Abstract
Systemic lupus erythematosus (SLE) is an autoimmune disease characterized by pathologic T cell–B cell interactions and autoantibody production. Defining the T cell populations that drive B cell responses in SLE may enable design of therapies that specifically target pathologic cell subsets. Here, we evaluated the phenotypes of CD4+ T cells in the circulation of 52 SLE patients drawn from multiple cohorts and identified a highly expanded PD-1hiCXCR5–CD4+ T cell population. Cytometric, transcriptomic, and functional assays demonstrated that PD-1hiCXCR5–CD4+ T cells from SLE patients are T peripheral helper (Tph) cells, a CXCR5– T cell population that stimulates B cell responses via IL-21. The frequency of Tph cells, but not T follicular helper (Tfh) cells, correlated with both clinical disease activity and the frequency of CD11c+ B cells in SLE patients. PD-1hiCD4+ T cells were found within lupus nephritis kidneys and correlated with B cell numbers in the kidney. Both IL-21 neutralization and CRISPR-mediated deletion of MAF abrogated the ability of Tph cells to induce memory B cell differentiation into plasmablasts in vitro. These findings identify Tph cells as a highly expanded T cell population in SLE and suggest a key role for Tph cells in stimulating pathologic B cell responses.
Keywords: Autoimmunity, Immunology
Keywords: Adaptive immunity, Lupus, T cells
Lupus patients have high frequencies of PD-1hi CXCR5- T peripheral helper cells, which stimulate B cell responses through MAF-dependent production of IL-21.
Introduction
Systemic lupus erythematosus (SLE) is a disease of broad autoimmunity, with breaches of tolerance in both T cells and B cells. Pathologic T cell–B cell interactions, B cell activation, and production of autoantibodies are hallmark features of SLE. Levels of anti–double stranded DNA (anti-dsDNA) autoantibodies rise with increased disease activity in SLE, such that this metric is often followed clinically in SLE patients (1). The frequency of circulating plasmablasts, which may serve as the source of rising antibody levels, also increase with disease activity (2, 3). In addition, a population of age-associated B cells (ABCs), which may differentiate into plasmablasts, is also highly expanded in SLE patients with active disease (4, 5). Expansion of these activated B cell populations in SLE generally requires help from T cells, suggesting that SLE disease activity involves ongoing T cell–B cell interactions (6, 7).
T follicular helper (Tfh) cells are often considered the primary T cell population capable of promoting the pathologic B cell response in SLE (8–10). Tfh cells are critical for stimulating B cell responses within follicles of secondary lymphoid organs in both infection and autoimmunity (11). Often identified as CXCR5+PD-1+CD4+ T cells, Tfh cells are recruited into lymphoid follicles by CXCL13, the ligand for CXCR5, and promote B cell maturation in germinal centers through production of IL-21, CXCL13, IL-4, and CD40L (11). Tfh cells have been observed to be expanded in cohorts of SLE patients (9, 10, 12, 13), and defective regulation of Tfh cells contributes to a lupus-like disease in multiple murine models (14–17).
There is increasing evidence that autoreactive T cell–B cell interactions in SLE also occur outside of germinal centers of secondary lymphoid organs. Murine SLE models have highlighted the role of extrafollicular CXCR4+CCR7+CD4+ T cells, which induce plasmablast development in splenic extrafollicular foci (18). T cell–B cell interactions occur within the interstitium of kidneys of lupus nephritis patients, and these interactions may drive B cell differentiation into plasma cells within the kidney (19, 20). In other autoimmune diseases, T cell–B cell interactions within peripheral inflamed sites can involve T cells with phenotypes distinct from germinal center Tfh cells, including the PD-1hiCXCR5– T peripheral helper (Tph) cell population in rheumatoid arthritis (RA) (21). Tph cells in RA synovium share several features with Tfh cells, including high expression of PD-1, ICOS, HLA-DR, and TIGIT; production of both CXCL13 and IL-21; and ability to drive B cell differentiation into plasma cells in vitro (22). However, Tph cells from RA synovium differ from Tfh cells in expression of migratory receptors and key transcriptional regulators. Specifically, Tph cells lack CXCR5, a defining marker for Tfh cells, and instead express chemokine receptors such as CCR2 and CCR5 that promote migration to sites of peripheral inflammation (22).
Here, we evaluated the T cell populations altered in the circulation of patients with SLE by high-dimensional mass cytometry. We identified a highly expanded population of PD-1hiCXCR5– T cells in SLE patients, including in new-onset SLE patients studied prior to initiation of strong immunosuppressive therapies. By phenotype, transcriptome, and function, these cells are Tph cells, which help B cells yet express migratory receptors that target peripheral tissues. Tph cell frequency correlated with features of active clinical disease and showed a specific correlation with CD11c+ ABCs in SLE patients. These studies reveal Tph cells as a highly expanded T cell population with pathologic potential in SLE.
Results
Expanded Tph cells in lupus nephritis patients.
We analyzed mass cytometry data on PBMCs from 27 lupus nephritis patients, 25 RA patients, and 25 noninflammatory controls that were generated by the Accelerating Medicines Partnership (AMP) RA/SLE Network, a multicenter study performing single cell analyses on blood and tissue samples from SLE and RA patients (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.130062DS1) (23–25). We focused the analysis on data that were generated with a panel designed to assess T cell phenotypes (Supplemental Table 2). In this dataset, the frequencies of CD4+ T cells and CD8+ T cells within PBMCs were comparable across diseases, and the frequency of memory (CD45RO+) cells within the CD4+ T cell subset was also similar (Supplemental Figure 1, A–C).
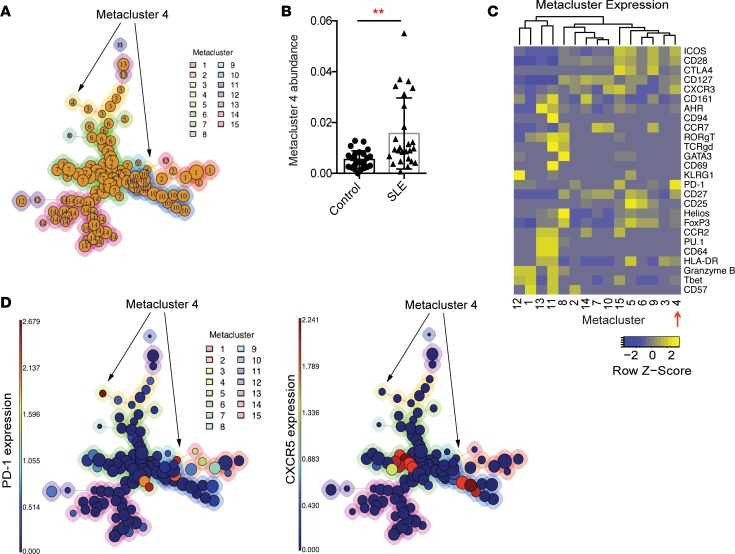
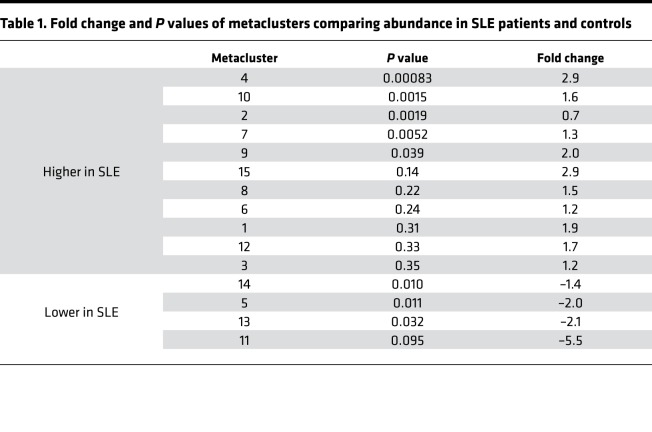
We used FlowSOM to evaluate memory CD4+ T cells in a multidimensional manner in order to identify subpopulations that are altered in SLE patients (26). FlowSOM was used to assign memory CD4+ T cells into 100 clusters, which were then aggregated into 15 metaclusters (Figure 1A). We compared the abundances of the metaclusters in SLE patient and control samples and identified 3 metaclusters with significantly increased abundance in SLE patients (metaclusters 2, 4, 7, Bonferroni adjusted P < 0.05) (Table 1 and Figure 1B). Of these, metacluster 4 also had a > 2-fold increase in abundance in SLE patients; therefore, we focused on this metacluster. Metacluster 4 contained cells with high expression of PD-1, as well as expression of ICOS and CXCR3 (Figure 1, C and D). Metacluster 4 was composed of 2 clusters, which mapped to distinct locations in the self-organizing map, suggesting heterogeneity of cells within the metacluster 4. A comparison of the 2 clusters that comprise metacluster 4 (cluster A and cluster B) demonstrated that these 2 clusters showed consistent expression of most markers, including expression of PD-1, ICOS, and CXCR3 (Supplemental Figure 2A). However, the 2 clusters differed in expression of HLA-DR, which was expressed in cluster A but not in cluster B (Supplemental Figure 2, A and B). Cluster B, which lacked HLA-DR, showed a larger expansion in SLE patients than did cluster A (Supplemental Figure 2C). Expression of CXCR5 was detected within clusters separate from metacluster 4 (Figure 1D). These results indicate that a population of PD-1hiCXCR5– T cells, with expression of ICOS and CXCR3 and variable expression of HLA-DR, is significantly expanded in the circulation of SLE patients.
Figure 1. Identification of an expanded CD4+ T cell population in the blood of SLE patients.
(A) FlowSOM analysis of AMP mass cytometry data gated on CD45RO+CD4+ T cells. Each circle represents an individual cluster. The aggregated metaclusters are indicated by the numbers within the circles and by the color around the circles. Circle size indicates the abundance of cells within the cluster. (B) Abundance of metacluster 4 in individual SLE patients (n = 26) and controls (n = 25). Error bars show mean ± SD. **P < 0.01 by Mann-Whitney U test. (C) Heatmap of row-normalized expression of mass cytometry markers in each metacluster. Markers with nonzero median expression in at least 1 metacluster are shown, excluding markers used for gating memory CD4+ T cells. (D) FlowSOM maps demonstrating level of expression of PD-1 and CXCR5 in the individual clusters. For A and D, arrows indicate location of metacluster 4.
Table 1. Fold change and P values of metaclusters comparing abundance in SLE patients and controls.
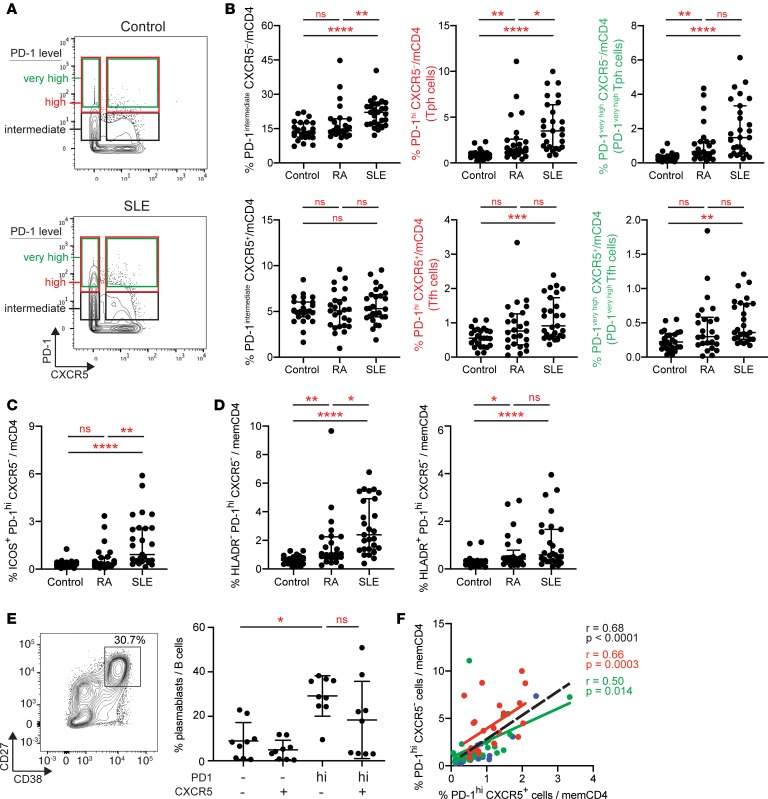
We confirmed the increased frequency of PD-1hiCXCR5– T cells in SLE patients through biaxial gating. The median MFI of PD-1 in metacluster 4 across all patients was 36; therefore, we focused our gating criteria on cells with high expression of PD-1 (MFI > 20, referred to as PD-1hi) to capture this population (Figure 2A and full gating shown in Supplemental Figure 1A). Using this gate, PD-1hiCXCR5– cells were highly expanded in SLE patients compared with noninflammatory controls (4.3-fold, P < 0.0001), and this expansion exceeded that observed in RA patients (Figure 2B). The frequency of PD-1hiCXCR5– cells in SLE patients was positively correlated with the frequency of cells in metacluster 4 (r = 0.6, P = 0.0012), suggesting that these 2 analyses capture a similar cell population. Quantification of CXCR5– cells with even higher PD-1 expression, requiring an MFI of 36 (PD-1very high cells), yielded similar results of a marked expansion in SLE patients compared with controls (P < 0.0001, 5.7-fold; Figure 2B). CXCR5– cells with intermediate PD-1 expression were also increased, but with a smaller relative expansion compared with controls (P < 0.0001, 1.7-fold) (Figure 2B). PD-1hiCXCR5– T cell populations that coexpress ICOS were also highly expanded in SLE patients (3.2-fold, P < 0.0001) (Figure 2C). By biaxial gating, both HLA-DR–PD-1hiCXCR5– and HLA-DR+PD-1hiCXCR5– cells were increased in SLE patients (Figure 2D, gated as shown in Supplemental Figure 2D).
Figure 2. Expanded PD-1hiCXCR5– Tph cells in the blood of SLE patients.
(A) Example of gating of memory CD4+ T cells with different levels of PD-1 expression in AMP mass cytometry data. (B) Quantification of CXCR5– and CXCR5+ memory CD4+ T cell populations with intermediate, high, or very high PD-1 expression as depicted in A in controls (n = 25), RA (n = 25), and SLE (n = 27) patients using AMP mass cytometry data. (C) Quantification of PD-1hiCXCR5–ICOS+ memory CD4+ T cells in AMP mass cytometry data as in B. (D) Quantification of PD-1hiCXCR5– cells that express or do not express HLA-DR as in B. (E) Example flow cytometry detection of plasmablasts in T cell–B cell cocultures and quantification of plasmablasts among B cells in cocultures of memory B cells with indicated CD4+ T cell subsets from SLE patients. Pooled data from 9 donors. Error bars show median ± interquartile range (B, C, D) or mean ± SD (E). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by Kruskal–Wallis with Dunn’s multiple comparisons test (B–E). (F) Correlation between PD-1hiCXCR5– and PD-1hiCXCR5+ cell frequencies in AMP mass cytometry data (red, SLE patients; green, RA patients; blue, controls; black line, all patients). Spearman correlation statistics shown.
This PD-1hiCXCR5– population with coexpression of ICOS and variable HLA-DR strongly resembles the Tph cell population, a PD-1hiCXCR5– B cell–helper T cell population abundant in RA synovium (22). To evaluate whether PD-1hiCXCR5– T cells in SLE patients have B cell–helper function consistent with a Tph cell identity, we performed in vitro T cell–B cell coculture assays. PD-1hiCXCR5– T cells sorted from PBMCs of SLE patients robustly induced differentiation of allogeneic memory B cells into CD38hiCD27+ plasmablasts, with activity comparable with that of PD-1hiCXCR5+ Tfh cells (Figure 2E). In contrast, PD-1– memory CD4+ T cells did not effectively induce plasmablasts. Together, these results identify a highly expanded PD-1hiCXCR5– T cell population in SLE patients that shows B cell helper activity; we refer to this population subsequently as Tph cells.
The absence of CXCR5 on Tph cells distinguishes these cells from Tfh cells, for which CXCR5 expression is considered a defining feature and an essential mediator of lymphoid follicle homing (11). Here, we define Tfh cells in the blood as PD-1hiCXCR5+CD4+ T cells; this population can be considered a circulating Tfh cell population with features that are similar but not identical to Tfh cells in lymphoid follicles (9, 27). Tfh cells were also significantly increased in the blood of SLE patients, but not RA patients, compared with controls (1.7-fold, P = 0.0013) (Figure 2B). Similar results were found when Tfh cells with high and very high PD-1 expression were analyzed. There was a positive correlation between Tph cell and Tfh cell frequencies in the total cohort overall — and specifically in SLE patients (Figure 2F). Notably, Tph cells substantially outnumbered Tfh cells in the circulation of SLE patients (median Tph 3.5% vs. Tfh 0.9% of memory CD4+ T cells), and the relative increase of Tph cells in SLE patients compared with controls (4.4-fold) exceeded that of Tfh cells (1.9-fold) (Figure 2B).
Clinical features of expanded Tph cells in SLE.
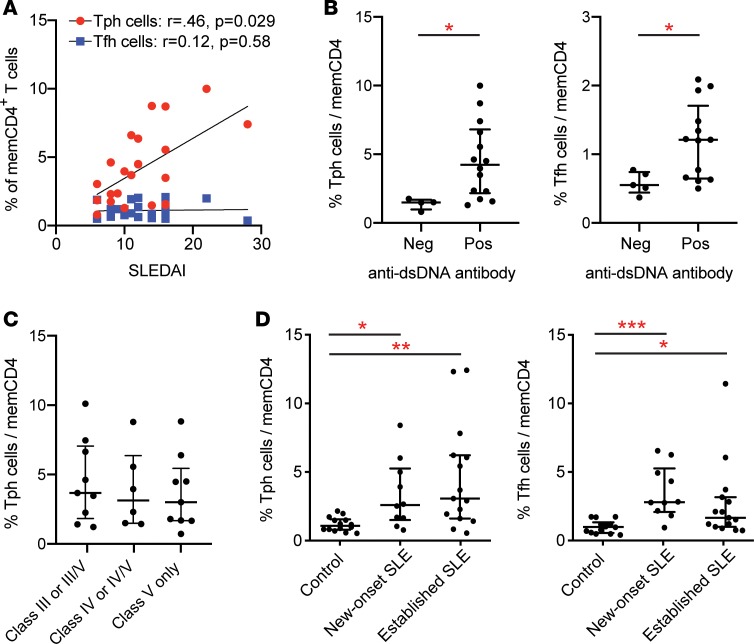
We next evaluated the relationship between Tph cell frequency and disease activity in the AMP cohort, in which all SLE patients had biopsy-demonstrated lupus nephritis. Among the patients who had a SELENA-SLEDAI score recorded upon enrollment (n = 21), Tph cell frequency was positively correlated with SELENA-SLEDAI score, while Tfh cell frequency did not show a positive correlation (Figure 3A). The frequency of Tph cells with very high PD-1 expression (MFI > 36) was also positively correlated with SLEDAI (r = 0.49, P = 0.018; Supplemental Figure 3), while PD-1intermediateCXCR5– cells did not show a significant correlation with SLEDAI (r = 0.38, P = 0.094). CXCR5+ cells showed no correlation with SLEDAI, regardless of the PD-1 expression threshold (Figure 3A and Supplemental Figure 3).
Figure 3. Clinical features of Tph cell expansion in SLE patients.
(A) Correlation between Tph cell (red) or Tfh cell (blue) frequency and disease activity by SELENA-SLEDAI in AMP lupus nephritis patients (n = 21). Spearman correlation statistics shown. (B) Frequency of Tph cells or Tfh cells in AMP lupus nephritis patients dichotomized based on anti-dsDNA antibody status. (C) Frequency of Tph cells in AMP lupus nephritis patients grouped according to histologic glomerulonephritis class. (D) Tph cell and Tfh cell frequencies in control (n = 10), new-onset SLE (n = 10), and established SLE (n = 15) patients in the BWH validation cohort. Error bars show median ± interquartile range (B–D). *P < 0.05, **P < 0.01, ***P < 0.001 by Mann-Whitney U test (B) and Kruskal–Wallis with Dunn’s multiple comparisons test (C and D).
Anti-dsDNA titers were available from a subset of the SLE patients in this cohort (n = 18). Antibody titers were measured at different institutions with varying assays, which limited the ability to assess linear correlations. When SLE patients were dichotomized based on anti-dsDNA antibody positivity, patients with a positive anti-dsDNA antibody test showed a 2.8-fold increase in the frequency of Tph cells compared with patients with a negative anti-dsDNA antibody test. Tfh cell frequency was also higher in patients with a positive anti-dsDNA antibody test (Figure 3B). Tph cell frequency was not significantly associated with histologic class of glomerulonephritis, patient race, or patient ethnicity, and it showed a negative association with patient age (Figure 3C and Supplemental Figure 4, A and B). These results indicate that Tph cell frequency is associated with high disease activity in lupus nephritis and shows a stronger association with clinical disease activity than does Tfh cell frequency.
To assess SLE patients with a wider range of clinical features, we analyzed an independent cohort of 15 SLE patients with established disease of varying manifestations (8 of 15 without a history of nephritis), as well as 10 patients with a new diagnosis of SLE prior to initiation of immunosuppressive therapy (Supplemental Table 3 and Supplemental Table 4). Mass cytometric analysis of these validation cohorts revealed increased frequencies of Tph cells both in patients with established disease (2.9-fold, P = 0.0017) and in patients with new-onset SLE (2.4-fold, P = 0.022) compared with noninflammatory controls (Figure 3D). These results, generated in an independent cohort of lupus patients analyzed by a separate cytometry analysis, confirm that Tph cells are highly expanded in SLE patients, including patients without nephritis. Further, expansion of Tph cells occurs early in the disease course and is not secondary to immunosuppressive therapy.
Potential Tph cells in lupus nephritis kidneys.
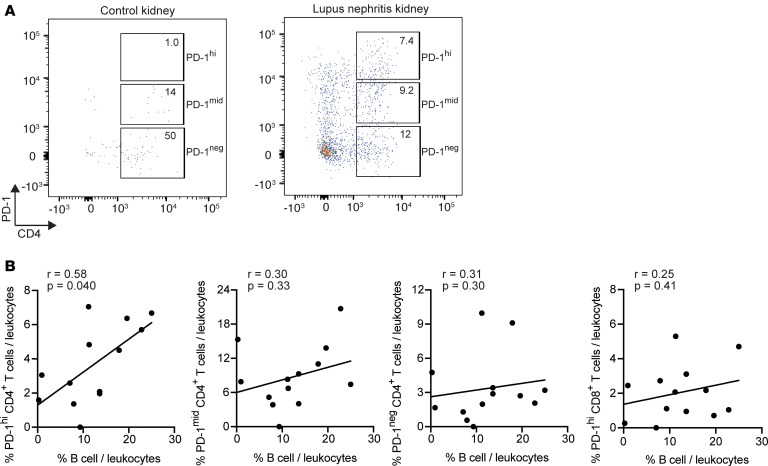
To interrogate the potential presence of Tph cells in the kidneys of patients with lupus nephritis, we analyzed flow cytometry data generated on dissociated kidney biopsy tissue from patients within the AMP Network (23). Viably cryopreserved kidney biopsies were dissociated into single cells and stained with a flow cytometry panel that included CD45, CD3, CD4, and PD-1, allowing for identification of PD-1hiCD4+ T cells, as well as CD19, to identify B cells. B cells were detected in lupus nephritis biopsies but not control kidney tissue in this cohort (23). Data were filtered to assess only kidney biopsies with > 100 leukocytes captured. Using these data, PD-1hiCD4+ T cells comprised > 5% of the total CD45+ leukocytes in 8 of 13 lupus nephritis biopsies (Figure 4, A and B). We identified a positive correlation between the frequency of PD-1hiCD4+ T cells and the frequency of B cells in the biopsy (Figure 4B). In contrast, there was no correlation between the frequency of B cells and CD4+ T cells with either negative or intermediate PD-1 expression, and there was no correlation between the frequency of B cells and PD-1hiCD8+ T cells. Because the flow cytometry data did not include chemokine receptors such as CXCR5, we cannot differentiate Tph cells from Tfh cells in these kidney samples. The specific association between PD-1hiCD4+ T cells and B cells is consistent with a B cell–helper function of these PD-1hiCD4+ T cells in lupus nephritis kidneys.
Figure 4. Potential Tph cells in lupus nephritis kidneys.
(A) Example of flow cytometry analysis of gated CD45+ leukocytes from control kidney tissue (left) or lupus nephritis kidney biopsy (right). (B) Correlation between the frequency of B cells and PD-1hi, PD-1intermediate, PD-1–CD4+ T cells or PD-1hiCD8+ T cells in lupus nephritis kidney biopsies (n = 13). Spearman correlation statistics shown.
Features of Tph cells in SLE.
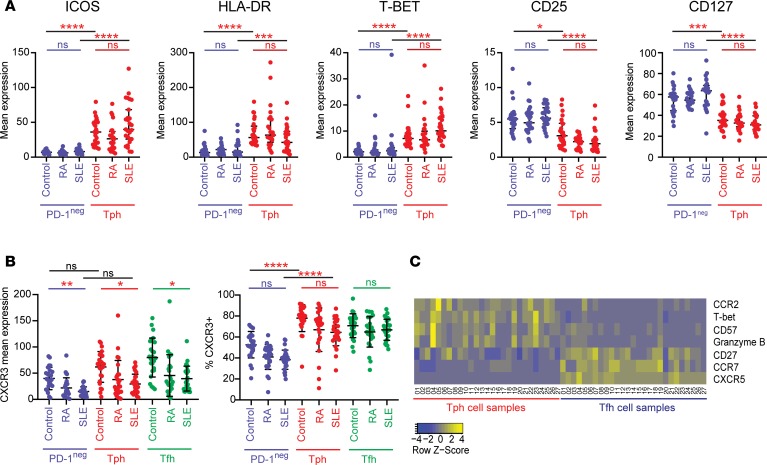
We next used the mass cytometry data to compare the phenotypes of Tph cells in blood from SLE, RA, and control donors within the AMP cohort. Tph cells from SLE patients appeared cytometrically similar to those from RA patients and controls, with features consistent with the prior description of Tph cells, including high expression of ICOS, HLA-DR, and T-bet and low expression of CD25 and CD127 (Figure 5A) (22). A broad analysis of all included cytometry markers identified 3 markers with significantly different expression on Tph cells from SLE patients compared with controls (PD-1, CCR2, and CXCR3; Bonferroni adjusted P < 0.05) (Supplemental Table 5). PD-1 expression was modestly increased in Tph cells from SLE patients compared with Tph cells from controls (1.18-fold, adjusted P = 0.039). Interestingly, the mean expression of chemokine receptor CXCR3 was lower on Tph cells from SLE patients compared with Tph cells from controls (–2.1-fold, P = 0.00048) (Figure 5B and Supplemental Table 5). This pattern was not specific to Tph cells, as CXCR3 expression was also significantly reduced in Tfh cells and PD-1– memory T cells from SLE patients (PD-1–: –2.6-fold, P < 0.0001; Tfh: –2.0-fold, adjusted P = 0.0004). The frequency of CXCR3+ cells as assessed by biaxial gating was less affected, suggesting that T cells from SLE patients may downregulate CXCR3 expression without entirely losing CXCR3 expression (Figure 5B). This pattern was also seen in the validation cohort, with SLE patients having a lower mean expression of CXCR3 compared with controls across Tph cells, Tfh cells, and PD-1– memory T cells (Supplemental Figure 5A). Both Tph cells and PD-1– cells from SLE patients also showed lower CCR2 expression compared with cells from controls, while mean CXCR5 expression was reduced in Tfh cells (gated to be PD-1hiCXCR5+) from SLE patients (Supplemental Figure 5, B and C). Lower mean expression of these chemokine receptors was also observed in the validation cohort (Supplemental Figure 5, D and E), suggesting that these changes are likely to reflect cell activation states characteristic of SLE rather than technical effects.
Figure 5. Cytometric features of Tph cells in SLE.
(A) Expression of characteristic proteins on Tph cells (red) and PD-1– memory CD4+ T cells (blue) in controls (n = 25), RA (n = 25), and lupus nephritis (SLE; n = 27) patients in the AMP cohort. (B) Mean expression of CXCR3 and frequency of CXCR3+ cells in PD-1– memory CD4+ T cells, Tph cells, and Tfh cells. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by Kruskal–Wallis with Dunn’s multiple comparisons test (A and B). Black bars indicate comparison of PD-1– and Tph cells either from controls or from SLE patients as indicated. Colored bars indicate comparison of control and SLE samples within the cell type. Error bars show mean ± SD. (C) Heatmap of row-normalized expression of 6 markers that are significantly differentially expressed comparing Tph cells and Tfh cells from lupus nephritis patients in the AMP cohort. CXCR5 is included for reference.
To characterize differences between Tph cells and Tfh cells from SLE patients, we compared the expression of each of the markers in the mass cytometry panel between these 2 populations. We identified 6 proteins, not including CXCR5, with significantly different expression on Tph cells and Tfh cells from SLE patients in the AMP cohort (Figure 5C). There was higher expression of T-bet, CCR2, CD57, and granzyme B and lower expression of CCR7 and CD27 on Tph cells compared with Tfh cells from SLE patients (Figure 5C and Supplemental Table 6, Bonferroni adjusted P < 0.05). These observations, in total, indicate that PD-1hiCXCR5– Tph cells from SLE patients are similar to those seen in RA patients and suggest that they represent the same T cell population.
Unique RNA-sequencing (RNA-seq) features of Tph cells in SLE patients.
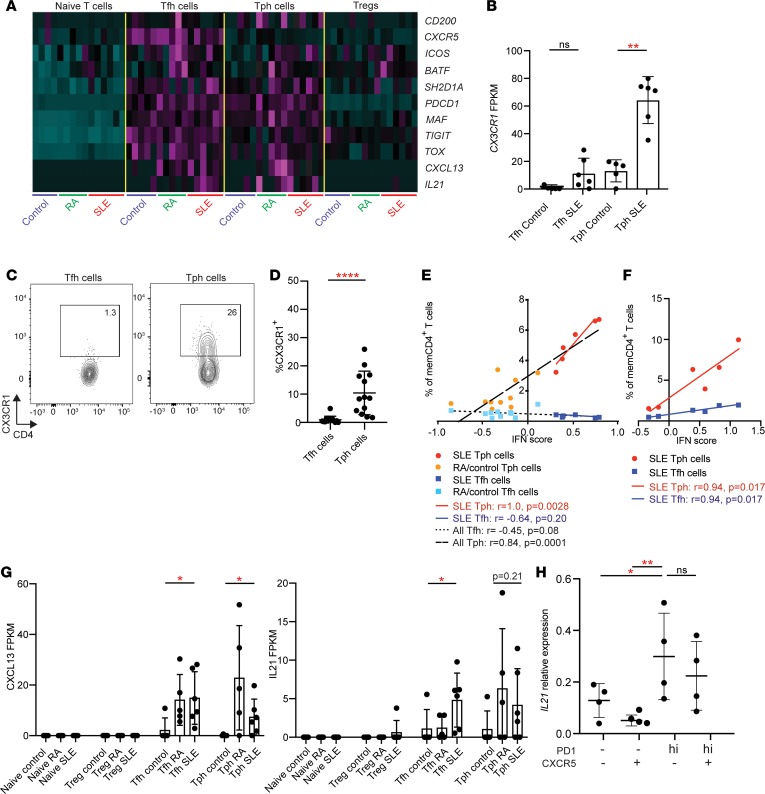
To further examine potential functional characteristics of Tph cells from SLE patients, we generated transcriptomes of Tph cells, Tfh cells, naive T cells, and Tregs sorted from PBMCs of SLE patients (n = 6), RA patients (n = 5), and noninflammatory controls (n = 5) by low-input RNA-seq (sorted as in Supplemental Figure 6A). We first evaluated the expression of a curated set of genes associated with Tfh cells and observed high expression of several Tfh-associated genes in both Tph cells and Tfh cells, including IL21, CXCL13, ICOS, MAF, BATF, and TOX (Figure 6A) (22, 28). In contrast, as expected, CXCR5 transcript was increased in Tfh cells but not Tph cells. These results confirm that PD-1hiCXCR5– Tph cells in SLE patients express a set of effector molecules and regulators associated with B cell–helper function.
Figure 6. RNA-seq features of Tph cells in SLE.
(A) Heatmap of row-normalized expression of Tfh-associated genes in T cell populations sorted from control, RA, and SLE patients. (B) Expression of CX3CR1 in RNA-seq data in Tfh and Tph cells from control and SLE patients. (C) Example flow cytometric detection of CX3CR1 in Tph cells and Tfh cells. (D) Quantification of CX3CR1 expression on Tph cells and Tfh cells in SLE patients in the AMP cohort. (E) Correlation between Tph or Tfh cell frequency and IFN score in the RNA-seq patient cohort (SLE, n = 6; RA, n = 5, control, n = 5). Black lines indicate overall Tph or Tfh correlations. Red line indicates correlation for SLE Tph cells. Blue line indicates correlation for SLE Tfh cells. (F) Correlation between Tph or Tfh cell frequency and IFN score in a subset of SLE patients from AMP cohort (n = 6). (G) Expression of CXCL13 and IL21 in T cell subsets in RNA-seq data. (H) IL21 expression by qPCR in PMA plus ionomycin–stimulated memory CD4+ T cell subsets from SLE patients (n = 4). Error bars show mean ± SD (B, G, and H) or median ± interquartile range (D). *P < 0.05, **P < 0.01, ****P < 0.0001 by Kruskal–Wallis with Dunn’s multiple comparisons test (B, D, and G), Mann-Whitney U test of Tph and Tfh control vs. SLE (G), and Spearman correlation statistics (E and F).
A comparison of the global transcriptomes of Tph cells and Tfh cells from SLE patients identified 229 genes with differential expression (P < 0.05, fold change > 2). Pathway analysis indicated pathways involving IL-2 (P = 3.6 × 10–10), T-bet (P = 1.9 × 10–8), CD28 (P = 3.4 × 10–7), and IL-21 (P = 1.1 × 10–6) as the top candidate upstream regulators with higher activity in Tph cells compared with Tfh cells. Notably, included in this gene list were several chemokine receptors with increased expression in Tph cells, including CX3CR1 (4.2-fold), CCR2 (4.4-fold), CXCR6 (10.4-fold), and CCR5 (4.1-fold) (Figure 6B and Supplemental Table 7) (21). CX3CR1 is of particular interest in SLE, as this receptor is commonly expressed on immune cells that infiltrate lupus nephritis kidneys (23). Using flow cytometry, we confirmed that a substantial portion of Tph cells, but not Tfh cells, expressed CX3CR1 (Figure 6, C and D).
We next explored differences in the transcriptomes of Tph cells from SLE patients compared with Tph cells from controls and identified 369 differentially expressed genes (P < 0.05, fold-change > 2) between the disease conditions (Supplemental Table 8). These genes were highly enriched in IFN-inducible genes; pathway analysis identified IFN signaling as the most enriched signature (P = 2.5 × 10–10), indicating a strong IFN signature in Tph cells from SLE patients. An IFN signature was similarly observed across other T cell populations from SLE patients (Supplemental Figure 6B). To further evaluate an IFN signature in Tph cells, we calculated an IFN signature score from the Tph cell transcriptomes and found that the IFN score was positively correlated with the frequency of Tph cells in circulation (Figure 6E). This positive correlation was observed across all patients studied and also present even among the SLE patients alone. To reproduce this observation in another cohort, we analyzed the correlation between Tph cell frequency and IFN score in a subset of SLE patients in the AMP cohort for whom IFN scores had been calculated using total PBMC transcriptomes (23). Here again, a positive correlation was observed between Tph cell frequency and IFN score (Figure 6F), suggesting a possible relationship between these 2 features of active SLE.
A focused assessment of the expression of cytokines associated with B cell–helper function detected both CXCL13 and IL21 in Tph cells and Tfh cells but not in Tregs or naive T cells. Expression of the B cell chemoattractant CXCL13 was significantly higher in both Tph cells and Tfh cells from SLE patients compared with cells from control donors (Figure 6G). IL21 also showed a trend toward higher expression in Tph cells from SLE patients compared with Tph cells from controls (Figure 6G). IFNG was also detected primarily in Tph cells and Tfh cells, while IL10 was detected variably across the T cell subsets (Supplemental Figure 7A). To confirm expression of IL21 in Tph cells from SLE patients, we analyzed IL21 by quantitative PCR (qPCR) in sorted Tph cells stimulated with PMA plus ionomycin. After stimulation, Tph cells expressed higher levels of IL21 than did PD-1– memory CD4+ T cell populations, with comparable expression in Tph cells and Tfh cells (Figure 6H). In contrast, IL10 expression did not significantly differ between Tph cells, Tfh cells, and PD-1– memory CD4+ T cells. (Supplemental Figure 7B). These results confirm that circulating Tph cells in SLE patients express IL21, IFNG, and CXCL13 and suggest that Tph cells in the circulation of SLE patients may be particularly active with mRNA expression of effector cytokines detectable directly ex vivo.
Tph cells are associated with plasmablasts and ABCs in lupus patients.
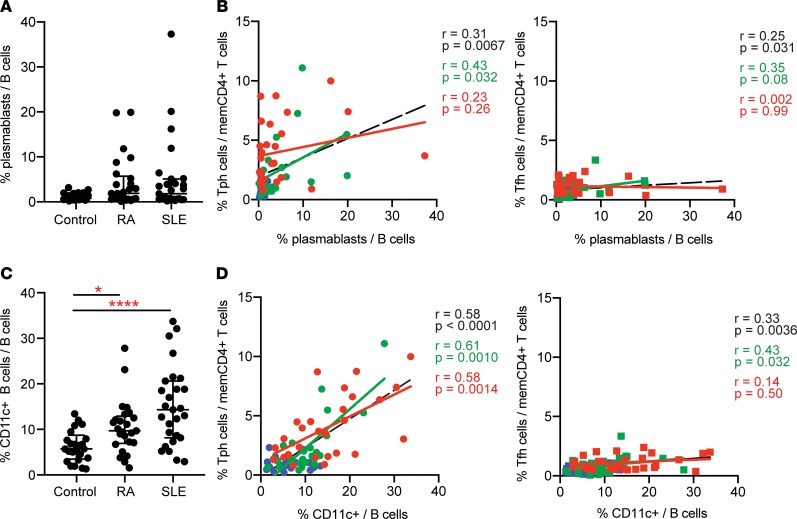
We next evaluated the correlation between Tph cells and different B cell populations in SLE patients. CD38hiCD27+ plasmablasts were increased in a subset of SLE patients in the AMP cohort, consistent with prior observations (Figure 7A, gated as shown in Supplemental Figure 8A) (29, 30). Across all patients and controls, the expansion of plasmablasts correlated positively with the frequency of both Tph cells and Tfh cells, although neither T cell population showed a strong correlation with plasmablast frequencies within the SLE cohort alone (Figure 7B).
Figure 7. Tph cells correlate with CD11c+ B cells and plasmablasts in SLE.
(A) Frequency of plasmablasts among B cells of controls (n = 25), RA (n = 25), and lupus nephritis (SLE; n = 27) patients in AMP mass cytometry dataset. (B) Correlation between the frequency of Tph cells or Tfh cells and plasmablasts in the AMP cohort (red, SLE patients; green, RA patients; blue, controls; black line, all patients). (C) Frequency of CD11c+ B cells as in A. (D) Correlation between Tph cells or Tfh cells and CD11c+ B cells as in B. Error bars show median ± interquartile range, with *P < 0.05, ****P < 0.0001 by Kruskal–Wallis with Dunn’s multiple comparisons test (A and C). Spearman correlation statistics shown in B and D.
Recently, a population of CD11c+CD21–CXCR5– B cells, sometimes referred to as ABCs, has been recognized as highly expanded in patients with active SLE (4). We quantified CD11c+ B cells using mass cytometry data generated with the AMP myeloid panel (Supplemental Table 2) and found that CD11c+ B cells composed, on average, about 15% of the B cells in lupus nephritis patients in the AMP cohort, 2.5-fold higher than controls (Figure 7C, gated as shown in Supplemental Figure 8B) (4). CD11c+ B cells were also increased in RA patients compared with controls, but to a lesser extent. The frequency of Tph cells correlated strongly with the frequency of CD11c+ B cells both across the overall cohort and specifically in SLE patients (Figure 7D). In contrast, Tfh cells showed a modest correlation with CD11c+ B cells overall and no correlation among SLE patients. In support of this observation, we also assessed the frequency of CD21–CXCR5– B cells, which could be identified in the AMP B cell panel. The frequency of CD11c+ B cells identified using the AMP myeloid mass cytometry panel correlates strongly with the frequency of CD21–CXCR5– B cells identified using the AMP B cell panel (Supplemental Figure 8, C and D), suggesting that these gating strategies identify highly overlapping cell populations, consistent with published reports (4, 31). Similar to observations with CD11c+ B cells, the frequency of CD21–CXCR5– B cells correlated positively with Tph cells but not Tfh cells (Supplemental Figure 8, C–E). These observations suggest a specific relationship between the expansion of Tph cells and CD11c+CD21– B cells in autoimmunity.
In murine models, generation of CD11c+ B cells requires T cell help and is enhanced by IL-21 and IFN-γ (6, 32). Because Tph cells express both IL-21 and IFN-γ, we hypothesized that Tph cells may stimulate the generation of CD11c+ B cells in vitro. Coculture of allogeneic healthy donor memory B cells with SLE donor T cells induced formation of a CD11c+ B cell population distinct from CD38hiCD27+ plasmablasts. Generation of CD11c+ B cells occurred comparably in cocultures of B cells with either Tph cells or Tfh cells (Supplemental Figure 8, F and G).
Tph cells help B cells in an IL-21– and MAF-dependent manner.
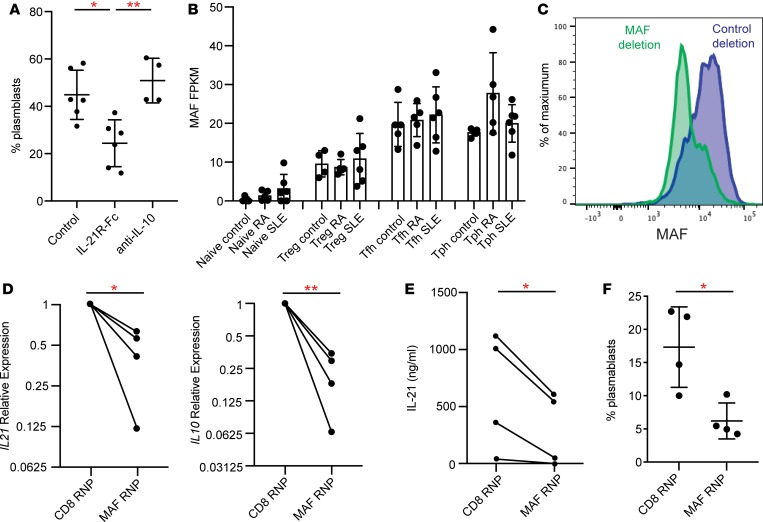
Our prior work indicated that the ability of Tph cells to induce B cell activation and differentiation into plasmablasts depends on production of IL-21 (22). Recently, a PD-1+CXCR5– population that coexpresses CXCR3 has been reported to help B activation and antibody secretion in an IL-10– but not IL-21–dependent manner (33). To determine whether Tph cells from SLE patients require IL-21 to stimulate plasmablasts, we evaluated the effect of neutralization of IL-21 in cocultures of Tph cells from SLE patients with allogeneic memory B cells. Blockade of IL-21 reduced the generation of plasmablasts in these cocultures by about 50%, while neutralization of IL-10 had no effect (Figure 8A).
Figure 8. Tph cells induce B cell responses in an IL-21– and MAF-dependent manner.
(A) Quantification of plasmablasts among B cells in cocultures of memory B cells from controls cocultured with Tph cells from SLE patients with neutralization of either IL-21 or IL-10 (n = 3 donors for IL-21R-Fc, n = 2 donors for anti–IL-10, 2 replicates per donor). (B) Expression of MAF in RNA-seq data in indicated T cell populations. (C) Flow cytometric detection of loss of MAF expression in CRISPR/Cas9-treated CD4+ cells. (D) Expression of IL21 and IL10 by qPCR in CD4+ T cells after treatment with MAF- or CD8-targeting Cas9 complexes. Values plotted were normalized to CD8 targeting control. (E) Expression of IL-21 by ELISA in CD4+ T cells after treatment with MAF- or CD8-targeting Cas9 complexes. (F) Plasmablast generation in cocultures of Tph cells treated with MAF- or CD8-targeting complexes cocultured with allogeneic memory B cells. Data pooled from 2 experiments with 2 different donors. Error bars show mean ± SD (A, B, and F). *P < 0.05, **P < 0.01. Paired t test statistics of normalized data in D and E. Unpaired t test statistics shown in F.
The regulation of IL-21 production differs between human and mouse T cells, and the transcriptional regulation of IL-21 production in human T cells remains incompletely defined (21). We noted that both circulating Tph cells and Tfh cells expressed high levels of the transcription factor MAF, a factor that has been implicated in IL-21 production from human Tfh cells (Figure 6A and Figure 8B) (34). In contrast, neither BCL6 nor SOX4 were elevated in Tph cells or Tfh cells from the circulation, and PRDM1 expression was higher in Tph cells than in Tfh cells, consistent with a low BCL6/PRDM1 ratio in Tph cells from both tissue and circulation (Supplemental Figure 9) (22). Given the coincident expression of MAF and IL-21 in both Tph cells and Tfh cells, we sought to directly evaluate the role of MAF in IL-21 production by human CD4+ T cells. We disrupted the gene encoding MAF by nucleofection of a CRISPR/Cas9 ribonucleoprotein complex in primary CD4+ T cells and in sorted Tph cells. We confirmed that the MAF-targeting construct reduced MAF expression by flow cytometry (Figure 8C). Deletion of MAF inhibited mRNA expression of both IL21 and IL10 in human CD4+ T cells and reduced secretion of IL-21 into culture media (Figure 8, D and E). In addition, CRISPR-mediated deletion of MAF in sorted Tph cells reduced the ability of Tph cells to induce B cell differentiation into plasmablasts in cocultures by over 50% (Figure 8F). Thus, Tph cells require the transcription factor MAF for IL-21–dependent B cell–helper function.
Discussion
Here, we have identified a markedly expanded Tph cell population in patients with SLE. Tph cells were initially described as a PD-1hiCXCR5–CD4+ T cell population abundant in the joints and expanded in the circulation of patients with seropositive RA (35). In this report, using multidimensional analysis of mass cytometry data, we found that PD-1hiCXCR5– T cells are highly expanded in the circulation of SLE patients, consistent with prior observations (12). We further demonstrated that these cells matched the Tph cell phenotype by multiple metrics: (a) high expression of PD-1, ICOS, HLA-DR, and TIGIT without CXCR5; (b) expression of cytokines associated with B cell recruitment and help (IL-21, CXCL13); and (c) ability to promote B cell differentiation into plasmablasts in an IL-21–dependent manner. By these criteria, the PD-1hiCXCR5– T cell population in SLE patients can be considered Tph cells.
The cytometric and transcriptomic similarities in Tph cells from RA and SLE patients help to define a core set of features that can be used to identify Tph cells across diseases, which includes expression of PD-1, ICOS, MAF, IL-21, and CXCL13 in the absence of CXCR5. Recently, antigen-specific T cells both in gut and blood of patients with celiac disease were found to have a phenotype that matches Tph cells, and cells with this phenotype were also increased in the circulation in small cohorts of SLE and systemic sclerosis patients (36). Tph cells are also increased in patients with type 1 diabetes — in particular, in patients with multiple autoantibodies (37). The expansion of Tph cells in multiple autoantibody-associated conditions, but not in seronegative RA or seronegative spondyloarthropathies, supports the idea that Tph cells represent a common driver of pathologic B cell activation in autoantibody-associated autoimmune diseases.
The correlations of Tph cell frequency with clinical and immunologic features of SLE suggest that Tph cells are important mediators of lupus immunopathology. While both Tph cells and Tfh cells were similarly increased in new-onset SLE patients, Tph cells substantially outnumbered Tfh cells in both established SLE and lupus nephritis patients, and only Tph cells showed a strong association with clinical disease activity by SLEDAI in lupus nephritis patients, suggesting that circulating Tph cell frequency may be a more robust marker of the activated T cell response than circulating Tfh cell frequency. However, given the highly heterogeneous nature of disease in SLE and the limitations of quantifying disease activity by SLEDAI, larger cohorts of SLE patients will be required to evaluate clinical correlations more definitively.
There is little doubt that Tfh cells are important in the pathogenesis of SLE, and these cells may be underrepresented in blood, given their localization in lymphoid follicles. However, both human and murine studies indicate that B cell–T cell interactions also occur outside of lymphoid follicles, both in extrafollicular foci of secondary lymphoid organs and within inflamed target tissues in autoimmunity (18–20, 22, 38–40). Tph cells appear well equipped to mediate these interactions, particularly in chronically inflamed peripheral tissues, where they may function to promote local accumulation of antibody-secreting cells (21). The ability of Tph cells to stimulate differentiation of memory, rather than naive, B cells in vitro is consistent with the idea that Tph cells may primarily interact with activated or memory B cells, which are more likely than naive B cells to traffic through inflamed tissues (41). The strong correlation between Tph cells, but not Tfh cells, and CD11c+ B cells in circulation further supports that idea that Tph cells may play a unique role in autoimmune pathology. Interestingly, CD11c+ B cells express little CXCR5 and also infiltrate target tissues, including lupus nephritis kidneys, raising the possibility that Tph cells and CD11c+ B cells may interact within peripheral tissues (4, 23, 31). This has not yet been experimentally tested.
We hypothesize that Tph cells likely migrate to inflamed tissues via expression of receptors such as CX3CR1, a chemokine receptor enriched on kidney-infiltrating leukocytes in lupus nephritis (23). By flow cytometry, we identified within lupus nephritis kidney biopsies CD4+ T cells with high PD-1 expression, which correlated with the frequency of infiltrating B cells. Published immunofluorescence microscopy studies further support the possibility of Tph cells in lupus nephritis kidneys, having demonstrated that PD-1+CD4+ T cells are in close contact with B cells within the interstitium of lupus nephritis kidneys (20). However, the chemokine receptor expression and essential effector functions of PD-1hi T cells in the kidney have not yet been demonstrated, and the extent of colocalization with specific B cell subsets, including CD11c+ B cells, requires further study.
Through in vitro coculture studies, we observed a critical role for IL-21 in the function of Tph cells from SLE patients. Tph cells sorted from SLE patients express IL-21, as well as CXCL13, and their ability to induce memory B cell differentiation into plasmablasts in vitro depends on IL-21, consistent with described effects of IL-21 on B cell differentiation in vivo (42). Production of IL-21 distinguishes Tph cells from a PD-1+CXCR5–CXCR3+ T cell population recently identified in pediatric SLE patients by Caielli and colleagues, which augments B cell responses via production of IL-10 and succinate (33). Importantly, we define the Tph cell population by requiring high expression of PD-1, which differs from the broader PD-1 expression used by Caielli and colleagues to identify IL-10/succinate–producing PD-1+CXCR5–CXCR3+ cells. Cytometric, transcriptomic, and functional analyses presented here indicate that the PD-1hiCXCR5– Tph cell population shows a strong resemblance to Tfh cells, including expression of IL-21 and CXCL13 ex vivo, consistent with prior observations of Tph cells in rheumatoid synovium (22). Given the key roles of IL-21 in autoreactive B cell activation, autoantibody production, and renal injury in murine models of lupus, we hypothesize that IL-21 production is an important aspect of Tph cell function, although other pathologic roles independent of B cell–helper function are also possible (43, 44). Subsequent studies will help to define the relationship between PD-1hi and PD-1+ T cells and the relationship between Tph cells and Tfh cells in SLE patients.
The transcriptional regulation of IL-21 production in human T cells remains incompletely understood. BCL6 loss or overexpression appears to have little effect on T cell production of IL-21 production (45). Overexpression of MAF, but not BCL6, in human T cells increased IL-21 production (34). Here, we found that Tph cells express high levels of MAF and that CRISPR-mediated deletion of MAF strongly inhibited IL-21 and IL-10 expression in T cells. Deletion of MAF also blunted the ability of Tph cells to stimulate plasmablast differentiation, indicating MAF as a key regulator of the ability of T cells to induce B cell differentiation.
In summary, we have found a highly expanded PD-1hiCXCR5– Tph cell population with IL-21–dependent B cell–helper function in SLE patients. We hypothesize that Tph cells may be important contributors to pathologic T cell–B cell interactions that occur outside of the follicles of secondary lymphoid organs. This work nominates Tph cells as a new therapeutic target in SLE and highlights that these cells should be considered in the development of strategies aimed at interrupting pathologic T cell–B cell interactions.
Methods
Patient cohorts.
Mass cytometry data was generated on PBMCs from patients enrolled in Phase I of the AMP RA/SLE Network (23, 24, 25). Patient characteristics and clinical assessments, including measurement of the modified SELENA-SLEDAI score and anti-dsDNA antibody assessment, were performed at each individual site. In the AMP cohort, all SLE patients met the 1997 ACR classification criteria for lupus and demonstrated class III, class IV, or class V glomerulonephritis on a clinically indicated kidney biopsy. RA patients met the 2010 ACR classification criteria for RA. Noninflammatory controls were screened for the absence of inflammatory diseases. For the validation cohorts, patients with SLE, seropositive RA (rheumatoid factor–positive and/or anti-CCP antibody–positive), and noninflammatory controls were enrolled at Brigham and Women’s Hospital with appropriate informed consent under IRB protocols approved by Partners HealthCare IRB. SLE patients met 1997 ACR classification criteria as assessed by the treating physician, who also determined the SLEDAI-2K score. The new-onset SLE cohort included patients with a diagnosis of SLE within the past 6 months prior to enrollment and without treatment with major immunosuppressive therapies. Treatment with hydroxychloroquine and prednisone ≤ 10mg was permitted. The established SLE cohort included patients with a diagnosis of SLE with varied treatments including major immunosuppressive therapies.
PBMC isolation and processing.
For the AMP cohort samples, blood was collected into lithium heparin tubes and PBMCs isolated by density centrifugation using SepMate tubes. For the BWH validation cohort, PBMCs were isolated by density centrifugation using Ficoll-Hypaque in 50-mL conical tubes. For both cohorts, PBMCs were washed in PBS and cryopreserved in a 10% DMSO–containing solution for batched analyses.
Generation of mass cytometry data.
For mass cytometry analysis of AMP samples, 79 PBMC samples included in the analysis were processed in a total of 5 batches, with the batches run within a span of 14 days. Batches had balanced numbers of samples from controls, RA patients, and SLE patients.
For mass cytometry analysis of validation cohort samples, 39 independent patient samples included in the analysis were processed in 3 batches run within a span of 8 days. Batches had balanced numbers of controls, established SLE, and new-onset SLE patient samples.
Cryopreserved PBMCs were thawed into RPMI Medium 1640 (Invitrogen, catalog 11875-085) supplemented with 5% heat-inactivated FBS (Invitrogen, catalog 16000044), 1 mM GlutaMAX (Invitrogen, catalog 35050079), antibiotic-antimycotic (Invitrogen, catalog 15240062), 2 mM MEM nonessential amino acids (Invitrogen, catalog 11140050), 10 mM HEPES (Invitrogen, catalog 15630080), 2.5 × 10–5 M 2-mercaptoethanol (MilliporeSigma, catalog M3148), 20 units/mL sodium heparin (MilliporeSigma, catalog H3393), and 25 units/mL benzonase nuclease (MilliporeSigma, catalog E1014). Cells were counted and 0.5 × 106 to 1 × 106 cells from each sample were transferred to a polypropylene plate for staining.
The samples were spun down and aspirated. A total of 5 μM of cisplatin viability staining reagent (Fluidigm, catalog 201064) was added for 2 minutes and then diluted with culture media. After centrifugation, Human TruStain FcX Fc receptor blocking reagent (BioLegend, catalog 422302) was used at a 1:100 dilution in CSB (PBS with 2.5 g BSA [MilliporeSigma, catalog A3059] and 100 mg of sodium azide [MilliporeSigma, catalog 71289]) for 10 minutes, followed by incubation with conjugated surface antibodies for 30 minutes. All antibodies were obtained from the Harvard Medical Area CyTOF Antibody Resource and Core.
A total of 16% stock paraformaldehyde (Thermo Fisher Scientific, catalog O4042-500) dissolved in PBS was used at a final concentration of 4% formaldehyde for 10 minutes in order to fix the samples before permeabilization with the FoxP3/Transcription Factor Staining Buffer Set (Thermo Fisher Scientific, catalog 00-5523-00). The samples were incubated with SCN-EDTA coupled palladium–based barcoding reagents for 15 minutes and then combined into a single sample. Conjugated intracellular antibodies were added into each tube and incubated for 30 minutes. Cells were then fixed with 1.6% formaldehyde for 10 minutes.
DNA was labeled for 20 minutes with an 18.75 μM iridium intercalator solution (Fluidigm, catalog 201192B). Samples were subsequently washed and reconstituted in Milli-Q filtered distilled water in the presence of EQ Four Element Calibration beads (Fluidigm, catalog 201078) at a final concentration of 1 × 106 cells/mL. Samples were acquired on a Helios CyTOF Mass Cytometer (Fluidigm). The raw FCS files were normalized to reduce signal deviation between samples over the course of multiday batch acquisitions, utilizing the bead standard normalization method established by Finck et al. (46). These normalized files were then deconvoluted into individual sample files using a single-cell–based debarcoding algorithm established by Zunder et al. (47). In the validation cohort, the normalized files were also compensated with a panel-specific spillover matrix to subtract cross-contaminating signals, utilizing the CyTOF-based compensation method established by Chevrier et al. (48).
In the AMP cohorts, PBMCs were stained with 3 CyTOF panels, all of which contributed data to the analyses presented (Supplemental Table 2). In the validation cohort, data from a T cell panel were used (Supplemental Table 4).
Mass cytometry data.
Mass cytometry data were first gated to exclude debris and identify DNA+ events. Nonviable cisplatin+ cells and equalization beads were excluded. Frequencies of Tph cells (PD-1hiCXCR5–CD4+ T cells), Tfh cells (PD-1hiCXCR5+CD4+ T cells), plasmablasts (CD19+CD20–CD38hiCD27+), CD11c+CD19+ B cells, and CD21loCD19+ B cells were quantified by manual gating, with uniform gates applied across all samples in each cohort. Two samples from the RA cohort were excluded due to their treatment with rituximab prior to sample collection, which precludes analysis of B cell populations. Gating for cell frequencies and expression intensity quantification were performed using FlowJo 10.4.2. FlowSOM analyses of the AMP mass cytometry data were performed using the implementation on Cytobank. Control (n = 25), RA (n = 24), and SLE (n = 26) samples were included; 1 RA sample and 1 SLE sample were excluded due to low memory CD4 T cell event counts. Excluding these 2 samples allowed for analysis of 5,296 gated memory CD4+ T cells per patient. Analysis was performed including all staining channels except the markers used to gate memory CD4+ T cells (CD45, CD4, CD8a, CD45RO, and CD45RA), assigning 100 clusters and 15 metaclusters with a random seed of 1559288433. Heatmaps show row-normalized median expression of markers in the metaclusters. Markers used to gate the analyzed input cell population and markers with a median expression of 0 in all clusters were excluded from heatmaps.
RNA-seq analysis.
CD3+ T cells from PBMCs from SLE patients (n = 6), RA patients (n = 5), and controls (n = 5) were isolated by positive selection using MACS columns, and CD3+ T cells were stained with a flow cytometry antibodies: CD3-BV510 (OKT3), CD4-PE/Cy7 (RPA-T4), CD45RA-BV605 (HI100), CD127-BV711 (A019D5), CD25-FITC (M-A251), TIGIT-PE (VSTM3), PD-1–APC/Cy7 (EH12.2H7), CXCR5-BV421 (J252D4) plus propidium iodide. Naive T cells, Tregs, Tph cells, and Tfh cells were sorted as indicated (Supplemental Figure 1). Cell sorting was performed on a Becton Dickinson FACSAria Fusion sorter using a 70 μm nozzle, with sort purity routinely > 98%. One thousand cells of each population were sorted directly into RLT lysis buffer with 1% β-mercaptoethanol (Qiagen). RNA was isolated using RNeasy Micro kits (Qiagen) and eluted in 12 μL of water.
A total of 5 μL of total RNA were transferred into wells of a 96-well plate, and RNA-seq libraries were prepared at Broad Technology Labs at the Broad Institute of Harvard and MIT (Cambridge, Massachusetts, USA) using the Illumina SmartSeq2 platform. Samples were sequenced on a HiSeq500 using 25 bp paired-end reads. Alignment and differential gene expression analysis was performed in Omicsoft Array Studio version 10.0.1.96. Briefly, cleaned reads were aligned to the human B38 genome reference by using the Omicsoft Aligner, with a maximum of 2 allowed mismatches. Gene level counts were determined by the OSA algorithm as implemented in Omicsoft Array Studio and using Ensembl.R86 gene models. Approximately 80%–90% of reads across all samples mapped to the human genome (corresponding to 1.4–16 million reads). Two samples with read counts less than 1 million were excluded. Differential gene expression analysis was performed by the DESeq2 algorithm as implemented in Omicsoft Array Studio, with samples from different groups serving as reference as indicated. A cutoff of 20 normalized counts in all replicate groups was applied when identifying a gene signature to remove genes with low expression. Pathway analyses were performed with Ingenuity Pathway Analysis (IPA; QIAGEN Inc.; www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/).
To calculate IFN scores for Tph cells transcriptomes, the expression of a set of IFN-stimulated genes identified as the union of 3 IFN-inducible gene modules described in Chiche et al. (23, 49). The IFN score was calculated as the average expression of this 124-gene set, using a scaled expression for each gene calculated by taking the difference from the mean and dividing by the SD in log scale. IFN scores for a subset of AMP patients (n = 6) for whom additional PBMCs were available was calculated in the same way using transcriptomes of total PBMC (23).
Analysis of flow cytometry of kidney biopsies.
Kidney biopsies from 24 lupus nephritis patients were analyzed by flow cytometry in the AMP Network as described (23). Cryopreserved kidney biopsies were thawed and dissociated into a single cell suspension using mechanical disruption and enzymatic digestion with Liberase TL. Single cell suspensions were stained with a flow cytometry panel including the following markers: anti–CD45-FITC (HI30), anti–CD19-PE (HIB19), anti–CD11c-PerCP/Cy5.5 (Bu15), anti–CD10-BV421 (HI10A), anti–CD14-BV510 (M5E2), anti–CD3-BV605 (UCHT1), anti–CD4-BV650 (RPA-T4), anti–CD8-BV711 (SK1), anti–CD31-AlexaFluor700 (WM59), anti–PD-1–APC (EH12.2H7) and propidium iodide (all from BioLegend). Data were acquired in a FASCAria Fusion, and flow cytometric quantification of cell populations was performed using FlowJo 10.4.2.
T and B cell cocultures.
Total B cells were isolated first from PBMCs from blood bank leukoreduction collars by magnetic bead positive selection using CD19 (Miltenyi Biotec). B cells were stained with anti–CD14-APC (M5E2), anti–CD3-PeCy7 (UCHT1), anti–CD27-FITC (O323), anti–IgD-BV421 (IA6-2), and propidium iodide antibodies (all from BioLegend), and memory B cells sorted as CD14–CD3–CD27+IgD– cells on a BD FACSAria Fusion to remove contaminating T cells and monocytes. To isolate T cell populations from lupus peripheral blood donors, PBMCs were stained directly with anti–CXCR5-BV421 (J252D4), anti–CD4-BV650 (RPA-T4), anti–PD-1–APC (EH12.2H7), anti–CD3-PE-Cy7 (UCHT1), anti–CD45RA-BV510 (HI100), and propidium iodide antibodies (all from BioLegend). T cell populations were sorted as follows: CD4+CD45RA–PD-1–CXCR5–, CD4+CD45RA–PD-1–CXCR5+, CD4+CD45RA–PD-1hiCXCR5– (Tph cells), and CD4+CD45RA–PD-1hiCXCR5+ (Tfh cells). Sorted T cell populations from SLE patients were cocultured with allogenic memory B cells from control donors at a ratio of 1:10 in 200 μL of RPMI/10% FBS and stimulated with LPS (5 μg/mL; Invitrogen) and SEB (1 μg/mL; Sigma-Aldrich) for 5–6 days. In indicated experiments, either recombinant IL-21R Fc chimera protein (R&D Systems, 20 μg mL–1) or anti–IL-10 antibody (BioLegend LEAF purified antibody, 10 μg mL–1) were added to cultures. Cells were harvested and analyzed by flow cytometry as described below.
Flow cytometry.
For CD38hiCD27+ plasmablasts and CXCR5–CD21–CD11c+ B cell quantification, cultured cells were washed once in PBS and stained in PBS/1% BSA with the following antibodies for 45 minutes: anti–CD27-FITC (O323), anti–PD-1–PE (EH12.2H7), anti–CD11c-BV510 (3.9), anti–CXCR5-BV605 (J252D4), anti–CD4-BV650 (RPA-T4), anti–CD21-APC (Bu32), anti–CD38-APC-Cy7(HB-7), anti–CD19-BV421 (HIB19), anti–CD138-PE-Cy7 (MI15), anti–CD278-PerCP/Cyanine5.5 (C398.4A) and propidium iodide. Cells were washed in cold PBS and data acquired on a BD Fortessa analyzer using FACSDiva software. Data were analyzed using FlowJo 10.4.2. A single set of gates for CD19, CD27, CD38, CXCR5, CD21, and CD11c were applied to all samples. The percentage of plasmablasts (CD19+CD38hiCD27+) among CD19+ B cells and percentage of atypical B cells (CD19+ nonplasmablast with CXCR5– CD21–CD11c+) among nonplasmablasts were calculated for indicated samples. Detection of CX3CR1 in Tph cells and Tfh cells was performed on the same PBMC samples used for mass cytometry of the BWH validation cohort using the following panel: anti–CD45RA-605 (HI100), anti–CD3-PE-Cy7 (UCHT1), anti–CD4-BV650 (RPA-T4), anti–PD-1–PE (EH12.2H7), anti–CXCR5-BV421(J252D4), anti–CX3CR1-FITC(2A9-1), anti–CXCR3-APC (G025H7), and propidium iodiden (all from BioLegend).
Reverse transcription PCR (RT-PCR) analyses.
RNA was isolated using RNeasy Micro Kits (Qiagen). cDNA was prepared using QuantiTect RT-PCR (Qiagen), and PCR was performed with Brilliant III SYBR Green on a AriaMx Real-Time PCR System. Primers used were as follows: RPL13A (forward: 5′ - CATAGGAAGCTGGGAGCAAG - 3′; reverse: 5′ - GCCCTCCAATCAGTCTTCTG - 3′), IL-10 (forward: 5′ - CGCATGTGAACTCCCTGG - 3′; reverse: 5′ - TAG ATGCCTTTCTCTTGGAGC - 3′), and IL-21 (forward: 5′ - AGGAAACCACCTTCCACAAA - 3′; reverse: 5′ - GAATCACATGAAGGGCATGTT - 3′). Expression levels relative to control gene RPL13A were calculated.
CRISPR/Cas9-mediated gene deletion.
Tph cells were flow sorted as described above and stimulated for 24 hours with anti-CD3/CD28 beads (1:5 bead/cell ratio). Single guide RNA-Cas 9 complexes were generated by first incubating MAF guide RNA (5′ - UGGAGAUCUCCUGCUUGAGGGUUUUAGAGCUAUGCU - 3′) with transactivating crRNA (tracrRNA) (IDT Technologies) in a 1:1 ratio for 2 minutes at 95°C and cooled at 37°C for 5 minutes. Afterward, the guide RNA and tracrRNA complex were incubated at 2:1 ratio with recombinant Cas9 (Macrolab) at 37°C for 1 hour. Sorted memory T cell subsets were stimulated overnight for 2 days with CD3/CD28 beads; they were then separated from the beads and washed with PBS. Cells were then electroporated with the single guide RNA-Cas9 complex at pulse EH-115 in a 4-D Nucleofector (Lonza) using P3 Primary cell 4D X Kit S (V4XP-3032, Lonza); they were then washed and resuspended in RPMI/10%FBS. Electroporated T cells were then cocultured on the same day with flow-sorted B cells as described above.
Data availability.
Mass cytometry data from the AMP SLE and RA cohorts datasets are available at the ImmPort repository (https://www.immport.org) from accessions SDY997 and SDY998. RNA-seq data from the BWH cohort dataset is available at the ImmPort repository from accession SDY1475. The data that support the findings of this study are available from the corresponding author upon request.
Statistics.
Statistical comparisons were performed in Prism as indicated in figure legends. Two-sided tests were used for all comparisons. For analysis of FlowSOM metaclusters in mass cytometry data, 2-sided Student’s t tests were calculated comparing metacluster abundances in SLE patients and controls, adjusted for multiple testing by Bonferroni correction. Significantly expanded clusters were considered to have adjusted P < 0.05 and fold change > 2. Comparison of mass cytometry marker expression on Tph cells and Tfh cells and comparison of expression on Tph cells from controls and SLE patients was performed using Student’s t tests with Bonferroni correction for multiple testing considering adjusted P < 0.05 as significant. All other analyses were performed using nonparametric tests, with Mann-Whitney U test used for comparison between 2 groups, and Kruskal-Wallis with Dunn’s multiple comparisons test was used for comparisons between 3 or more groups, except as indicated.
Study approval.
Human subjects research was performed according to the IRBs at sites participating in the AMP RA/SLE Network via approved protocols with appropriate informed consent (23, 24, 25). Patients recruited at Brigham and Women’s Hospital were enrolled with written informed consent under protocols 2014P002558 and 2016P001660 approved by the Partners HealthCare IRB.
Author contributions
GK, MFG, KHC, MP, JPB, DW, and CP recruited patients and collected and analyzed clinical data. AVB, VSW, and MFG performed the functional experiments. JAL and JK performed the mass cytometry data acquisition. AVB, DAR, CYF, YQ, and JAL analyzed the mass cytometry data. ZJL sorted and processed samples for RNA-seq. MFM, MBB, SEA, and DAR supervised analyses of mass cytometry and RNA-seq. ESM, GW, KXZ, AA, YC, and DAR analyzed the RNA-seq data. BD, JHA, MP, JPB, DW, CP, JAJ, JMG, and PAN participated in the design of the study and interpretation of the data. AVB and DAR wrote the initial draft; VFW, JPB, JAJ, KHC, BD, JMG, and PAN edited it; and all authors participated in writing the final manuscript, with the exception of MFG, who died prior to manuscript completion.
Supplementary Material
Acknowledgments
This work was supported, in part, by funding from the Lupus Research Alliance, Rheumatology Research Foundation, Tobe and Stephen E. Malawista, MD Endowment in Academic Rheumatology, Burroughs Wellcome Fund Career Award in Medical Sciences, and NIAMS K08 AR072791 to DAR. AVB was supported by a Rheumatology Research Foundation and Medical Graduate Student Preceptorship. AVB and PAN were supported by the Fundación Bechara, and PAN was supported by funding from the Lupus Research Alliance, NIAMS P30 AR070253, and NIAMS R01 AR065538. We thank Adam Chicoine and the BWH Human Immunology Center Flow Cytometry Core for cell sorting assistance. We thank Jason Gilliland, Sophia Zarakhovich, and Kimberly Kerr from Merck & Co. Inc. for their assistance in RNA-seq data processing.
Version 1. 09/19/2019
In-Press Preview
Version 2. 10/17/2019
Electronic publication
Version 3. 10/17/2019
The affiliation for the AMP Network was updated.
Footnotes
Conflict of interest: DAR and MBB are coinventors on patent WO2017213695A1 submitted on Tph cells. SA, EM, YQ, and GW are employees of Merck & Co. Inc. KZ was an employee of Merck & Co. Inc., and MM was an employee of Merck Sharp & Dohme Corp., a subsidiary of Merck & Co. Inc., during participation in this manuscript.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(20):e130062.https://doi.org/10.1172/jci.insight.130062.
Contributor Information
Alexandra V. Bocharnikov, Email: abocharnikov@bwh.harvard.edu.
Joshua Keegan, Email: jkeegan@bwh.harvard.edu.
Vanessa S. Wacleche, Email: vwacleche@bwh.harvard.edu.
Ye Cao, Email: ycao13@bwh.harvard.edu.
Chamith Y. Fonseka, Email: cfonseka@broadinstitute.org.
Guoxing Wang, Email: guoxing.wang@merck.com.
Kelvin X. Zhang, Email: kelvinxizhang@gmail.com.
Arnon Arazi, Email: arnonar@broadinstitute.org.
Gregory Keras, Email: gkeras@bwh.harvard.edu.
Zhihan J. Li, Email: zjli@bwh.harvard.edu.
Yujie Qu, Email: yujie_qu@merck.com.
Michael F. Gurish, Email: mgurish@rics.bwh.harvard.edu.
Michelle Petri, Email: mpetri@jhmi.edu.
Jill P. Buyon, Email: Jill.Buyon@nyulangone.org.
Chaim Putterman, Email: chaim.putterman@einstein.yu.edu.
David Wofsy, Email: david.wofsy@ucsf.edu.
Judith A. James, Email: Judith-James@omrf.org.
Joel M. Guthridge, Email: Joel-Guthridge@omrf.org.
Betty Diamond, Email: bdiamond@nshs.edu.
Jennifer H. Anolik, Email: jennifer_anolik@urmc.rochester.edu.
Matthew F. Mackey, Email: matthew.f.mackey@gsk.com.
Stephen E. Alves, Email: stephen_alves@merck.com.
Peter A. Nigrovic, Email: pnigrovic@bwh.harvard.edu.
Karen H. Costenbader, Email: KCOSTENBADER@BWH.HARVARD.EDU.
Michael B. Brenner, Email: mbrenner@rics.bwh.harvard.edu.
James A. Lederer, Email: jlederer@rics.bwh.harvard.edu.
Deepak A. Rao, Email: darao@bwh.harvard.edu.
References
- 1.Kavanaugh AF, Solomon DH, American College of Rheumatology Ad Hoc Committee on Immunologic Testing Guidelines Guidelines for immunologic laboratory testing in the rheumatic diseases: anti-DNA antibody tests. Arthritis Rheum. 2002;47(5):546–555. doi: 10.1002/art.10558. [DOI] [PubMed] [Google Scholar]
- 2.Arce E, Jackson DG, Gill MA, Bennett LB, Banchereau J, Pascual V. Increased frequency of pre-germinal center B cells and plasma cell precursors in the blood of children with systemic lupus erythematosus. J Immunol. 2001;167(4):2361–2369. doi: 10.4049/jimmunol.167.4.2361. [DOI] [PubMed] [Google Scholar]
- 3.Jacobi AM, et al. Correlation between circulating CD27high plasma cells and disease activity in patients with systemic lupus erythematosus. Arthritis Rheum. 2003;48(5):1332–1342. doi: 10.1002/art.10949. [DOI] [PubMed] [Google Scholar]
- 4.Wang S, et al. IL-21 drives expansion and plasma cell differentiation of autoreactive CD11chiT-bet+ B cells in SLE. Nat Commun. 2018;9(1):1758. doi: 10.1038/s41467-018-03750-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naradikian MS, Hao Y, Cancro MP. Age-associated B cells: key mediators of both protective and autoreactive humoral responses. Immunol Rev. 2016;269(1):118–129. doi: 10.1111/imr.12380. [DOI] [PubMed] [Google Scholar]
- 6.Manni M, et al. Regulation of age-associated B cells by IRF5 in systemic autoimmunity. Nat Immunol. 2018;19(4):407–419. doi: 10.1038/s41590-018-0056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malkiel S, Barlev AN, Atisha-Fregoso Y, Suurmond J, Diamond B. Plasma Cell Differentiation Pathways in Systemic Lupus Erythematosus. Front Immunol. 2018;9:427. doi: 10.3389/fimmu.2018.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanco P, Ueno H, Schmitt N. T follicular helper (Tfh) cells in lupus: Activation and involvement in SLE pathogenesis. Eur J Immunol. 2016;46(2):281–290. doi: 10.1002/eji.201545760. [DOI] [PubMed] [Google Scholar]
- 9.He J, et al. Circulating precursor CCR7(lo)PD-1(hi) CXCR5(+) CD4(+) T cells indicate Tfh cell activity and promote antibody responses upon antigen reexposure. Immunity. 2013;39(4):770–781. doi: 10.1016/j.immuni.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Simpson N, et al. Expansion of circulating T cells resembling follicular helper T cells is a fixed phenotype that identifies a subset of severe systemic lupus erythematosus. Arthritis Rheum. 2010;62(1):234–244. doi: 10.1002/art.25032. [DOI] [PubMed] [Google Scholar]
- 11.Crotty S. Follicular helper CD4 T cells (TFH) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 12.Choi JY, et al. Circulating follicular helper-like T cells in systemic lupus erythematosus: association with disease activity. Arthritis Rheumatol. 2015;67(4):988–999. doi: 10.1002/art.39020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, et al. Circulating CXCR5+CD4+helper T cells in systemic lupus erythematosus patients share phenotypic properties with germinal center follicular helper T cells and promote antibody production. Lupus. 2015;24(9):909–917. doi: 10.1177/0961203314567750. [DOI] [PubMed] [Google Scholar]
- 14.Vinuesa CG, et al. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature. 2005;435(7041):452–458. doi: 10.1038/nature03555. [DOI] [PubMed] [Google Scholar]
- 15.Yi W, et al. The mTORC1-4E-BP-eIF4E axis controls de novo Bcl6 protein synthesis in T cells and systemic autoimmunity. Nat Commun. 2017;8(1):254. doi: 10.1038/s41467-017-00348-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu YL, Metz DP, Chung J, Siu G, Zhang M. B7RP-1 blockade ameliorates autoimmunity through regulation of follicular helper T cells. J Immunol. 2009;182:1421–1428. doi: 10.4049/jimmunol.182.3.1421. [DOI] [PubMed] [Google Scholar]
- 17.Kim CJ, et al. The Transcription Factor Ets1 Suppresses T Follicular Helper Type 2 Cell Differentiation to Halt the Onset of Systemic Lupus Erythematosus. Immunity. 2018;49(6):1034–1048.e8. doi: 10.1016/j.immuni.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 18.Odegard JM, et al. ICOS-dependent extrafollicular helper T cells elicit IgG production via IL-21 in systemic autoimmunity. J Exp Med. 2008;205(12):2873–2886. doi: 10.1084/jem.20080840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang A, et al. In situ B cell-mediated immune responses and tubulointerstitial inflammation in human lupus nephritis. J Immunol. 2011;186(3):1849–1860. doi: 10.4049/jimmunol.1001983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liarski VM, et al. Cell distance mapping identifies functional T follicular helper cells in inflamed human renal tissue. Sci Transl Med. 2014;6(230):230ra46. doi: 10.1126/scitranslmed.3008146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao DA. T Cells That Help B Cells in Chronically Inflamed Tissues. Front Immunol. 2018;9:1924. doi: 10.3389/fimmu.2018.01924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao DA, et al. Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature. 2017;542(7639):110–114. doi: 10.1038/nature20810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arazi A, et al. The immune cell landscape in kidneys of patients with lupus nephritis. Nat Immunol. 2019;20(7):902–914. doi: 10.1038/s41590-019-0398-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang F, et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat Immunol. 2019;20(7):928–942. doi: 10.1038/s41590-019-0378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Der E, et al. Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat Immunol. 2019;20(7):915–927. doi: 10.1038/s41590-019-0386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Gassen S, et al. FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A. 2015;87(7):636–645. doi: 10.1002/cyto.a.22625. [DOI] [PubMed] [Google Scholar]
- 27.Vella LA, et al. T follicular helper cells in human efferent lymph retain lymphoid characteristics. J Clin Invest. 2019;129(8):3185–3200. doi: 10.1172/JCI125628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kenefeck R, et al. Follicular helper T cell signature in type 1 diabetes. J Clin Invest. 2015;125(1):292–303. doi: 10.1172/JCI76238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banchereau R, et al. Personalized Immunomonitoring Uncovers Molecular Networks that Stratify Lupus Patients. Cell. 2016;165(3):551–565. doi: 10.1016/j.cell.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tipton CM, et al. Diversity, cellular origin and autoreactivity of antibody-secreting cell population expansions in acute systemic lupus erythematosus. Nat Immunol. 2015;16(7):755–765. doi: 10.1038/ni.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenks SA, et al. Distinct Effector B Cells Induced by Unregulated Toll-like Receptor 7 Contribute to Pathogenic Responses in Systemic Lupus Erythematosus. Immunity. 2018;49(4):725–739.e6. doi: 10.1016/j.immuni.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naradikian MS, et al. Cutting Edge: IL-4, IL-21, and IFN-gamma Interact To Govern T-bet and CD11c Expression in TLR-Activated B Cells. J Immunol. 2016;197(4):1023–1028. doi: 10.4049/jimmunol.1600522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caielli S, et al. A CD4+ T cell population expanded in lupus blood provides B cell help through interleukin-10 and succinate. Nat Med. 2019;25(1):75–81. doi: 10.1038/s41591-018-0254-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kroenke MA, et al. Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. J Immunol. 2012;188(8):3734–3744. doi: 10.4049/jimmunol.1103246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rao DA, et al. T peripheral helper cells are expanded in the circulation of active SLE patients and correlate with CD21low B cells [abstract]. Arthritis Rheumatol. 2018; 70 (suppl 10). https://acrabstracts.org/abstract/t-peripheral-helper-cells-are-expanded-in-the-circulation-of-active-sle-patients-and-correlate-with-cd21low-b-cells/ Accessed September 23, 2019.
- 36.Christophersen A, et al. Distinct phenotype of CD4+ T cells driving celiac disease identified in multiple autoimmune conditions. Nat Med. 2019;25(5):734–737. doi: 10.1038/s41591-019-0403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ekman I, et al. Circulating CXCR5–PD-1hi peripheral T helper cells are associated with progression to type 1 diabetes. Diabetologia. 2019;62(9):1681–1688. doi: 10.1007/s00125-019-4936-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheel T, Gursche A, Zacher J, Häupl T, Berek C. V-region gene analysis of locally defined synovial B and plasma cells reveals selected B cell expansion and accumulation of plasma cell clones in rheumatoid arthritis. Arthritis Rheum. 2011;63(1):63–72. doi: 10.1002/art.27767. [DOI] [PubMed] [Google Scholar]
- 39.Vu Van D, et al. Local T/B cooperation in inflamed tissues is supported by T follicular helper-like cells. Nat Commun. 2016;7:10875. doi: 10.1038/ncomms10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sweet RA, Ols ML, Cullen JL, Milam AV, Yagita H, Shlomchik MJ. Facultative role for T cells in extrafollicular Toll-like receptor-dependent autoreactive B-cell responses in vivo. Proc Natl Acad Sci USA. 2011;108(19):7932–7937. doi: 10.1073/pnas.1018571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oh JE, et al. Migrant memory B cells secrete luminal antibody in the vagina. Nature. 2019;571(7763):122–126. doi: 10.1038/s41586-019-1285-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zotos D, et al. IL-21 regulates germinal center B cell differentiation and proliferation through a B cell-intrinsic mechanism. J Exp Med. 2010;207(2):365–378. doi: 10.1084/jem.20091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi JY, et al. Disruption of Pathogenic Cellular Networks by IL-21 Blockade Leads to Disease Amelioration in Murine Lupus. J Immunol. 2017;198(7):2578–2588. doi: 10.4049/jimmunol.1601687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang M, et al. Interleukin-21 receptor blockade inhibits secondary humoral responses and halts the progression of preestablished disease in the (NZB x NZW)F1 systemic lupus erythematosus model. Arthritis Rheumatol. 2015;67(10):2723–2731. doi: 10.1002/art.39233. [DOI] [PubMed] [Google Scholar]
- 45.Nurieva RI, et al. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325(5943):1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Finck R, et al. Normalization of mass cytometry data with bead standards. Cytometry A. 2013;83(5):483–494. doi: 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zunder ER, et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat Protoc. 2015;10(2):316–333. doi: 10.1038/nprot.2015.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chevrier S, Crowell HL, Zanotelli VRT, Engler S, Robinson MD, Bodenmiller B. Compensation of Signal Spillover in Suspension and Imaging Mass Cytometry. Cell Syst. 2018;6(5):612–620.e5. doi: 10.1016/j.cels.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chiche L, et al. Modular transcriptional repertoire analyses of adults with systemic lupus erythematosus reveal distinct type I and type II IFN signatures. Arthritis Rheumatol. 2014;66(6):1583–1595. doi: 10.1002/art.38628. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass cytometry data from the AMP SLE and RA cohorts datasets are available at the ImmPort repository (https://www.immport.org) from accessions SDY997 and SDY998. RNA-seq data from the BWH cohort dataset is available at the ImmPort repository from accession SDY1475. The data that support the findings of this study are available from the corresponding author upon request.