4C). We observed an upregulated permeability of the colon in the RictorLyz2-Cre mice on day 8 (Figure 4D) that was accompanied by an increased infiltration of CD45+ leukocytes consisting of CD4+ T cells, CD8+ T cells, and CD19+ B cells into their colon (Figure 4, E and F, and Supplemental Figure 4). Hence, mTORC2 activity in macrophages contributes to the maintenance of gut homeostasis and limits inflammation during epithelial damage.
Figure 4. RictorLyz2-Cre mice are more susceptible to DSS-induced acute colitis.
(A–F) Rictorfl/fl and RictorLyz2-Cre mice were challenged with 5% DSS in drinking water for 5 days, followed by normal water for 9 (A) or 3 (B–F) days. Survival (A) and weight (B) was monitored daily. One representative of 2 independent experiments is shown. (A) Data represent survival of 15 Rictorfl/fl and 11 RictorLyz2-Cre mice. P value was determined by log-rank test. (B) Data represent the mean ± SEM (n = 9–11). (C) Colon length on day 8. Data represent the mean ± SEM (n = 9–11). (D) Colon permeability on day 8 by FITC-Dextran gavage and subsequent measurement of fluorescence intensity in the serum. Data represent the mean ± SEM (n = 9–10). (E) Representative dot plots for the leukocyte population (CD45+) of Rictorfl/fl and RictorLyz2-Cre lamina propria cells are shown. (F) Lamina propria cells of the colon were isolated on day 8 and flow cytometric analysis of immune cell populations was performed (Supplemental Figure 2). Data represent the mean ± SEM (n = 12). P values for B–D and F were determined by unpaired 2-tailed Student’s t test. *P < 0.05; **P < 0.01.
mTORC2 regulates metabolic control of macrophage polarization in the colon.
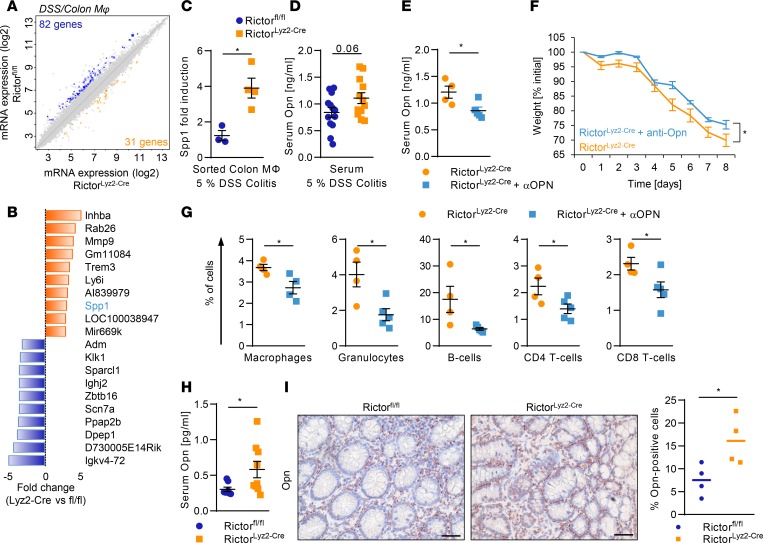
To identify functions that may mediate colon homeostasis by mTORC2 in colonic macrophages during DSS-induced acute colitis, we sorted CD11b+CD64+MHCII+Ly6C– macrophages from the colon at the peak of the inflammation on day 8. Microarray analysis revealed that 113 genes were more than 2-fold differentially expressed between the colonic macrophages of RictorLyz2-Cre and control mice (Figure 5A). Gene set enrichment analysis (GSEA) showed that antiinflammatory M2-like macrophage polarization was reduced in RictorLyz2-Cre compared with Rictorfl/fl colonic macrophages during acute DSS-induced colitis (Supplemental Figure 5A). Similarly, peritoneal macrophages (Supplemental Figure 5, B and C) and BM-derived macrophages (BMDM) (Supplemental Figure 5, D and E) showed decreased M2 marker gene expression, while M1 signature genes were increased (Supplemental Figure 5F). Hence, mTORC2 is important for efficient antiinflammatory polarization of macrophages in vitro and in vivo and prevents overactivation of proinflammatory polarization genes.
Figure 5. mTORC2 in macrophages suppresses Opn to diminish colitis.
(A) Scatterplot of global gene expression profiles of sorted Rictorfl/fl and RictorLyz2-Cre colonic macrophages on day 8 during DSS-induced colitis is shown. (B) The 10 most highly differentially regulated genes from the microarray analysis in the RictorLyz2-Cre and Rictorfl/fl colonic macrophages are shown. (C) mRNA levels of Spp1 in sorted colonic macrophages on day 8 during 5% DSS-induced colitis. Data represent the mean ± SEM (n = 3–4). (D) Opn levels in serum on day 8 of Rictorfl/fl and RictorLyz2-Cre mice during 5% DSS-induced colitis model are shown. Data represent the mean ± SEM (n = 11). (E–G) RictorLyz2-Cre mice were challenged with 5% DSS in drinking water for 5 days, followed by normal water for 3 days. Mice were i.p. injected with 650 μg/kg Opn-neutralizing antibody or placebo on days 1, 3, and 6. (E) Serum Opn levels in blood on day 8. Data represent the mean ± SEM (n = 5). (F) Weight was monitored daily. Data represent the mean ± SEM (n = 5). (G) Lamina propria cells of the colon were isolated on day 8, and flow cytometric analysis was performed. Data represent the mean ± SEM (n = 4). *P < 0.05. (H) Opn levels in serum of Rictorfl/fl and RictorLyz2-Cre mice after the AOM/DSS model are shown. Data represent the mean ± SEM (n = 12–13). (I) IHC of Opn in colorectal tumor sections of AOM/DSS-treated mice. Scale bars: 40 μm. Right panel, number, and means of Opn+ cells evaluated from 4 IHC samples. P values were determined by unpaired 2-tailed Student’s t test. *P < 0.05.
mTORC2 regulates gut homeostasis by restraining osteopontin expression.
We then investigated in more mechanistic detail how mTORC2 maintains homeostasis of the gut. Microarray analysis during acute DSS-induced colitis revealed that secreted phosphoprotein 1 (Spp1), which encodes osteopontin (Opn) (42, 43), was one of the most highly upregulated genes in RictorLyz2-Cre colonic macrophages (Figure 5B), and this was confirmed by real-time PCR analysis (Figure 5C). Interestingly, SPP1 tended to be increased in the serum of RictorLyz2-Cre mice during acute DSS-induced colitis, as well (Figure 5D). Opn has been functionally implicated in different aspects of colitis in the mouse (44, 45). Treatment of RictorLyz2-Cre mice with an Opn-neutralizing antibody diminished the serum levels of Opn in these animals (Figure 5E) and reduced the weight loss as well as immune cell infiltration into the colon of the mice significantly (Figure 5, F and G). In summary, the exacerbated inflammation in the RictorLyz2-Cre mice during the acute DSS-induced colitis could be reverted by the neutralization of Opn.
SPP1 correlates with poor survival in human CRC patients.
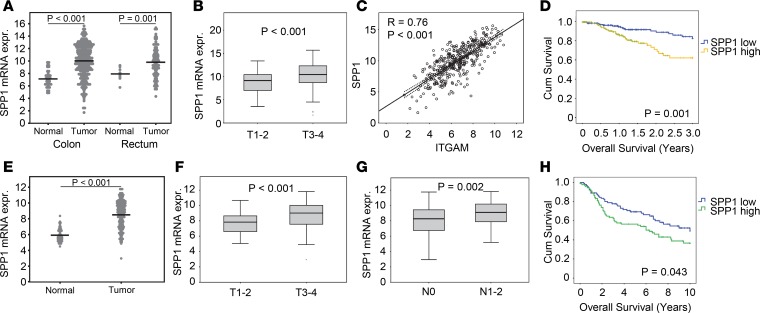
We asked whether Opn might also play a role in the tumorigenic process of CRC (46). We found significantly increased serum levels of Opn in the RictorLyz2-Cre mice (Figure 5H). Moreover, histological assessment of CAC tumors showed that Opn expression in the tumor stroma was higher in tumors of RictorLyz2-Cre mice (Figure 5I). Next, we investigated whether SPP1 is relevant for human CRC and analyzed 433 samples with colorectal adenocarcinoma (COAD) from The Cancer Genome Atlas (TCGA) (47). Expression of SPP1 was significantly higher in primary tumor samples compared with adjacent normal tissues both in the colon and rectum (Figure 6A). Moreover, SPP1 expression was found to be significantly higher in more advanced tumors of stages T3 and T4 (Figure 6B), and SPP1 was also increased in primary tumors prone to lymph node metastasis (Supplemental Figure 6A). SPP1 strongly correlated with expression of the monocyte/macrophage markers CD11b (ITGAM) (Figure 6C) and CD68 (Supplemental Figure 6B). Importantly, expression of SPP1 in primary tumors was significantly associated with lower overall survival rates after 3 years (P = 0.001) (Figure 6D) and 10 years (P = 0.013). ITGAM was not associated with overall survival (Supplemental Figure 6C). These results were replicated in a second CRC dataset (ref. 48 and Figure 6, E–H).
Figure 6. SPP1 correlates with tumor size and poor survival in human colorectal cancer patients.
(A) SPP1 mRNA expression of colon primary tumor (n = 286) compared with normal colon tissue (n = 41) and rectum primary tumor (n = 9) compared with normal rectum tissue (n = 94) of the TCGA data. Lines represent the means. (B) Box blots comparing SPP1 mRNA expression in colorectal primary tumor samples of low (T1-2, n = 69) and high (T3-4, n = 300) T stage. (C) Scatter plot showing correlations between SPP1 and ITGAM (n = 380). Regression lines with corresponding 95% CI are presented within each graph. (D) Kaplan-Meier analysis of colorectal primary tumor samples distributed into low (n = 181) and high (n = 180) SPP1 expression. (E) SPP1 mRNA expression of colon primary tumor (n = 186) compared with normal colon tissue (n = 54) of the GSE41258 data. Lines represent the means. (F and G) Box plots comparing SPP1 mRNA expression in colorectal primary tumor samples of low (T1-2, n = 38) and high (T3-4, n = 144) T stage (F) and negative (N0, n = 93) and positive (N1-2, n = 89) N stage (G). (H) Kaplan-Meier analysis of colorectal primary tumor samples distributed into low (n = 91) and high (n = 91) SPP1 expression. Data are displayed as log2 mRNA expression intensity. Unpaired 2-tailed t test was used to calculate P values. To determine P values for the Kaplan-Meier analysis, we performed log-rank test.
Myeloid mTORC2 negatively associates with outcome in human CRC.
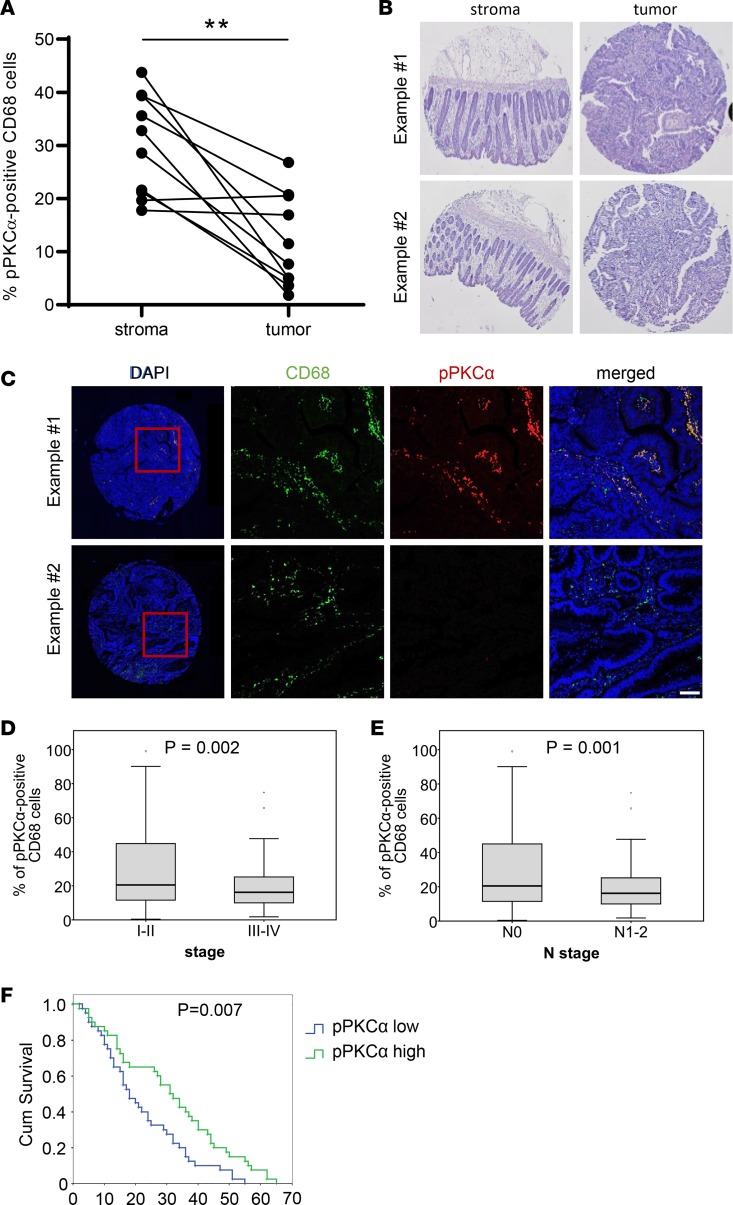
To more directly test whether mTORC2 in macrophages affects outcome in human CRC, we analyzed 180 tissue samples of CRC patients. mTORC2 activity was measured by phosphorylation of PKCα serine 657 in CD68+ macrophages. We confirmed that mTORC2 activity was downregulated in tumor areas compared with nontumorigenic regions in 10 randomly selected paired patient samples (Figure 7, A and B). mTORC2 activity in tumor areas showed patient-to-patient variability (Figure 7, A and C). mTORC2 activity in macrophages was negatively associated with tumor stage (Figure 7D) and lymph node metastasis (Figure 7E). While mTORC2 activity was not associated with overall survival (data not shown), low mTORC2 activity in macrophages in tumors was significantly associated with lower survival time in the dying population (Figure 7F). Collectively, these data suggest that inactivation of mTORC2 in macrophages is associated with a worse clinical outcome in human CRC patients.
Figure 7. mTORC2 activity correlates with tumor stage and survival in human colorectal cancer patients.
(A) Percentage of pPKCα+CD68+ macrophages in tumor and associated stroma of the corresponding patient. (B) Representative H&E staining of stroma and tumor samples from A. Magnification, 2×. (C) Tumor samples from B showing high (upper panel) and low (lower panel) pPKCα (red) staining and CD68+ macrophages (green). Scale bar: 50 µm. (D and E) Box plots comparing pPKCα+CD68+ macrophages in colorectal primary tumor samples of low (I–II, n = 63) and high (III–IV, n = 117) stage (D) and negative (N0, n = 111) and positive (N1-2, n = 69) N stage (E). (F) Kaplan-Meier analysis of colorectal primary tumor samples distributed into low (n = 83) and high (n = 83) pPKCα expression. Unpaired 2-tailed t test was used to calculate P values. To determine P values for the Kaplan-Meier analysis, we performed log-rank test.
Pharmacological inhibition of mTORC2 exacerbates experimental CAC.
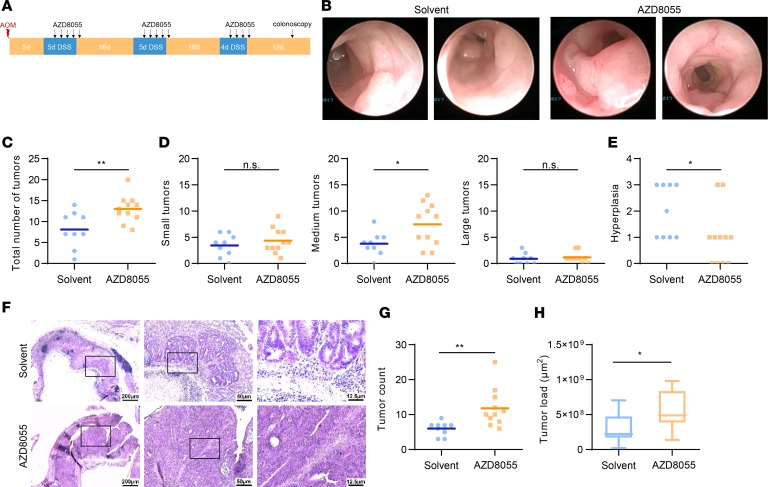
Our results at this point showed that low mTORC2 activity in macrophages was associated with a poor outcome in CAC and thus raised the possibility that pharmacological inhibition of mTORC2 might actually increase colorectal tumorigenesis. Therefore, we subjected WT mice to the AOM/DSS model and treated them with the second-generation mTOR inhibitor AZD8055 (Figure 8A). On day 6 after the last DSS cycle, we performed colonoscopy and observed significantly increased tumor numbers in the AZD8055-treated animals (Figure 8, B and C). This enhancement in tumor burden was due to an increased presence of medium-sized tumors (Figure 7D). Hyperplasia was decreased in the inhibitor-treated mice (Figure 8E). Histological analysis of colon tissue confirmed increased tumor size in animals treated with AZD8055 (Figure 8, F–H).
Figure 8. A second-generation mTOR inhibitor exacerbates tumorigenesis in DSS-induced cancer.
(A) Scheme of the AOM/DSS-induced colitis–associated cancer model with the treatment of AZD8055. Day 0 corresponds to the AOM administration. A colonoscopy was performed on day 57. Mice were sacrificed on day 67. (B) Representative pictures of the colonoscopy. (C–E) Total (C) and size-divided (D) tumor count during the colonoscopy, as well as hyperplasia (E). (F) Representative picture of the H&E stainings in 3 different magnifications. From left to right: ×2.5 (scale bar: 200 μm), ×10 (scale bar: 50 μm), ×40 (scale bar: 12.5 μm). (G and H) Tumor count (G) and tumor load (H) analyzed by the H&E stainings. P values were determined by unpaired 2-tailed Student’s t test.
Discussion
In this work, we have shown that blockade of mTORC2 in macrophages leads to aggravated disease of colitis-associated CRC in mice and humans. This was partly due to the protective role of mTORC2 during colitis to dampen inflammatory damage, a well-known driving force for the development of CRC. The exacerbated inflammation during acute colitis seen with loss of Rictor in macrophages was mediated via SPP1/Opn, as blockade of this inflammatory mediator reverted the phenotype. Deletion of Rictor in macrophages or global inhibition of mTORC1/mTORC2 increased tumor development and growth in a colitis-associated CRC model. While our findings in mice are limited to the AOM/DSS model, the reduced activity of mTORC2 was confirmed in human CRC samples. In those samples, high SPP1 expression and low mTORC2 activity in macrophages were negatively associated with a good prognostic outcome.
The recognition that mTORC2 is involved in the regulation of proliferation, apoptosis, and metabolic reprogramming of cancer cells has made this pathway a new promising target in cancer therapy (33). Anticancer targets are usually identified in vitro under growth-promoting conditions in 2-D cultures. It is important to emphasize that mTORC2 activity could not be detected in mouse and human tumor cells in vivo. Whereas mTORC2 (and mTORC1) activation is generally observed in tumor cell lines upon in vitro culture, the activation of this ubiquitous signaling pathway could not be found in human CRC cells in situ. A recent report showed that mTORC2 activity of CRC cell lines observed in 2-D cultures in vitro is lost upon 3-D culture conditions (49), signifying that a 3-D culture can better mimic the in vivo conditions concerning mTOR pathway activation. We found mTORC2 activity confined to stromal tissue macrophages during conditions of disturbed homeostasis, such as DSS-induced intestinal damage. This mTORC2 activity of macrophages was lost in tumor-infiltrating cells, suggesting that tumor cells induce inactivation of this pathway by yet-to-be-defined signals. Hence, mTORC2 is mainly active in stromal macrophages of the tissue to promote repair mechanisms and to regain tissue homeostasis. Inactivation of mTORC2 activity in the colon and in the tumor stroma, thus, enhances tumorigenesis and metastasis.
mTORC2 is implicated in the regulation of innate immune cell functions (5). In DCs, deletion of Rictor augments stimulation of T cells and the production of IL-6, TNF-α, and IL-12 (50, 51). Moreover, myeloid Rictor deficiency reduces antiinflammatory polarization potential of macrophages, while M1 polarization is similar or augmented (23–26). We found enhanced M1 and reduced M2 polarization in BMDMs and in colonic macrophages during colitis in vivo when looking at macrophages deficient for functional mTORC2 activity. Our data show that myeloid mTORC2 is a critical regulator of gut homeostasis during acute colitis and colitis-associated CRC, likely due to the reduced antiinflammatory polarization of Rictor-deficient macrophages. Interestingly, in a noninflammatory mouse model of melanoma, mTORC2 deficiency in macrophages is beneficial (25), while our data show that myeloid mTORC2 deficiency in CRC promotes tumorigenesis. Hence, mTORC2 in the innate immune system plays distinct roles in different types of cancer.
Our data suggest Opn can mediate the exacerbated colitis in the RictorLyz2-Cre mice. Opn is involved in several chronic inflammatory conditions including colitis, where it has multifaceted functions (43–45, 52–56). Opn mRNA and protein levels are overexpressed in several other types of cancer — for example, in lung (57), breast (58), or prostate cancer (46). Moreover, Opn is involved in the regulation of nearly all hallmarks of cancer (42), such as angiogenesis in melanomas (59) or metastasis in bladder cancer (60). Recently, Opn deficiency was found to suppress intestinal tumor development in a genetic mouse model (61). Additionally, OPN has been correlated with decreased patient survival (62). Our data now show that high OPN derived from mTORC2-inhibited macrophages is associated with poor survival in 2 cohorts of CRC patients.
A series of ATP-competitive inhibitors that block mTORC1 and mTORC2 at the same time (35) have already entered clinical trials (30). We now describe a suppressive role of mTORC2 in macrophages for an inflammation-based tumor model of CRC. Similarly, mTORC1 has antiinflammatory properties by stimulating production of IL-10 in DCs that protect the colon from chronic inflammation (22). Hence, global inhibition of mTORC1 and mTORC2 in cancer patients could have adverse inflammatory effects on the immune system by promoting tumorigenesis, as seen with a second-generation mTOR inhibitor in our mouse model. Thus, altering the innate immune system and its inflammatory response, as shown here, can be driving forces of cancer development. The potential unfavorable activities of second-generation mTOR inhibitors might especially be important for inflammation-associated tumors, where mTORC2 activation in macrophages seems to reestablish tissue homeostasis after colon damage. Instead, our work indicates activation of mTORC2 in macrophages as a therapeutic possibility in CRC.
Methods
Mouse strains.
B6 Rictorfl/fl mice were described previously (63). Lyz2-Cre mice were originally described by Clausen et al. (64). Rictorfl/fl mice were crossed to Lyz2Cre/+ mice to obtain Rictorfl/fl Lyz2Cre/+ (denoted RictorLyz2-Cre) or Rictorfl/fl Lyz2+/+ (denoted Rictorfl/fl) littermates. Male and female mice were used similarly for the experiments. Mice were bred at the Division of Biomedical Research, Medical University of Vienna, and housed with constant temperature and a 12-hour day/night cycle. Genotyping of the mice was performed with the DirectPCR-Tail lysis reagent (Peqlab). PCR was performed with the following primers: Rictor: 5′-ACT GAA TAT GTT CAT GGT TGT G-3′; 5′-GAA GTT ATT CAG ATG GCC CAG C-3′; Lyz2-Cre: 5′-CCC AGA AAT GCC AGA TTA CG-3′; 5′-CTT GGG CTG CCA GAA TTT CTC-3′; 5′-TTA CAG TCG GCC AGG CTG AC-3′.
Acute DSS-induced colitis.
Cohoused mice of both genotypes were used between 6 and 9 weeks of age. Mice were given 5% DSS (36,000–50,000 molecular weight, MP Biomedicals) in their drinking water for 5 days. Afterward, DSS-solution was replaced by normal water. Body weight of the mice was monitored daily. Mice surviving the experiment were anesthetized with ketamine/xylazine (Livisto and Ceva, respectively), and blood was collected. Mice were euthanized by cervical dislocation, colon length was measured with a ruler, and lamina propria cells were isolated and stained for flow cytometry.
Colitis-associated CRC model.
Cohoused mice of both genotypes were used between 6 and 9 weeks of age. Mice were injected i.p. with 10 mg/kg body weight AOM (MilliporeSigma). 5 days after the injection, mice were given 2% DSS in the drinking water for 5 days. This was followed by 16 days of normal water. On day 26, mice were again given a 2% DSS solution for 5 days, followed by 16 days of normal water. The last 2% DSS cycle was started on day 47 and lasted 4 days. Ten days after the last DSS cycle, mice were anesthetized with ketamine/xylazine, and blood was collected. Afterward, mice were euthanized by cervical dislocation, and colon was fixed in 4% paraformaldehyde overnight for H&E staining. Tumor number and size were determined microscopically after H&E staining.
Treatment with AZD8055.
Female C57BL/6 mice (obtained from the Department of Laboratory Animal Science and Genetics at the Medical University of Vienna) at the age of 5 weeks (average weight on day 0 was 17.4 g) were subjected to the AOM/DSS cancer model. Each DSS cycle was accompanied — with a delay of 1 day — by oral gavage of 10 mg/kg AZD8055 (Selleckchem). AZD8055 was dissolved in DMSO and stored at –20°C at a stock solution of 50 mg/ml. Mice were treated with 10 mg/kg by preparing a solution of 5 mg/ml for the gavaging procedure by adding 30% PEG 300 (MilliporeSigma) and ddH2O, with the final concentration of DMSO being 4%. This was further diluted by adding H2O to obtain a suitable volume for gavaging (~200 μl).
Colonoscopy.
Twenty-four hours before the intervention, mice received a rehydration solution consisting of 1.35% glucose, 0.29% sodium citrate, 0.26% trisodium citrate, dihydrate, 0.15% potassium chloride, and 3.45% PEG between 6,000–35,000 molecular weight (MilliporeSigma), and food was eliminated. The colonoscope (Karl Storz) was inserted, and the colon was inflated via the connection to an air pump (Eheim). A video was recorded based on a hyperplasia score, defined by the granularity of the surface was established in a blinded manner. During colonoscopy, tumors were counted and sized.
Treatment with Opn-neutralizing antibody.
During the acute DSS-induced colitis model, RictorLyz2-Cre mice were i.p. injected with 650 μg/kg body weight Opn-neutralizing antibody (R&D, AF808) or IgG control (R&D, AB-108-C) on day 1, 3, and 6.
Analysis of intestinal permeability in mice.
Analysis of intestinal permeability was carried out according to a previously described protocol (65). Mice were starved of water and food overnight. The next day, mice were gavaged with 440 mg/kg body weight FD4 FITC-dextran (MilliporeSigma). After 4 hours, mice were anesthetized with ketamine/xylazine, and blood was collected into Microvette CB 300 LH (Sarstedt). Blood was centrifuged at 2,000 g for 5 minutes at room temperature. Serum was collected and diluted in an equal volume of PBS. Diluted serum (100 μl) was added to a 96-well plate, and fluorescence was measured at an excitation wavelength of 485 nm and an emission wavelength of 528 nm on a spectrophotofluorometer (BioTek Synergy HT). Serially diluted FITC‑dextran was used to create a standard curve.
Cell culture.
Cells were cultured in a humidified incubator at 37°C and 5% CO2. BM from the femur and tibia was isolated, cultivated, and differentiated for 6 days on petri dishes (Greiner Bio-One) in MΦ‑differentiation media, which consisted of DMEM high glucose (Thermo Fisher Scientific), 10% low endotoxin FBS (Thermo Fisher Scientific), 2 mM L-glutamine (Lonza), 100 U/ml penicillin (MilliporeSigma), 100 μg/ml streptomycin (MilliporeSigma), 50 μg/ml β-mercaptoethanol (Thermo Fisher Scientific), and 15% L929 conditioned media. After the 6-day differentiation process, macrophages were seeded in 10% MΦ-media (without L929 supplementation) and allowed to rest for at least 4 hours before stimulation. Peritoneal macrophages were flushed with cold PBS from the peritoneal cavity, seeded in 10% MΦ-media onto petri dishes, and allowed to rest overnight before stimulation. In vitro stimulations were carried out by supplementing the macrophage culture with the following agents and concentrations for the indicated times: 100 ng/ml lipopolysaccharide from Escherichia coli 0111:B4 (LPS) (MilliporeSigma), 20 ng/ml IFN-γ (Peprotech), 10 ng/ml IL-4 (Peprotech).
Three-dimensional collagen gel cultures.
Human peripheral blood mononuclear cells (PBMCs) were isolated from EDTA-blood samples of healthy donors by density centrifugation using Ficoll-Paque Plus (density 1.077 g/ml, GE Healthcare). PBMCs were transferred into nontissue culture treated petri dishes in plain RPMI (Thermo Fisher Scientific) and 2 mM L-glutamine. After 2 hours of monocyte adhesion, all other cells were removed by washing 3 times with plain RPMI. Monocytes were differentiated into macrophages by cultivation in RPMI supplemented with 10% heat-inactivated FBS (Thermo Fisher Scientific), 2 mM L-glutamine, and 20 ng/ml recombinant human M-CSF (Peprotech) for 6 days. A total of 2 × 105 MDMs alone or in coculture with 500 freshly trypsinized colon cancer cells (DLD-1) were resuspended in 300 μl of a collagen I solution consisting of 2 mg/ml collagen I (rat tail, Corning), DMEM supplemented with 5% FBS, 10% of 10× PBS, and 1 M NaOH for neutralization as described previously (66). After solidification, the collagen gels were cultivated for 8 days in DMEM, 5% FBS, until tumor cells formed spheres within the gels. Medium was exchanged every other day. Gels were fixed in 4% paraformaldehyde (Roth) and embedded in paraffin.
Flow cytometry.
The colon was cut into small pieces, and epithelial cells were removed by two 20-minute sessions of incubation with HBSS (Thermo Fisher Scientific) supplemented with 10% FBS, 5 mM EDTA (MilliporeSigma), and 1% penicillin/streptomycin at 37°C with shaking at 200 rpm. Supernatant was removed and tissue was digested by 50 minutes incubation with RPMI supplemented with 10% FBS, 1% penicillin/streptomycin, 2 mM glutamine, 1 mg/ml Collagenase VIII (MilliporeSigma), and 2 mg/ml DNase (Worthington) at 37°C with shaking at 200 rpm. Supernatant was collected, and cells were filtered with a 70-μm cell strainer. Cells were centrifuged at 500 g for 8 minutes at 4°C. Lamina propria cells were washed with cold PBS and resuspended in FACS buffer (PBS, 2% FBS, 2 mM EDTA). Cells were blocked with TruStainfcX (BioLegend) for 20 minutes on ice and, afterward, incubated for 30 minutes with directly labeled antibodies. Cells were washed and taken up in FACS buffer for analysis on a FACSCalibur (BD Biosciences), FACSCanto II (BD Biosciences), FACSARIA I (BD Biosciences), or LSRFortessa (BD Biosciences). Data were analyzed by FlowJo (Tree Star Inc.). Antibodies from eBioscience were CD11b (clone M1/70) and CD3e (clone eBio500A2). CD45.2 (clone 104), I-Ab (clone AF6-120.1), CD64 (clone X54-5/7.1), CD8 (catalog 53-6.7), and CD4 (clone RM4-5) were obtained from BioLegend, and CD19 (clone 1D3) was purchased from BD Biosciences.
Glucose uptake assay.
Macrophages were seeded on 6-well plates and treated as indicated. After incubation, cells were collected and incubated with 100 μM 2-NBDG (Invitrogen) in PBS for 10 minutes at 37°C. Uptake was stopped by adding cold PBS. After a washing step, cells were taken up in FACS buffer and analyzed on a FACSCalibur (BD Biosciences).
Immunoblot.
Cells were pretreated in 6- or 12-well culture plates (CytoOne) as indicated. After stimulation, cells were washed once with PBS and taken up in 100 μl/well Triton X-100 lysis buffer consisting of 10 mM HEPES pH 7.9 (MilliporeSigma), 70 mM NaCl (Merck), 1 mM EDTA (MilliporeSigma), 1% Triton X-100 (MilliporeSigma), protease inhibitor cocktail (Roche Diagnostics), 10 mM NaF (Merck), 2 mM Na4P2O7 (Merck), 2 mM glycerophosphat (MilliporeSigma), and 200 μM Na3VO4 (MilliporeSigma). Cells were sonicated 3 times for 10 seconds and kept on ice for 30 minutes. Protein concentration was measured by using the Protein Assay Dye Reagent Concentrate (Bio-Rad) according to the manufacturer’s protocol. Protein concentration was adjusted with lysis buffer and 4× SDS reducing sample buffer containing 250 mM Tris-HCl (pH 6.8), 40% glycerol (Merck), 8% SDS (MilliporeSigma), 400 mM dithiothreitol (MilliporeSigma), and bromphenolblue (Merck) was added. Samples were boiled for 5 minutes, resolved on 12% SDS-PAGE, and transferred to a nitrocellulose membrane (Peqlab). Membranes were blocked in 4% dry milk for 1 hour at room temperature. The primary antibody was added overnight at 4°C. The secondary antibody (Bethyl, A120-101P), conjugated with horseradish peroxidase, was added for 45 minutes at room temperature in 4% dry milk. Proteins were detected by chemiluminescence using the Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific). The reaction was visualized using Amersham Hyperfilm ECL (GE Healthcare) and the Medical X-ray Processor 2000 (Kodak). Primary antibodies were Rictor (clone 53A2), pAkt S473 (clone D9E), pan-Akt (clone 40D4), histon H3 (clone D1H2), pAkt T308 (clone D25E6), pNDRG1 T346 (clone D98G11), pPRAS40 T246 (clone C77D7), and GAPDH (all from Cell Signaling Technologies).
Histology.
Colons of the mice were removed, washed, fixed in 4% paraformaldehyde, dehydrated (isopropanol/xylene/paraffin), embedded in paraffin, and sectioned with a microtome (Micros). H&E staining was performed, and slides were mounted with Eukitt.
IHC.
Tissues were fixed, dehydrated, and embedded in paraffin. Paraffin was removed using xylol, isopropanol, and an alcohol gradient in descending order (all from Carl Roth). For antigen retrieval, the slides were incubated in modified citrate buffer (pH 6) (Dako) at 120°C. Slides were incubated in 1% H2O2 and permeabilized with 0.1% Tween 20. Primary antibodies, Ki-67 (AB9260, Chemicon), Opn (AF808, R&D Systems) and pS6 (2215, Cell Signaling Technology), were incubated overnight at 4°C. After washing with PBS, a biotinylated secondary antibody was added for 45 minutes, and slides were incubated with Streptavidin-HRP complexes conjugated with peroxidase. To visualize positive staining, sections were incubated with aminoethyl carbazole for 20 minutes, and hematoxylin was used for counterstaining the nuclei. Staining intensities were quantified using ImageJ (NIH).
Immunofluorescence.
Paraffin embedded colon disease spectrum tissue arrays (US Biomax Inc.) containing, in total, 180 cores from colon cancer patients were used for immunofluorescence stainings. Rehydration and antigen retrieval was performed as described for IHC stainings. After blocking with PBS containing 1% BSA and 0.3% Triton, sections were incubated overnight at 4°C with primary antibodies against pAKT Ser473 (Cell Signaling Technologies), pPKCα Ser657 (Santa Cruz Biotechnology Inc., sc-12356), and F4/80 (eBioscience, 123101) for mouse macrophages or CD68 (Santa Cruz Biotechnology Inc., 137002) for human macrophages. The next day, fluorescently labeled secondary antibodies and DAPI were added for 1 hour. Slides were scanned using the tile scan function of the Leica confocal microscope TCS SP8. ImageJ software was used to analyze staining percentages where pPKCα or pAKT positivity was calculated in relation to macrophage numbers.
mRNA expression analysis.
BMDM or peritoneal macrophages were pretreated as indicated and lysed directly in TRI Reagent (MilliporeSigma). RNA was isolated by phenol-chloroform extraction, and cDNA was generated with the GoScript Reverse Transcription system (Promega). For real-time PCR, GoTaq qPCR Master Mix (Promega) was used, and analysis was done on a StepOnePlus Real-Time PCR System. Expression levels of target mRNAs were normalized to β-actin.
Microarray analysis.
Colonic macrophages were sorted (CD45+CD11b+CD64+MHCII+Ly6C–) from the lamina propria cells, and RNA was isolated by the Quiagen RNeasy Plus Micro Kit. Microarray analysis was performed with the Affymetrix GeneChip Mouse Gene 2.0 ST array. Normalization of the data was done with the Affymetrix Expression Console. Analysis was performed with the Affymetrix Transcriptome Analysis Console (TAC) v3.0. Differential expression of genes between Rictorfl/fl and RictorLyz2-Cre colonic macrophages was defined by more than 2-fold difference and a significance value of smaller than 0.05. Microarray data have been deposited under GEO accession number GSE96525.
GSEA.
The GSEA tool from the Broad Institute website was used for the identification of enriched gene signatures. Normalized gene expression data obtained from sorted Rictorfl/fl (n = 3) and RictorLyz2-Cre (n = 4) colonic macrophages was used for comparison. We screened the gene set “CLASSSICALY ACTIVATED VS TYPE 2 ACTIVATED MACROPHAGE DN”.
Statistics.
For bioinformatic analysis of human CRC patients, we used the RNA sequencing gene expression file for CRC generated by the TCGA research network (47) and the study from Sheffer et al. (GSE41258) (48). Statistical analysis was performed with SPSS v21. Two-tailed unpaired (independent) t test was used to calculate P values. Levene’s test with a threshold of 0.05 was performed to choose the appropriate P value for the t test. Pearson correlation analysis was applied to calculate corresponding R and P values for scatter plots. Kaplan-Meier plots were generated by classifying the samples into groups of low and high target gene expression according to the median gene expression value. To test for the significance of Kaplan-Meier analysis, we performed the log-rank test. P ≤ 0.05 was considered to be statistically significant. Pairwise comparisons were analyzed using 1-way ANOVA and Bonferroni correction.
All mouse results are expressed as mean ± SEM. Unpaired student t test (2-tailed) was used to detect statistical significance. For the survival analysis shown in the Kaplan-Meier plots, log-rank test was used for the comparison of the groups. P values smaller than 0.05 were defined as statistically significant.
Study approval.
All animal studies were approved by the official Austrian ethics committee for animal experiments (GZ BMWF-66.009/0028-II/3b/2014 and GZ BMWFW-66.009/0255-WF/V/3b/2015).
Author contributions
TW conceptualized and supervised the study. Investigation was performed by KK, BS, SDF, M. Hirtl, DS, ML, NS, JMM, DU, and M. Herac. AS, AB, GS, MB, HD, M. Herac, MM, MAM, and TW supplied resources. KK and TW wrote the first draft, and all authors participated in review and edits.
Supplementary Material
Acknowledgments
We thank Astrid Fabry, Theresia Lengheimer, and Roberto Plasenzotti for support in mouse breeding. TW is supported by grants from the Austrian Science Fund (FWF) P27701-B20 and P30857-B28, by the Vienna Science and Technology Fund (WWTF) LS18-058, and by the Foundation for Sarcoidosis Research. MM is supported by the Austrian Science Fund (FWF) grant FWF-P25336-B13.
Version 1. 10/17/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(20):e124164.https://doi.org/10.1172/jci.insight.124164.
Contributor Information
Karl Katholnig, Email: karl.katholnig@gmx.at.
Birgit Schütz, Email: birgit.schuetz@meduniwien.ac.at.
David Schörghofer, Email: david.schoerghofer@startmail.com.
Monika Linke, Email: monika.linke.gruber@gmail.com.
Daniela Unterleuthner, Email: daniela.unterleuthner@meduniwien.ac.at.
Martin Hirtl, Email: hirtl.martin@gmail.com.
Michaela Lang, Email: lang@microbial-ecology.net.
Merima Herac, Email: merima.herac@meduniwien.ac.at.
Andreas Spittler, Email: andreas.spittler@meduniwien.ac.at.
Andreas Bergthaler, Email: ABergthaler@cemm.oeaw.ac.at.
Gernot Schabbauer, Email: gernot.schabbauer@meduniwien.ac.at.
Helmut Dolznig, Email: helmut.dolznig@meduniwien.ac.at.
Markus Hengstschläger, Email: markus.hengstschlaeger@meduniwien.ac.at.
Mario Mikula, Email: mario.mikula@meduniwien.ac.at.
Thomas Weichhart, Email: thomas.weichhart@meduniwien.ac.at.
References
- 1.Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;168(6):960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cornu M, Albert V, Hall MN. mTOR in aging, metabolism, and cancer. Curr Opin Genet Dev. 2013;23(1):53–62. doi: 10.1016/j.gde.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Linke M, Fritsch SD, Sukhbaatar N, Hengstschläger M, Weichhart T. mTORC1 and mTORC2 as regulators of cell metabolism in immunity. FEBS Lett. 2017;591(19):3089–3103. doi: 10.1002/1873-3468.12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weichhart T, Hengstschläger M, Linke M. Regulation of innate immune cell function by mTOR. Nat Rev Immunol. 2015;15(10):599–614. doi: 10.1038/nri3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimobayashi M, Hall MN. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol. 2014;15(3):155–162. doi: 10.1038/nrm3757. [DOI] [PubMed] [Google Scholar]
- 7.Oh WJ, Jacinto E. mTOR complex 2 signaling and functions. Cell Cycle. 2011;10(14):2305–2316. doi: 10.4161/cc.10.14.16586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikenoue T, Inoki K, Yang Q, Zhou X, Guan KL. Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. EMBO J. 2008;27(14):1919–1931. doi: 10.1038/emboj.2008.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guri Y, et al. mTORC2 Promotes Tumorigenesis via Lipid Synthesis. Cancer Cell. 2017;32(6):807–823.e12. doi: 10.1016/j.ccell.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 10.Foley TM, et al. Dual PI3K/mTOR Inhibition in Colorectal Cancers with APC and PIK3CA Mutations. Mol Cancer Res. 2017;15(3):317–327. doi: 10.1158/1541-7786.MCR-16-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu Y, et al. mTORC2 Regulates Amino Acid Metabolism in Cancer by Phosphorylation of the Cystine-Glutamate Antiporter xCT. Mol Cell. 2017;67(1):128–138.e7. doi: 10.1016/j.molcel.2017.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masui K, Cavenee WK, Mischel PS. mTORC2 in the center of cancer metabolic reprogramming. Trends Endocrinol Metab. 2014;25(7):364–373. doi: 10.1016/j.tem.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kühl AA, Erben U, Kredel LI, Siegmund B. Diversity of Intestinal Macrophages in Inflammatory Bowel Diseases. Front Immunol. 2015;6:613. doi: 10.3389/fimmu.2015.00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347(6):417–429. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- 15.Kaplan GG. The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol. 2015;12(12):720–727. doi: 10.1038/nrgastro.2015.150. [DOI] [PubMed] [Google Scholar]
- 16.Bouma G, Strober W. The immunological and genetic basis of inflammatory bowel disease. Nat Rev Immunol. 2003;3(7):521–533. doi: 10.1038/nri1132. [DOI] [PubMed] [Google Scholar]
- 17.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long AG, Lundsmith ET, Hamilton KE. Inflammation and Colorectal Cancer. Curr Colorectal Cancer Rep. 2017;13(4):341–351. doi: 10.1007/s11888-017-0373-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zigmond E, Jung S. Intestinal macrophages: well educated exceptions from the rule. Trends Immunol. 2013;34(4):162–168. doi: 10.1016/j.it.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Saleh M, Trinchieri G. Innate immune mechanisms of colitis and colitis-associated colorectal cancer. Nat Rev Immunol. 2011;11(1):9–20. doi: 10.1038/nri2891. [DOI] [PubMed] [Google Scholar]
- 21.Katholnig K, Linke M, Pham H, Hengstschläger M, Weichhart T. Immune responses of macrophages and dendritic cells regulated by mTOR signalling. Biochem Soc Trans. 2013;41(4):927–933. doi: 10.1042/BST20130032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohtani M, Hoshii T, Fujii H, Koyasu S, Hirao A, Matsuda S. Cutting edge: mTORC1 in intestinal CD11c+ CD11b+ dendritic cells regulates intestinal homeostasis by promoting IL-10 production. J Immunol. 2012;188(10):4736–4740. doi: 10.4049/jimmunol.1200069. [DOI] [PubMed] [Google Scholar]
- 23.Festuccia WT, Pouliot P, Bakan I, Sabatini DM, Laplante M. Myeloid-specific Rictor deletion induces M1 macrophage polarization and potentiates in vivo pro-inflammatory response to lipopolysaccharide. PLoS ONE. 2014;9(4):e95432. doi: 10.1371/journal.pone.0095432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hallowell RW, et al. mTORC2 signalling regulates M2 macrophage differentiation in response to helminth infection and adaptive thermogenesis. Nat Commun. 2017;8:14208. doi: 10.1038/ncomms14208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang SC, et al. Metabolic Reprogramming Mediated by the mTORC2-IRF4 Signaling Axis Is Essential for Macrophage Alternative Activation. Immunity. 2016;45(4):817–830. doi: 10.1016/j.immuni.2016.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Babaev VR, Huang J, Ding L, Zhang Y, May JM, Linton MF. Loss of Rictor in Monocyte/Macrophages Suppresses Their Proliferation and Viability Reducing Atherosclerosis in LDLR Null Mice. Front Immunol. 2018;9:215. doi: 10.3389/fimmu.2018.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh MH, et al. mTORC2 Signaling Selectively Regulates the Generation and Function of Tissue-Resident Peritoneal Macrophages. Cell Rep. 2017;20(10):2439–2454. doi: 10.1016/j.celrep.2017.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feldman ME, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7(2):e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu K, et al. Biochemical, cellular, and in vivo activity of novel ATP-competitive and selective inhibitors of the mammalian target of rapamycin. Cancer Res. 2009;69(15):6232–6240. doi: 10.1158/0008-5472.CAN-09-0299. [DOI] [PubMed] [Google Scholar]
- 30.Chiarini F, Evangelisti C, McCubrey JA, Martelli AM. Current treatment strategies for inhibiting mTOR in cancer. Trends Pharmacol Sci. 2015;36(2):124–135. doi: 10.1016/j.tips.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 31.Motzer RJ, et al. Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet. 2008;372(9637):449–456. doi: 10.1016/S0140-6736(08)61039-9. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Kim SG, Blenis J. Rapamycin: one drug, many effects. Cell Metab. 2014;19(3):373–379. doi: 10.1016/j.cmet.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou Z, Chen J, Yang J, Bai X. Targeted Inhibition of Rictor/mTORC2 in Cancer Treatment: A New Era after Rapamycin. Curr Cancer Drug Targets. 2016;16(4):288–304. doi: 10.2174/1568009616666151113120830. [DOI] [PubMed] [Google Scholar]
- 34.Schenone S, Brullo C, Musumeci F, Radi M, Botta M. ATP-competitive inhibitors of mTOR: an update. Curr Med Chem. 2011;18(20):2995–3014. doi: 10.2174/092986711796391651. [DOI] [PubMed] [Google Scholar]
- 35.Shor B, Gibbons JJ, Abraham RT, Yu K. Targeting mTOR globally in cancer: thinking beyond rapamycin. Cell Cycle. 2009;8(23):3831–3837. doi: 10.4161/cc.8.23.10070. [DOI] [PubMed] [Google Scholar]
- 36.Zhang H, et al. mTOR ATP-competitive inhibitor INK128 inhibits neuroblastoma growth via blocking mTORC signaling. Apoptosis. 2015;20(1):50–62. doi: 10.1007/s10495-014-1066-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Q, et al. Development of ATP-competitive mTOR inhibitors. Methods Mol Biol. 2012;821:447–460. doi: 10.1007/978-1-61779-430-8_29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka T, Kohno H, Suzuki R, Yamada Y, Sugie S, Mori H. A novel inflammation-related mouse colon carcinogenesis model induced by azoxymethane and dextran sodium sulfate. Cancer Sci. 2003;94(11):965–973. doi: 10.1111/j.1349-7006.2003.tb01386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chassaing B, Aitken JD, Malleshappa M, Vijay-Kumar M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr Protoc Immunol. 2014;104:Unit 15.25. doi: 10.1002/0471142735.im1525s104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thaker AI, Shaker A, Rao MS, Ciorba MA. Modeling colitis-associated cancer with azoxymethane (AOM) and dextran sulfate sodium (DSS) J Vis Exp. 2012;(67):4100. doi: 10.3791/4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sayer B, Lu J, Green C, Söderholm JD, Akhtar M, McKay DM. Dextran sodium sulphate-induced colitis perturbs muscarinic cholinergic control of colonic epithelial ion transport. Br J Pharmacol. 2002;135(7):1794–1800. doi: 10.1038/sj.bjp.0704633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shevde LA, Samant RS. Role of osteopontin in the pathophysiology of cancer. Matrix Biol. 2014;37:131–141. doi: 10.1016/j.matbio.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lund SA, Giachelli CM, Scatena M. The role of osteopontin in inflammatory processes. J Cell Commun Signal. 2009;3(3-4):311–322. doi: 10.1007/s12079-009-0068-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toyonaga T, et al. Osteopontin Deficiency Accelerates Spontaneous Colitis in Mice with Disrupted Gut Microbiota and Macrophage Phagocytic Activity. PLoS ONE. 2015;10(8):e0135552. doi: 10.1371/journal.pone.0135552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong J, Eckhardt ER, Oz HS, Bruemmer D, de Villiers WJ. Osteopontin deficiency protects mice from Dextran sodium sulfate-induced colitis. Inflamm Bowel Dis. 2006;12(8):790–796. doi: 10.1097/00054725-200608000-00015. [DOI] [PubMed] [Google Scholar]
- 46.Thalmann GN, et al. Osteopontin: possible role in prostate cancer progression. Clin Cancer Res. 1999;5(8):2271–2277. [PubMed] [Google Scholar]
- 47.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheffer M, et al. Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proc Natl Acad Sci USA. 2009;106(17):7131–7136. doi: 10.1073/pnas.0902232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riedl A, et al. Comparison of cancer cells in 2D vs 3D culture reveals differences in AKT-mTOR-S6K signaling and drug responses. J Cell Sci. 2017;130(1):203–218. doi: 10.1242/jcs.188102. [DOI] [PubMed] [Google Scholar]
- 50.Brown J, Wang H, Suttles J, Graves DT, Martin M. Mammalian target of rapamycin complex 2 (mTORC2) negatively regulates Toll-like receptor 4-mediated inflammatory response via FoxO1. J Biol Chem. 2011;286(52):44295–44305. doi: 10.1074/jbc.M111.258053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raïch-Regué D, Rosborough BR, Watson AR, McGeachy MJ, Turnquist HR, Thomson AW. mTORC2 Deficiency in Myeloid Dendritic Cells Enhances Their Allogeneic Th1 and Th17 Stimulatory Ability after TLR4 Ligation In Vitro and In Vivo. J Immunol. 2015;194(10):4767–4776. doi: 10.4049/jimmunol.1402551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Inoue M, Shinohara ML. Intracellular osteopontin (iOPN) and immunity. Immunol Res. 2011;49(1-3):160–172. doi: 10.1007/s12026-010-8179-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clemente N, et al. Osteopontin Bridging Innate and Adaptive Immunity in Autoimmune Diseases. J Immunol Res. 2016;2016:7675437. doi: 10.1155/2016/7675437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sato T, et al. Osteopontin/Eta-1 upregulated in Crohn’s disease regulates the Th1 immune response. Gut. 2005;54(9):1254–1262. doi: 10.1136/gut.2004.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kourepini E, Aggelakopoulou M, Alissafi T, Paschalidis N, Simoes DC, Panoutsakopoulou V. Osteopontin expression by CD103- dendritic cells drives intestinal inflammation. Proc Natl Acad Sci USA. 2014;111(9):E856–E865. doi: 10.1073/pnas.1316447111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heilmann K, et al. Osteopontin as two-sided mediator of intestinal inflammation. J Cell Mol Med. 2009;13(6):1162–1174. doi: 10.1111/j.1582-4934.2008.00428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chambers AF, Wilson SM, Kerkvliet N, O’Malley FP, Harris JF, Casson AG. Osteopontin expression in lung cancer. Lung Cancer. 1996;15(3):311–323. doi: 10.1016/0169-5002(95)00595-1. [DOI] [PubMed] [Google Scholar]
- 58.Tuck AB, et al. Osteopontin expression in a group of lymph node negative breast cancer patients. Int J Cancer. 1998;79(5):502–508. doi: 10.1002/(SICI)1097-0215(19981023)79:5<502::AID-IJC10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 59.Kale S, Raja R, Thorat D, Soundararajan G, Patil TV, Kundu GC. Osteopontin signaling upregulates cyclooxygenase-2 expression in tumor-associated macrophages leading to enhanced angiogenesis and melanoma growth via α9β1 integrin. Oncogene. 2014;33(18):2295–2306. doi: 10.1038/onc.2013.184. [DOI] [PubMed] [Google Scholar]
- 60.Ahmed M, et al. An Osteopontin/CD44 Axis in RhoGDI2-Mediated Metastasis Suppression. Cancer Cell. 2016;30(3):432–443. doi: 10.1016/j.ccell.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ishigamori R, Komiya M, Takasu S, Mutoh M, Imai T, Takahashi M. Osteopontin Deficiency Suppresses Intestinal Tumor Development in Apc-Deficient Min Mice. Int J Mol Sci. 2017;18(5):E1058. doi: 10.3390/ijms18051058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klement JD, et al. An osteopontin/CD44 immune checkpoint controls CD8+ T cell activation and tumor immune evasion. J Clin Invest. 2018;128(12):5549–5560. doi: 10.1172/JCI123360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shiota C, Woo JT, Lindner J, Shelton KD, Magnuson MA. Multiallelic disruption of the rictor gene in mice reveals that mTOR complex 2 is essential for fetal growth and viability. Dev Cell. 2006;11(4):583–589. doi: 10.1016/j.devcel.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 64.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8(4):265–277. doi: 10.1023/A:1008942828960. [DOI] [PubMed] [Google Scholar]
- 65.Gupta JN. Analysis of Intestinal Permeability in Mice. Bio-protocol. 2014;4(22):e1289 [Google Scholar]
- 66.Dolznig H, et al. Modeling colon adenocarcinomas in vitro a 3D co-culture system induces cancer-relevant pathways upon tumor cell and stromal fibroblast interaction. Am J Pathol. 2011;179(1):487–501. doi: 10.1016/j.ajpath.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.