Abstract
It is proposed that the impaired sympathoadrenal response to hypoglycemia induced by recurrent insulin-induced hypoglycemia (RH) is an adaptive phenomenon induced by specific changes in microRNA expression in the ventromedial hypothalamus (VMH). To test this hypothesis, genome-wide microRNAomic profiling of the VMH by RNA-sequencing was performed in control rats and rats treated for RH. Differential expression analysis identified microRNA-7a-5p and microRNA-665 as potential mediators of this phenomenon. To further test this hypothesis, experiments were conducted consisting of targeted lentiviral-mediated overexpression of microRNA-7a-5p and downregulation of microRNA-665 in the VMH. Hyperinsulinemic hypoglycemic clamp experiments demonstrated that targeted overexpression of microRNA-7a-5p (but not downregulation of microRNA-665) in the VMH of RH rats restored the epinephrine response to hypoglycemia. This restored response to hypoglycemia was associated with a restoration of GABAA receptor gene expression. Finally, a direct interaction of microRNA-7a-5p with the 3′-UTR of GABAA receptor α1-subunit (Gabra1) gene was demonstrated in a luciferase assay. These findings indicate that (a) the impaired sympathoadrenal response RH induces is associated with changes in VMH microRNA expression and (b) microRNA-7a-5p, possibly via direct downregulation of GABA receptor gene expression, may serve as a mediator of the altered sympathoadrenal response to hypoglycemia.
Keywords: Endocrinology
Keywords: Noncoding RNAs
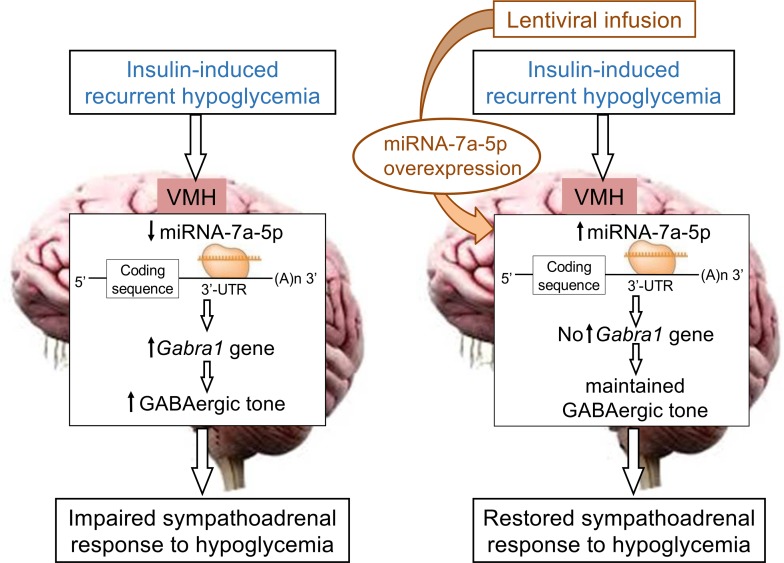
MicroRNA-7a-5p regulates GABAergic tone, which plays an important role in the ventromedial hypothalamus to mediate the sympathoadrenal response to hypoglycemia.
Introduction
Hypoglycemia is a major obstacle in achieving tight glycemic control in insulin-treated patients with diabetes (1). It is well documented that recurrent episodes of insulin-induced hypoglycemia (RH) induce a maladaptive response (termed hypoglycemia-associated autonomic failure; HAAF) in which defective brain glucose sensing and impaired glucose counterregulatory response (CRR) increase the risk for severe hypoglycemia (2). Several mechanisms have been proposed to explain how RH induces HAAF, including (a) the actions of counterregulatory hormones released during these episodes (cortisol, epinephrine, opioids) (3), (b) enhanced cerebral glucose metabolism transport/phosphorylation/oxidation (2, 4), (c) increased cerebral storage of glycogen (5), (d) increased GABAergic tone (6), and (e) the use of alternative fuels (e.g., β-hydroxybutyrate, acetate, lactate) to support brain metabolism (7). However, the underlying basis of this pathophysiological maladaptation in the brain remains unknown.
Studies have demonstrated that hypoglycemia induces changes in the expression of genes (and mRNA) involved in hypothalamic glucose metabolism/sensing (8, 9) that may influence the brain’s adaptive response to hypoglycemia. Of these gene products, γ-aminobutyric acid (GABA), the most ubiquitously expressed inhibitory neurotransmitter in the mammalian central nervous system, has been shown to play an important role in regulation of the glucose sensing and hypoglycemic counterregulation (6, 10). Studies have reported that RH-induced increased GABAergic inhibitory tone at the level of the GABAA receptor in the ventromedial hypothalamus (VMH) acts to blunt the CRRs to hypoglycemia (11). We speculated that these RH-induced maladaptive changes occurring at the level of gene expression could be mediated by microRNAs. MicroRNAs are endogenously expressed, ~22-nucleotide-long, single-stranded, noncoding RNAs that negatively regulate the expression of genes (or mRNA) either by mRNA degradation or by translational repression, mainly via base-pairing interactions with the 3′ untranslated region (3′-UTR) of target genes in both normal physiological (12) and disease contexts (13).
Studies have shown that a relatively large number of known microRNAs are abundantly expressed in the mammalian brain in regional and distinct patterns to regulate area-specific functions (14, 15). Within the brain, microRNAs expressed or enriched in the hypothalamus have been shown to be important regulators of vital bodily functions (16, 17). Because microRNAs can target several mRNAs, microRNAs are now considered proximal posttranscriptional regulators affecting multiple parallel biological pathways (18, 19). In recent studies, microRNAs have been demonstrated to be a key regulator of glucose homeostasis (20–22) and glucose sensing within pancreatic islets (23, 24). In addition, microRNAs have been shown to be associated with sympathetic nervous system regulation (25). Therefore, we postulated that in response to RH, altered microRNA expression in key glucose-sensing areas of the brain could explain, at least in part, the regulation and changes of mRNA (gene) expression involved in glucose metabolism or sensing and the maladaptive downregulation of the sympathoadrenal response to hypoglycemia. Thus, the present study sought to investigate the role of altered microRNA expression in the VMH as a potentially novel mechanistic basis of RH-induced HAAF.
Results
RH induces the genome-wide microRNAomic changes in the VMH.
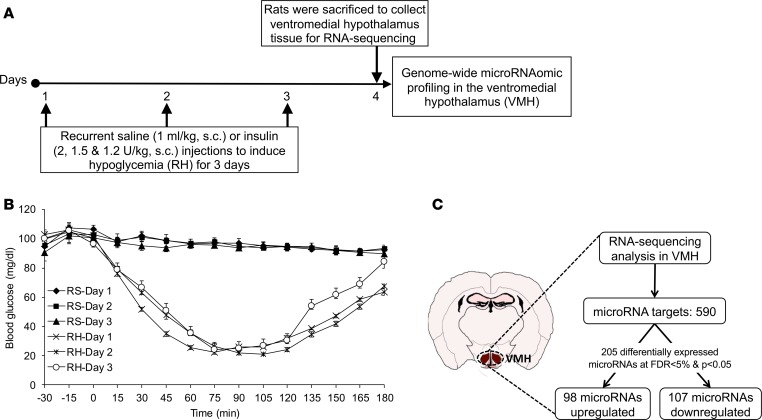
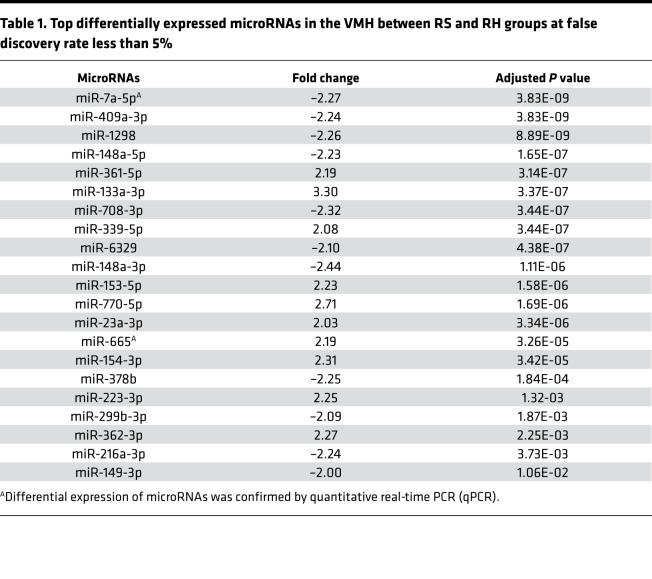
To evaluate the extent to which RH induces microRNA changes in the brain, RNA-sequencing was performed in the VMH of recurrent saline (RS) control and RH rats (Figure 1A). In the RH group, the levels of blood glucose remained at 25–50 mg/dL for about 90 minutes after insulin injection over the 3 days of hypoglycemic episodes. However, in the RS control group, blood glucose levels were near or at normal levels over the 3 days of saline injections (Figure 1B). High-throughput RNA-sequencing data identified 590 microRNAs in the VMH of control and RH rats, of which 205 microRNAs were significantly different between treatment groups (see Supplemental Data; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.130521DS1). It was noted that out of the 205 microRNAs, 98 were upregulated and 107 were downregulated in RH group as compared with controls (Figure 1C), of which 21 microRNAs displayed larger than 2-fold expression change (i.e., top differentially expressed microRNAs; Table 1) at an FDR of less than 5%.
Figure 1. RH leads to differential microRNA expression in the VMH.
(A) Male Sprague-Dawley rats were injected with regular human insulin subcutaneously (s.c.) for 3 consecutive days to induce RH (2 U/kg on day 1, 1.5 U/kg on day 2, and 1.2 U/kg on day 3). For the control group, rats were injected with saline (1 mL/kg, s.c.) for 3 consecutive days (RS). Following overnight fasting on day 4, both RS (i.e., control) and RH (i.e., recurrent insulin–treated rats) groups were sacrificed to collect VMH brain tissues to determine genome-wide microRNA profiling by RNA-sequencing. (B) Blood glucose levels (mg/dL) in RS or RH groups during 3-day protocol. (C) Differential expression of microRNAs in the VMH between control and RH groups at false discovery rate (FDR) less than 5% and P less than 0.05. Data are expressed as mean ± SEM (n = 6/group).
Table 1. Top differentially expressed microRNAs in the VMH between RS and RH groups at false discovery rate less than 5%.
Potential microRNAs and their expressions.
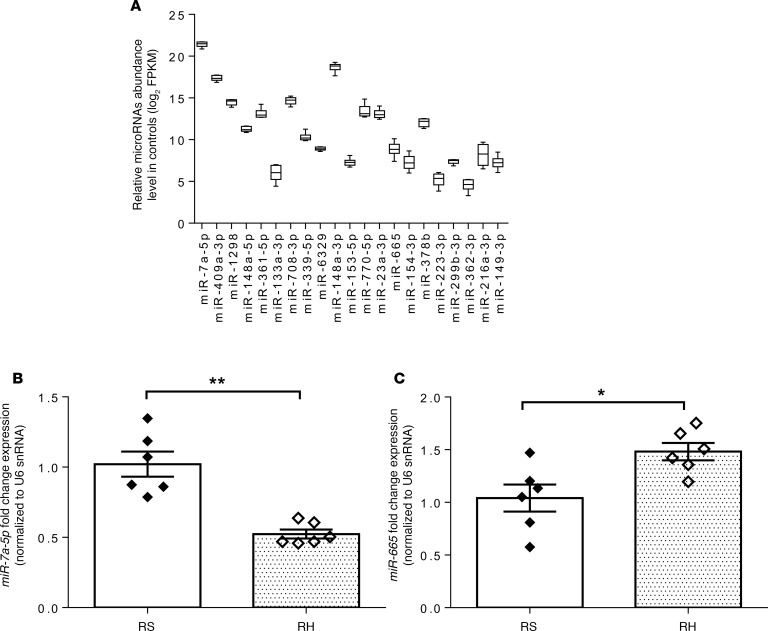
Because RH leads to impaired CNS-mediated CRR, we targeted microRNAs that regulate the genes most likely involved in regulating RH-induced impaired brain glucose sensing or metabolism by using Ingenuity Pathway Analysis (IPA) web-based software (QIAGEN Inc.; www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/), which uses data from TargetScan, TarBase, miRecords, and Ingenuity expert findings (26). We identified microRNA-7a-5p and microRNA-665 as potential targets for impaired brain glucose sensing or metabolism in RH. These microRNAs have been reported to alter the posttranscriptional regulation of MAPK-interacting serine/threonine-protein kinase 2 (23) and thioredoxin-like 1 (27) genes, which have been shown to regulate pancreatic β cells’ proliferation and cellular response to glucose deprivation, respectively. From the top differentially expressed microRNAs, microRNA-7a-5p, which has been reported as a brain-enriched microRNA (16, 28), was highly abundant in the VMH (Figure 2A). qPCR analysis of microRNA-7a-5p and microRNA-665, using total RNA isolated from VMH of control and RH rats, supported the sequencing findings as indicated by a significant decrease in microRNA-7a-5p expression and a significant increase in microRNA-665 expression in RH rats as compared with controls (Figure 2, B and C). The fold change expression in these microRNAs assessed by qPCR was highly consistent with the fold change expression observed by RNA-sequencing (see Table 1).
Figure 2. Expression of key microRNAs by quantitative real-time PCR confirms the differential microRNA expression observed by RNA-sequencing.
(A) Abundance level of top differentially expressed microRNAs in the VMH of control rats. (B) MicroRNA-7a-5p and (C) microRNA-665 expression by quantitative real-time PCR (qPCR) in the VMH of control (RS) and RH rats. MicroRNA expression levels are represented as relative fold change expression after normalization with reference to the expression of U6 small nuclear RNA (U6 snRNA) endogenous control. Data are expressed as mean ± SEM (n = 6/group). *P < 0.05, and **P < 0.01 vs. RS group; unpaired 2-tailed Student’s t test. MicroRNA abundance levels are expressed as log2 of FPKM. FPKM, fragments per kilobase of transcript per million reads mapped to microRNAs.
Lentiviral-mediated microRNA manipulation in the VMH.
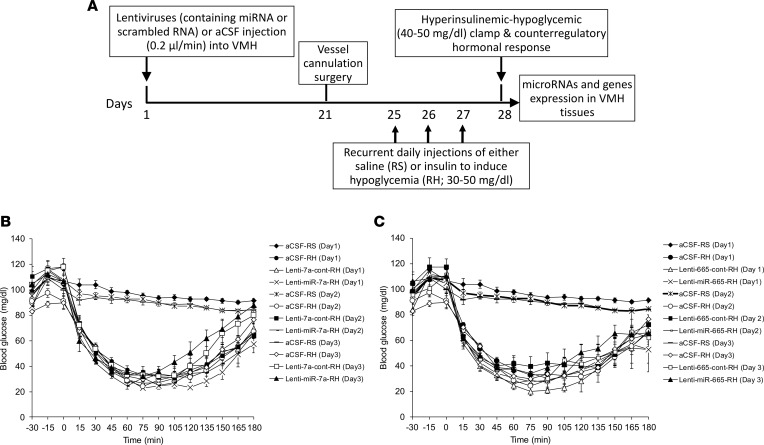
To more definitively explore a potential causal relationship of decreased microRNA-7a-5p expression or increased microRNA-665 expression in mediating the RH-induced impaired sympathoadrenal response to hypoglycemia, experiments were conducted to determine whether targeted reversal of these RH-induced microRNA changes in the VMH could restore the blunted sympathoadrenal response to hypoglycemia (Figure 3A). To test this hypothesis, we selectively (a) overexpressed microRNA-7a-5p and (b) downregulated expression of microRNA-665 in the VMH by lentiviral injections, expressing either microRNA-7a (lenti-miR-7a) or anti–microRNA-665 (lenti-miR-665), respectively, into the VMH of RH rats. Control lentiviral vectors (lenti-7a-cont or lenti-665-cont) were infused into RH rats as respective controls (see Supplemental Methods). There was no change observed in body weight and blood glucose of any of the RH group following 4 weeks of lentiviral delivery into the VMH (data not shown). In the RH group, the levels of blood glucose remained at 25–50 mg/dL for about 90 minutes after insulin injection over the 3 days of hypoglycemic episodes. However, in the aCSF-treated RS group, blood glucose levels were near or at normal levels over the 3 days of saline injections (Figure 3, B and C).
Figure 3. Schematic timeline representing study design for lentiviral-mediated microRNA manipulation in the VMH and hypoglycemic clamps.
(A) Male Sprague-Dawley rats were subjected to either artificial cerebrospinal fluid (aCSF) or lentivirus (containing either control or targeted microRNA sequence) injection into the VMH by using a stereotaxic frame. Three weeks following intra-VMH injection, all rats subsequently had vascular catheters implanted under ketamine anesthesia. Four days following vascular surgery, rats that received aCSF into the VMH were subjected to either RS or RH for 3 days, while rats that received lentiviruses into the VMH were subjected to the RH only. Following overnight fasting (i.e., after 4 weeks of intra-VMH injection), all rats were subjected to the hyperinsulinemic (20 mU/kg/min) hypoglycemic (40–50 mg/dL for 90 minutes) clamps for assessing counterregulatory hormonal response in plasma. (B and C) Blood glucose levels (mg/dL) during the 3-day protocol in RS rats treated with intra-VMH aCSF-RS and RH rats treated with either aCSF-RH or control lentivirus (lenti-7a-cont-RH or lenti-665-cont-RH) or microRNA-encoding lentivirus (lenti-miR-7a-RH or lenti-miR-665-RH). Data are expressed as mean ± SEM (n = 6–7/group).
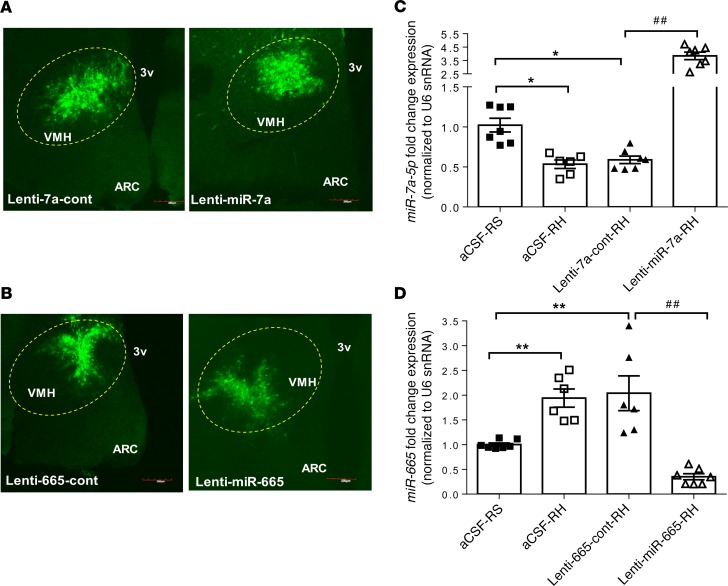
The targeted delivery of lentiviruses was confirmed by GFP immunofluorescence staining in the VMH of the control rats treated with control and with targeted microRNA-encoding lentiviruses (lenti-miR-7a or lenti-miR-665) (Figure 4, A and B). The lentiviruses’ efficacy was measured by the expressions of the microRNAs in the VMH using microRNA TaqMan real-time PCR assay (Figure 4, C and D). The decreased expression of microRNA-7a-5p induced by RH was reversed in the VMH of lenti-miR-7a–treated rats as noted by increased microRNA-7a-5p expression (Figure 4C). Similarly, the overexpression of microRNA-665 RH induced was decreased in the VMH of lenti-miR-665–treated rats (Figure 4D). Rats treated with intra-VMH respective control lentivirus had microRNA levels similar to control intra-VMH aCSF-treated rats.
Figure 4. Lentiviruses’ delivery in the VMH modulates the microRNA expression.
(A and B) GFP immunostaining in the VMH of rats treated with either control lentiviruses (lenti-7a-cont or lenti-665-cont) or lentiviruses containing targeted microRNA sequence (lenti-miR-7a or lenti-miR-665) in the VMH. Scale bars: 200 μm. VMH, ventromedial hypothalamus; ARC, arcuate nucleus of the hypothalamus; 3v, third ventricle. (C) MicroRNA-7a-5p and (D) microRNA-665 expression by qPCR in the VMH of RS rats treated with intra-VMH aCSF-RS and recurrent hypoglycemic rats treated with aCSF-RH, control lentiviruses (lenti-7a-cont or lenti-665-cont), or microRNA-encoding lentiviruses (lenti-miR-7a-RH or lenti-miR-665-RH). MicroRNA expression levels are represented as relative fold change after normalization with reference to the expression of U6 snRNA endogenous control. Data are expressed as mean ± SEM (n = 6–7/group). *P < 0.05, and **P < 0.01 vs. aCSF-RS group; ##P < 0.01 vs. lenti-7a-cont-RH or lenti-665-cont group; 1-way ANOVA followed by Tukey’s test.
Restoration of microRNA-7a-5p expression in the VMH normalizes the RH-induced impaired sympathoadrenal response to hypoglycemia.
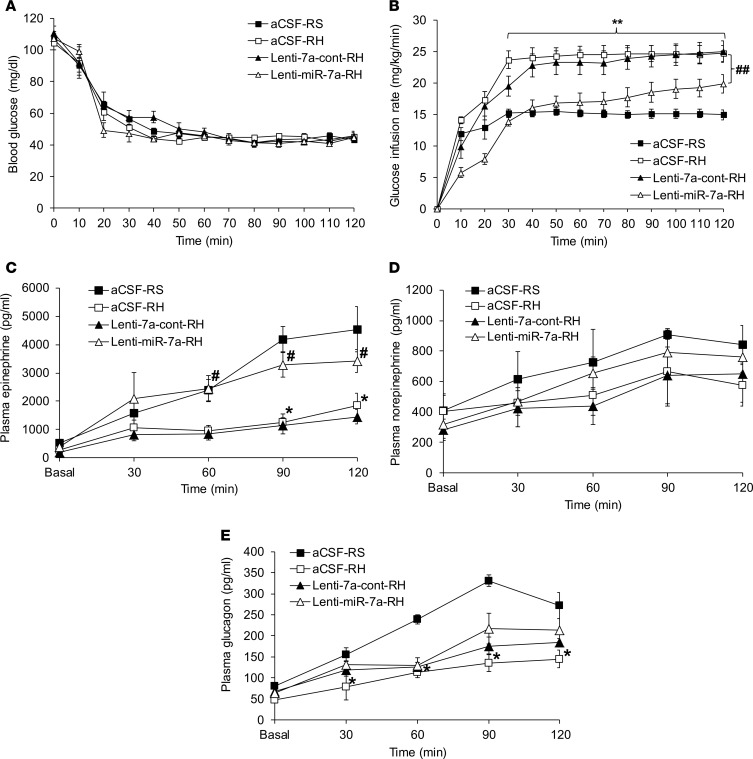
Four weeks following aCSF or lentivirus infusion into the VMH and following RH or RS protocols, hyperinsulinemic hypoglycemic clamps were performed in all groups. Blood glucose levels were matched in all groups (~45 mg/dL) for 90 minutes (Figure 5A). Consistent with the expected blunting of the CRR in the RH group, the exogenous glucose requirement was higher as compared with the control RS group (aCSF-RH vs. aCSF-RS, P < 0.01; Figure 5B). The GIR in the RH group receiving either aCSF or control lentiviruses in the VMH was similar (lenti-miR-7a-cont-RH vs. aCSF-RH, P = NS; Figure 5B). Consistent with an improvement in counterregulatory hormone response, RH rats treated with intra-VMH infusion of microRNA-7a–expressing lentiviruses were noted to have a significantly lowered GIR as compared with the RH rats that received control lentiviruses (lenti-miR-7a-RH vs. lenti-7a-cont-RH, P < 0.01; Figure 5B).
Figure 5. Targeted reversal of microRNA-7a-5p expression in the VMH restores the RH-induced impaired sympathoadrenal response to hypoglycemia.
(A) Blood glucose levels (mg/dL) and (B) glucose infusion rates (GIRs; mg/kg/min) during hyperinsulinemic (20 mU/kg/min) hypoglycemic (40–50 mg/dL for 90 minutes) clamps. Plasma (C) epinephrine (pg/mL), (D) norepinephrine (pg/mL), and (E) glucagon (pg/mL) levels during basal (at 0 minutes) and hypoglycemic periods (30–120 minutes) of hyperinsulinemic hypoglycemic clamps in RS rats treated with intra-VMH aCSF-RS and recurrent hypoglycemic rats treated with either aCSF-RH or control lentiviruses (lenti-7a-cont-RH) or microRNA-7a–encoding lentiviruses (lenti-miR-7a-RH). Data are expressed as mean ± SEM (n = 6–7/group). *P < 0.05, and **P < 0.01 vs. aCSF-RS group; #P < 0.05, and ##P < 0.01 vs. Lenti-miR-7a-RH group; 2-way repeated-measures ANOVA followed by Tukey’s test.
Plasma epinephrine levels increased robustly during hypoglycemia in control rats. However, as expected, the epinephrine response was blunted by 60% in RH rats (aCSF-RH vs. aCSF-RS, P < 0.01; Figure 5C). The delivery of control lentiviruses in the VMH of RH rats did not alter the epinephrine response as compared to the aCSF-treated RH group (lenti-7a-cont-RH vs. aCSF-RH, P = NS; Figure 5C); however, the RH rats treated with VMH lenti-miR-7a showed an increased epinephrine response to hypoglycemia (lenti-miR-7a-RH vs. lenti-7a-cont-RH, P < 0.01; Figure 5C) to levels not different from control rats. Peak epinephrine levels achieved during hypoglycemia were positively correlated with microRNA-7a-5p expression levels measured by qPCR (r = 0.41; P < 0.05; data not shown). Plasma norepinephrine levels during hypoglycemia increased to a similar extent in all groups (Figure 5D).
Plasma glucagon levels increased during hypoglycemia in the control rats, and this effect was blunted by 57% in RH rats (aCSF-RH vs. aCSF-RS, P < 0.01; Figure 5E). The infusion of neither control lentiviruses nor microRNA-7a–containing lentiviruses significantly improved this blunted glucagon response to hypoglycemia (lenti-miR-7a-RH vs. lenti-7a-cont-RH, P = NS; Figure 5E).
MicroRNA-665 downregulation in the VMH does not normalize the RH-induced impaired CRR to hypoglycemia.
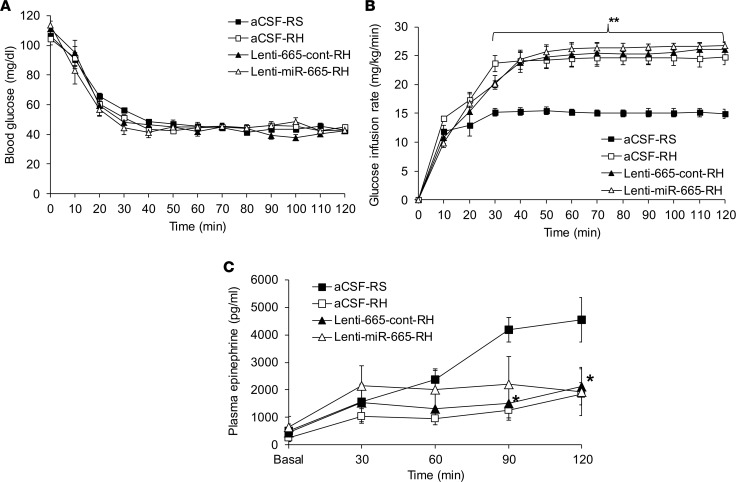
The CRR to hypoglycemia was assessed by experiments in which the RH-induced upregulation of microRNA-665 in the VMH was downregulated with lentivirus-mediated microRNA-665. During the hypoglycemic clamp, blood glucose levels were matched in all groups (~45 mg/dL) for 90 minutes (Figure 6A). As expected, the GIRs were similar between aCSF- and control lentivirus–treated RH rats. MicroRNA-665 knockdown in the VMH resulted in similar GIRs between lenti-665-cont–treated and lenti-miR-665–treated RH rats (lenti-miR-665-RH vs. lenti-665-cont-RH, P = NS; Figure 6B). These results were consistent with the lack of effect of microRNA-665 knockdown on the RH-induced impaired epinephrine response (lenti-miRZip-665-RH vs. lenti-665-cont-RH, P = NS; Figure 6C).
Figure 6. Lentiviral-mediated microRNA-665 downregulation in the VMH does not normalize the RH-induced impaired CRR to hypoglycemia.
(A) Blood glucose levels (mg/dL), (B) GIRs (mg/kg/min), and (C) plasma epinephrine (pg/mL) levels during hyperinsulinemic (20 mU/kg/min) hypoglycemic (40–50 mg/dL for 90 minutes) clamps in RS rats treated with intra-VMH aCSF-RS and recurrent hypoglycemic rats treated with either aCSF-RH or control lentiviruses (lenti-665-cont-RH) or lentiviruses encoding anti–microRNA-665 (lenti-miR-665-RH). Data are expressed as mean ± SEM (n = 6–7/group). *P < 0.05 vs. aCSF-RS group, and **P < 0.01 vs. aCSF-RS group; 2-way repeated-measures ANOVA followed by Tukey’s test.
Overexpression of the microRNA-7a-5p in the VMH counteracts the metabolic gene expression changes induced by RH.
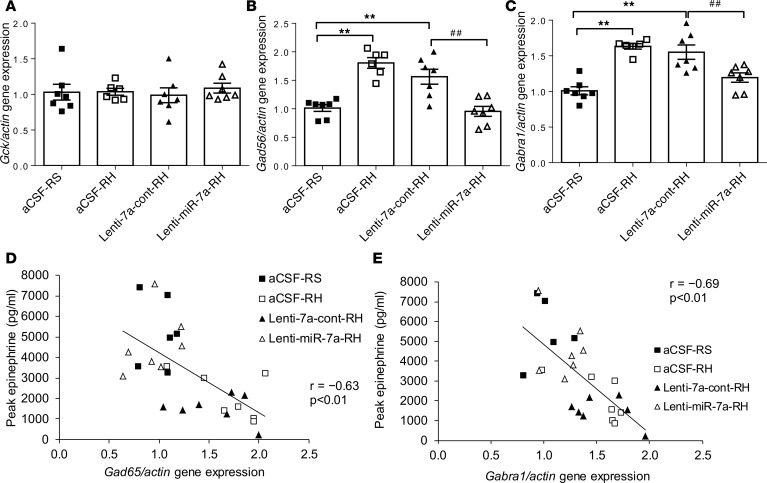
We measured the expression of genes in the VMH that are known to be involved in the regulation of the CRR to hypoglycemia. Real-time PCR of VMH micropunches demonstrated that mRNA levels for both glutamic acid decarboxylase 65 (Gad65 or Gad2: a neuronal enzyme involved in the synthesis of the inhibitory neurotransmitter, GABA), and the GABAA receptor α1-subunit (Gabra1: an ionotropic receptor for GABA), were increased by 78% and 61%, respectively, in RH rats (aCSF-RH vs. aCSF-RS, P < 0.01; Figure 7, B and C). Glucokinase (Gck) mRNA levels, however, were not significantly altered by RH (Figure 7A). Notably, experiments involving the overexpression of microRNA-7a-5p resulted in a restoration of the RH-induced increased Gad65 and Gabra1 (lenti-miR-7a vs. lenti-7a-cont, P < 0.01; Figure 7, B and C) whereas control lentiviruses in RH rats did not alter gene expression. Peak epinephrine levels achieved during hypoglycemia were highly correlated with Gad65 (r = –0.63; P < 0.01; Figure 7D) and Gabra1 (r = –0.69; P < 0.01; Figure 7E) gene expression levels measured by qPCR.
Figure 7. Restoration of the microRNA-7a-5p in the VMH counteracts the metabolic gene expression changes induced by RH.
(A) Glucokinase (Gck), (B) glutamic acid decarboxylase 65 (Gad65), (C) GABAA receptor α1-subunit (Gabra1) gene expression by qPCR and correlation of (D) Gad65 or (E) Gabra1 fold change expression with peak epinephrine levels in the VMH of RS rats treated with intra-VMH aCSF-RS and recurrent hypoglycemic rats treated with either aCSF-RH or control lentiviruses (lenti-7a-cont-RH) or microRNA-7a–encoding lentiviruses (lenti-miR-7a-RH). Gene expression levels are represented as relative fold change after normalization with reference to the expression of β-actin endogenous control. Data are expressed as mean ± SEM (n = 6–7/group). **P < 0.01 vs. aCSF-RS group; ##P < 0.01 vs. lenti-7a-cont-RH or lenti-665-cont group; 1-way ANOVA followed by Tukey’s test.
Overexpression of the microRNA-7a-5p negatively regulates the expression of Gabra1 gene by binding to its 3′-UTR region.
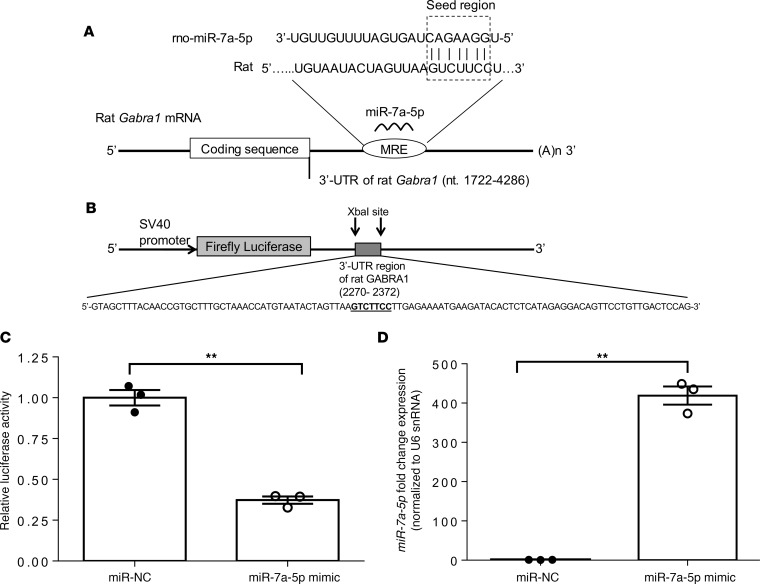
Based on the prediction algorithms of TargetScan, microRNA-7a-5p is predicted to bind to the Gabra1 3′-UTR with its seed sequence (5′-GGAAGAC-3′) complementary to nucleotides 2316–2322. Although the above result indicated an association between microRNA-7a-5p and GABA receptor expression, whether this association is direct or indirect remained unknown. To test whether microRNA-7a-5p directly targets expression of Gabra1, human embryonic kidney 293T (HEK293T) cells were cotransfected with either microRNA-7a-5p mimic or microRNA-7a-5p negative control, renilla luciferase and firefly luciferase reporters containing the 3′-UTR of Gabra1 gene (Figure 8, A and B). Results indicated that microRNA-7a-5p significantly decreased the relative luciferase activity by 63% via direct binding to the Gabra1 3′-UTR region in the luciferase construct (Figure 8C). Next, the expression of microRNA-7a-5p was measured in HEK293T-cotransfected cells by qPCR. Results indicated that the endogenous level of microRNA-7a-5p in HEK293T cells transfected with microRNA-7a-5p negative control was low, while the transfection of HEK293T cells with microRNA-7a-5p mimic led to a ~400-fold increase in microRNA-7a-5p expression (Figure 8D). Overall, these results indicate that microRNA-7a-5p binds to the 3′-UTR region of Gabra1 gene that negatively regulates Gabra1 gene expression at a posttranscriptional level.
Figure 8. Restoration of the microRNA-7a-5p negatively regulates the expression of Gabra1 gene by binding to its 3′-UTR region.
(A) Schematic diagram of rat Gabra1 mRNA containing the predicted conserved target site of microRNA-7a-5p at microRNA regulatory element (MRE) on 3′-UTR. The seed region is indicated in the box. (B) Schematic diagram of SV-40–driven firefly luciferase reporter construct containing a DNA insert at XbaI site (nt 1934), ~100-bp oligonucleotide sequence from the 3′-UTR region of rat Gabra1 gene, containing predicted rno-miR-7a-5p target site. (C) Relative luciferase activity and (D) microRNA-7a-5p expression in HEK293T cells, cotransfected with firefly luciferase reporter construct (shown in B), renilla luciferase plasmid (transfection control), and with either microRNA mimic for rat microRNA-7a-5p (miR-7a-5p mimic) or microRNA mimic negative control (miR-NC). Relative luciferase activity (firefly normalized to renilla) values of miR-7a-5p were normalized to miR-NC–transfected controls. MicroRNA-7a-5p expression was measured by qPCR and represented as relative fold change expression after normalization with reference to the expression of U6 snRNA endogenous control. Data are expressed as mean ± SEM of 3 independent experiments. **P < 0.01 for difference between miR-NC– and miR-7a-5p mimic–treated samples; unpaired 2-tailed Student’s t test.
MicroRNA-7a-5p expression in both neuronal and non-neuronal cell types.
To determine the cell types involved in microRNA-7a–mediated action, control rats receiving microRNA-7a lentivirus in the VMH were tested for phenotypic expression of GFP with either NeuN (neuronal marker) or glial fibrillary acidic protein (GFAP) as astrocytic marker after 4 weeks of lentiviral injection. GFP was colocalized with NeuN as well as with GFAP (white arrows, Supplemental Figure 1). Because the lentivirus expresses GFP, the colocalization of GFP with NeuN or GFAP indicates that lentiviral-mediated microRNA-7a overexpression occurs in both neuronal and non-neuronal cell types.
Discussion
To prevent episodes of severe hypoglycemia in people with diabetes, understanding the basic mechanism by which RH induces a maladaptive response (i.e., HAAF) remains a fundamental unanswered question in the field. Consistent with previous studies demonstrating a role for microRNAs in regulating glucose homeostasis (20, 21) and glucose sensing within pancreatic islets (22, 24), this study notes the change in expression of several microRNAs induced by RH. We further demonstrate that microRNA-7a-5p is a fundamental regulator of the adrenomedullary response to hypoglycemia. MicroRNA-7a-5p was highly enriched in the VMH and markedly downregulated by RH. We demonstrate that targeted overexpression of microRNA-7a-5p in the VMH with a lentiviral approach restored the sympathoadrenal response to hypoglycemia. This restored response to hypoglycemia was associated with a restoration of GABAA receptor gene expression. It is further demonstrated that there was direct binding of the microRNA-7a-5p to the 3′-UTR of Gabra1 gene, which is consistent with repressed Gabra1 gene expression. Thus, these results suggest that microRNA-7a-5p in the VMH plays an important role in regulation of brain glucose sensing and sympathoadrenal response by regulating the GABAergic system and therefore may be an appropriate therapeutic target for the regulation of HAAF in people with diabetes.
According to our results, repeated episodes of insulin-induced hypoglycemia markedly alter the microRNA profile in the VMH, indicating that the pathophysiological maladaptation RH induces is associated with specific genomic changes in the brain. The microRNA profile changes in the VMH allowed us to identify the top differentially expressed microRNAs, of which microRNA-7a-5p and microRNA-665 were selected as potential candidates responsible for the maladaptive response in RH. Previous studies have shown that microRNA-7a is a brain-enriched microRNA that is abundantly expressed in hypothalamic nuclei (16, 28). Consistent with this finding, our RNA-sequencing data in the VMH indicated that microRNA-7a-5p is highly abundant as compared with other top differentially expressed microRNAs, which led us to speculate that microRNA-7a-5p plays an important role in mediating this adaptive response to hypoglycemia. In agreement with these findings, lentiviral-mediated overexpression of microRNA-7a-5p in the VMH enhanced the epinephrine response to hypoglycemia and reduced the GIR during hypoglycemic clamp (both results consistent with an improved CRR). Interestingly, this restored sympathoadrenal response to hypoglycemia did not coincide with a restoration of the pancreatic α cell (i.e., glucagon) response. Because the glucagon response to hypoglycemia is regulated by several factors, including altered intraislet insulin secretion, adrenergic signaling, and sympathetic innervation, the lack of glucagon response restoration with VMH microRNA-7a-5p overexpression in RH indicates that systemic catecholamines are qualitatively less important than other factors in mediating the glucagon response to hypoglycemia. Previous studies demonstrated that the acute administration of the GABAA receptor antagonist bicuculline methiodide into the VMH increased the glucagon response to hypoglycemia of RH-treated rats (6). It remains unclear to what extent differences in experimental approaches (acute versus chronic model; pharmacological versus genomic approaches; post-RH versus ante-RH treatments) accounts for these differential effects on glucagon responses.
The proposed concept that HAAF is mediated by changes in VMH microRNA does not conflict with previously postulated mechanisms for the development of HAAF (4). Indeed, a role for increased GABAergic tone in the VMH contributing to the impaired CRR to hypoglycemia induced by RH has been suggested (6). It was interesting that glutamic acid decarboxylase 65 (Gad65 or Gad2, a neuronal enzyme involved in the synthesis of the inhibitory neurotransmitter, GABA) as well as Gabra1 (an ionotropic receptor for GABA) expression were coordinately increased by RH and coordinately normalized by experiments in which microRNA-7a-5p was overexpressed, leading to a restoration of sympathoadrenal response to hypoglycemia. These findings are highlighted by the data shown in Figure 7, indicating a correlation between both Gabra1 and Gad65 expression with the epinephrine response to hypoglycemia. These findings suggest that microRNA-7a-5p in the VMH regulates the sympathoadrenal response to hypoglycemia by regulating GABAergic tone in the VMH.
Because microRNAs negatively regulate the expression of a gene by binding to its 3′-UTR, we sought to determine whether the microRNA-7a-5p–mediated regulation of sympathoadrenal response occurs via direct interaction with GABAergic system genes. Based on TargetScan prediction, microRNA-7a-5p does not bind to the 3′-UTR sequence of Gad65. Thus, it is speculated that the restoration of RH-induced changes in Gad65 expression observed by intra-VMH microRNA-7a-5p overexpression is not the result of direct microRNA-mRNA interaction but is likely mediated through indirect processes. However, the TargetScan tool indicated that microRNA-7a-5p is predicted to bind to the 3′-UTR sequence of Gabra1 gene. To determine the direct binding of microRNA-7a with Gabra1 gene, luciferase assays were performed in which Gabra1 3′-UTR DNA sequence was ligated into the luciferase reporter constructs. Results from the luciferase assay demonstrated that microRNA-7a-5p directly binds to the 3′-UTR sequence and induces repression of Gabra1 gene, which is associated with GABAergic inhibition. By identifying Gabra1 gene as a direct target of microRNA-7a-5p, our data raise the hypothesis that altered regulation of microRNA-7a-5p (and consequently expression of their target genes) may predispose to impaired CNS glucose sensing and blunted sympathoadrenal response to hypoglycemia.
MicroRNA-7 is an evolutionarily highly conserved small RNA and is considered a prototypical neuroendocrine microRNA, being expressed at high levels in neurons and neuroendocrine organs (29, 30). Although previous studies demonstrated that microRNA-7 is highly expressed in neurons as compared with astrocytes (31), in the current study, lentivirus-mediated overexpression of microRNA-7a occurred in both neuronal and astrocytic cells (Supplemental Figure 1). Thus, although it remains unclear whether the observed microRNA-7a-5p–mediated improvements in the sympathoadrenal response are mediated by neuronal or astrocytic cells in the VMH, our data are consistent with the notion that these effects occur (at least in part) via actions in VMH cells expressing Gabra1 (as indicated by the direct interaction of microRNA-7a-5p with Gabra1 3′-UTR).
In conclusion, these findings indicate a potentially novel role for microRNAs in mediating the RH-induced, maladaptive, impaired CRR to hypoglycemia. These findings specifically identify microRNA-7a-5p as a potential novel therapeutic target that can restore the sympathoadrenal response in HAAF.
Methods
Animals.
Male Sprague-Dawley rats (SASCO SD, strain code: 400; Charles River Laboratories, Inc.) weighing 250–300 g (~9 weeks old) were maintained under standard housing conditions (room temperature 22–24°C with 12-hour light/12-hour dark cycle) in the Comparative Medicine Center at the University of Utah. The animals had free access to food (2920X Teklad global rodent chow diet, Envigo Corp.) and water and were acclimatized to handling for 1 week before experimental manipulation.
Recurrent hypoglycemia.
Initially, we evaluated the extent to which RH could induce microRNA changes in the VMH. In brief, male Sprague-Dawley rats were subjected to 3 days’ RS injections or RH and then euthanized on day 4 to study microRNA profiling in the VMH by RNA-sequencing (Figure 1A). Rats were injected with regular human insulin (2 U/kg on day 1, 1.5 U/kg on day 2, and 1.2 U/kg on day 3; Humulin R, Lilly) s.c. to induce recurrent hypoglycemia for 3 consecutive days. Food was withheld for 3 hours daily, and blood glucose levels were measured from a tail nick every 15 minutes using a glucometer (Ascensia Contour BG, Bayer HealthCare). Following insulin injection, blood glucose levels were allowed to drop to 25–50 mg/dL for 90 minutes. At the end of this period, the rats were given free access to food again. Control rats received a daily injection of normal saline (RS; 1 mL/kg, s.c.) for 3 consecutive days. On day 4, following overnight fasting, both RS (n = 6) and RH (n = 6) rats were euthanized to collect brains on dry ice, which were stored at –80°C (Figure 1).
Genome-wide microRNAomic profiling in the VMH.
To determine the effect of RH on genome-wide microRNAomic profiles, RNA-sequencing was performed from punch biopsies (0.5-mm diameter; Stoelting Co.) of the VMH, which were dissected from 250-μm frozen brain sections (32, 33) of RS (n = 6) and RH (n = 6) rats. In brief, total RNA was extracted and purified from the VMH using miRNeasy Mini Kit (QIAGEN Inc.), and the quality of the RNA was determined by Agilent 2200 TapeStation (Agilent Technologies). Small RNA libraries were prepared using the NEBNext Multiplex Small RNA (New England Biolabs). Sequencing was performed using HiSeq 50 Cycle Single-Read Sequencing version 4 reagents (Illumina). Differential expression analysis of microRNA sequence data was performed with the Bioconductor package DESeq2 (34). Differential expression was evaluated by fitting a negative binomial generalized linear model and then adjusting the P value for multiple testing using the Benjamini-Hochberg correction with an FDR of less than 5%. Putative gene targets of differentially expressed microRNAs were identified in the brain using IPA software, which uses data from TargetScan, TarBase, miRecords, and Ingenuity expert findings, and were restricted to experimentally observed or highly confident predictions. The changes in key microRNAs observed by RNA-sequencing were confirmed by real-time PCR using TaqMan microRNA assays as described below. RNA-sequencing data were deposited into the Gene Expression Omnibus database (GSE138472).
Lentiviral-mediated microRNA manipulation.
Based on the findings that RH altered expression of specific microRNAs in the VMH, lentivirus microinjection rescue experiments were conducted to determine whether targeted reversal of the noted RH-induced microRNAs changes in the VMH could restore the sympathoadrenal response to hypoglycemia. In brief, all rats were subjected to either aCSF or lentivirus (containing either control or targeted microRNA sequences) injection into the VMH by stereotaxic (Benchmark, NeuroLab) surgery. The lentiviruses were injected into the VMH 4 weeks before the hypoglycemic clamp to allow the optimal microRNA expression (35). Three weeks following intra-VMH surgery, all rats had vascular catheters implanted under anesthesia. Four days after vascular surgery, rats were randomized to 1 of 4 treatment groups (n = 6–7/group): (a) intra-VMH aCSF + RS (control), (b) intra-VMH aCSF + RH, (c) intra-VMH lentivirus control + RH, or (c) intra-VMH lentiviruses containing targeted microRNA sequence + RH (see Figure 3A). Following 3 days of RS or RH, and after an overnight fast, all rats were subjected to a hyperinsulinemic (20 mU/kg/min) hypoglycemic (40–50 mg/dL for 90 minutes) clamp and then euthanized to harvest the brain tissues to test the gene and microRNA expression in the VMH by real-time PCR using TaqMan assays.
Lentivirus injection into the VMH.
The details of microRNA lentivector constructs and lentivirus production are provided in supplemental materials. In brief, rats were anesthetized with an intraperitoneal injection of ketamine/xylazine mixture and mounted in a stereotaxic apparatus (Benchmark Scientific). To inject lentiviruses into the VMH, a bilateral osmotic infusion cannula (28-gauge, 1.2 mm, length 9.2 mm; Plastics One) was inserted at the coordinates from bregma (anteroposterior −2.6 mm, mediolateral ±0.6 mm, and dorsoventral −9.2 mm) using a stereotaxic frame (Benchmark Scientific). The lentiviruses (2 × 108 to 1 × 109 infectious units/mL) were injected into the VMH using a bilateral infusion cannula (Plastics One) connected to a 1-mL syringe at a rate of 0.2 μL/min for 10 minutes, and the cannula was left in the VMH for an additional 5 minutes to avoid backflow.
Vascular surgery.
Three weeks following VMH neurosurgery, rats were anesthetized with an intraperitoneal injection of ketamine/xylazine. As previously described (36), a microrenathane catheter (MRE 033, Braintree Scientific Inc.) was inserted into the left carotid artery (for blood glucose and hormone sampling), and 2 catheters (MRE 025, Braintree Scientific Inc.) were implanted into the right jugular vein (for insulin and glucose infusions).
Hyperinsulinemic hypoglycemic clamp and hormone analysis.
Hyperinsulinemic (20 mU/kg/min) hypoglycemic clamps were performed in both RS and RH rats, during which blood glucose was lowered to 40–50 mg/dL using variable 50% dextrose infusion for 90 minutes (37). Blood samples were collected during the basal and every 30 minutes of hypoglycemia for plasma catecholamine ELISA (Abnova) and glucagon (MilliporeSigma) assays. At the end of clamps, all rats were sacrificed to collect brains to test the microRNAs and gene expression in the VMH by real-time PCR using TaqMan assays as described below.
MicroRNA expression by qPCR.
In brief, total RNA extracted from frozen VMH tissues (miRNeasy Mini Kit from QIAGEN Inc.) was reverse-transcribed and then amplified by PCR using TaqMan microRNA assay kits (Applied Biosystems), which contain a microRNA-specific stem-loop reverse transcription primer and the microRNA-specific PCR primers with TaqMan probe. TaqMan microRNA assay for the microRNA-7a-5p (dme-miR-7; assay ID 000268), microRNA-665 (mmu-miR-665; assay ID 001948), and U6 snRNA (assay ID 001973) were used.
Gene expression by qPCR.
The total cellular RNA isolated from the VMH after the hypoglycemic clamp was also used to assess the gene expression by real-time PCR using TaqMan gene assays for Gad65 (Gad2; assay ID Rn00561244_m1), Gabra1 (assay ID: Rn00788315_m1), Gck (assay ID Rn00561265_m1), and the housekeeping gene β-actin (Actb; assay ID Rn00667869_m1).
Immunofluorescence.
To confirm targeted delivery of lentiviruses, a separate cohort of control rats receiving either control or targeted microRNA lentiviruses (both for microRNA-7a and microRNA-665) into the VMH were tested for the GFP immunofluorescence. A subgroup of rats receiving microRNA-7a lentivirus into the VMH was subjected to the coimmunofluorescent staining of GFP with either NeuN (neuronal marker) or GFAP (astrocytic marker) to assess whether the lentivirus-mediated microRNA-7a overexpression occurs via neuronal or non-neuronal cell types. In brief, 4 weeks following intra-VMH control or targeted microRNA lentivirus injections, rats were perfused with 4% paraformaldehyde under ketamine/xylazine anesthesia to collect brains for immunofluorescence staining. Serial coronal brain sections (25 μm) through the VMH were cut on a cryostat and incubated with mouse monoclonal anti-GFP (1:200; sc-9996, Santa Cruz Biotechnology) and rabbit monoclonal anti-NeuN (1:200; MABN140, MilliporeSigma) or rabbit monoclonal anti-GFAP (1:200; ab33922, Abcam) in 10% goat serum at 4°C overnight. The following day, the sections were washed thoroughly with 1× PBS 3 times and incubated with goat anti-mouse Alexa Fluor 488 (1:1000, A11029, Life Technologies Corporation) and goat anti-rabbit Alexa Fluor 546 (1:1000, A11035, Life Technologies Corporation) in 10% goat serum for 1 hour at room temperature. The staining was visualized under fluorescence microscope (Olympus Fluoview FV1000) using the Fluoview software (Olympus FV10-ASW, version 4.2b).
Prediction of potential microRNA-7a-5p targets using TargetScan.
We used the TargetScan software (release 7.2) (26) to identify the microRNA-7a-5p targets. TargetScan is a web-based tool for sequence-based prediction of biological targets of microRNAs that considers matches to the 3′-UTR region and their orthologs, as defined by UCSC Genome Browser. TargetScan is reported to be a robust tool because it enables a more complete search at isoform level, it penalizes the less conserved interactions, and its databases are up-to-date (38).
Luciferase reporter constructs containing microRNA-7a-5p target site.
The 3′-UTR region of rat Gabra1 gene (GenBank XM_017597179.1; nt 1722–4286) was identified using the UCSC Genome Browser. The target site for rno-miR-7a-5p within the rat Gabra1 3′-UTR region (nt 2301–2323) was predicted using the TargetScan software (release 7.2). A ~100-bp DNA fragment (nt 2270–2372) of the 3′-UTR of Gabra1 gene containing predicted rno-miR-7a-5p target site was amplified from rat brain VMH genomic DNA using AccuPrime pfx SuperMix (Invitrogen) and primers designed with XbaI restriction site (tctaga) appended at the 5′ end (forward: 5′-CGCtctagaGTAGCTTTACAACCGTGCTTTG-3′; and reverse: 5′-CGCtctagaCTGGAGTCAACAGGAACTGTC-3′; synthesized by DNA/Peptide Synthesis core facility at the University of Utah). The purified DNA fragment was ligated into the XbaI site (nt 1934) located downstream of the luciferase gene in the SV-40–driven firefly pGL3-promoter vector (Promega; 5010 bp) to construct a luciferase reporter plasmid. The sequence and orientation of the DNA insert were tested by Sanger sequencing using this primer for sense oligonucleotides: 5′-CCTCATAAAGGCCAAGAAGGG-3′ (Integrated DNA Technologies).
Luciferase assays.
HEK293T cells (a gift from S. Swaminathan’s lab) were maintained at 37°C and 5% CO2 in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum and 100 U/mL of penicillin/streptomycin (Life Technologies Corporation). In brief, HEK293T cells were plated in 6-well plates (Corning Inc.), and the next day cells were cotransfected with (a) firefly luciferase reporter plasmid DNA (300 ng) containing Gabra1 3′-UTR fragment, (b) pcDNA3 vector as filler DNA (200 ng), (c) a control pRL-CMV renilla luciferase reporter plasmid (30 ng; Promega), and (d) mirVana microRNA mimic (100 nM; Ambion), corresponding to rat miR-7a-5p or an mirVana miR-NC (100 nM; Ambion) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Cells were harvested 24 hours after transfection and assayed using a Dual-Luciferase Reporter Assay System (Promega). Luciferase reporter activities were measured by using Turner Biosystems 20/20 Luminometer. For data analysis, firefly luciferase activity was firstly normalized to renilla (transfection control), and then rno-miR-7a-5p was normalized to miR-NC–transfected cells.
Statistics.
The results are represented as mean ± SEM. Data for body weight, blood glucose, and microRNA and mRNA expressions were analyzed by 1-way ANOVA. Results for blood glucose, GIR, catecholamine, and glucagon were analyzed by 2-way repeated-measures ANOVA. Post hoc analyses were conducted by Tukey’s multiple-comparison tests. Pearson’s correlation analysis was performed on individual samples to evaluate the correlation between variables. A P value less than 0.05 was considered statistically significant.
Study approval.
All procedures were performed in accordance with the US National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011) and approved by the Institutional Animal Care and Use Committee of the University of Utah.
Author contributions
RA designed and performed experiments, analyzed and interpreted the results, and wrote the manuscript. GD and DV performed experiments and reviewed the manuscript. MM, AVDA, and CT performed experiments. SS reviewed the manuscript. SJF designed the study, interpreted the results, and edited the manuscript.
Supplementary Material
Acknowledgments
This work was supported by funding from National Institutes of Health R01 NS070235-01A1 (to SJF) and R01DK118082 (to SJF), the Juvenile Diabetes Research Foundation 2-SRA-2014-270-M-R (to SJF), and the University of Utah’s Diabetes and Metabolism Research Center. We are grateful for support from the High-throughput Genomics and Bioinformatic Analysis core facility of Huntsman Cancer Institute and the Genomics, DNA/Peptide Synthesis, and Fluorescence Microscopy core facilities of Heath Sciences cores at the University of Utah. The authors have declared that no conflict of interest exists.
Version 1. 10/17/2019
Electronic publication
Version 2. 11/14/2019
Updated issue information.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(20):e130521.https://doi.org/10.1172/jci.insight.130521.
Contributor Information
Rahul Agrawal, Email: rahul19agrawal@gmail.com.
Griffin Durupt, Email: griffindurupt@gmail.com.
Dinesh Verma, Email: dineshpgi@gmail.com.
Michael Montgomery, Email: u1043989@utah.edu.
Casey Taylor, Email: casey.j.taylor4@gmail.com.
Sankar Swaminathan, Email: sankar.swaminathan@hsc.utah.edu.
Simon J. Fisher, Email: simon.fisher@u2m2.utah.edu.
References
- 1.Cryer PE. The barrier of hypoglycemia in diabetes. Diabetes. 2008;57(12):3169–3176. doi: 10.2337/db08-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cryer PE. Mechanisms of hypoglycemia-associated autonomic failure and its component syndromes in diabetes. Diabetes. 2005;54(12):3592–3601. doi: 10.2337/diabetes.54.12.3592. [DOI] [PubMed] [Google Scholar]
- 3.McGregor VP, Banarer S, Cryer PE. Elevated endogenous cortisol reduces autonomic neuroendocrine and symptom responses to subsequent hypoglycemia. Am J Physiol Endocrinol Metab. 2002;282(4):E770–E777. doi: 10.1152/ajpendo.00447.2001. [DOI] [PubMed] [Google Scholar]
- 4.Chan O, Sherwin R. Influence of VMH fuel sensing on hypoglycemic responses. Trends Endocrinol Metab. 2013;24(12):616–624. doi: 10.1016/j.tem.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herzog RI, Chan O, Yu S, Dziura J, McNay EC, Sherwin RS. Effect of acute and recurrent hypoglycemia on changes in brain glycogen concentration. Endocrinology. 2008;149(4):1499–1504. doi: 10.1210/en.2007-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan O, et al. Increased GABAergic tone in the ventromedial hypothalamus contributes to suppression of counterregulatory responses after antecedent hypoglycemia. Diabetes. 2008;57(5):1363–1370. doi: 10.2337/db07-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borg MA, Tamborlane WV, Shulman GI, Sherwin RS. Local lactate perfusion of the ventromedial hypothalamus suppresses hypoglycemic counterregulation. Diabetes. 2003;52(3):663–666. doi: 10.2337/diabetes.52.3.663. [DOI] [PubMed] [Google Scholar]
- 8.Vavaiya KV, Paranjape SA, Briski KP. Testicular regulation of neuronal glucose and monocarboxylate transporter gene expression profiles in CNS metabolic sensing sites during acute and recurrent insulin-induced hypoglycemia. J Mol Neurosci. 2007;31(1):37–46. doi: 10.1007/BF02686116. [DOI] [PubMed] [Google Scholar]
- 9.Mastaitis JW, Wurmbach E, Cheng H, Sealfon SC, Mobbs CV. Acute induction of gene expression in brain and liver by insulin-induced hypoglycemia. Diabetes. 2005;54(4):952–958. doi: 10.2337/diabetes.54.4.952. [DOI] [PubMed] [Google Scholar]
- 10.Chan O, et al. Increased GABAergic output in the ventromedial hypothalamus contributes to impaired hypoglycemic counterregulation in diabetic rats. Diabetes. 2011;60(5):1582–1589. doi: 10.2337/db10-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan O, Zhu W, Ding Y, McCrimmon RJ, Sherwin RS. Blockade of GABA(A) receptors in the ventromedial hypothalamus further stimulates glucagon and sympathoadrenal but not the hypothalamo-pituitary-adrenal response to hypoglycemia. Diabetes. 2006;55(4):1080–1087. doi: 10.2337/diabetes.55.04.06.db05-0958. [DOI] [PubMed] [Google Scholar]
- 12.Jacovetti C, Matkovich SJ, Rodriguez-Trejo A, Guay C, Regazzi R. Postnatal β-cell maturation is associated with islet-specific microRNA changes induced by nutrient shifts at weaning. Nat Commun. 2015;6:8084. doi: 10.1038/ncomms9084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grasso M, Piscopo P, Confaloni A, Denti MA. Circulating miRNAs as biomarkers for neurodegenerative disorders. Molecules. 2014;19(5):6891–6910. doi: 10.3390/molecules19056891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olsen L, Klausen M, Helboe L, Nielsen FC, Werge T. MicroRNAs show mutually exclusive expression patterns in the brain of adult male rats. PLoS ONE. 2009;4(10):e7225. doi: 10.1371/journal.pone.0007225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meister B, Herzer S, Silahtaroglu A. MicroRNAs in the hypothalamus. Neuroendocrinology. 2013;98(4):243–253. doi: 10.1159/000355619. [DOI] [PubMed] [Google Scholar]
- 16.Herzer S, Silahtaroglu A, Meister B. Locked nucleic acid-based in situ hybridisation reveals miR-7a as a hypothalamus-enriched microRNA with a distinct expression pattern. J Neuroendocrinol. 2012;24(12):1492–1504. doi: 10.1111/j.1365-2826.2012.02358.x. [DOI] [PubMed] [Google Scholar]
- 17.Taouis M. MicroRNAs in the hypothalamus. Best Pract Res Clin Endocrinol Metab. 2016;30(5):641–651. doi: 10.1016/j.beem.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13(4):271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 19.Felekkis K, Touvana E, Stefanou Ch, Deltas C. microRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia. 2010;14(4):236–240. [PMC free article] [PubMed] [Google Scholar]
- 20.Liu W, et al. Hepatic miR-378 targets p110α and controls glucose and lipid homeostasis by modulating hepatic insulin signalling. Nat Commun. 2014;5:5684. doi: 10.1038/ncomms6684. [DOI] [PubMed] [Google Scholar]
- 21.Dooley J, et al. The microRNA-29 family dictates the balance between homeostatic and pathological glucose handling in diabetes and obesity. Diabetes. 2016;65(1):53–61. doi: 10.2337/db15-0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu X, et al. MicroRNA-26a regulates insulin sensitivity and metabolism of glucose and lipids. J Clin Invest. 2015;125(6):2497–2509. doi: 10.1172/JCI75438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Liu J, Liu C, Naji A, Stoffers DA. MicroRNA-7 regulates the mTOR pathway and proliferation in adult pancreatic β-cells. Diabetes. 2013;62(3):887–895. doi: 10.2337/db12-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Latreille M, et al. MicroRNA-7a regulates pancreatic β cell function. J Clin Invest. 2014;124(6):2722–2735. doi: 10.1172/JCI73066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson KL, et al. A novel interaction between sympathetic overactivity and aberrant regulation of renin by miR-181a in BPH/2J genetically hypertensive mice. Hypertension. 2013;62(4):775–781. doi: 10.1161/HYPERTENSIONAHA.113.01701. [DOI] [PubMed] [Google Scholar]
- 26.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiménez A, Pelto-Huikko M, Gustafsson JA, Miranda-Vizuete A. Characterization of human thioredoxin-like-1: potential involvement in the cellular response against glucose deprivation. FEBS Lett. 2006;580(3):960–967. doi: 10.1016/j.febslet.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 28.Adlakha YK, Saini N. Brain microRNAs and insights into biological functions and therapeutic potential of brain enriched miRNA-128. Mol Cancer. 2014;13:33. doi: 10.1186/1476-4598-13-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bravo-Egana V, et al. Quantitative differential expression analysis reveals miR-7 as major islet microRNA. Biochem Biophys Res Commun. 2008;366(4):922–926. doi: 10.1016/j.bbrc.2007.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci USA. 2009;106(31):13052–13057. doi: 10.1073/pnas.0906277106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palkovits M. Punch sampling biopsy technique. Meth Enzymol. 1983;103:368–376. doi: 10.1016/s0076-6879(83)03025-6. [DOI] [PubMed] [Google Scholar]
- 33.Segal JP, et al. Use of laser-capture microdissection for the identification of marker genes for the ventromedial hypothalamic nucleus. J Neurosci. 2005;25(16):4181–4188. doi: 10.1523/JNEUROSCI.0158-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Åkerblom M, et al. Visualization and genetic modification of resident brain microglia using lentiviral vectors regulated by microRNA-9. Nat Commun. 2013;4:1770. doi: 10.1038/ncomms2801. [DOI] [PubMed] [Google Scholar]
- 36.Puente EC, et al. Recurrent moderate hypoglycemia ameliorates brain damage and cognitive dysfunction induced by severe hypoglycemia. Diabetes. 2010;59(4):1055–1062. doi: 10.2337/db09-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishihara KK, Haywood SC, Daphna-Iken D, Puente EC, Fisher SJ. Brain insulin infusion does not augment the counterregulatory response to hypoglycemia or glucoprivation. Metab Clin Exp. 2009;58(6):812–820. doi: 10.1016/j.metabol.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riffo-Campos ÁL, Riquelme I, Brebi-Mieville P. Tools for sequence-based miRNA target prediction: what to choose? Int J Mol Sci. 2016;17(12):E1987. doi: 10.3390/ijms17121987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.