Abstract
Chromatin modifiers act to coordinate gene expression changes critical to neuronal differentiation from neural stem/progenitor cells (NSPCs). Lysine-specific methyltransferase 2D (KMT2D) encodes a histone methyltransferase that promotes transcriptional activation and is frequently mutated in cancers and in the majority (>70%) of patients diagnosed with the congenital, multisystem intellectual disability disorder Kabuki syndrome 1 (KS1). Critical roles for KMT2D are established in various non-neural tissues, but the effects of KMT2D loss in brain cell development have not been described. We conducted parallel studies of proliferation, differentiation, transcription, and chromatin profiling in KMT2D-deficient human and mouse models to define KMT2D-regulated functions in neurodevelopmental contexts, including adult-born hippocampal NSPCs in vivo and in vitro. We report cell-autonomous defects in proliferation, cell cycle, and survival, accompanied by early NSPC maturation in several KMT2D-deficient model systems. Transcriptional suppression in KMT2D-deficient cells indicated strong perturbation of hypoxia-responsive metabolism pathways. Functional experiments confirmed abnormalities of cellular hypoxia responses in KMT2D-deficient neural cells and accelerated NSPC maturation in vivo. Together, our findings support a model in which loss of KMT2D function suppresses expression of oxygen-responsive gene programs important to neural progenitor maintenance, resulting in precocious neuronal differentiation in a mouse model of KS1.
Keywords: Genetics, Neuroscience
Keywords: Adult stem cells, Epigenetics, Neurological disorders
Loss of chromatin modifier KMT2D perturbs adult-born hippocampal neural stem/progenitor cell maintenance through transcriptional suppression of hypoxia-responsive gene expression.
Introduction
Trithorax group proteins promote chromatin accessibility by exerting antagonistic functions against Polycomb group transcriptional suppressors to activate gene expression (1). Fine-tuning of cell type transitions during neuronal development from neural stem/progenitor cells (NSPCs) depends critically on this duality, as evidenced by severe neurodevelopmental defects caused by variants in numerous chromatin-modifying genes (2). Loss-of-function variants in genes encoding 2 such enzymes, lysine-specific methyltransferase 2D (KMT2D) and lysine-specific demethylase 6A (KDM6A/UTX) cause the intellectual disability (ID) disorder Kabuki syndrome (KS1 and KS2, respectively) (3, 4). Up to 74% (5) of KS cases result from mutations in KMT2D (KS1), which encodes a major histone H3 lysine 4 (H3K4) methyltransferase, which catalyzes chromatin-opening modifications at context-specific targets. Developmental requirements for KMT2D in cardiac precursors (6), B cells (7, 8), muscle and adipose (9), and epithelial tissues (10) have been linked, respectively, to KMT2D-associated cardiac, immunological, and oncogenic phenotypes (11), yet the effects of KMT2D deficiency in neurodevelopment are not yet understood.
We previously described a mouse model of KS1, Kmt2d+/βgeo [Mll2Gt(RRt024)Byg], demonstrating characteristic features including craniofacial abnormalities and visuospatial memory impairments, associated with decreased adult-born hippocampal NSPCs in the dentate gyrus (DG) (12). Decreased DG gray matter volume was subsequently observed in patients with KS1 (13). The continual birth and integration of new neurons makes adult neurogenesis the most potent form of lifelong plasticity in the mammalian brain (14), though recent studies have disagreed on its extent in humans (15–17). During late embryonic stages, a subset of multipotent NSPCs persists in the DG (18), becoming subject to an array of intrinsic and extrinsic factors affecting NSPC maintenance, i.e., self-renewal, proliferation, and neuronal differentiation, throughout adult life. Mounting evidence tightly links metabolic rewiring (19) and hypoxic states in the DG (20, 21) to cell-intrinsic regulation of NSPC maintenance.
Here, we find that KMT2D deficiency strongly suppresses metabolic gene expression and leads to reduced proliferation, abnormal hypoxia responses, and precocious neuronal maturation in multiple KS1 model systems. Importantly, these phenotypes were validated in vivo in a KS1 mouse model, supporting a role for these abnormalities in the pathogenesis of KS1-associated ID.
Results
Genetic ablation of the Kmt2d Su(var)3-9, enhancer-of-zeste and trithorax methyltransferase domain disrupts proliferation and cell cycle in a cell-autonomous manner.
We first selected the HT22 mouse hippocampal neuronal cell line (22) for analysis of KMT2D catalytic function in a neuronal context. The DNA sequence encoding the Su(var)3-9, enhancer-of-zeste and trithorax (SET) methyltransferase domain was deleted by CRISPR/Cas9 with an upstream small guide RNA (sgRNAup) in exon 52, and either sgRNA1 (exon 54) or sgRNA2 (intron 54), resulting in deletions of 565 bp (Kmt2dΔ1) or 654 bp (Kmt2dΔ2), respectively, as verified by Sanger DNA sequencing, in silico translation, and PCR (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.129375DS1). Targeted cells were clonally expanded to establish heterozygous (Kmt2d+/Δ) and homozygous (Kmt2dΔ/Δ) cell lines for comparison against the parental wild-type line (Kmt2d+/+). Both biological replicate alleles, Kmt2dΔ1 and Kmt2dΔ2, were represented in the present studies; thus, the combined data are denoted hereafter simply as Kmt2d+/Δ or Kmt2dΔ/Δ. Kmt2d mRNA encoded within the targeted region was about 50% decreased in Kmt2d+/Δ cells and absent in Kmt2dΔ/Δ cells, while Kmt2d mRNA from exons upstream of the deletion site was unaffected (Supplemental Figure 1C). Immunofluorescence against KMT2D, detecting a peptide sequence upstream of deletions (Supplemental Figure 1D), demonstrated distinctly nuclear KMT2D distribution in Kmt2d+/+ cells but more diffuse distribution in Kmt2d+/Δ and Kmt2dΔ/Δ cells, and we observed uniformly nuclear expression of a neuronal nuclear marker, RNA binding protein fox-1 homolog 3 (RBFOX3), independent of genotype (Figure 1A).
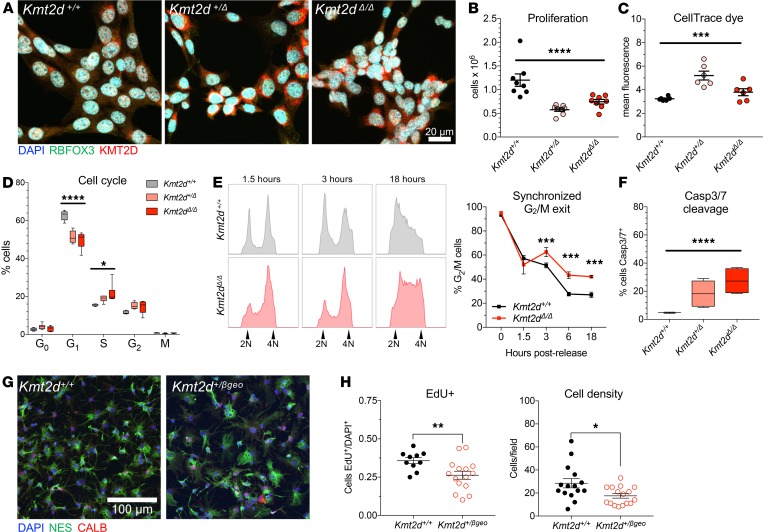
Figure 1. Genetic ablation of the Kmt2d SET methyltransferase domain disrupts proliferation and cell cycle in a cell-autonomous manner.
(A) Representative immunostaining against KMT2D and RBFOX3 in Kmt2d+/+, Kmt2d+/Δ, and Kmt2dΔ/Δ HT22 cells. (B) Decreased proliferation in Kmt2d-inactivated cells quantified 72 hours after equal density plating. One-way ANOVA. (C) Generational tracking reveals fewer cell divisions, i.e., reduced dye dilution, of CellTrace Violet in Kmt2d+/Δ and Kmt2dΔ/Δ cells at 72 hours. One-way ANOVA. (D) Flow cytometric quantification of cell cycle phases using marker Ki-67 (KI67) and DAPI fluorescence. One-way ANOVA for each cycle phase, independently. (E) Kmt2d+/+ and Kmt2dΔ/Δ cells synchronized and released for analysis of G2/M exit, by DNA content, up to 18 hours after release, and quantification of cells in G2/M (technical triplicates per time point). Bars indicate mean ± SEM. Two-way ANOVA (P < 0.0001) with post hoc multiple comparisons correction. (F) Flow cytometric quantification of early apoptotic cells by caspase-3/7 fluorescence. One-way ANOVA. (G) Confocal images of nestin (NES) and calbindin (CALB) expressing primary hippocampal NSPCs from Kmt2d+/+ and Kmt2d+/βgeo mice, and (H) quantified proliferation. One-tailed Student’s t test. Bars indicate mean ± SEM. Boxes indicate mean ± interquartile range; whiskers indicate minima and maxima. (*P < 0.05, **P < 0.01, ***P < 0.001; ****P < 0.0001). Scale bars: 20 μm (A), 100 μm (G).
Proliferation analysis after equal-density plating revealed cell densities approximately 52% lower in Kmt2d+/Δ cells and approximately 39% lower in Kmt2dΔ/Δ cells, compared with wild-type (Figure 1B). This defect was supported by dye-based generational tracking, detecting modestly reduced dilution of a fluorescent tracer, i.e., fewer cell divisions, in Kmt2d+/Δ and Kmt2dΔ/Δ daughter cells compared with wild-type (Figure 1C and Supplemental Figure 1E), while initial dye uptake in parental cells was genotype independent. Flow cytometric analysis of cell cycle occupancy, using marker of proliferation KI67 (KI67) and a DNA label, revealed that Kmt2d+/Δ cells and Kmt2dΔ/Δ cells were enriched for S and G2 phase, compared with wild-type (Figure 1D and Supplemental Figure 1F). To characterize temporal dynamics of cell cycle progression, we synchronized cells in G2/M phase and analyzed DNA content at time points after release (Figure 1E). Wild-type cells exited G2/M phase at higher rates than Kmt2dΔ/Δ cells, at 3 hours and up to 18 hours after release. Cell death was profiled by flow cytometric detection of caspase-3/7 substrate cleavage to distinguish early apoptotic cells. Compared with wild-type, apoptotic cell proportions were greater in both Kmt2d+/Δ cells (~287%) and Kmt2dΔ/Δ cells (~478%) (Figure 1F).
To examine proliferation in primary hippocampal progenitors, we isolated NSPCs from microdissected DG of Kmt2d+/βgeo mice and wild-type littermates. NSPCs exhibited characteristic expression of NSPC marker nestin (NES), with a minority of cells expressing mature neuron marker calbindin (CALB) (Figure 1G). Cells were plated at equal density and pulsed with cell division marker 5-ethynyl-2′-deoxyuridine (EdU), then quantified by confocal microscopy. Compared with wild-type, Kmt2d+/βgeo NSPCs demonstrated lower proliferation rates as measured by EdU incorporation and cell density (Figure 1H).
Findings of proliferation defects, G2/M cell cycle delay, and increased apoptosis in hippocampal cells bearing Kmt2d inactivation by SET domain deletion, together with proliferation defects in primary Kmt2d+/βgeo hippocampal NSPCs, support a cell-intrinsic role for KMT2D activity in neurodevelopmental contexts.
Suppressed transcription of KMT2D-regulated hypoxia response genes upon loss of the KMT2D SET methyltransferase domain.
We performed high-coverage RNA-Seq comparing 3 Kmt2dΔ/Δ clones against the parental Kmt2d+/+ line, each in technical triplicate, followed by differential expression analysis. Libraries clustered robustly by genotype with clear separation of Kmt2dΔ/Δ cells from Kmt2d+/+ by principal component analysis, yielding 575 significantly differentially expressed genes (DEGs) at a false discovery rate (FDR) of 0.05 in Kmt2dΔ/Δ cells compared with Kmt2d+/+ (Figure 2A, Supplemental Figure 2, A and B, and Supplemental Table 1). Approximately 76% of DEGs (436 genes) were downregulated in Kmt2dΔ/Δ cells, including known KMT2D targets, such as Krueppel-like factor 10 (Klf10) (12), revealing strong global transcriptional suppression from Kmt2d inactivation. Overrepresentation analysis revealed significant enrichment of gene networks among Kmt2dΔ/Δ downregulated DEGs, including glycolysis and hypoxia-inducible factor 1A (HIF1A) signaling, while Kmt2dΔ/Δ upregulated DEGs were enriched in fewer networks (Figure 2B).
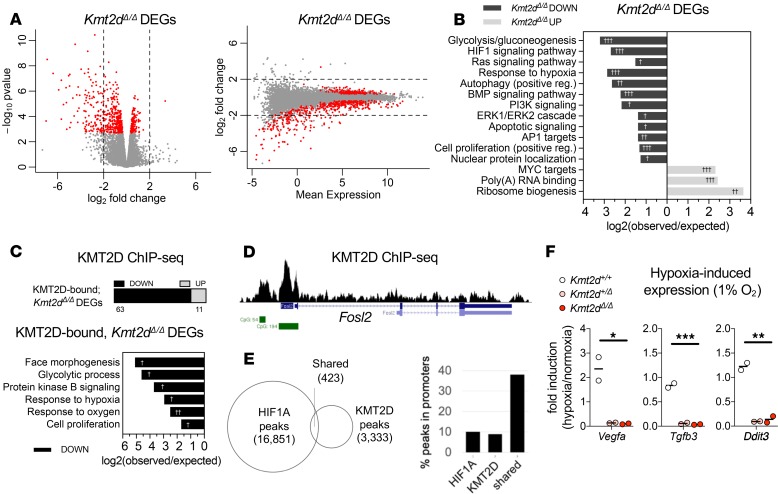
Figure 2. Suppressed transcription of KMT2D-regulated hypoxia response genes upon loss of the Kmt2d SET methyltransferase domain in neuronal cells.
(A) Expression analysis by RNA-Seq in HT22 cells revealed 575 significant differentially expressed genes (DEGs) in Kmt2dΔ/Δ clones (3 biological replicates) relative to Kmt2d+/+ cells, each in technical triplicate. Fold changes in expression indicate the most significant Kmt2dΔ/Δ DEGs (~76%, red dots) are downregulated in Kmt2dΔ/Δ cells, plotted against P value and mean expression. (B) Gene networks significantly enriched among down- or upregulated Kmt2dΔ/Δ DEGs. (C) Kmt2dΔ/Δ DEGs that are also KMT2D bound, as determined by ChIP-Seq chromatin profiling in Kmt2d+/+ HT22 cells, and gene networks significantly enriched among KMT2D-bound, Kmt2dΔ/Δ DEGs. (D) Representative ChIP-Seq track of a KMT2D-bound, Kmt2dΔ/Δ DEG depicting KMT2D binding peaks (shown in black), RefSeq gene annotations (shown in blue), and CpG islands (shown in green). (E) Overlapping loci of observed KMT2D-ChIP peaks in HT22 cells and HIF1A-ChIP peaks in embryonic hearts (26). Overlapping KMT2D/HIF1A peak regions, compared with individually bound regions, are enriched at gene promoters. (F) Reverse transcription quantitative PCR (RT-qPCR) analysis of hypoxia-induced gene expression in Kmt2d+/+, Kmt2d+/Δ, and Kmt2dΔ/Δ cells, following 72 hours in normoxia (21% O2) or hypoxia (1% O2), with fold induction of target gene mRNA. Two biological replicates per genotype, each in technical triplicate. One-way ANOVA (*P < 0.05, **P < 0.01, and ***P < 0.001). Fisher’s exact test (†FDR < 0.05, ††FDR < 0.01, and †††FDR < 0.001).
We reasoned that among Kmt2dΔ/Δ DEGs, a subset of genes found to also bind KMT2D itself in wild-type cells would more likely represent direct transcriptional consequences of Kmt2d inactivation, whereas unbound DEGs could reflect secondary effects. We performed chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-Seq) using a previously validated ChIP-grade KMT2D antibody (9) in Kmt2d+/+ HT22 cells. We identified 3756 KMT2D binding peaks significantly enriched over input (Supplemental Table 2), of which approximately 10% occurred inside promoters, approximately 33% (1235 peaks) occurred within 5 kb of a transcription start site (TSS ± 5 kb), and approximately 25% occurred within 2 kb (Supplemental Figure 2, C–F). To account for promoter and enhancer interactions (9, 10, 23), we reasoned that TSS ± 5 kb peaks, compared with more distal peaks, are more likely to reflect KMT2D cis-regulatory functions on proximal genes, so we refer to these as KMT2D-bound genes. The 1463 observed KMT2D-bound genes (Supplemental Table 3) were significantly enriched in mRNA 3′-UTR binding, rho GTPase signaling, circadian clock, translation, oxidative stress, HIF1A signaling, and other pathways (Supplemental Figure 2G).
We then intersected KMT2D-bound genes with Kmt2dΔ/Δ DEGs to reveal 74 putative direct target genes (Supplemental Table 3), of which ~85% (63 genes) were downregulated (Figure 2C), including insulin-like growth factor 1 (Igf1) and fos-like antigen 2 (Fosl2). At least 20 observed KMT2D-bound, Kmt2dΔ/Δ DEGs were previously described as KMT2D targets in other tissues (7, 24). KMT2D-bound, Kmt2dΔ/Δ downregulated DEGs were most significantly enriched for pathways including face morphogenesis, glycolysis, hypoxia response, and proliferation, and surprisingly, 29 of these 63 genes are also HIF1A regulated (25). Although craniofacial features associate with KS1, enrichment of face morphogenesis genes in HT22 cells likely reflects pleiotropic gene functions. KMT2D ChIP-Seq peaks on HIF1A-regulated genes clustered at promoters and enhancers, often overlapping CpG islands in genes such as Fosl2, with others clustering at alternative TSSs, as in retinoic acid receptor–α (Rara), or in enhancer-like peaks, as in DNA damage-inducible transcript 4 (Ddit4) (Figure 2D and Supplemental Figure 2H).
A large fraction of KMT2D-bound, Kmt2dΔ/Δ DEGs control oxygen-responsive metabolism, warranting interrogation of shared KMT2D and HIF1A binding sites. We first intersected KMT2D peaks with HIF1A peaks previously found in embryonic hearts (26), finding 423 overlapped regions (Figure 2E). Like KMT2D, HIF1A showed approximately 10% of peaks located inside promoters, but among shared KMT2D/HIF1A–bound peaks this fraction approached approximately 40%, supporting cooperative regulatory activity (Supplemental Figure 2I). We identified 289 TSS ± 5 kb genes, as defined above, for these overlapped KMT2D/HIF1A–bound peaks, including 8 Kmt2dΔ/Δ DEGs (Supplemental Table 3).
To check whether KMT2D/HIF1A–bound genes generalize to other tissues, we next interrogated independent gene sets having experimentally validated, hypoxia-induced HIF1A binding in the promoter (27). Of 86 validated genes, 5 were KMT2D bound, Kmt2dΔ/Δ downregulated DEGs, 23.3-fold more than expected by chance (Fisher’s exact test, P = 4.74e-6) (Supplemental Table 3). Of 81 genes validated in 3 or more tissues, 3 were KMT2D bound, Kmt2dΔ/Δ downregulated DEGs: Klf10, Rara, and Ddit4 (Fisher’s exact test, P = 0.002).
Given the prevalence of oxygen response genes among Kmt2dΔ/Δ downregulated DEGs and shared KMT2D/HIF1A targets, we hypothesized there is a positive regulatory role for KMT2D in transcriptional responses to hypoxia in HT22 cells. We subjected Kmt2d+/+, Kmt2d+/Δ, and Kmt2dΔ/Δ cells to normoxia (21% O2) or hypoxia (1% O2), and measured hypoxia-induced gene expression responses. Analysis of canonical HIF1A targets, vascular endothelial growth factor A (Vegfa), Bcl2/adenovirus E1B 19-KD protein-interacting protein 3 (Bnip3), Ddit3, and cyclin-dependent kinase inhibitor 1A (Cdkn1a), in Kmt2d+/+ cells revealed robust upregulation upon hypoxic exposure; in contrast, Kmt2d+/Δ and Kmt2dΔ/Δ cell lines failed to induce these genes to comparable levels (Figure 2F and Supplemental Figure 2J). In hypoxic conditions, stabilized HIF1A undergoes nuclear translocation, i.e., activation. We therefore quantified nucleus-localized HIF1A fluorescence under normoxia (21% O2) and hypoxia (1% O2) (Supplemental Figure 2K). Unexpectedly, in normoxia, Kmt2dΔ/Δ cells exhibited more than 2-fold greater HIF1A activation than Kmt2d+/+ cells. Upon hypoxic exposure, HIF1A activation doubled in wild-type cells but failed to respond in Kmt2d+/Δ cells and Kmt2dΔ/Δ cells.
Taken together, our data suggest that KMT2D plays an important role in positively regulating HIF1A-inducible, oxygen-responsive metabolic gene programs in neuronal cells.
KS1 patient–derived cells recapitulate KMT2D-associated defects in proliferation and cell cycle.
We reprogrammed skin biopsy fibroblasts to generate induced pluripotent stem cells (iPSCs) from a previously described female KS1 patient (KS1-1) bearing a heterozygous nonsense KMT2D mutation (c.7903C>T:p.R2635*) with characteristic facial features, congenital heart disease, and visuospatial memory impairments (28). We selected KS1 iPSCs bearing normal 46,XX karyotype (Supplemental Figure 3A) and characteristic morphology (Figure 3A) for comparison against previously described iPSC lines from unrelated healthy controls (C1-2, C3-1) (29). KMT2D mRNA quantification in KS1 iPSCs confirmed decreased message compared with controls, as expected because of haploinsufficiency (Supplemental Figure 3, B and C). Quantification after EdU pulse demonstrated lower proliferation rates (~25%) in KS1 iPSCs compared with controls (Figure 3B), accompanied by a shift in cell cycle occupancy (Figure 3C, Supplemental Figure 3D) favoring G2/M phase (24% more cells).
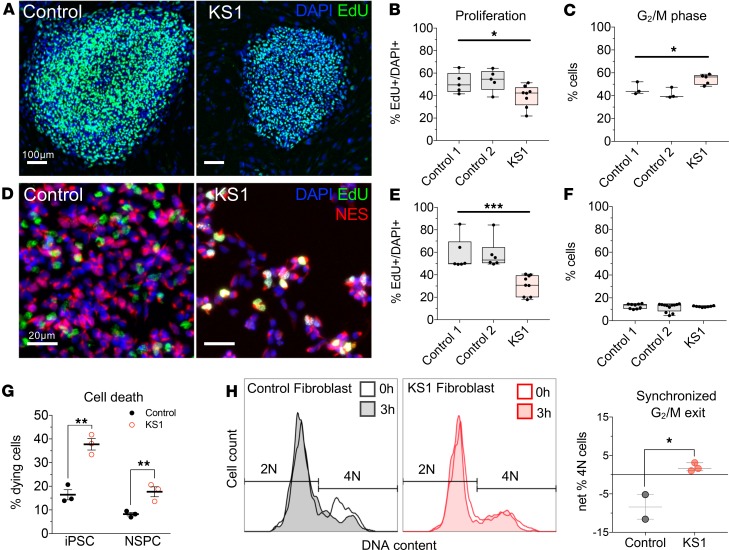
Figure 3. KS1 patient–derived cells recapitulate KMT2D-associated defects in proliferation and cell cycle.
(A) Representative immunostaining of iPSCs derived from a KMT2D+/– KS1 patient (c.7903C>T:p.R2635*) and healthy controls. (B) Proliferating cells were pulsed with EdU for 30 minutes and quantified by flow cytometry. One-way ANOVA. (C) Cell cycle analysis in iPSCs, discriminating 2N and 4N DNA content (G1/G0 and G2/M, respectively) by flow cytometry using DAPI fluorescence. One-way ANOVA. (D) Representative immunostaining of NES-expressing NSPCs induced from iPSCs of KS1 patient and controls. (E) EdU pulse assay quantified by flow cytometry. One-way ANOVA. (F) Cell cycle defect analysis in NSPCs. One-way ANOVA. (G) Quantification of dying cells by flow cytometric scatter profiles in KS1 patient and control cells. One-tailed Student’s t test. (H) Synchronized G2/M exit analysis by flow cytometry in fibroblasts from KS1 patients (KS1-1, KS1-2, KS1-3) and healthy controls (controls 3 and 4), in triplicate per cell line. Cells were enriched for G2/M phase using nocodazole and analyzed by DAPI fluorescence to quantify G2/M phase cell fractions at 0 and 3 hours after release. One-tailed Student’s t test. Bars indicate mean ± SEM. Boxes indicate mean ± interquartile range; whiskers indicate minima and maxima. (*P < 0.05, **P < 0.01, and ***P < 0.001.) Scale bars: 100 μm (A), 20 μm (D).
We next generated NES-expressing NSPCs through parallel differentiation of KS1 and control iPSCs, using an established small molecule inhibition protocol (30). RT-qPCR confirmed decreased KMT2D in KS1 NSPCs (Supplemental Figure 3E), and cells displayed normal morphology independent of genotype (Figure 3D, Supplemental Figure 3F). EdU incorporation rates revealed KS1 NSPCs had a marked proliferation defect (~47% reduced, Figure 3E) and fewer mitotic divisions (Supplemental Figure 3G). KS1 NSPCs did not display a cell cycle defect (Figure 3F, Supplemental Figure 3H), suggesting either cell type dependence or loss of this phenotype during in vitro differentiation. Flow cytometry indicated higher proportions of dying cells in KS1 samples compared with controls among both iPSCs (~130%) and NSPCs (~115%) (Figure 3G, Supplemental Figure 3, I and J).
To determine whether G2/M bias, seen in KS1 iPSCs, occurred in unmanipulated primary cells from additional KS1 patients, we analyzed fibroblasts from 3 molecularly confirmed KS1 patients (KS1-1, KS1-2, KS1-3) and healthy controls. Fibroblasts were synchronized in G2/M phase followed by flow cytometric analysis of DNA content. At 3 hours after release, control cells had exited G2/M phase, in contrast to KS1 cells, which remained in G2/M (Figure 3H). Thus, delayed G2/M exit was consistent in primary, nonreprogrammed cells from 3 patients with KS1.
Transcriptional suppression of metabolic genes in cycling cells and precocious neuronal differentiation in KS1 patient–derived NSPCs.
To interrogate transcriptional consequences of KMT2D loss in the context of neuronal differentiation, we performed single-cell RNA sequencing (scRNA-Seq) in iPSCs and NSPCs from the KS1 patient and controls (Supplemental Figure 4A). By inspecting expression of cell-type markers, we confirmed that libraries segregated into clusters reflecting distinct cell identities of the expected lineages (Supplemental Figure 4, B–D).
First, differential expression analysis in iPSCs and NSPCs identified genes downregulated or upregulated in the KS1 patient relative to healthy controls (Supplemental Figure 4E). KS1 iPSCs displayed strong transcriptional suppression among 421 DEGs, with 372 genes downregulated and 49 genes upregulated (Supplemental Table 4). NSPCs showed less directional bias, having 346 significant DEGs, among which 147 genes were downregulated and 199 genes were upregulated (Supplemental Table 5). Intersection of KS1 iPSC and NSPC DEG lists showed that 40 genes were shared (downregulated) and 10 genes were shared (upregulated) (Supplemental Figure 4, F and G, and Supplemental Table 6). Shared downregulated genes included glycolysis genes, aldehyde dehydrogenase 7 family member A1 (ALDH7A1), enolase 1 (ENO1), and triosephosphate isomerase 1 (TPI1), as well as factors important to stem cell maintenance, including proliferation-associated protein 2G4 (PA2G4) and lin-28 homolog A (LIN28A). As in Kmt2dΔ/Δ HT22 cells, downregulated genes in KS1 patient iPSCs and NSPCs were significantly enriched for HIF1A direct targets, genes containing the hypoxia-responsive element 5′-RCGTG-3′ motif, and known hypoxia response genes (Supplemental Figure 4H).
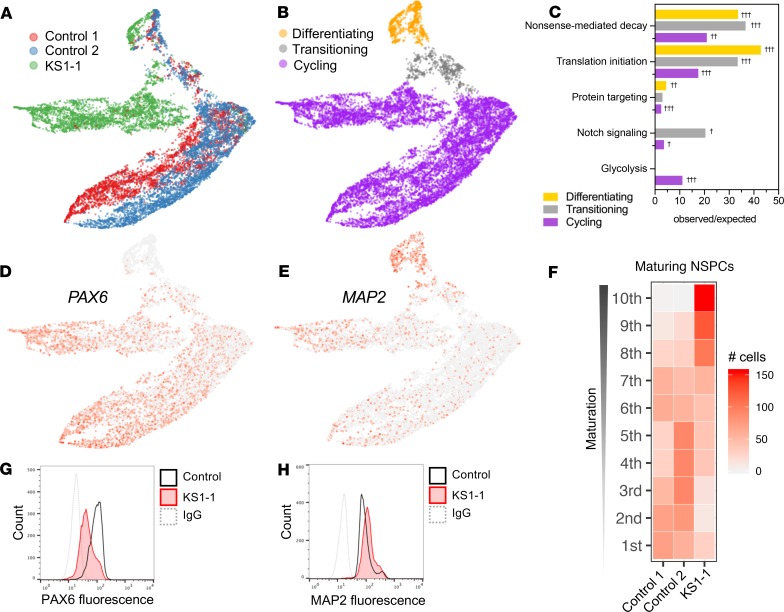
We next focused on NSPCs from KS1 and controls to interrogate transcriptional effects during neuronal differentiation. We used Uniform Manifold Approximation and Projection (UMAP) to visualize single cells in a manner that displays high-dimensionality data while preserving both local and global relationships (31). Control NSPCs were tightly clustered, indicating similar expression profiles, in contrast to a distinct separation of KS1 cells, which gradually lessens in a subset of cells that more closely resemble controls (Figure 4A, top). We then partitioned single-cell libraries into developmentally informative subsets as follows. First, we verified that differences in cell cycle phase composition did not account for KS1-associated differential gene expression in NSPCs (Supplemental Figure 5A and Supplemental Table 7). Next, we partitioned cells by stage-specific marker expression to define a differentiation trajectory consisting of early or “cycling” NSPCs, “transitioning” NSPCs, and “differentiating” NSPCs (Figure 4B). Cycling cells comprised the majority of NSPCs analyzed and exhibited the greatest KS1-associated expression differences, while expression profiles of transitioning and differentiating NSPCs showed gradual convergence of gene expression. We analyzed DEGs exclusively within cycling, transitioning, and differentiating NSPC subsets to determine whether particular gene networks drive transcriptional differences in a stage-specific manner (Figure 4C and Supplemental Table 7). KS1 DEGs in transitioning NSPCs, and to a lesser extent cycling NSPCs, showed enrichment of genes comprising the Notch signaling pathway, including delta-like protein 3 (DLL3), protein jagged-1 (JAG1), transcription factor HES-5 (HES5), and cyclin D1 (CCND1). Cycling NSPCs had DEGs enriched in glycolysis pathways.
Figure 4. Transcriptional suppression of metabolic genes in cycling cells and precocious neuronal differentiation in KS1 patient–derived NSPCs.
(A) scRNA-Seq profiling in patient and healthy control iPSC-derived NSPCs (~5000 cells per patient), with Uniform Manifold Approximation Projection (UMAP) to visualize gene expression differences between cells. (B) NSPCs partitioned by maturation stage as defined by stage-specific marker expression and (C) enriched gene networks, analyzed exclusively among DEGs for each NSPC subset (cycling, transitioning, and differentiating). (D and E) Representative UMAPs annotated by relative expression intensities of NSPC markers, revealing the maturation trajectory from early NSPCs (PAX6+) to differentiating NSPCs (MAP2+). (F) Heatmap comparing density of NSPCs along the maturation trajectory, defined by binned marker expression from earliest (first) to most differentiated (10th) deciles, with KS1 cells disproportionately occupying the most mature bins. (G and H) Protein-level experimental validation of marker expression differences by flow cytometry in NSPCs from KS1 patient and controls, plotting fluorescence intensities of PAX6 and MAP2. Fisher’s exact test (†FDR < 0.05, ††FDR < 0.01, and †††FDR < 0.001).
Apart from increased rates of KS1 cell death (Figure 3G), another possible factor in the observed decrease of proliferative KS1 NSPCs (Figure 3E) could be a change in cellular differentiation, such as precocious cell maturation, resulting in depletion of cycling precursors. To explore this by scRNA-Seq, we examined markers ranging from immature cells (paired box protein Pax-6–positive, PAX6+) to the most differentiated cells (microtubule-associated protein 2–positive, MAP2+) (Figure 4, D and E). We further restricted analysis to the transitioning and differentiating, i.e., “maturing” NSPC subset (Supplemental Figure 5B), defining a trajectory that enabled parsing of cells into binned deciles of increasing maturation (Supplemental Figure 5, C–H). Quantification of cell densities revealed strong bias of KS1 NSPCs in the most matured bins relative to controls (Figure 4F), i.e., greater representation of mature NSPCs from KS1 than controls. These transcriptional signatures were corroborated experimentally at the protein level; KS1 NSPCs had increased MAP2 fluorescence and decreased PAX6 fluorescence relative to control based on flow cytometry (Figure 4, G and H).
Together, these results link transcriptional suppression of metabolic gene pathways to cell-autonomous proliferation defects in KMT2D-deficient KS1 patient–derived stem cell models, and scRNA-Seq data suggest that precocious differentiation could contribute to KS1-associated neurodevelopmental defects.
In vivo defects of neurogenesis and NSPC differentiation in a Kmt2d+/βgeo mouse model of KS1.
Finally, we asked whether proliferative defects, transcriptional suppression, and precocious differentiation phenotypes could be validated in vivo, using an established KS1 mouse model. Kmt2d+/βgeo mice, bearing a Kmt2d-truncating mutation, were previously found to exhibit visuospatial memory impairments and fewer doublecortin-positive (DCX+) NSPCs in the DG subgranular zone (SGZ) (12, 32), but NSPC lineage progression in Kmt2d+/βgeo mice has not been characterized.
We conducted cell cycle and RNA-Seq analysis in Kmt2d+/βgeo mice, using an EdU pulse paradigm to label adult-born cells. We sampled microdissected DG within 1 cell cycle (16 hours) to capture the full complement of dividing NSPCs (Figure 5A), then purified EdU+ nuclei by FACS (Figure 5B and Supplemental Figure 6A). DNA content analysis revealed enrichment of G2/M phase in Kmt2d+/βgeo EdU+ DG nuclei (Figure 5C). We next profiled transcription by RNA-Seq in purified EdU+ DG nuclei, yielding 827 DEGs (Supplemental Figure 6, B and C, and Supplemental Table 8). The 416 downregulated genes in Kmt2d+/βgeo nuclei were enriched for misfolded protein binding, tricarboxylic acid cycle, proteasome complex, oxygen response, and poly(A) RNA–binding genes. Given the observed downregulation of poly(A) RNA–binding genes, we considered the possibility that improper 3′-UTR–mediated mRNA metabolism could lead to accumulation of transcripts influencing NSPC maturation. Indeed, despite little overall bias toward up- or downregulation in Kmt2d+/βgeo DG nuclei, interrogating positive regulators of neuronal differentiation revealed a marked predominance of proneural transcripts upregulated, having only 3 genes downregulated but 14 genes upregulated, including copine 1 (Cpne1), focal adhesion kinase 1 (Ptk2), RAS-related protein RAB11A (Rab11A), and retinoblastoma-associated protein 1 (Rb1). Interestingly, KS1 patient NSPCs also showed upregulated proneural genes, such as nuclear receptor subfamily 2, group F, member 1 (NR2F1) and proneural transcription factor HES-1 (HES1), while Kmt2dΔ/Δ HT22 cells had upregulated brain-derived neurotrophic factor (Bdnf) and neuron-specific microtubule component (Tubb3/Tuj1). Such proneural gene expression observed across KS1 models raises the possibility that NSPC differentiation rates could be altered in Kmt2d+/βgeo mice.
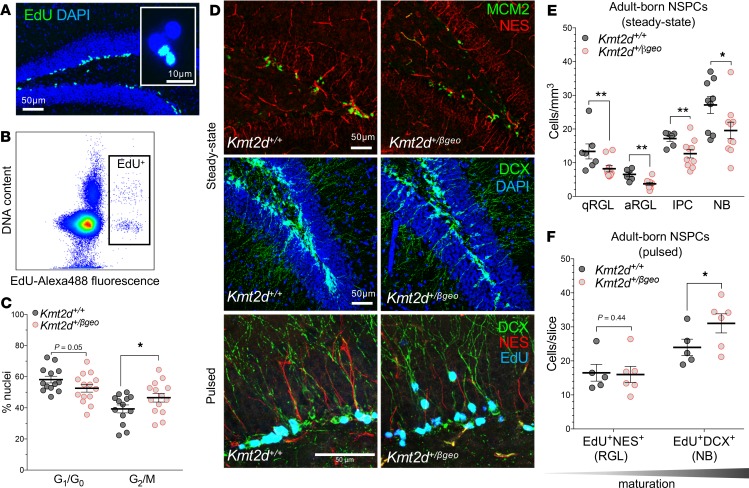
Figure 5. In vivo defects of neurogenesis and NSPC differentiation in a Kmt2d+/βgeo mouse model of KS1.
(A) Immunostaining images of dividing (EdU-pulsed) dentate gyrus (DG) NSPCs and nuclei purified from microdissected DG by fluorescence-activated cell sorting (FACS) (B) of labeled nuclei. (C) Cell cycle analysis in purified EdU+ DG nuclei from Kmt2d+/+ and Kmt2d+/βgeo mice sampled 16 hours after pulse, using DAPI fluorescence (13–14 mice per genotype, 200–500 nuclei per mouse). (D) Representative confocal immunostaining of neurogenesis markers in the DG of adult Kmt2d+/+ and Kmt2d+/βgeo mice at steady state (6–10 mice per genotype, 7–10 Z-stack images per mouse) or after EdU pulse (5–6 mice per genotype, 10 Z-stack images per mouse). NES+ radial glia-like (RGL) NSPCs, in either quiescent (minichromosome maintenance complex component 2–negative, MCM2–) or activated (MCM2+) states (qRGL and aRGL, respectively), MCM2+ NES946 intermediate progenitor cells (IPCs), and DCX+ neuroblasts (NBs) were quantified. (E and F) Quantification of stage-specific NSPC densities (qRGL, aRGL, IPC, and NB) in adult Kmt2d+/+ and Kmt2dβ/geo mice at steady state (E) or after EdU pulse-chase (2 weeks) to birth date, differentiating NSPCs (F). Bars indicate mean ± SEM. One-tailed Student’s t test (*P < 0.05, **P < 0.01, and ***P < 0.001). Scale bars: 50 μm, unless otherwise specified (A, inset, 10 μm).
To examine NSPC lineage progression in vivo, we analyzed stage-specific cell abundances both at steady state and after birth dating of adult-born NSPCs by EdU pulse, comparing Kmt2d+/βgeo mice to sex- and age-matched Kmt2d+/+ littermates (Figure 5D and Supplemental Figure 6D).
At steady state, we observed significantly fewer NSPCs in Kmt2d+/βgeo mice compared with Kmt2d+/+ mice at all stages (Figure 5E). The cell division marker, MCM2, distinguished NES+ NSPCs in the quiescent (MCM2–) or activated (MCM2+) state. Importantly, qRGL (NES+MCM2–) NSPCs were approximately 39% less numerous in Kmt2d+/βgeo mice, indicating a baseline paucity in the stem cell pool. aRGL (NES+MCM2+) NSPCs were approximately 43% less numerous, and IPC (NES–MCM2+) NSPCs were approximately 26% fewer. We confirmed prior observations (12, 32) of fewer NB (DCX+) NSPCs, finding a 28% decrease in Kmt2d+/βgeo mice. By stratifying analysis along the septotemporal axis of the DG, we observed that aRGL NSPC reductions in Kmt2d+/βgeo mice were more pronounced in the septal DG than the temporal region (Supplemental Figure 6E), congruous with spatial memory defects (12) localized to the septal DG (33). Because DCX+ NSPCs migrate radially during maturation, we compared radial distances of DCX+ cell bodies from the SGZ plane and observed increased distances in Kmt2d+/βgeo mice (Supplemental Figure 6F). Finally, despite diminished NSPC populations in Kmt2d+/βgeo mice, we observed no numeric differences among mature neurons (RBFOX3+) (Supplemental Figure 6G), nor were gross anatomical differences seen by MRI volumetric analysis (Supplemental Figure 6H, Supplemental Table 9).
From these data, we then calculated a lineage progression index to approximate the expansion potential of each successive neurogenic cell type. Although Kmt2d+/βgeo mice showed fewer total NSPCs of each type at steady state, the lineage progression index at each cell type transition did not differ significantly (Supplemental Figure 6I), suggesting that particular cell type transition impairments are not responsible for the adult neurogenesis defect. However, we did note substantially higher variance of RGL activation rates in Kmt2d+/βgeo mice, suggesting impaired coordination of NSPC mitotic entry (Supplemental Figure 6J).
Pulse labeling with marker-based imaging enables precise measurement of birth dates, i.e., mitotic division, of specific cell types. To resolve temporal dynamics of NSPC differentiation in Kmt2d+/βgeo and wild-type mice, we pulsed adult mice with EdU for 2 weeks, during which a subset of labeled DG cells is expected to reach a late NSPC (NB) stage, characterized by radial extension of a DCX+ neuronal process. In contrast, another subset of pulsed cells, bearing an NES+ qRGL-like process, represents NSPCs that remain in a stem-like state. Thus, by quantifying EdU-labeled cells exhibiting either a DCX+ neuronal process (EdU+DCX+) or an NES+ qRGL-like process (EdU+NES+) (Figure 5F and Supplemental Figure 7), one can compare relative differentiation status, where a higher proportion of EdU+DCX+ cells would indicate early or precocious maturation. Indeed, though steady state cell numbers again showed fewer total NES+ and DCX+ NSPCs in Kmt2d+/βgeo mice compared with wild-type, among pulsed cells the Kmt2d+/βgeo mice exhibited a significantly greater fraction of EdU+DCX+ immature neurons (Figure 5F). In other words, Kmt2d+/βgeo DG NSPCs born within the preceding 2 weeks had achieved a more advanced differentiation state than wild-type cells born in the same window.
Together, studies of adult neurogenesis dynamics in Kmt2d+/βgeo mice suggest that in vivo neurodevelopmental effects of KMT2D loss recapitulate many phenotypes observed initially in vitro using mouse HT22 cells and KS1 patient–derived cells. Although comparison of gene expression profiles across these KS1 models revealed few individual genes with shared dysregulation among all models, at the network level we observed high enrichment of HIF1A regulatory and RNA metabolism pathways in a comparison of all DEGs in these KS1 models (Supplemental Figure 8, A–D).
Precocious differentiation and reduced hypoxia responses in Kmt2d+/βgeo primary hippocampal NSPCs.
Cellular oxygen availability has previously been directly linked to maintenance and differentiation of embryonic (33) and adult DG (19) NSPCs. Primary hippocampal NSPCs of Kmt2d+/βgeo mice showed increased HIF1A activation compared with wild-type NSPCs, and both genotypes showed increased HIF1A activation upon treatment by HIF1A-stabilizing agent dimethyloxaloylglycine (DMOG) for 12 hours (Supplemental Figure 9). We then subjected NSPCs to a standard in vitro neuronal differentiation protocol, quantifying cell marker expression between 0 and 8 days (Supplemental Figure 10, A and B). Before differentiation (day 0), wild-type NSPCs expressed low levels of a mature DG neuron marker, prospero-related homeobox 1 (PROX1), while Kmt2d+/βgeo NSPCs, surprisingly, showed an increase (Supplemental Figure 10C). By measuring expression of a proneural transcription factor, achaete-scute homolog 1 (ASCL1), we observed a baseline decrease (day 0) in Kmt2d+/βgeo NSPCs compared with wild-type (Supplemental Figure 10D). In contrast, after 2 days in differentiation conditions, Kmt2d+/βgeo NSPCs responded with greater ASCL1 expression compared with wild-type, an effect sustained at 4 and 8 days. DMOG treatment increased ASCL1 levels in both genotypes, though to greater magnitude in wild-type than Kmt2d+/βgeo NSPCs. Together, these data are consistent with a link between cellular hypoxia response and neuronal differentiation in hippocampal NSPCs (20).
Discussion
The ID disorder KS1 is caused by mutations in the histone methyltransferase KMT2D, but mechanistic links to neurodevelopmental and cognitive consequences in patients are not yet clear. KS1 diagnoses are typically made after birth, but the inherent reversibility of chromatin modifications raises the possibility that a detailed understanding of KMT2D activity in neuronal cells could identify molecular targets for postnatal interventions in KS1-associated ID.
Here, we report that KMT2D-deficient human and mouse neurodevelopment models, in vitro and in vivo, demonstrate similar patterns of transcriptional suppression, proliferative defects, and precocious cellular differentiation. These phenotypes were cell autonomous in vitro, suggesting that (a) chromatin and gene expression studies in neurogenic cell types could yield disease-relevant KMT2D targets and (b) these cellular models provide platforms for screening of novel therapeutic strategies or targeted manipulations. We performed transcriptomic and KMT2D profiling in these models and observed systematic suppression of hypoxia response pathways, particularly among HIF1A-regulated genes that are also directly KMT2D bound in neuronal cells. Physically overlapping KMT2D- and HIF1A-bound genomic loci were observed across tissues, approximately 40% of these at promoters, raising the possibility of shared etiologies in embryonically distinct KS1-affected organ systems. Furthermore, KMT2D-deficient neuronal cells, in contrast to isogenic wild-type cells, were unable to mount characteristic hypoxia-inducible gene activation responses when exposed to low-oxygen conditions, demonstrating oxygen response defects in KS1 models.
The implication of hypoxia response defects in KS1 suggests clinical relevance of recent findings in neurodevelopmental regulation. First, the adult hippocampal NSPC niche harbors locally hypoxic, but dynamic, microenvironments, and the hypoxic state positively influences NSPC survival (20, 21). Thus, compromised hypoxia responses could render cells particularly vulnerable to changes in oxygen levels as experienced by maturing NSPCs as they migrate from DG SGZ vasculature. Second, NSPC maturation is coupled to a metabolic rewiring from glycolysis in early NSPCs, to oxidative phosphorylation in maturing neurons. Zheng and colleagues (19) recently found this metabolic switch, marked by suppression of glycolytic genes, to be essential for neuronal maturation. In KS1 neural models we observed suppression of hypoxia-responsive glycolytic genes accompanied by upregulation of proneuronal differentiation genes, and demonstrated precocious maturation of DG NSPCs by in vivo pulsing of adult Kmt2d+/βgeo mice, as well as in vitro differentiation of Kmt2d+/βgeo primary DG NSPCs. Future studies could determine whether targeted chromatin opening at hypoxia response loci normalizes differentiation dynamics in KS1 NSPCs.
Analogous findings regarding premature activation of terminal differentiation genes, reduced proliferation, and precocious maturation in KMT2D-depleted keratinocytes were recently linked to disorganized epidermal stratification (10). Furthermore, in KMT2D-deficient cardiomyocytes, loss of H3K4me2 at KMT2D-bound hypoxia response genes was associated with cell cycle and proliferative defects in heart development (6). In contrast, KMT2D deletion in B cells conferred proliferative advantage and impaired cell maturation, despite significant upregulation of differentiation genes (8, 9). Thus, while KMT2D’s role in enhancer-mediated gene expression during differentiation is well established (11), phenotypic manifestations appear to be cell type and stage dependent. We now extend KMT2D-associated phenotypes of transcriptional perturbation, hypoxia response, cell cycle, proliferation, and premature differentiation to neuronal contexts. Phenotypic concordance across tissues of disparate embryonic origin suggests that KMT2D targets important to KS1 phenotypes support basic cellular homeostatic functions related to housekeeping, energy production, and cell cycle progression, rather than genes with purely brain-specific function. Furthermore, we report concordant phenotypes both from nonsense KMT2D mutations (patient iPSCs and NSPCs) and mutations limited to the KMT2D SET domain (HT22 cells, Kmt2d+/βgeo mice), indicating that loss of either gene dosage or catalytic function of KMT2D can be pathogenic.
Present results indicate that adult hippocampal neurogenesis defects, which we previously found to associate with visuospatial memory defects in Kmt2d+/βgeo mice, are observable at all stages examined, including fewer quiescent NSPCs in the DG, which could indicate either postnatal depletion or altered niche development in the embryo. Despite having fewer total NSPCs, by pulse-chase experiments we observed the Kmt2d+/βgeo NSPC population to achieve a more advanced maturation stage than that of wild-type littermates. Interestingly, adult-born NSPCs wield a disproportionately strong influence on DG circuitry and visuospatial learning during younger, but not older, neuronal maturation stages (34). This stage-dependent coupling of NSPC maturation with cognitive outcomes increases the likelihood that accelerated neuronal differentiation rates could negatively affect visuospatial memory acquisition. Furthermore, multispecies comparisons demonstrate that measured decreases in neurogenesis rates are consistent with accelerated neuronal maturation rates across the life span (35).
The apparent paradox of increased HIF1A activation, despite blunted hypoxia-responsive expression in Kmt2d+/Δ and Kmt2dΔ/Δ neuronal cells, raises 2 possibilities. First, chronic HIF1A activity could result in cellular compensatory efforts to downregulate hypoxia response genes. In this case, heterochromatin environments at HIF1A-binding genes could prevent induction. Alternatively, cellular oxygen sensing could be coupled to gene expression through chromatin states in an HIF1A-independent manner. Independent studies recently discovered direct oxygen sensing by KDM6A/UTX (the H3K27 demethylase lost in KS2 patients) as well as the H3K4/H3K36 demethylase 5A (KDM5A), which controlled chromatin states and cell differentiation in an HIF1A-independent manner (36, 37). These findings link hypoxia-induced histone methylation at H3K4, H3K27, H3K9, and H3K36 directly with control of maturation in multiple cell types, further supporting the notion that KS-associated transcriptional suppression, in the adult DG context, could affect NSPC stage-dependent learning (34) via metabolic dysregulation. Hypoxia-upregulated H3K4me3 peaks (37) were enriched in HIF1A target gene promoters, where we presently observed high overlaps in KMT2D/HIF1A–bound loci. Strikingly, loss of KDM5A, whose activity opposes that of KMT2D at H3K4 sites, caused upregulation of hypoxia-responsive genes (37), i.e., an effect opposite to the present KS1-associated suppression of hypoxia response genes, such as Klf10 and Bcl2/adenovirus E1B 19-KD protein-interacting protein 3–like (Bnip3l). Several histone demethylases, and at least 33 chromatin modifiers in total, have been shown to affect hypoxia response genes, 11 of these associating with developmental disorders or cancers, yet KMT2D and other histone methyltransferases have not yet been implicated (38).
In summary, our findings suggest that KMT2D deficiency disrupts neurogenesis by negatively affecting NSPC maintenance functions, including cell cycle, proliferation, and survival, accompanied by decreased adult NSPC numbers and precocious neuronal differentiation. Chromatin and transcriptome profiling identified KMT2D- and HIF1A-regulated gene programs suppressed across KS1 model systems, implicating previously described roles for hypoxia responses in regulating neuronal differentiation. Indeed, we functionally demonstrate KMT2D-dependent HIF1A activation and target gene induction in KS1 neural models and diminished response to hypoxic conditions during in vitro neuronal differentiation in Kmt2d+/βgeo NSPCs. Together, these findings are consistent with an etiological model for KS1-associated developmental changes in which KMT2D loss transcriptionally suppresses oxygen response programs critical to early NSPC maintenance, favoring precocious cellular differentiation during hippocampal neurogenesis.
Methods
Media and reagents are listed (Supplemental Table 10).
Animals.
The Kmt2d+/βgeo allele [Mll2Gt(RRt024)Byg] was generated by Bay Genomics (University of California) through the random insertion of a gene trap vector. Kmt2d+/βgeo mice were fully backcrossed to a C57BL/6J background (Jackson Laboratories) over more than 10 generations. Animals were housed in a 14-hour light/10-hour dark cycle with free access to food and water. Experiments compared age- and sex-matched littermates. Genotyping by PCR has been described (12).
Primary hippocampal NSPCs.
Female C57BL/6J mice (Jackson Laboratories) were mated to Kmt2d+/βgeo males and sacrificed for embryo harvest at embryonic day 18. Microdissected DG from Kmt2d+/βgeo and Kmt2d+/+ littermate embryos was processed for NSPC isolation and in vitro differentiation as described previously (39).
Patient-derived iPSCs, NSPCs, and fibroblasts.
Skin biopsy fibroblasts were cultured from molecularly confirmed KS1 patients (KS1-1, KS1-2, KS1-3). KS1-1 fibroblasts were reprogrammed using nonintegrating Sendai virus vectors (CytoTune-iPS 2.0). Five days after induction, cells were transferred to mouse embryonic fibroblast (MEF) feeder plates in iPSC medium. Twenty-one days after induction, high-quality colonies were manually selected for propagation and karyotyping by G-banding. Generation of healthy control lines (C3-1 and C1-2) was previously described (29). Feeder MEFs from E13.5 CF-1 mice were mitotically inactivated by irradiation. iPSCs were enzymatically passaged every 4–8 days using collagenase. NSPCs were induced from iPSCs as previously described (30), briefly, by inhibiting glycogen synthase kinase 3, transforming growth factor–β, γ-secretase, and Notch signaling pathways using small molecules CHIR99021 (4 μM), SB431542 (3 μM), (3 μM), and Compound E (0.1 μM), in the presence of human leukemia inhibitory factor (10 ng/mL) and Rho-associated, coiled-coil containing protein kinase inhibitor (5 μM) for 7 days. NSPCs were split with Accutase and propagated in neural induction medium on a Matrigel.
CRISPR/Cas9 deletions in HT22 cells.
HT22 mouse hippocampal cells are commercially available but were a gift of the Goff Laboratory and maintained in HT22 medium. sgRNAs targeting 2 loci spanning the Kmt2d SET domain–encoding region, with cut sites in exon 52 and either exon 54 (Kmt2dΔ1) or intron 54 (Kmt2dΔ2), were integrated into Cas9 plasmid (pSpCas9BB-2A-puro v2.0) and delivered to cells at 20% confluence using Lipofectamine 2000 according to the manufacturer’s protocol. After puromycin selection, cells were identified by PCR (primers listed in Supplemental Table 10) and clonally expanded. Following Sanger sequencing, a subset of clones appearing to be heterozygous based on PCR, but found to bear strand invasion, were removed from analyses.
RNA-Seq in HT22 cells: library preparation.
Cells were plated at equal density and sampled at 60% confluence. Total RNA was isolated from 3 biological replicates of Kmt2dΔ/Δ clones and Kmt2d+/+ parental cells using Direct-Zol RNA MicroPrep, and libraries were constructed in technical triplicate using NEBNext Poly(A) Magnetic Isolation Module and NEBNext Ultra II RNA Library Prep Kit for Illumina, with size selection by AMPure XP beads, according to manufacturers’ protocols. Library quantification and quality checks were done using KAPA Library Quantification Kit for Illumina, High Sensitivity DNA Kit on BioAnalyzer, and Qubit dsDNA HS Assay. Paired-end 50-bp reads were obtained for pooled libraries using Illumina HiSeq 2500.
RNA-Seq in HT22 cells: data analysis.
We first obtained a FASTA file with all mouse cDNA sequences (Mus_musculus.GRCm38.cdna.all.fa.gz) from Ensembl (http://uswest.ensembl.org/Mus_musculus/Info/Index, version 91, downloaded January 2018). Then, sequencing reads were pseudoaligned to this FASTA file, and transcript abundances were subsequently quantified, using Salmon (40). We then used the tximport R package (41) to convert the transcript abundances into normalized gene-level counts, by setting the “countsFromAbundance” parameter equal to “lengthScaledTPM.” Next, we used the edgeR (42, 43) and limma (44) R packages to log2 transform these gene-level counts and normalize each of the samples with the “voom” function using the effective library size (that is, the product of the library size and the normalization factors, the latter of which we computed with the “calcNormFactors” function provided in edgeR). Subsequently, we estimated the mean-variance relationship and calculated weights for each observation. To account for the correlation between technical replicates of the same clone when performing the differential analysis, we fit a mixed linear model, using the function “duplicateCorrelation” from the statmod R package (45) to block on clone. The differential analysis was then performed using the limma R package. DEGs were called with 0.05 as the cutoff for the FDR.
When performing the principal component analysis, transcript abundances were first converted into gene-level counts using the tximport R package, with the “countsFromAbundance” parameter equal to “no.” Then, we applied a variance-stabilizing transformation to these gene-level counts using the “vst” function from the DESeq2 R package (46), with the parameter “blind” set to “TRUE,” and subsequently estimated the principal components (without scaling the expression matrix) using the 1000 most variable genes.
scRNA-Seq: library preparation.
NSPCs were induced in parallel from each iPSC line (KS1-1, C1-2, C3-1) under identical conditions and passaged 3 times before sampling. iPSCs were detached from MEF feeders using collagenase (200 units/mL). iPSCs and NSPCs were dissociated to single-cell suspension using Accutase. Cell counts and viability were analyzed using Countess II. scRNA-Seq libraries were created with Chromium Single Cell 3′ Library & Gel Bead Kit v2 (10x Genomics) according to the manufacturer’s protocol. Targeted cell recovery for each sample was 5000 cells. Sufficient cDNA for library construction was obtained using 20 amplification cycles for iPSC libraries and 16 cycles for NSPC libraries. Sample indexing was achieved using 11 PCR cycles for iPSC libraries and 5 cycles for NSPC libraries. scRNA-Seq libraries were sequenced using Illumina NextSeq 500.
scRNA-Seq: data analysis.
Sequencing output was processed through the Cell Ranger 2.1.0 preprocessing pipeline using default parameters, with the exception of --expect-cells = 5000 for “cellranger count” and --normalize = none for “cellranger aggr.” Reads were quantified against hg19 using the 10x reference genome and transcriptome builds (refdata-cellranger-GRCh38-1.2.0). The aggregated raw count matrix was then used as input for the Monocle 2 scRNA-Seq framework. DEG analysis was performed on all NSPCs and iPSCs with respect to genotype (KS1 patient vs. healthy control) and was performed using the Monocle 2 (47) likelihood ratio test (0.1% FDR, Monocle 2 likelihood ratio test, Benjamini-Hochberg corrected) with “num_genes_expressed” added as a nuisance parameter to both the full and reduced models. The directionality of the differential gene test was determined by calculating the mean gene expression across all KS1 patient–derived and healthy control cells, evaluating the relative fold change. High-variance genes were selected as those with a positive residual to the estimated dispersion fit and a mean number of reads per cell greater than or equal to 0.0005. Cell cycle stage was determined by profiling cell cycle–associated genes across all cells and assigning cell cycle state using the R/Bioconductor package scran (48). Dimensionality reduction and visualization were performed via UMAP (31) on the log10(counts + 1) of the high-variance genes in the NSPC data set. The first 10 principal components were used as input for UMAP using the default parameters of the R/CRAN package umap. Cells were assigned to clusters using Monocle 2’s implementation of the louvain community detection algorithm. Learned clusters were then aggregated by hand based on marker gene expression into 3 clusters (“differentiating,” “transitioning,” “cycling”). Differential gene expression analysis within clusters and between genotypes was performed as described above. The “differentiating” cluster was then segregated, and a smooth line was fitted using a linear regression. This line was determined to represent the direction of differentiation by examination of marker genes (Supplemental Figure 5, C–H). The residuals of this fit were then plotted, and deciles were calculated containing an equal number of cells along the axis of differentiation. The number of cells in each decile was then counted with respect to genotype.
ChIP-Seq: library preparation.
Kmt2d+/+ HT22 cells were sampled at 70% confluence and processed for pull down with ChIP-grade KMT2D antibody (MilliporeSigma) according to Encyclopedia of DNA Elements (ENCODE) guidelines. Sonicated, reverse–cross-linked chromatin served as input control. Briefly, approximately 300 million cells per cell line were cross-linked in 1% formaldehyde and quenched with 0.125 M glycine, and cell lysate supernatants were collected for immediate processing or snap-frozen for storage at –80°C. Nuclei (20 million/sample) were diluted in 1 mL RIPA buffer and were sonicated using a Bioruptor for 6 cycles of 5 minutes (60 s on/30 s off) in ice-cold water bath. Supernatants containing sheared chromatin were precleared with Protein A Dynabeads and incubated overnight at 4°C with 8 μg KMT2D antibody. ChIP DNA was recovered by Dynabead incubation (overnight at 4°C plus 6 hours at room temperature) before 6 sequential salt washes of increasing stringency, then eluted and reverse–cross-linked overnight at 65°C. DNA was purified using DNA Clean and Concentrator (Zymo Research) and quantified using High Sensitivity DNA Kit on BioAnalyzer and Qubit dsDNA HS Assay. DNA libraries were constructed using NEBNext Ultra II DNA Library Prep Kit for Illumina and quantified using KAPA Library Quantification Kit for Illumina. Paired-end 75-bp reads were obtained for pooled libraries using Illumina HiSeq 2500.
ChIP-Seq: data analysis.
Sequencing reads were aligned to the mouse reference genome (mm10) using Bowtie2 (49). Then, duplicate reads were removed with the function MarkDuplicates from Picard (http://broadinstitute.github.io/picard/). Peaks were subsequently called using model-based analysis of ChIP-Seq 2 (50), with the “keep-dup” parameter equal to “all.” After peak calling, we excluded all peaks that overlapped with blacklisted regions provided by ENCODE (51). As a quality metric, using the resulting list of peaks, we computed the fraction of reads in peaks (frip) with the “featureCounts” function in the Rsubread package (52), with the “requireBothEndsMapped” parameter equal to “TRUE,” and the “countChimericFragments” and “countMultiMappingReads” parameters equal to “FALSE.” We found frip to be 2.9%, which is within the typically encountered range of values for a point-source factor (53).
To identify genes likely to be regulated in cis by KMT2D, we first obtained the coordinates of 10-kb regions centered on the TSS for each gene, using the “promoters” function from the EnsDb.Mmusculus.v79 R package (54), with the “filter” parameter equal to “TxBiotypeFilter(“protein_coding”)” and the “upstream” and “downstream” parameters both equal to 5000. Subsequently, we selected those genes whose extended promoter (± 5 kb from the TSS) overlapped with at least 1 KMT2D peak, using the “findOverlaps” function in the GenomicRanges R package (55).
Purification of EdU+ nuclei.
Mice were given 150 mg/kg EdU by intraperitoneal injection and sampled after 16 hours. DG was microdissected in ice-cold PBS immediately following sacrifice by halothane inhalation. Total nuclei were purified as described (56) with the addition of RNase inhibitor to all buffers. Briefly, DG was dounce-homogenized in 1 mL lysis buffer and layered above a sucrose gradient for ultracentrifugation at 139,800 g (max) for 2 hours at 4°C. Nuclei were resuspended in Click-iT EdU Alexa Fluor 488 with RNAse inhibitor and incubated 30 minutes at room temperature. Samples were passed through a 40-μm filter (Thermo Fisher Scientific), stained with 1 μg/mL DAPI, and kept on ice before sorting. Lysates processed identically from non–EdU-injected mice served as negative controls during sorting with Beckman Coulter MoFlo Cell Sorter. Cell cycle analysis by DNA content was performed with gates discriminating 2N and 4N cells by DAPI fluorescence.
RNA-Seq: EdU+ nuclei.
Purified EdU+ nuclei from 3 Kmt2d+/βgeo and 3 wild-type littermate female mice (500 nuclei pooled per genotype) were sorted into Smart-Seq 2 lysis buffer (2 μL Smart-Seq2 lysis buffer with RNase inhibitor, 1 μL oligo-dT primer, and 1 μL deoxynucleoside triphosphate; briefly spun by tabletop microcentrifuge; and snap-frozen on dry ice. Nuclei were processed according to a modified Smart-seq2 protocol (57). Briefly, lysates were thawed to 4°C, heated to 72°C for 5 minutes, and immediately placed on ice. Template-switching first-strand cDNA synthesis was performed using a 5′-biotinylated template switch oligo. cDNAs were amplified using 20 cycles of KAPA HiFi PCR and 5′-biotinylated in situ PCR primer. Amplified cDNA was cleaned using a 1:1 ratio of Ampure XP beads, and approximately 250 pg was input to a one-quarter–sized Nextera XT tagmentation reaction. Tagmented fragments were amplified for 12 enrichment cycles, and dual indexes were added to each well to uniquely label each library. Concentrations were assessed with Quant-iT PicoGreen dsDNA Reagent (Invitrogen, Thermo Fisher Scientific), and samples were diluted to approximately 2 nM and pooled. Paired-end 105-bp reads were obtained for pooled libraries using Illumina HiSeq 2500. Paired-end reads were aligned to mm10 using HISAT2 (58) with default parameters except –p = 8. Aligned reads from individual samples were quantified against a reference genome (GENCODE vM8) using cuffquant (59). Normalized expression estimates across all samples were obtained using cuffnorm with default parameters (60).
RT-qPCR.
Total RNA was isolated by RNeasy Mini Kit (QIAGEN), and cDNA libraries were constructed with High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to manufacturers’ protocols. Experiments were performed in technical triplicate, with biological replicates as indicated. Probes were from TaqMan.
Immunostaining, confocal imaging, and processing.
Coronal brain sections of 30 μm (every sixth slice) were analyzed in serial order. Briefly, adult brains were PFA-fixed by transcardial perfusion and postfixed for 12 hours before cryoprotection by 30% sucrose in phosphate buffer. Brains were sectioned by cryostat (Leica), directly mounted to charged slides, and stored at –80°C. Antigen retrieval (DakoCytomation) was performed at 95°C for 20 minutes. Overnight incubation at 4°C in primary antibodies (Supplemental Table 10) preceded incubation in Alexa Fluor–conjugated secondary antibody (1:500). Tiled, Z-stacked images were acquired using Zeiss LSM780 FCS AxioObserver confocal microscope and Zen software (Zeiss) to encompass the entire DG structure. Images were quantified using Imaris (BitPlane) by experimenters blinded to genotype. Cell counts were corrected by DG area multiplied by Z-thickness and expressed as cells/mm3. For pulse-label experiments, mice were injected with 150 mg/kg EdU in saline every 48 hours and sampled as above. DCX+ neuroblast distance from the SGZ plane was measured in Fiji (NIH). Patient-derived cell imaging used EVOS FL Cell Imaging System with analysis in Fiji.
FACS and analysis.
Flow cytometry analysis with FACSverse and FACSsuite (BD Biosciences) and sorting by Beckman Coulter MoFlo Cell Sorter were done with proper gate settings and doublet discrimination (Supplemental Figure 3J, Supplemental Figure 6A). Runs of 10,000 or more cells were analyzed from technical triplicate culture wells and analyzed in FlowJo v10 (TreeStar Inc). Unstained and secondary-only samples served as controls. Cells were sampled after a 30-minute pulse of EdU (10 μM) using Click-iT EdU Flow Cytometry Assay (Thermo Fisher Scientific). CellTrace Violet and CellEvent caspase-3/7 reagent (Thermo Fisher Scientific) were used according to the manufacturer’s protocols. For cycle synchronization, 250 ng/mL nocodazole (MilliporeSigma) was applied for 18 hours before release.
MRI.
3D T2-weighted MRI (9.4 T) was performed on PFA-perfused brains of Kmt2d+/βgeo (n = 3) and Kmt2d+/+ (n = 3) female mice aged 4 months. Atlas-based, volume-corrected analysis was performed in 25 brain regions (DtiStudio).
Data availability.
High-throughput data are publicly available: RNA-Seq and ChIP-Seq in Gene Expression Omnibus (GEO) database (GSE126167) and scRNA-Seq in GEO (GSE126027). Scripts for scRNA-Seq analysis are available at https://github.com/Jaugust7/Kabuki-Syndrome-scRNA-analysis
Statistics.
For high-throughput experiments, see Methods. For cellular assays, see figure legends. Statistical analyses with multiple-comparisons correction were done with GraphPad Prism (version 7.0b). Gene set enrichments were determined according to WebGestalt (61) or by Fisher’s exact test in R version 3.5.2 as indicated. P values below 0.05 were considered significant and are denoted as indicated in the legends (*P < 0.05, **P < 0.01, and ***P < 0.001).
Study approval.
All mouse experiments were performed using protocols approved by the Animal Care and Use Committee of Johns Hopkins University (JHU) School of Medicine and are in accordance with NIH guidelines. Informed consent regarding KS1 patient samples was obtained according to institutional IRB and Institutional Stem Cell Research Oversight protocols approved by JHU.
Author contributions
GAC and HTB conceived the study; GAC and HTB wrote the manuscript; GAC, HNN, GHC, JDR, BLW, and LZ performed experiments; and GAC, LB, JJA, KDH, and LAG analyzed data.
Supplementary Material
Acknowledgments
HTB is funded through an Early Independence Award from the NIH (DP5OD017877), the Icelandic Research Fund (195835-051), and the Louma G. Foundation. Imaging was performed with NIH support (S10OD016374). The karyotype facility was supported by the National Institute of Child Health and Human Development (1U54 HD079123-01A1). FACS was performed at the Bloomberg School of Public Health. We thank Michael Sherman for assistance with image quantification and Manisha Aggarwal for MRI. Schematics were created by Mark Sandusky or with Biorender.com. Hongjun Song and Kai Ge provided critical reagents and advice. Hal Dietz and Gregg Semenza provided conceptual guidance.
Version 1. 08/29/2019
In-Press Preview
Version 2. 10/17/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(20):e129375.https://doi.org/10.1172/jci.insight.129375.
Contributor Information
Giovanni A. Carosso, Email: gcaross1@jhmi.edu.
Leandros Boukas, Email: lboukas1@jhmi.edu.
Jonathan J. Augustin, Email: jaugust7@jhmi.edu.
Ha Nam Nguyen, Email: namoadida@gmail.com.
Briana L. Winer, Email: Brianaleigh2013@aol.com.
Gabrielle H. Cannon, Email: ghcannon14@gmail.com.
Johanna D. Robertson, Email: jojorobertson@comcast.net.
Li Zhang, Email: lzhang32@jhmi.edu.
Kasper D. Hansen, Email: khansen@jhsph.edu.
Loyal A. Goff, Email: loyalgoff@jhmi.edu.
Hans T. Bjornsson, Email: hbjorns1@jhmi.edu.
References
- 1.Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G. Genome regulation by Polycomb and Trithorax: 70 years and counting. Cell. 2017;171(1):34–57. doi: 10.1016/j.cell.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Bjornsson HT. The Mendelian disorders of the epigenetic machinery. Genome Res. 2015;25(10):1473–1481. doi: 10.1101/gr.190629.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ng SB, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42(9):790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyake N, et al. KDM6A point mutations cause Kabuki syndrome. Hum Mutat. 2013;34(1):108–110. doi: 10.1002/humu.22229. [DOI] [PubMed] [Google Scholar]
- 5.Hannibal MC, et al. Spectrum of MLL2 (ALR) mutations in 110 cases of Kabuki syndrome. Am J Med Genet A. 2011;155A(7):1511–1516. doi: 10.1002/ajmg.a.34074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ang SY, et al. KMT2D regulates specific programs in heart development via histone H3 lysine 4 di-methylation. Development. 2016;143(5):810–821. doi: 10.1242/dev.132688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ortega-Molina A, et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat Med. 2015;21(10):1199–1208. doi: 10.1038/nm.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med. 2015;21(10):1190–1198. doi: 10.1038/nm.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JE, et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife. 2013;2:e01503. doi: 10.7554/eLife.01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin-Shiao E, et al. KMT2D regulates p63 target enhancers to coordinate epithelial homeostasis. Genes Dev. 2018;32(2):181–193. doi: 10.1101/gad.306241.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Froimchuk E, Jang Y, Ge K. Histone H3 lysine 4 methyltransferase KMT2D. Gene. 2017;627:337–342. doi: 10.1016/j.gene.2017.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bjornsson HT, et al. Histone deacetylase inhibition rescues structural and functional brain deficits in a mouse model of Kabuki syndrome. Sci Transl Med. 2014;6(256):256ra135. doi: 10.1126/scitranslmed.3009278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boisgontier J, et al. Anatomical and functional abnormalities on MRI in Kabuki syndrome. Neuroimage Clin. 2019;21:101610. doi: 10.1016/j.nicl.2018.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonçalves JT, Schafer ST, Gage FH. Adult neurogenesis in the hippocampus: from stem cells to behavior. Cell. 2016;167(4):897–914. doi: 10.1016/j.cell.2016.10.021. [DOI] [PubMed] [Google Scholar]
- 15.Moreno-Jiménez EP, et al. Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat Med. 2019;25(4):554–560. doi: 10.1038/s41591-019-0375-9. [DOI] [PubMed] [Google Scholar]
- 16.Tobin MK, et al. Human hippocampal neurogenesis persists in aged adults and Alzheimer’s disease patients. Cell Stem Cell. 2019;24(6):974–982.e3. doi: 10.1016/j.stem.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sorrells SF, et al. Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature. 2018;555(7696):377–381. doi: 10.1038/nature25975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berg DA, et al. A common embryonic origin of stem cells drives developmental and adult neurogenesis. Cell. 2019;177(3):654–668.e15. doi: 10.1016/j.cell.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng X, et al. Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. Elife. 2016;5:e13374. doi: 10.7554/eLife.13374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mazumdar J, et al. O2 regulates stem cells through Wnt/β-catenin signalling. Nat Cell Biol. 2010;12(10):1007–1013. doi: 10.1038/ncb2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chatzi C, Schnell E, Westbrook GL. Localized hypoxia within the subgranular zone determines the early survival of newborn hippocampal granule cells. Elife. 2015;4:e08722. doi: 10.7554/eLife.08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morimoto BH, Koshland DE. Induction and expression of long- and short-term neurosecretory potentiation in a neural cell line. Neuron. 1990;5(6):875–880. doi: 10.1016/0896-6273(90)90347-I. [DOI] [PubMed] [Google Scholar]
- 23.Wang C, et al. Enhancer priming by H3K4 methyltransferase MLL4 controls cell fate transition. Proc Natl Acad Sci U S A. 2016;113(42):11871–11876. doi: 10.1073/pnas.1606857113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo C, et al. Global identification of MLL2-targeted loci reveals MLL2’s role in diverse signaling pathways. Proc Natl Acad Sci U S A. 2012;109(43):17603–17608. doi: 10.1073/pnas.1208807109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Semenza GL. HIF-1: mediator of physiological and pathophysiological responses to hypoxia. J Appl Physiol. 2000;88(4):1474–1480. doi: 10.1152/jappl.2000.88.4.1474. [DOI] [PubMed] [Google Scholar]
- 26.Guimarães-Camboa N, et al. HIF1α represses cell stress pathways to allow proliferation of hypoxic fetal cardiomyocytes. Dev Cell. 2015;33(5):507–521. doi: 10.1016/j.devcel.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benita Y, Kikuchi H, Smith AD, Zhang MQ, Chung DC, Xavier RJ. An integrative genomics approach identifies Hypoxia Inducible Factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucleic Acids Res. 2009;37(14):4587–4602. doi: 10.1093/nar/gkp425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanz JH, Lipkin P, Rosenbaum K, Mahone EM. Developmental profile and trajectory of neuropsychological skills in a child with Kabuki syndrome: implications for assessment of syndromes associated with intellectual disability. Clin Neuropsychol. 2010;24(7):1181–1192. doi: 10.1080/13854046.2010.506198. [DOI] [PubMed] [Google Scholar]
- 29.Wen Z, et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature. 2014;515(7527):414–418. doi: 10.1038/nature13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li W, et al. Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc Natl Acad Sci U S A. 2011;108(20):8299–8304. doi: 10.1073/pnas.1014041108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McInnes L, Healy J, Saul N, Großberger L. UMAP: uniform manifold approximation and projection. J Open Source Softw. 2018;3(29):861. doi: 10.21105/joss.00861. [DOI] [Google Scholar]
- 32.Benjamin JS, et al. A ketogenic diet rescues hippocampal memory defects in a mouse model of Kabuki syndrome. Proc Natl Acad Sci U S A. 2017;114(1):125–130. doi: 10.1073/pnas.1611431114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris AM, Churchwell JC, Kesner RP, Gilbert PE. Selective lesions of the dentate gyrus produce disruptions in place learning for adjacent spatial locations. Neurobiol Learn Mem. 2012;97(3):326–331. doi: 10.1016/j.nlm.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhuo JM, et al. Young adult born neurons enhance hippocampal dependent performance via influences on bilateral networks. Elife. 2016;5:e22429. doi: 10.7554/eLife.22429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snyder JS. Recalibrating the relevance of adult neurogenesis. Trends Neurosci. 2019;42(3):164–178. doi: 10.1016/j.tins.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 36.Chakraborty AA, et al. Histone demethylase KDM6A directly senses oxygen to control chromatin and cell fate. Science. 2019;363(6432):1217–1222. doi: 10.1126/science.aaw1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Batie M, Frost J, Frost M, Wilson JW, Schofield P, Rocha S. Hypoxia induces rapid changes to histone methylation and reprograms chromatin. Science. 2019;363(6432):1222–1226. doi: 10.1126/science.aau5870. [DOI] [PubMed] [Google Scholar]
- 38.Luo W, Wang Y. Epigenetic regulators: multifunctional proteins modulating hypoxia-inducible factor-α protein stability and activity. Cell Mol Life Sci. 2018;75(6):1043–1056. doi: 10.1007/s00018-017-2684-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernas S, Leiter O, Walker T, Kempermann G. Isolation, culture and differentiation of adult hippocampal precursor cells. Bio Protoc. 2017;7(21):e2603. doi: 10.21769/BioProtoc.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14(4):417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40(10):4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Giner G, Smyth GK. Statmod: probability calculations for the inverse Gaussian distribution. Cornell University website. https://arxiv.org/abs/1603.06687 Updated July 28, 2016. Accessed September 19, 2019.
- 46.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lun AT, McCarthy DJ, Marioni JC. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 2016;5:2122. doi: 10.12688/f1000research.9501.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amemiya HM, Kundaje A, Boyle AP. The ENCODE blacklist: identification of problematic regions of the genome. Sci Rep. 2019;9(1):9354. doi: 10.1038/s41598-019-45839-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liao Y, Smyth GK, Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019;47(8):e47. doi: 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22(9):1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. doi: 10.18129/B9.bioc.EnsDb.Mmusculus.v79. Rainer J. EnsDb.Mmusculus.v79: Ensembl based annotation package. R package version 2.99.0. Bioconductor. Accessed September 19, 2019. [DOI]
- 55.Lawrence M, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9(8):e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rizzardi LF, et al. Neuronal brain-region-specific DNA methylation and chromatin accessibility are associated with neuropsychiatric trait heritability. Nat Neurosci. 2019;22(2):307–316. doi: 10.1038/s41593-018-0297-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Picelli S, Björklund ÅK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10(11):1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 58.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mudge JM, Harrow J. Creating reference gene annotation for the mouse C57BL6/J genome assembly. Mamm Genome. 2015;26(9-10):366–378. doi: 10.1007/s00335-015-9583-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017;45(W1):W130–W137. doi: 10.1093/nar/gkx356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
High-throughput data are publicly available: RNA-Seq and ChIP-Seq in Gene Expression Omnibus (GEO) database (GSE126167) and scRNA-Seq in GEO (GSE126027). Scripts for scRNA-Seq analysis are available at https://github.com/Jaugust7/Kabuki-Syndrome-scRNA-analysis